Significance
Microorganisms persisting in hydraulically fractured shales must maintain osmotic balance in hypersaline fluids, gain energy in the absence of electron acceptors, and acquire carbon and nitrogen to synthesize cell building blocks. We provide evidence that that cofermentation of amino acids (Stickland reaction) meets all of these organismal needs, thus functioning as a keystone metabolism in enriched and natural microbial communities from hydraulically fractured shales. This amino acid-based metabolic network can be rationally designed to optimize biogenic methane yields and minimize undesirable chemistries in this engineered ecosystem. Our proposed ecological framework extends to the human gut and other protein-rich ecosystems, where the role of Stickland fermentations and their derived syntrophies play unrecognized roles in carbon and nitrogen turnover.
Keywords: hydraulic fracturing, metaproteomics, Stickland reaction, methanogenesis, metagenomics
Abstract
Hydraulic fracturing is one of the industrial processes behind the surging natural gas output in the United States. This technology inadvertently creates an engineered microbial ecosystem thousands of meters below Earth’s surface. Here, we used laboratory reactors to perform manipulations of persisting shale microbial communities that are currently not feasible in field scenarios. Metaproteomic and metabolite findings from the laboratory were then corroborated using regression-based modeling performed on metagenomic and metabolite data from more than 40 produced fluids from five hydraulically fractured shale wells. Collectively, our findings show that Halanaerobium, Geotoga, and Methanohalophilus strain abundances predict a significant fraction of nitrogen and carbon metabolites in the field. Our laboratory findings also exposed cryptic predatory, cooperative, and competitive interactions that impact microorganisms across fractured shales. Scaling these results from the laboratory to the field identified mechanisms underpinning biogeochemical reactions, yielding knowledge that can be harnessed to potentially increase energy yields and inform management practices in hydraulically fractured shales.
In 2016, natural gas became the main source of electricity in the United States—the first time in history that a natural resource other than coal has provided a bulk of the nation’s power (1). Sixty percent of the natural gas produced in the United States comes from hydraulically fractured shales, a majority of which is generated in Ohio, West Virginia, and Pennsylvania (1). Hydraulic fracturing (HF) is the high-pressure injection of water, chemical additives, and proppant into the Earth’s subsurface to fracture hydrocarbon-bearing shales, thereby releasing economically important trapped natural gases. This process unintentionally creates a new microbial ecosystem, where a subset of surface-derived microorganisms proliferate in shales more than 2,500 m below the Earth’s surface.
Recent research suggests that microbial life in shales may impact gas and oil production efficiencies (2, 3). For instance, the persistence of methanogens in these ecosystems may contribute to increased biogenic methane formation by Methanohalophilus, while negative impacts, such as corrosion and sulfidogenesis (“souring”), are associated with other prevalent microbial community members including Halanaerobium (2–9). To grow in fractured shales, microorganisms must adapt to increased salinities and reduced chemical conditions where fermentative metabolisms prevail (2). Given these selective pressures, persisting shale-hosted microbial communities are constrained to several halotolerant members, including Halanaerobium and Methanohalophilus, which co-occur across every fractured shale sampled to date (2). Metagenomic and metabolite analyses from a single well suggested that glycine betaine (GB), an amino acid derivative, may play an important role as an osmoprotectant and as an energy source for these co-occurring shale organisms (8). However, the GB-supported metabolisms employed across geographically and geologically distinct fractured shales remain unknown.
Here, we use a combination of field investigations and detailed laboratory microcosm experiments to define the metabolic network supported by GB. First, we sample GB prevalence and concentration in the field using temporally collected fluid samples collected from Utica and Marcellus fractured shale wells. We then established laboratory microcosm reactors with Utica-produced fluids collected 96 d after HF and used proteomics to define the impacts of GB on persisting shale microbial communities. To extend these laboratory-discovered processes back to the field scale, we conducted a paired metagenome and metabolome analysis from over 40 samples collected across five fractured shale wells located in the Appalachian basin. This comprehensive dataset offers unique insight into previously cryptic amino acid-based metabolisms that may sustain life in these economically important ecosystems.
Constructing Model Shale Microbial Communities in the Laboratory
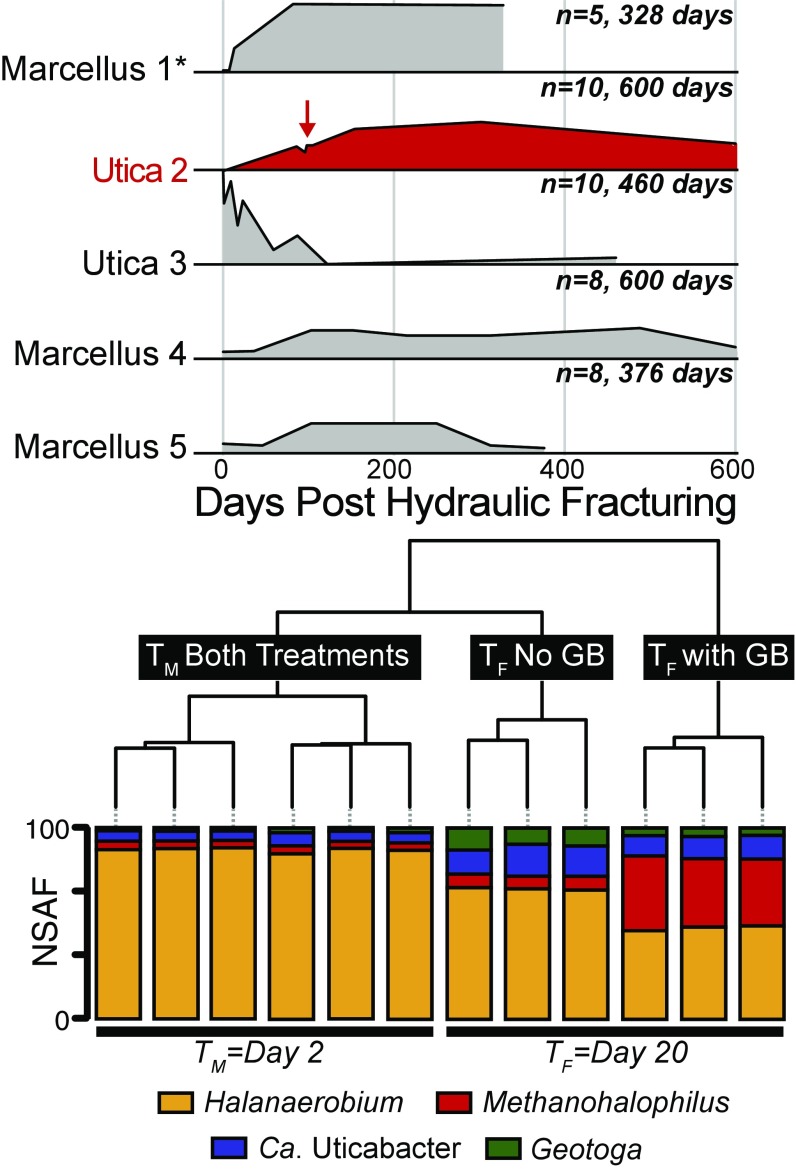
To understand the broader importance of GB across geographically distinct fractured shales, we profiled GB concentrations in input (fluids injected during HF) and produced fluids from five shale wells sampled up to 600 d after HF (Fig. 1 and Dataset S1). GB was present in all hydraulically fractured shale wells, with two of the five wells showing a trend where GB is not detected in the input fluids, but is then produced and maintained in situ (Marcellus 1 and Utica 2). For these two wells, GB was positively correlated to salinity (Pearson, R = 0.87, P < 0.001), corroborating our prior hypothesis that this metabolite is likely microbially synthesized in situ to support microbial adaptation to brine-level salinities (8). In the other three wells, GB was detected in the input fluids, albeit at low concentrations (>0.8 μM). This could be a result of operators using recycled produced fluids as input fluids or the exogenous addition of GB as a surfactant amended to input fluids (10) (https://fracfocus.org/chemical-use). GB dynamics in these wells hint at both microbial utilization and production; however, it is also possible that GB is leached from the dissolution of shale rock (Fig. 1).
Fig. 1.
Area plots show relative GB concentration trends through time across five HF wells in two shale formations: (Marcellus and Utica), with the number of samples denoted and plotted on an identical y axis. Inocula for microcosm experiments were obtained from the well shown in red at the time point indicated by the red arrow (96 d after HF). Data from one well was previously reported (8) and is indicated by an asterisk (*). Exact GB concentrations range from below detect to 8.1 μM and are provided in Dataset S1. Hierarchal clustering of metaproteomic data are shown from day 2 (TM) and day 20 (TF) postinoculation of the laboratory microcosm experiment. Stacked bars represent the relative abundance [normalized spectral abundance factor (NSAF)] of proteins from each organism indicated by color within each sample. Time point and microcosm treatment are indicated in black boxes with white text.
