Abstract
Obesity has evolved into a global pandemic that constitutes a major threat to public health. The majority of obesity-related health care costs are due to cardiometabolic complications, such as insulin resistance, dyslipidemia, and hypertension, which are risk factors for Type 2 diabetes and cardiovascular disease. However, many obese individuals, often called metabolically healthy obese (MHO), seem to be protected from these cardiometabolic complications. Conversely, there is a group of individuals who suffer from cardiometabolic complications despite being of normal weight; a condition termed metabolically obese normal weight (MONW). Recent large-scale genomic studies have provided evidence that a number of genetic variants show an association with increased adiposity but a favorable cardiometabolic profile, an indicator for the genetic basis of the MHO and MONW phenotypes. Many of these loci are located in or near genes that implicate pathways involved in adipogenesis, fat distribution, insulin signaling, and insulin resistance. It has been suggested that a threshold for subcutaneous adipose tissue expandability may be at play in the manifestation of MHO and MONW, where expiry of adipose tissue storage capacity could lead to ectopic lipid accumulation in non-adipose tissues such as liver, muscle, heart, and pancreatic beta cells. Understanding the genetic aspects of the mechanisms that underpin MHO and MONW is crucial to define appropriate public health action points and to develop effective intervention measures.
Keywords: adiposity, cardiometabolic disease, genomics, metabolically healthy obesity, metabolically obese normal weight
INTRODUCTION
Obesity has been described as a global pandemic (43, 45, 58). The incidence of obesity has more than doubled worldwide since 1980 (66), posing a severe burden on the health care system. The majority of obesity-related health care costs are due to cardiometabolic comorbidities, such as insulin resistance, dyslipidemia, and hypertension, which are risk factors for Type 2 diabetes (T2D) and cardiovascular disease (61, 70).
Interestingly, some obese individuals appear to be protected against obesity-related cardiometabolic complications. This condition, first described in the 1980s (2, 54), is referred to as metabolically healthy obesity (MHO). Conversely, there are also individuals who have an elevated cardiometabolic risk that is similar to that of an unhealthy obese population, while having a body weight within the normal range. These individuals are referred to as metabolically obese normal weight (MONW) (46, 47).
Age, sex, ancestry, diet, smoking, and physical activity are known demographic and environmental factors contributing to MHO and MONW (11, 13, 28, 29, 42, 44). There are no estimates available for the heritability of these two conditions in current literature. Nevertheless, recent large-scale genomic studies have provided evidence that a number of genetic variants implicate an inverse relationship between increased adiposity and an unfavorable cardiometabolic profile, indicative of a genetic basis of the MHO and MONW phenotypes (25, 32, 33, 50, 67, 68). In this review, we will consolidate the increasing evidence of a genetic predisposition for MHO and MONW, provide comparison between studies, and unify shared hypotheses for the underlying mechanisms.
DEFINITIONS OF MHO AND MONW
Despite the simple notion, there is much debate on the definition of MHO (22, 23, 42, 57, 64). There are no universally accepted criteria for MHO (22, 23, 42, 57, 64), but it has been broadly defined as obesity, diagnosed by body mass index (BMI) ≥30 kg/m2, in the absence of cardiometabolic comorbidities (6, 22, 23, 42, 57, 64). The prevalence of MHO has been estimated to be 20–69% in the obese population (4, 7, 18, 21, 23, 37, 44, 55, 64). MHO is characterized by relatively high insulin sensitivity and healthy glycemic, lipid, inflammation, blood pressure, and hormonal profiles, with low incidence of cardiometabolic diseases, despite high adiposity (1, 8, 22, 24, 37).
Similar to MHO, various criteria have been used to define the MONW status. The condition is often described as normal weight (BMI < 25 kg/m2) with several cardiometabolic abnormalities typically seen in obese individuals (4, 5, 13, 17, 37). MONW is characterized by elevated triglycerides (TG), glycemic, and C-reactive protein (CRP) levels, reduced high-density lipoprotein cholesterol (HDL-C) and adiponectin levels, increased insulin resistance, and increased risk of T2D and cardiovascular disease (4, 5, 13, 17, 19, 37). The prevalence of MONW among normal weight individuals is estimated to be 4–25% (4, 7, 11, 18, 23, 37, 55, 64).
CURRENT EVIDENCE ON THE GENETIC BASIS OF MHO AND MONW
Despite the lack of consensus on the definitions of MHO and MONW, recent genomic studies have provided novel insights into the genetic basis of these two phenotypes by examining associations of genome-wide variants with adiposity and cardiometabolic traits. In two large-scale genome-wide association studies (GWAS) of body fat percentage (BFP), several variants were identified to cause a predisposition to increased adiposity but reduced cardiometabolic risk, representative of the MHO phenotype (25, 33). Other studies examined aggregate scores of insulin resistance loci and uncovered a link between genetic predisposition to insulin resistance and decreased adiposity, which implicates the MONW phenotype (32, 50, 67, 68). It is important to note that these two conditions are flipsides of the same coin, depending on which of the two alleles for the variant is considered to be the effect allele. In the following, we will discuss recent findings on the genetic determinants of these two paradoxical conditions.
GWAS of BFP identifies a favorable adiposity locus near the IRS1 gene.
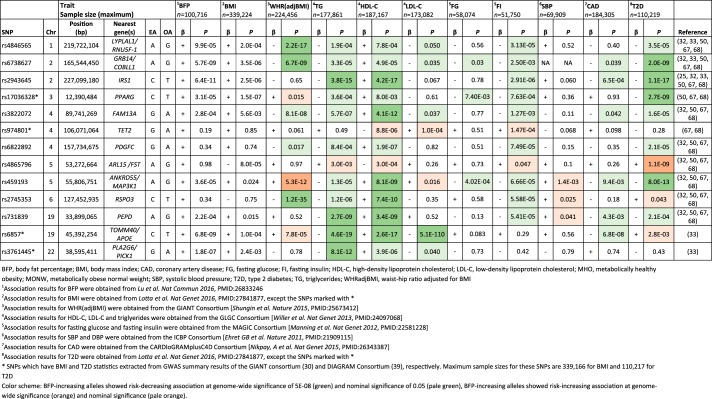
In 2011, the first genome-wide meta-analysis of BFP was published, including up to 76,202 individuals of European and Indian Asian descent from 26 GWAS (25). Three loci were found to be associated with BFP, including a locus near the previously established FTO obesity gene (14, 51, 60, 65), and two novel loci, near SPRY2 and IRS1.
Opposite to what would be expected based on the known association between lower BFP and a favorable metabolic profile, the BFP-decreasing allele at the IRS1 locus was associated with lower HDL-C and higher TG levels and increased insulin resistance. The effect size of the IRS1 locus on BFP was significantly larger in men than in women, with each allele decreasing BFP by 0.20 and 0.06%, respectively. The association with HDL-C and TG was also greater in men than in women, whereas the association with insulin resistance was similar between men and women.
The BFP-decreasing allele of the IRS1 locus showed a significant reduction in subcutaneous adipose tissue (SAT) in men, but not in women. No association with visceral adipose tissue (VAT) was found in either sex. Consequently, the fat-decreasing allele at this locus was associated with a higher VAT/SAT ratio in men, but not in women. The BFP-decreasing allele at the IRS1 locus was also associated with lower adiponectin levels in men, but not in women, whereas no association was found for circulating leptin levels. Leptin and adiponectin are both hormones secreted by adipocytes, of which the former regulates appetite and food intake (27), whereas the latter glucose and lipid levels (69). Leptin levels are known to correlate positively with body fatness, whereas the correlation between body fatness and adiponectin levels is inverse (27, 69). Interestingly, studies have shown that transgenic overexpression of adiponectin permits healthy expansion of SAT in leptin-deficient (ob/ob) mice. Consequently, this prevents accumulation of lipids in the liver and retains insulin sensitivity (26). Hence, it was hypothesized that lower adiponectin level could be associated with reduced ability to expand SAT in men with BFP-decreasing allele of the IRS1 locus. This could then lead to a flux of lipids into liver, which may increase insulin resistance through lipotoxicity (63).
The variant most strongly associated with BFP near the IRS1 locus was located 500 kb upstream of the gene. In expression quantitative trait locus (eQTL) analyses, the BFP-decreasing allele exhibited reduced IRS1 expression in SAT and VAT, whereas no effect was seen in blood or liver. The reduced expression in SAT and VAT was found to be more significant in men than in women. In analyses for basal IRS1 gene expression, adipocytes of female mice exhibited higher Irs1 expression than those of male mice in both SAT and VAT, while in humans, women had higher expression of IRS1 in VAT than men, but no sex difference was observed in SAT. The higher basal levels of IRS1 in adipose tissue of women could be beneficial in buffering a modest level of impairment in IRS1 gene expression. Women also have a stronger drive to store fat subcutaneously than men, which could overcome a defect in IRS-1 function (25).
The IRS1 gene is likely to be involved in the regulation of insulin and insulin-like growth factor-1 action. Previous studies have shown that a single nucleotide polymorphism (SNP) (rs2943641) at the same locus is associated with reduced expression of IRS-1 protein and reduced insulin-induced phosphatidylinositol 3-OH kinase activity in skeletal muscle, which take part in insulin signal transduction (48). Knockout mice of Irs1 are insulin resistant and hyperinsulinemic, despite being lean (3, 59), consistent with the association between the BFP-decreasing IRS1 allele and an unfavorable cardiometabolic profile in humans.
