Significance
Here, we capture and study a reactive iron porphyrin carbene (IPC) intermediate in the heme binding pocket of an engineered cytochrome c protein. IPCs have never before been directly characterized in a protein, although they are thought to be the key catalytic intermediate common to an array of abiological but synthetically useful carbene transfer reactions catalyzed by wild-type and engineered heme proteins. Our work provides insight into how a “carbene transferase” acquired its new-to-nature function as well as how it facilitates efficient and selective transfer of the carbene to a second substrate. Knowledge gained by studying this versatile intermediate provides a foundation for studying the mechanisms of carbene transfer reactions and will facilitate the engineering of carbene transfer enzymes.
Keywords: carbene, reactive intermediate, heme, metalloenzyme, carbene transferase
Abstract
Recently, heme proteins have been discovered and engineered by directed evolution to catalyze chemical transformations that are biochemically unprecedented. Many of these nonnatural enzyme-catalyzed reactions are assumed to proceed through a catalytic iron porphyrin carbene (IPC) intermediate, although this intermediate has never been observed in a protein. Using crystallographic, spectroscopic, and computational methods, we have captured and studied a catalytic IPC intermediate in the active site of an enzyme derived from thermostable Rhodothermus marinus (Rma) cytochrome c. High-resolution crystal structures and computational methods reveal how directed evolution created an active site for carbene transfer in an electron transfer protein and how the laboratory-evolved enzyme achieves perfect carbene transfer stereoselectivity by holding the catalytic IPC in a single orientation. We also discovered that the IPC in Rma cytochrome c has a singlet ground electronic state and that the protein environment uses geometrical constraints and noncovalent interactions to influence different IPC electronic states. This information helps us to understand the impressive reactivity and selectivity of carbene transfer enzymes and offers insights that will guide and inspire future engineering efforts.
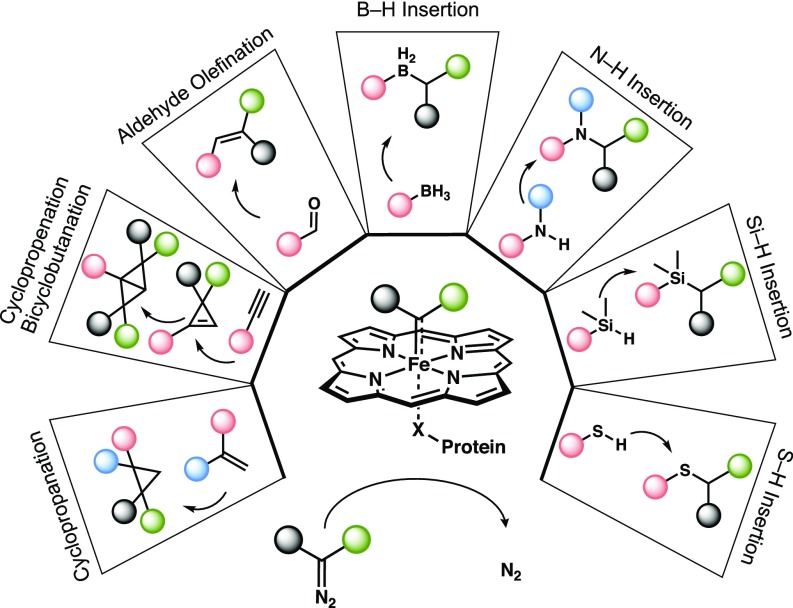
Carbenes are formally neutral, divalent carbon species with two nonbonded electrons. They are typically reactive due to their incomplete octet electron configuration (1). Highly stabilized (“persistent”) carbenes exist in biological systems, most notably as the deprotonated thiazolium group of thiamine in pyruvate oxidases (2). In contrast, reactive metal-carbenes have not been found in nature, despite their versatility and utility in chemical synthesis (3–5). Recent protein engineering efforts have revealed that heme proteins can catalyze a myriad of formal carbene transfer reactions, including alkene cyclopropanation (6–9), alkyne cyclopropenation and bicyclobutanation (10), and carbonyl olefination (11) as well as carbon−boron (12), –nitrogen (13), –silicon (14), and –sulfur (15) bond formation (Fig. 1). The proteins enable the heme center to make products that iron porphyrin alone does not (10, 12), and their genetically encoded structures direct selectivity that can be switched by evolution (6, 8–10, 12, 14), making it possible to create whole new biocatalytic pathways to important molecules.
Fig. 1.
The scope of laboratory-evolved biocatalytic carbene transfer reactions. Engineered heme proteins catalyze a broad range of abiological reactions with diazo reagents. These reactions are assumed to proceed through a catalytic IPC intermediate. X indicates porphyrin proximal ligand, usually serine or histidine. All published examples of heme-dependent carbene transfer enzymes accept diazo reagents having one or more electron-withdrawing groups (e.g., COOR, CF3) as carbene precursors. The active resting state of the catalyst is Fe(II)-porphyrin.
By analogy to known transition metal-catalyzed reactions that use diazo reagents as carbene precursors (3–5), the emerging enzymatic carbene transfer activities of heme proteins are all thought to involve a reactive iron porphyrin carbene (IPC) intermediate; transfer of the carbene to a second substrate and product release regenerate the protein catalyst (6–15). Our current understanding of enzyme-catalyzed carbene transfer reactions comes primarily from experimental characterizations of model IPC compounds (16–21) and their computational studies (22–27). Determining the structures and properties of reactive intermediates is central to deciphering how enzymes enhance reaction rates and dictate selectivity (28), and recent mechanistic studies of cyclopropanation enzymes (24, 26, 27) have also underscored the importance of the IPC intermediate and the need to characterize it within a protein scaffold.
Results and Discussion
Structural Characterization of Engineered Carbene Transferase, Rhodothermus marinus cytochrome c V75T M100D M103E.
