Significance
To date, the importance of liver X receptors (LXRs) in atherosclerosis development has been gleaned from their pharmacological or genetic manipulation. Here, we show that altering LXRα phosphorylation can shape proatherogenic responses to fat-rich diets, uncovering previously unrecognized mechanisms. Disrupting LXRα phosphorylation in myeloid cells triggers global changes in gene expression in macrophages, including the up-regulation of proliferation-promoting factors, consistent with increased proliferation of lesion-resident cells. This leads to an enhanced atherosclerotic plaque burden and plaques with altered phenotypic features. Notably, novel LXRα-regulated targets revealed by impaired LXRα phosphorylation are markedly distinct from those promoted by LXR ligand activation. Overall, this work reveals LXRα phosphorylation as an important determinant of atherosclerosis development. This could be exploited for the design of novel antiatherosclerotic strategies.
Keywords: liver X receptor, atherosclerosis, macrophages, proliferation, FoxM1
Abstract
Macrophages are key immune cells for the initiation and development of atherosclerotic lesions. However, the macrophage regulatory nodes that determine how lesions progress in response to dietary challenges are not fully understood. Liver X receptors (LXRs) are sterol-regulated transcription factors that play a central role in atherosclerosis by integrating cholesterol homeostasis and immunity. LXR pharmacological activation elicits a robust antiatherosclerotic transcriptional program in macrophages that can be affected by LXRα S196 phosphorylation in vitro. To investigate the impact of these transcriptional changes in atherosclerosis development, we have generated mice carrying a Ser-to-Ala mutation in myeloid cells in the LDL receptor (LDLR)-deficient atherosclerotic background (M-S196ALdlr-KO). M-S196ALdlr-KO mice fed a high-fat diet exhibit increased atherosclerotic plaque burden and lesions with smaller necrotic cores and thinner fibrous caps. These diet-induced phenotypic changes are consistent with a reprogramed macrophage transcriptome promoted by LXRα-S196A during atherosclerosis development. Remarkably, expression of several proliferation-promoting factors, including the protooncogene FoxM1 and its targets, is induced by LXRα-S196A. This is consistent with increased proliferation of plaque-resident cells in M-S196ALdlr-KO mice. Moreover, disrupted LXRα phosphorylation increases expression of phagocytic molecules, resulting in increased apoptotic cell removal by macrophages, explaining the reduced necrotic cores. Finally, the macrophage transcriptome promoted by LXRα-S196A under dietary perturbation is markedly distinct from that revealed by LXR ligand activation, highlighting the singularity of this posttranslational modification. Overall, our findings demonstrate that LXRα phosphorylation at S196 is an important determinant of atherosclerotic plaque development through selective changes in gene transcription that affect multiple pathways.
Atherosclerosis is a chronic inflammatory process and the major pathology responsible for cardiovascular disease, which is now the leading cause of global mortality (1). This pathology results from the accumulation of lipids, immune cells, and extracellular matrix within arterial walls, causing flow limitation (2). Atherosclerotic lesions progress and may rupture and thrombose, occluding the vessel and leading to myocardial infarcts or strokes. Macrophages are immune cells involved in most key pathways for the development of atherosclerosis, including uptake of oxidized LDL, cholesterol efflux, foam cell and fatty streak formation, local proliferation, apoptosis, programmed removal of dead cells or efferocytosis, necrotic core formation, and contribution to plaque stability (2).
Liver X receptors (LXRs) are ligand-activated transcription factors that play vital roles in cholesterol homeostasis (3) and inflammation (4). LXRs are expressed as two isotypes, LXRα and LXRβ, which display 78% sequence homology yet vary in their tissue expression and regulation (5). Both LXRs are endogenously activated by oxidized metabolites of cholesterol (6) and intermediates of the cholesterol biosynthesis pathway (7), as well as by various synthetic ligands (8). Pharmacological activation of these receptors has been demonstrated to modulate a range of lipid and inflammatory disorders (9). With regard to atherosclerosis, activating LXRs attenuate atherosclerosis progression (3) via promotion of cholesterol efflux through lipid-laden macrophages present in the atherosclerotic lesions and inhibition of vascular inflammation (4), and possibly by affecting other aspects of lipid metabolism (10). Additionally, ligand-activated LXR promotes CCR7-dependent plaque regression (11). Functional studies in macrophages further indicate that LXRα is required for a robust antiatherosclerotic response to LXR ligands and LXRα plays a selective role in limiting atherosclerosis in response to hyperlipidemia (12).
LXRα transcriptional activity can be modulated by several posttranslational modifications (5), including phosphorylation at serine (S) 198 in the human sequence, corresponding to S196 in the mouse ortholog. We have demonstrated that modulation of LXRα phosphorylation significantly modifies its target gene repertoire in macrophage cell lines overexpressing the receptor, thereby altering pathways known to be relevant to the development of atherosclerosis (13, 14). Interestingly, we previously showed phosphorylated S196-LXRα is present in progressive atherosclerotic lesions (14), suggesting LXRα phosphorylation at this residue could be important for the development of atherosclerotic plaques. However, the specific contribution of myeloid LXRα phosphorylation to atherosclerosis development remains unknown.
To investigate this, we have generated a mouse model specifically expressing a Ser-to-Ala phosphorylation mutant of LXRα in myeloid cells (M-S196A) in the LDL receptor (LDLR)-deficient (Ldlr-KO) atherosclerotic background (M-S196ALdlr-KO). Disrupting LXRα phosphorylation in myeloid cells, including macrophages, promotes plaque burden yet modulates plaque phenotype to acquire distinctive characteristics, such as smaller necrotic cores and thinner fibrous caps encapsulating the lesions. These phenotypic changes are consistent with a reprogrammed macrophage transcriptome. Notably, cell cycle progression and proliferation pathways are markedly induced in M-S196ALdlr-KO macrophages, specifically the expression of the FoxM1 transcription factor and several of its targets. This is associated with increased lesion-resident cell proliferation in the LXRα phosphomutant mice. In addition, changes in the expression of various phagocytic molecules result in enhanced macrophage efferocytosis, thus explaining the reduced necrotic cores present in M-S196ALdlr-KO mice. Interestingly, most of the phosphorylation-sensitive genes identified are not subject to LXR ligand regulation, and we show that global transcriptional changes in response to impaired LXRα phosphorylation under dietary perturbation are markedly distinct from those revealed by ligand activation. Overall, these findings demonstrate LXRα phosphorylation at S196 determines atherosclerotic plaque progression by promoting changes in local cell proliferation, efferocytosis, and necrotic core formation.
Results
Impaired Myeloid LXRα Phosphorylation Promotes Atherosclerosis.
