Abstract
Analysis of skeletal muscle cross sections is an important experimental technique in muscle biology. Many aspects of immunohistochemistry and fluorescence microscopy can now be automated, but most image quantification techniques still require extensive human input, slowing progress and introducing the possibility of user bias. MyoVision is a new software package that was developed to overcome these limitations. The software improves upon previously reported automatic techniques and analyzes images without requiring significant human input and correction. When compared with data derived by manual quantification, MyoVision achieves an accuracy of ≥94% for basic measurements such as fiber number, fiber type distribution, fiber cross-sectional area, and myonuclear number. Scientists can download the software free from www.MyoVision.org and use it to automate the analysis of their own experimental data. This will improve the efficiency and consistency of the analysis of muscle cross sections and help to reduce the burden of routine image quantification in muscle biology.
NEW & NOTEWORTHY Scientists currently analyze images of immunofluorescently labeled skeletal muscle using time-consuming techniques that require sustained human supervision. As well as being inefficient, these techniques can increase variability in studies that quantify morphological adaptations of skeletal muscle at the cellular level. MyoVision is new software that overcomes these limitations by performing high-content analysis of muscle cross sections with minimal manual input. It is open source and freely available.
Keywords: automation software, cell morphology, high-content microscopy, image analysis, skeletal muscle
INTRODUCTION
Skeletal muscle is primarily composed of elongated, multinucleated cells, called fibers, which adapt to external stimuli and changing functional demands. Hormones, exercise, physical inactivity, spaceflight, denervation, aging, acute and chronic diseases/infections, genetic defects, and metabolic disorders all modulate skeletal muscle fibers in ways that are visually discernible using immunofluorescence microscopy (1, 2, 27, 31, 32, 35, 36). Specifically, fiber cross-sectional area (CSA), fiber type proportion [i.e., percent slow-twitch, fast-twitch, and hybrid fibers, based on myosin heavy-chain (MyHC) expression], myonuclear number, and muscle stem cell (i.e., satellite cell) number can change profoundly in the aforementioned conditions. As a result, numerous research groups seek to measure tissue- and cellular-level morphology as part of their analyses.
Image quantification is often the most time-consuming part of this process. In muscle biology laboratories at the University of Kentucky, samples are typically sectioned and stained in batches. The experimental protocols (including lengthy immunohistochemical procedures) are normally completed within a few days. In contrast, before the development of MyoVision, the software described in this manuscript, quantification of experimental data sets typically required weeks to months of intensive human effort.
Several groups, including the current authors, have described theoretical frameworks for automating the analysis of muscle cross sections without making software available for general use (13, 15, 16, 19, 21, 22, 24). Other groups have published their computer code. For example, Bergmeister et al. (3) shared a plug-in for ImageJ that can measure fiber number, size, and type. SMASH is software, written by Smith and Barton, that can semiautomatically expedite analyses, mainly as a free alternative to expensive commercial software packages (30). These programs have attractive features but require extensive user interaction and/or optimization. For example, in SMASH, the user has to set a segmentation filter interactively for each image in order for the software to produce useful results. This makes it difficult to automate the analysis and introduces the potential for user bias.
MyoVision overcomes most of these limitations. MyoVision provides high-content quantification of muscle features, including fiber number, CSA, minimum Feret diameter, myonuclear number, and fiber type distribution, without requiring human supervision. Data presented on the following pages show that it is also more reliable and more accurate than available alternative techniques and packages. It is available for free download from www.MyoVision.org.
METHODS
Animals and Tissue Preparation
All of the images used for estimating MyoVision accuracy were obtained during previous studies of mouse plantaris tissue (12, 14). No mice were killed specifically for the development and validation of the software. Each of the images showed a complete plantaris cross section and was formed by stitching together multiple fields of view (see Image Acquisition and Quantification). One cross section per mouse was analyzed in all of the studies. The cell detection and counting comparisons used images from six plantaris cross sections labeled with anti-laminin antibody (total of 5,800 fibers). The CSA analyses included >14,000 muscle fibers from cross sections labeled with antidystrophin antibody (four groups of n = 4 mice per group subjected to sham, 3-day, 7-day, or 14-day synergist ablation surgery). 3,300 muscle fibers from six cross sections labeled with anti-laminin and isoform-specific anti-MyHC antibodies were analyzed for the fiber-type distribution comparisons. Finally, the myonuclear counting analysis was based on 3,800 fibers from six plantaris cross sections stained with antidystrophin antibodies and DAPI.
Immunohistochemistry
Immunohistochemical procedures were carried out as previously described by Fry et al. (10). A detailed, recommended protocol for both mouse and human muscle cross sections is included in the appendix. In summary, for fiber typing, unfixed sections (7 µm) were incubated for 90 min at room temperature with antibodies to MyHC types I, IIA, and IIB (1:100; cat. no. BA.D5, SC.71, and BF.F3, respectively, University of Iowa Developmental Studies Hybridoma Bank, Iowa City, IA), in addition to rabbit anti-laminin IgG (1:100, L9393; Sigma-Aldrich, St. Louis, MO). MyHC type IIX expression was inferred from unstained fibers. Fluorescence-conjugated secondary antibodies were applied to different mouse immunoglobulin subtypes for 1 h to visualize MyHC expression and laminin. Sections were postfixed in absolute methanol before mounting.
For dystrophin identification, muscle sections were rehydrated with PBS and blocked in mouse-on-mouse blocking reagent (Vector Laboratories, Burlingame, CA). After washing, incubation with antidystrophin antibody (1:50; cat. no. VPD505; Vector Laboratories) overnight was followed by incubation for 75 min with goat anti-mouse biotinylated secondary antibody (1:1,000, 115-065-205; Jackson ImmunoResearch, West Grove, PA). Sections were washed again, incubated 30 min in SA-FITC (1:150, no. SA-5001; Vector Laboratories), and postfixed in 4% paraformaldehyde before mounting using Vectashield fluorescent mounting medium with DAPI (Vector Laboratories).
Image Acquisition and Quantification
All images were acquired using an upright microscope at ×20 magnification (AxioImager M1, Zen 2.3 Imaging Software; Zeiss, Göttingen, Germany), which automatically acquires consecutive fields in multiple channels. These fields (~35 per plantaris) were then optionally stitched together into a single mosaic image.
Manual Analysis for Fiber-Type Distribution
Manual analyses were performed first by a blinded operator using ImageJ followed by visual confirmation by a second researcher (28). Briefly, multiple channels were background subtracted, normalized for their intensities, false colored, and merged into a single image using Zen 2 Lite or Pro. The different fiber types were visually identified on the basis of color differences in the merged image using the cell counter tool. Fibers were sequentially counted as Type I (Cy5, pink), Type IIA (FITC, green), and Type IIB (TRITC, red). Fibers that were counted as negative under all three channels were classified as Type IIX. Classification of fiber type required roughly 1 h per cross section.
Semimanual analysis using Zen.
Muscle-fiber CSA was determined by a single operator using Zen 2 Pro (Zeiss) image-processing software. The image analysis macro was designed using preexisting tools within the Zen 2 Pro software. Images were preprocessed to optimize contrast for the membrane immunofluorescence. In the image analysis macro, the DAPI filter was selected with the entire cross section of muscle being used as the region of interest (ROI). The input image was smoothed with a Gaussian filter (sigma 1.5) without sharpening and a minimum 485–500, and a maximum >1000 intensity threshold was used for the watershed separation of fibers. A predetermined minimum CSA exclusion of 150 µm2 and maximum 5,000 µm2 was employed (representing the smallest and largest fibers found in a mouse plantaris cross section) (10). After the initial automatic segmentation, the manual cutting and merging tools were used to correct segmentation errors. For an 800–1,000 fiber cross section of whole muscle, this process generally took 1.5–2 h. Fiber identification (ID) and CSA were selected as the output variables, so that fiber number and average CSA could be determined. Visual verification of appropriate fiber sizes was then carried out, and all merged fibers that were not manually separated (generally <2% of fibers) were excluded from the analysis.
SMASH.