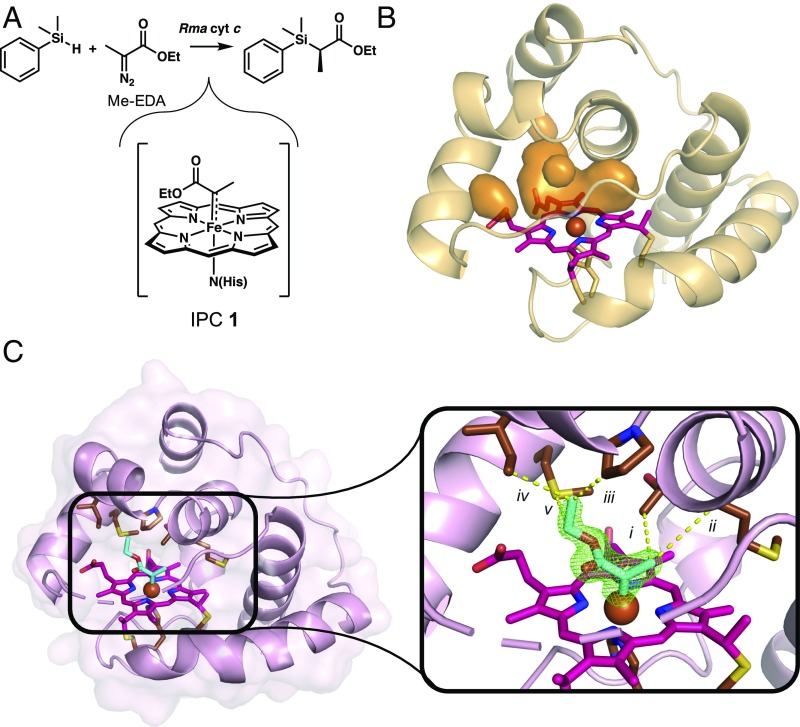
Rhodothermus marinus (Rma) cytochrome c (cyt c) is an electron transfer protein [Protein Data Bank (PDB) ID code 3CP5] (29) previously engineered to catalyze the formation of carbon−silicon (14) and carbon−boron (12) bonds by inserting a reactive iron-carbene into the corresponding Si–H and B–H bonds of a silane or borane substrate. We set out to capture and characterize the IPC intermediate in Rma cyt c by turning to Rma cyt c V75T M100D M103E (TDE), a variant previously engineered to catalyze silylation reactions with high efficiency (14). Rma TDE was discovered after three rounds of site saturation mutagenesis and screening for enhanced activity, which resulted in amino acid substitutions V75T, M100D, and M103E. This protein is capable of catalyzing thousands of rounds of carbene transfer using ethyl 2-diazopropanoate (Me-EDA) (Fig. 2A) as a carbene precursor, presumably through the intermediacy of IPC 1 (Fig. 2A). To visualize the active site for carbene transfer, we obtained an X-ray crystal structure of Rma TDE at 1.47 Å (PDB ID code 6CUK) (SI Appendix, Table S1A). This structure reveals a pocket on the distal face of the heme created by the three mutations (Fig. 2B). In Rma TDE, the native distal methionine has been mutated to an aspartate (M100D), the α-carbon of which is displaced by 4.8 Å relative to its position in the wild-type structure (SI Appendix, Fig. S1). This displacement produces a cavity in the space previously occupied by the M100 sidechain and allows a water molecule (or hydroxide ion) to coordinate to the heme iron in the enzyme resting state. Overall, directed evolution created an entirely new active site in Rma TDE, where substrates bind, the carbene forms, and catalysis takes place.
Fig. 2.
Carbene transfer silylation reaction and structures of resting and carbene-bound Rma TDE. (A) The C–Si bond-forming reaction catalyzed by Rma cytochrome c proceeds via intermediate IPC 1. (B) Mutations V75T, M100D, and M103E (Rma TDE) in Rma cytochrome c increased its carbene transfer activity (14). The crystal structure of Rma TDE with no Me-EDA bound (1.47 Å; PDB ID code 6CUK) reveals a cavity distal to the heme (orange surfaces) that is not present in wild-type Rma cyt c. (C) The crystal structure of carbene-bound Rma TDE (1.29 Å; PDB ID code 6CUN), with the carbene species in cyan. Residues D100, T101, and D102 are unresolved. (Inset) Omit map in green (FO–FC) of the carbene electron density contoured at 3σ. Interactions between the carbene and amino acid residues are shown with yellow lines. (i) T75, 3.7 Å. (ii) M76, 4.3 Å. (iii) P79, 3.5 Å. (iv) I83, 3.6 Å. (v) M89, 3.5 Å.
Crystallographic Observation of an IPC in Rma TDE.
To observe an IPC in the protein scaffold, crystals of Rma TDE were soaked with Me-EDA at room temperature for 30 min under air, reduced with sodium dithionite, and flash frozen in liquid nitrogen before diffraction. Under these conditions, we determined an X-ray crystal structure at 1.29 Å that shows a species bound to the iron porphyrin (PDB ID code 6CUN) that was not present in our previous structure of Rma TDE (PDB ID code 6CUK) (SI Appendix, Table S1A). We observed contiguous electron density from the iron porphyrin to an active site ligand, consistent with an enzymatic IPC intermediate, rather than iron-bound Me-EDA (Fig. 2C). The observed electron density is in agreement with a single carbene species in a single conformation (SI Appendix, section IIH), suggesting that this structure represents the dominant form of the IPC intermediate in solution during catalysis. The high occupancy of the carbene indicates that the rate of carbene formation greatly exceeds the rate of carbene decay in the absence of a second substrate, making the direct characterization of an enzymatic IPC intermediate possible.