To investigate the impact of macrophage LXRα phosphorylation on the development of atherosclerosis, we generated a mouse model expressing a serine-to-alanine mutation at residue 196 in LXRα in myeloid cells (M-S196A) on a proatherosclerotic (LDLR-deficient or Ldlr-KO) background (M-S196ALdlr-KO) (SI Appendix, Fig. S1A). Effective expression of a Cre-driven targeting construct introducing S196A knock-in in the sense strand was demonstrated in M-S196ALdlr-KO mice compared with WTLdrl-KO control littermates (SI Appendix, Fig. S1B). Mice were fed a fat-rich Western diet (WD) to accelerate plaque progression. LXRα expression was similar in WTLdlr-KO and M-S196ALdlr-KO mice (SI Appendix, Fig. S1 C and D). M-S196ALdlr-KO mice developed normally, and no change in body weight before, during, or after WD feeding was observed (SI Appendix, Fig. S1 E–G). There were no detectable changes in basal metabolic characteristics, including total cholesterol, HDL, or LDL/very-LDL levels and the amounts of triglycerides, free fatty acids, and insulin in the plasma of M-S196ALdlr-KO mice compared with WTLdlr-KO mice (SI Appendix, Fig. S2). Interestingly, M-S196ALdlr-KO mice showed a significant increase in atherosclerosis plaque burden in their aortas as measured by en face Oil Red O staining (Fig. 1A) and aortic root plaque coverage (Fig. 1 B and C). However, this was not associated with changes in the levels of CD68+ cells in the lesions (Fig. 1D).
Fig. 1.
M-S196ALdlr-KO mice develop increased atherosclerosis on a WD. (A, Left) Representative images of en face Oil Red O-stained whole aortas (n = 8–11 per group). (Original magnification: 8×.) (A, Right) Quantification of stained areas as percent plaque coverage for each genotype. (B) H&E-stained aortic roots (n = 9–10 per group). (Scale bars: 500 μm.) (C) Quantification of stained areas as percent plaque coverage for each genotype. (D) CD68 staining of aortic roots (n = 7–8 per group). Data are mean ± SEM (*P < 0.05, relative to WTLdlr-KO mice). NS, not significant.
Changes in LXRα Phosphorylation at Ser196 Reprogram Global Macrophage Gene Expression in the Context of Diet-Induced Atherosclerosis.
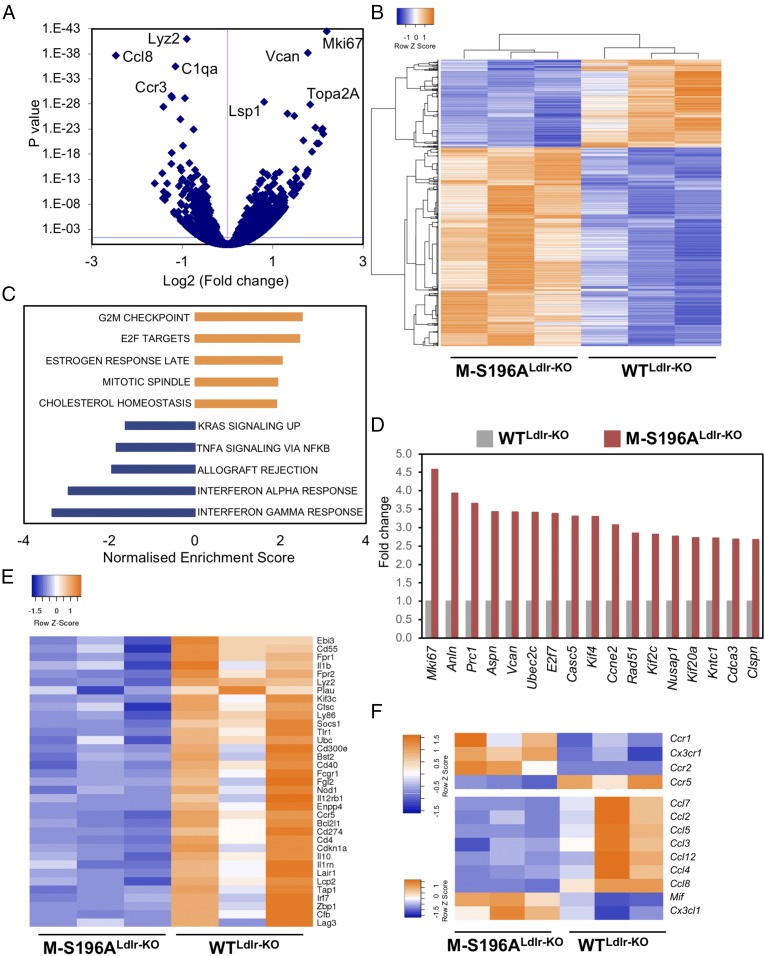
To explore in more detail the pathways underlying the changes observed in atherosclerosis development, we investigated the transcriptomic profiles of macrophages differentiated from the bone marrow of mice exposed to the WD. RNA-sequencing (RNA-seq) analysis revealed significant genome-wide changes in transcript levels (Fig. 2 A and B). LXRα-S196A significantly induced (n = 460) or reduced (n = 210) gene expression. The impact of LXRα-S196A on the basal macrophage expression of well-established LXR targets varied: While Srebf1 and Abcg1 levels were not affected, other targets, such as Abca1 and Apoe, were significantly reduced (SI Appendix, Fig. S3A). Hallmark pathway analysis identified G2/M checkpoint and E2F targets to be markedly enriched, indicating cell cycle and cell proliferation pathways are induced in the mutant-expressing macrophages (Fig. 2C and SI Appendix, Fig. S4 A and B). This was further confirmed by reactome pathway analysis (SI Appendix, Table S1). Several genes involved in these processes were regulated over twofold, including cell proliferation marker Mik67 or Ki67 (4.58-fold, P = 3.46E-43) (Fig. 2D). Concomitant to these changes in cell proliferation genes, there was a substantial reduction in the expression of genes associated with the immune response (Fig. 2 C and E and SI Appendix, Fig. S4C and Table S2). We observed opposing changes in the expression of chemokine receptors involved in monocyte trafficking to atherosclerotic lesions and some of the chemokines they bind to (15) (Fig. 2F). For example, expression of the chemokine receptors Ccr1 (1.43-fold, P = 0.004), Ccr2 (1.73-fold, P = 3.8 × 10−15), and Cx3cr1 (1.8-fold, P = 0.0008) was increased, whereas Ccr5 (0.55-fold, P = 6.19 × 10−17) expression was diminished in LXRα-S196A macrophages compared with WT macrophages. Such differential expression of chemokine receptors and their ligands may explain the lack of change in the overall number of CD68+ cells retained in the plaques of M-S196ALdlr-KO mice. Furthermore, there was no difference in the number of circulating monocytes between genotypes (SI Appendix, Fig. S1H).
Fig. 2.