Detailed descriptions of the SMASH software have been published (30). When using the SMASH software for the analyses described in this study, the initial segmentation was performed on all laminin images with parameters set to blue (fiber outline color), 0.323 (pixel size – μm/pixel), and 8 (segmentation filter – low is more segmentation). The segmentation filter was selected by empirically testing values from 5 to 12 and visually inspecting results for the first image. Subsequent fiber filter parameters were set to default values. Although SMASH was designed to be semiautomatic, no manual correction was performed for either the initial segmentation or the fiber filter to compare with the fully automatic analyses of MyoVision.
ImageJ plug-in.
The ImageJ plug-in was downloaded according to instructions described by Bergmeister et al. (3), and the intensity threshold for each laminin image was set to 600, which was within the range set by the manual analysis of the same images. The ROI was set to the entire muscle cross section. The same CSA filters (a minimum of 150 µm2 and a maximum 5,000 µm2) were applied to all analyzed images.
MyoVision Software Implementation
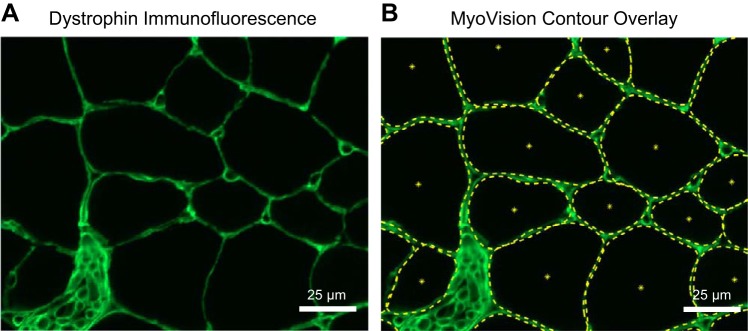
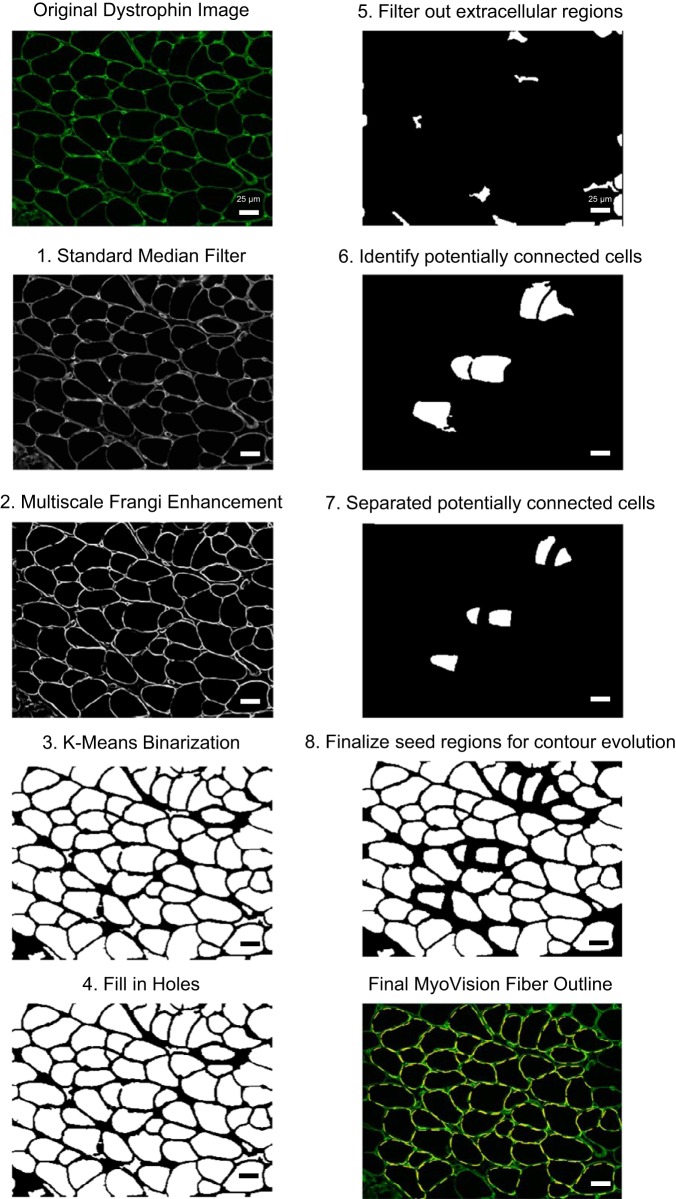
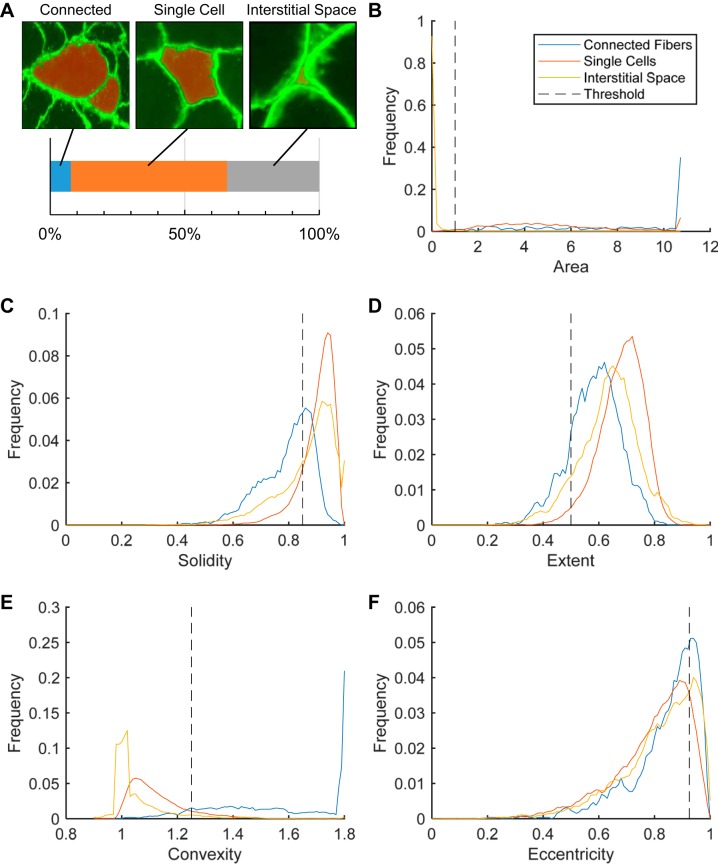
For a detailed description of the algorithm rationale and implementation, please see the appendix. Figure 1A shows an example of a portion of skeletal muscle cross section stained for dystrophin. Figure 1B shows the same image after processing in the MyoVision software. The algorithm workflow is also diagrammed in Fig. 2. Briefly, skeletal muscle cross sections labeled with anti-laminin or -dystrophin immunofluorescence were filtered and enhanced for line and edge structures. The enhanced image was segregated into membrane and cytoplasmic regions using Euclidian Distance K-means clustering. Each separated cytoplasmic region was designated as a seed region, which belongs to one of three classifications 1) multiple connected cells, 2) a single cell, or 3) interstitial space (Fig. 3A; red regions). Using shape descriptors (Fig. 3, B–F), seed regions that were more likely to be connected cells were selected for watershed separation. The contour of each processed seed region was fine-tuned by spline-evolution until convergence. Contour evolution was carried out over an energy field calculated using the Chan Vese – Vector Field Convolution. For fiber type and myonuclear analyses, fiber outlines generated from the laminin/dystrophin reference image were combined and converted into a mask overlay. This mask was subsequently applied to each additional single-channel immunofluorescence image, indicating nuclei or a particular MyHC isoform.
Fig. 1.
MyoVision fiber outline. A: dystrophin-labeled immunofluorescence intensity image of a mouse plantaris muscle cross section, false colored in green. B: same image as in A with MyoVision cell outline dotted in yellow. Scale bar = 25 μm.
Fig. 2.
MyoVision workflow. Major steps in the MyoVision cell detection and outlining algorithm. For detailed descriptions, please see the appendix. Steps 1 and 2: filtering to prepare for initial segmentation. Steps 3 and 4: convert the image to foreground and background and then perform morphological operations. Steps 5 and 6: calculate shape descriptors for each seed region and identify potentially connected seeds. Step 7: distance-based watershed transformation to separate the potentially connected fibers. Step 8: combine separated fibers with single fibers and generate parametric splines to prepare for contour evolution. Scale bar = 25 μm.
