Significance
This work identifies tRNAiMet precursors as a direct target of tumor suppressor miR-34a, and indicates that targeted suppression of tRNAiMet levels attenuates cell proliferation while inducing cell cycle arrest and apoptosis. The tRNAiMet may act as an oncogene and contribute significantly to tumorigenesis. Our findings provide conceptual and mechanistic insights into biology and cancer epigenetics and might have significant therapeutic implications.
Keywords: miR-34a, tRNAiMet, proliferation, cell cycle, apoptosis
Abstract
It remains unknown whether microRNA (miRNA/miR) can target transfer RNA (tRNA) molecules. Here we provide evidence that miR-34a physically interacts with and functionally targets tRNAiMet precursors in both in vitro pulldown and Argonaute 2 (AGO2) cleavage assays. We find that miR-34a suppresses breast carcinogenesis, at least in part by lowering the levels of tRNAiMet through AGO2-mediated repression, consequently inhibiting the proliferation of breast cancer cells and inducing cell cycle arrest and apoptosis. Moreover, miR-34a expression is negatively correlated with tRNAiMet levels in cancer cell lines. Furthermore, we find that tRNAiMet knockdown also reduces cell proliferation while inducing cell cycle arrest and apoptosis. Conversely, ectopic expression of tRNAiMet promotes cell proliferation, inhibits apoptosis, and accelerates the S/G2 transition. Moreover, the enforced expression of modified tRNAiMet completely restores the phenotypic changes induced by miR-34a. Our results demonstrate that miR-34a directly targets tRNAiMet precursors via AGO2-mediated cleavage, and that tRNAiMet functions as an oncogene, potentially representing a target molecule for therapeutic intervention.
In humans, the most well-defined tumor suppressor micro RNAs (miRNAs) are members of the miR-34 family, including miR-34a, miR-34b, and miR-34c. These miRNAs are encoded by two different genes; miR-34a is encoded by its own transcript, whereas miR-34b and miR-34c share a common primary transcript (1). The loci harboring these two genes are located in regions associated with fragile sites of the genome that have been shown to be frequently altered in cancer (2). Interestingly, recent findings suggest that members of the miR-34 family are direct transcriptional targets of the tumor suppressor p53 (3–7), which is essential for miR-34 expression induced by DNA damage and oncogenic stress. Therefore, miR-34a may be a key player in the p53 network, mediating the biological function of p53 by regulating the expression of various genes. The inactivation of miR-34a attenuates p53-mediated apoptosis in response to genotoxic stress (6), whereas the ectopic expression of miR-34a causes a dramatic reprogramming of gene expression and induces cell cycle arrest, apoptosis, and senescence (3–7).
The down-regulation or loss of miR-34a expression has been linked to the development of numerous types of cancer, including glioblastomas and malignant peripheral nerve sheath tumors as well as breast, colon, ovarian, pancreatic, and prostate cancers (3, 8–13). Both genetic and epigenetic mechanisms contribute to the reduction of miR-34a expression in malignant cells, including the inactivation of p53 and CpG methylation of the miR-34a promoter (11, 14). miR-34a has been shown to directly target the 3′ UTRs of numerous oncogene mRNAs, including Bcl-2, SIRT1, Fra-1, E2F, c-Met, Notch1, Notch2, CDK4/6, VEGF, ARAF, PIK3R2, cyclin D3, cyclin E2, and PLK1 (4, 7, 9, 10, 13, 15, 16), which may contribute to its tumor-suppressive role. However, whether miR-34a or other miRNAs can target tRNA molecules remains unknown.
Results
miR-34a Functionally Targets tRNAiMet.
We recently demonstrated that ionizing radiation induces miR-34a expression in human mammary epithelial cells (HMECs) (17). At the same time, miR-34a has been reported to be down-regulated in breast cancer samples (9). Since overexpression of tRNAs has been demonstrated in breast cancer (18), we analyzed the sequences of miR-34a and mature tRNA molecules and realized that tRNAiMet could be a potential target of miR-34a. Sequence analysis revealed that the mature miR-34a and tRNAiMet sequences in humans, rats, and mice share 100% identity, and a potential binding motif for miR-34a and tRNAiMet has been identified (Fig. 1 A and B). The predicted minimum free energy of the interaction between miR-34a and tRNAiMet was −15.9 kcal/mol (Fig. 1B), whereas the minimum free energy of disruption of the tRNAiMet acceptor arm was −15.2 kcal/mol.