The crystal structure of carbene-bound Rma TDE displays some similarities to previously studied small molecule IPC complexes. The Fe–C bond length of 1.9 Å is consistent with the previously determined structure of the IPC 1-methylimidazole-ligated mesotetrakis(pentafluorophenyl)porphyrin (TPFPP) diphenylcarbene, where the reported Fe–C bond length is 1.83 Å (20). The Fe–N (imidazole) bond length of 2.1 Å in the protein is also consistent with that in the small molecule structure (2.17 Å) (20). Out-of-plane distortions, particularly ruffling distortions, are observed in both the imidazole-ligated TPFPP diphenylcarbene structure and the carbene-bound Rma TDE structure. Whereas the mean out-of-plane deviation for TPFPP diphenylcarbene was 1.03 Å (SI Appendix, Table S2), it is smaller for carbene-bound Rma TDE (0.6 Å; the deviation for Rma TDE alone is 0.7 Å). The difference in distortion between both Rma TDE structures is small, and both values are within the range reported for c-type cytochrome proteins (0.3–1.2 Å) (30). Ruffling is known to be the main distortion in these proteins and is thought to be induced by covalent attachment of the heme to the protein (31).
In contrast to what we observed in Rma TDE in the absence of Me-EDA, amino acid residues D100, T101, and D102 are unresolved in the carbene-bound Rma TDE structure, indicating that this loop region is flexible and the carbene is solvent accessible (Fig. 2C). The crystal structure also reveals close, nonpolar contacts between the iron-carbene and five amino acid sidechains (T75, M76, P79, I83, and M89 are 3.5–4.3 Å from the carbene) (Fig. 2C), suggesting that the active site is strongly hydrophobic, despite its solvent accessibility.
IPC Electronic Structure and Spectroscopy.
When first proposed as a reactive enzyme intermediate, the IPC in cytochrome P450 was presented as analogous to oxoferryl intermediates found in the natural P450 catalytic cycle (6). This assignment was supported by the Fe(IV)-like Mössbauer parameters obtained for small molecule IPC complexes (20, 21, 32). However, Zhang and coworkers (24, 26, 27) have argued that IPCs are better described as Fe(II) closed shell singlets, lower in energy than the triplet states. Most recently, Shaik and coworkers (23) studied a model IPC complex and concluded that the lowest energy state for their IPC was an open shell singlet, best described as an S = 1/2 Fe(III) that is antiferromagnetically coupled to a carbene radical. Although the IPC open shell singlet is similar to the Fe(III)-superoxide electronic structure in heme oxygen binding (33), no experimental evidence has been published to support the relevance of an open shell singlet configuration in IPCs. Computational studies from both the Zhang group and the Shaik group suggest that the energy differences between electronic states are small (22–24, 27), and thus, the open shell singlet, closed shell singlet, and triplet states should all be considered plausible ground states for the IPC intermediate in the environment of an enzyme active site.
Encouraged by our success in capturing the IPC in the enzyme crystals, we sought to use spectroscopy to characterize the electronic structure of the IPC in Rma TDE. Under conditions similar to those where Rma TDE is catalytically active, we prepared solution samples of both Rma TDE and Me-EDA–treated Rma TDE and analyzed them with ultraviolet-visible (SI Appendix, Fig. S2), electron paramagnetic resonance (EPR), and Mössbauer spectroscopy. Neither sample gave EPR signals in either parallel (10 mW) or perpendicular (2 mW) mode at 4 K (SI Appendix, Fig. S3). While the absence of EPR signals under these conditions does not prove that the samples are diamagnetic, these results are consistent with our assignment of the resting enzyme and carbene-bound enzyme as low-spin species (vide infra). Mössbauer spectra of Rma TDE at 80 K were best fit as a single species with an isomer shift (∂Fe) of 0.45 mm·s−1 and a quadrupole splitting (|∆EQ|) of 1.18 mm·s−1 (SI Appendix, Fig. S4), consistent with an S = 0 low-spin Fe(II) resting-state species. This observation is in contrast to other Fe(II) hemes in proteins lacking an endogenous distal ligand to iron, which have been shown to favor high-spin (S = 2) states (34, 35). Mössbauer spectra of Me-EDA–treated Rma TDE at 80 K indicated the presence of a major species (88% abundance) and a minor species (12% abundance), the latter of which was assigned as ferrous Rma TDE. The major species was assigned as carbene-bound Rma TDE based on the Mössbauer parameters obtained (∂Fe = 0.18 mm·s−1, |∆EQ| = 1.83 mm·s−1) and the lack of observed EPR signal (SI Appendix, Figs. S3 and S4). Thus, this reactive intermediate can be made in high yields and studied in both freeze-quenched solutions and crystals.
Computational Models of the IPC.
