Significance
The majority of mutations residing within the rod domain of myosin have been associated with skeletal myopathy with or without cardiomyopathy. However, the molecular mechanisms underlying the variation in clinical and pathological phenotypes of myopathies associated with the MYH7 mutation are still poorly understood. We used CRISPR/Cas9-mediated genome engineering to develop a fly model for Laing distal myopathy to investigate the pathological mechanisms of the recurrent L1729del MYH7 mutation. This study unveils structural and functional phenotypes associated with this mutation in skeletal and heart muscles, and identifies a mechanism that alleviates the pathological phenotype, suggesting that E3-ligase modifier gene activity may reduce or enhance the impact of this myosin mutation in patients.
Keywords: myosin myopathy, Laing distal myopathy, myosin, Drosophila, Abba/Thin
Abstract
Myosin is a molecular motor indispensable for body movement and heart contractility. Apart from pure cardiomyopathy, mutations in MYH7 encoding slow/β-cardiac myosin heavy chain also cause skeletal muscle disease with or without cardiac involvement. Mutations within the α-helical rod domain of MYH7 are mainly associated with Laing distal myopathy. To investigate the mechanisms underlying the pathology of the recurrent causative MYH7 mutation (K1729del), we have developed a Drosophila melanogaster model of Laing distal myopathy by genomic engineering of the Drosophila Mhc locus. Homozygous MhcK1728del animals die during larval/pupal stages, and both homozygous and heterozygous larvae display reduced muscle function. Flies expressing only MhcK1728del in indirect flight and jump muscles, and heterozygous MhcK1728del animals, were flightless, with reduced movement and decreased lifespan. Sarcomeres of MhcK1728del mutant indirect flight muscles and larval body wall muscles were disrupted with clearly disorganized muscle filaments. Homozygous MhcK1728del larvae also demonstrated structural and functional impairments in heart muscle, which were not observed in heterozygous animals, indicating a dose-dependent effect of the mutated allele. The impaired jump and flight ability and the myopathy of indirect flight and leg muscles associated with MhcK1728del were fully suppressed by expression of Abba/Thin, an E3-ligase that is essential for maintaining sarcomere integrity. This model of Laing distal myopathy in Drosophila recapitulates certain morphological phenotypic features seen in Laing distal myopathy patients with the recurrent K1729del mutation. Our observations that Abba/Thin modulates these phenotypes suggest that manipulation of Abba/Thin activity levels may be beneficial in Laing distal myopathy.
Myosin is a molecular motor, converting chemical energy into mechanical force, and is indispensable for body movement and heart contractility. Striated muscle myosin heavy chain (MyHC) isoforms are encoded by a multigene family, with individual genes being differentially expressed and having different functional properties (1). In adult human limb skeletal muscle, slow/β-cardiac MyHC (MyHC I), encoded by MYH7, is expressed in type 1 muscle fibers and in the heart ventricles (2).
Slow/β-cardiac MyHC (MYH7) was the first striated muscle MyHC isoform associated with disease in humans (3). Hundreds of different dominant mutations in MYH7, mainly located within the globular head of MYH7 and some in the rod region, have been associated with hypertrophic and dilated cardiomyopathy (4). Apart from pure cardiomyopathy, mutations in MYH7 can also cause skeletal muscle disease with or without cardiac involvement (5). A vast majority of these mutations are located at the elongated α-helical coiled-coil C-terminal rod domain (light meromyosin, LMM) of MYH7. These mutations have been associated with Laing distal myopathy and myosin storage myopathy (MSM), two skeletal myopathies with distinct morphological and clinical phenotypes (5, 6).
Laing myopathy is a rare distal myopathy that was reported for the first time in 2004, when five different heterozygous mutations were identified in six families with a distinct clinical feature (7). In most cases, symptoms present early in childhood but can arise within an age range from congenital to 50 y of age. The clinical symptoms in typical cases begin with weakness of ankle dorsiflexors and a “hanging big-toe” sign. Cardiac involvement is not usually a feature of Laing distal myopathy, although association with cardiomyopathy has been reported (8–11). The muscle pathological changes are variable, but a common finding is a predominance of type 1 fibers that are numerous and small and may appear as fiber-type disproportion and core-like lesions (5, 12). The biological effects of a subset of Laing distal myopathy mutations have been assessed in muscle and nonmuscle cells and in transgenic Caenorhabditis elegans (13, 14). Although these studies have provided new insights into the pathogenesis, data interpretation is confounded by time limitations and extra chromosomal arrays. Consequently, no consensus has been reached regarding disease mechanism.
The K1729del MYH7 mutation associated with Laing distal myopathy has been reported in several unrelated families, with the largest cohort of patients originating from the Safor region in Spain (12). The patients display clinical variability with or without cardiac involvement (7, 12, 15). All affected individuals presented with weakness of great toe/ankle dorsiflexors and a majority showed proximal and axial muscle involvement and weakness of neck flexor, finger extensor, and mild facial weakness. The most relevant morphological alterations in affected individuals in this family were muscle fiber-type disproportion, core-like lesions, and mitochondrial abnormalities (12). Many of the molecular mechanisms that govern sarcomerogenesis and muscle development are conserved between the fruit fly Drosophila melanogaster and humans. In Drosophila all MyHC isoforms are encoded by a single Mhc gene through alternative RNA splicing (16), enabling identification of direct effects of mutant alleles on myofibril formation and stability.
Here, we used CRISPR/Cas9 genome engineering to develop a fly model for Laing distal myopathy to investigate the pathobiological mechanisms of the recurrent K1729del MYH7 mutation. We show that the corresponding Drosophila MhcK1728del allele expressed alone, or in trans to a wild-type allele, causes profound effects on muscle structure and function performance and lifespan. The associated defects in sarcomere structure and myofibril damage resemble those observed in Laing distal myopathy. Larvae homozygous for MhcK1728del also display structural and functional changes in the heart muscle. Overexpression of the Drosophila protein Abba/Thin, which has essential roles in maintaining sarcomeric integrity (17, 18) and is homologous to human Trim32 (human limb-girdle muscular dystrophy type 2H), was sufficient to suppress the phenotypes associated with heterozygous MhcK1728del expression. Collectively, the MhcK1728del mutant phenotype recapitulated certain muscle morphological phenotypes manifest in Laing distal myopathy patients carrying the K1729del myh7 mutation, and was rescued by increased expression of abba/thin.
Materials and Methods
Generation of the MhcK1728del Allele.
The MhcK1728del allele (corresponding to K1729 in humans, S1) was generated by CRISPR/Cas9-mediated genome editing. The targeting sequence (CTCGGACTCCAGCTTCCTCT) was cloned into pCFD3-dU6:3gRNA plasmid and coinjected with a 177-base-long single-stranded oligodeoxynucleotide (ssODNs) in fly embryos expressing Cas9 during oogenesis (y1 M{vas-Cas9}ZH2A w1118). The ssODN donor encompassed the K1728del mutation and a silent mutation located at position 5295 (NM_165191) (CCAAG > CTAAG) to introduce a DdeI enzyme digestion site. To identify mutant flies, the genomic region of interest was amplified by PCR from whole-fly extracts and digested with DdeI. The K1728del mutation was confirmed by sequencing of exons over the entire Mhc locus. The homozygous lethal MhcK1728del allele was maintained as a stock over a CyO balancer chromosome carrying Deformed > YFP.
Drosophila Genetics and Lines.