Taken together, the BFP-decreasing allele at the locus near IRS1 exhibited features of the MONW phenotype. It was associated with an increased VAT/SAT ratio, adverse lipid profile, insulin resistance, reduced adiponectin levels, and increased risk of T2D and coronary artery disease (CAD) (Fig. 1). Conversely, aligning the results according to the BFP-increasing allele, the association pattern exhibits favorable adiposity and hence, is reminiscent of the MHO phenotype (Fig. 2). In contrast to IRS1, the BFP-increasing alleles of the loci in FTO and near SPRY2 showed associations with cardiometabolic traits that were consistent with the known association between increased adiposity and an unfavorable cardiometabolic profile (25). The discovery of the IRS1 locus implicated the first clear genetic link between increased adiposity and favorable cardiometabolic profile and suggested an interplay between subcutaneous fat expandability and improved insulin sensitivity as evidenced by epidemiological studies, eQTL analyses, and mouse studies. Amid the relatively modest sample size, as superseded by subsequent studies, it provides credible support to the genetic basis of MHO at multiple levels.
Fig. 1.
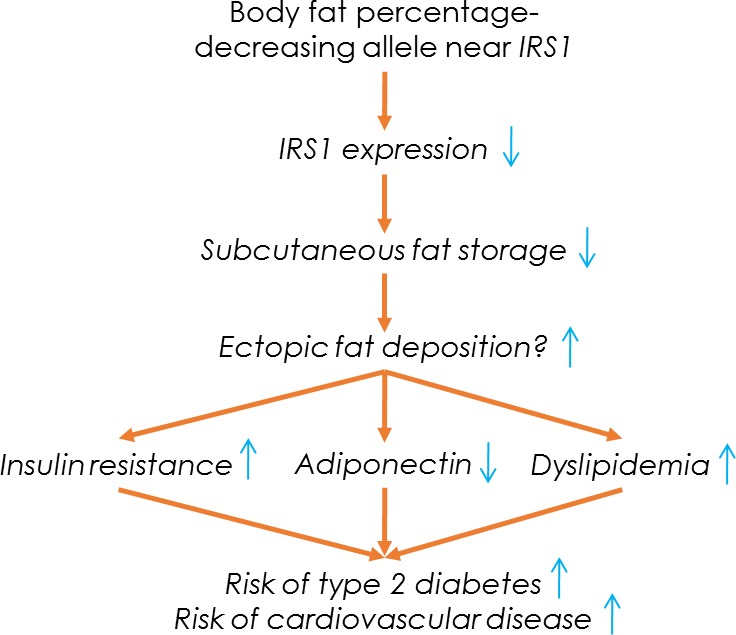
Possible pathways and mechanisms involved at the locus near the IRS1 gene.
Fig. 2.
Loci implicated in metabolically healthy obesity (MHO) and metabolically obese normal weight (MONW) and their association with cardiometabolic traits in meta-analyses of genome-wide association studies.
An extended GWAS of BFP identifies additional loci linked to favorable adiposity.
In 2016, an extended genome-wide meta-analysis of BFP, including up to 100,716 individuals from 56 studies, mainly of European ancestry, confirmed the associations for the IRS1, FTO, and SPRY2 loci and identified nine novel BFP-associated loci (33). Four of the novel loci, in or near MC4R, TMEM18, TUFM/SH2B1, and SEC16B, showed stronger association with BMI than BFP. The remaining five loci, in or near GRB14/COBLL1, TOMM40, IGF2BP1, PLA2G6/PICK1, and CRTC1, displayed stronger association with BFP than BMI (Fig. 3). As BMI represents the sum of fat and lean mass, whereas BFP is a ratio of body fat mass to body weight, the stronger association with BFP may indicate that the loci affect adiposity in particular.
Fig. 3.
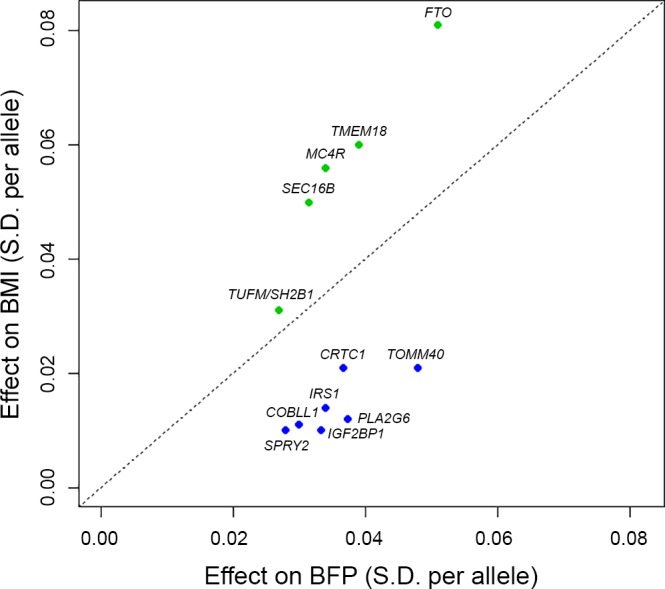
Comparison of effects of 12 body fat percentage (BFP) increasing loci on BFP and body mass index (BMI). Loci that have larger effects on BMI than BFP (above diagonal) are colored green, whereas loci with larger effects on BFP than BMI (below diagonal) are colored blue. The effect sizes are indicated in units of SD of inverse normally transformed traits (mean = 0, SD = 1). Adapted from Ref. (33).
The four novel loci that were associated more strongly with BMI than BFP had all been previously discovered in GWAS for BMI and were shown to influence body weight through their role in the central nervous system (10, 14, 31, 51, 56, 60, 65). As their BMI-increasing alleles were typically associated with an unfavorable cardiometabolic profile, we will not discuss them here. Of the five novel loci that were more strongly associated with BFP than BMI, the BFP-increasing alleles in three loci, near GRB14/COBLL1, TOMM40, and PLA2G6/PICK1, showed cardiometabolically protective effects (Fig. 2). The BFP-increasing allele at the locus near GRB14/COBLL1 was associated with an overall favorable cardiometabolic profile, including higher HDL-C and lower TG levels, and reduced risk of T2D and CAD. These protective associations may have been mediated by a favorable fat distribution, as the BFP-increasing allele was also associated with lower waist-to-hip ratio adjusted for body mass index (WHRadjBMI). In eQTL analyses, the BFP-increasing allele was associated with reduced adipose tissue expression of GRB14, which encodes a protein that binds directly to the insulin receptor. Grb14-deficient mice show improved glucose homeostasis and enhanced insulin action through increased phosphorylation of IRS1 in the liver and skeletal muscle (12). An increase in GRB14 expression has been found in adipose tissue from insulin-resistant mice and obese patients with T2D (9).
The BFP-increasing allele at the locus near a mitochondrial membrane protein-encoding TOMM40 gene showed favorable associations with some cardiometabolic traits, including a favorable lipid profile and reduced risk of CAD, while showing unfavorable associations with other traits, including higher WHRadjBMI, VAT, and liver fat and increased risk of T2D. The BFP-increasing variant is in modest linkage disequilibrium with two variants in the nearby APOE gene found associated with HDL-C, TG, and LDL-C (3). Conditional analyses were carried out to distinguish between the TOMM40 and APOE signals by conditioning the TOMM40 variant’s associations with BFP, HDL-C, TG, and LDL-C on the lipid-associated SNPs in APOE, and vice versa. After conditioning, the associations of the TOMM40 locus with BFP, HDL-C and TG were attenuated but remained significant, whereas the association with LDL-C disappeared (33). Conversely, there was no association between the APOE variants and BFP after conditioning on the TOMM40 variant, whereas the HDL-C, TG, and LDL-C associations remained significant after conditioning. These findings suggest that the BFP and lipid associations of the TOMM40 locus are partially independent on the APOE signal (33).
The BFP-increasing allele in the locus near PLA2G6/PICK1 was associated with lower TG levels in men and women, and with lower insulin levels and risk of T2D particularly in men. However, it was also associated with higher VAT in men. This locus harbors several genes that may drive the phenotypic associations. One strong candidate is the adipose tissue-expressed PICK1 gene: mice deficient in the gene show increased body fat and reduced lean mass, reduced TG levels, and increased insulin sensitivity, compensating for impaired insulin secretion (20).
To conclude, in addition to the previously identified IRS1 locus, this study identified three BFP-associated loci that displayed cross-phenotype association signatures that are reminiscent of the MHO phenotype. The BFP-increasing allele of the locus near GRB14/COBLL1 showed an overall favorable metabolic profile, similar to the previously identified IRS1 locus (25), whereas the loci near TOMM40/APOE and PLA2G6/PICK1 showed favorable associations with some cardiometabolic traits and unfavorable or no associations with others. The inconsistency of the BFP-increasing allele at the TOMM40/APOE and PLA2G6/PICK1 loci in inducing positive cardiometabolic outcomes is troubling for the interpretation of the functional role of these genes in metabolic health. Nevertheless, the identification of these three novel loci is a major step forward as it highlights the complexity of mechanisms underlying MHO and MONW, suggesting that there is a need for a more inclusive and less homogeneous interpretation of the underlying mechanisms implicated by loci related to MHO and MONW.