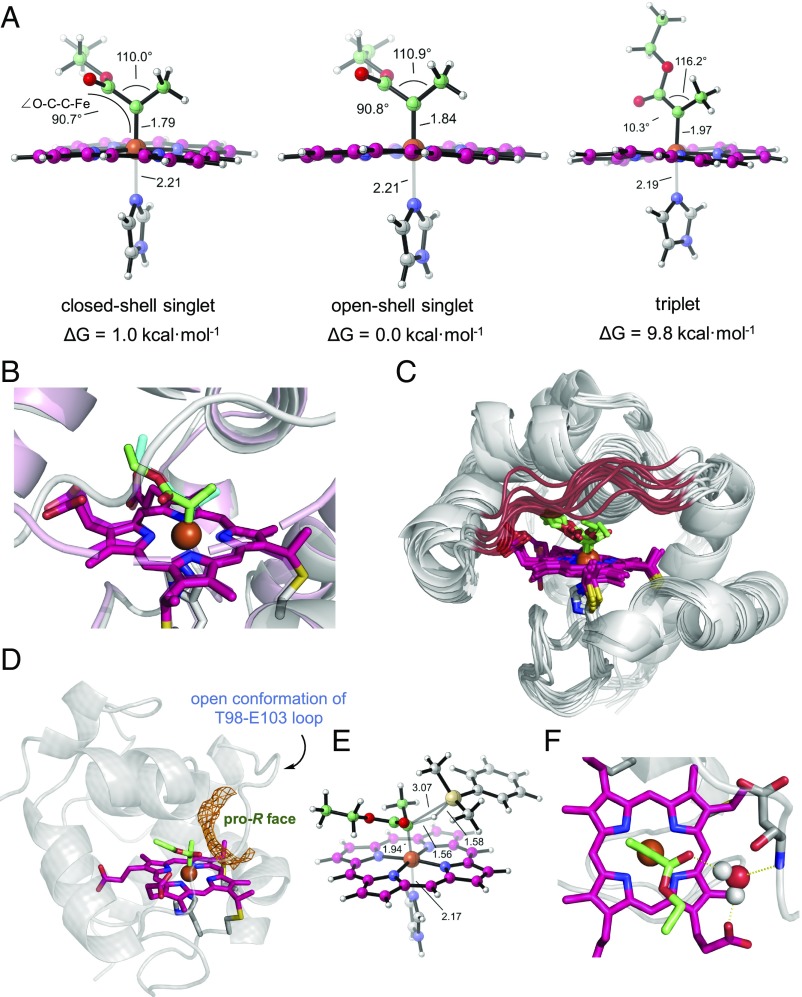
The symmetric Mössbauer line shape of Me-EDA–treated Rma TDE and its lack of EPR signal suggested that this is an S = 0 species or an S = 1 species with a large zero-field splitting. To further differentiate these electronic states, we used density functional theory (DFT) to characterize model structures of IPC 1 in three possible electronic states. Our calculations showed that the closed shell singlet (S = 0) state was 8.8 kcal·mol−1 more stable than the triplet state (S = 1) and 1.0 kcal·mol−1 less stable than the open shell singlet (Fig. 3A and SI Appendix, Fig. S5). The geometrical features of the DFT-optimized structures also suggested that the IPC in Rma TDE exists as a singlet: the orientation of the ester group in the carbene-bound Rma TDE crystal structure is similar to that observed in the optimized closed shell and open shell singlet structures but distinctly different from the triplet state (Fig. 3A). The electronic structure of cytochrome proteins is known to correlate with heme ruffling (31, 36), and the singlet ground state is further supported by heme ruffling analysis (SI Appendix, Table S2): the ruffling out-of-plane distortion observed in the crystal structure of carbene-bound Rma TDE (0.5 Å) is more similar to that observed in the DFT-optimized closed shell singlet (0.401 Å) and open shell singlet (0.421 Å) states than in the triplet state (0.784 Å), where the heme is more distorted. While our DFT calculations suggested that the triplet state was energetically and geometrically unfavorable, the open shell and closed shell singlet states are very close in energy and cannot be unambiguously assigned to IPC 1 (SI Appendix, Fig. S5). We also simulated the Mössbauer parameters of the open shell and closed shell singlet states but found that their predicted Mössbauer parameters were within experimental error of the observed values (SI Appendix, Fig. S4E). Spectroscopy and theory indicate that carbene-bound Rma TDE is a ground-state singlet.
Fig. 3.
Computational models of carbene-bound Rma TDE. (A) DFT optimized structures of model IPC 1 in the closed shell singlet, open shell singlet, and triplet electronic states. DFT calculations predict that the singlet state is 8.8 kcal·mol−1 more stable than the triplet state, which is consistent with previously characterized small molecule IPCs. The calculated Fe–C bond lengths are 1.79, 1.84, and 1.97 Å for the closed shell singlet, open shell singlet, and triplet, respectively. The Fe–N (imidazole) bond lengths are 2.21, 2.21, and 2.19 Å, respectively. These numbers are within experimental error of the lengths observed in the carbene-bound Rma TDE crystal structure, where the Fe–C bond length is 1.9 Å and the Fe–N bond length is 2.1 Å. (B) Overlay of the carbene-bound Rma TDE crystal structure with a QM/MM-optimized structure of carbene-bound Rma TDE. QM/MM structure: protein scaffold is gray, and carbene is green. Crystal structure (PDB ID code 6CUN): protein scaffold is light pink, and carbene is cyan. The preferred carbene conformation determined by MD simulations and QM/MM calculations is almost identical to that determined by X-ray crystallography. (C) Overlay of 10 representative snapshots obtained from 1,000 ns of MD simulation on the carbene-bound Rma TDE. Simulations show that the active site front loop (residues 98–103; highlighted in red) is highly flexible, switching between “open” and “closed” conformations. Despite the front loop’s high flexibility, the carbene is stabilized in a single major conformation (SI Appendix, Fig. S7). (D) Snapshot obtained from an MD trajectory on carbene-bound Rma TDE with the front loop in the open conformation, showing that the carbene pro-R face is solvent exposed. Solvent-accessible area is showed in orange. (E) DFT-optimized structure of the lowest-energy pro-R transition state (closed shell singlet state) for Si–H insertion with IPC 1 (SI Appendix, Fig. S8). (F) A representative view of an MD snapshot on carbene-bound Rma TDE showing interactions between the bridging water with the heme carboxylate, the carbene carbonyl, and the amide bond of residue D100. Hydrogen-bonding interactions with the carbene have been shown to affect the stability of IPCs (25, 27, 37).
The DFT models suggest that the observed end-on carbene is thermodynamically more stable than the alternative (Fe, N)-bridged carbene, which also requires a large distortion of the porphyrin that is difficult to attain for the covalently ligated heme in cyt c proteins (SI Appendix, Fig. S6). This is in contrast to the thiolate-ligated IPC computationally studied by Shaik and coworkers (23), where the (Fe, N)-bridged mode was found to be thermodynamically favored. Small molecule (Fe, N)-bridged IPCs have been synthesized and crystallized (17–19, 25); their experimental characterizations are distinct from that of end-on IPCs. As far as we know, (Fe, N)-bridged carbenes are not active intermediates in carbene transfer reactions (37).