Fig. 1.
miR-34a targets tRNAiMet. (A) Sequence of the mature miR-34a (Upper) and a proposed target sequence in tRNAiMet (Lower) in humans, rats, and mice. (B) Location of the binding motif in mature tRNAiMet. The indicated energy was calculated with RNAhybrid; the red arrow indicates the predicated AGO2 cleavage site. (C) 15 μg of DNA-free total RNA isolated from HCC1806 cells was subjected to an in vitro pulldown assay with the indicated concentration of biotin-labeled hsa-miR-34a or negative control probes. The enrichment of tRNAiMet was determined by qRT-PCR with total tRNAiMet primers and normalized to total input. (D) 15 μg of DNA-free total RNA isolated from HCC1806 cells was subjected to an in vitro pulldown assay with 1 μM biotin-labeled hsa-miR-34a or negative control probes. The enrichment of immature tRNAiMet was determined by qRT-PCR with immature tRNAiMet primers and normalized to total input. (E) At 10 d after Dox treatment, the Dox-inducible hsa-miR-34a-expressing HCC1806 cells were cross-linked and immunoprecipitated with ChIP-grade anti-Ago2, and the levels of miR-34a and immature tRNAiMet were detected by qRT-PCR. (F) 1 μM or 2 μM hsa-miR-34a or negative control siRNA was preincubated with 100 ng or 200 ng of human wild-type AGO2 or mutant AGO2 at 37 °C for 30 min, then incubated with 2.5 μM heat-denatured tRNAiMet (sequences, modification sites, and mutations showed at the Lower) at 37 °C for 2 h. Cleavage products were resolved by 15% denaturing polyacrylamide gel electrophoresis. (G) The cloverleaf structure of tRNAiMet shows endogenously modified bases, artificially modified bases in synthetic modified tRNAiMet used in this study (purple), and scrambled ones (arrow-denoted red bases) in the scrambled control. (H) Whole-cell lysates prepared from NIH 3T3 AGO2+/+ and AGO2−/− cells were subjected to SDS/PAGE and Western blot analysis using antibody against mouse AGO2 (mAGO2). Total RNA isolated from NIH 3T3 AGO2+/+ and AGO2−/− cells was subjected to qRT-PCR using primers for mm-miR-34a and mouse immature tRNAiMet. (I) Whole-cell lysates prepared from NIH 3T3 AGO2−/− cells stably expressing either mAGO2 or GFP were subjected to SDS/PAGE and Western blot analysis using antibody against mAGO2; total RNA isolated from NIH 3T3 AGO2−/− cells stably expressing either mAGO2 or GFP was subjected to qRT-PCR using primers for mm-miR-34a and mouse immature tRNAiMet. Dox(−) served as the control. D, dihydrouridine; m1A, 1-methyladenosine; m1G, 1-methylguanosine; m2G, N2-methylguanpsine; m5C, 5-methylcytidine; m7G, 7-methylguanosine; mActin, mouse actin; mGAPDH, mouse GAPDH; MT-AGO, mutant AGO2 (D597N); NC siRNA, negative control siRNA; t6A, N6-threonylcarbamoyladenosine; tRF, tRNAiMet fragment; WT-AGO2, wild-type AGO2. *P < 0.05; **P < 0.03.
We then studied the interaction of miR-34a with tRNAiMet molecules and precursors in cell-free extracts using various pulldown assays. Biotinylated Homo sapiens miR-34a (hsa-miR-34a) was used as a probe to pull down its binding partners in vitro, and quantitative real-time RT-PCR (qRT-PCR) was used to quantify the tRNAiMet species captured by the miR-34a probe. The amount of captured tRNAiMet species was increased in a dose-dependent fashion. Up to 2.5% of the total cellular tRNAiMet could be captured from the cell-free extracts, while a nonspecific control probe showed no dose-dependent capture of tRNAiMet (Fig. 1C). In principle, the primers used here would not distinguish the mature and immature tRNAiMet molecules, precursor tRNAiMet (pretRNAiMet) and primary transcript of tRNAiMet (pri-tRNAiMet); however, in vitro experiments with pure species showed that unmodified tRNAiMet was amplified 30-fold more efficiently than the mature modified species (SI Appendix, Fig. S1).