Changes in LXRα phosphorylation reprogram macrophage gene expression. (A) Volcano plot of log2 ratio vs. P value of differentially expressed genes comparing 12-wk-old, WD-fed M-S196ALdlr-KO and WTLdlr-KO bone marrow-derived macrophages (n = 3 per group). The blue line indicates an adjusted P value threshold of 0.04 (Wald test for logistic regression). (B) Clustered heat map of RNA-seq gene counts in WD-fed macrophages (n = 3 mice per group). (C) Gene set enrichment analysis showing enriched pathways in M-S196ALdlr-KO macrophages derived from hallmark gene sets. (D) Fold change of RNA-seq gene counts in M-S196ALdlr-KO compared with WTLdlr-KO macrophages (set as 1) (n = 3 per genotype) for the top induced genes (≥twofold expression, P ≤ 0.01) involved in cell proliferation. (E) Heat map of RNA-seq gene counts of immune response genes down-regulated by S196A in WD-fed macrophages (n = 3 mice per group). (F) Heat map of RNA-seq gene counts of chemokine receptor (Top) and chemokine ligand genes (Bottom) showing differentially expressed genes in S196A WD-fed macrophages (n = 3 mice per group). For all heat maps, blue and orange depict up-regulated and down-regulated genes, respectively, and only genes showing ≥1.3-fold change with P ≤ 0.01 are shown.
M-S196A Induces Expression of FoxM1 and Lesion-Resident Cells.
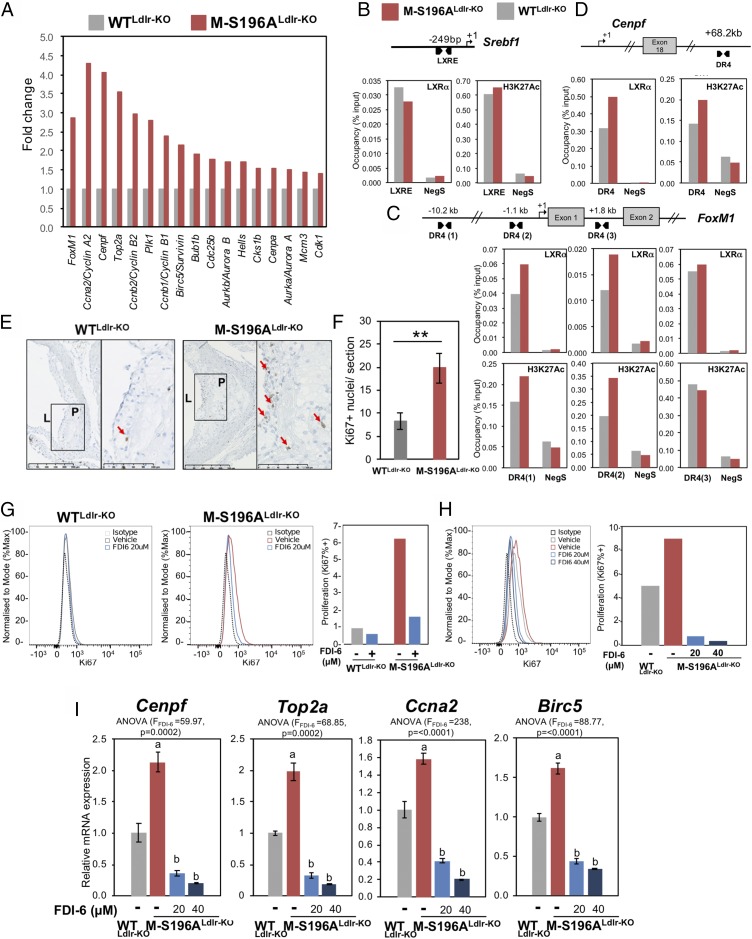
Examination of the RNA-seq datasets revealed LXRα-S196A cells expressed almost threefold (P = 7.76E-14) more protooncogene FoxM1 compared with macrophages expressing WT LXRα (Fig. 3A and SI Appendix, Fig. S4D). This was also the case for several FoxM1 target genes (16) (Fig. 3A and SI Appendix, Fig. S4D). While LXRα activation was previously shown to inhibit cell proliferation via inhibition of FoxM1 in hepatic carcinoma cells (17), its regulation in macrophages has never been documented. LXRs modulate gene transcription by heterodimerizing with the retinoid X receptor (RXR) and binding to specific DNA sequences termed LXR response elements (LXREs) in the transcriptional regulatory regions of their target genes (18). Similar to the well-known Srebf1 LXR target (Fig. 3B), specific LXRα occupancy was observed at the FoxM1 gene in macrophages at different sites (Fig. 3D), further indicating that FoxM1 is an LXRα target in these cells. These sites were initially identified by ChIP-sequencing analysis (SI Appendix, Fig. S5A) and in silico by sequence similarity to reported LXREs. We also confirmed that, unlike FoxM1, genes upstream of this gene were not significantly influenced by the expression of LXRα-S196A in macrophages (SI Appendix, Fig. S5B). Contrary to Srebf1, which is not affected by LXRα-S196A expression, LXRα occupancy, as well as H3K27 acetylation, at FoxM1 and at one of its targets, Cenpf, was modestly enhanced in macrophages expressing the phosphomutant receptor, suggesting this may be one of the mechanisms by which LXRα-S196A influences the expression of these genes. The enhanced levels of several promitotic genes suggested cell proliferation could be altered in M-S196ALdlr-KO macrophages. Indeed, macrophages expressing the LXRα-S196A mutant showed about a 20% increase in proliferation in culture measured as Ki67 levels (SI Appendix, Fig. S5 C and D). Recent studies have highlighted the important role local macrophage proliferation plays in lesion development (19). Consistent with a significant increase in the regulation of FoxM1 and other genes involved in cell cycle pathways, increased proliferation of lesion-resident cells as measured by Ki67 staining was observed in the atherosclerotic plaques of M-S196ALdlr-KO mice (Fig. 3B), which was associated with increased nuclei content (SI Appendix, Fig. S6B) compared with WTLdlr-KO mice. Notably, pharmacological inhibition of FoxM1 using the specific inhibitor FDI-6 (20, 21) reduced the enhanced M-S196ALdlr-KO macrophage proliferation observed (Fig. 3 G and H) and consistently reduced FoxM1 target gene expression (Fig. 3I), confirming that FoxM1 up-regulation mediates, at least in part, some of the increased macrophage proliferation observed in M-S196ALdlr-KO mice. This strongly suggests that FoxM1-dependent enhanced local proliferation in the plaques could contribute to increased plaque size exhibited by M-S196ALdlr-KO mice, as has been postulated (22–24).
Fig. 3.