Computational Modeling of the IPC Enzymatic Intermediate.
We next used molecular dynamics (MD) to understand how the protein scaffold of Rma TDE affects the conformation and properties of the IPC intermediate. MD simulations were performed on singlet IPC 1 docked into Rma TDE (PDB ID code 6CUK) using various IPC 1 conformations as starting points and explicit water as solvent. MD trajectories revealed that, regardless of the initial carbene orientation, the IPC converges to a single well-defined orientation (Fig. 3B and SI Appendix, Fig. S7). This conformation is remarkably similar to that observed in the carbene-bound Rma cyt c crystal structure, indicating the catalytic relevance of the latter in solution (Fig. 3B). Furthermore, MD simulations highlighted the flexibility of the active site front loop (T98-E103) (Fig. 3C), favoring an “open” conformation where the IPC is solvent exposed and accessible to substrates with pro-R facial selectivity, while keeping the pro-S face inaccessible (Fig. 3D). This high degree of loop flexibility is consistent with the unresolved residues (D100–D102) observed in the carbene-bound Rma TDE crystal structure (Fig. 2C). To further investigate the protein’s enantiopreference, we performed DFT calculations to characterize the intrinsic geometric features of the pro-R transition state for carbene Si−H insertion using a truncated computational model and dimethylphenylsilane as substrate (Fig. 3E and SI Appendix, Fig. S8). When comparing the lowest energy-optimized pro-R transition state with the most populated structure of carbene-bound Rma TDE, the space made available when Rma TDE is in its open conformation provides a perfect fit for the silane substrate (Fig. 3 D and E and SI Appendix, Fig. S8). However, the pro-S transition state could only take place when the IPC is rotated away from its preferred conformation, which is disfavored by the active site of Rma TDE. Thus, in Rma TDE-catalyzed carbon−silicon bond-forming reactions, the protein dictates substrate approach to the IPC intermediate from the pro-R face, which on carbene transfer to the Si−H bond of the silane substrate, gives R-configured organosilicon products exclusively as observed experimentally (>99% enantiomeric excess) (14).
MD simulations also revealed the presence of a water molecule that forms persistent hydrogen bonds bridging one of the heme carboxylate groups and the carbonyl group of the carbene (Fig. 3E). This water molecule is highly conserved, with its position further stabilized by polar contacts with the front loop residues. In the carbene-bound Rma TDE crystal structure, this bridging water was not observed but was likely obscured due to its proximity to the unresolved residues D100–D102. Hydrogen bonding is known to affect the electronic structure and stability of IPC intermediates (25, 27, 37). In view of this, we designed a truncated computational model of an IPC–water complex, and through DFT calculations, we found that the H-bonding water molecule stabilizes the carbene complex by about 7 kcal·mol−1 in its closed shell singlet state. This stabilizing effect is more substantial on the closed shell singlet than on the open shell singlet or triplet states, contributing to determining the preferred conformation adopted by the IPC in the protein (SI Appendix, Fig. S9). Hybrid quantum mechanics/molecular mechanics (QM/MM) calculations showed that, within Rma TDE, the singlet–triplet energy gap of the hydrogen-bonded IPC increases by 2.9 kcal·mol−1 (8.8 kcal·mol−1 in the truncated model, 11.7 kcal·mol−1 when considering the full system) (SI Appendix, Fig. S10). This significant destabilization of the radical triplet state is largely due to conformational restraints imposed by the protein active site and the bridging water: to minimize steric clash with the P79 and I83 sidechains, the carbene ester is forced to adopt a conformation parallel to the heme, which is intrinsically preferred by the closed shell and open shell singlet states but disfavors the radical triplet state, as this conformation cannot stabilize the unpaired electron in the carbene p orbital by the adjacent ester. Thus, mediated by amino acid residues and an ordered water, the active site of Rma TDE imposes multiple conformational restraints on the IPC intermediate to stabilize the closed shell singlet state and disfavor the presence of carbene radicals.
Summary and Conclusions
In conclusion, characterization of a catalytically active IPC intermediate inside the active site of an enzyme has provided a foundation from which to study enzyme-catalyzed carbene transfer reactions and explain the reactivity and selectivity of Rma TDE-catalyzed carbene transfer. Structural characterization of the Rma TDE enzyme showed that the three mutations introduced during directed evolution led to dramatic changes in a loop distal to the heme and generated a protein structure preorganized for substrate binding and IPC formation. The experimental IPC structure and computational modeling showed that Rma TDE stabilizes a single conformation of the IPC that gives the enzyme complete stereoselectivity control during carbene transfer. The conformational restraints imposed by the enzyme also favor the formation of a singlet iron-carbene. This convergent approach to characterizing the emerging family of carbene transferases enables informed engineering that will enhance the generality and utility of these powerful catalysts.
Materials and Methods
Detailed experimental methods are presented in SI Appendix.
Cloning, Expression, and Purification of Rma TDE.
The gene encoding Rma TDE was obtained as a gBlock and cloned into pET22b(+). Expression was performed in HyperBroth (AthenaES) using Escherichia coli BL21 E. cloni EXPRESS (Lucigen). 57Fe-labeled protein was produced using defined media supplemented with 57Fe. Rma TDE was purified via a HisTrap HP column (GE Healthcare).
Crystallography.
Crystals of Rma TDE were grown using the sitting drop vapor diffusion method and were cryoprotected before diffraction at the Stanford Synchrotron Radiation Lightsource on beamline 12–2. Carbene-bound crystals of Rma TDE were prepared by soaking preformed crystals with concentrated solutions of Me-EDA and sodium dithionite. Structures were determined by molecular replacement, and models were built using standard procedures.
EPR Spectroscopy.