Fly culture, crosses, and analyses were performed on standard fly food and at room temperature (22 °C) unless otherwise stated. The w1118 (w−) line was used as a wild-type genetic background. Transgenic lines carrying MhcK1728del/+ were generated on the w− background (BestGene). As control, the w−;Sco/CyO, Deformed-YFP fly line was used (Bloomington Stock Center). Flies carrying the loss-of-function allele Mhc1 or the amorphic Mhc10 allele that results in undetectable levels of Mhc in the indirect flight muscle (IFM) and jump muscle (19), were kindly provided by S. I. Bernstein, San Diego State University, San Diego. HandC-GFP (expressing GFP in the heart muscle) was kindly provided by A. Paulalat, University of Osnabrück, Germany. Mef2-Gal4 (expressing Gal4 in muscle lineages) was obtained from the Bloomington Stock Center (stock no. 27390). Flies carrying UAS-Abba on the third chromosome (w; UAS-Abba) and AbbaMJO0348, which is a loss-of-function allele, were kindly provided by H. Nguyen, University of Erlangen-Nuremberg, Germany. For rescue assays, w;MhcK1728del/+;UAS-Abba/Mef2-Gal4, w;MhcK1728del/Mhc10;UAS-Abba/Mef2-Gal4, or w;MhcK1728del/AbbaMJO0348 were analyzed. MhcK1728del/Mhc1,UAS-Mhc;Mef2-Gal4/+ was analyzed to assess the impact of overexpression of the wild-type Mhc allele in rescue of lethality in MhcK1728del animals heterozygote with a null Mhc allele (Mhc1). The phenotypic effect of the MhcK1728del allele was further studied by overexpression of either the mutated or wild-type Mhc (Mhc1,UAS-MhcK1728del/+;Mef2-Gal4/+ or Mhc1,UAS-Mhc/+;Mef2-Gal4/+). For construction of UAS-MhcK1728del, see SI Appendix. The genotypes used in this study are listed in a table (SI Appendix, Table S1).
Larval Motility Assays.
The crawling assay was performed with third-instar larvae, as previously described (20). Briefly, single larvae were transferred to 9-cm apple-juice plates and the animal’s position was recorded for 1 min to trace the movements. At least 20 animals of each genotype were analyzed. In the larval turning assay (21), third-instar larvae were placed on apple-juice plates and gently rolled ventral side up. The time taken for larvae to return to dorsal side up position and continue their forward movement was recorded.
Jumping and Flight Assays in Adult Flies.
Four-day-old, 2-wk-old, and 5-wk-old flies were evaluated for jump and flight abilities. The jump assay was essentially as previously described by Swank et al. (22); jump ability was defined as the horizontal distance a fly was able to jump from a 7-cm-high platform. Wings were removed before jump-testing. Twenty flies per genotype were assessed in 10 replicates. Flight ability was assessed for 20 min at room temperature in a transparent vial (9-cm high, internal diameter 2.6 cm) with a light source at the top of the vial to encourage flies to fly. The vial was agitated every 5 min. Flies that were not able to perform flight motion within 20 min were considered flightless. Twenty flies per genotype were assessed in five replicates.
Viability and Lifespan of Adult Flies.
Ten flies of the same sex were kept on 12-mL standard fly food. Flies were regularly transferred to new food vials and dead flies were recorded every 3 d until day 30 (0, 3, 6, 9, 12, 15, 18, 21, 24, 27, and 30). Day 0 denotes eclosion. Fly lifespan analysis was performed in four replicates.
Analysis of Third-Instar Larval Heart Rate.
Heart rates were measured using fluorescence microscopy images of the beating heart in third-instar larvae carrying HandC-GFP (GFP expressed in the heart), as previously described (23). Manual heart-rate counting was performed using a cell counter. Twenty larvae of each genotype were analyzed in triplicates.
Immunofluorescence and Confocal Microscopy of Larval, Pupa, and Adult Flies.
Dissected body wall and heart muscles from third-instar larvae and IFM from pupae and adult flies (4-d-old and 2- and 5-wk-old flies) were fixed with 4% formaldehyde for 10 min. Samples were permeabilized in PBS containing 2% BSA and 1% Triton X-100 for 30 min and then incubated overnight at 4 °C with primary antibodies. Primary antibodies were: IgG mouse anti-myosin (1:100), kindly provided by J. D. Saide, Boston University, Boston, Massachusetts; IgG1 Rat anti-Titin/Kettin MAC155 (1:100; Abcam); IgG Rabbit anti-Obscurin (1:100), kindly provided by B. Bullard, University of York, York, United Kingdom). Secondary antibodies were: goat IgG anti-mouse Cy3 #115-165-146 (1:1,000), Donkey IgG anti-rabbit Alexa Fluor 647 #711-605-152 (1:600), and Donkey IgG ant-Rat Alexa Fluor 647 #712-605-150 (1:200), all from Jackson ImmunoResearch. Samples were mounted in Fluoromount-G and imaged with a Confocal LSM800 microscope (Zeiss), using 63× objective. Images were processed using Photoshop (Adobe).
Ultrastructural Analysis.
Preparation of samples for transmission electron microscopy of IFMs in adult flies, was as previously described (24). Samples were viewed on a TEM LEO 912 Omega (Zeiss) or a JEOL 1011 electron microscope and images were captured with a VELETA Olympus digital CCD camera or with a Gatan 782 camera.
Statistical Analysis.
Graphs and statistical comparisons were generated with IBM SPSS 20 Statistics (IBM Corp.). Statistical data are presented as mean ± SEM. Survival data were analyzed with Kaplan–Meier, and statistical comparisons were made with log-rank pairwise analysis. Statistical significance for heart rate and locomotor effects were determined by general linear model univariate ANOVA, followed by Bonferroni post hoc. The mean difference was considered to be statistically significant at the 95% confidence level. Results were considered as not significant (ns) when P > 0.05, very significant when 0.01 < P < 0.05 (*), and extremely significant when P < 0.001 (**). Figures were assembled with Adobe Photoshop and Illustrator CC 2015.5 (Adobe Systems).
Supplementary Information Appendix.
A detailed description of additional methods is provided in the SI Appendix.
Results
The MhcK1728del Allele Impairs Fitness and Causes Homozygous Lethality.
Alignment of the MyHC from human (MYH7) and Drosophila (Mhc) indicates that K1729 in MYH7 is conserved and corresponds to K1728 in the Drosophila Mhc locus (SI Appendix, Fig. S1). The K1729del mutation, located in the LMM of MYH7, is predicted to disrupt the heptad repeats of the α-helical coiled-coil C-terminal rod domain of myosin. To generate a fly model for this mutation, we used targeted genome editing using CRISPR/Cas9. The resulting MhcK1728del allele was lethal homozygous and lethal in trans with the Mhc1 loss-of-function allele and this lethality could be rescued by muscle-specific expression of wild-type Mhc (Materials and Methods). Heterozygous MhcK1728del animals (MhcK1728del/+) were viable but exhibited a shorter mean lifespan than control flies (Sco/CyO) and flies heterozygous for the loss-of-function Mhc1 allele at 25 °C (Fig. 1A). In addition, they displayed defects in wing posture similar to those reported for Mhc10 mutants that selectively lack Mhc protein in fast-twitch IFMs and jump muscles (25, 26). The penetrance of wing-posture defects was increased in flies expressing MhcK1728del in trans to Mhc10 (MhcK1728del/Mhc10). No homozygous MhcK1728del adult flies (MhcK1728del/MhcK1728del) were obtained. Some MhcK1728del homozygous animals died during embryogenesis, while the majority died during larval stages. A few homozygous animals developed to third-instar larval stages and underwent pupation; however, they never developed into flies and the time needed to reach pupal stage was approximately three times longer than that for heterozygous siblings. Thus, the MhcK1728del allele has an impact on animal fitness with the homozygous condition being lethal.