Impaired macrophage LXRα phosphorylation induces FoxM1 expression and increases plaque cell proliferation. (A) Fold-change of RNA-seq gene counts in WD-fed M-S196ALdlr-KO compared with WD-fed WTLdlr-KO macrophages (set as 1) for FoxM1 and FoxM1 target genes with ≥1.3-fold expression (P ≤ 0.01; n = 3 per genotype). LXR occupancy and H3K27 acetylation (HK27Ac) at the Srebf1 reported LXRE (B) and FoxM1 (C) and Cenpf (D) identified DR4 sequences (LXR-binding sites) in bone marrow-derived macrophages from M-S196ALdlr-KO and WTLdlr-KO WD-fed mice are shown. Data shown are normalized to input compared with a region within in a gene desert (Neg S) as a negative control. One representative experiment of three (each using n = 2 mice per genotype) is shown. (E) Representative images of plaques exhibiting Ki67+ nuclei. L, lumen; P, plaque. (Scale bars: 250 μm.) (F) Quantification of Ki67+ nuclei in WD-fed WTLdlr-KO and M-S196ALdlr-KO plaques (n = 6–10 mice per group). **P ≤ 0.01. (G) Flow cytometry histograms of Ki67 expression in F4/80+ macrophages (Left) and bar chart of percentage of Ki67+ cells in WTLdlrKO and M-S196ALdlr-KO macrophages exposed to 20 μM FDI-6 inhibitor (Right). Data shown are representative of three independent experiments (n = 2 mice per genotype each). (H) Histogram (Left) and bar chart of percentage of Ki67+ M-S196ALdlr-KO macrophages in response to indicated concentrations of FDI-6 (Right). Data shown are representative of three independent experiments (n = 2 mice per genotype each). (I) RT-qPCR analysis of FoxM1 target genes in WTLdlr-KO and M-S196ALdlr-KO macrophages. Normalized data are shown relative to WTLdlr-KO as mean ± SD (n = 3) compared with WT-DMSO (a) and with M-S196A-DMSO (b).
M-196ALdlr-KO Mice Display Phenotypic Changes in Their Necrotic Cores and Fibrous Caps.
The observed changes in gene expression suggest a complex interaction of pathways involved in the progression of atherosclerosis. Unexpectedly, despite their larger size, size-matched atherosclerotic lesions in M-S196ALdlr-KO mice display smaller necrotic cores (Fig. 4A). Programmed cell removal or efferocytosis has been shown to strongly impact the formation of necrotic cores in advanced plaques (25). In agreement with this, macrophage engulfment of apoptotic cells was significantly increased (Fig. 4B and SI Appendix, Fig. S6C). Further interrogation of the LXRα-S196A–regulated transcriptome showed differential expression of several pro- and antiphagocytic molecules (Fig. 4C and SI Appendix, Fig. S3B). These include Ccr2 (1.73-fold, P = 3.8 × 10−15), Gpr132 (1.64-fold, P = 1.73 × 10−10), Itgb3 (1.59-fold, P = 0.007), and Mfge8 (1.37-fold, P = 7.46 × 10−5), which are known to promote efferocytosis (25), as well as molecules known to render apoptotic cells resistant to efferocytosis, such as Cd47 (26) (0.75-fold, P = 0.0001) and Tnf (0.71-fold, P = 0.0004) in M-S196ALdlr-KO macrophages. This is consistent with the enhanced efferocytosis observed in these cells. Another important morphological feature of atherosclerotic lesions influenced by macrophages is the thinning of the protective collagenous scar surrounding them or fibrous caps (2). Interestingly, M-S196ALdlr-KO lesions show a reduced fibrous cap thickness with overall smaller fibrous cap areas (SI Appendix, Fig. S6 D and E). This could result from the diminished expression of several collagen genes, including Col1a1 (0.6-fold, P = 9.3 × 10−04), Col1a2 (0.7-fold, P = 9.9 × 10−03), Col3a1 (0.6-fold, P = 4.0 × 10−03), Col5a1 (0.6-fold, P = 3.4 × 10−03), and Col6a1 (0.5-fold, P = 4.2 × 10−05), and increased levels of matrix-degrading molecules, such as Mmp8 (1.57-fold, P = 6.1 × 10−10) and Mmp12 (1.48-fold, P = 8.8 × 10−6). Overall, these data highlight the complex phenotypic changes present in atherosclerotic lesions resulting from changes in LXRα phosphorylation.
Fig. 4.
M-S196ALdlr-KO mice show decreased plaque necrotic cores and increased efferocytosis capacity. (A, Left) H&E-stained mature plaques depict necrotic core (NC; n = 4–6 per group); representative images are shown. (A, Right) Quantification of H&E-stained areas for each genotype. (Scale bars: 250 μm.) (B) Engulfment of apoptotic Jurkat cells (*P ≤ 0.05; n = 6 per group). (C) Fold change of RNA-seq gene counts in WD-fed M-S196ALdlr-KO compared with WD-fed WTLdlr-KO macrophages (set as 1) for pro- and antiphagocytic genes (n = 3 per genotype).
Disrupted LXRα Phosphorylation at Ser196 Alters Ligand Responses in Macrophages.
Our findings indicate that in the context of an atherogenic diet, changes in LXRα phosphorylation modulate the macrophage transcriptome and promote the atherosclerotic plaque burden. To further understand the magnitude of the transcriptional changes imposed by the LXRα phosphorylation mutant, we next examined whether WT- and S196A-expressing macrophages respond differently to an LXR ligand and explored the differences in global transcript changes between ligand activation and reduced LXRα phosphorylation. RNA-seq analysis was performed on bone marrow-derived macrophages from M-196ALdlr-KO mice exposed to the WD for 12 wk and cultured in the presence of vehicle or LXR ligand GW3965. GW ligand activation promoted changes in macrophage gene expression that were different in cells expressing the S196A mutant compared with WT macrophages (SI Appendix, Fig. S7 A–C). Gene set enrichment analysis revealed the pathways subject to changes in LXRα phosphorylation in the presence of the LXR ligand are similar to those seen in the absence of GW (SI Appendix, Fig. S7D). For instance, genes involved in nuclear division and cell cycle regulation remained strongly induced by LXRα-S196A, further emphasizing the importance of LXRα phosphorylation in the modulation of these pathways. Remarkably, it became apparent that while a small subset of genes was differentially regulated by the mutant only in the context of the ligand (94 induced and 50 reduced compared with WT cells), most differences in gene expression were observed in the absence of ligand (Fig. 5 A and B). Additionally, our datasets showed that ligand responses (either induction or repression) were similar in both WT- and mutant-expressing macrophages for a subset of genes, but not for others (Fig. 5C and SI Appendix, Fig. S8). Further analysis revealed that both the magnitude of the response and the identity of the genes were strikingly different between the response to the ligand (regulation by GW in either WT or S196A cells) and to phosphorylation (modulation by LXRα-S196A compared with WT cells) (SI Appendix, Fig. S7 E and F). This highlights the significance of S196 phosphorylation in rewiring the LXR-modulated transcriptome.
Fig. 5.