EPR spectra were recorded on a Bruker EMX X-band CW-EPR spectrometer using Bruker Win-EPR software (version 3.0). Data were collected in parallel mode with a power of 10 mW and in perpendicular mode with a power of 2 mW, both at 4 K. Samples of Rma TDE were prepared under anaerobic conditions in the presence of sodium dithionite before flash freezing with liquid nitrogen.
Mössbauer Spectroscopy.
Mössbauer spectra were recorded using a spectrometer from SEE Co., and data analysis was performed using the WMOSS program. Solution samples of 57Fe-labeled Rma TDE were prepared under anaerobic conditions in the presence of sodium dithionite before flash freezing with liquid nitrogen. Samples were stored under liquid nitrogen until being mounted on the cryostat, and data were recorded at 77 K.
DFT Calculations.
DFT calculations were carried out using Gaussian09 (38) and Gaussian16 (39) using a truncated model. Geometry optimizations and frequency calculations were performed using (U)B3LYP (40–42) functional with the SDD basis set for iron and 6–31G(d) on all other atoms and within the conductor-like polarizable continuum model (CPCM) (diethyl ether, ε = 4) (43, 44) to have an estimation of the dielectric permittivity in the enzyme active site. Enthalpies and entropies were calculated for 1 atm and 298.15 K. A correction to the harmonic oscillator approximation was also applied to the entropy calculations by raising all frequencies below 100 cm–1 to 100 cm–1 (45, 46). Single-point energy calculations were performed using the dispersion-corrected functional (U)B3LYP-D3(BJ) (47, 48) with the Def2TZVP basis set on all atoms and with CPCM (diethyl ether, ε = 4). All energy values discussed in the manuscript correspond to the quasiharmonic corrected Gibbs energies (∆G-qh) at (U)B3LYP-D3(BJ)/Def2TZVP/PCM(diethyl ether)//(U)B3LYP/6–31G(d)+SDD(Fe)/PCM(diethyl ether) level if not otherwise noted. Further descriptions and details of the methodology are provided in SI Appendix.
Hybrid QM/MM Calculations.
QM/MM calculations within the ONIOM approach (49, 50) were carried out using Gaussian09 (38). Geometry optimizations were performed using (U)B3LYP (40–42) functional in combination with the SDD basis set for iron and 6–31G(d) on all other atoms and AmberFF14Sb force field (51) using a mechanical embedding scheme. Stationary points were verified as minima by a vibrational frequency analysis, thermal corrections were calculated for 1 atm and 298.15 K, and a correction to the harmonic oscillator approximation was included as described above. Single-point energy calculations were performed at the (U)B3LYP/Def2TZVP:AmberFF14Sb level and using an electrostatic embedding scheme. Snapshots for QM/MM calculations were obtained from classical MD trajectories. Further descriptions and details of the methodology are provided in SI Appendix.
MD Simulations.
MD simulations were performed using the GPU code (pmemd) (52) of the AMBER 16 package (53). Parameters for the carbene-bound IPC were generated within the antechamber and MCPB.py (54) modules in AMBER16 package using the general AMBER force field (gaff) (55), with partial charges set to fit the electrostatic potential generated at the B3LYP/6–31G(d) level by the RESP model (56). Protonation states of protein residues were predicted using H++ server. Each protein was immersed in a preequilibrated truncated cuboid box with a 10-Å buffer of TIP3P (57) water molecules using the leap module, and the systems were neutralized by addition of explicit counterions (Na+ and Cl−). All subsequent calculations were done using the widely tested Stony Brook modification of the Amber14 force field (ff14sb) (58). After minimization, heating, and equilibration of the system (SI Appendix has a full description of the protocol used), production trajectories were then run for 1,000 ns (1 µs). The obtained trajectories were processed and analyzed using the cpptraj (58) module from Ambertools utilities.
Supplementary Material
Acknowledgments
We thank J. M. Bollinger Jr., K. Chen, X. Huang, C. Krebs, C. J. Pollock, and R. K. Zhang for helpful discussions and A. Tang for experimental assistance. This work was supported by National Science Foundation Division of Chemistry Grant CHE-1361104 (to K.N.H.); the Rothenberg Innovation Initiative (RI2) Program (S.B.J.K. and F.H.A.); the Jacobs Institute for Molecular Engineering for Medicine at Caltech (S.B.J.K. and F.H.A.); National Science Foundation Division of Molecular and Cellular Biosciences Grant MCB-1513007 (to F.H.A.); and Office of Chemical, Bioengineering, Environmental and Transport Systems SusChEM Initiative Grant CBET-1403077 (to F.H.A.). R.D.L. is supported by NIH National Research Service Award Training Grant 5 T32 GM07616. M.G.-B. thanks the Ramón Areces Foundation for a postdoctoral fellowship. M.J.C. thanks the Center for Environmental Microbial Interactions at Caltech for a fellowship. Crystallography experiments were supported by J. Kaiser and the Caltech Molecular Observatory. EPR experiments were performed with the assistance of P. Oyala and supported by National Science Foundation Grant NSF-1531940 (to the Caltech EPR Facility). Computational resources were provided by the University of California, Los Angeles Institute for Digital Research and Education and the Extreme Science and Engineering Discovery Environment, which is supported by National Science Foundation Grant OCI-1053575.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6CUK and 6CUN).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807027115/-/DCSupplemental.