Follow-up of insulin resistance-associated loci reveals a genetic link to MONW.
Instead of approaching the evidence for MHO and MONW by screening adiposity-increasing loci for favorable effects on insulin resistance and other cardiometabolic traits, one can also approach this evidence by taking insulin resistance or other cardiometabolic traits as the starting point and screening for favorable adiposity effects. Indeed, a study published in 2014 examined whether GWAS-identified insulin resistance loci could be associated with a common, “lipodystrophy-like” phenotype (68). In monogenic lipodystrophy, patients have partial or complete lack of subcutaneous fat in combination with severe insulin resistance and dyslipidemia (16, 52, 53).
The associations of 19 known insulin resistance-increasing loci (50) with eight lipodystrophy-related traits were examined using results from published GWAS for BMI, VAT/SAT ratio, HDL-C, TG, hepatic steatosis and liver enzyme alanine transaminase (ALT), and circulating levels of adiponectin and sex-hormone binding globulin (SHBG). Hierarchical clustering was performed to group the 19 loci based on their associations with the eight traits to identify co-regulated and functionally related genes (68). Two clusters were observed. The first cluster contained 11 loci, in or near IRS1, GRB14, ARL15, FAM13A, LYPLAL1, PEPD, PDGFC, RSPO3, PPARG, TET2, and ANKRD55 (Fig. 2). A genetic risk score built using the insulin resistance-increasing alleles at these 11 loci was associated with lower BMI, HDL-C, adiponectin, and SHBG levels; higher TG, ALT, and VAT/SAT ratio; and greater hepatic steatosis. The second cluster contained five loci and was not associated with any of the tested traits, except with higher BMI, and the three remaining insulin resistance loci did not cluster with any other variants. Therefore, we will only focus on the cluster of 11 loci in this review.
The genetic score of the 11 loci was examined for associations with six additional cardiometabolic diseases and disease-related outcomes for which the risk was known to be increased in monogenic lipodystrophy, including T2D, CAD, systolic blood pressure (SBP), diastolic blood pressure (DBP), and carotid intima media thickness (cIMT) and carotid plague. In these analyses, including up to 69,828 individuals of European ancestry, the genetic score was associated with an increased risk of T2D and CAD and higher levels of SBP and DBP, but there was no association with either cIMT or carotid plaque.
As the score of 11 loci showed association with lower BMI, but higher VAT/SAT ratio, and elevated cardiometabolic risk, it was suggested that these loci may play a role in the development of a MONW-like phenotype. A separate meta-analysis of five studies, including up to 18,565 individuals of European descent, replicated the associations of the same 11 loci, except TET2, with lower adiposity but higher risk of T2D, hypertension, and heart disease (50). In 2016, the associations of the 11-locus score were confirmed in a larger sample size, including up to 164,609 individuals of European descent (67), showing an association pattern of the insulin resistance increasing alleles with lower adiposity yet an unfavorable cardiometabolic profile, resembling the MONW phenotype.
The hierarchical clustering of the loci associated with surrogate measures of insulin resistance suggests co-regulated pathways implicated within clusters, which is helpful in elucidating crucial aspects of the intertwined regulatory mechanisms at play. While the interpretation of the 11-locus cluster will require further pathway analyses and experimental model studies to be substantiated, this study opened up the possibility that an interplay between subcutaneous fat storage capacity and insulin resistance may contribute to the MONW phenotype.
Novel insulin resistance loci highlight a role for peripheral adipose storage in MONW.
Multiple novel insulin resistance loci were identified in 2016 through a screening approach that combined published GWAS summary results for fasting insulin, TG, and HDL-C levels, which are hallmarks of insulin resistance. More specifically, GWAS summary results from up to 188,577 individuals of European ancestry were screened for loci showing association with higher fasting insulin (P < 0.005), higher TG (P < 0.005), and lower HDL-C levels (P < 0.005) (32). This approach identified 53 insulin resistance loci that were subsequently combined into a genetic score. The 53-locus score was associated with lower BFP and BMI, lower hip circumference and gynoid and leg fat mass, and higher waist circumference and risk of T2D. The authors proposed that the association with smaller hip circumference and gynoid and leg fat could indicate an impaired ability to expand the peripheral fat compartment (32). Furthermore, as the genetic score was associated with higher levels of ALT and γ-glutamyltransferase, it was hypothesized that the score may be associated with hepatic lipid deposition due to failure of lipid storage in SAT. Analyses in women with familial partial lipodystrophy type 1 (FPLD1) showed a higher burden of the alleles at these 53 loci, suggesting that the polygenic predisposition of these 53 loci may contribute to the FPLD1 phenotype. An overlap was also found with regulatory regions of lipodystrophy in adipose tissues, and pathway analysis and cellular models indicated the involvement of some of the insulin resistance loci with adipocyte gene expression and lipid accumulation.
This study on insulin resistance harvested a large number of loci due to an increase in sample size and a novel analysis strategy focusing on a combination of fasting insulin, triglycerides, and HDL-cholesterol. The association of the identified insulin resistance loci as a genetic score with peripheral fat storage capacity was inferred through concurrent measures of fat distribution and liver markers in the studied population. These epidemiological observations were partly supported by results from analyses of pathway enrichment and from experimental studies in cell cultures. Hence, this study provides strong evidence supporting a link between subcutaneous adipose tissue expandability and insulin resistance as previously suggested by rodent models.
DISCUSSION
With the increasing evidence of a genetic link between increased adiposity and a favorable cardiometabolic profile, or conversely, between decreased adiposity and an unfavorable cardiometabolic profile, a genetic basis of the MHO and MONW phenotypes is substantiated. This review provides a summary of the findings on genetic loci that contribute to MHO and MONW reported so far.
A number of physiological pathways, including adipogenesis, angiogenesis, adipose tissue dysfunction and expandability, adipose inflammation, macrophage infiltration and activation, lipid oxidative capacity, fat distribution, ectopic fat accumulation, and impaired mitochondrial function, have been suggested to contribute to the metabolic heterogeneity seen among obese and normal-weight individuals (8, 15, 35, 36, 40, 63). These findings have been largely based on studies conducted in transgenic rodent models (15) or human cell lines (35, 36), and some in obesity-discordant monozygotic twins (49). The genomic studies published so far have highlighted the involvement of four main mechanisms in MHO and MONW: insulin signaling, insulin resistance, adipogenesis, and fat distribution. The role of insulin signaling and insulin resistance was implicated by the favorable cardiometabolic effects of adiposity-increasing loci that have important roles in the insulin receptor signaling pathway, such as IRS1 and GRB14/COBLL1 (25, 33), as well as findings on the relationship between genetic predisposition to insulin resistance and decreased adiposity (32, 50, 68). The link between insulin resistance and decreased adiposity was hypothesized to reflect a common, low-penetrant “lipodystrophy-like” syndrome that may share underlying mechanisms with monogenic lipodystrophies (32, 50, 67, 68). The role of fat distribution was implicated by the association of aggregate scores of insulin resistance-increasing loci with reduced peripheral SAT (25, 32, 33, 50, 67, 68). The importance of adipogenesis was shown in pathway analyses and knockdown experiments for some of the implicated genes, where adipocyte gene expression or lipid storage capacity was affected (32, 38, 62).
One hypothesis proposed to underlie the link between genetic predisposition to insulin resistance and MONW in the studies we reviewed is that insulin resistance-increasing loci may be involved in determining an individual threshold for SAT expandability (63). As subcutaneous fat storage space expires, lipid starts to accumulate ectopically in nonadipose tissues, such as the liver and skeletal muscle. This may disrupt insulin signaling in liver and muscle, leading to whole-body insulin resistance and dyslipidemia (25, 48) through a lipotoxic mechanism (63). Adipogenesis allows healthy expansion of SAT, preventing ectopic lipid deposition (25, 32, 33, 50, 67, 68). These intertwined mechanisms may play an important role in the development of MHO and MONW phenotypes. However, causality between subcutaneous fat storage capacity and insulin resistance could not be deduced from the studies performed so far (32, 50, 68). Furthermore, we noted that some loci implicated in MHO are associated with decreased WHRadjBMI, while others are associated with increased WHRadjBMI (Fig. 2). This suggests the presence of other mechanisms than fat distribution and subcutaneous fat expansion in mediating the paradoxical link between increased adiposity and a favorable cardiometabolic profile. Some identified SNPs in linkage disequilibrium with other SNPs in nearby genes might also exhibit heterogeneous effects, as seen for the locus near TOMM40/APOE (33), or show pleiotropic effects on adiposity and cardiometabolic traits, which could provide an alternative explanation for the observed results. Extended gene discovery studies focusing on specific combinations of adiposity and cardiometabolic traits, along with analyses of the enrichment of loci representing such association signatures in biological pathways, will be needed to unravel the mechanisms underlying the heterogeneous association signatures of loci implicated in MHO and MONW. Follow-up analyses using more refined human phenotypes and studies in experimental models will be required to validate such novel mechanistic insights.