To understand the possible sources and metabolic roles of this prevalent metabolite, we generated laboratory microcosms using produced fluids collected 96 d post-HF (Utica well 2, Fig. 1, red arrow). To identify the microbial sources of GB, these reactors were established without shale rock. Triplicate anoxic microcosms were amended with and without GB in a chemically undefined medium containing yeast extract (see Materials and Methods for recipe) and incubated for 20 d, with three time points chosen for metabolite, metagenome, and metaproteome analyses. Abiotic controls showed no metabolite changes through the experiment (SI Appendix, Fig. S1). Time points were collected at the beginning (T0), at maximum cell density on day 2 (TM), and upon substantial methane production (1.5 log fold increase from T0) on day 20 (TF) (Dataset S2). Metagenomic sequencing facilitated the reconstruction of four draft genomes belonging to the genera Halanaerobium, Methanohalophilus, Geotoga, and a novel genus within the Clostridiales (SI Appendix, Figs. S2 and S3 and Dataset S3). The organisms from which these genomes were reconstructed were the only members of the microbial community in both the GB and non-GB enrichment cultures at all time points (SI Appendix, Fig. S4). This enrichment reflects the low genus-level diversity previously reported in late-produced fluids from Utica and Marcellus shales (2, 8, 11).
Genomes reported here were estimated to be greater than 93% complete, with less than 2% contamination, and contained full-length 16S rRNA genes (Dataset S3). Based on the recently proposed Genomic Standards Consortium standards (12), the genomes recovered here would be considered high quality. The unassigned Clostridiales genome is most closely related to Dethiosulfatibacter by 16S rRNA gene analysis (∼90% identity, SILVA) and Dethiosulfatibacter aminovorans by average nucleotide identity at the genome level (73.1%) (SI Appendix, Figs. S2 and S3). Following the naming convention for genomes assembled from metagenomes (13), we propose the genus name Candidatus Uticabacter based on the shale formation from which this genome was recovered. 16S rRNA gene fragments (V4 region) were identical to the near-complete 16S rRNA gene in our Candidatus Uticabacter genome, suggesting that members of this genus have been previously detected in a hydraulically fractured shale well in the Sichuan Basin in China [National Center for Biotechnology Information (NCBI) SRR2094439.12567.1] (9). Beyond Ca. Uticabacter, the other members recovered in our laboratory genomic analyses are routinely reported in studies from fractured shales across the United States (14–16). For instance, 16S rRNA genes corresponding to Halanaerobium and Methanohalophilus are recovered from in all but 1 of these 17 studies (8). Together, these findings demonstrate that the microorganisms detected in our microcosms, and likely their metabolic interactions, are relevant to fractured shale ecosystems.
Next, we used metagenome-resolved metaproteomics to uncover the active metabolisms assigned to each genus. A total of 555,973 unique peptides were recovered from 15 metaproteomes, with an average of 37,046 unique peptides per microcosm sample (Dataset S4). Across all time points and treatments, a majority of the proteins analyzed were from the genus Halanaerobium (63%). Proteins from other members of the microbial community were also detected, with 15% of total proteins from Methanohalophilus, 11% from Ca. Uticabacter, and 7% from Geotoga (Fig. 1). Interestingly, overall protein content and taxonomic assignment could not be statistically differentiated between the GB and non-GB treatment at the middle time point, largely driven by the dominance and conserved metabolism of Halanaerobium across the two treatments. The final time point (TF) was statistically differentiated by treatment, with proteins from Methanohalophilus enriched in the GB microcosm where methane was produced in high amounts, while proteins assigned to Ca. Uticabacter and Geotoga were more enriched in the non-GB microcosm that produced significantly less methane.
Osmoprotection Mechanisms Enabling Salinity Adaptation in Laboratory Reactors
Given the hypersaline conditions observed in late (>40 d post-HF)-produced fluids (Dataset S1), we profiled microcosm metaproteomic data for evidence of osmoprotection strategies. While it has been well documented by our group, and others, that these organisms encode versatile osmoprotection strategies (8, 17, 18), the preferred mechanisms and how they change with extracellular availability of an osmoprotectant were unknown. Consistent with production and consumption patterns of GB across wells (Fig. 1), all organisms in the microcosm have the potential to uptake GB, with Methanohalophilus exclusively utilizing the compatible solute strategy through uptake and synthesis (SI Appendix, Fig. S5). From glycine, Methanohalophilus can produce GB through sarcosine and N-N-dimethylglycine intermediates (SI Appendix). Notably, this pathway is expressed regardless of GB amendment, signifying that GB may be key to biogenic methane production in fractured shales.
Collectively, metaproteomic data indicate that Ca. Uticabacter and Halanaerobium likely use the salt-in strategy through sodium/proton antiporters, while Methanohalophilus and Geotoga are reliant on the osmolyte strategy (SI Appendix, Fig. S5). Inferring osmoprotectant function from metaomics is complicated by the fact that many transporters are nonspecific and often these compounds can play other roles in cellular assimilation or energy production. Despite these challenges, our findings expand upon prior reports that Halanaerobium solely uses a salt-in strategy and provides proteomic evidence for the use of choline uptake for osmoprotection (SI Appendix, Fig. S5). Given that GB has multiple assimilatory (osmoprotection, nitrogen, and carbon source) and dissimilatory (energy generation) uses, and is prevalent in fractured shales (Fig. 1), we suggest GB may be a keystone metabolite. Here, we used GB and non-GB amended laboratory microcosms to investigate the ecological interactions, including predation, mutualism, and competition, present in fractured-shale microbial communities.
Viral Predation and Resistance Is Ongoing in Laboratory Reactors
To elucidate predator–prey interactions in these microcosms, we identified viral genomes, linked these viruses to hosts, and measured their activity. Fifty-four assembled viral contigs were recovered and clustered into 16 unique populations, 25% of which were affiliated with the Order Caudovirales, while the remaining majority (75%) were taxonomically novel. Viral dynamics were coordinated to their host, and notably not impacted by GB amendment (SI Appendix, Fig. S6). Two of the viral populations found in this microcosm were also previously reported (8) in Marcellus well 1 (Fig. 1 and Dataset S3). This finding demonstrates the relevance of these laboratory-enriched viruses to the shale ecosystem.
We detected 326 unique viral peptides from 13 of the 16 viral populations (Datasets S3 and S4). Most of the viral peptides were identified as proteins with unknown function (36%); however, peptides involved in virion production (e.g., terminase and head proteins) and viral integration into host genomes (e.g., resolvase and recombinase) were also detected (SI Appendix, Fig. S6). These expression data show that a majority of the microcosm viruses are active, and these include both temperate and lytic infections. Thus, fractured-shale microbial communities are likely evolving under strong constraints exerted by a diverse set of viruses.
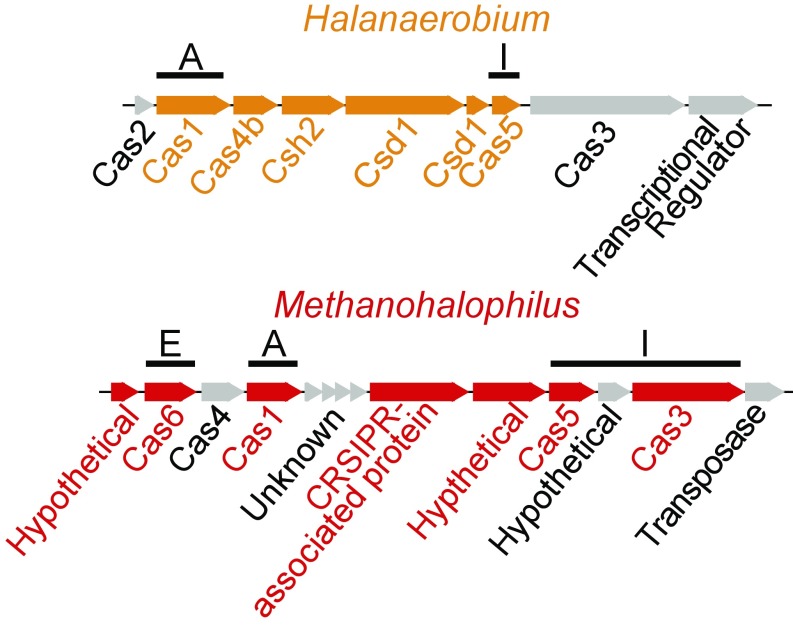
Previously, we detected spacer incorporation in a Halanaerobium genome over time from field-produced fluids (Marcellus well 1) (8). Here, we provide evidence for the activity of the CRIPSR-Cas system from deep biosphere microbial communities. Cas proteins for all three functional stages of adaptive immunity were expressed (adaptation, expression, interference; Fig. 2) (19). Of particular importance, both Methanohalophilus and Halanaerobium expressed adaptive proteins for incorporating spacers into CRISPR loci (Cas1), as well as interference proteins for producing cognate RNAs (Cas5) that bind to and cleave the viral DNA (Cas 3, Methanohalophilus only) (SI Appendix). The congruence between laboratory and field viral populations and evidence of CRISPR-Cas activity demonstrate that the strong viral predation captured in our laboratory microcosms reflects ongoing viral–host interactions maintained at the ecosystem scale.
Fig. 2.
Halanaerobium and Methanohalophilus CRISPR-Cas system genes are shown, with corresponding peptides detected in proteomics highlighted in orange and red, respectively. Genes for adaptive immunity are denoted by functional stage, with adaptation (A), expression (E), and interference (I) stages all represented in metaproteomic data.