Fig. 3.
Empirically determined thresholds for shape descriptors. A: representative images of seed regions (red), dystrophin overlay (green), and their respective manual classifications. Shape descriptors are calculated for a total of 6,781 seed regions (520 connected, 3,939 single cells, and 2,322 interstitial spaces). B–F: shape descriptors and their respective frequency distributions (black dotted line represents the threshold selected for the MyoVision algorithm).
Accuracy Measurements
Accuracy for fiber and myonuclei counting and for analyses of cross-sectional areas was defined by Eq. 1:
| (1) |
The reference value was defined as the value obtained by trained muscle researchers using semiautomated or fully manual methods (Fig. 4A, solid line), except for the case of cross-sectional area. Because manual techniques typically underestimated fiber cross-sectional area (see Fig. 5), the reference cross-sectional areas were calculated as A0 × (manual value) + A1, where A0 was the slope and A1 was the y intercept of a regression line fitted to the algorithm values (Fig. 6A, dashed lines).
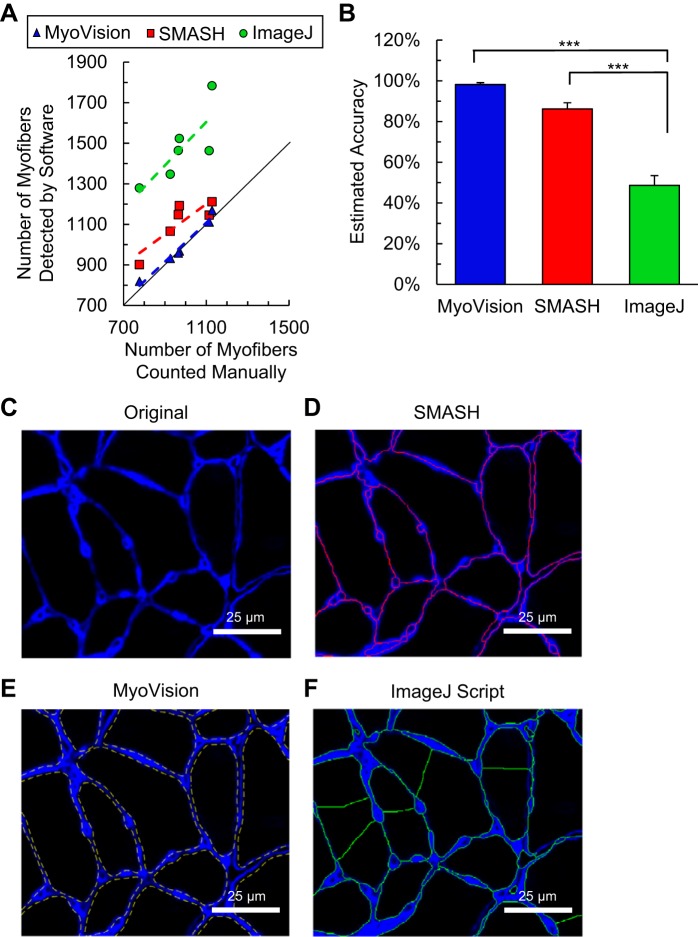
Fig. 4.
Fiber detection and counting. A: comparison of MyoVision (blue), SMASH (red), and ImageJ plug-in (green) with manual fiber counting for six mouse plantaris muscles. MyoVision counts lie directly on the line of identity (black). Markers above the line of identity represent over-segmentation. B: estimated accuracy of each method is expressed as percent difference from manual counts (*** denotes P < 0.001). C–F: representative images of the segmentation for each algorithm compared with the original laminin-labeled (blue) image. Significant oversegmentation (a single fiber is broken up into multiple fibers) can be readily observed for the ImageJ plug-in. Scale bar = 25 μm.
Fig. 5.
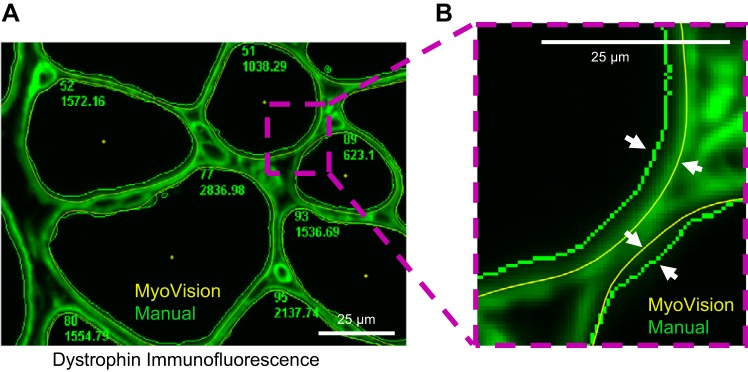
MyoVision versus semimanual outlines. A: MyoVision (yellow) and manual (green) cell outlines for the same dystrophin-labeled image. B: zoomed-in view of the region from A boxed in purple dotted line. White arrows highlight the difference between the MyoVision and the manual outlines. MyoVision outlines are closer to the dystrophin staining, therefore producing fiber CAS measurements that are consistently larger than the manual measurements (Fig. 6A). Scale bar = 25 μm.
Fig. 6.
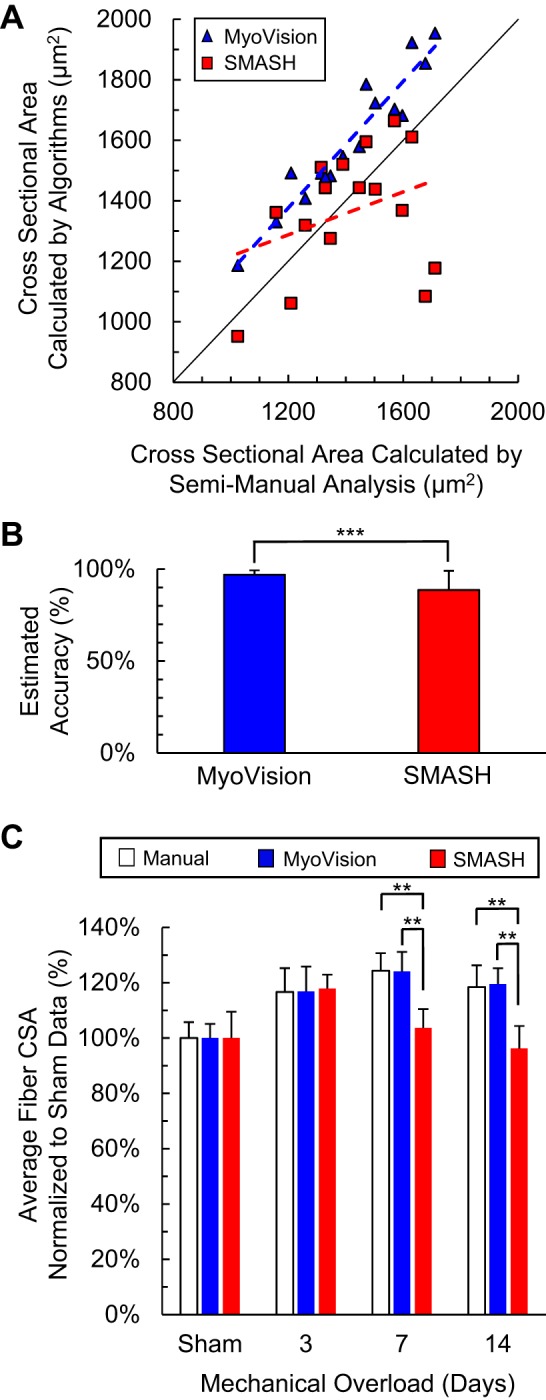
Average fiber cross-sectional area (CSA). A: comparison of MyoVision (blue) and SMASH (red) cell size measurements with manual measurements for 16 mouse plantaris cross sections with and without mechanical overload. The line of identity is shown in black. MyoVision measurements are bigger than semimanual measurements for every sample. Dashed line denotes linear regression line. B: estimated accuracy for MyoVision and SMASH measurements of fiber CSA (***P < 0.001). C: relative increase in fiber CSA (hypertrophy) over the mechanical overload time course. SMASH demonstrates significant underestimation of fiber CSA as growth increases (* and ** denote P < 0.05 and 0.01, respectively).