Understanding the underlying mechanisms that lead to MHO and MONW is important to explain why not all obese individuals develop metabolic impairments and to find new ways to prevent development of cardiometabolic disease among those who are already obese. Identifying genetic determinants of MHO and MONW may open up new avenues for drug development and for defining public health action points. For example, the thiazolidinedione class of antidiabetic drugs has been shown to be capable of stimulating adipose tissue differentiation by activating PPARG, which leads to increased body weight but improved insulin sensitivity at the same time (41). If individuals can be identified with a genetic predisposition to either MHO or MONW, tailored medical intervention such as these could be selectively used for prevention and treatment.
GRANTS
This work was supported by Danish Council for Independent Research (DFF) Grant 6110-00183 and Novo Nordisk Foundation Grant NNF17OC0026848.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
L.O.H. prepared figures; L.O.H. and T.O.K. drafted manuscript; L.O.H., R.J.F.L., and T.O.K. edited and revised manuscript; L.O.H., R.J.F.L., and T.O.K. approved final version of manuscript; R.J.F.L. and T.O.K. conceived and designed research.
REFERENCES
- 1.Aguilar-Salinas CA, García EG, Robles L, Riaño D, Ruiz-Gomez DG, García-Ulloa AC, Melgarejo MA, Zamora M, Guillen-Pineda LE, Mehta R, Canizales-Quinteros S, Tusie Luna MT, Gómez-Pérez FJ. High adiponectin concentrations are associated with the metabolically healthy obese phenotype. J Clin Endocrinol Metab 93: 4075–4079, 2008. doi: 10.1210/jc.2007-2724. [DOI] [PubMed] [Google Scholar]
- 2.Andres R. Effect of obesity on total mortality. Int J Obes 4: 381–386, 1980. [PubMed] [Google Scholar]
- 3.Araki E, Lipes MA, Patti ME, Brüning JC, Haag B III, Johnson RS, Kahn CR. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature 372: 186–190, 1994. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- 4.Arnlöv J, Sundström J, Ingelsson E, Lind L. Impact of BMI and the metabolic syndrome on the risk of diabetes in middle-aged men. Diabetes Care 34: 61–65, 2011. doi: 10.2337/dc10-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aung K, Lorenzo C, Hinojosa MA, Haffner SM. Risk of developing diabetes and cardiovascular disease in metabolically unhealthy normal-weight and metabolically healthy obese individuals. J Clin Endocrinol Metab 99: 462–468, 2014. doi: 10.1210/jc.2013-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blüher M. The distinction of metabolically ‘healthy’ from ‘unhealthy’ obese individuals. Curr Opin Lipidol 21: 38–43, 2010. doi: 10.1097/MOL.0b013e3283346ccc. [DOI] [PubMed] [Google Scholar]
- 7.Bradshaw PT, Monda KL, Stevens J. Metabolic syndrome in healthy obese, overweight, and normal weight individuals: the Atherosclerosis Risk in Communities Study. Obesity (Silver Spring) 21: 203–209, 2013. doi: 10.1002/oby.20248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brochu M, Tchernof A, Dionne IJ, Sites CK, Eltabbakh GH, Sims EA, Poehlman ET. What are the physical characteristics associated with a normal metabolic profile despite a high level of obesity in postmenopausal women? J Clin Endocrinol Metab 86: 1020–1025, 2001. doi: 10.1210/jcem.86.3.7365. [DOI] [PubMed] [Google Scholar]
- 9.Cariou B, Capitaine N, Le Marcis V, Vega N, Béréziat V, Kergoat M, Laville M, Girard J, Vidal H, Burnol AF. Increased adipose tissue expression of Grb14 in several models of insulin resistance. FASEB J 18: 965–967, 2004. doi: 10.1096/fj.03-0824fje. [DOI] [PubMed] [Google Scholar]
- 10.Chambers JC, Elliott P, Zabaneh D, Zhang W, Li Y, Froguel P, Balding D, Scott J, Kooner JS. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet 40: 716–718, 2008. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 11.Conus F, Allison DB, Rabasa-Lhoret R, St-Onge M, St-Pierre DH, Tremblay-Lebeau A, Poehlman ET. Metabolic and behavioral characteristics of metabolically obese but normal-weight women. J Clin Endocrinol Metab 89: 5013–5020, 2004. doi: 10.1210/jc.2004-0265. [DOI] [PubMed] [Google Scholar]
- 12.Cooney GJ, Lyons RJ, Crew AJ, Jensen TE, Molero JC, Mitchell CJ, Biden TJ, Ormandy CJ, James DE, Daly RJ. Improved glucose homeostasis and enhanced insulin signalling in Grb14-deficient mice. EMBO J 23: 582–593, 2004. doi: 10.1038/sj.emboj.7600082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckel N, Mühlenbruch K, Meidtner K, Boeing H, Stefan N, Schulze MB. Characterization of metabolically unhealthy normal-weight individuals: Risk factors and their associations with type 2 diabetes. Metabolism 64: 862–871, 2015. doi: 10.1016/j.metabol.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316: 889–894, 2007. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gavrilova O, Marcus-Samuels B, Graham D, Kim JK, Shulman GI, Castle AL, Vinson C, Eckhaus M, Reitman ML. Surgical implantation of adipose tissue reverses diabetes in lipoatrophic mice. J Clin Invest 105: 271–278, 2000. doi: 10.1172/JCI7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George S, Rochford JJ, Wolfrum C, Gray SL, Schinner S, Wilson JC, Soos MA, Murgatroyd PR, Williams RM, Acerini CL, Dunger DB, Barford D, Umpleby AM, Wareham NJ, Davies HA, Schafer AJ, Stoffel M, O’Rahilly S, Barroso I. A family with severe insulin resistance and diabetes due to a mutation in AKT2. Science 304: 1325–1328, 2004. doi: 10.1126/science.1096706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hadaegh F, Bozorgmanesh M, Safarkhani M, Khalili D, Azizi F. “Predictability of body mass index for diabetes: affected by the presence of metabolic syndrome?”. BMC Public Health 11: 383, 2011. doi: 10.1186/1471-2458-11-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamer M, Stamatakis E. Metabolically healthy obesity and risk of all-cause and cardiovascular disease mortality. J Clin Endocrinol Metab 97: 2482–2488, 2012. doi: 10.1210/jc.2011-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinnouho GM, Czernichow S, Dugravot A, Nabi H, Brunner EJ, Kivimaki M, Singh-Manoux A. Metabolically healthy obesity and the risk of cardiovascular disease and type 2 diabetes: the Whitehall II cohort study. Eur Heart J 36: 551–559, 2015. doi: 10.1093/eurheartj/ehu123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holst B, Madsen KL, Jansen AM, Jin C, Rickhag M, Lund VK, Jensen M, Bhatia V, Sørensen G, Madsen AN, Xue Z, Møller SK, Woldbye D, Qvortrup K, Huganir R, Stamou D, Kjærulff O, Gether U. PICK1 deficiency impairs secretory vesicle biogenesis and leads to growth retardation and decreased glucose tolerance. PLoS Biol 11: e1001542, 2013. doi: 10.1371/journal.pbio.1001542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iacobellis G, Ribaudo MC, Zappaterreno A, Iannucci CV, Leonetti F. Prevalence of uncomplicated obesity in an Italian obese population. Obes Res 13: 1116–1122, 2005. doi: 10.1038/oby.2005.130. [DOI] [PubMed] [Google Scholar]
- 22.Karelis AD. Metabolically healthy but obese individuals. Lancet 372: 1281–1283, 2008. doi: 10.1016/S0140-6736(08)61531-7. [DOI] [PubMed] [Google Scholar]
- 23.Karelis AD, Brochu M, Rabasa-Lhoret R. Can we identify metabolically healthy but obese individuals (MHO)? Diabetes Metab 30: 569–572, 2004. doi: 10.1016/S1262-3636(07)70156-8. [DOI] [PubMed] [Google Scholar]
- 24.Karelis AD, Faraj M, Bastard JP, St-Pierre DH, Brochu M, Prud’homme D, Rabasa-Lhoret R. The metabolically healthy but obese individual presents a favorable inflammation profile. J Clin Endocrinol Metab 90: 4145–4150, 2005. doi: 10.1210/jc.2005-0482. [DOI] [PubMed] [Google Scholar]