Mutualistic Interactions Sustain Biogenic Methane Production in Laboratory Reactors
Consistent with our prior metagenome findings and physiological characterizations of the genus (8, 20, 21), Methanohalophilus is inferred to be an obligate methylotrophic methanogen, lacking the capacity to utilize hydrogen or acetate. Additionally, this genome lacked the genes necessary to directly use quaternary amines like choline and GB (22–24). Halanaerobium appears to be an obligate fermenter, as the genome lacks an electron transport chain and terminal oxidase or reductase genes (4, 8). We have previously suggested based solely on metagenomic inferences that the fermentation of the amino acid derivative GB will yield products sustaining methylotrophic methanogens in fractured shales (8).
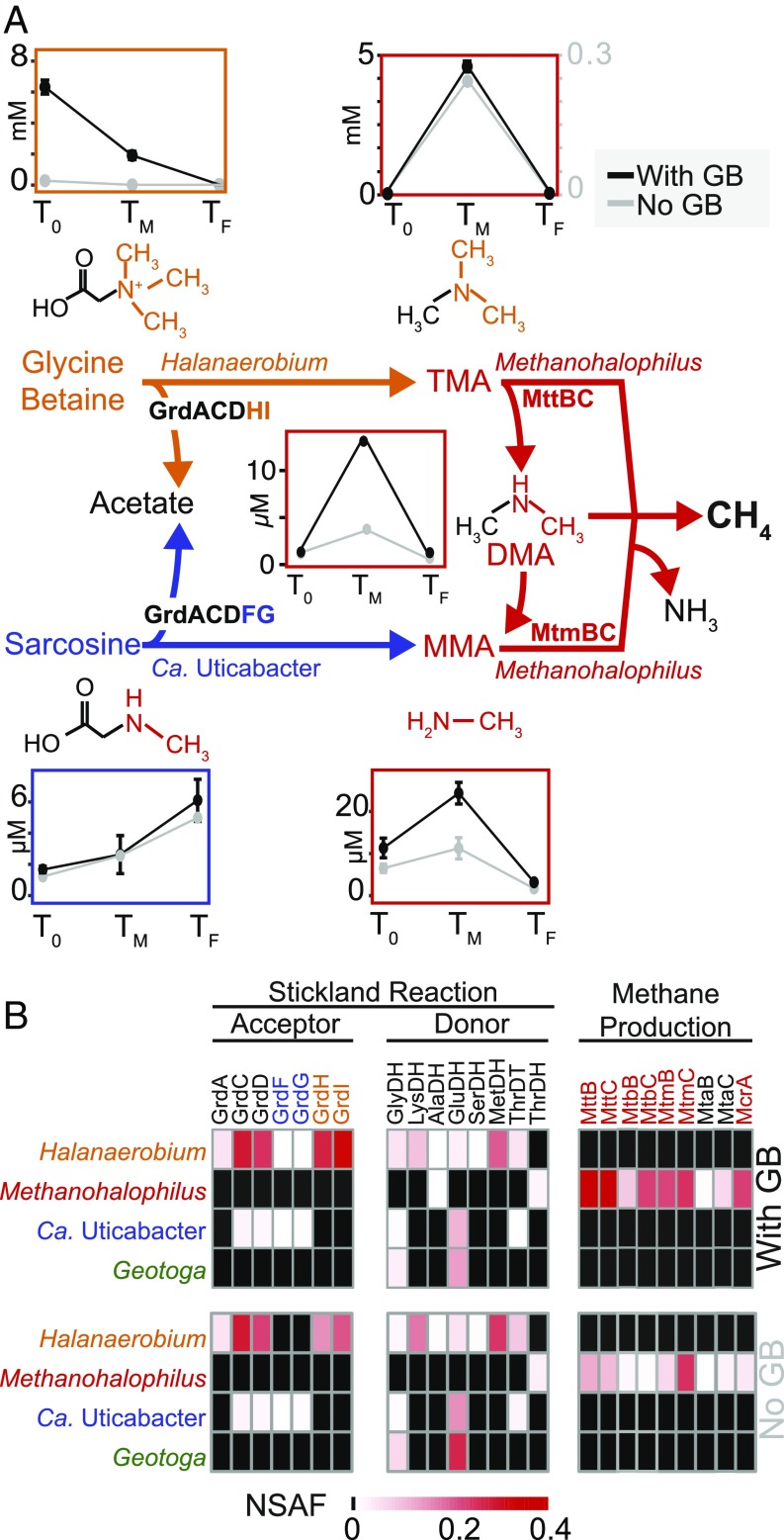
To better elucidate this metabolic cross-feeding, we used linear discriminant analysis to identify and report the significant metabolisms occurring at different stages of biogenic methane production [LEfSe (25); Dataset S2]. Our proteomics data revealed that GB was fermented by Halanaerobium to yield trimethylamine (TMA) at the middle time point, which sustained methanogenesis at the later time point. The proteins necessary for this metabolic symbiosis (Halanaerobium GrdHI and Methanohalophilus MttB) were discriminating features of the middle and final time points, respectively, and we failed to identify any other sources for TMA production (Fig. 3). Other possible sources of methane include methanol (MtaB), monomethylamine (MtmB), and dimethylamine (MtbB), but not acetate, as corresponding proteins were detected for methylotrophic substrates only. Our findings are consistent with prior reports where methylotrophic methanogenesis is more prevalent in saline ecosystems, likely because this methanogenesis pathway (rather than hydrogenotrophic or acetoclastic alternatives) generates higher energy yields that are needed to sustain the increased cost of osmoprotectant synthesis (2, 26).
Fig. 3.
(A) Center colored pathway shows Stickland reactions from GB and sarcosine to TMA and methylamine (MMA), respectively, fuel methanogenesis with pathways colored by organism. Chemical structures are shown, with cleaved products colored. Corresponding line graphs shows average metabolite concentrations with SD of triplicate samples through time colored by treatment (black, GB; gray, no GB). Note that TMA is reported with a dual y axis and all dynamics of methanogenesis substrates (TMA, DMA, and MMA) are shown in red boxes. Acetate concentrations over time can be found in Fig. 4B. Geotoga is not represented here because it does not have potential to carry out a Stickland reaction. (B) Heat maps display NSAF values for proteins detected by metaproteomics in GB amended (Top) and no-GB (Bottom) microcosms at the TF timepoint.
Metabolite analysis supported the metaproteomic results, revealing that 90% of GB consumed in the first 2 d was recovered as TMA. Ninety-five percent of this TMA was subsequently converted to methane by the last time point. Interestingly in the non-GB reactors, the proteomic and metabolomic patterns are similar but less prominent, with 60% of Halanaerobium-produced TMA converted to methane. The fact that GB metabolism occurs regardless of experimental manipulations has ramifications for in situ processes, as the substrate concentrations in the non-GB microcosm were similar to field conditions (Fig. 1). Moreover, the synthesis of GB in the shale-free laboratory microcosm supports our supposition that increased GB over time in the field-derived produced fluids was due to microbial synthesis (Fig. 1). The ubiquity of Methanohalophilus across fractured shales (2) and the high efficiency of methane production demonstrated here indicate that methylamine methanogenesis may be active and important to shale natural gas production. Supporting our findings, a prior study predicted that biogenic methane accounted for 12% of methane produced in a shale-gas well lifetime (3). Our findings leave open the possibility that the augmentation of fractured shales with exogenous methyl-C1 compounds could enhance biogenic methane production down well, analogous to acetate amendment techniques currently employed in coal-bed methane recovery (27).
We next examined the capacity for other Stickland fermentations that support methanogenesis. Similar to Halanaerobium, Ca. Uticabacter expressed proteins to ferment sarcosine (sarcosine reductase, GrdFG) (28), yielding monomethylamine that Methanohalophilus utilizes for methane production (Fig. 3 and SI Appendix, Fig. S7). Monomethylamine concentrations and necessary enzymes (MtmB) followed the same pattern as TMA but were significantly lower (Fig. 3 and SI Appendix, Fig. S1). Unlike GB, sarcosine does not decrease with monomethylamine formation but rather increases over time in both biological treatments, suggesting microbial sarcosine production exceeds its removal (SI Appendix). We show that mutualistic exchange of methylamines produces biogenic methane in fractured-shale microbial communities.
Untangling the Stickland Fermentation Network Revealed Substrate Partitioning and Competition in Laboratory Reactors
While our field and laboratory studies indicated that GB is readily reduced to TMA by the prevalent and highly dominant shale bacterium Halanaerobium (2, 7), the amino acid electron donor for this fermentation was unknown. Our laboratory study illuminated the genomic potential for utilizing known Stickland electron donors and acceptors in a shale-derived microbial community, with the reactions and key functional genes for these metabolisms summarized in Table 1.
Table 1.