Accuracy for analysis of fiber-type distributions was defined by Eq. 2 to prevent division by zero errors when muscle cross sections did not contain any Type I fibers.
| (2) |
Statistics
Reported values represent means ± SE. Statistical analyses were performed using GraphPad Prism version 7.0 for Windows, GraphPad Software, La Jolla CA, www.graphpad.com. P values less than 0.05 were considered to be statistically significant. One-way ANOVA followed by Tukey multiple-comparisons test was performed for fiber counting accuracy measurements. Paired, two-tailed Student’s t-tests were performed for accuracy measurements of fiber CSA and myonuclear counting. Repeated-measures two-way ANOVA followed by Bonferroni multiple-comparisons tests were performed for CSA changes with mechanical overload and for fiber-typing accuracy measurements.
RESULTS
Fiber Counting
The individual fibers are identified and outlined in yellow. One of the key features of MyoVision is that seed regions are prefiltered using shape descriptors before the conventional watershed transformation is applied. Such a prefiltering approach minimizes oversegmentation and improves the accuracy of fiber counts. Figure 4 shows that the number of fibers counted by MyoVision is 101.4 ± 1.0% of the number counted by manual analysis, which corresponds to an accuracy of 98.2 ± 0.9% (see Eq. 1) if manual counts are accepted as true values. In contrast, SMASH (a program that implements metric-based filtering after the watershed transformation) overestimates the number of fibers (113.8 ± 3.0%) and has an accuracy of only 86.2 ± 3.0%. The plug-in for ImageJ described by Bergmeister et al. (3) oversegments (151.4 ± 4.8%) the fibers (Fig. 4F) and had an accuracy of only 48.6 ± 4.8%. This plug-in was not considered further in this work because of its poor performance in these initial tests.
Fiber Cross-Sectional Area
A direct assessment of the cytoplasmic outlines from MyoVision and semimanual analyses demonstrates that MyoVision outlines lie closer to the membrane than the semimanual outlines (Fig. 5). When comparing the CSAs calculated by the MyoVision and SMASH algorithms to the reference values obtained by semimanual analysis, MyoVision produces higher CSA measurements than the semimanual method (Fig. 6A). This is because users typically choose a more conservative threshold (smaller cytoplasmic regions) in the semimanual method to reduce the number of manual corrections needed to separate connected cells. Figure 6B shows that the areas calculated by the MyoVision software are more accurate than those produced by the SMASH algorithm (P < 0.001).
Figure 6C shows that MyoVision-derived estimates for the relative increases in fiber CSA in response to a hypertrophic stimulus are indistinguishable from those determined by manual analysis (P > 0.05). This contrasts with the relative growths calculated by SMASH, which differ at 7 and 14 days of mechanical muscle overload from manual measurements and from MyoVision-derived values (all P values are less than 0.05). These statistical tests show that MyoVision-based estimates of muscle fiber hypertrophy are more consistent with manual measurements than those obtained using SMASH.
Fiber-Type Classification
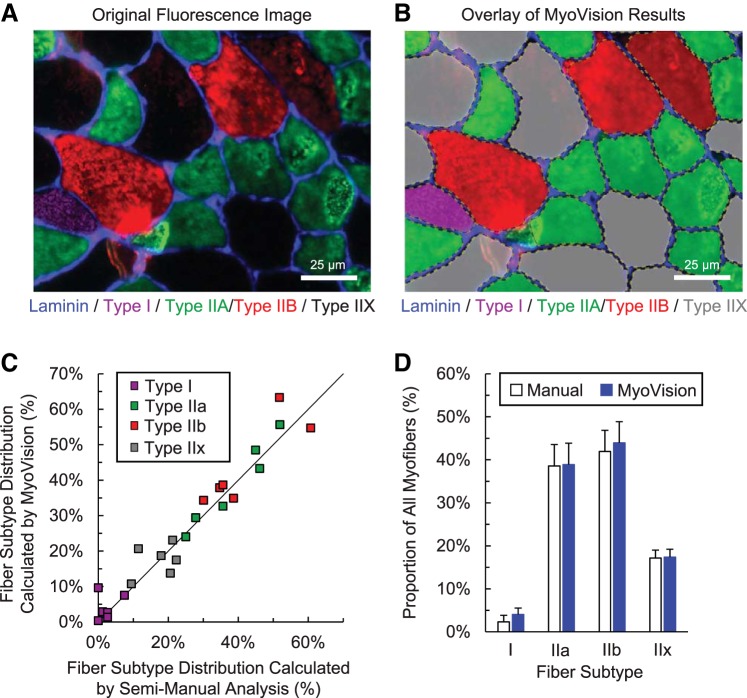
Figure 7A shows a representative image of muscle fibers immunofluorescently labeled for various MyHC isoforms and delineated by the basal lamina. Staining is shown in arbitrary false color: blue for laminin, magenta for Type I, green for Type IIA, and red for Type IIB. Type IIX is inferred from a lack of MyHC immunofluorescence. MyoVision uses the laminin channel to detect and outline the muscle fibers and then classifies fiber types on the basis of the intensities detected for the different MyHC antibodies. Figure 7B shows an overlay of the MyoVision fiber type classification results. MyoVision results are linearly and positively correlated to manual counts (Pearson, R = 0.97), and a two-way ANOVA shows no statistical difference between the proportions of each fiber type measured by hand and by MyoVision. The accuracy of MyoVision fiber type analysis is estimated to be 96.5 ± 0.6% compared with manual analysis.
Fig. 7.
Fiber-type classification. A: plantaris cross section image immunofluorescently stained for three different myosin heavy-chain subtypes and labeled for laminin. B: MyoVision outline and classification of fiber types in A. C: comparison of MyoVision fiber type distribution results with manual counts for six murine plantaris muscles. The line of identity is shown in black. D: comparison of average fiber type distribution for all mice shows no significant difference between MyoVision and manual analyses. Scale bar = 25 μm.
Myonuclear Number
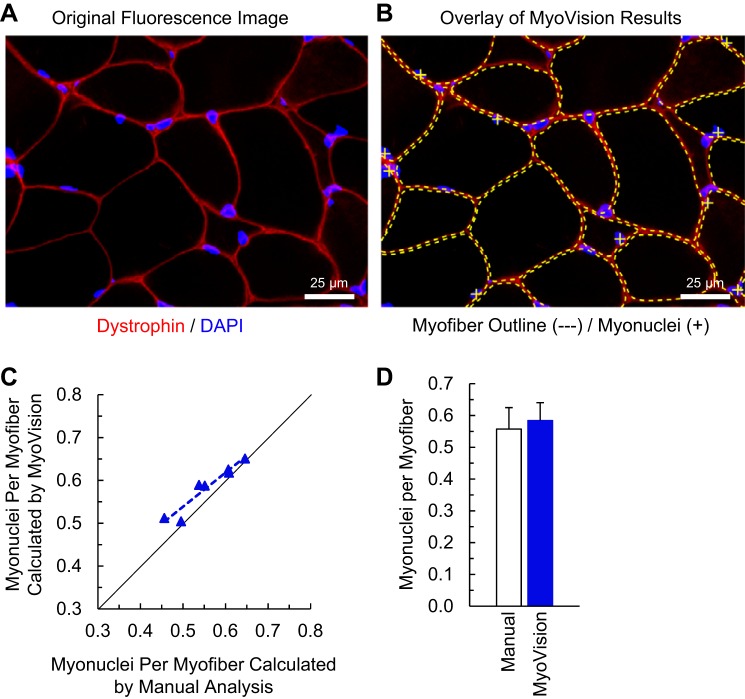
Myonuclei are differentiated from interstitial nuclei because of their location under the sarcolemma. Automatic counting of myonuclei, thus, requires accurate delineation of the sarcolemma. Figure 8A shows dystrophin immunofluorescence demarcating the sarcolemma in red and DAPI-stained nuclear DNA in blue. Figure 8B shows the results from MyoVision myonuclear detection, where the sarcolemma is outlined by yellow dotted lines and the myonuclei (defined as any nuclear region having its centroid and greater than 50% of its area inside the sarcolemma) are indicated by yellow plus signs. MyoVision myonuclear counts are linearly correlated with manual counts (Pearson, R = 0.96), and a Student’s t-test shows no statistical difference between the manual and MyoVision myonuclear counts. MyoVision myonuclear counting accuracy is estimated to be 94.9 ± 1.7% relative to manual analysis.