Macrophage transcriptional reprogramming in response to changes in LXRα phosphorylation is fundamentally different from ligand activation responses. (A and B) RNA-seq analysis on M-S196ALdlr-KO and WTLdlr-KO macrophages from WD-fed mice exposed to 1 μM GW3965 (GW) (n = 3 per group). Venn diagrams of genes induced (A) or reduced (B) in S196A compared with WT cells are shown. (C) Comparison of ligand responses in M-S196ALdlr-KO and WTLdlr-KO macrophages. (Middle) Venn diagram of genes induced (UP) or reduced (DOWN) by GW in WT LXRα cells compared with differentially expressed genes (DEGs) in S196A vs. WT in vehicle-treated (DMSO) conditions. Bar graphs show fold changes in gene expression for genes induced (Top) or reduced (Bottom) by GW that are also differentially regulated by S196A. #, not significant. (D) Clustered heat map of RNA-seq gene counts for FoxM1 and its target genes in macrophages from WD-fed mice (n = 3 per group) treated as indicated. (E) RT-qPCR analysis of FoxM1 and its target genes in WD-fed WTLdlr-KO and M-S196ALdlr-KO macrophages. Normalized data are shown relative to WTLdlr-KO macrophages as mean ± SD (n = 3) [P ≤ 0.001 compared with WT-DMSO (a), P ≤ 0.001 compared with S196A-DMSO (b), and P ≤ 0.001 compared with WT-GW (c)]. (F) mRNA expression of FoxM1 regulators. Data and statistical analysis are as in E.
Finally, we investigated whether this dichotomy between ligand- and phosphorylation-induced responses was apparent in the regulation of the phosphorylation-sensitive gene FoxM1 and some of its target genes. FoxM1 was not significantly affected by exposure to the LXR ligand in WTLdlr-KO cells (Fig. 5 D and E). By contrast, GW3965 activation markedly reduced FoxM1 mRNA levels in M-S196ALdlr-KO macrophages (Fig. 5 D and E). This regulatory pattern was recapitulated by most FoxM1 targets examined. In addition, established transcriptional regulators of FoxM1 were strongly (Top2a, Rad51, and Check2) or moderately (Melk) induced in M-S196ALdlr-KO cells in unstimulated conditions compared with WTLdlr-KO cells (Fig. 5F). Consistent with the known antiproliferative effects of LXR ligands, the expression of these genes was strongly attenuated by GW3965, although this occurred preferentially in the mutant cells. Other LXRα phosphorylation-sensitive genes implicated in cell cycle progression mimic this mode of regulation (SI Appendix, Fig. S7G).
Overall, our findings suggest that LXRα phosphorylation at Ser196 is a powerful means of regulating LXRα transcriptional activity that has important consequences for macrophage biology and for the progression of a metabolic, inflammatory, and proliferative disease, such as atherosclerosis.
Discussion
The macrophage regulatory nodes that determine how the atherosclerotic lesion progresses in response to dietary challenges are not fully understood. LXRs have key roles in the regulation of macrophage lipid homeostasis and inflammation, and, as such, they strongly modulate the progression of metabolic diseases, such as atherosclerosis (3). The importance of these receptors in disease development has been mainly gleaned from studies evaluating the consequences of their pharmacological or genetic manipulation. However, it remained unknown whether alternative modulation of the activity of these receptors, for instance, by altering posttranslational modifications of the receptor, could shape the proatherogenic responses of fat-rich diets, thus altering disease development. We previously showed LXRα is phosphorylated in cholesterol-loaded macrophages and in progressive atherosclerotic plaques (13). We have now explored the impact of LXRα phosphorylation on atherosclerosis development by expressing an LXRα Ser196-to-Ala mutant, previously shown to disrupt LXRα phosphorylation (13), specifically in myeloid cells on the LDLR-null background (M-S196ALdlr-KO).
LXRα-S196A expression in cells of the myeloid lineage, including macrophages, increases atherosclerotic plaque burden (Fig. 1 and SI Appendix, Fig. S9). This is consistent with an enhanced number of proliferating lesion-resident cells (Fig. 3E) in M-S196ALdlr-KO mice and the up-regulation of genes driving cell cycle progression in macrophages, particularly at the G2/M checkpoint, accounting for up to 15% of the total changes in gene expression exerted by the phosphorylation mutant (Fig. 2C). During the past decade, established paradigms of atherosclerotic plaque formation and progression have been revisited, and local proliferation of macrophages has been demonstrated to be an important driver of atherosclerosis development in advanced atherosclerotic plaques (22, 24, 27). However, the specific players modulating macrophage proliferation in the context of atherosclerosis remain poorly understood. Proliferation of lesional macrophages has been linked to up-regulation of the scavenger receptor Msr1 (22, 28), but a defined mechanism has remained elusive. Our findings now indicate that modulation of LXRα phosphorylation plays an important role in this process.
LXRs are known modulators of cell proliferation in other disease conditions where cell proliferation is critical, including cancer (29). For instance, LXR activation inhibits proliferation of B and T cells and macrophages (30–32) as well as several cancer cell lines, including prostate (LNCaP) (33), breast (MCF7) (34), and colon (HTC111) (35). Identified antiproliferative mechanisms by classic LXR agonists appear to be independent of the lipogenic activity of LXR (34) but, rather, linked to β-catenin activity (35), cyclins (33), and sterol metabolism (32). In contrast to these inhibitory effects, LXR activation was recently reported to enhance proliferation of neural progenitor cells in a MAPK/extracellular signal-regulated kinase pathway-dependent manner (36). Inverse agonism of LXR with a novel synthetic inverse agonist has also shown promise as a potential cancer treatment through inhibition of lipogenesis, glycolysis, and by regulating the expression of key glycolytic and lipogenic genes (29). However, in our study, LXR is shown to target cell cycle promoting factors in an atherosclerotic context.
The underlying mechanisms explaining the reported LXR antiproliferative actions may be cell-specific. The inhibitory effects of LXR ligands on macrophage colony-stimulating factor–stimulated macrophage proliferation involve down-regulation of the cyclin-dependent kinase regulators cyclin D1 (Ccnd1) and B1 (Ccnb1) (31). Both cyclins are inhibited by LXRα through the transcriptional repression of FoxM1 in hepatic carcinoma cells (17). Conversely, LXR antagonism with a sulfated oxysterol promotes hepatic proliferation, in part, through the induction of FoxM1 (37). FoxM1 is an essential proliferation-associated transcription factor found overexpressed in numerous solid tumors (38–41). Its expression is restricted to actively dividing cells, and it is regulated in a cell cycle-dependent manner by a wide range of proliferative signals (42). FoxM1 levels are induced in the G1 phase, maintained throughout the S phase, and reach maximum expression in the G2/M phase (43–46). We now demonstrate that chronic disruption of LXRα phosphorylation in macrophages enhances FoxM1 expression and several of FoxM1-associated regulators and targets driving cell cycle progression (16) (Fig. 3A and SI Appendix, Fig. S4D). We have now identified multiple LXRα-binding sites at the FoxM1 locus, confirming FoxM1 is an LXR target in macrophages (Fig. 3C). LXRα occupancy and H3K27 acetylation are modestly enhanced at some of these sites in cells expressing the LXRα phosphomutant, indicating this is likely one of the mechanisms underlying enhanced FoxM1 expression in these cells. It is possible that expression of the S-to-A mutant could affect the structure/function of LXRα unrelated to its impact on the phosphorylation of the receptor. Nevertheless, we have previously shown by molecular modeling that phosphorylated and nonphosphorylated receptors can adopt different structures that could influence protein–protein interactions, ultimately modulating LXRα function (14). Importantly, induced FoxM1 levels are associated with increased lesional cell proliferation (Fig. 3E) and cell content (SI Appendix, Fig. S6B), consistent with larger atherosclerotic lesions. Among FoxM1 regulators, Bub1b [or Bub receptor 1 (BubR1), increased 1.9-fold, P = 1.26 × 10−6 in the LXRα-S196A–expressing macrophages] has been shown to alter atherosclerosis, as impaired expression of BubR1 results in decreased macrophage proliferation and attenuated atherogenesis (47). Furthermore, LXRα-S196A macrophages show decreased mRNA expression of the FoxM1 inhibitor SASH1 (48) (0.8-fold, P = 1.33 × 10−4), which has been implicated in the development of atherosclerosis in people who smoke, through its upregulation in monocytes. Notably, we now provide evidence that specific inhibition of FoxM1 reduces proliferation of LXRα-S196A macrophages, confirming the relevance of this pathway. Overall, our findings further highlight the importance of this set of molecules in atherosclerosis progression.