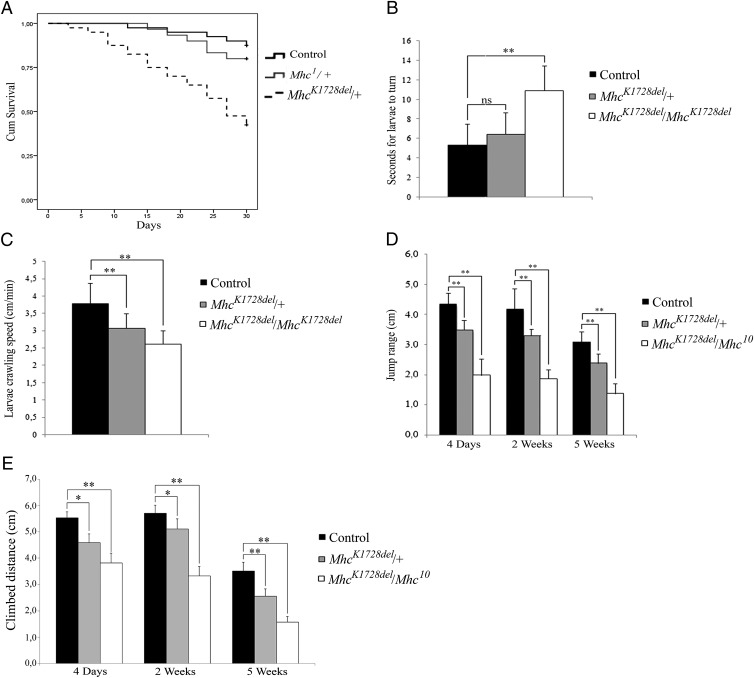
Fig. 1.
Impaired muscle function in MhcK1728del mutants. (A) Life span: Heterozygous MhcK1728del/+ adult flies show mean lifespan of 23 d vs. 29 d for control flies (P < 0.001 log-rank test) and vs. 28 d for heterozygous Mhc1/+ flies (P = 0.002 log-rank test). (B) Larval turning assay: Homozygous MhcK1728del/MhcK1728del larvae show a significant increase in time needed to turn belly-down when placed on their backs (P < 0.001), compared with controls. No significant increase in time is seen for the heterozygous MhcK1728del/+ larvae. (C) Larval crawling speed: Both heterozygotes and homozygotes show significant reduction in crawling ability (P < 0.0001) at third-instar larval stage. (D) Adult jump ability at 4 d, 2 wk, and 5 wk of age: jump ability of MhcK1728del/+ and MhcK1728del/Mhc10 flies is reduced, compared with controls (P < 0.0001 for all age groups). For a given genotype, there is no significant difference in jump ability between 4-d-old and 2-wk-old flies, but ability significantly declines between 2- and 5-wk-old flies (P < 0.0001). (E) Adult climbing ability at 4 d, 2 wk, and 5 wk of age: climbing ability of MhcK1728del/+ and MhcK1728del/Mhc10 flies is severely impaired compared with control flies (P < 0.0003) at all time points measured. There is no significant difference in climbing ability between 4-d-old and 2-wk-old flies, as observed with jump ability, but the deterioration is highly significant between 2 and 5 wk for all genotypes (P < 0.0001). *P < 0.05; **P < 0.001.
Reduced Motility in Animals Heterozygous and Homozygous for MhcK1728del.
Larval turning and crawling assays were used to evaluate muscle function in heterozygous and homozygous MhcK1728del larvae (MhcK1728del/MhcK1728del). In larval turning assays, animals homozygous for MhcK1728del showed a twofold increase in time needed to turn over and resume crawling on the ventral side compared with control animals (homozygous MhcK1728del P < 0.001 vs. controls) (Fig. 1B). Furthermore, homozygous and heterozygous MhcK1728del larvae exhibited an extremely significant reduction in crawling ability compared with the control animals (homozygous MhcK1728del: 2.61 ± 0.39 cm, P < 0.0001 vs. 3.79 ± 0.59 cm, and heterozygous MhcK1728del: 3.06 ± 0.43, P < 0.0001 vs. 3.79 ± 0.59 cm) (Fig. 1C). There was also a significant reduction in crawling ability between homozygous and heterozygous MhcK1728del animals (P = 0.014) (Fig. 1C).
Because homozygous MhcK1728del animals do not survive to adulthood, we employed the Mhc10 allele to genetically produce adult flies in which only the MhcK1728del mutant protein is expressed in flight and jump muscles. Jump, climbing, and flight ability was subsequently analyzed in 4-d-old, and 2- and 5-wk-old MhcK1728del/Mhc10 trans-heterozygous adult flies. Jump ability of MhcK1728del/Mhc10 was severely impaired, with a jump range less than half of the control flies (P ≤ 0.0001) at all time points measured. There was no significant difference in jump ability between 4-d-old and 2-wk-old flies, but jump ability significantly decreased in both mutant and control flies between 2 and 5 wk (P < 0.0001) (Fig. 1D). Heterozygous MhcK1728del flies also showed a significant reduction in jump ability, but to a lesser extent than the MhcK1728del/Mhc10 trans-heterozygotes (Fig. 1D). Rapid iterative negative geotaxis (RING) assessment of MhcK1728del/Mhc10 indicated severely impaired climbing ability that was significantly reduced compared with controls (P < 0.0003) (Fig. 1E) at all time points measured. As with jump ability, there was no significant difference in climbing ability between 4-d-old and 2-wk-old flies, but climbing ability significantly decreased in both mutant and control flies between 2 and 5 wk (P < 0.0001) (Fig. 1E). A significant reduction in climbing ability was also observed in heterozygous MhcK1728del flies, but to a lesser extent than in the MhcK1728del/Mhc10 trans-heterozygotes (Fig. 1E). In the flight assay, control flies were able to leave the open vial within 20 min. In contrast, MhcK1728del/Mhc10 trans-heterozygotes and the heterozygous MhcK1728del flies remained in the vial and failed to beat their wings, indicating a complete lack of flight ability. Jump, climbing, and flight ability was also analyzed in flies overexpressing either wild-type Mhc or MhcK1728del in muscle with the Mef2-Gal4 driver. Jump ability was significantly impaired in Mef2 > MhcK1728del flies compared with Mef2 > Mhc flies at both 4 d and 2 wk (P < 0.038 at 4-d-old and P < 0.003 at 2-wk-old flies) (SI Appendix, Fig. S2A), as was climbing ability at both time points (P < 0.004 at 4-d-old and P < 0.0001 at 2-wk-old flies) (SI Appendix, Fig. S2B). Mef2-Gal4–driven expression of MhcK1728del, but not of Mhc, also caused loss of flight ability. Taken together, these results indicate that the expression of MhcK1728del results in impairment of muscle function.
Muscle Morphology.