Fig. 8.
Myonuclear number. A: plantaris cross-section image immunofluorescently labeled with DAPI for nuclei and colabeled for dystrophin. B: MyoVision outlines (dotted yellow lines) and classification of the myonuclei (yellow plus signs) in A. C: comparison of MyoVision myonuclear counting results with human counts for six mouse plantaris muscles. The line of identity is shown in black. D: comparison of average myonuclei per fiber for all mice shows no significant difference between MyoVision and human analysis. Scale bar = 25 μm.
Software and Interface
The software (including source code) along with instructions and demonstration videos are freely available at www.MyoVision.org.
DISCUSSION
High-content microscopy is a vital tool for muscle biology research. Routine image-based techniques require significant manual optimization and continuous human supervision. ImageJ plug-ins can differ significantly among laboratories, and without standardization, indiscriminate usage of ImageJ plug-ins may exacerbate problems with reproducibility. SMASH is a well-designed software package that acts as a free version of commercially available image analysis systems, but it requires varying amounts of manual correction to ensure accuracy. The primary goal for MyoVision was to decrease the demand for human guidance without sacrificing analytical accuracy and precision.
MyoVision incorporates several algorithms that have been used in previous analysis programs (4, 6, 9, 13, 15, 17, 19, 21–26, 30, 33, 37) but streamlines the workflow to enhance computational efficiency and robustness. In particular, most previous algorithms used one of two options: the watershed transformation or the active contour model (snake algorithm). In contrast, MyoVision invokes both algorithms in sequence. The hybrid “watersnake” approach first separates connected muscle fibers using the efficient watershed transformation and then fine-tunes the cytoplasmic boundaries using the active contour method.
The data presented in this work show that MyoVision measures fiber number, CSA, fiber type, and myonuclear number with high accuracy (98.2%, 96.9%, 96.5%, and 94.9%, respectively). While these measurements can be performed by carefully trained investigators, manual analysis is often subjective and time consuming, especially for myonuclear counting. As described in methods, completing all of the above measurements using manual techniques might require hours of sustained human effort per cross section. MyoVision requires a few minutes to produce statistically equivalent results. It also eliminates interindividual variation, which may facilitate data sharing among laboratories throughout the muscle research community.
The biggest limitation for MyoVision is its sensitivity to the quality of the input image. Because fiber detection and outlining are performed entirely on the reference image, degraded fluorescence signals can detrimentally impact the software’s accuracy. Patches of weak staining within a cross section will prevent accurate fiber detection. Irregular noise levels, especially high-intensity punctate noise and/or holes within the fiber myofibrillar regions, also cause problems for fiber detection. Currently, incorrect outlines can be eliminated through the user interface by defining regions of interest. However, the software does not yet allow for manual addition of fiber outlines and/or modification of fiber boundaries. High-quality input images are, therefore, required for accurate results. Detailed protocols for immunohistochemical processing of both mouse and human skeletal muscle are provided in the appendix for reference. These protocols should help investigators to produce high-quality images that can be processed quickly and accurately using MyoVision.
GRANTS
This work was made possible by grants from the National Institutes of Health to J. J. McCarthy (AR061939), to C. A. Peterson and J. J. McCarthy (AR060701), and to K. A. Murach (AR071753), with salary support (UL1TR001998) to K. S. Campbell.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Y.W., C.A.P., J.J.M., and K.S.C. conceived and designed research; Y.W., K.A.M., C.D.V., and K.S.C. performed experiments; Y.W., K.A.M., I.J.V., and C.D.V. analyzed data; Y.W., K.A.M., I.J.V., C.S.F., C.A.P., J.J.M., and K.S.C. interpreted results of experiments; Y.W. prepared figures; Y.W., K.A.M., and C.S.F. drafted manuscript; Y.W., K.A.M., I.J.V., C.S.F., C.A.P., J.J.M., and K.S.C. edited and revised manuscript; Y.W., K.A.M., I.J.V., C.S.F., C.A.P., J.J.M., and K.S.C. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank Dr. Janna Jackson for her discussions and insights.
APPENDIX
Part 1. Detailed Description of MyoVision Software Implementation
MyoVision draws from multiple previously reported algorithms from as early as 1998. During the early stages of development, it became apparent very quickly that different groups may develop similar algorithms to analyze skeletal muscle, but default parameters tend to differ, and preprocessing and postprocessing methods vary as well. The following sections provide a brief review of the literature, software design rationale, and detailed information of how MyoVision analyzes immunofluorescence images of skeletal muscle cross sections.
Program development considerations.
Several published automatic algorithms for muscle fiber segmentation utilized the active contour model (GVF snake) and demonstrated very high levels of accuracy and consistency (13, 15, 19, 22, 25). In 1998, Klemenčič et al. (15) reported for the first time reasonable success using a spline-based active contour model to detect fiber boundaries and measure cross-sectional area. However, their implementation was not fully automatic because it involved a manual initiation step in which a trained researcher selected the center point for each fiber region to be analyzed. Brox et al. (4) and Kim et al. (13) improved upon the classic edge-based active contour approach by extending it with region-based intensity and texture components and implementing an automatic initialization procedure. They showed the accuracy for fiber counting and cross-sectional area measurements to be greater than 98%; however, the algorithm was developed for hematoxylin-and-eosin-stained images. Recognizing the advantages of the automated active contour model, Sertel et al. (29) further developed the algorithm to automatically analyze ATPase-stained muscle cross sections and for the first time demonstrated an 89% accuracy for metabolic fiber typing. Mula et al. (22) subsequently implemented a slightly different automatic initialization protocol and applied the active contour model to muscle cross sections enhanced by immunofluorescent labeling in 2013. A common theme among these approaches was the inclusion of a robust line detection algorithm, known as multiscale vessel enhancement filtering (9). Despite improvements afforded by the gradient vector flow method of energy calculation, noise and concavities can still present problems to the GVF snake. Improvements in the form of the vector field convolution (VFC) method dramatically increased the capture range and noise resistance of the parametric snake (17). As demonstrated by Brox et al. (4) and Kim et al. (13), incorporating region-based information significantly improves accuracy. The most widely used region-based active contour model is the classic active contour without edges, first described by Chan and Vese in 2001 (6). A recent study in which the VFC-based energy calculation is combined with a modified version of the Chan-Vese algorithm, named VFCCV, highlighted the benefits of using both line- and region-based information (33).
Historically, one of the major weaknesses of the active contour model was its high computational demand. To avoid such computational costs, several other automatic implementations for the segmentation of skeletal muscle fiber cross sections employed the more computationally efficient watershed transformation (3, 26). This was the same fundamental approach to muscle fiber segmentation used by the semiautomatic programs as well (20, 30). One of the known drawbacks of the watershed transformation is oversegmentation, which occurs when single regions are erroneously separated into multiple smaller ones (8). As such, local minima suppression and marker-controlled approaches were introduced to minimize oversegmentation, but boundary accuracy remained an area of concern (34). Precision is vital for consistently applying the rules necessary for classifying a myonucleus, because even a slight overestimation of the myofiber boundaries can result in significant variations in myonuclear counts (11). Additionally, muscle tissue from different species can vary in the degrees of membrane thicknesses and interfiber networks, which made it imperative that automated myofiber segmentation programs take the local changes in membrane staining into strict consideration (20).
In 2012, Sáez et al. directly benchmarked the watershed transformation against the active contour model in immunofluorescently stained skeletal muscle cross sections and showed that the watershed transformation is up to six times faster than the active contour model with little difference in accuracy (26). However, Sáez et al. (26) did not report any optimization in terms of their active contour initialization and evolution conditions. This can be a significant step because, in other applications, such as live cell tracking, the active contour model demonstrates higher accuracy than the watershed transformation, and given improved initialization conditions, the active contour model is not significantly slower than the watershed transformation (7). For MyoVision, using VFC instead of GVF for the external energy calculation improves upon the capture range, which indirectly optimizes contour initialization by allowing faster convergence.