Importantly, we show evidence that reduced LXRα phosphorylation during atherosclerosis development reprograms the macrophage transcriptome (Fig. 2). Global gene expression analysis revealed that in these cells, most genes are sensitive to the expression of the LXRα phosphomutant in the absence of LXR ligand stimulation, with only an additional 82 genes being modulated by the mutant in the presence of the GW3965 ligand compared with the 670 LXRα phosphorylation-sensitive genes under basal conditions (SI Appendix, Fig. S7 E and F). This suggests that modulation by LXRα-S196A expression is different from the regulation by ligand activation of the receptor. Indeed, comparison of the various datasets evidenced that the transcriptomic rewiring in response to impaired LXRα phosphorylation in the mutant cells is remarkably distinct and does not merely phenocopy ligand responses (Fig. 5C and SI Appendix, Fig. S8 C and D), thus highlighting the importance of this posttranslational modification in modulating the activity of LXRα in the context of a metabolic and inflammatory disease. An example of this is the regulation of FoxM1 expression and its regulated pathways, which are induced in LXRα-S196A–expressing cells (Fig. 3A and SI Appendix, Fig. S4D). Previous reports have linked LXRα activation with FoxM1 repression in hepatic carcinoma cells (17). We observed that FoxM1 inhibition by GW3965 is recapitulated in macrophages from mice exposed to a high-fat diet that are developing atherosclerotic plaques (Fig. 5 D and E). However, this is preferentially observed in the LXRα phosphorylation mutant-expressing cells. In fact, most FoxM1 targets and modulators, as well as other factors involved in the G2/M cell cycle transition, mirror this pattern of regulation (Fig. 5 E and F and SI Appendix, Fig. S7G). This suggests that in macrophages, the antiproliferative effects of LXR ligands are enhanced when LXRα phosphorylation is disrupted. Intriguingly, these transcriptomic changes are observed in cells that have been differentiated and cultured in vitro from precursor cells exposed to the proatherosclerotic environment in the bone marrow of WD-fed mice and could be the result of epigenetic changes influenced by the diet. Indeed, there are reports showing a distinct metabolic environment (e.g., in type 2 diabetes) can epigenetically imprint bone marrow progenitor cells that can derive into a “preprogrammed” macrophage state associated with changes in gene expression (49). Outside the scope of this study, future investigations will help establish changes in the epigenome of LXRα-S196A–expressing cells and how they affect metabolic, proliferative, and inflammatory pathways.
The observed changes in gene expression are likely to result from a combination of factors, including differential binding to DNA (as shown in Fig. 3D) and possibly variations in the interaction with cofactors. Indeed, we have previously described that differential NCoR binding to the LXR phosphomutant is important for the variation in the expression of certain targets (13). In addition, using molecular modeling, we have shown how LXRα in its unphosphorylated state may impact cofactor interactions by exposing an alternative surface for protein interaction (14). Thus, a mutation disrupting phosphorylation would be expected to mimic these interactions in vivo, which could also partly explain the observed changes in gene expression. Based on this molecular modeling, it is currently unclear how the change in LXR phosphorylation at this serine residue could impact DNA binding. The S198 residue does not contact directly with DNA or the RXRα DNA-binding domain. However, this was examined for one particular LXRE, and it may likely vary at different LXRE/DNA sequences, thus explaining the gene-specific effects observed when the phosphomutant is expressed. Future investigations beyond the scope of this study comparing global WT and phosphomutant LXR DNA-binding profiles will be able to provide further insight into the molecular mechanisms behind the observed differences in gene expression.
Beyond changes in cell proliferation, the enhanced plaque burden in M-S196ALdlr-KO mice is likely the result of the complex modulation of additional pathways relevant for atherosclerosis development. LXRα promotes the expression of factors important for macrophage efferocytotic capacity, such as the MerTK receptor for apoptotic cells (50), which can influence the formation of the necrotic core and plaque stability (51). Despite the overall increase in atherosclerosis, we found that size-matched advanced plaques from M-S196ALdlr-KO mice exhibited significantly reduced necrotic cores (Fig. 4A) consistent with the increased capacity of M-S196ALdlr-KO macrophages to engulf apoptotic cells (Fig. 4B and SI Appendix, Fig. S6C). Transcriptomic profiling revealed that in addition to the reduced expression of Mertk and its ligand Gas6, several genes known to promote phagocytosis were significantly up-regulated in M-S196ALdlr-KO cells, including the prophagocytotic receptors Ccr2 (52), Gpr132 (53), and Itgb3 (54) and the bridging molecule Mfge8 (55) (Fig. 4C and SI Appendix, Fig. S3B). This differential expression could explain the increased phagocytic ability of the LXRα phosphomutant-expressing macrophages (Fig. 4B).
With regard to the signals that may regulate LXRα phosphorylation in the context of atherosclerosis progression, we previously showed that LXRα is phosphorylated at this same residue both in response to cholesterol loading in cultured macrophages and in foam cells within atherosclerotic plaques from animal models fed a high-fat diet (13). A possible candidate could be desmosterol (a cholesterol precursor and the last intermediate in the Bloch biosynthesis pathway), which accumulates in macrophage foam cells and atherosclerotic plaques (7) and acts as an endogenous LXR ligand modulating its target gene expression (7, 56). Desmosterol promotes phosphorylation of LXRα in macrophages in culture (SI Appendix, Fig. S10) and could be one of the many signals that affect LXRα phosphorylation in an atherosclerotic environment, although it is likely that a combination of them is responsible for ultimately promoting this modification. It is possible that endogenous cues preventing LXRα phosphorylation, thus mimicking the Ser-to-Ala mutation, may alter cell proliferation and phagocytosis in a similar manner. However, they are also likely to exert other (similar or opposing) effects to other transcription factors or signaling molecules. Future studies will aim to further identify endogenous signals modulating LXRα phosphorylation in the context of atherosclerosis.