- 25.Kilpeläinen TO, Zillikens MC, Stančákova A, Finucane FM, Ried JS, Langenberg C, Zhang W, Beckmann JS, Luan J, Vandenput L, Styrkarsdottir U, Zhou Y, Smith AV, Zhao JH, Amin N, Vedantam S, Shin SY, Haritunians T, Fu M, Feitosa MF, Kumari M, Halldorsson BV, Tikkanen E, Mangino M, Hayward C, Song C, Arnold AM, Aulchenko YS, Oostra BA, Campbell H, Cupples LA, Davis KE, Döring A, Eiriksdottir G, Estrada K, Fernández-Real JM, Garcia M, Gieger C, Glazer NL, Guiducci C, Hofman A, Humphries SE, Isomaa B, Jacobs LC, Jula A, Karasik D, Karlsson MK, Khaw KT, Kim LJ, Kivimäki M, Klopp N, Kühnel B, Kuusisto J, Liu Y, Ljunggren O, Lorentzon M, Luben RN, McKnight B, Mellström D, Mitchell BD, Mooser V, Moreno JM, Männistö S, O’Connell JR, Pascoe L, Peltonen L, Peral B, Perola M, Psaty BM, Salomaa V, Savage DB, Semple RK, Skaric-Juric T, Sigurdsson G, Song KS, Spector TD, Syvänen AC, Talmud PJ, Thorleifsson G, Thorsteinsdottir U, Uitterlinden AG, van Duijn CM, Vidal-Puig A, Wild SH, Wright AF, Clegg DJ, Schadt E, Wilson JF, Rudan I, Ripatti S, Borecki IB, Shuldiner AR, Ingelsson E, Jansson JO, Kaplan RC, Gudnason V, Harris TB, Groop L, Kiel DP, Rivadeneira F, Walker M, Barroso I, Vollenweider P, Waeber G, Chambers JC, Kooner JS, Soranzo N, Hirschhorn JN, Stefansson K, Wichmann HE, Ohlsson C, O’Rahilly S, Wareham NJ, Speliotes EK, Fox CS, Laakso M, Loos RJ. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet 43: 753–760, 2011. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JY, van de Wall E, Laplante M, Azzara A, Trujillo ME, Hofmann SM, Schraw T, Durand JL, Li H, Li G, Jelicks LA, Mehler MF, Hui DY, Deshaies Y, Shulman GI, Schwartz GJ, Scherer PE. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J Clin Invest 117: 2621–2637, 2007. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klok MD, Jakobsdottir S, Drent ML. The role of leptin and ghrelin in the regulation of food intake and body weight in humans: a review. Obes Rev 8: 21–34, 2007. doi: 10.1111/j.1467-789X.2006.00270.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee K. Metabolically obese but normal weight (MONW) and metabolically healthy but obese (MHO) phenotypes in Koreans: characteristics and health behaviors. Asia Pac J Clin Nutr 18: 280–284, 2009. [PubMed] [Google Scholar]
- 29.Li L, Yin J, Cheng H, Wang Y, Gao S, Li M, Grant SF, Li C, Mi J, Li M. Identification of genetic and environmental factors predicting metabolically healthy obesity in children: data from the BCAMS Study. J Clin Endocrinol Metab 101: 1816–1825, 2016. doi: 10.1210/jc.2015-3760. [DOI] [PubMed] [Google Scholar]
- 30.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan J, Mägi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Zhao JH, Zhao W, Chen J, Fehrmann R, Hedman AK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg-Gresham JL, Buyske S, Demirkan A, Deng G, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Leach IM, Medina-Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stančáková A, Strawbridge RJ, Sung YJ, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Isaacs A, Albrecht E, Ärnlöv J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Blüher M, Böhringer S, Bonnycastle LL, Böttcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Chen YI, Clarke R, Daw EW, de Craen AJM, Delgado G, Dimitriou M, Doney ASF, Eklund N, Estrada K, Eury E, Folkersen L, Fraser RM, Garcia ME, Geller F, Giedraitis V, Gigante B, Go AS, Golay A, Goodall AH, Gordon SD, Gorski M, Grabe HJ, Grallert H, Grammer TB, Gräßler J, Grönberg H, Groves CJ, Gusto G, Haessler J, Hall P, Haller T, Hallmans G, Hartman CA, Hassinen M, Hayward C, Heard-Costa NL, Helmer Q, Hengstenberg C, Holmen O, Hottenga JJ, James AL, Jeff JM, Johansson Å, Jolley J, Juliusdottir T, Kinnunen L, Koenig W, Koskenvuo M, Kratzer W, Laitinen J, Lamina C, Leander K, Lee NR, Lichtner P, Lind L, Lindström J, Lo KS, Lobbens S, Lorbeer R, Lu Y, Mach F, Magnusson PKE, Mahajan A, McArdle WL, McLachlan S, Menni C, Merger S, Mihailov E, Milani L, Moayyeri A, Monda KL, Morken MA, Mulas A, Müller G, Müller-Nurasyid M, Musk AW, Nagaraja R, Nöthen MM, Nolte IM, Pilz S, Rayner NW, Renstrom F, Rettig R, Ried JS, Ripke S, Robertson NR, Rose LM, Sanna S, Scharnagl H, Scholtens S, Schumacher FR, Scott WR, Seufferlein T, Shi J, Smith AV, Smolonska J, Stanton AV, Steinthorsdottir V, Stirrups K, Stringham HM, Sundström J, Swertz MA, Swift AJ, Syvänen AC, Tan ST, Tayo BO, Thorand B, Thorleifsson G, Tyrer JP, Uh HW, Vandenput L, Verhulst FC, Vermeulen SH, Verweij N, Vonk JM, Waite LL, Warren HR, Waterworth D, Weedon MN, Wilkens LR, Willenborg C, Wilsgaard T, Wojczynski MK, Wong A, Wright AF, Zhang Q, LifeLines Cohort Study, Brennan EP, Choi M, Dastani Z, Drong AW, Eriksson P, Franco-Cereceda A, Gådin JR, Gharavi AG, Goddard ME, Handsaker RE, Huang J, Karpe F, Kathiresan S, Keildson S, Kiryluk K, Kubo M, Lee JY, Liang L, Lifton RP, Ma B, McCarroll SA, McKnight AJ, Min JL, Moffatt MF, Montgomery GW, Murabito JM, Nicholson G, Nyholt DR, Okada Y, Perry JRB, Dorajoo R, Reinmaa E, Salem RM, Sandholm N, Scott RA, Stolk L, Takahashi A, Tanaka T, van ’t Hooft FM, Vinkhuyzen AAE, Westra HJ, Zheng W, Zondervan KT, StudyADIPOGen Consortium, AGEN-BMI Working Group, CARDIOGRAMplusC4D Consortium, CKDGen Consortium, GLGC, ICBP, MAGIC Investigators, MuTHER Consortium, MIGen Consortium, PAGE Consortium, ReproGen Consortium, GENIE Consortium; International Endogene Consortium, Heath AC, Arveiler D, Bakker SJL, Beilby J, Bergman RN, Blangero J, Bovet P, Campbell H, Caulfield MJ, Cesana G, Chakravarti A, Chasman DI, Chines PS, Collins FS, Crawford DC, Cupples LA, Cusi D, Danesh J, de Faire U, den Ruijter HM, Dominiczak AF, Erbel R, Erdmann J, Eriksson JG, Farrall M, Felix SB, Ferrannini E, Ferrières J, Ford I, Forouhi NG, Forrester T, Franco OH, Gansevoort RT, Gejman PV, Gieger C, Gottesman O, Gudnason V, Gyllensten U, Hall AS, Harris TB, Hattersley AT, Hicks AA, Hindorff LA, Hingorani AD, Hofman A, Homuth G, Hovingh GK, Humphries SE, Hunt SC, Hyppönen E, Illig T, Jacobs KB, Jarvelin MR, Jöckel KH, Johansen B, Jousilahti P, Jukema JW, Jula AM, Kaprio J, Kastelein JJP, Keinanen-Kiukaanniemi SM, Kiemeney LA, Knekt P, Kooner JS, Kooperberg C, Kovacs P, Kraja AT, Kumari M, Kuusisto J, Lakka TA, Langenberg C, Marchand LL, Lehtimäki T, Lyssenko V, Männistö S, Marette A, Matise TC, McKenzie CA, McKnight B, Moll FL, Morris AD, Morris AP, Murray JC, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, Madden PAF, Pasterkamp G, Peden JF, Peters A, Postma DS, Pramstaller PP, Price JF, Qi L, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ridker PM, Rioux JD, Ritchie MD, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schunkert H, Schwarz PEH, Sever P, Shuldiner AR, Sinisalo J, Stolk RP, Strauch K, Tönjes A, Trégouët DA, Tremblay A, Tremoli E, Virtamo J, Vohl MC, Völker U, Waeber G, Willemsen G, Witteman JC, Zillikens MC, Adair LS, Amouyel P, Asselbergs FW, Assimes TL, Bochud M, Boehm BO, Boerwinkle E, Bornstein SR, Bottinger EP, Bouchard C, Cauchi S, Chambers JC, Chanock SJ, Cooper RS, de Bakker PIW, Dedoussis G, Ferrucci L, Franks PW, Froguel P, Groop LC, Haiman CA, Hamsten A, Hui J, Hunter DJ, Hveem K, Kaplan RC, Kivimaki M, Kuh D, Laakso M, Liu Y, Martin NG, März W, Melbye M, Metspalu A, Moebus S, Munroe PB, Njølstad I, Oostra BA, Palmer CNA, Pedersen NL, Perola M, Pérusse L, Peters U, Power C, Quertermous T, Rauramaa R, Rivadeneira F, Saaristo TE, Saleheen D, Sattar N, Schadt EE, Schlessinger D, Slagboom PE, Snieder H, Spector TD, Thorsteinsdottir U, Stumvoll M, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Walker M, Wallaschofski H, Wareham NJ, Watkins H, Weir DR, Wichmann HE, Wilson JF, Zanen P, Borecki IB, Deloukas P, Fox CS, Heid IM, O’Connell JR, Strachan DP, Stefansson K, van Duijn CM, Abecasis GR, Franke L, Frayling TM, McCarthy MI, Visscher PM, Scherag A, Willer CJ, Boehnke M, Mohlke KL, Lindgren CM, Beckmann JS, Barroso I, North KE, Ingelsson E, Hirschhorn JN, Loos RJF, Speliotes EK. Genetic studies of body mass index yield new insights for obesity biology. Nature 518: 197–206, 2015. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M, Freathy RM, Attwood AP, Beckmann JS, Berndt SI, Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial, Jacobs KB, Chanock SJ, Hayes RB, Bergmann S, Bennett AJ, Bingham SA, Bochud M, Brown M, Cauchi S, Connell JM, Cooper C, Smith GD, Day I, Dina C, De S, Dermitzakis ET, Doney AS, Elliott KS, Elliott P, Evans DM, Sadaf Farooqi I, Froguel P, Ghori J, Groves CJ, Gwilliam R, Hadley D, Hall AS, Hattersley AT, Hebebrand J, Heid IM, KORA, Lamina C, Gieger C, Illig T, Meitinger T, Wichmann HE, Herrera B, Hinney A, Hunt SE, Jarvelin MR, Johnson T, Jolley JD, Karpe F, Keniry A, Khaw KT, Luben RN, Mangino M, Marchini J, McArdle WL, McGinnis R, Meyre D, Munroe PB, Morris AD, Ness AR, Neville MJ, Nica AC, Ong KK, O’Rahilly S, Owen KR, Palmer CN, Papadakis K, Potter S, Pouta A, Qi L, Nurses’ Health Study, Randall JC, Rayner NW, Ring SM, Sandhu MS, Scherag A, Sims MA, Song K, Soranzo N, Speliotes EK, Diabetes Genetics Initiative, Syddall HE, Teichmann SA, Timpson NJ, Tobias JH, Uda M, SardiNIA Study, Vogel CI, Wallace C, Waterworth DM, Weedon MN, Wellcome Trust Case Control Consortium; Willer CJ, FUSION, Wraight, Yuan X, Zeggini E, Hirschhorn JN, Strachan DP, Ouwehand WH, Caulfield MJ, Samani NJ, Frayling TM, Vollenweider P, Waeber G, Mooser V, Deloukas P, McCarthy MI, Wareham NJ, Barroso I, Jacobs KB, Chanock SJ, Hayes RB, Lamina C, Gieger C, Illig T, Meitinger T, Wichmann HE, Kraft P, Hankinson SE, Hunter DJ, Hu FB, Lyon HN, Voight BF, Ridderstrale M, Groop L, Scheet P, Sanna S, Abecasis GR, Albai G, Nagaraja R, Schlessinger D, Jackson AU, Tuomilehto J, Collins FS, Boehnke M, Mohlke KL. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet 40: 768–775, 2008. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lotta LA, Gulati P, Day FR, Payne F, Ongen H, van de Bunt M, Gaulton KJ, Eicher JD, Sharp SJ, Luan J, De Lucia Rolfe E, Stewart ID, Wheeler E, Willems SM, Adams C, Yaghootkar H, Forouhi NG, Khaw KT, Johnson AD, Semple RK, Frayling T, Perry JR, Dermitzakis E, McCarthy MI, Barroso I, Wareham NJ, Savage DB, Langenberg C, O’Rahilly S, Scott RA; EPIC-InterAct Consortium; Cambridge FPLD1 Consortium . Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat Genet 49: 17–26, 2017. doi: 10.1038/ng.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, Day FR, Gustafsson S, Buchkovich ML, Na J, Bataille V, Cousminer DL, Dastani Z, Drong AW, Esko T, Evans DM, Falchi M, Feitosa MF, Ferreira T, Hedman AK, Haring R, Hysi PG, Iles MM, Justice AE, Kanoni S, Lagou V, Li R, Li X, Locke A, Lu C, Mägi R, Perry JR, Pers TH, Qi Q, Sanna M, Schmidt EM, Scott WR, Shungin D, Teumer A, Vinkhuyzen AA, Walker RW, Westra HJ, Zhang M, Zhang W, Zhao JH, Zhu Z, Afzal U, Ahluwalia TS, Bakker SJ, Bellis C, Bonnefond A, Borodulin K, Buchman AS, Cederholm T, Choh AC, Choi HJ, Curran JE, de Groot LC, De Jager PL, Dhonukshe-Rutten RA, Enneman AW, Eury E, Evans DS, Forsen T, Friedrich N, Fumeron F, Garcia ME, Gärtner S, Han BG, Havulinna AS, Hayward C, Hernandez D, Hillege H, Ittermann T, Kent JW, Kolcic I, Laatikainen T, Lahti J, Mateo Leach I, Lee CG, Lee JY, Liu T, Liu Y, Lobbens S, Loh M, Lyytikäinen LP, Medina-Gomez C, Michaëlsson K, Nalls MA, Nielson CM, Oozageer L, Pascoe L, Paternoster L, Polašek O, Ripatti S, Sarzynski MA, Shin CS, Narančić NS, Spira D, Srikanth P, Steinhagen-Thiessen E, Sung YJ, Swart KM, Taittonen L, Tanaka T, Tikkanen E, van der Velde N, van Schoor NM, Verweij N, Wright AF, Yu L, Zmuda JM, Eklund N, Forrester T, Grarup N, Jackson AU, Kristiansson K, Kuulasmaa T, Kuusisto J, Lichtner P, Luan J, Mahajan A, Männistö S, Palmer CD, Ried JS, Scott RA, Stancáková A, Wagner PJ, Demirkan A, Döring A, Gudnason V, Kiel DP, Kühnel B, Mangino M, Mcknight B, Menni C, O’Connell JR, Oostra BA, Shuldiner AR, Song K, Vandenput L, van Duijn CM, Vollenweider P, White CC, Boehnke M, Boettcher Y, Cooper RS, Forouhi NG, Gieger C, Grallert H, Hingorani A, Jørgensen T, Jousilahti P, Kivimaki M, Kumari M, Laakso M, Langenberg C, Linneberg A, Luke A, Mckenzie CA, Palotie A, Pedersen O, Peters A, Strauch K, Tayo BO, Wareham NJ, Bennett DA, Bertram L, Blangero J, Blüher M, Bouchard C, Campbell H, Cho NH, Cummings SR, Czerwinski SA, Demuth I, Eckardt R, Eriksson JG, Ferrucci L, Franco OH, Froguel P, Gansevoort RT, Hansen T, Harris TB, Hastie N, Heliövaara M, Hofman A, Jordan JM, Jula A, Kähönen M, Kajantie E, Knekt PB, Koskinen S, Kovacs P, Lehtimäki T, Lind L, Liu Y, Orwoll ES, Osmond C, Perola M, Pérusse L, Raitakari OT, Rankinen T, Rao DC, Rice TK, Rivadeneira F, Rudan I, Salomaa V, Sørensen TI, Stumvoll M, Tönjes A, Towne B, Tranah GJ, Tremblay A, Uitterlinden AG, van der Harst P, Vartiainen E, Viikari JS, Vitart V, Vohl MC, Völzke H, Walker M, Wallaschofski H, Wild S, Wilson JF, Yengo L, Bishop DT, Borecki IB, Chambers JC, Cupples LA, Dehghan A, Deloukas P, Fatemifar G, Fox C, Furey TS, Franke L, Han J, Hunter DJ, Karjalainen J, Karpe F, Kaplan RC, Kooner JS, McCarthy MI, Murabito JM, Morris AP, Bishop JA, North KE, Ohlsson C, Ong KK, Prokopenko I, Richards JB, Schadt EE, Spector TD, Widén E, Willer CJ, Yang J, Ingelsson E, Mohlke KL, Hirschhorn JN, Pospisilik JA, Zillikens MC, Lindgren C, Kilpeläinen TO, Loos RJ. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat Commun 7: 10495, 2016. doi: 10.1038/ncomms10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, Rybin D, Liu CT, Bielak LF, Prokopenko I, Amin N, Barnes D, Cadby G, Hottenga JJ, Ingelsson E, Jackson AU, Johnson T, Kanoni S, Ladenvall C, Lagou V, Lahti J, Lecoeur C, Liu Y, Martinez-Larrad MT, Montasser ME, Navarro P, Perry JR, Rasmussen-Torvik LJ, Salo P, Sattar N, Shungin D, Strawbridge RJ, Tanaka T, van Duijn CM, An P, de Andrade M, Andrews JS, Aspelund T, Atalay M, Aulchenko Y, Balkau B, Bandinelli S, Beckmann JS, Beilby JP, Bellis C, Bergman RN, Blangero J, Boban M, Boehnke M, Boerwinkle E, Bonnycastle LL, Boomsma DI, Borecki IB, Böttcher Y, Bouchard C, Brunner E, Budimir D, Campbell H, Carlson O, Chines PS, Clarke R, Collins FS, Corbatón-Anchuelo A, Couper D, de Faire U, Dedoussis GV, Deloukas P, Dimitriou M, Egan JM, Eiriksdottir G, Erdos MR, Eriksson JG, Eury E, Ferrucci L, Ford I, Forouhi NG, Fox CS, Franzosi MG, Franks PW, Frayling TM, Froguel P, Galan P, de Geus E, Gigante B, Glazer NL, Goel A, Groop L, Gudnason V, Hallmans G, Hamsten A, Hansson O, Harris TB, Hayward C, Heath S, Hercberg S, Hicks AA, Hingorani A, Hofman A, Hui J, Hung J, Jarvelin MR, Jhun MA, Johnson PC, Jukema JW, Jula A, Kao WH, Kaprio J, Kardia SL, Keinanen-Kiukaanniemi S, Kivimaki M, Kolcic I, Kovacs P, Kumari M, Kuusisto J, Kyvik KO, Laakso M, Lakka T, Lannfelt L, Lathrop GM, Launer LJ, Leander K, Li G, Lind L, Lindstrom J, Lobbens S, Loos RJ, Luan J, Lyssenko V, Mägi R, Magnusson PK, Marmot M, Meneton P, Mohlke KL, Mooser V, Morken MA, Miljkovic I, Narisu N, O’Connell J, Ong KK, Oostra BA, Palmer LJ, Palotie A, Pankow JS, Peden JF, Pedersen NL, Pehlic M, Peltonen L, Penninx B, Pericic M, Perola M, Perusse L, Peyser PA, Polasek O, Pramstaller PP, Province MA, Räikkönen K, Rauramaa R, Rehnberg E, Rice K, Rotter JI, Rudan I, Ruokonen A, Saaristo T, Sabater-Lleal M, Salomaa V, Savage DB, Saxena R, Schwarz P, Seedorf U, Sennblad B, Serrano-Rios M, Shuldiner AR, Sijbrands EJ, Siscovick DS, Smit JH, Small KS, Smith NL, Smith AV, Stančáková A, Stirrups K, Stumvoll M, Sun YV, Swift AJ, Tönjes A, Tuomilehto J, Trompet S, Uitterlinden AG, Uusitupa M, Vikström M, Vitart V, Vohl MC, Voight BF, Vollenweider P, Waeber G, Waterworth DM, Watkins H, Wheeler E, Widen E, Wild SH, Willems SM, Willemsen G, Wilson JF, Witteman JC, Wright AF, Yaghootkar H, Zelenika D, Zemunik T, Zgaga L, Wareham NJ, McCarthy MI, Barroso I, Watanabe RM, Florez JC, Dupuis J, Meigs JB, Langenberg C; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Multiple Tissue Human Expression Resource (MUTHER) Consortium . A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 44: 659–669, 2012. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mantzaris MD, Tsianos EV, Galaris D. Interruption of triacylglycerol synthesis in the endoplasmic reticulum is the initiating event for saturated fatty acid-induced lipotoxicity in liver cells. FEBS J 278: 519–530, 2011. doi: 10.1111/j.1742-4658.2010.07972.x. [DOI] [PubMed] [Google Scholar]
- 36.McLaughlin T, Deng A, Yee G, Lamendola C, Reaven G, Tsao PS, Cushman SW, Sherman A. Inflammation in subcutaneous adipose tissue: relationship to adipose cell size. Diabetologia 53: 369–377, 2010. doi: 10.1007/s00125-009-1496-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meigs JB, Wilson PW, Fox CS, Vasan RS, Nathan DM, Sullivan LM, D’Agostino RB. Body mass index, metabolic syndrome, and risk of type 2 diabetes or cardiovascular disease. J Clin Endocrinol Metab 91: 2906–2912, 2006. doi: 10.1210/jc.2006-0594. [DOI] [PubMed] [Google Scholar]
- 38.Miki H, Yamauchi T, Suzuki R, Komeda K, Tsuchida A, Kubota N, Terauchi Y, Kamon J, Kaburagi Y, Matsui J, Akanuma Y, Nagai R, Kimura S, Tobe K, Kadowaki T. Essential role of insulin receptor substrate 1 (IRS-1) and IRS-2 in adipocyte differentiation. Mol Cell Biol 21: 2521–2532, 2001. doi: 10.1128/MCB.21.7.2521-2532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J, Lindgren CM, Müller-Nurasyid M, Pechlivanis S, Rayner NW, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S, Johnson AD, Dimas AS, Loos RJ, Vedantam S, Chen H, Florez JC, Fox C, Liu CT, Rybin D, Couper DJ, Kao WH, Li M, Cornelis MC, Kraft P, Sun Q, van Dam RM, Stringham HM, Chines PS, Fischer K, Fontanillas P, Holmen OL, Hunt SE, Jackson AU, Kong A, Lawrence R, Meyer J, Perry JR, Platou CG, Potter S, Rehnberg E, Robertson N, Sivapalaratnam S, Stančáková A, Stirrups K, Thorleifsson G, Tikkanen E, Wood AR, Almgren P, Atalay M, Benediktsson R, Bonnycastle LL, Burtt N, Carey J, Charpentier G, Crenshaw AT, Doney AS, Dorkhan M, Edkins S, Emilsson V, Eury E, Forsen T, Gertow K, Gigante B, Grant GB, Groves CJ, Guiducci C, Herder C, Hreidarsson AB, Hui J, James A, Jonsson A, Rathmann W, Klopp N, Kravic J, Krjutškov K, Langford C, Leander K, Lindholm E, Lobbens S, Männistö S, Mirza G, Mühleisen TW, Musk B, Parkin M, Rallidis L, Saramies J, Sennblad B, Shah S, Sigurðsson G, Silveira A, Steinbach G, Thorand B, Trakalo J, Veglia F, Wennauer R, Winckler W, Zabaneh D, Campbell H, van Duijn C, Uitterlinden AG, Hofman A, Sijbrands E, Abecasis GR, Owen KR, Zeggini E, Trip MD, Forouhi NG, Syvänen AC, Eriksson JG, Peltonen L, Nöthen MM, Balkau B, Palmer CN, Lyssenko V, Tuomi T, Isomaa B, Hunter DJ, Qi L, Wellcome Trust Case Control Consortium, Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators, Genetic Investigation of ANthropometric Traits (GIANT) Consortium, Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Shuldiner AR, Roden M, Barroso I, Wilsgaard T, Beilby J, Hovingh K, Price JF, Wilson JF, Rauramaa R, Lakka TA, Lind L, Dedoussis G, Njølstad I, Pedersen NL, Khaw KT, Wareham NJ, Keinanen-Kiukaanniemi SM, Saaristo TE, Korpi-Hyövälti E, Saltevo J, Laakso M, Kuusisto J, Metspalu A, Collins FS, Mohlke KL, Bergman RN, Tuomilehto J, Boehm BO, Gieger C, Hveem K, Cauchi S, Froguel P, Baldassarre D, Tremoli E, Humphries SE, Saleheen D, Danesh J, Ingelsson E, Ripatti S, Salomaa V, Erbel R, Jöckel KH, Moebus S, Peters A, Illig T, de Faire U, Hamsten A, Morris AD, Donnelly PJ, Frayling TM, Hattersley AT, Boerwinkle E, Melander O, Kathiresan S, Nilsson PM, Deloukas P, Thorsteinsdottir U, Groop LC, Stefansson K, Hu F, Pankow JS, Dupuis J, Meigs JB, Altshuler D, Boehnke M, McCarthy MI,. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 44: 981–990, 2012. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naukkarinen J, Heinonen S, Hakkarainen A, Lundbom J, Vuolteenaho K, Saarinen L, Hautaniemi S, Rodriguez A, Frühbeck G, Pajunen P, Hyötyläinen T, Orešič M, Moilanen E, Suomalainen A, Lundbom N, Kaprio J, Rissanen A, Pietiläinen KH. Characterising metabolically healthy obesity in weight-discordant monozygotic twins. Diabetologia 57: 167–176, 2014. doi: 10.1007/s00125-013-3066-y. [DOI] [PubMed] [Google Scholar]
- 41.Nichols GA, Gomez-Caminero A. Weight changes following the initiation of new anti-hyperglycaemic therapies. Diabetes Obes Metab 9: 96–102, 2007. doi: 10.1111/j.1463-1326.2006.00580.x. [DOI] [PubMed] [Google Scholar]
- 42.Phillips CM, Dillon C, Harrington JM, McCarthy VJ, Kearney PM, Fitzgerald AP, Perry IJ. Defining metabolically healthy obesity: role of dietary and lifestyle factors. PLoS One 8: e76188, 2013. doi: 10.1371/journal.pone.0076188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popkin BM, Adair LS, Ng SW. Global nutrition transition and the pandemic of obesity in developing countries. Nutr Rev 70: 3–21, 2012. doi: 10.1111/j.1753-4887.2011.00456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Primeau V, Coderre L, Karelis AD, Brochu M, Lavoie ME, Messier V, Sladek R, Rabasa-Lhoret R. Characterizing the profile of obese patients who are metabolically healthy. Int J Obes 35: 971–981, 2011. doi: 10.1038/ijo.2010.216. [DOI] [PubMed] [Google Scholar]
- 45.Roth J, Qiang X, Marbán SL, Redelt H, Lowell BC. The obesity pandemic: where have we been and where are we going? Obes Res 12, Suppl 2: 88S–101S, 2004. doi: 10.1038/oby.2004.273. [DOI] [PubMed] [Google Scholar]
- 46.Ruderman NB, Berchtold P, Schneider S. Obesity-associated disorders in normal-weight individuals: some speculations. Int J Obes 6, Suppl 1: 151–157, 1982. [PubMed] [Google Scholar]
- 47.Ruderman NB, Schneider SH, Berchtold P. The “metabolically-obese,” normal-weight individual. Am J Clin Nutr 34: 1617–1621, 1981. doi: 10.1093/ajcn/34.8.1617. [DOI] [PubMed] [Google Scholar]