To address the effects of the MhcK1728del mutation on sarcomere organization, we dissected and labeled larval body muscles from third-instar larvae and IFMs from late pupae and adult flies for sarcomeric proteins. Sallimus (also known as Kettin or Titin) is a large protein that localizes to the Z-disks, while Obscurin (Unc-89) (27) links myosin filaments at the M-band of the sarcomere. Double-immunofluorescence analyses with antibodies against Mhc and Kettin/Titin or Mhc and Obscurin reveal a severely disrupted sarcomere structure in homozygous MhcK1728del larvae. Both the Z-disks and M-bands appear reduced and discontinuous, while Mhc fails to concentrate in a distinct A-band at either side of the Z-disk and instead occupies nearly the entire space between the Z-disks (Fig. 2 A and B). A similar sarcomeric disruption phenotype is evident in late pupal IFMs (Fig. 2 C and D). Sarcomeric irregularities are evident also in heterozygous MhcK1728del animals, which exhibit less defined Mhc-containing A-bands (Fig. 2) and slightly wider and jagged Z-disks and M-bands in the larval body wall (Fig. 2 A and B). We also analyzed IFMs from 4-d-old and 2-wk-old adult flies overexpressing either wild-type Mhc (Mef2 > Mhc) or mutated Mhc (Mef2 > MhcK1728del). Mef2 > MhcK1728del flies displayed a severely disrupted sarcomeric structure, including less-well separated A-bands and severe undefined structure of Z-disks and M-bands, compared with Mef2 > Mhc flies (SI Appendix, Fig. S2C).
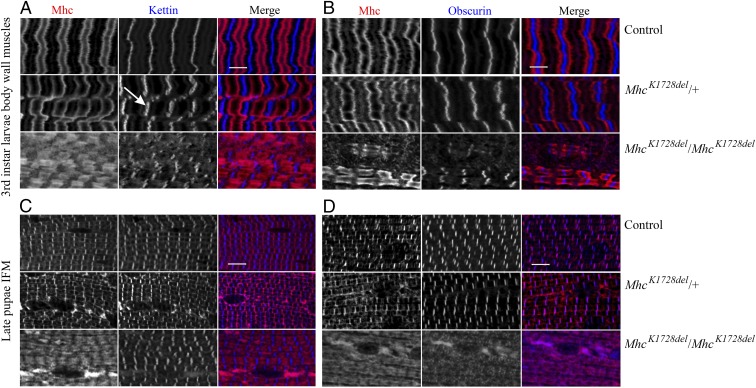
Fig. 2.
Muscle morphology in MhcK1728del mutant larvae and pupae. Larval body wall muscles and pupal IFM were labeled for myosin (Mhc) and Kettin/Titin (A and C) or Mhc and Obscurin (B and D). Each panel shows control animals (Top), MhcK1728del/+ animals (Middle), and MhcK1728del/MhcK1728del animals (Bottom). (A and B) Heterozygous MhcK1728del larvae show less-defined sarcomeric striations than control animals, Kettin/Titin is detected in slightly broad and jagged stripes at the Z-disks (arrow in A) and Obscurin as jagged stripes at M-bands (B). Homozygous larvae show disruptions in sarcomere and immature striations with myosin-containing A-bands appearing unstructured and extensively stretched (A). Z-disks (Kettin/Titin) and M-bands (Obscurin) appear reduced and discontinuous (A and B). (C and D) The IFMs of heterozygous pupae show less defined sarcomeric striated patterns. Homozygous MhcK1728del pupae display severely disrupted sarcomere structure and widely overextended myosin-containing A-bands. Staining with Obscurin revealed unstructured and immature M-band development (D). Depositions of myosin are detected in both heterozygous and homozygous MhcK1728del pupae, and were greater in number and size in homozygotes (C and D). (Scale bars, 5 µm.)
To explore potential progressive effects of the MhcK1728del allele on sarcomere structure, we analyzed IFMs of adult flies at 4 d old, and 2 and 5 wk old. In 4-d-old MhcK1728del/Mhc10 flies that exclusively express MhcK1728del in indirect flight and jump muscles, Mhc is detected in the distinct A-bands; however, the bands generally appear less separated and Mhc is only faintly detected among the muscle fibers. Notably, some areas showed increased myosin immunofluorescence, probably indicating myosin accumulation. These undefined subcellular structures diminished with time, accompanied by more extensive areas of sarcomeric disruption (evident from mislocalization of Mhc, Kettin, and Obscurin) and regions of myofibril atrophy in 5-wk-old flies (Fig. 3). Similar but less-severe structural defects were observed in IFMs of heterozygous MhcK1728del flies, with areas of increased myosin immunofluorescence at day 4. Defects in sarcomere organization were less severe and there was no atrophy evident in these animals. Thus, MhcK1728del appears to result in a progressively worsening adult muscle phenotype, as has been reported in patients with Laing distal myopathy.
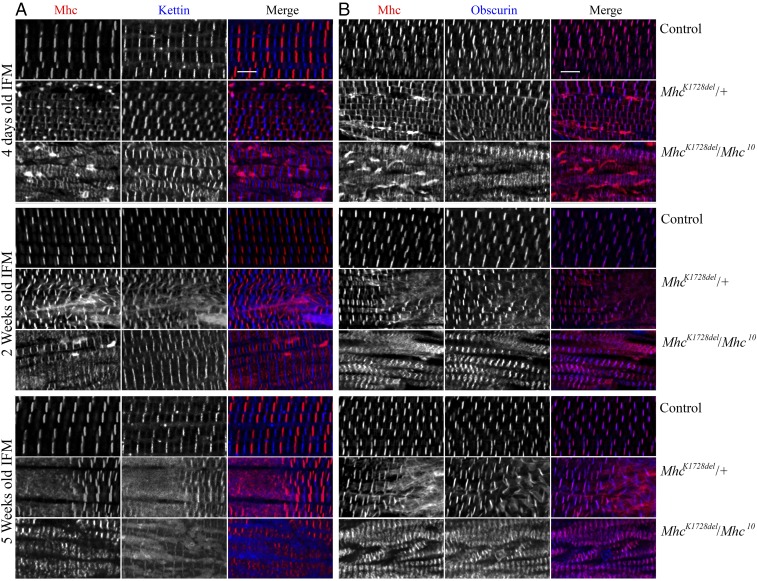
Fig. 3.
Muscle morphology in IFMs of MhcK1728del mutant adult flies. IFMs were labeled for Mhc and Kettin/Titin (A) or Mhc and Obscurin (B) in 4-d-old, and 2- and 5-wk-old adult flies. Each panel shows control animals (Top), MhcK1728del/+ animals (Middle), and MhcK1728del/Mhc10 animals (Bottom) for each age group. Control flies show parallel periodic striations across the IFMs at all ages. Heterozygous MhcK1728del/+ and MhcK1728del/Mhc10 animals show progressive disruptions in sarcomeric structure with age. The 4-d-old MhcK1728del/Mhc10 flies show distinct but less spaced A-bands and areas with increased myosin intensity, probably indicating myosin accumulation, which diminished with time. The 2- and 5-wk-old adult myofibrils of heterozygous MhcK1728del/+ and MhcK1728del/Mhc10 flies display sarcomere fragmentation and dissolution of Z-disks. The 5-wk-old MhcK1728del/Mhc10 flies show myofibril atrophy. (Scale bars, 5 µm.)
Ultrastructural analysis of the IFM was performed on 7-d-old adult flies. Well-organized sarcomeres with preserved Z-disk and M-bands were observed in IFMs of control flies (Fig. 4 A–C). In IFMs of heterozygous MhcK1728del flies, fibers displayed focal thick filament loss (Fig. 4D) and sarcomeric disorganization, with Z-disk streaming spanning more than one sarcomere (Fig. 4 E, G, and H) and accumulation of Z-disk–derived material (Fig. 4F). Very occasionally, some mitochondrial aggregates were observed (Fig. 4I). Myofibrils of MhcK1728del/Mhc10 trans-heterozygous flies that express only the MhcK1728del mutant protein in indirect flight and jump muscles, showed large areas of severe thick filament loss, with pale Z-disk and Z-disk fragmentation, as well as areas containing small electron-dense granular material (Fig. 4 J–L).