Summary of Stickland half-reactions shown in Fig. 4
| Donors/acceptors | Compound | Relevant gene | Half-reaction | Refs. |
| Acceptor | GB | GB reductase (grdHI) | GB ⇒ Acetyl-P + TMA | 28, 32, 33 |
| Acceptor | Sarcosine | Sarcosine reductase (grdFG) | Sarcosine ⇒ Acetyl-P + Monomethylamine | 28, 32, 33 |
| Acceptor | Glycine | Glycine reductase (grdBE) | Glycine ⇒ Acetyl-P + Ammonium | 28, 32, 33 |
| Donor | Lysine | 3,5-Diaminohexanoate dehydrogenase (kdd) | Lysine + NAD+ ⇒ ⇒ 5-Amino-3-oxohexanoate + NADH | 29, 31, 32 |
| Donor | Threonine | Threonine dehydrogenase (tdh) | Threonine + NAD+ ⇒ l-2-Amino-3-oxobutanoate + NADH | 29, 30, 32 |
| Donor | Glycine | Glycine dehydrogenase (gvcD) | Glycine + NAD+ ⇒ Ammonium + CO2 + NADH | 29, 32, 33 |
Relevant gene abbreviations are noted (shown in parentheses).
Based on coupled metaomic data from the GB enrichment, we conclude that lysine is likely the primary electron donor used by Halanaerobium to reduce GB to TMA (Fig. 4). Using the enzyme 3,5-diaminohexanoate dehydrogenase (22), Halanaerobium is the only bacterium to oxidize lysine to acetate, butyrate, and ammonia through crotonyl-CoA in the microcosm (SI Appendix). The pattern of expression for this enzyme was significantly correlated to that of GB reductase (P < 0.01), and metabolite stoichiometry demonstrated that 93% of the lysine was oxidized in the first 2 d during primary GB reduction. Of the other possible Stickland electron donors (29–31), lysine was in the greatest concentration, accounting for up to 17% of GB reduction, while other Halanaerobium Stickland donors implicated by proteomics and metabolomics included serine (7.2%), methionine (6.7%), glycine (4.1%), and threonine (3.8%) (Datasets S1 and S4). Given that only a little more than a third of GB reduction can be accounted for from known amino acid reductants in the Stickland reaction, Halanaerobium also uses hydrogen or other currently unknown reductants as the electron donor for GB reduction.
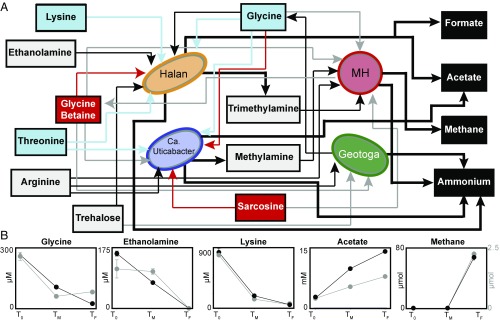
Fig. 4.
Metabolic network of interactions revealed by metaproteomics and metabolite analyses. (A) Network of Halanaerobium (orange), Methanohalophilus (red), Ca. Uticabacter (blue), and Geotoga (green) shows the interconnected metabolisms of shale organisms. Arrows pointing toward and away from microbes show utilization and production, respectively. Arrow line color denotes substrate utilization: red (oxidant in the Stickland reaction), blue (reductant in the Stickland reaction), and gray (osmoprotectant). Bold black lines indicate the production of substrates, and terminal end products are noted in black boxes. (B) Line graph shows average with SD of triplicate metabolite concentrations through time colored by treatment (black, GB; gray, no GB). Abiotic control metabolite concentrations did not change significantly over time but showed glycine was added from media, not produced fluids (Materials and Methods and SI Appendix, Fig. S5, and Dataset S1). Note, methane is shown on a dual y axis.
Unlike lysine, which represents a noncompetitive substrate for Halanaerobium, other Stickland electron donors are more widely used by members of the community. In addition to Halanaerobium, Ca. Uticabacter can also compete for glycine as a Stickland electron donor to support sarcosine reduction, or may use glycine as both a donor and acceptor simultaneously (28, 32, 33) (SI Appendix). Glycine is consumed at all time points except at the last time point of the no-GB amendment (Fig. 4). From proteomic and metabolite analysis, we infer that Geotoga is responsible for this glycine production, via operation of the glycine cleavage system in reverse (Fig. 4 and SI Appendix, Fig. S8), using ethylene glycol as an oxidant. Overall, glycine is the most interconnected metabolite based on its variety of uses in the microcosm community (Fig. 4).
In summary, the laboratory microcosms demonstrated that GB and glycine have both adaptive and metabolic roles in fractured-shale communities. For instance, GB is synthesized and used as an osmoprotectant by Methanohalophilus, while Halanaerobium utilizes GB to produce energy, providing Methanohalophilus with substrates. Similarly, Methanohalophilus uses glycine to synthesize GB for osmoprotection, while Ca. Uticabacter uses glycine to reduce sarcosine to the methanogenic substrate, monomethylamine. In addition to amino acids, sugars like trehalose and maltose can also be used as an energy source (Halanaerobium) and an osmoprotectant (Geotoga). Overall, our study focuses on the multiple uses for amino acids (and their derivatives) in facilitating microorganism growth and maintenance in up to 2,500-m-deep fractured shales. Also hinting at the importance of organic nitrogen to rock-hosted systems, Lloyd et al. (34) demonstrated the significance of detrital proteins to supporting life in deep marine sediments. It is intriguing to speculate that these nitrogen transformations may be a conserved metabolism in the deep biosphere.
Noncompetitive Substrates and End Products Uncovered in Laboratory Reactors
In addition to predation, mutualism, and competition, we identified noncompetitive substrates that provide energy for a single organism. Our proteomics data showed Halanaerobium uniquely fermented ethanolamine (EutBCEGH) and trehalose (TrePP) (SI Appendix, Figs. S1 and S9), with the former substrate likely relevant to shale where ethanolamine is provided exogenously as a corrosion inhibitor and endogenously through biomass turnover of cell membranes (35). Collectively, the interconnected amino acid and sugar fermentations result in the buildup of methane, ammonium, formate, and acetate (Fig. 4). Acetate was the most abundant produced metabolite, with the greatest production rate occurring before TM (Fig. 4). As expected, Halanaerobium-mediated GB reduction was responsible for the increased concentration of acetate between the two treatments, accounting for 97% of the difference between amended and nonamended GB microcosms. Congruent with the observed accumulation of acetate in microcosm studies, Geotoga, Ca. Uticabacter, and Halanaerobium all expressed genes for acetate production, with a fourfold to ninefold greater expression of acetate kinase from Halanaerobium. Other carbon sources supporting acetate production include trehalose and ethanolamine fermentation by Halanaerobium and ethylene glycol fermentation by Halanaerobium and Geotoga, which together could explain a third of the acetate produced in the nonamended reactors (Dataset S1). The fermentation of ethylene glycol may be important to fractured shales in the field, where this compound is commonly added to input fluids for use as a gelling agent in HF (https://fracfocus.org/chemical-use).
Extending Laboratory Reactor Findings to the Field Scale: Microcosm-Generated Hypotheses Are Validated in Appalachian Basin-Produced Fluids
Batch-operated laboratory microcosms more readily permitted the quantification of gas and metabolic waste products generated by the shale microbial consortia (Fig. 4), allowing mass-balance calculations that are currently not feasible at the field scale. Key outcomes from our laboratory microcosms included (i) deciphering trade-offs between osmoprotectant and energy use, (ii) unveiling the pervasive Stickland fermentation network, and (iii) discovering new interconnected metabolites that may be essential to the shale metabolic economy (Fig. 4). Specifically, we demonstrated that GB is a keystone metabolite that not only is vital to salinity adaptation but also is fermented to TMA and acetate by Halanaerobium, a metabolism that subsequently fuels methane production by Methanohalophilus. Glycine was the most connected metabolite, with proteomics indicating use as an energy source for Halanaerobium Stickland reactions, transportation into the cell for osmoprotectant generation by Methanohalophilus, and intracellular synthesis for assimilatory purposes by Geotoga.