- 48.Rung J, Cauchi S, Albrechtsen A, Shen L, Rocheleau G, Cavalcanti-Proença C, Bacot F, Balkau B, Belisle A, Borch-Johnsen K, Charpentier G, Dina C, Durand E, Elliott P, Hadjadj S, Järvelin MR, Laitinen J, Lauritzen T, Marre M, Mazur A, Meyre D, Montpetit A, Pisinger C, Posner B, Poulsen P, Pouta A, Prentki M, Ribel-Madsen R, Ruokonen A, Sandbaek A, Serre D, Tichet J, Vaxillaire M, Wojtaszewski JF, Vaag A, Hansen T, Polychronakos C, Pedersen O, Froguel P, Sladek R. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet 41: 1110–1115, 2009. [Erratum in Nat Genet 41: 1156, 2009] doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 49.Samocha-Bonet D, Dixit VD, Kahn CR, Leibel RL, Lin X, Nieuwdorp M, Pietiläinen KH, Rabasa-Lhoret R, Roden M, Scherer PE, Klein S, Ravussin E. Metabolically healthy and unhealthy obese—the 2013 Stock Conference report. Obes Rev 15: 697–708, 2014. doi: 10.1111/obr.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott RA, Fall T, Pasko D, Barker A, Sharp SJ, Arriola L, Balkau B, Barricarte A, Barroso I, Boeing H, Clavel-Chapelon F, Crowe FL, Dekker JM, Fagherazzi G, Ferrannini E, Forouhi NG, Franks PW, Gavrila D, Giedraitis V, Grioni S, Groop LC, Kaaks R, Key TJ, Kühn T, Lotta LA, Nilsson PM, Overvad K, Palli D, Panico S, Quirós JR, Rolandsson O, Roswall N, Sacerdote C, Sala N, Sánchez MJ, Schulze MB, Siddiq A, Slimani N, Sluijs I, Spijkerman AM, Tjonneland A, Tumino R, van der A DL, Yaghootkar H, McCarthy MI, Semple RK, Riboli E, Walker M, Ingelsson E, Frayling TM, Savage DB, Langenberg C, Wareham NJ; RISC study group; EPIC-InterAct consortium . Common genetic variants highlight the role of insulin resistance and body fat distribution in type 2 diabetes, independent of obesity. Diabetes 63: 4378–4387, 2014. doi: 10.2337/db14-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orrú M, Usala G, Dei M, Lai S, Maschio A, Busonero F, Mulas A, Ehret GB, Fink AA, Weder AB, Cooper RS, Galan P, Chakravarti A, Schlessinger D, Cao A, Lakatta E, Abecasis GR. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet 3: e115, 2007. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Semple RK, Chatterjee VK, O’Rahilly S. PPAR gamma and human metabolic disease. J Clin Invest 116: 581–589, 2006. doi: 10.1172/JCI28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Semple RK, Savage DB, Cochran EK, Gorden P, O’Rahilly S. Genetic syndromes of severe insulin resistance. Endocr Rev 32: 498–514, 2011. doi: 10.1210/er.2010-0020. [DOI] [PubMed] [Google Scholar]
- 54.Sims EA. Characterization of the syndromes of obesity, in Diabetes Mellitus and Obesity (Brodoff BN, Bleicher SJ, editors). Baltimore: Williams & Wilkins, 1982, p. 219–226. [Google Scholar]
- 55.Song Y, Manson JE, Meigs JB, Ridker PM, Buring JE, Liu S. Comparison of usefulness of body mass index versus metabolic risk factors in predicting 10-year risk of cardiovascular events in women. Am J Cardiol 100: 1654–1658, 2007. doi: 10.1016/j.amjcard.2007.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Mägi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segrè AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpeläinen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proença C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grässler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jørgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, König IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaløy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimäki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O’Donnell CJ, O’Rahilly S, Ong KK, Oostra B, Paré G, Parker AN, Perola M, Pichler I, Pietiläinen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstråle M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tönjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kähönen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Grönberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ MAGIC; Procardis Consortium . Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 42: 937–948, 2010. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stefan N, Kantartzis K, Machann J, Schick F, Thamer C, Rittig K, Balletshofer B, Machicao F, Fritsche A, Häring HU. Identification and characterization of metabolically benign obesity in humans. Arch Intern Med 168: 1609–1616, 2008. doi: 10.1001/archinte.168.15.1609. [DOI] [PubMed] [Google Scholar]
- 58.Swinburn BA, Sacks G, Hall KD, McPherson K, Finegood DT, Moodie ML, Gortmaker SL. The global obesity pandemic: shaped by global drivers and local environments. Lancet 378: 804–814, 2011. doi: 10.1016/S0140-6736(11)60813-1. [DOI] [PubMed] [Google Scholar]
- 59.Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, Hayakawa T, Terauchi Y, Ueki K, Kaburagi Y, Satoh S, Sekihara H, Yoshioka S, Horikoshi H, Furuta Y, Ikawa Y, Kasuga M, Yazaki Y, Aizawa S. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature 372: 182–186, 1994. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 60.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 41: 18–24, 2009. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 61.Trasande L, Elbel B. The economic burden placed on healthcare systems by childhood obesity. Expert Rev Pharmacoecon Outcomes Res 12: 39–45, 2012. doi: 10.1586/erp.11.93. [DOI] [PubMed] [Google Scholar]
- 62.Tseng YH, Kriauciunas KM, Kokkotou E, Kahn CR. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol Cell Biol 24: 1918–1929, 2004. doi: 10.1128/MCB.24.5.1918-1929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Virtue S, Vidal-Puig A. It's not how fat you are, it's what you do with it that counts. PLoS Biol 6: e237, 2008. doi: 10.1371/journal.pbio.0060237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wildman RP, Muntner P, Reynolds K, McGinn AP, Rajpathak S, Wylie-Rosett J, Sowers MR. The obese without cardiometabolic risk factor clustering and the normal weight with cardiometabolic risk factor clustering: prevalence and correlates of 2 phenotypes among the US population (NHANES 1999-2004). Arch Intern Med 168: 1617–1624, 2008. doi: 10.1001/archinte.168.15.1617. [DOI] [PubMed] [Google Scholar]
- 65.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, McArdle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O’Rahilly S, Purmann C, Rees MG, Ridderstråle M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YC, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Waterworth DM, Watkins N, Witteman JC, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JN; Wellcome Trust Case Control Consortium; Genetic Investigation of ANthropometric Traits Consortium . Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 41: 25–34, 2009. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.World Health Organization. Obesity and overweight Fact sheet. 2016.
- 67.Yaghootkar H, Lotta LA, Tyrrell J, Smit RA, Jones SE, Donnelly L, Beaumont R, Campbell A, Tuke MA, Hayward C, Ruth KS, Padmanabhan S, Jukema JW, Palmer CC, Hattersley A, Freathy RM, Langenberg C, Wareham NJ, Wood AR, Murray A, Weedon MN, Sattar N, Pearson E, Scott RA, Frayling TM. Genetic evidence for a link between favorable adiposity and lower risk of type 2 diabetes, hypertension, and heart disease. Diabetes 65: 2448–2460, 2016. doi: 10.2337/db15-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yaghootkar H, Scott RA, White CC, Zhang W, Speliotes E, Munroe PB, Ehret GB, Bis JC, Fox CS, Walker M, Borecki IB, Knowles JW, Yerges-Armstrong L, Ohlsson C, Perry JR, Chambers JC, Kooner JS, Franceschini N, Langenberg C, Hivert MF, Dastani Z, Richards JB, Semple RK, Frayling TM. Genetic evidence for a normal-weight “metabolically obese” phenotype linking insulin resistance, hypertension, coronary artery disease, and type 2 diabetes. Diabetes 63: 4369–4377, 2014. doi: 10.2337/db14-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamauchi T, Kamon J, Minokoshi Y, Ito Y, Waki H, Uchida S, Yamashita S, Noda M, Kita S, Ueki K, Eto K, Akanuma Y, Froguel P, Foufelle F, Ferre P, Carling D, Kimura S, Nagai R, Kahn BB, Kadowaki T. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat Med 8: 1288–1295, 2002. doi: 10.1038/nm788. [DOI] [PubMed] [Google Scholar]
- 70.Yang Z, Hall AG. The financial burden of overweight and obesity among elderly Americans: the dynamics of weight, longevity, and health care cost. Health Serv Res 43: 849–868, 2008. doi: 10.1111/j.1475-6773.2007.00801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]