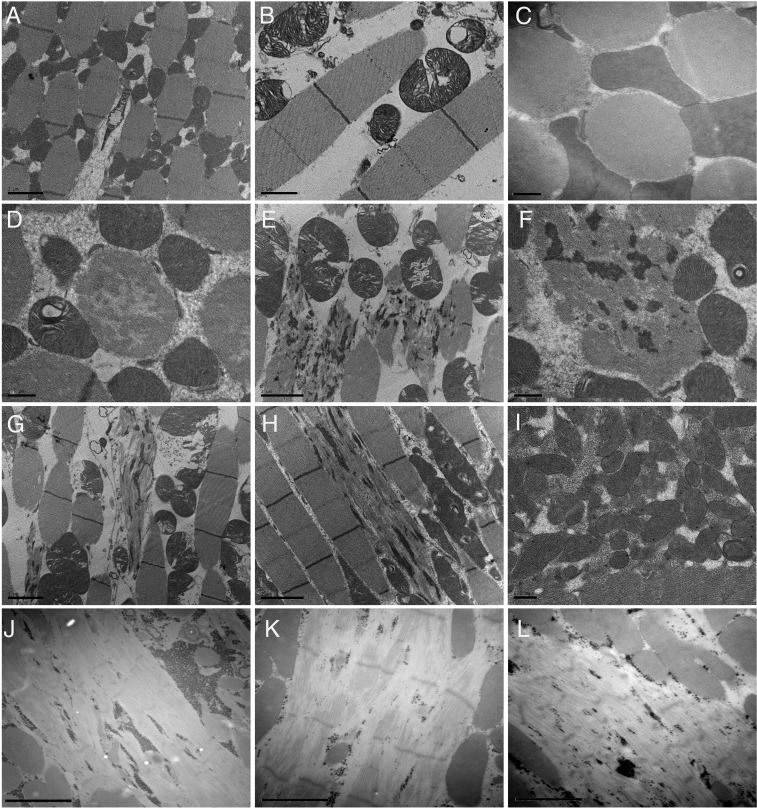
Fig. 4.
Transmission electron micrographs of IFMs in MhcK1728del flies. Transmission electron micrographs of IFMs in 7-d-old control (A–C), heterozygous MhcK1728del/+ (D–I), and MhcK1728del/Mhc10 flies (J–L). (A–C) Control flies show regular sarcomere structure with well-defined Z-disk and M-bands (A and B) and dense fibers (C, transverse section). (D and F) Transverse sections of heterozygous MhcK1728del/+ flies reveal fibers with focal thick filament loss (D) and accumulation of Z-disk–derived material (F). (E, G, and H) Longitudinal sections reveal fibers with Z-disk streaming and extensions leading to myofibrillar disorganization. (I) Collections of mitochondria with some electron-dense inclusions. (J–L) IFMs of MhcK1728del/Mhc10 flies revealed severe thick filament loss, pale Z-disks and accumulation of electron dense granular material. [Scale bars: 2 µm (A, E, G, H, and J–L), 1 µm (B and C), and 0.5 µm (D, F, and I).]
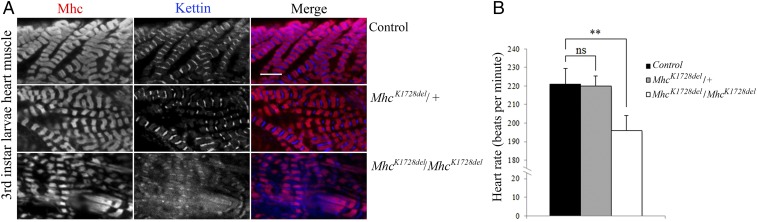
Homozygous MhcK1728del Allele Causes Reduced Heart Rate and Morphological Defects.
To assess the effects of MhcK1728del mutation on cardiac muscle structure and function, morphological and heart-rate analyses were assessed in the hearts of the heterozygous and homozygous MhcK1728del third-instar larvae. Immunofluorescence analyses with antibodies detecting Mhc and Kettin/Titin reveal a severely disrupted sarcomere structure in the heart of homozygous MhcK1728del larvae. Kettin/Titin fails to concentrate in distinct Z-disks and instead occupies nearly the entire space between the sarcomeres. Mhc-containing A-bands appear extended and less defined. In contrast, no obvious defect is observed in the heart muscle of heterozygous MhcK1728del larvae (Fig. 5A). Expression of the homozygous MhcK1728del allele resulted in a significant 10% reduction in heart rate (P = 0.002) (Fig. 5B) compared with control animals. There was no difference in heart rate between heterozygous MhcK1728del and control animals.
Fig. 5.
Functional and structural impairment in homozygous MhcK1728del larval heart muscles. (A) Heart muscles of third-instar larvae were labeled for Mhc and Kettin/Titin. Heart muscle of heterozygous MhcK1728del/+ mutant (Middle) shows normal sarcomeric striated patterns, compared with control (Top). Homozygous MhcK1728del/MhcK1728del larval heart muscle (Bottom) exhibits disturbed sarcomere structures with disorganised Z-disks and diffuse Kettin-staining. (Scale bar, 5 µm.) (B) Heart rate assessment: Heterozygous MhcK1728del/+ animals show no significant changes in heart rate compared with controls. Homozygous MhcK1728del/MhcK1728del larvae show significantly lower heart rate compared with control and heterozygous MhcK1728del/+ animals (P < 0.0001). Twenty larvae of each group were analyzed in triplicate. ns, P > 0.05; **P < 0.001.
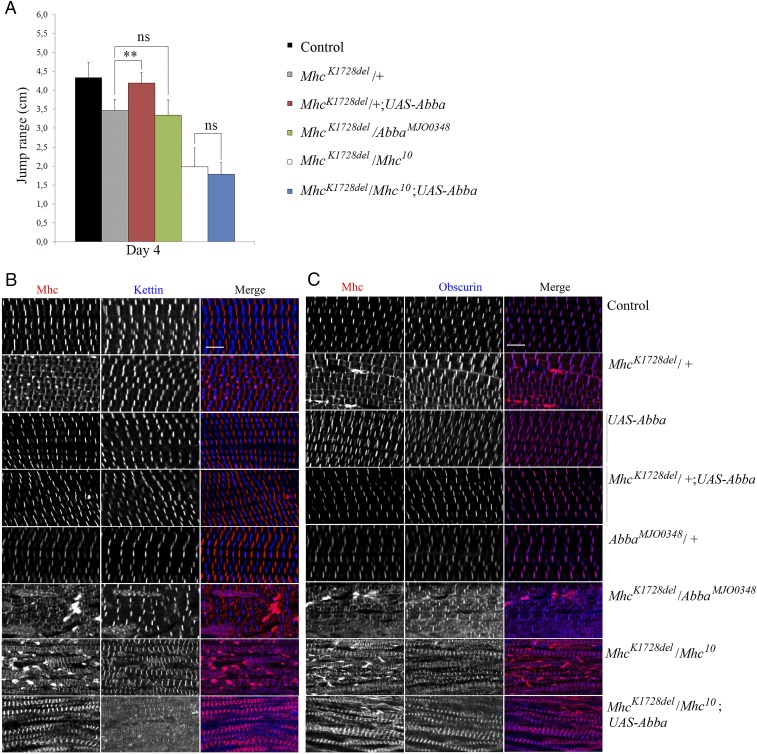
MhcK1728del-Induced Muscle Defects Are Suppressed by Overexpression of Abba/Thin.