References
- 1.Bourissou D, Guerret O, Gabbaï FP, Bertrand G. Stable carbenes. Chem Rev. 2000;100:39–92. doi: 10.1021/cr940472u. [DOI] [PubMed] [Google Scholar]
- 2.Meyer D, Neumann P, Ficner R, Tittmann K. Observation of a stable carbene at the active site of a thiamin enzyme. Nat Chem Biol. 2013;9:488–490. doi: 10.1038/nchembio.1275. [DOI] [PubMed] [Google Scholar]
- 3.Kornecki KP, et al. Direct spectroscopic characterization of a transitory dirhodium donor-acceptor carbene complex. Science. 2013;342:351–354. doi: 10.1126/science.1243200. [DOI] [PubMed] [Google Scholar]
- 4.Doyle MP. Catalytic methods for metal carbene transformations. Chem Rev. 1986;86:919–939. doi: 10.1021/cr940066a. [DOI] [PubMed] [Google Scholar]
- 5.Ford A, et al. Modern organic synthesis with α-diazocarbonyl compounds. Chem Rev. 2015;115:9981–10080. doi: 10.1021/acs.chemrev.5b00121. [DOI] [PubMed] [Google Scholar]
- 6.Coelho PS, Brustad EM, Kannan A, Arnold FH. Olefin cyclopropanation via carbene transfer catalyzed by engineered cytochrome P450 enzymes. Science. 2013;339:307–310. doi: 10.1126/science.1231434. [DOI] [PubMed] [Google Scholar]
- 7.Bordeaux M, Tyagi V, Fasan R. Highly diastereoselective and enantioselective olefin cyclopropanation using engineered myoglobin-based catalysts. Angew Chem Int Ed Engl. 2015;54:1744–1748. doi: 10.1002/anie.201409928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gober JG, et al. Mutating a highly conserved residue in diverse cytochrome P450s facilitates diastereoselective olefin cyclopropanation. ChemBioChem. 2016;17:394–397. doi: 10.1002/cbic.201500624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knight AM, et al. Diverse engineered heme proteins enable stereodivergent cyclopropanation of unactivated alkenes. ACS Cent Sci. 2018;4:372–377. doi: 10.1021/acscentsci.7b00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen K, Huang X, Kan SBJ, Zhang RK, Arnold FH. Enzymatic construction of highly strained carbocycles. Science. 2018;360:71–75. doi: 10.1126/science.aar4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weissenborn MJ, et al. Enzyme-catalyzed carbonyl olefination by the E. coli protein YfeX in the absence of phosphines. ChemCatChem. 2016;8:1636–1640. [Google Scholar]
- 12.Kan SBJ, Huang X, Gumulya Y, Chen K, Arnold FH. Genetically programmed chiral organoborane synthesis. Nature. 2017;552:132–136. doi: 10.1038/nature24996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang ZJ, Peck NE, Renata H, Arnold FH. Cytochrome P450-catalyzed insertion of carbenoids into N-H bonds. Chem Sci (Camb) 2014;5:598–601. doi: 10.1039/C3SC52535J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kan SBJ, Lewis RD, Chen K, Arnold FH. Directed evolution of cytochrome c for carbon-silicon bond formation: Bringing silicon to life. Science. 2016;354:1048–1051. doi: 10.1126/science.aah6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyagi V, Bonn RB, Fasan R. Intermolecular carbene S-H insertion catalysed by engineered myoglobin-based catalysts. Chem Sci (Camb) 2015;6:2488–2494. doi: 10.1039/c5sc00080g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansuy D, et al. Dichlorocarbene complexes of iron(II)-porphyrins-crystal and molecular structure of Fe(TPP)(CCl2)(H2O) Angew Chem Int Ed Engl. 1978;17:781–782. [Google Scholar]
- 17.Chevrier B, Weiss R, Lange M, Chottard JC, Mansuy D. An iron(III)-porphyrin complex with a vinylidene group inserted into an iron-nitrogen bond: Relevance to the structure of the active oxygen complex of catalase. J Am Chem Soc. 1981;103:2899–2901. [Google Scholar]
- 18.Olmstead MM, Cheng RJ, Balch AL. X-ray crystallographic characterization of an iron porphyrin with a vinylidene carbene inserted into an iron-nitrogen bond. Inorg Chem. 1982;21:4143–4148. [Google Scholar]
- 19.Artaud I, Gregoire P, Leduc P, Mansuy D. Formation and fate of iron-carbene complexes in reactions between a diazoalkane and iron-porphyrins: Relevance to the mechanism of formation of N-substituted hemes in cytochrome P-450 dependent oxidation of sydnones. J Am Chem Soc. 1990;112:6899–6905. [Google Scholar]
- 20.Li Y, Huang JS, Zhou ZY, Che CM, You XZ. Remarkably stable iron porphyrins bearing nonheteroatom-stabilized carbene or (alkoxycarbonyl)carbenes: Isolation, X-ray crystal structures, and carbon atom transfer reactions with hydrocarbons. J Am Chem Soc. 2002;124:13185–13193. doi: 10.1021/ja020391c. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, et al. Electronic configuration and ligand nature of five-coordinate iron porphyrin carbene complexes: An experimental study. J Am Chem Soc. 2017;139:5023–5026. doi: 10.1021/jacs.7b01722. [DOI] [PubMed] [Google Scholar]
- 22.Khade RL, et al. Iron porphyrin carbenes as catalytic intermediates: Structures, Mössbauer and NMR spectroscopic properties, and bonding. Angew Chem Int Ed Engl. 2014;53:7574–7578. doi: 10.1002/anie.201402472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharon DA, Mallick D, Wang B, Shaik S. Computation sheds insight into iron porphyrin carbenes’ electronic structure, formation, and N-H insertion reactivity. J Am Chem Soc. 2016;138:9597–9610. doi: 10.1021/jacs.6b04636. [DOI] [PubMed] [Google Scholar]
- 24.Wei Y, Tinoco A, Steck V, Fasan R, Zhang Y. Cyclopropanations via heme carbenes: Basic mechanism and effects of carbene substituent, protein axial ligand, and porphyrin substitution. J Am Chem Soc. 2018;140:1649–1662. doi: 10.1021/jacs.7b09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatsumi K, Hoffmann R. Metalloporphyrins with unusual geometries. 2. Slipped and skewed bimetallic structures, carbene and oxo complexes, and insertions into metal-porphyrin bonds. Inorg Chem. 1981;20:3771–3784. [Google Scholar]