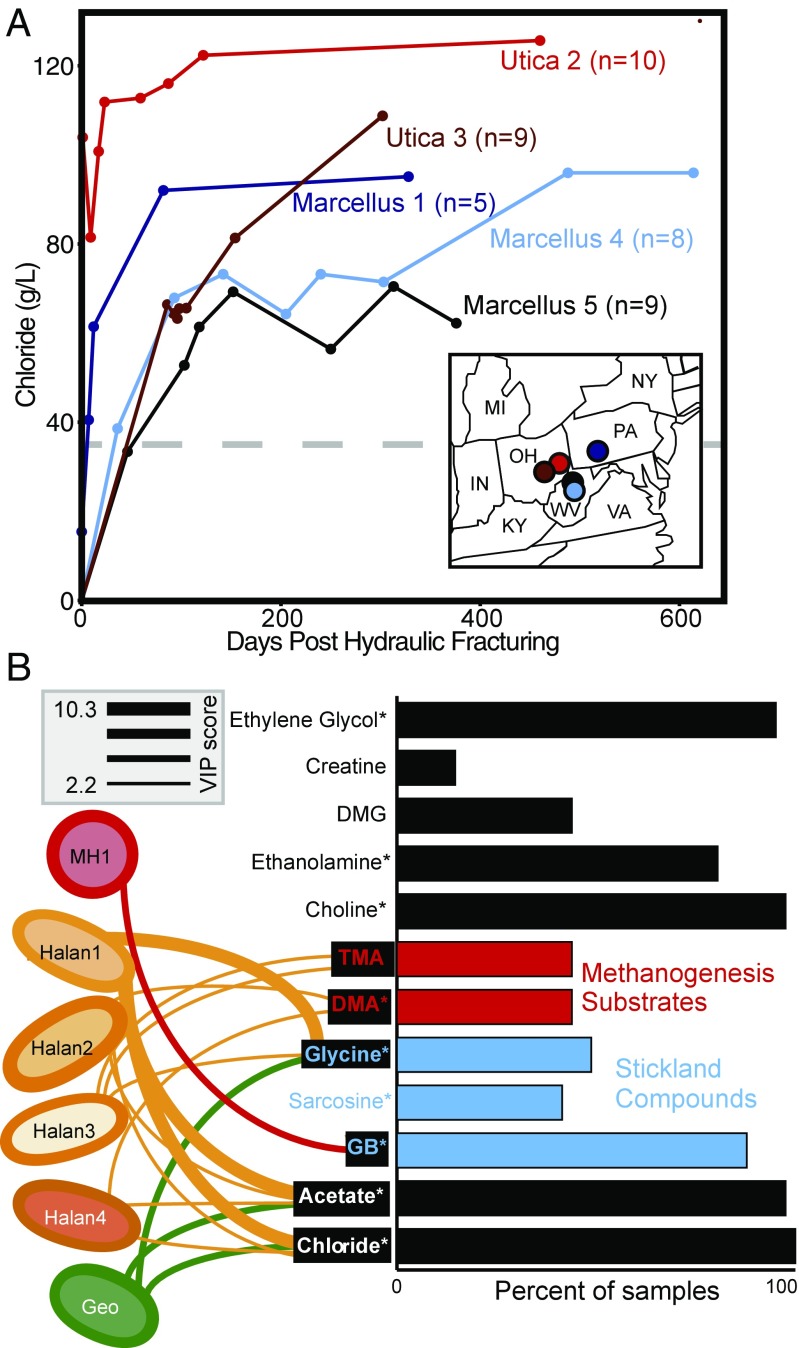
To quantify the relevance of these laboratory-identified processes at the field scale, we analyzed input and produced fluid metagenomic and metabolite data. This includes samples from one previously published well (n = 5; ref. 8) and 36 samples from four additional wells reported here. For each well, samples span input fracture fluids to produced fluids collected up to at least 300 d after HF. Along this timescale, fluids transition from freshwater to hypersaline (>35 chloride g/L) (Fig. 5). From our field metagenome data, we defined microbial strain membership and relative abundance across the samples using a single copy, conserved marker gene, RpsC (Materials and Methods and Dataset S5). Across these produced fluid metagenomes, we identified multiple strains of Halanaerobium and Methanohalophilus (nine and three strains, respectively) and a single strain of Geotoga. Ca. Uticabacter was removed from this analysis due to detection in less than five samples. For comparison, reliance on 16S rRNA gene diversity would have only resolved a single sequence type for each of these genera, showing this strain-level resolution better captured the genotypic microdiversity previously observed in shale fluids (8).
Fig. 5.
(A) Chloride concentrations for field samples with paired metabolites and metagenomes are shown (n = 41), with color denoting well. Above the dashed line, hypersaline conditions are indicated. Circles on the Inset map show each well’s geographic location. (B) Bar graph shows the prevalence of key metabolites uncovered by laboratory experiments across 41 input and produced fluid samples from five wells. Substrates are colored by metabolism (red, methanogenesis substrates; blue, Stickland reaction substrates). Asterisks signify metabolites detected in at least one of the input fluids described here. Concentration of field metabolites that could be significantly predicted (sPLS regression, R2 > 0.1) by the field relative abundance of microorganisms are denoted with black boxes. Taxa from microcosm experiments that were significant variables (VIP values >2) in metabolite prediction are shown by connections between metabolites and Halanaerobium (Halan), Methanohalophilus (MH), and Geotoga (Geo), with the thickness of the line denoting variable importance. The top three predictions are shown for each strain, with Halanaerobium strains numbered 1–4.
Consistent with the laboratory reactor data, metabolites related to osmoprotection were highly correlated in produced fluids across shale wells, with GB, choline, sarcosine, and creatine positively correlated to chloride (SI Appendix, Fig. S10). Of these compounds, GB is generally regarded as the most potent osmoprotectant (17), and thus it is possible that sarcosine and creatine may instead support GB biosynthesis as outlined in SI Appendix. Given that several of these metabolites are detected in the input fluids and are known additives in the fracturing process (https://fracfocus.org/chemical-use) (Fig. 5 and Dataset S1), this finding provides further evidence that chemicals added during HF support life in this man-made ecosystem.
Similar to our laboratory microcosms, field metabolite analyses showed that Stickland reaction substrates have significant coordinated associations. GB was positively correlated to TMA across produced fluids from fractured shales, supporting the notion that this osmoprotectant can be fermented to yield methanogenic substrates (Fig. 3 and SI Appendix, Fig. S10). Additionally, another Stickland electron acceptor identified in our microcosm, sarcosine, was removed concomitant with the production of acetate, signifying that methylamine fermentation may contribute to acetate buildup in shales (Fig. 4). Using both our laboratory-based proteomic findings and field metabolite data, we conclude that GB fermentation is likely mediated with threonine, leucine, and glycine as possible electron donors in the field. Lysine was not detected in produced fluids, which may suggest rapid consumption in the field. Similarly, hydrogen may also represent an important electron donor that cannot be accurately measured in the field. Alternatively, we must consider the absence of measured Stickland donors in the field may signify that electron donors could be an important constraint on microbial methane production in situ. Collectively, our field metabolite and metagenome data signify the ubiquity of the Stickland reaction across shale well microbial communities.
Across the field-produced fluid samples, microbial communities converge at late time points (>40 d after HF), despite initial differences in inoculum, well operator, or location (SI Appendix, Fig. S10). Thus, we next examined whether the relative abundance of these produced fluid microbial communities were predictive of metabolites in the shale produced fluids. Partial least-squares (PLS) regressions demonstrated that the produced fluid microbial community composition predicted the concentration of seven metabolites in field-derived fluids. These predicted metabolites included acetate, glycine, TMA, DMA chloride, ethanolamine, and GB (Fig. 5), many of which were integral metabolites identified in our laboratory microcosms (Fig. 4). However, ethanolamine was not included in Fig. 5 or these remaining analyses because the correlations supporting this prediction in the field data may be spurious (SI Appendix, Fig. S11).
To better resolve the microbial strains associated with shale chemistries, we ranked the organisms’ contribution to metabolite prediction using a value importance in projection (VIP) score to define significance (>2). A single Halanaerobium strain contributed to the predictions of all seven metabolites, with the top five highest VIP scores linking one strain to predictions of chloride, acetate, glycine, TMA, and GB. This finding is consistent with our laboratory reactor inferences suggesting that, in saline fluids (chloride), Halanaerobium uses glycine to reduce GB, generating TMA and acetate. Additionally, the other three Halanaerobium were each predictive of different metabolite profiles, suggesting niche partitioning at the strain level may occur in this ecosystem.
Other genera identified in our laboratory microcosms also had predictive value at the field scale. For instance, for Geotoga, the highest predictive score was for glycine concentrations, consistent with our laboratory proteomic evidence for glycine production via the glycine cleavage system. Methanohalophilus, which is only detected in low abundance in persisting shale microbial communities, had a strain that was predictive of GB concentrations. This is supported by our laboratory proteomics data showing the osmoprotection by these methanogens may represent a microbial source for this keystone metabolite in shales. Alternatively, this relationship to GB could be explained by the dependency of Methanohalophilus on GB fermentation for the synthesis of methylamine substrates. Collectively, this regression-based modeling of the field-collected chemical and biological data revealed a near-perfect congruence between metabolisms active in our laboratory microcosm and field-scale biogeochemistry across geographically and geologically distinct fractured shales.
Conclusion
This study demonstrates how cultivation-based investigations, coupled to high-resolution metaomics in both the laboratory and field, can help establish paradigms for microorganisms influencing terrestrial microbial ecology and biogeochemistry. Laboratory microcosms minimized many of the physical, chemical, and biological confounding factors that prevent elucidation of metabolic interactions in the field. Results from these reactors enabled us to tease apart the complex intertwined metabolisms and trade-offs that underpin even a “simple” microbial community (Fig. 4). Using regression-based modeling, we show that the relative abundance of the few bacterial taxa identified in our microcosms can predict a significant fraction of the carbon and nitrogen variability in hydraulically fractured shales. The Stickland reactions identified in this study are critical to microbial persistence, providing gene targets for other protein-rich environments including the human gut (36) and soils (37), where the importance of this amino acid metabolism is largely unrealized. Since our laboratory results retain their applicability at the field scale, they provide a conceptual framework to better understand or even manipulate desired biogeochemical processes in the deep terrestrial biosphere.
Materials and Methods
Experimental Design and Sample Collection.
HF input fluids and shale-produced fluids were collected from well heads and gas–fluid separators. These fluids were collected from five wells in the Utica and Marcellus shales, in Ohio (n = 2), West Virginia (n = 2), and Pennsylvania (n = 1). Our earlier study temporally characterized geochemical and microbiological signatures of produced fluids collected from Marcellus well 1 (8). This study contributes geochemical and metagenomic data from four additional wells in the Utica and Marcellus shales (Datasets S1 and S3).