In humans, members of the TRIM/RBCC superfamily of E3-ligases [the human muscle-specific RING finger (MuRFs)] proteins can regulate the degradation of MyHCs. We previously reported that MuRF1 deficiency combined with deleterious MuRF3 mutation leads to subsarcolemmal accumulation of myosin (28). In Drosophila, the TRIM/RBCC protein family member Abba/Thin (Another B-box Affiliate) is required for sarcomeric integrity during muscle formation and function, and mutants display abnormal accumulation of Mhc globular structures (17, 18). We therefore investigated whether levels of Abba/Thin expression have any effect on the muscle defects associated with MhcK1728del, using the muscle-specific Mef2-Gal4 driver to overexpress Abba (17). In flies trans-heterozygous for MhcK1728del and Mhc10, overexpression of Abba did not result in significant rescue of either jump (MhcK1728del/Mhc10;Mef2 > Abba) (Fig. 6A) or flight ability. However, morphological analyses of the IFM revealed an apparent reduction in aberrant myosin accumulation, although no improvement in sarcomeric organization were seen (Fig. 6 B and C). Thus, in the absence of wild-type myosin, the muscle defects caused by MhcK1728del cannot be improved by increased levels of Abba. In contrast, overexpression of Abba/Thin was able to improve muscle functions of heterozygous MhcK1728del animals (MhcK1728del/+;Mef2 > Abba). Jump and flight abilities were evaluated in 4-d-old MhcK1728del heterozygous adult flies. Jump ability of MhcK1728del heterozygotes was restored to that of the wild-type upon muscle-specific expression of Abba (Fig. 6A), and flies also regained flight ability. To assess the effects of Abba/Thin overexpression on the morphology of the IFMs in animals heterozygous for MhcK1728del, we again used antibodies against Mhc and Kettin/Titin or Mhc and Obscurin. Immunofluorescence analyses revealed an organized sarcomeric structure in these flies with distinct Z-disks, M-bands, and Mhc-containing thick filaments. Moreover, the distinct areas of increased myosin immunofluorescence that were observed in 4-d-old heterozygous MhcK1728del flies were not detected in heterozygous MhcK1728del flies that overexpressed Abba (Fig. 6 B and C). These findings indicate that elevated levels of Abba are able to alleviate muscle defects associated with the MhcK1728del allele in the presence of wild-type Mhc.
Fig. 6.
Rescue of 4-d-old heterozygous MhcK1728del/+ fly muscle function and morphology with overexpression of abba/thin. (A) Adult jump ability at 4 d of age: jump ability of MhcK1728del/+ and MhcK1728del/Mhc10 flies is significantly reduced, compared with controls. Overexpression of Abba in flies with MhcK1728del in trans to Mhc10 (MhcK1728del/Mhc10;UAS-Abba/Mef2-Gal4), did not cause any significant rescue of jump ability. Jump ability of heterozygous MhcK1728del/+ flies was restored upon overexpression of Abba (MhcK1728del/+;UAS-Abba/Mef2-Gal4) to levels comparable with controls (P < 0.0001). Flies heterozygous for both MhcK1728del and the loss-of-function allele abbaMJO0348 (MhcK1728del/abbaMJO0348) did not show significant alteration in jump ability compared with flies heterozygous only for MhcK1728del (MhcK1728del/+). ns, P > 0.05; **P < 0.001. (B and C) IFMs of 4-d-old adult flies were labeled for Mhc and Kettin/Titin (B) or Mhc and Obscurin (C). Control flies, flies with overexpression of Abba (UAS-Abba/Mef2-Gal4), and flies heterozygous for the loss of function Abba allele (abbaMJO0348) show parallel periodic sarcomeric striations in the IFMs. Less-defined sarcomeric structures are observed in the IFM of heterozygous MhcK1728del/+ flies. Structured periodic sarcomeric striations are restored in heterozygous MhcK1728del animals that overexpress Abba (MhcK1728del/+;UAS-Abba/Mef2-Gal4). Overexpression of Abba in flies that carry MhcK1728del in trans to Mhc10 (MhcK1728del/Mhc10;UAS-Abba/Mef2-Gal4) does not improve sarcomeric organization, but results in an apparent reduction in aberrant myosin accumulation seen in flies with MhcK1728del in trans to Mhc10 (MhcK1728del/Mhc10). Flies heterozygous for MhcK1728del and abbaMJO0348 (MhcK1728del/abbaMJO0348) show IFMs with fragmentation of sarcomeres and amorphous myosin, Z-disks and M-bands observed. (Scale bars, 5 µm.)
We next addressed if reduced Abba levels had any effect on the muscle phenotypes displayed by heterozygous MhcK1728del flies. Thus, 4-d-old animals heterozygous for both MhcK1728del and the loss-of-function allele abbaMJO0348 were subjected to functional and structural analyses. Although removal of one wild-type copy of Abba (MhcK1728del/abbaMJO0348) did not cause a significant alteration in jump ability compared with MhcK1728del heterozygotes controls (Fig. 6A), flies exhibited a more severe muscle phenotype, with unstructured Mhc-containing thick filaments and areas with increased myosin immunofluorescence, indicating myosin accumulation (Fig. 6 B and C). Moreover, Obscurin also failed to concentrate in M-bands and the Z-disks appeared less structured in MhcK1728del/abbaMJO0348 than in heterozygous MhcK1728del flies (Fig. 6 B and C). Neither overexpression of Abba alone nor abbaMJO0348 heterozygosity caused detectable morphological changes in muscle or sarcomere organization (Fig. 6 B and C). Collectively, these data show that Abba expression levels modulate the muscle phenotype observed in heterozygous MhcK1728del animals, with elevated Abba levels restoring muscle function and morphology, while reduced Abba levels enhance the severity of muscle phenotypes.
Discussion
In the present study we have established a Laing distal myopathy model in D. melanogaster, using CRISPR/Cas9 genome editing to incorporate specific amino acid changes in the Drosophila Mhc locus. This model recapitulates certain muscle morphological features seen in Laing distal myopathy patients carrying the recurrent K1729del mutation, and offers a valuable system for understanding the underlying pathology and testing therapeutic approaches.
Many mutations have been reported to cause Laing distal myopathy, including amino acid deletions in the LMM region, such as K1617del and K1729del, and are able to cause different morphological and clinical features. Initially, it was postulated that the various morphological and clinical phenotypes are dictated by the position of the mutation within the slow/β-cardiac MyHC protein. However, the intrafamilial variation of clinical and pathological phenotypes (12), and the overlap of clinical and pathologic phenotypes associated with various mutated residues (5), means general conclusions are hard to make. As tissue biopsy obtained from patients is limited, it has been difficult to assess the mechanism of pathogenicity and to follow the course of muscle damage during disease development. Moreover, patients with myopathy often show compensatory changes in protein expression that can obscure the primary effect of the mutation. This is further hampered by the presence of more than one MyHC isoform in skeletal muscle. Establishing a simplified disease model in D. melanogaster, which contains a single Mhc gene, allows controlled experimental interrogation of disease mechanisms (29–31). The sarcomeric structure of myofibrils is largely conserved between human and flies and the IFM is particularly amenable to functional, genetic, and molecular analyses.