On the one hand, the active contour (snake) algorithm is very good at using local image information to evolve contours to the edges of the cells. On the other hand, the watershed transformation is extremely efficient in separating connected objects. It became reasonably obvious that these two algorithms can complement each other, and implementing one algorithm does not preclude the implementation of the second. Therefore, MyoVision was developed as a hybrid “watersnake” algorithm. It uses a watershed transformation to separate connected myofibers and then optimizes the boundaries of these fibers using an active contour algorithm.
Myofiber detection.
The analytical workflow is shown in Fig. 2. Briefly, a single-channel image of a muscle cross section was immunofluorescently labeled for laminin (Fig. 2A). The membrane reference image labeled with laminin or dystrophin immunofluorescence is processed using a 3 × 3 median spatial filter (Fig. 2B). Edges are subsequently enhanced by calculating the multiscale Hessian-based edge probability map as described by Frangi et al. (9) using scales from σmin = 1 to σmax = 4 because Sertel et al. (29) empirically determined that these scales produced the best edge enhancement results for skeletal muscle cross sections. Each pixel of the edge probability map is classified using an automatic K-means clustering algorithm where the number of clusters is determined on the basis of minimizing the average pixel to cluster center distances. The lowest cluster is considered background/cytoplasm, therefore, creating a binary image where the membrane is white and everything else is black (Fig. 2C).
After clearing away regions touching the edge of the image, shape descriptors (extent, convexity, solidity, and area) of each cytoplasmic seed region are calculated. The extent of a region is defined as the ratio of the area of the region to the area of the smallest-area rectangle containing the region. Convexity is defined as the ratio of the perimeter of the region to the perimeter of the convex hull of the region, where the convex hull is defined as the smallest convex polygon that can contain the region. Solidity is defined as the ratio between the area of the region and the area of the convex hull of the region.
To determine the optimal threshold for shape descriptors, a preliminary calibration study was performed. 6,781 seed regions were generated from dystrophin-labeled images of skeletal muscle cross sections and manually classified as “connected fibers,” “single cell,” or “noncellular space” (Figure 3A). Regions where multiple fibers were inadequately separated represented less than 8% of all generated seeds, whereas interstitial spaces represented almost 35% of all of the seeds. This suggested that typical algorithms applied watershed segmentation over 100% of all seed regions, whereas only 8% of the regions actually required watershed separation.
Seed region area (expressed as a percentage of the maximum area of all seed regions) was the best predictor of noncellular or interstitial space (Fig. 3B). There appeared to be very little distinction between connected and unconnected seed regions in terms of eccentricity; therefore, this measure is not included as a differentiation factor (Fig. 3F). Seed regions with extent values below 0.5, solidity less than 0.85, and convexity greater than 1.25 (Fig. 3, C–E) were identified as potentially connected myofibers and were selected for further processing via a single watershed segmentation. Distances greater than 5 μm were suppressed during this process to further minimize oversegmentation. MyoVision, thus, restricted the watershed analysis to the regions that were most likely to be connected fibers based on the described combinations of shape descriptor variables. The utilization of shape descriptors helped to minimize the problems typically associated with indiscriminate use of watershed methods.
The contours of the seed regions were subsequently refined using the parametric spline-based active contour model. Image forces were calculated by convolving a vector field kernel (width = 64 pixels, type = power, α = 2.6) with the gradient magnitude of the multiscale enhanced edge map generated in the second step (for a detailed description of vector field convolution, see Ref. 17). Additionally, image region information was incorporated as the difference between the average fluorescence intensities inside and outside of the evolving contour, similar to the algorithm described in the VFCCV approach (33). Contour evolution was set to stop upon convergence (when the area stopped changing per iteration).
Myofiber-type classification and myonuclear counting.
For fiber-typing analyses, MyoVision used K-means clustering to classify each pixel as foreground (myosin heavy-chain staining) or background based on fluorescence intensity. Then, using the myofiber outline as detected in the reference (laminin/dystrophin) image, MyoVision classified the myofibers with greater than 25% foreground area as positive for the particular myosin heavy-chain isoform. For myonuclear counting analyses, all nuclei were detected using the fluorescence channel stained for DNA content (DAPI for our images). Initial binarization was achieved using the automatic threshold algorithm described by Otsu (23). The detected nuclear regions were subsequently subjected to a marker-controlled watershed segmentation to separate connected nuclei. Nuclei were classified as myonuclear if both the center of mass for the nucleus and at least 50% of the total area of that nucleus fell within the myofiber, as outlined using the reference image, in accordance with previously published counting methods (5, 19).
Part 2. Determining Myosin Heavy-Chain (MyHC) Fiber-Type Distribution, Muscle Fiber Cross-Sectional Area (CSA), Fiber Type-Specific CSA, and Myonuclear Density in Mouse and Human Skeletal Muscle via Immunohistochemistry
Tissue processing.
mouse.
-
After sacrifice, coat the muscle liberally with Tissue-Tek OCT (Sakura Finetek, Torrance, CA) and freeze the muscle in liquid-nitrogen-cooled isopentane.
-
•
Muscle should be completely covered in OCT, as insufficient coverage can affect the quality of tissue preservation during freezing and also make it more challenging to obtain complete muscle cross sections when sectioning.
-
•
When the isopentane begins to freeze around the periphery of the freezing vessel, it is at the correct temperature to freeze the muscle.
-
•
Muscle should be pinned to a cork covered in aluminum foil at resting length. The muscle can be frozen directly on the aluminum, and after the isopentane has evaporated (~5 min on dry ice), it can be easily transferred to the storage container.
-
•
For small muscles, such as the plantaris and soleus, ~15–20 s is sufficient to fully freeze the muscle, whereas larger muscles, such as the gastrocnemius, may take longer (~30 s).
-
•
-
Remove enough muscle with a blade or scalpel, so that cutting is initiated at or near the midbelly, and then mount the tissue upright in a bolus of OCT using freeze-spray.
-
•
Use the freeze-spray to ensure that the warm OCT does not cause freeze damage, and all manipulations should be performed in the cryostat.
-
•
Let the tissue equilibrate to the temperature of the cryostat (23–24°C) for 20–30 min, and then cut 7-µm-thick sections from the mounted tissue onto charged slides.
human.
-
Following the biopsy procedure, identify a portion of muscle (~50 mg) where fiber orientation can be determined. Mount the muscle on a small piece of cork with the base coated in tragacanth gum. The muscle biopsy is mounted upright on the cork, such that the fibers are perpendicular to the surface of the cork.
-
•
Properly orienting the muscle biopsy in regard to fiber direction is needed for cross-sectional assessment of the sample.
-
•
A “base” of tragacanth gum can be created on the cork, so that one end of the muscle sample can be partially submerged in the tragacanth gum to stand the sample upright on the cork. This will prevent the muscle sample from folding over during the freezing process.
-
•
The same isopentane preparation for the freezing of mouse muscle is applied to the human sample. Once the isopentane is sufficiently cold, the muscle can be placed “muscle-side down” in the isopentane by holding the cork with forceps.
-
•
The human muscle should remain in the chilled isopentane for ~15–20 s to ensure that the entire sample is frozen.
-
•
Human muscle samples frozen in tragacanth gum can be cut from their cork base and then mounted upright using OCT and freeze-spray in a similar fashion to mouse muscle.
As with mouse muscle, the mounted human muscle will need to equilibrate to the temperature of the cryostat (23–24°C) for 20–30 min before cutting 7-µm-thick sections from the mounted tissue onto charged slides.
Mouse and human slide processing.
-
Let frozen cross sections air-dry at room temperature for 1–6 h.
-
• It is important that the sections are completely dry before moving them to −20°C for storage or beginning a staining protocol. Moisture in the tissue will cause bubbling of the sections, resulting in poor staining quality.
-
• Sections can be stored in a sealed box at −20°C for several months before staining.
-
• If using sections from the −20°C, allow air-drying for at least 5 min on the bench.
-
• For best results, begin the staining protocol on the day of sectioning (at least an hour after the sections have been drying at room temperature). Staining quality declines the longer the samples are stored at −20°C.
-
•
Prior to initiating the staining protocol, draw a circle around the section with a PAP pen (no. H-4000; Vector Laboratories, Burlingame, CA).
Tissue staining.
mouse.
Rehydrate the sections in PBS for 5 min.
-
Block the sections for 1 h with mouse-on-mouse blocking kit (1 drop/1 ml PBS) (MKB-2213; Vector Laboratories).