In summary, we have shown that disrupting LXRα phosphorylation in cells of the myeloid lineage affects the development of atherosclerosis that could be explained through altered cell proliferation and efferocytosis. We also show that chronically modulating LXRα phosphorylation reprograms gene regulation in macrophages under basal conditions and significantly affects their response to ligand stimulation. These findings add to our fundamental knowledge of how LXRα activity can be regulated and introduce functional consequences of its modification by phosphorylation, which should be heeded to manipulate these receptors for the design of novel cardiovascular therapies.
Materials and Methods
Analysis of Murine Atherosclerosis Development.
WTLdlr-KO and M-S196ALdlr-KO mice were generated as described in SI Appendix, SI Materials and Methods. All animal procedures and experimentation were approved by the UK’s Home Office under the Animals (Scientific Procedures) Act 1986, PPL 1390 (70/7354). Eight-week-old WTLdlr-KO and M-S196ALdlr-KO male mice were fed a WD ad libitum (20% fat, 0.15% cholesterol; no. 5342 AIN-7A; Test Diet Limited) for 12 wk. Murine atherosclerosis and lesion characterization were performed as described in SI Appendix, SI Materials and Methods.
Cell Culture and in Vitro Experiments.
Bone marrow-derived macrophages were prepared as reported by Pineda-Torra et al. (57). Peritoneal macrophages and Jurkat cells were prepared as described in SI Appendix, SI Materials and Methods. An efferocytosis assay and flow cytometry were performed as described in SI Appendix, SI Materials and Methods.
Gene Expression Analysis.
RNA quantification, RT-qPCR, and RNA sequencing and analysis were performed as described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Edward Fisher and Prof. Michael Garabedian (New York University School of Medicine) for their insightful discussions and Theresa Leon for providing Jurkat cells. This work was supported by Medical Research Council New Investigator Grant G0801278 (to I.P.-T.), British Heart Foundation (BHF) Project Grant PG/13/10/30000 (to I.P.-T.), a University College of London Grand Challenges PhD Studentship (to I.P.-T.), BHF Studentship FS/12/59/30649 (to I.P.-T.), the Royal Free Charity PhD Program in Medicine (I.P.-T.), and the University College London Division of Medicine (I.P.-T.). It was also supported by Spanish Ministry of Economy and Competitiveness Grants SAF2014-56819-R (to A.C.) and SAF2015-71878-REDT (to A.C.). T.H.T. was funded by the State São Paulo Research Foundation Grant 2017/12314-0.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.D.M. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE112213).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721245115/-/DCSupplemental.
References
- 1.Roth GA, et al. Global and regional patterns in cardiovascular mortality from 1990 to 2013. Circulation. 2015;132:1667–1678. doi: 10.1161/CIRCULATIONAHA.114.008720. [DOI] [PubMed] [Google Scholar]
- 2.Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: A dynamic balance. Nat Rev Immunol. 2013;13:709–721. doi: 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee SD, Tontonoz P. Liver X receptors at the intersection of lipid metabolism and atherogenesis. Atherosclerosis. 2015;242:29–36. doi: 10.1016/j.atherosclerosis.2015.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pascual-García M, Valledor AF. Biological roles of liver X receptors in immune cells. Arch Immunol Ther Exp (Warsz) 2012;60:235–249. doi: 10.1007/s00005-012-0179-9. [DOI] [PubMed] [Google Scholar]
- 5.Becares N, Gage MC, Pineda-Torra I. Posttranslational modifications of lipid-activated nuclear receptors: Focus on metabolism. Endocrinology. 2017;158:213–225. doi: 10.1210/en.2016-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXR α. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 7.Spann NJ, et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins JL, et al. Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J Med Chem. 2002;45:1963–1966. doi: 10.1021/jm0255116. [DOI] [PubMed] [Google Scholar]
- 9.Hong C, Tontonoz P. Liver X receptors in lipid metabolism: Opportunities for drug discovery. Nat Rev Drug Discov. 2014;13:433–444. doi: 10.1038/nrd4280. [DOI] [PubMed] [Google Scholar]
- 10.Ito A, et al. LXRs link metabolism to inflammation through Abca1-dependent regulation of membrane composition and TLR signaling. eLife. 2015;4:e08009. doi: 10.7554/eLife.08009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feig JE, et al. LXR promotes the maximal egress of monocyte-derived cells from mouse aortic plaques during atherosclerosis regression. J Clin Invest. 2010;120:4415–4424. doi: 10.1172/JCI38911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bischoff ED, et al. Non-redundant roles for LXRalpha and LXRbeta in atherosclerosis susceptibility in low density lipoprotein receptor knockout mice. J Lipid Res. 2010;51:900–906. doi: 10.1194/jlr.M900096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torra IP, et al. Phosphorylation of liver X receptor alpha selectively regulates target gene expression in macrophages. Mol Cell Biol. 2008;28:2626–2636. doi: 10.1128/MCB.01575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C, et al. Modulation of macrophage gene expression via LXRα serine 198 phosphorylation. Mol Cell Biol. 2015;35:2024–2034. doi: 10.1128/MCB.00985-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley K, Pramod AB, Croft M, Ravichandran KS, Ting JP. How mouse macrophages sense what is going on. Front Immunol. 2016;7:204. doi: 10.3389/fimmu.2016.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaiswal N, Chakraborty S, Nag A. Biology of FOXM1 and its emerging role in cancer therapy. J Proteins Proteomics. 2014;5:1–24. [Google Scholar]
- 17.Hu C, et al. LXRα-mediated downregulation of FOXM1 suppresses the proliferation of hepatocellular carcinoma cells. Oncogene. 2014;33:2888–2897. doi: 10.1038/onc.2013.250. [DOI] [PubMed] [Google Scholar]
- 18.Teboul M, et al. OR-1, a member of the nuclear receptor superfamily that interacts with the 9-cis-retinoic acid receptor. Proc Natl Acad Sci USA. 1995;92:2096–2100. doi: 10.1073/pnas.92.6.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang J, et al. Inhibiting macrophage proliferation suppresses atherosclerotic plaque inflammation. Sci Adv. 2015;1:e1400223. doi: 10.1126/sciadv.1400223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gormally MV, et al. Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nat Commun. 2014;5:5165. doi: 10.1038/ncomms6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsico G, Gormally MV. Small molecule inhibition of FOXM1: How to bring a novel compound into genomic context. Genom Data. 2014;3:19–23. doi: 10.1016/j.gdata.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robbins CS, et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat Med. 2013;19:1166–1172. doi: 10.1038/nm.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang J, et al. Inhibiting macrophage proliferation suppresses atherosclerotic plaque inflammation. Sci Adv. 2015;1:e1400223. doi: 10.1126/sciadv.1400223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lhoták Š, et al. Characterization of proliferating lesion-resident cells during all stages of atherosclerotic growth. J Am Heart Assoc. 2016;5:e003945. doi: 10.1161/JAHA.116.003945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kojima Y, Weissman IL, Leeper NJ. The role of efferocytosis in atherosclerosis. Circulation. 2017;135:476–489. doi: 10.1161/CIRCULATIONAHA.116.025684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kojima Y, et al. CD47-blocking antibodies restore phagocytosis and prevent atherosclerosis. Nature. 2016;536:86–90. doi: 10.1038/nature18935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenkins SJ, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakai M, et al. The scavenger receptor serves as a route for internalization of lysophosphatidylcholine in oxidized low density lipoprotein-induced macrophage proliferation. J Biol Chem. 1996;271:27346–27352. doi: 10.1074/jbc.271.44.27346. [DOI] [PubMed] [Google Scholar]