Our MhcK1728del allele, corresponding to the recurrent K1729del mutation in slow/β-cardiac MyHC, represents a disease model for one of the most common Laing distal myopathy-causing mutations (7, 12, 15). Although the severity of myopathy is age-unrelated, it is progressive with time and is characterized by muscle fiber-type disproportion, core-like lesions in the muscle fibers, and mitochondrial abnormalities (7, 12, 15). Our analyses of heterozygous MhcK1728del Drosophila muscle revealed large areas of sarcomeric disorganization with Z-disk streaming and extensions, reminiscent of the core and mini core-like lesions observed in muscle biopsies from patients carrying the heterozygous K1729del mutation. An increased severity of the morphological changes was observed with age, supporting progressive myopathy. The expression of the mutated allele in an otherwise wild-type situation also caused sarcomere disruption, indicating that expression of the mutant allele is the underlying cause of the phenotype.
This muscle pathology differs from the abnormalities observed in a Drosophila model of the E706K myosin heavy chain IIa myopathy, which shows abnormal packing of thin and thick filaments, rimmed vacuoles, cylindrical spirals, and abnormal membrane invaginations as the main ultrastructural features (29). We occasionally observed mitochondrial abnormalities, a feature that has been found in some, but not in all patients carrying the heterozygous K1729del mutation (12).
Consistent with their aberrant muscle architecture, MhcK1728del flies also display defects in flight, jumping, and climbing ability, as well as a reduced lifespan. Homozygosity of the MhcK1728del allele is not compatible with fly viability. Impaired larval muscle functions, such as turning from belly-up to belly-down, were observed in homozygous MhcK1728del mutants during the first- and second-larval instar, and most animals died during larval stages, although a few made it to pupariation but failed to eclose. The lethal phenotype is consistent with the demonstrated severe sarcomere abnormalities, with disintegration of sarcomeric structure and mislocalization of core components, such as Kettin/Titin and M-bands, indicating a marked impact of the mutated myosin on muscle function. Notably, all Laing distal myopathy patients reported so far have a dominant mode of inheritance being heterozygous for the mutation. Our findings may indicate that homozygous MYH7 mutation associated with Laing distal myopathy is not compatible with a normal life expectancy.
MyHC is also the main component of the sarcomeric thick filaments in both human and Drosophila heart muscle (32). Development of cardiomyopathy is an uncommon clinical phenotype in Laing distal myopathy patients carrying a heterozygous mutation in MYH7. However, cardiomyopathy is a clinical feature in patients with MSM caused by homozygous MYH7 mutations (33). Our Drosophila larval cardiac assessment revealed alterations of heart rate in homozygous, but not heterozygous MhcK1728del animals, which correlated with sarcomeric structural defects in the heart. This might indicate an underlying dose-dependent mechanism for sarcomeric disruption in the heart and, hence, development of cardiomyopathy.
The Mhc K1728del mutant phenotype, including altered muscle function and morphology, was mimicked in flies overexpressing the MhcK1728del allele (SI Appendix), further supporting that the K1728del Mhc mutation is the underlying cause of the phenotypes observed in the Drosophila model.
In Drosophila, the Abba/Thin protein has a fundamental role in maintaining myofibril bundling and sarcomeric integrity during muscle development and usage (17, 18). Abba localizes to Z-disks in muscle and loss of Abba causes progressive muscle degeneration and accumulation of Mhc in globular structures (17). The closest human homolog to Abba/Thin is the TRIM32 E3 ubiquitin ligase (17, 18), which belongs to the TRIM/RBCC superfamily. Mutations in TRIM32 are associated with autosomal recessive limb-girdle muscular dystrophy 2H, a muscle-wasting disease with variable clinical spectrum ranging from almost asymptomatic to wheelchair-bound patients (34). The TRIM/RBCC family also includes the MuRF E3 ligases, which regulate degradation of MyHCs (35, 36). Patients with compound MuRF1 (TRIM63) deficiency and a deleterious MuRF3 (TRIM54) missense mutation develop cardiomyopathy and skeletal myopathy associated with subsarcolemmal myosin accumulation and aggregates of fragmented sarcomeres (28). Similar phenotypes were observed in double-knockout mice for MuRF1 and MuRF3 (37), and mice lacking MuRF1 are resistant to skeletal muscle atrophy (38, 39). The phenotypes described for patients with mutations in TRIM/RBCC family members resemble those caused by Abba/Thin deficiency in flies (17, 18), including reduced muscle function, myofiber fragmentation, and subsarcolemmal accumulation of myosin (17, 18). Here, we show that overexpression of Abba/Thin can restore muscle function and morphology in flies with MhcK1728del mutation. Abba overexpression fully suppressed the phenotypic effects of heterozygous MhcK1728del, including reduced jump ability, lack of flight ability, and the myopathy of IFMs. In contrast, reduced Abba levels in animals heterozygous for AbbaMJO0348 enhances the muscle defects associated with the heterozygous MhcK1728del condition, showing that Abba functions to counteract the deleterious effects of the MhcK1728del. This could be explained by a proposed E3-ligase function of Abba that is able to clear the accumulation of malfunctioning MyHC. Thus, an intriguing question is whether the observed variation in clinical and pathologic phenotypes of Lys1729del mutation may in part be due to the genetic background related to the existence of E3-ligase modifier genes that may reduce or enhance the impact of the mutation. This will be important to explore in future studies.
In conclusion, we have generated a Laing distal myopathy model in Drosophila, and show that the phenotypes associated with the heterozygous MhcK1728del condition are suppressed by overexpression of the TRIM family protein Abba/Thin.
Supplementary Material
Acknowledgments
The study was supported by grants from the European Union’s Seventh Framework Programme for research, technological development, and demonstration under Grant 608473 (to H.T.), the Swedish Research Council (H.T. and R.H.P.), the Swedish Cancer Foundation (A.E.U. and R.H.P.), and Fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III (ISCIII PI14/00738) and FEDER funds “a way to achieve Europe” (M.O.). The funders had no role in the design of the study and collection, analysis, decision to publish, interpretation of data, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800727115/-/DCSupplemental.