In this study, a single sample from the Utica well 2 time series was used to build microcosms to assess microbial interactions among shale microorganisms. The single produced fluid sample was collected from a gas–fluid separator in October 2014 (day 96 post-HF) from an oil-gas well in Ohio, United States. The microcosm experiment consisted of three treatments: (i) 5 mM GB and produced fluid, (ii) no GB and produced fluid, and (iii) 5 mM GB and no produced fluid. Each treatment was done in triplicate and consisted of 10% anoxic, produced fluid (day 96) and 90% sterile modified DSMZ 479 media dispensed in Balch tubes sealed with butyl rubber stoppers and aluminum crimps under an atmosphere of N2/CO2 [80:20 (vol/vol)]. Before mixing with produced fluids, the modified DSMZ medium (per liter) included 87 g of sodium chloride, 1.5 g of potassium chloride, 6.0 g of magnesium chloride, 0.4 g of calcium chloride, 1.0 g of ammonium chloride, 2.0 g of yeast extract, 2.0 g of trypticase peptone, 0.2 g of coenzyme M, 0.2 g of sodium sulfide, and 4.0 g of sodium bicarbonate, and was brought to a pH of 7.2 using 1 mM NaOH. This undefined medium was selected for two reasons: (i) to facilitate sufficient biomass production necessary for proteomics measurements and (ii) to try to capture the undefined nature of many of the compounds used in the fracturing process (https://fracfocus.org/chemical-use). Growth curves were done in triplicate for each treatment, using optical density measurements at 600 nm as an analog for microbial growth (Dataset S2). Microcosm methane production was quantified at every microcosm time point that growth was measured using a Shimadzu (GC-2014) gas chromatograph equipped with a thermal conductivity detector using helium as a carrier gas at 100 °C. All GC measurements are included in Dataset S2. Samples for metagenomics, metabolites, and proteomics were taken at the beginning (T0), at maximum cell density on day 2 (TM), and upon substantial methane production on day 20 (TF) (Dataset S2). To reflect the natural salinity gradient established in hydraulically fractured wells, for example, chloride ranges from 8.3 mg/L in the input to 95 g/L over the 328 d of well sampling, our microcosms were established with a salinity of ∼94 g/L chloride.
Microcosm and Field Fluid Chemistry Analysis.
Twenty-one fluid samples from microcosm experiments and 40 samples from Utica and Marcellus produced fluids were filtered (0.2 μm) at time of collection and sent to the Pacific Northwest National Laboratory for metabolite analysis by NMR. Samples were diluted by 10% (vol/vol) with 5 mM 2,2-dimethyl-2-silapentane-5-sulfonate-d6 as an internal standard. All NMR spectra were collected using a Varian Direct Drive 600-MHz NMR spectrometer equipped with a 5-mm triple resonance salt-tolerant cold probe. The 1D 1H NMR spectra of all samples were processed, assigned, and analyzed using Chenomx NMR Suite 8.3 with quantification based on spectral intensities relative to the internal standard. Candidate metabolites present in each of the complex mixtures were determined by matching the chemical shift, J-coupling, and intensity information of experimental NMR signals against the NMR signals of standard metabolites in the Chenomx library. The 1D 1H spectra were collected following standard Chenomx data collection guidelines (38), using a 1D NOESY presaturation (TNNOESY) experiment with 65,536 complex points and at least 512 scans at 298 K. Additionally, 2D spectra (including 1H–13C heteronuclear single-quantum correlation spectroscopy, 1H-1H total correlation spectroscopy) were acquired on most of the fluid samples, aiding in the 1D 1H assignments of acetate, ethanol, ethylene glycol, methanol, and methylamine (MMA). Biological triplicates had similar metabolite pools, with all data reported (Dataset S1). Fluid samples from the no-cell control were done in single and showed consistent metabolite concentrations throughout the experiment. NMR metabolite methods and analyses of Marcellus 1 and Utica 2 produced fluids were reported previously in Daly et al. (8). Here, we reanalyzed the same produced fluids to search for important compounds outlined by proteomics in the two wells presented in Daly et al. (e.g., lysine) and analyzed produced fluids from three additional wells (Dataset S1). Chloride concentrations from produced fluids were obtained using a Thermo Scientific Dionex ICS-2100 ion chromatograph and are included in Dataset S1.
Metagenomic Sequencing and Assembly.
Total nucleic acids were extracted from five microcosm samples [inoculum (T0), GB + cells at TM, GB + cells at TF, no GB + cells at TM, and no GB + cells at TF] using the PowerSoil DNA Isolation kit (MoBio), eluted in 100 μL, and stored at −20 °C until sequencing. DNA for the microcosm inoculum (T0) was submitted for sequencing at the Genomics Shared Resource facility at The Ohio State University. Libraries were prepared with the Nextera XT Library System in accordance with the manufacturer’s instructions. Genomic DNA was sheared by sonication, and fragments were end-repaired. Sequencing adapters were ligated, and library fragments were amplified with five cycles of PCR before solid-phase reversible immobilization size selection, library quantification, and validation. Libraries were sequenced on the Illumina HiSeq 2500 platform, and paired-end reads of 113 cycles were collected. The other four metagenomes were sequenced at the Joint Genome Institute. Briefly, libraries were created and quantified using an Illumina Library creation kit (KAPA Biosystems) with solid-phase reversible 402 immobilization size selection. Libraries were then sequenced on the Illumina HiSeq 2500 sequencing platform utilizing a TruSeq Rapid paired-end 404 cluster kit. DNA was extracted and sequenced from all produced and input fluids as outlined previously (8). All raw reads from microcosms, produced fluids, and input fluids were trimmed from both the 5′ and 3′ ends with Sickle, and then each sample was assembled individually with IDBA-UD (8, 39–41) using default parameters. Metagenome statistics including amount of sequencing are noted in Dataset S3.
Metagenome Binning and Annotation for Proteomics Database.
All scaffolds ≥2.5 kb were included when binning genomes from the metagenomic assembly. Scaffolds were annotated as described previously (8). Briefly, ORFs were predicted with MetaProdigal (42), and sequences were compared with USEARCH (43) to KEGG, UniRef90, and InterProScan (44) with single and reverse best-hit matches of >60 bases reported. We obtained near-complete, curated draft (>93% estimated completion, <1% overages) genome resolved bins using a combination of phylogenetic signal, coverage, and GC content, for a Halanaerobium, Ca. Uticabacter, Methanohalophilus, and Geotoga (Dataset S3). As described previously (8, 39), genome completion was estimated based on the presence of core gene sets (Bacteria, 31 genes, and Archaea, 104 genes), using Amphora2 (45). Contamination (gene copies >1 per bin) indicating potential misbins, along with GC and phylogeny, were used to manually remove potential contamination from the bins. Given the dominance and high strain variation in some samples, highly abundant genomes (>400×, bacterial and viral) often failed to assemble. To recover these genomes, subassemblies were performed to reconstruct the dominant genomes, using 10%, 5%, and 1% of the reads (8). Given the high strain variation, we were able to recover only a single near-complete Halanaerobium bin from the most abundant strain using a 1% subassembly. However, we know there were at least two other strains of Halanaerobium in the microcosm. To capture the most proteomic signal, we binned Halanaerobium as a whole from the inoculum to create a community Halanaerobium bin. This allowed us to see the activity of Halanaerobium as a whole in the microcosm; thus, here we refer to Halanaerobium at the genus level. All genome statistics including 16S rRNA gene presence, completion, and length are reported in Dataset S3. Fasta files of nucleotide and amino acid sequences for each genome bin are included in Datasets S6 and S7, respectively.
Near–full-length ribosomal 16S rRNA gene sequences were reconstructed from unassembled Illumina reads from microcosms and input and produced fluids using EMIRGE (46). To reconstruct 16S rRNA gene sequences, we followed the protocol with trimmed paired-end reads where both reads were at least 20 nt used as inputs and 50 iterations. EMIRGE sequences were chimaera checked before phylogenetic gene analyses. EMIRGE abundances for the microcosm experiment are shown in SI Appendix, Fig. S4. Necessary scripts and analyses to perform metagenome assembly, EMIRGE, annotation, and single-copy genes can be accessed from github (https://github.com/TheWrightonLab/metagenome_analyses).
Viral genomes were identified from all subassemblies using VirSorter (47, 48) hosted on the CyVerse discovery environment (49) (Dataset S3). VirSorter was run with default parameters using the virome database, retaining viruses and prophage with category 1 and 2 status. Viral genomes were then clustered using GenomeCluster hosted on the CyVerse discovery environment with 95% average nucleotide identity over 80% of the smallest contig (48). We combined the four microbial and 16 unique viral genome bins to build the metagenomic database for proteomic assessment.
Metaproteomic Extraction, Spectral Analysis, and Data Acquisition.