- 29.Flaveny CA, et al. Broad anti-tumor activity of a small molecule that selectively targets the Warburg effect and lipogenesis. Cancer Cell. 2015;28:42–56. doi: 10.1016/j.ccell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solt LA, Kamenecka TM, Burris TP. LXR-mediated inhibition of CD4+ T helper cells. PLoS One. 2012;7:e46615. doi: 10.1371/journal.pone.0046615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pascual-García M, et al. Liver X receptors inhibit macrophage proliferation through downregulation of cyclins D1 and B1 and cyclin-dependent kinases 2 and 4. J Immunol. 2011;186:4656–4667. doi: 10.4049/jimmunol.1000585. [DOI] [PubMed] [Google Scholar]
- 32.Bensinger SJ, et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134:97–111. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukuchi J, Kokontis JM, Hiipakka RA, Chuu CP, Liao S. Antiproliferative effect of liver X receptor agonists on LNCaP human prostate cancer cells. Cancer Res. 2004;64:7686–7689. doi: 10.1158/0008-5472.CAN-04-2332. [DOI] [PubMed] [Google Scholar]
- 34.Vedin L-L, Lewandowski SA, Parini P, Gustafsson J-Å, Steffensen KR. The oxysterol receptor LXR inhibits proliferation of human breast cancer cells. Carcinogenesis. 2009;30:575–579. doi: 10.1093/carcin/bgp029. [DOI] [PubMed] [Google Scholar]
- 35.Uno S, et al. Suppression of β-catenin signaling by liver X receptor ligands. Biochem Pharmacol. 2009;77:186–195. doi: 10.1016/j.bcp.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Wang JZ, Fang Y, Ji WD, Xu H. LXR agonists promote the proliferation of neural progenitor cells through MEK-ERK pathway. Biochem Biophys Res Commun. 2017;483:216–222. doi: 10.1016/j.bbrc.2016.12.163. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, et al. Cholesterol metabolite, 5-cholesten-3β-25-diol-3-sulfate, promotes hepatic proliferation in mice. J Steroid Biochem Mol Biol. 2012;132:262–270. doi: 10.1016/j.jsbmb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teh M-T, et al. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 2002;62:4773–4780. [PubMed] [Google Scholar]
- 39.Chan DW, et al. Over-expression of FOXM1 transcription factor is associated with cervical cancer progression and pathogenesis. J Pathol. 2008;215:245–252. doi: 10.1002/path.2355. [DOI] [PubMed] [Google Scholar]
- 40.Curtis C, et al. METABRIC Group The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang I-C, et al. Transgenic expression of the forkhead box M1 transcription factor induces formation of lung tumors. Oncogene. 2008;27:4137–4149. doi: 10.1038/onc.2008.60. [DOI] [PubMed] [Google Scholar]
- 42.Wierstra I. The transcription factor FOXM1 (Forkhead box M1): Proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles. Adv Cancer Res. 2013;118:97–398. doi: 10.1016/B978-0-12-407173-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 43.Korver W, Roose J, Wilson A, Clevers H. The winged-helix transcription factor Trident is expressed in actively dividing lymphocytes. Immunobiology. 1997;198:157–161. doi: 10.1016/S0171-2985(97)80036-8. [DOI] [PubMed] [Google Scholar]
- 44.Korver W, et al. The human TRIDENT/HFH-11/FKHL16 gene: Structure, localization, and promoter characterization. Genomics. 1997;46:435–442. doi: 10.1006/geno.1997.5065. [DOI] [PubMed] [Google Scholar]
- 45.Ye H, et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17:1626–1641. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Down CF, Millour J, Lam EWF, Watson RJ. Binding of FoxM1 to G2/M gene promoters is dependent upon B-Myb. Biochim Biophys Acta. 2012;1819:855–862. doi: 10.1016/j.bbagrm.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka S, et al. BubR1 insufficiency results in decreased macrophage proliferation and attenuated atherogenesis in apolipoprotein E-deficient mice. J Am Heart Assoc. 2016;5:e004081. doi: 10.1161/JAHA.116.004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weidmann H, et al. SASH1, a new potential link between smoking and atherosclerosis. Atherosclerosis. 2015;242:571–579. doi: 10.1016/j.atherosclerosis.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 49.Gallagher KA, et al. Epigenetic changes in bone marrow progenitor cells influence the inflammatory phenotype and alter wound healing in type 2 diabetes. Diabetes. 2015;64:1420–1430. doi: 10.2337/db14-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A-Gonzalez N, et al. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity. 2009;31:245–258. doi: 10.1016/j.immuni.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thorp E, Cui D, Schrijvers DM, Kuriakose G, Tabas I. Mertk receptor mutation reduces efferocytosis efficiency and promotes apoptotic cell accumulation and plaque necrosis in atherosclerotic lesions of apoe-/- mice. Arterioscler Thromb Vasc Biol. 2008;28:1421–1428. doi: 10.1161/ATVBAHA.108.167197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka T, Terada M, Ariyoshi K, Morimoto K. Monocyte chemoattractant protein-1/CC chemokine ligand 2 enhances apoptotic cell removal by macrophages through Rac1 activation. Biochem Biophys Res Commun. 2010;399:677–682. doi: 10.1016/j.bbrc.2010.07.141. [DOI] [PubMed] [Google Scholar]
- 53.Bolick DT, et al. G2A deficiency in mice promotes macrophage activation and atherosclerosis. Circ Res. 2009;104:318–327. doi: 10.1161/CIRCRESAHA.108.181131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanayama R, et al. Identification of a factor that links apoptotic cells to phagocytes. Nature. 2002;417:182–187. doi: 10.1038/417182a. [DOI] [PubMed] [Google Scholar]
- 55.Ait-Oufella H, et al. Lactadherin deficiency leads to apoptotic cell accumulation and accelerated atherosclerosis in mice. Circulation. 2007;115:2168–2177. doi: 10.1161/CIRCULATIONAHA.106.662080. [DOI] [PubMed] [Google Scholar]
- 56.Muse ED, et al. Cell-specific discrimination of desmosterol and desmosterol mimetics confers selective regulation of LXR and SREBP in macrophages. Proc Natl Acad Sci USA. 2018;115:E4680–E4689. doi: 10.1073/pnas.1714518115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pineda-Torra I, Gage M, de Juan A, Pello OM. Isolation, culture, and polarization of murine bone marrow-derived and peritoneal macrophages. Methods Mol Biol. 2015;1339:101–109. doi: 10.1007/978-1-4939-2929-0_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.