This blocking step allows for the use of mouse primary antibodies on mouse tissue.
Wash 3 × 5 min in PBS.
-
Incubate the sections in 1°Ab for 90 min at room temperature (RT) or rocking overnight at 4°C (produces comparable results). Concentrates are diluted into supernatants:
MyHC Type 1 fibers: BA.D5 IgG2b concentrate (1:100) (BA.D5, from Developmental Studies Hybridoma Bank (DHSB), Iowa City, IA)
MyHC Type 2a fibers: SC.71 IgG1 supernatant (DHSB)
-
MyHC Type 2b fibers: BF.F3 IgM supernatant or Type 2x fibers: 6H1 IgM supernatant (DHSB)
In our experience, BF.F3 (MyHC 2b) generally produces better results than 6H1 (MyHC 2x) in mouse tissue.
-
Laminin (fiber borders): Rabbit antilaminin (1:150) (L9393; Sigma)
SC.71 and BF.F3 supernatants are combined in a 1:1 ratio with BA.D5 and laminin concentrate added at a 1:100 and 1:150 dilution, respectively, i.e., for 100 µl of antibody, combine 50 µl SC.71 with 50 µl 6H1 and add 1 µl BA.D5 and 0.75 µl laminin.
Wash 3 × 5 min in PBS.
-
Incubate the sections in 2°Ab for 60 min at RT, diluted in PBS.
-
•
MyHC Type 1 fibers: Goat anti-mouse IgG2b, Alexa Fluor 647-conjugated 2°Ab (1:250) (A21242; Invitrogen, Carlsbad, CA). Type 1 fibers appear pink.
-
•
MyHC Type 2a fibers: Goat anti-mouse IgG1, Alexa Fluor 488-conjugated 2°Ab (1:500) (Invitrogen, A21121). Type 2a fibers appear green.
-
•
MyHC Type 2x fibers: Goat anti-mouse IgM, Alexa Flour 555-conjugated 2°Ab (1:250) (Invitrogen, A21426). Type 2x fibers appear red.
-
•
Fiber borders: Goat anti-rabbit IgG, AMCA-conjugated 2°Ab (1:150) (Vector, Cl−1000). Laminin appears blue.
-
•
Mount with Vectashield mounting media (H-1000; Vector Laboratories).
human.
Rehydrate the sections in PBS for 5 min.
-
Incubate the sections in 1°Ab for 90 min at room temperature (RT) or rocking overnight at 4°C (produces comparable results). Concentrates are diluted into supernatants:
-
•
MyHC Type 1 fibers: BA.D5 IgG2b concentrate (1:100) (BA.D5 from DHSB)
-
•
MyHC Type 2a fibers: SC.71 IgG1 supernatant (DHSB)
-
•
Type 2x fibers: 6H1 IgM supernatant (DHSB)
6H1 (MyHC 2x) works well in human muscle sections.
-
•
Laminin (fiber borders): Rabbit anti-laminin (1:150) (L9393; Sigma)
SC.71 and 6H1 supernatants are combined in a 1:1 ratio with BA.D5 and laminin concentrate added at a 1:100 and 1:150 dilution, respectively, i.e., for 100 µl of antibody, combine 50 µl SC.71 with 50 µl 6H1 and add 1 µl BA.D5 and 0.75 µl laminin.
-
•
Wash 3 × 5 min in PBS.
-
Incubate the sections in 2°Ab for 60 min at RT, diluted in PBS.
-
•
MyHC Type 1 fibers: Goat anti-mouse IgG2b, Alexa Fluor 647-conjugated 2°Ab (1:250) (Invitrogen, A21242). Type 1 fibers appear pink.
-
•
MyHC Type 2a fibers: Goat anti-mouse IgG1, Alexa Fluor 488-conjugated 2°Ab (1:500) (A21121; Invitrogen). Type 2a fibers appear green.
-
•
MyHC Type 2x fibers: Goat anti-mouse IgM, Alexa Flour 555-conjugated 2°Ab (1:250) (Invitrogen, A21426). Type 2x fibers appear red.
-
•
Laminin (fiber borders): Goat anti-rabbit IgG, AMCA-conjugated 2°Ab (1:150) (Vector Laboratories, Cl−1000). Laminin appears blue.
-
•
Wash 3 × 5 min in PBS.
Mount with Vectashield mounting media (H-1000; Vector).
Dystrophin staining.
Dystrophin, as opposed to laminin, can also be used for muscle fiber CSA determination. One dystrophin antibody that we use in our laboratory is a mouse IgG1, which means MyHC 2a fibers (SC.71) cannot be stained simultaneously. If excluding MyHC 2a fibers, all other fiber types can be costained with dystrophin using the protocols above (substituting dystrophin for laminin). Another dystrophin that works well is raised in rabbit and is compatible with all fiber-typing antibodies. Total muscle fiber CSA can be determined (with or without fiber type) using dystrophin or laminin.
-
1.
Dystrophin 1° Ab: Mouse anti-dystrophin IgG1 (1:100) (VP-D505; Vector) (mouse-on-mouse block for 1 h before 1° Ab if staining mouse sections) or Rabbit anti-dystrophin (1:200) (SC-15376; Santa Cruz Biotechnology)
-
2.
2° Ab: Goat anti-mouse IgG1, Alexa Fluor 488-conjugated 2° Ab (1:500) (A21121; Invitrogen) or goat anti-rabbit IgG, AMCA-conjugated 2°Ab (1:150) (Vector, Cl−1000)
Dystrophin and DAPI staining for myonuclear counts.
Utilizing the same procedures as above:
-
1.
Dystrophin 1° Ab: Mouse anti-dystrophin IgG1 (1:100) (VP-D505; Vector Laboratories) (mouse-on-mouse block for 1 h before 1° Ab if staining mouse sections) or rabbit anti-dystrophin (1:200) (SC-15376; Santa Cruz Biotechnology, Santa Cruz, CA) in PBS for 90 min at RT or ON rocking at 4°C.
-
2.
2° Ab: Goat anti-mouse IgG1, Alexa Fluor 488-conjugated 2° Ab (1:500) (A21121; Invitrogen) or goat anti-rabbit IgG Alexa Fluor 555 (A-21429; Invitrogen) for 60 min in PBS at RT. Dystrophin appears green or red.
Nuclei can be identified via DAPI staining before applying coverslips. Incubate 5 min in DAPI (1:10,000 diluted in PBS at RT) (D35471; Invitrogen), then coverslip with Vectashield mounting media (Vector, H-1000). Alternatively, slides can be coverslipped using Vectashield with DAPI (H-1200; Vector).