To see whether immature tRNAiMet species could actually be captured from whole-cell lysates, we designed primers to amplify selectively any immature tRNAiMet species captured by the hsa-miR-34a pulldown. Analysis showed that immature tRNAiMet molecules (pretRNAiMet and/or pri-tRNAiMet) were also enriched by the wild-type miR-34a probe (Fig. 1D), although the efficiency of the pulldown of the longer species was only approximately one-quarter of that seen with shorter amplicons. Taken together, these findings appear to show that miR-34a can target precursors of tRNAiMet, but whether mature species are also targeted is not clear.
miRNAs regulate gene expression through translational inhibition or transcript degradation via Argonaute 2 (AGO2)-catalyzed cleavage (19). To determine if AGO2 is capable of miR-34a–directed tRNAiMet cleavage, we stably transfected HCC1806 cells with a doxycycline (Dox)-inducible hsa-miR-34a plasmid (SI Appendix, Fig. S2). AGO2 was then precipitated from whole-cell lysates by a ChIP-grade antibody. Real-time RIP-PCR indicated that hsa-miR-34a was significantly enriched by anti-AGO2 antibodies (2.6-fold; P = 0.043) (Fig. 1E, Upper), as expected. Immature tRNAiMet was also significantly enriched (3.5-fold; P = 0.015) (Fig. 1E, Lower). To test the AGO2-mediated tRNAiMet cleavage hypothesis directly, we performed an in vitro cleavage assay (Fig. 1F) by incubating human AGO2 protein with miR-34a and either synthetic unmodified tRNAiMet or a synthetic modified tRNAiMet that contained three of the posttranscriptional modifications present in mature tRNAiMet or a synthetic scrambled version of the tRNAiMet sequence (Fig. 1G). This assay showed that wild-type AGO2 could catalyze miR-34a–mediated cleavage of unmodified tRNAiMet, but not the modified tRNAiMet or the scrambled tRNAiMet (Fig. 1F, Left). A control siRNA and mutant AGO2 (D597N) produced no cleavage of any tRNA species (Fig. 1F, Middle and Right).
The proposed model of interaction between miR-34a and tRNAiMet (Fig. 1B) suggests that two tRNAiMet fragments (approximately 18 nt and 16 nt; lane 4; Fig. 1F, Left) cleaved by AGO2 may have been derived from the miR-34a–targeted half of the amino acid arm and half of the T stem/loop (16 nt) and half of the T stem/loop and half of the anticodon arm (18 nt), and a 31-nt fragment from half of the anticodon arm and D stem/loop and half of the amino acid arm, although other possibilities are indicated in Fig. 1B based on the types of AGO2 cleavages seen in other systems (20). To test our hypothesis in vivo, this model was then validated by RNA-Seq analysis using enforced miR-34a expression HCC1806 cell line. We also note that the unmodified tRNAiMet sample that we used displays two bands in the gel (lanes 2–4, Fig. 1F, Left), and only the top, more intense one appeared to be cleaved by AGO2 (lane 4, Fig. 1F, Left). Mass spectrometry of the sample (Dharmacon), suggested that the top, more intense band is the bona fide unmodified tRNAiMet, while the lower, less intense band is a degradation product (SI Appendix, Fig. S3).
Interestingly, miR-34a did not target modified tRNAiMet (Fig. 1F, Left). AGO2 cleavage of the miR-34a/tRNAiMet complex in vitro occurs efficiently with unmodified tRNAiMet, presumably due to the lack of modified ribonucleotides. This suggests that in vivo, miR-34a modulates the pool of mature functional tRNAiMet primarily by depleting its precursors. Additional controls showed that miR-34a alone could not directly cleave tRNAiMet in either RNA-RNA binding buffer or DNAzyme cleavage buffer (21, 22); such cleavage occurred only when AGO2 was present in the AGO2-specific buffer (SI Appendix, Fig. S4).
To further confirm the key role of AGO2 in miR-34a–guided tRNAiMet degradation, we then used mouse embryonic fibroblast NIH 3T3 AGO2+/+ and AGO2−/− lines as a model system to explore the effect of AGO2 on the expression of immature tRNAiMet. We found that a marked reduction of AGO2 led to a modest down-regulation of mm-miR-34a and profound overexpression of immature tRNAiMet (Fig. 1H). In contrast, restored expression of AGO2 in NIH 3T3 AGO2−/− cells resulted in a modest up-regulation of mm-miR-34a and a remarkable reduction of the immature tRNAiMet (Fig. 1I). Taken together, these results suggest that miR-34a can bind to tRNAiMet both in vitro and in vivo and functionally direct tRNAiMet degradation, at least in part, through AGO2-catalyzed cleavage of unmodified tRNAiMet molecules.
miR-34a Expression Levels Are Inversely Correlated with tRNAiMet in Ionizing Radiation-Exposed HMECs and Breast Cancer Cells.