Liquid culture (1.2 mL) from each microcosm sample was collected anaerobically, centrifuged for 15 min at 10,000 × g, separated from the supernatant, and stored at −80 °C until shipment to Pacific Northwest National Laboratory. Proteins in the pellet were precipitated and washed twice with acetone. Then the pellet was lightly dried under nitrogen. Filter-aided sample preparation kits were used for protein digestion according to the manufacturer’s instructions. Resultant peptides were snap-frozen in liquid N2, digested again overnight, and concentrated to ∼30 μL using a SpeedVac (Labconco). Final peptide concentrations were determined using a bicinchoninic acid assay. All mass-spectrometric data were acquired using a Q-Exactive Plus (Thermo Scientific) connected to an nanoACQUITY UPLC M-Class liquid chromatography system (Waters) via in-house 70-cm column packed using Phenomenex Jupiter 3-μm C18 particles and in-house built electrospray apparatus. MS/MS spectra were compared with the predicted protein collections using the search tool MSGF+ (50). Contaminant proteins typically observed in proteomics experiments were also included in the protein collections searched. The searches were performed using ±20-ppm parent mass tolerance, parent signal isotope correction, partially tryptic enzymatic cleavage rules, and variable oxidation of methionine. In addition, a decoy sequence approach (51) was employed to assess false-discovery rates. Data were collated using an in-house program, imported into a SQL server database, filtered to ∼1% false-discovery rate (peptide to spectrum level), and combined at the protein level to provide unique peptide count (per protein) and observation count (that is, spectral count) data. Spectral count data for each identified protein was normalized using normalized spectral abundance frequency (NSAF) calculations, accounting for protein length and proteins per sample (Dataset S4). Note that metaproteomics were not done on produced fluid samples from the field.
Microcosm Metabolic, Phylogenetic, and Statistical Analyses.
Proteins for osmoprotection (SI Appendix, Fig. S5), the Stickland reaction, and other metabolisms discussed were mined from the amino acid annotation files of binned genomes using BLASTp with a bit score cutoff of 60 (a technical homolog) and cross-checked in metaproteomics data. For each metabolism discussed, scaffold and gene location for genes of interest are included (Dataset S3). If >75% of proteins required for a multisubunit enzyme were detected in the proteomics, we gave it the status of detected in the proteome.
Significance of activity reported was based on linear discriminant analysis effect size (LEfSe) (25, 52). LEfSe analysis was performed between time points (e.g., TM to TF in GB) and treatments (e.g., TM of GB to TM of no GB) to find features (proteins) differentially active. LEfSe combines the standard tests for statistical significance (Kruskal–Wallis test and pairwise Wilcoxon test) with linear discriminant analysis (25). It ranks features by effect size, which puts features that explain most of the biological difference at top. LEfSe analysis was performed at the α value of 0.05 for the Kruskal–Wallis test and the threshold of 2 on the logarithmic linear discriminant analysis score for discriminative features. All error bars shown here are indicative of 1 SD from the mean, and all significance statements refer to a P value of less than 0.05.
Phylogenetic analyses were performed for genome bins and metagenomes using ribosomal S3 protein amino acid sequences (genomes and metagenomes) and 16S rRNA genes (genomes only). 16S rRNA genes recovered from microcosm genomes and their nearest neighbors (SILVA database; ref. 53) were aligned using MUSCLE version 3.8.31. The resulting alignment was manually curated and a phylogenetic tree was constructed with RAxML 7.2.9 (GTR Gamma nucleotide model, 100 bootstrap replicates). For the S3 protein tree, amino acid sequences were pulled from the microcosm bins and augmented with sequences mined from National Center for Biotechnology Information and Joint Genome Institute–Integrated Microbial Genomes databases. Sequences were aligned using MUSCLE, version 3.8.31, and run through ProtPipeliner, a Python script developed in-house for generation of phylogenetic trees (https://github.com/TheWrightonLab). A maximum-likelihood phylogeny for the alignment of S3 ribosomal proteins and 16S rRNA genes was conducted using RAxML, version 8.3.1, under the LG+α+γ model of evolution with 100 bootstrap replicates. All phylogenetic trees were visualized in iTOL and can be found in SI Appendix (SI Appendix, Figs. S2 and S3).
Phylogenetic and Statistical Analysis of Field Data.
Ribosomal S3 proteins were used to track strain resolved abundance patterns across the HF dataset (Dataset S5). First, all annotated ribosomal S3 proteins from 41 input and produced fluid metagenomes were pulled to build an S3 database. Then, using Bowtie 2 (54), metagenomic reads were competitively mapped by sample to the S3 database using zero mismatches. Strain resolved relative abundance was obtained by quantifying the percentage of total reads that mapped divided by the length of the sequence and then normalizing to within each sample (https://github.com/TheWrightonLab). Strains included in this analysis had to have 95% of the S3 sequence covered with mapped reads. Ribosomal protein tree with all amino acid sequences used in this analysis was obtained using methods outlined above and is shown in Dataset S8.
To predict fluid metabolites from the microcosm microbial community, we used sparse PLS (sPLS) (55, 56), as implemented in the R package mixOmics (57). In other words, this approach enabled us to model a relationship between microbial abundance and fluid chemistry traits. In addition, the predictors were ranked according to their VIP (58). The VIP measure of a predictor estimates its contribution in the PLS regression. The predictors having high VIP values are assumed important for the PLS prediction of the response variable. The VIP values of the prokaryotic functional subnetworks are provided in Dataset S9. All R scripts for modeling of HF datasets are included in Dataset S10.
Viral Analyses.
We used two methods to link viral contigs to microbial hosts. First, as described previously, CRISPR arrays were identified in each genome bin by using the CRISPR recognition tool plugin in Geneious R8 (8). To link microbial hosts and viruses, we used BLASTn to identify viral contigs with matching spacer sequences. All matches were manually confirmed as perfect matches by aligning sequences in Geneious R8. Second, we used the d2S hexamer frequency dissimilarity measure (59) between viral contigs and host genomes to predict viral–host associations. Analyses were run with five microcosm genomes and 16 viral population representatives. In all cases, the d2S dissimilarity measure predictions were congruent with CRISPR spacer array linkages.
In SI Appendix, Fig. S6, expressed viral proteins are divided into seven categories: DNA/replication, lysogeny, structure, lysis, hypothetical, transposase, and other. DNA/replication category referred to amino acid sequences associated with DNA metabolism such as DNA methyltransferases and helicases. Lysogeny refers to the viral lysogenic cycle and was made up of recombinases and resolvases. The structural category included tail sheath proteins, terminases, and phage tail tape measures. The transposase category was only made up of transposase-associated amino acid sequences, while hypothetical referred to proteins of unknown function or hypothetical distinction.
Supplementary Material
Acknowledgments
The authors would like to thank the anonymous reviewers, as well as Duncan J. Kountz for their insightful comments. M.A.B. is partially funded by Faye Fellowship from the Ohio State University Environmental Science Graduate Program. The following PIs (K.C.W., P.J.M., D.R.C., S.S., and M.J.W.) and their respective affiliates are partially supported by funding from the National Sciences Foundation Dimensions of Biodiversity (Award 1342701). Samples for this research were provided by the Marcellus Shale Energy and Environment Laboratory funded by Department of Energy’s National Energy Technology Laboratory Grant DE FE0024297. Metagenomic sequencing for this research was performed by the Joint Genome Institute via a large-scale sequencing award (Award 1931) (to K.C.W.). The work conducted by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the US Department of Energy under Contract DE-AC02-05CH11231. The NMR data and MS-proteomics data in this work was collected via a general proposal (ID 49615) using instrumentation at the Environmental Molecular Science Laboratory, a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory. Pacific Northwest National Lab is operated by Battelle for the DOE under Contract DE-AC05-76RL01830.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genomes resolved from the microcosm dataset reported in this paper have been deposited in National Center for Biotechnology Information BioProject (accession number PRJNA308326; biosample numbers SAMN05172267, SAMN05172290, SAMN06343770, and SAMN06343770). All of the metagenomic nucleotide files reported in this paper have been deposited in the Joint Genome Institute Genome Portal database, genome.jgi.doe.gov/, or with the National Center for Biotechnology Information (see Dataset S3 for accession numbers). All mass spectrometry proteomics data from all microcosm experiments reported in this paper have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (dataset identifiers PXD008490 and 10.6019/PXD008490).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800155115/-/DCSupplemental.
References
- 1.US Energy Information Administration 2017 Electricity explained: Electricity in the United States (US Department of Energy, Washington, DC). Available at https://www.eia.gov/energyexplained/index.php?page=electricity_in_the_united_states. Accessed June 12, 2017.
- 2.Mouser PJ, Borton M, Darrah TH, Hartsock A, Wrighton KC. Hydraulic fracturing offers view of microbial life in the deep terrestrial subsurface. FEMS Microbiol Ecol. 2016;92:fiw166. doi: 10.1093/femsec/fiw166. [DOI] [PubMed] [Google Scholar]
- 3.Cokar M, Ford B, Kallos MS, Gates ID. New gas material balance to quantify biogenic gas generation rates from shallow organic-matter-rich shales. Fuel. 2013;104:443–451. [Google Scholar]
- 4.Booker AE, et al. Sulfide generation by dominant Halanaerobium microorganisms in hydraulically fractured shales. MSphere. 2017;2:e00257-17. doi: 10.1128/mSphereDirect.00257-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akob DM, Cozzarelli IM, Dunlap DS, Rowan EL, Lorah MM. Organic and inorganic composition and microbiology of produced waters from Pennsylvania shale gas wells. Appl Geochem. 2015;60:116–125. [Google Scholar]
- 6.Liang R, et al. Metabolic capability of a predominant Halanaerobium sp. in hydraulically fractured gas wells and its implication in pipeline corrosion. Front Microbiol. 2016;7:988. doi: 10.3389/fmicb.2016.00988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipus D, et al. Predominance and metabolic potential of Halanaerobium spp. in produced water from hydraulically fractured Marcellus shale wells. Appl Environ Microbiol. 2017;83:e02659-16. doi: 10.1128/AEM.02659-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly RA, et al. Microbial metabolisms in a 2.5-km-deep ecosystem created by hydraulic fracturing in shales. Nat Microbiol. 2016;1:16146. doi: 10.1038/nmicrobiol.2016.146. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Yu Z, Zhang H, Thompson IP. Microbial distribution and variation in produced water from separators to storage tanks of shale gas wells in Sichuan Basin, China. Environ Sci Water Res Technol. 2017;3:340–351. [Google Scholar]
- 10.Xiao Z, Samuel M, Tibbles R, Moussa O. 2005. Hydraulic fracturing method. US Patent Appl US20050003965A1.