References
- 1.Schiaffino S, Reggiani C. Myosin isoforms in mammalian skeletal muscle. J Appl Physiol (1985) 1994;77:493–501. doi: 10.1152/jappl.1994.77.2.493. [DOI] [PubMed] [Google Scholar]
- 2.Smerdu V, Karsch-Mizrachi I, Campione M, Leinwand L, Schiaffino S. Type IIx myosin heavy chain transcripts are expressed in type IIb fibers of human skeletal muscle. Am J Physiol. 1994;267:C1723–C1728. doi: 10.1152/ajpcell.1994.267.6.C1723. [DOI] [PubMed] [Google Scholar]
- 3.Geisterfer-Lowrance AA, et al. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 4.Walsh R, Rutland C, Thomas R, Loughna S. Cardiomyopathy: A systematic review of disease-causing mutations in myosin heavy chain 7 and their phenotypic manifestations. Cardiology. 2010;115:49–60. doi: 10.1159/000252808. [DOI] [PubMed] [Google Scholar]
- 5.Tajsharghi H, Oldfors A. Myosinopathies: Pathology and mechanisms. Acta Neuropathol. 2013;125:3–18. doi: 10.1007/s00401-012-1024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamont PJ, et al. Novel mutations widen the phenotypic spectrum of slow skeletal/β-cardiac myosin (MYH7) distal myopathy. Hum Mutat. 2014;35:868–879. doi: 10.1002/humu.22553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meredith C, et al. Mutations in the slow skeletal muscle fiber myosin heavy chain gene (MYH7) cause laing early-onset distal myopathy (MPD1) Am J Hum Genet. 2004;75:703–708. doi: 10.1086/424760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Udd B. 165th ENMC International Workshop: Distal myopathies 6-8th February 2009 Naarden, The Netherlands. Neuromuscul Disord. 2009;19:429–438. doi: 10.1016/j.nmd.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Homayoun H, et al. Novel mutation in MYH7 gene associated with distal myopathy and cardiomyopathy. Neuromuscul Disord. 2011;21:219–222. doi: 10.1016/j.nmd.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Overeem S, et al. Symptomatic distal myopathy with cardiomyopathy due to a MYH7 mutation. Neuromuscul Disord. 2007;17:490–493. doi: 10.1016/j.nmd.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Darin N, Tajsharghi H, Ostman-Smith I, Gilljam T, Oldfors A. New skeletal myopathy and cardiomyopathy associated with a missense mutation in MYH7. Neurology. 2007;68:2041–2042. doi: 10.1212/01.wnl.0000264430.55233.72. [DOI] [PubMed] [Google Scholar]
- 12.Muelas N, et al. MYH7 gene tail mutation causing myopathic profiles beyond Laing distal myopathy. Neurology. 2010;75:732–741. doi: 10.1212/WNL.0b013e3181eee4d5. [DOI] [PubMed] [Google Scholar]
- 13.Buvoli M, Buvoli A, Leinwand LA. Effects of pathogenic proline mutations on myosin assembly. J Mol Biol. 2012;415:807–818. doi: 10.1016/j.jmb.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahl-Halvarsson M, Pokrzywa M, Rauthan M, Pilon M, Tajsharghi H. Myosin storage myopathy in C. elegans and human cultured muscle cells. PLoS One. 2017;12:e0170613. doi: 10.1371/journal.pone.0170613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedera P, Petty EM, Bui MR, Blaivas M, Fink JK. The second kindred with autosomal dominant distal myopathy linked to chromosome 14q: Genetic and clinical analysis. Arch Neurol. 2003;60:1321–1325. doi: 10.1001/archneur.60.9.1321. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein SI, Mogami K, Donady JJ, Emerson CP., Jr Drosophila muscle myosin heavy chain encoded by a single gene in a cluster of muscle mutations. Nature. 1983;302:393–397. doi: 10.1038/302393a0. [DOI] [PubMed] [Google Scholar]
- 17.Domsch K, Ezzeddine N, Nguyen HT. Abba is an essential TRIM/RBCC protein to maintain the integrity of sarcomeric cytoarchitecture. J Cell Sci. 2013;126:3314–3323. doi: 10.1242/jcs.122366. [DOI] [PubMed] [Google Scholar]
- 18.LaBeau-DiMenna EM, et al. Thin, a Trim32 ortholog, is essential for myofibril stability and is required for the integrity of the costamere in Drosophila. Proc Natl Acad Sci USA. 2012;109:17983–17988. doi: 10.1073/pnas.1208408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collier VL, Kronert WA, O’Donnell PT, Edwards KA, Bernstein SI. Alternative myosin hinge regions are utilized in a tissue-specific fashion that correlates with muscle contraction speed. Genes Dev. 1990;4:885–895. doi: 10.1101/gad.4.6.885. [DOI] [PubMed] [Google Scholar]
- 20.Nichols CD, Becnel J, Pandey UB. Methods to assay Drosophila behavior. J Vis Exp. 2012:3795. doi: 10.3791/3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Estes PS, et al. Wild-type and A315T mutant TDP-43 exert differential neurotoxicity in a Drosophila model of ALS. Hum Mol Genet. 2011;20:2308–2321. doi: 10.1093/hmg/ddr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swank DM, et al. The myosin converter domain modulates muscle performance. Nat Cell Biol. 2002;4:312–316. doi: 10.1038/ncb776. [DOI] [PubMed] [Google Scholar]
- 23.Novak SM, Joardar A, Gregorio CC, Zarnescu DC. Regulation of heart rate in Drosophila via fragile X mental retardation protein. PLoS One. 2015;10:e0142836. doi: 10.1371/journal.pone.0142836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burkart C, et al. Modular proteins from the Drosophila sallimus (sls) gene and their expression in muscles with different extensibility. J Mol Biol. 2007;367:953–969. doi: 10.1016/j.jmb.2007.01.059. [DOI] [PubMed] [Google Scholar]
- 25.O’Donnell PT, Collier VL, Mogami K, Bernstein SI. Ultrastructural and molecular analyses of homozygous-viable Drosophila melanogaster muscle mutants indicate there is a complex pattern of myosin heavy-chain isoform distribution. Genes Dev. 1989;3:1233–1246. doi: 10.1101/gad.3.8.1233. [DOI] [PubMed] [Google Scholar]
- 26.Nongthomba U, Cummins M, Clark S, Vigoreaux JO, Sparrow JC. Suppression of muscle hypercontraction by mutations in the myosin heavy chain gene of Drosophila melanogaster. Genetics. 2003;164:209–222. doi: 10.1093/genetics/164.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katzemich A, et al. The function of the M-line protein obscurin in controlling the symmetry of the sarcomere in the flight muscle of Drosophila. J Cell Sci. 2012;125:3367–3379. doi: 10.1242/jcs.097345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olivé M, et al. New cardiac and skeletal protein aggregate myopathy associated with combined MuRF1 and MuRF3 mutations. Hum Mol Genet. 2015;24:3638–3650. doi: 10.1093/hmg/ddv108. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, et al. Expression of the inclusion body myopathy 3 mutation in Drosophila depresses myosin function and stability and recapitulates muscle inclusions and weakness. Mol Biol Cell. 2012;23:2057–2065. doi: 10.1091/mbc.E12-02-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viswanathan MC, et al. Myosin storage myopathy mutations yield defective myosin filament assembly in vitro and disrupted myofibrillar structure and function in vivo. Hum Mol Genet. 2017;26:4799–4813. doi: 10.1093/hmg/ddx359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suggs JA, et al. A Drosophila model of dominant inclusion body myopathy type 3 shows diminished myosin kinetics that reduce muscle power and yield myofibrillar defects. Dis Model Mech. 2017;10:761–771. doi: 10.1242/dmm.028050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cammarato A, et al. A mighty small heart: The cardiac proteome of adult Drosophila melanogaster. PLoS One. 2011;6:e18497. doi: 10.1371/journal.pone.0018497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tajsharghi H, Oldfors A, Macleod DP, Swash M. Homozygous mutation in MYH7 in myosin storage myopathy and cardiomyopathy. Neurology. 2007;68:962. doi: 10.1212/01.wnl.0000257131.13438.2c. [DOI] [PubMed] [Google Scholar]
- 34.Schoser BG, et al. Commonality of TRIM32 mutation in causing sarcotubular myopathy and LGMD2H. Ann Neurol. 2005;57:591–595. doi: 10.1002/ana.20441. [DOI] [PubMed] [Google Scholar]
- 35.Freemont PS. RING for destruction? Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 36.Glass D, Roubenoff R. Recent advances in the biology and therapy of muscle wasting. Ann N Y Acad Sci. 2010;1211:25–36. doi: 10.1111/j.1749-6632.2010.05809.x. [DOI] [PubMed] [Google Scholar]
- 37.Fielitz J, et al. Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J Clin Invest. 2007;117:2486–2495. doi: 10.1172/JCI32827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 39.Lecker SH, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.