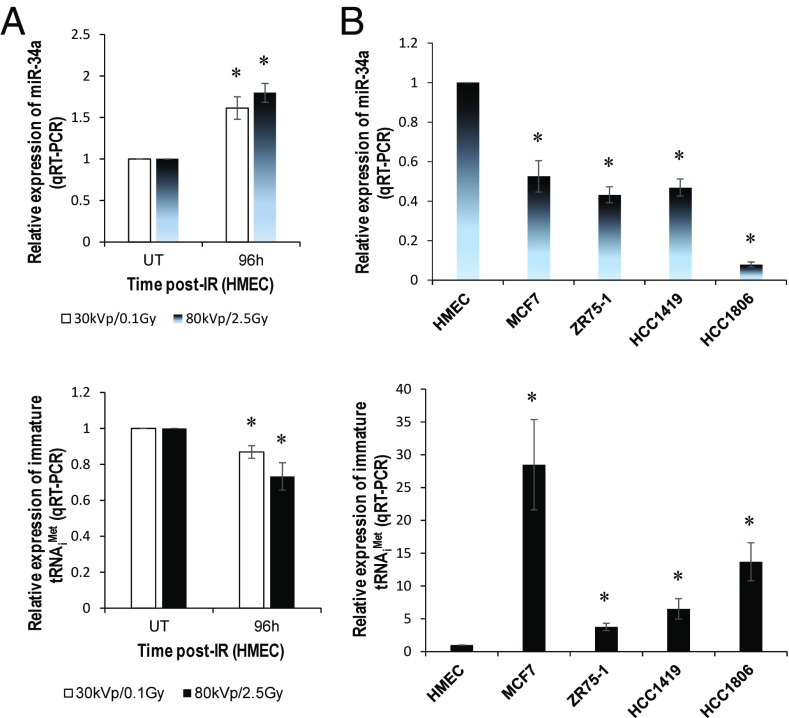
Ionizing radiation (IR)-induced miR-34a expression was inversely correlated with the level of immature tRNAiMet (Fig. 2A). miR-34a was down-regulated in all of the breast cancer cell lines examined (Fig. 2B, Upper), consistent with recent reports (9, 13). Interestingly, the levels of immature tRNAiMet were elevated in these breast cancer cell lines (Fig. 2B, Lower) and were generally inversely correlated with the levels of miR-34a (Fig. 2B, Upper), suggesting that miR-34a may be a significant regulator of tRNAiMet level. To establish the ratio of endogenous miR-34a to tRNAiMet molecules, we generated standard curves with two variants of Ct value and molecule number (SI Appendix, Fig. S5 A and B), and then calculated and analyzed the numbers of both molecules in IR-exposed HMECs and breast cancer cell lines (SI Appendix, Fig. S6 A and B). The ratio of miR-34a to total tRNAiMet molecules ranged from 1:17 to 1:19 in IR-exposed HMECs, but from 1:7.3 × 103 to 1:1.7 × 105 in breast cancer cell lines. We found that total tRNAiMet was also down-regulated in the IR-exposed HMECs and overexpressed in all breast cancer cell lines examined (SI Appendix, Fig. S7 A and B).
Fig. 2.
miR-34a is inversely correlated with immature tRNAiMet. (A) Total RNA samples isolated from HMECs exposed to the indicated doses of ionizing radiation (IR) were subjected to qRT-PCR using miR-34a (Upper) and immature tRNAiMet (Lower) primers. (B) Total RNA isolated from the indicated normal and breast cancer cell lines was subjected to qRT-PCR using miR-34a (Upper) and immature tRNAiMet (Lower) primers. *P < 0.05.
miR-34a Suppresses Cell Proliferation and Induces Cell Cycle Arrest and Apoptosis.