- 11.Mohan AM, Bibby KJ, Lipus D, Hammack RW, Gregory KB. The functional potential of microbial communities in hydraulic fracturing source water and produced water from natural gas extraction characterized by metagenomic sequencing. PLoS One. 2014;9:e107682. doi: 10.1371/journal.pone.0107682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowers RM, et al. Genome Standards Consortium Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol. 2017;35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konstantinidis KT, Rosselló-Móra R. Classifying the uncultivated microbial majority: A place for metagenomic data in the Candidatus proposal. Syst Appl Microbiol. 2015;38:223–230. doi: 10.1016/j.syapm.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Murali Mohan A, et al. Microbial community changes in hydraulic fracturing fluids and produced water from shale gas extraction. Environ Sci Technol. 2013;47:13141–13150. doi: 10.1021/es402928b. [DOI] [PubMed] [Google Scholar]
- 15.Davis JP, Struchtemeyer CG, Elshahed MS. Bacterial communities associated with production facilities of two newly drilled thermogenic natural gas wells in the Barnett Shale (Texas, USA) Microb Ecol. 2012;64:942–954. doi: 10.1007/s00248-012-0073-3. [DOI] [PubMed] [Google Scholar]
- 16.Cluff MA, Hartsock A, MacRae JD, Carter K, Mouser PJ. Temporal changes in microbial ecology and geochemistry in produced water from hydraulically fractured Marcellus shale gas wells. Environ Sci Technol. 2014;48:6508–6517. doi: 10.1021/es501173p. [DOI] [PubMed] [Google Scholar]
- 17.Oren A. Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev. 1999;63:334–348. doi: 10.1128/mmbr.63.2.334-348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipus D, Vikram A, Ross DE, Bibby K. Draft genome sequence of Methanohalophilus mahii strain DAL1 reconstructed from a hydraulic fracturing-produced water metagenome. Genome Announc. 2016;4:e00899-16. doi: 10.1128/genomeA.00899-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathrani IM, Boone DR, Mah RA, Fox GE, Lau PP. Methanohalophilus zhilinae sp. nov., an alkaliphilic, halophilic, methylotrophic methanogen. Int J Syst Bacteriol. 1988;38:139–142. doi: 10.1099/00207713-38-2-139. [DOI] [PubMed] [Google Scholar]
- 21.Paterek JR, Smith PH. Methanohalophilus mahii gen. nov., sp. nov., a methylotrophic halophilic methanogen. Int J Syst Evol Microbiol. 1988;38:122–123. [Google Scholar]
- 22.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci USA. 2012;109:21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watkins AJ, Roussel EG, Webster G, Parkes RJ, Sass H. Choline and N,N-dimethylethanolamine as direct substrates for methanogens. Appl Environ Microbiol. 2012;78:8298–8303. doi: 10.1128/AEM.01941-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ticak T, Kountz DJ, Girosky KE, Krzycki JA, Ferguson DJ., Jr A nonpyrrolysine member of the widely distributed trimethylamine methyltransferase family is a glycine betaine methyltransferase. Proc Natl Acad Sci USA. 2014;111:E4668–E4676. doi: 10.1073/pnas.1409642111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oren A. Thermodynamic limits to microbial life at high salt concentrations. Environ Microbiol. 2011;13:1908–1923. doi: 10.1111/j.1462-2920.2010.02365.x. [DOI] [PubMed] [Google Scholar]
- 27.Park SY, Liang Y. Biogenic methane production from coal: A review on recent research and development on microbially enhanced coalbed methane (MECBM) Fuel. 2016;166:258–267. [Google Scholar]
- 28.Andreesen JR. Glycine reductase mechanism. Curr Opin Chem Biol. 2004;8:454–461. doi: 10.1016/j.cbpa.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Barker HA. Amino acid degradation by anaerobic bacteria. Annu Rev Biochem. 1981;50:23–40. doi: 10.1146/annurev.bi.50.070181.000323. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz AC, Schäfer R. New amino acids, and heterocyclic compounds participating in the Stickland reaction of Clostridium sticklandii. Arch Mikrobiol. 1973;93:267–276. doi: 10.1007/BF00412026. [DOI] [PubMed] [Google Scholar]
- 31.Stadtman TC, White FH., Jr Tracer studies on ornithine, lysine, and formate metabolism in an amino acid fermenting Clostridium. J Bacteriol. 1954;67:651–657. doi: 10.1128/jb.67.6.651-657.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fonknechten N, et al. Clostridium sticklandii, a specialist in amino acid degradation: Revisiting its metabolism through its genome sequence. BMC Genomics. 2010;11:555. doi: 10.1186/1471-2164-11-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andreesen JR. Glycine metabolism in anaerobes. Antonie Van Leeuwenhoek. 1994;66:223–237. doi: 10.1007/BF00871641. [DOI] [PubMed] [Google Scholar]
- 34.Lloyd KG, et al. Predominant archaea in marine sediments degrade detrital proteins. Nature. 2013;496:215–218. doi: 10.1038/nature12033. [DOI] [PubMed] [Google Scholar]
- 35.White DA. Form and Function of Phospholipids. Elsevier Science; Amsterdam: 1973. The phospholipid composition of mammalian tissues; pp. 441–482. [Google Scholar]
- 36.Fischbach MA, Sonnenburg JL. Eating for two: How metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zindel U, et al. Eubacterium acidaminophilum sp. nov., a versatile amino acid-degrading anaerobe producing or utilizing H2 or formate. Arch Microbiol. 1988;150:254–266. [Google Scholar]
- 38.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: Quantitative analysis of 1H NMR metabolomics data. Anal Chem. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 39.Solden LM, et al. New roles in hemicellulosic sugar fermentation for the uncultivated Bacteroidetes family BS11. ISME J. 2017;11:691–703. doi: 10.1038/ismej.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wrighton KC, et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science. 2012;337:1661–1665. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- 41.Angle JC, et al. Methanogenesis in oxygenated soils is a substantial fraction of wetland methane emissions. Nat Commun. 2017;8:1567. doi: 10.1038/s41467-017-01753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics. 2012;28:2223–2230. doi: 10.1093/bioinformatics/bts429. [DOI] [PubMed] [Google Scholar]
- 43.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 44.Quevillon E, et al. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005;33(Suppl_2):W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu M, Scott AJ. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics. 2012;28:1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 46.Miller CS, Baker BJ, Thomas BC, Singer SW, Banfield JF. EMIRGE: Reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biol. 2011;12:R44. doi: 10.1186/gb-2011-12-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roux S, Enault F, Hurwitz BL, Sullivan MB. VirSorter: Mining viral signal from microbial genomic data. PeerJ. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roux S, et al. Ecogenomics of virophages and their giant virus hosts assessed through time series metagenomics. Nat Commun. 2017;8:858. doi: 10.1038/s41467-017-01086-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Devisetty UK, Kennedy K, Sarando P, Merchant N, Lyons E. Bringing your tools to CyVerse discovery environment using Docker. F1000Res. 2016;5:1442. doi: 10.12688/f1000research.8935.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 52.Borton MA, et al. Chemical and pathogen-induced inflammation disrupt the murine intestinal microbiome. Microbiome. 2017;5:47. doi: 10.1186/s40168-017-0264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guidi L, et al. Tara Oceans Coordinators Plankton networks driving carbon export in the oligotrophic ocean. Nature. 2016;532:465–470. doi: 10.1038/nature16942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen H, Huang JZ. Sparse principal component analysis via regularized low rank matrix approximation. J Multivar Anal. 2008;99:1015–1034. [Google Scholar]
- 57.Lê Cao K-A, Rossouw D, Robert-Granié C, Besse P. A sparse PLS for variable selection when integrating omics data. Stat Appl Genet Mol Biol. 2008;7:35. doi: 10.2202/1544-6115.1390. [DOI] [PubMed] [Google Scholar]
- 58.Chong I-G, Jun C-H. Performance of some variable selection methods when multicollinearity is present. Chemom Intell Lab Syst. 2005;78:103–112. [Google Scholar]
- 59.Ahlgren NA, Ren J, Lu YY, Fuhrman JA, Sun F. Alignment-free $d_2^*$ oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res. 2017;45:39–53. doi: 10.1093/nar/gkw1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.