- 26.Khade RL, Zhang Y. Catalytic and biocatalytic iron porphyrin carbene formation: Effects of binding mode, carbene substituent, porphyrin substituent, and protein axial ligand. J Am Chem Soc. 2015;137:7560–7563. doi: 10.1021/jacs.5b03437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khade RL, Zhang Y. C-H insertions by iron porphyrin carbene: Basic mechanism and origin of substrate selectivity. Chemistry. 2017;23:17654–17658. doi: 10.1002/chem.201704631. [DOI] [PubMed] [Google Scholar]
- 28.Rittle J, Green MT. Cytochrome P450 compound I: Capture, characterization, and C-H bond activation kinetics. Science. 2010;330:933–937. doi: 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]
- 29.Stelter M, et al. A novel type of monoheme cytochrome c: Biochemical and structural characterization at 1.23 A resolution of rhodothermus marinus cytochrome c. Biochemistry. 2008;47:11953–11963. doi: 10.1021/bi800999g. [DOI] [PubMed] [Google Scholar]
- 30.Graves AB, Graves MT, Liptak MD. Measurement of heme ruffling changes in MhuD using UV-vis spectroscopy. J Phys Chem B. 2016;120:3844–3853. doi: 10.1021/acs.jpcb.6b01497. [DOI] [PubMed] [Google Scholar]
- 31.Kleingardner JG, Bren KL. Biological significance and applications of heme c proteins and peptides. Acc Chem Res. 2015;48:1845–1852. doi: 10.1021/acs.accounts.5b00106. [DOI] [PubMed] [Google Scholar]
- 32.English DR, Hendrickson DN, Suslick KS. Mössbauer spectra of oxidized iron porphyrins. Inorg Chem. 1983;22:367–368. [Google Scholar]
- 33.Wilson SA, et al. X-ray absorption spectroscopic investigation of the electronic structure differences in solution and crystalline oxyhemoglobin. Proc Natl Acad Sci USA. 2013;110:16333–16338. doi: 10.1073/pnas.1315734110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ran Y, et al. Spectroscopic identification of heme axial ligands in HtsA that are involved in heme acquisition by Streptococcus pyogenes. Biochemistry. 2010;49:2834–2842. doi: 10.1021/bi901987h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engler N, Prusakov V, Ostermann A, Parak FG. A water network within a protein: Temperature-dependent water ligation in H64V-metmyoglobin and relaxation to deoxymyoglobin. Eur Biophys J. 2003;31:595–607. doi: 10.1007/s00249-002-0255-x. [DOI] [PubMed] [Google Scholar]
- 36.Liptak MD, Wen X, Bren KL. NMR and DFT investigation of heme ruffling: Functional implications for cytochrome c. J Am Chem Soc. 2010;132:9753–9763. doi: 10.1021/ja102098p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dzik WI, Xu X, Zhang XP, Reek JNH, de Bruin B. ‘Carbene radicals’ in Co(II)(por)-catalyzed olefin cyclopropanation. J Am Chem Soc. 2010;132:10891–10902. doi: 10.1021/ja103768r. [DOI] [PubMed] [Google Scholar]
- 38.Frisch MJ, et al. 2009. Gaussian 09, Revision A.02 (Gaussian Inc., Wallingford, CT)
- 39.Frisch MJ, et al. 2016. Gaussian 16, Revision B.01 (Gaussian, Inc., Wallingford, CT)
- 40.Becke AD. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A Gen Phys. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 41.Becke AD. Density‐functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 42.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B Condens Matter. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 43.Barone V, Cossi M. Quantum calculation of molecular energies and energy gradients in solution by a conductor solvent model. J Phys Chem A. 1998;102:1995–2001. [Google Scholar]
- 44.Cossi M, Rega N, Scalmani G, Barone V. Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J Comput Chem. 2003;24:669–681. doi: 10.1002/jcc.10189. [DOI] [PubMed] [Google Scholar]
- 45.Ribeiro RF, Marenich AV, Cramer CJ, Truhlar DG. Use of solution-phase vibrational frequencies in continuum models for the free energy of solvation. J Phys Chem B. 2011;115:14556–14562. doi: 10.1021/jp205508z. [DOI] [PubMed] [Google Scholar]
- 46.Zhao Y, Truhlar DG. Computational characterization and modeling of buckyball tweezers: Density functional study of concave-convex pi...pi interactions. Phys Chem Chem Phys. 2008;10:2813–2818. doi: 10.1039/b717744e. [DOI] [PubMed] [Google Scholar]
- 47.Grimme S, Ehrlich S, Goerigk L. Effect of the damping function in dispersion corrected density functional theory. J Comput Chem. 2011;32:1456–1465. doi: 10.1002/jcc.21759. [DOI] [PubMed] [Google Scholar]
- 48.Grimme S, Antony J, Ehrlich S, Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J Chem Phys. 2010;132:154104. doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- 49.Dapprich S, Komáromi I, Byun KS, Morokuma K, Frisch MJ. A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J Mol Struct THEOCHEM. 1999;461–462:1–21. [Google Scholar]
- 50.Chung LW, et al. The ONIOM method and its applications. Chem Rev. 2015;115:5678–5796. doi: 10.1021/cr5004419. [DOI] [PubMed] [Google Scholar]
- 51.Maier JA, et al. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J Chem Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 53.Case DA, et al. Amber 16. University of California; San Francisco: 2016. [Google Scholar]
- 54.Li P, Merz KM., Jr MCPB.py: A python based metal center parameter builder. J Chem Inf Model. 2016;56:599–604. doi: 10.1021/acs.jcim.5b00674. [DOI] [PubMed] [Google Scholar]
- 55.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 56.Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- 57.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 58.Roe DR, Cheatham TE., 3rd PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.