miR-34a has been suggested to play an important role in cancer cell proliferation, apoptosis, migration, and invasion (3, 8–13). Thus, we further examined the significance of miR-34a in breast carcinogenesis using the Dox-inducible miR-34a–expressing HCC1806 cell line as a model system. We found that miR-34a induced by Dox significantly suppressed breast cancer cell proliferation (Fig. 3 A and B). Ectopic miR-34a caused a profound reduction in immature tRNAiMet (SI Appendix, Fig. S8A) and substantially induced S-phase cell cycle arrest and apoptosis in the HCC1806 cells (Fig. 3 C and D). We also noted that the copy number of miR-34a is higher in HCC1806 cells expressing miR-34a [1.5-fold higher for Dox(−) and twofold higher for Dox(+)] compared with the parental cells (SI Appendix, Fig. S8B). Furthermore, application of 800 ng/mL doxycycline (the same dose used for inducible miR-34a expression in HCC1806 cells expressing miR-34a) had no effect on HCC1806 cell proliferation (SI Appendix, Fig. S8C), although a transient reduction in the expression of total tRNAiMet was found at 24 h after treatment (SI Appendix, Fig. S8D).
Fig. 3.
Ectopic miR-34a suppresses breast cancer cell proliferation while inducing apoptosis and cell cycle arrest at S phase. (A) At 10 d after Dox treatment, HCC1806 cells expressing miR-34a were replated to determine the effect of ectopic miR-34a expression on cell proliferation. (B) At 10 d after Dox exposure, the levels of miR-34a were detected by qRT-PCR. (C and D) The effects of ectopic miR-34a on apoptosis (C) and the cell cycle (D) were determined at 10 d after Dox exposure; Dox(−) served as the control. *P < 0.05.
To further validate our findings, we evaluated the effect of ectopic miR-34a expression on apoptosis and cell cycle in another breast cancer cell line, MCF7. Flow cytometry analyses showed a modest but significant induction of apoptosis and G2 arrest in MCF7 cells transfected with miR-34a (SI Appendix, Fig. S9 A–C).
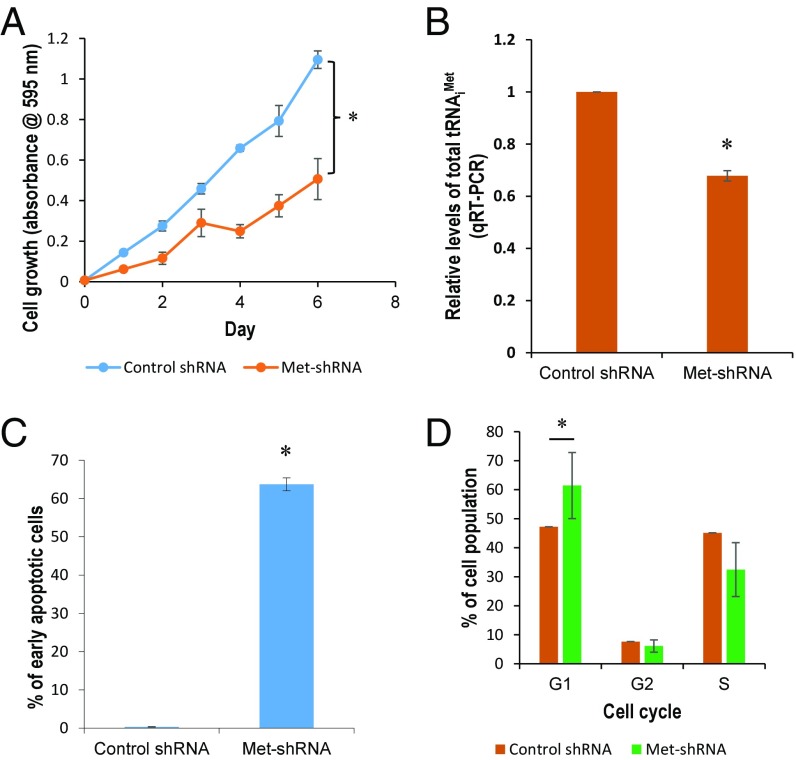
Knockdown of tRNAiMet Attenuates Cell Proliferation and Induces Apoptosis and G1-Phase Arrest.