REFERENCES
- 1.Ansved T, Larsson L. Effects of denervation on enzyme-histochemical and morphometrical properties of the rat soleus muscle in relation to age. Acta Physiol Scand 139: 297–304, 1990. doi: 10.1111/j.1748-1716.1990.tb08927.x. [DOI] [PubMed] [Google Scholar]
- 2.Bagley JR, Murach KA, Trappe SW. Microgravity-induced fiber type shift in human skeletal muscle. Gravit Space Res 26: 34–40, 2012. [Google Scholar]
- 3.Bergmeister KD, Gröger M, Aman M, Willensdorfer A, Manzano-Szalai K, Salminger S, Aszmann OC. Automated muscle fiber type population analysis with ImageJ of whole rat muscles using rapid myosin heavy chain immunohistochemistry. Muscle Nerve 54: 292–299, 2016. doi: 10.1002/mus.25033. [DOI] [PubMed] [Google Scholar]
- 4.Brox T, Kim Y-J, Weickert J, Feiden W. Fully-automated analysis of muscle fiber images with combined region and edge-based active contours. In: Bildverarbeitung für die Medizin 2006: Algorithmen Systeme Anwendungen Proceedings des Workshops vom 19 – 21 März 2006 in Hamburg, edited by Handels H, Ehrhardt J, Horsch A, Meinzer H-P, Tolxdorff T. Berlin: Springer, 2006, p. 86–90. doi: 10.1007/3-540-32137-3_18. [DOI] [Google Scholar]
- 5.Bruusgaard JC, Egner IM, Larsen TK, Dupre-Aucouturier S, Desplanches D, Gundersen K. No change in myonuclear number during muscle unloading and reloading. J Appl Physiol (1985) 113: 290–296, 2012. doi: 10.1152/japplphysiol.00436.2012. [DOI] [PubMed] [Google Scholar]
- 6.Chan TF, Vese LA. Active contours without edges. IEEE Trans Image Process 10: 266–277, 2001. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- 7.Degerman J, Faijerson J, Althoff K, Thorlin T, Rodriguez J, Gustavsson TA. A comparative study between level set and watershed image segmentation for tracking stem cells in time-lapse microscopy. https://pdfs.semanticscholar.org/e6dc/f8e64f9e8ef6429394d6d0d99e26a41f17ed.pdf [retrieved 1 June, 2017]. [Google Scholar]
- 8.El Allaoui A. Medical image segmentation by marker-controlled watershed and mathematical morphology. Int J Multimedia Appl 4: 1–9, 2012. doi: 10.5121/ijma.2012.4301. [DOI] [Google Scholar]
- 9.Frangi AF, Niessen WJ, Vincken KL, Viergever MA. Multiscale vessel enhancement filtering. In: Medical Image Computing and Computer-Assisted Interventation—MICCAI’98. New York: Springer, 1998, p. 130–137. doi: 10.1007/BFb0056195. [DOI] [Google Scholar]
- 10.Fry CS, Lee JD, Jackson JR, Kirby TJ, Stasko SA, Liu H, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Regulation of the muscle fiber microenvironment by activated satellite cells during hypertrophy. FASEB J 28: 1654–1665, 2014. doi: 10.1096/fj.13-239426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gundersen K. Muscle memory and a new cellular model for muscle atrophy and hypertrophy. J Exp Biol 219: 235–242, 2016. doi: 10.1242/jeb.124495. [DOI] [PubMed] [Google Scholar]
- 12.Jackson JR, Kirby TJ, Fry CS, Cooper RL, McCarthy JJ, Peterson CA, Dupont-Versteegden EE. Reduced voluntary running performance is associated with impaired coordination as a result of muscle satellite cell depletion in adult mice. Skelet Muscle 5: 41, 2015. doi: 10.1186/s13395-015-0065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim YJ, Brox T, Feiden W, Weickert J. Fully automated segmentation and morphometrical analysis of muscle fiber images. Cytometry A 71A: 8–15, 2007. doi: 10.1002/cyto.a.20334. [DOI] [PubMed] [Google Scholar]
- 14.Kirby TJ, Patel RM, McClintock TS, Dupont-Versteegden EE, Peterson CA, McCarthy JJ. Myonuclear transcription is responsive to mechanical load and DNA content but uncoupled from cell size during hypertrophy. Mol Biol Cell 27: 788–798, 2016. doi: 10.1091/mbc.E15-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klemenčič A, Kovačič S, Pernuš F. Automated segmentation of muscle fiber images using active contour models. Cytometry 32: 317–326, 1998. doi:. [DOI] [PubMed] [Google Scholar]
- 16.Kostrominova TY, Reiner DS, Haas RH, Ingermanson R, McDonough PM. Automated methods for the analysis of skeletal muscle fiber size and metabolic type. Int Rev Cell Mol Biol 306: 275–332, 2013. doi: 10.1016/B978-0-12-407694-5.00007-9. [DOI] [PubMed] [Google Scholar]
- 17.Li B, Acton ST. Active contour external force using vector field convolution for image segmentation. IEEE Trans Image Process 16: 2096–2106, 2007. doi: 10.1109/TIP.2007.899601. [DOI] [PubMed] [Google Scholar]
- 19.Liu F, Fry CS, Mula J, Jackson JR, Lee JD, Peterson CA, Yang L. Automated fiber-type-specific cross-sectional area assessment and myonuclei counting in skeletal muscle. J Appl Physiol (1985) 115: 1714–1724, 2013. doi: 10.1152/japplphysiol.00848.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meunier B, Picard B, Astruc T, Labas R. Development of image analysis tool for the classification of muscle fibre type using immunohistochemical staining. Histochem Cell Biol 134: 307–317, 2010. doi: 10.1007/s00418-010-0733-7. [DOI] [PubMed] [Google Scholar]
- 21.Miazaki M, Viana MP, Yang Z, Comin CH, Wang Y, da F Costa L, Xu X. Automated high-content morphological analysis of muscle fiber histology. Comput Biol Med 63: 28–35, 2015. doi: 10.1016/j.compbiomed.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mula J, Lee JD, Liu F, Yang L, Peterson CA. Automated image analysis of skeletal muscle fiber cross-sectional area. J Appl Physiol (1985) 114: 148–155, 2013. doi: 10.1152/japplphysiol.01022.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otsu N. A threshold selection method from gray-level histograms. Automatica 11: 23–27, 1975. [Google Scholar]
- 24.Pertl C, Eblenkamp M, Pertl A, Pfeifer S, Wintermantel E, Lochmüller H, Walter MC, Krause S, Thirion C. A new web-based method for automated analysis of muscle histology. BMC Musculoskelet Disord 14: 26, 2013. doi: 10.1186/1471-2474-14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Röhrle O, Köstler H, Loch M. Segmentation of Skeletal Muscle Fibres for Applications in Computational Skeletal Muscle Mechanics. In: Computational Biomechanics for Medicine: Soft Tissues and the Musculoskeletal System, edited by Wittek A, Nielsen MFP, Miller K. New York, NY: Springer, 2011, p. 107–117. doi: 10.1007/978-1-4419-9619-0_12. [DOI] [Google Scholar]
- 26.Sáez A, Rivas E, Montero-Sánchez A, Paradas C, Acha B, Pascual A, Serrano C, Escudero LM. Quantifiable diagnosis of muscular dystrophies and neurogenic atrophies through network analysis. BMC Med 11: 77, 2013. doi: 10.1186/1741-7015-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev 91: 1447–1531, 2011. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 28.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675, 2012. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sertel O, Dogdas B, Chiu CS, Gurcan MN. Microscopic image analysis for quantitative characterization of muscle fiber type composition. Comput Med Imaging Graph 35: 616–628, 2011. doi: 10.1016/j.compmedimag.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Smith LR, Barton ER. SMASH—semi-automatic muscle analysis using segmentation of histology: a MATLAB application. Skelet Muscle 4: 21, 2014. doi: 10.1186/2044-5040-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stec MJ, Kelly NA, Many GM, Windham ST, Tuggle SC, Bamman MM. Ribosome biogenesis may augment resistance training-induced myofiber hypertrophy and is required for myotube growth in vitro. Am J Physiol Endocrinol Metab 310: E652–E661, 2016. doi: 10.1152/ajpendo.00486.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suga T, Kimura E, Morioka Y, Ikawa M, Li S, Uchino K, Uchida Y, Yamashita S, Maeda Y, Chamberlain JS, Uchino M. Muscle fiber type-predominant promoter activity in lentiviral-mediated transgenic mouse. PLoS One 6: e16908, 2011. doi: 10.1371/journal.pone.0016908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J, Ray N, Zhang H.. VFCCV snake: A novel active contour model combining edge and regional information. In: 2014 IEEE International Conference on Image Processing. Piscataway, NJ: IEEE, 2014, p. 927–931. [Google Scholar]
- 34.Wang D, Vallotton P.. Improved marker-controlled watershed segmentation with local boundary priors. In: 25th International Conference of Image and Vision Computing New Zealand. Piscataway, NJ: IEEE, 2010, p. 1–6. [Google Scholar]
- 35.Wang JF, Forst J, Schröder S, Schröder JM. Correlation of muscle fiber type measurements with clinical and molecular genetic data in Duchenne muscular dystrophy. Neuromuscul Disord 9: 150–158, 1999. doi: 10.1016/S0960-8966(98)00114-X. [DOI] [PubMed] [Google Scholar]
- 36.White JR, Confides AL, Moore-Reed S, Hoch JM, Dupont-Versteegden EE. Regrowth after skeletal muscle atrophy is impaired in aged rats, despite similar responses in signaling pathways. Exp Gerontol 64: 17–32, 2015. doi: 10.1016/j.exger.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu C, Prince JL. Generalized gradient vector flow external forces for active contours. Signal Process 71: 131–139, 1998. doi: 10.1016/S0165-1684(98)00140-6. [DOI] [Google Scholar]