We next generated a stable tRNAiMet-knockdown HCC1806 cell line to explore the role of tRNAiMet in breast cancer and to establish whether the biological effects of miR-34a are mediated, at least in part, by the targeting of tRNAiMet. As expected, knockdown of tRNAiMet attenuated cell proliferation (Fig. 4 A and B) and induced apoptosis and G1-phase cell cycle arrest (Fig. 4 C and D). Apoptotic HCC1806 cells stably expressing tRNAiMet small hairpin RNA (shRNA) carried more copies of tRNAiMet shRNA than nonapoptotic ones (SI Appendix, Fig. S10A). Interestingly, aneuploid cells were identified in the tRNAiMet-knockdown cell line by both fluorescent microscopy (SI Appendix, Fig. S10B) and flow cytometry. To exclude potential off-target effects, we then transiently transfected HCC1806 cells with either wild-type tRNAiMet dsiRNA (Dicer substrate short-interfering RNA) or scrambled tRNAiMet dsiRNA. Untransfected cells served as a control. Analysis by qRT-PCR showed that wild-type tRNAiMet dsiRNA significantly reduced the levels of total tRNAiMet at the indicated time points, while the scrambled tRNAiMet dsiRNA had no significant effect (SI Appendix, Fig. S10C). The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay indicated a significant reduction in cell proliferation induced by wild-type tRNAiMet dsiRNA, while this reduction was completely abolished by scrambled tRNAiMet dsiRNA (SI Appendix, Fig. S10D). These results indicate the specificity of wild-type tRNAiMet dsiRNA function.
Fig. 4.
tRNAiMet knockdown inhibits breast cancer cell proliferation while inducing apoptosis and G1 cell cycle arrest. (A) The effect of tRNAiMet knockdown on cell proliferation was evaluated in HCC1806 cells expressing tRNAiMet-shRNA. (B) The levels of total tRNAiMet were measured in HCC1806 cells expressing tRNAiMet-shRNA by qRT-PCR. (C and D) The effect of tRNAiMet knockdown on apoptosis (C) and cell cycle (D) was determined in HCC1806 cells expressing tRNAiMet-shRNA. *P < 0.05.
We stably transfected MCF7 cells with the same shRNA vectors and found similar reductions in cell proliferation (SI Appendix, Fig. S11 A and B), induction of G1 arrest (SI Appendix, Fig. S11C), and induction of apoptosis (SI Appendix, Fig. S11D).
Ectopic tRNAiMet Expression Restores the Phenotypic Changes Induced by miR-34a.
To dissect the cellular roles of tRNAiMet and further analyze its oncogenic potential, we generated a stable tRNAiMet -overexpressing HCC1806 cell line. qRT-PCR revealed a higher copy number of tRNAiMet in HCC1806 cells expressing tRNAiMet (SI Appendix, Fig. S12). The expression of ectopic tRNAiMet enhanced cell growth, inhibited apoptosis, and accelerated the S/G2 transition (SI Appendix, Fig. S13). To validate in vivo that miR-34a–induced phenotypic alterations are mediated, at least in part by targeting tRNAiMet, we then performed rescue experiments by transient transfection of HCC1806 cells with either miR-34 alone or in combination with the modified tRNAiMet. As expected, miR-34a–induced suppression of proliferation and induction of apoptosis (Fig. 5 A and C) were completely restored by transfection with modified tRNAiMet (Fig. 5 D and F).
Fig. 5.
Enforced expression of tRNAiMet restores the phenotypic changes induced by miR-34a. (A) At 24 h after transfection, HCC1806 cells transiently transfected with either 30 nM miR-34a mimic or negative control siRNA were replated in 96-well plates for the MTT assay. The experiments were done in triplicate. (B) At 72 h after transfection, miR-34a levels were determined by qRT-PCR. (C) At 24 h and 96 h after transfection, the cells were harvested, and apoptosis was analyzed. The experiments were done in duplicate. (D) At 24 h after transfection, the HCC1806 cells transiently transfected with either 30 nM miR-34a mimic or in combination with modified tRNAiMet (tRNAiMet rescue) were replated in 96-well plates for the MTT assay. The experiments were done in triplicate. (E) At 72 h after transfection, the levels of total tRNAiMet were measured by qRT-PCR. (F) At 72 h after transfection, the cells were harvested, and apoptosis was analyzed. The experiments were done in duplicate. *P < 0.05.
Discussion
Although thousands of miRNA targets have been discovered to date, no evidence that miRNAs can target tRNA molecules has been reported. This study reveals that the tumor suppressor miR-34a can directly target unmodified tRNAiMet, leading to tRNAiMet degradation via AGO2-mediated cleavage, and suggests a conceptually and mechanistically novel process in miRNA-mediated posttranscriptional regulation. We demonstrate a physical and functional interaction between these two molecules in in vitro pulldown and AGO2 cleavage assays. We also provide evidence that tRNAiMet may act as an oncogene.
As a key player in miRNA-mediated mRNA degradation, AGO2 may also play a pivotal role in miR-34a–directed tRNAiMet cleavage, since miR-34a failed to cleave unmodified tRNAiMet in the presence of mutant AGO2 and occurred only in the presence of wild-type AGO2. Although we show that miR-34a selectively targets unmodified tRNAiMet, most likely in the nucleus, we cannot exclude the possibility that miR-34a could also somehow target fully mature tRNAiMet in the cytoplasm, since the modified tRNAiMet used in our experiments was not fully modified due to technical restrictions. Even though we have proposed a model to display the possible cleavage sites based on the data from in vitro AGO2 cleavage assay and RNA-Seq analysis, it may not fully represent the in vivo situation. Although the role of AGO2 in miR-34a–induced tRNAiMet degradation has not been reported previously, AGO2 has been shown to bind selectively to tRNAMet in human HEK293 cells (23). Sequence analysis predicts that miR-34a may also target other tRNAs in addition to tRNAiMet, such as tRNALeu(CAG), tRNAGlu(TTC), and tRNAArg(CCT). The existence and potential biological repercussions of these interactions require future study.
Our findings are strengthened because the sequences of mature tRNAiMet are mostly identical among humans, rats, and mice, including the 3′ terminal sequence that we believe to be involved in binding to miR-34a (SI Appendix, Table S1). The canonical mechanism of miRNA-mediated mRNA cleavage requires perfect or nearly perfect complementarity, which is not the case for miR-34a:tRNAiMet pairs. However, the data we present here demonstrate that the binding motif between miR-34a and tRNAiMet appears to be more stable than the tRNAiMet arm. Furthermore, there is strong evidence that in the case of miRNA-directed mRNA degradation in mammals, the complementarity between miRNA and mRNA is largely imperfect (24, 25), although the mechanisms involved are unclear and require further investigation.
The data presented here highlight that miR-34a could directly target unmodified immature tRNAiMet (Fig. 1 D–F). We are not sure whether miR-34a can interact in vivo with mature tRNAiMet. One previously unsuspected role of tRNAiMet modifications may be to protect this tRNA against miR-34a–mediated AGO2 cleavage; since modified nucleotides m1G9 and m5C47 both locate at a structurally important junction of tRNAiMet, they may increase tRNA thermal stability, consequently preventing this tRNA from degradation (26). Although it is unclear why miR-34a does not target cleavage of modified tRNAiMet, we suggest that tRNAiMet modifications may inhibit binding of miR-34a and AGO2 to tRNA.
Even though our findings suggest a direct interaction between miR-34a and unmodified immature tRNAiMet, tRNA molecules are highly structured; their 3′ ends generally form a double-stranded structure with their 5′ ends in which it is termed the amino acyl arm. How the double-stranded structures are unwound to permit recognition by miR-34a remains unclear. In addition to Dicer, a core component of the RICS complex that mediates duplex unwinding (27), conformational alterations of RICS during assembly may also contribute to unwinding the double-stranded structure (28). Furthermore, a recent study has indicated an involvement of RNA helicase A in RNA duplex unwinding (29). Here we show that an interaction between miR34a and highly structured tRNAiMet may be mediated via AGO2. To date, few studies have taken into consideration secondary structures of 3′ UTRs that interact with miRNAs. Among those, it was recently shown that miR-26b targets highly structured EphA2 mRNA (30, 31), with this interaction mediated by a classical RISC complex. In the future, it would be interesting to conduct an in-depth analysis of all participating members of the RISC complex and any associated proteins.
For several years, deregulated protein synthesis has been associated with oncogenic transformation and tumorigenesis (32). Indeed, cancer cells typically exhibit an increased growth rate, which may be due, at least in part, to the global up-regulation of tRNA molecules (33). Although tRNA levels are elevated in human malignancies such as breast cancer (18), the underlying mechanism remains poorly understood. Here we provide evidence that miR-34a plays an important role in governing the levels of tRNAiMet, and that this interaction may affect levels of cellular proliferation and apoptosis. Taken together, our findings may serve as an important roadmap for future analyses of interactions of miRNAs and other noncoding RNAs, their roles in regulation of key biological processes, and their contributions to carcinogenesis.
Materials and Methods
All cell lines used in this study, including HMECs and MCF7, ZR75-1, HCC1419, and HCC1806 cells, were cultured in appropriate media at 37 °C in a humidified atmosphere containing 5% CO2. Levels of hsa-miR-34a and tRNAiMet were detected by qRT-PCR. Physical and functional interactions between miR-34 and tRNAiMet were analyzed using in vitro pulldown and AGO2 cleavage assays. Stable cell lines were generated using conventional methods in combination with cell sorting. Cell proliferation was determined with the MTT assay (34). Cell cycle and apoptosis were analyzed by flow cytometry. Detailed information is provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Leemor Joshua-Tor, Dr. Elad Elkayam, and Ms. Amanda R. Epstein (Watson School of Biological Sciences, Cold Spring Harbor Laboratory) for kindly providing the human AGO2 D597A mutant protein; Prof. Dónal O’Carroll (MRC Centre for Regenerative Medicine, University of Edinburgh), Prof. Phillip Zamore (Howard Hughes Medical Institute, University of Massachusetts Medical School), and Dr. Karina Zhuravleva (Howard Hughes Medical Institute, University of Massachusetts Medical School) for providing the MEF Ago2−/− and MEF Ago2−/− Ago2-overexpression cell lines; Dr. U. Wieden-Kothe for helping to generate the standard curves for calculating miR-34a and tRNAiMet molecular numbers; and Dr. Andrey Golubov for helpful discussions. This work was funded by the Canadian Breast Cancer Foundation, Canadian Institutes for Health Research, and Alberta Cancer Foundation/Alberta Innovates-Health Solutions.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703029115/-/DCSupplemental.
References
- 1.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 2.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumamoto K, et al. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008;68:3193–3203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 7.Tarasov V, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 8.Corney DC, et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, et al. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med. 2013;13:109–117. doi: 10.1007/s10238-012-0186-5. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian S, et al. Genome-wide transcriptome analyses reveal p53 inactivation-mediated loss of miR-34a expression in malignant peripheral nerve sheath tumours. J Pathol. 2010;220:58–70. doi: 10.1002/path.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang S, et al. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2013;32:4294–4303. doi: 10.1038/onc.2012.432. [DOI] [PubMed] [Google Scholar]
- 14.Vogt M, et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 15.Bernardo BC, et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci USA. 2012;109:17615–17620. doi: 10.1073/pnas.1206432109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lal A, et al. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011;7:e1002363. doi: 10.1371/journal.pgen.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang B, Li D, Kovalchuk O. p53 Ser15 phosphorylation and histone modifications contribute to IR-induced miR-34a transcription in mammary epithelial cells. Cell Cycle. 2013;12:2073–2083. doi: 10.4161/cc.25135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavon-Eternod M, et al. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 20.Xu K, Lin J, Zandi R, Roth JA, Ji L. MicroRNA-mediated target mRNA cleavage and 3′-uridylation in human cells. Sci Rep. 2016;6:30242. doi: 10.1038/srep30242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romero-López C, Berzal-Herranz A. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA. 2009;15:1740–1752. doi: 10.1261/rna.1680809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khachigian LM, et al. c-Jun regulates vascular smooth muscle cells growth and neointima formation after arterial injury. J Biol Chem. 2002;277:22985–22991. doi: 10.1074/jbc.M200977200. [DOI] [PubMed] [Google Scholar]
- 23.Maniataki E, Mourelatos Z. Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA. 2005;11:849–852. doi: 10.1261/rna.2210805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim LP, et al. Microarray analysis shows that some microRNAs down-regulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 26.Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- 27.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, function and role in cancer. Curr Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 30.Wu N, et al. Role of microRNA-26b in glioma development and its mediated regulation on EphA2. PLoS One. 2011;6:e16264. doi: 10.1371/journal.pone.0016264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winter J, et al. Comparative 3' UTR analysis allows identification of regulatory clusters that drive Eph/ephrin expression in cancer cell lines. PLoS One. 2008;3:e2780. doi: 10.1371/journal.pone.0002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berns A. A tRNA with oncogenic capacity. Cell. 2008;133:29–30. doi: 10.1016/j.cell.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 33.Tuller T. The effect of dysregulation of tRNA genes and translation efficiency mutations in cancer and neurodegeneration. Front Genet. 2012;3:201. doi: 10.3389/fgene.2012.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berridge MV, Herst PM, Tan AS. Tetrazolium dyes as tools in cell biology: New insights into their cellular reduction. Biotechnol Annu Rev. 2005;11:127–152. doi: 10.1016/S1387-2656(05)11004-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.