Significance
Virophages are parasites of giant viruses within protists. They reduce giant virus production and increase host cell survival. They provide a defense system for protists against giant viruses in diverse environments, likely with ecological relevance for protist populations. To understand the remarkable virophage life cycle, it is crucial to investigate how they assemble into infectious particles and which processes require interactions with giant virus and host. We examined the marine virophage mavirus to show that its major and minor capsid proteins assemble into virus-like particles in the absence of specific host or viral factors. Subsequently, the virophage-encoded protease processes the major capsid protein to prepare virions for infection.
Keywords: virus assembly, capsid maturation, viral protease, crystallography, electron microscopy
Abstract
Virophages have the unique property of parasitizing giant viruses within unicellular hosts. Little is understood about how they form infectious virions in this tripartite interplay. We provide mechanistic insights into assembly and maturation of mavirus, a marine virophage, by combining structural and stability studies on capsomers, virus-like particles (VLPs), and native virions. We found that the mavirus protease processes the double jelly-roll (DJR) major capsid protein (MCP) at multiple C-terminal sites and that these sites are conserved among virophages. Mavirus MCP assembled in Escherichia coli in the absence and presence of penton protein, forming VLPs with defined size and shape. While quantifying VLPs in E. coli lysates, we found that full-length rather than processed MCP is the competent state for capsid assembly. Full-length MCP was thermally more labile than truncated MCP, and crystal structures of both states indicate that full-length MCP has an expanded DJR core. Thus, we propose that the MCP C-terminal domain serves as a scaffolding domain by adding strain on MCP to confer assembly competence. Mavirus protease processed MCP more efficiently after capsid assembly, which provides a regulation mechanism for timing capsid maturation. By analogy to Sputnik and adenovirus, we propose that MCP processing renders mavirus particles infection competent by loosening interactions between genome and capsid shell and destabilizing pentons for genome release into host cells. The high structural similarity of mavirus and Sputnik capsid proteins together with conservation of protease and MCP processing suggest that assembly and maturation mechanisms described here are universal for virophages.
Virion morphogenesis is crucial for the viral life cycle. Dynamic stages of the virus, including host infection, replication, and virion formation, were emphasized after the discovery of giant viruses, which spurred discussions to define the intracellular virion factory as actual “viral self” (1, 2). The interplay between giant virus and host cell became even more complex when virophages were discovered as parasites of giant viruses (3). Giant viruses of the family Mimiviridae reproduce within cytoplasmic virion factories (reviewed in ref. 4) that are exploited by virophages of the family Lavidaviridae, likely for transcription (5). Research on virophages so far mainly focused on their isolation, cultivation, and genetic characterization, which led to the description of Sputnik (3, 6, 7), Zamilon (8), and mavirus (9). They infect freshwater and marine protozoa, but metagenomic data suggest that virophages also infect algae (10–12). Their ∼70-nm icosahedral capsids enclose a linear or circular dsDNA genome of 17–19 kbp, carrying 20–21 ORFs. It was further revealed that virophages can integrate into genomes of both giant viruses (6) and host cells (13, 14); the latter is proposed to be a defense strategy against giant viruses, potentially with ecological relevance for protist populations (3, 9, 13, 14).
Although dozens of virophage genomes are described, little is understood about how they form infectious particles and which processes rely on interactions with the giant virus and the host cell. Interestingly, the four genes conserved among virophages are all proposed to be involved in virion morphogenesis (5, 12). They code for a double jelly-roll (DJR) major capsid protein (MCP), a single jelly-roll (SJR) minor capsid protein called penton protein, a clan CE cysteine protease, and an FtsK/HerA-type ATPase (12, 15). Cryo-electron microscopy (EM) uncovered the structure of the mature Sputnik particle with triangulation number T = 27, 260 trimeric MCP capsomers, and 12 pentameric penton proteins (“pentons”) (16, 17). MCP capsomers with quasi-sixfold symmetry form the capsid’s facets, and pentons show pentagonal symmetry to build the vertices as observed in other DJR viruses of the PRD1-adenovirus lineage (16, 18). The JR fold consists of two sandwiched β-sheets, each composed of four antiparallel strands labeled B to I, generating sheets BIDG and CHEF (19). Notably, the Sputnik MCP was found to be 87 residues shorter at the C terminus than predicted from the gene sequence (16). It was proposed that MCP processing occurs during virophage assembly, but experimental evidence is lacking.
Although the structure of the Sputnik capsid revealed the arrangement of capsid proteins in the mature virion, little is known about how they assembled, whether assembly and processing involved giant virus or host factors, and how the conserved protease and ATPase are involved in morphogenesis. Here, we explored virophage capsid assembly and maturation processing, including effects on capsid stability and implications for host infection. Insights into these potentially conserved processes are crucial to understand the remarkable virophage life cycle and how it interacts with giant virus and host cell. We studied the marine virophage mavirus that infects the widespread nanoflagellate Cafeteria roenbergensis and exploits the Cafeteria roenbergensis virus (CroV) for replication (9). Its ability to integrate into host cell genomes and to act as a defense system against CroV has been proposed to be of ecological importance for zooplankton populations (14).
Results
Mavirus Protease Processes MCP at Multiple Sites in Vitro.
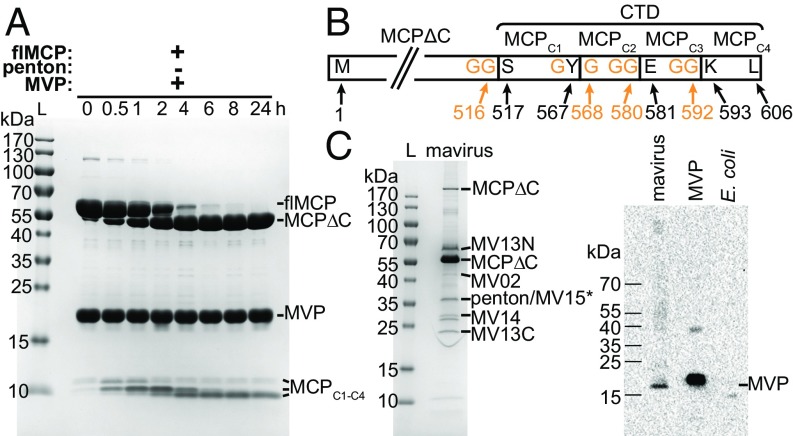
The virophage protease is suggested to be involved in capsid maturation, as observed for other eukaryotic viruses of the PRD1-adenovirus lineage (15, 20). To investigate its function in the virophage mavirus, we recombinantly expressed the mavirus cysteine protease (MVP), MCP, and penton protein. Purified MVP was present as a dimer, MCP as a trimeric capsomer, and penton protein as a pentamer (SI Appendix, Fig. S1 A–C). MVP cleaved recombinant MCP at multiple sites but not recombinant penton protein (Fig. 1A and SI Appendix, Fig. S1E). We used electrospray-ionization quadrupole time-of-flight mass spectrometry (MS) to identify MCP cleavage fragments from this in vitro turnover (SI Appendix, Fig. S1F and Table S1). The largest MCP fragment comprised residues (res.) 1–516 of full-length MCP (flMCP), which we termed MCPΔC (Fig. 1B). The C-terminal domain (CTD) of flMCP (res. 517–606) seems to be cleaved from flMCP first in this in vitro experiment and was further processed at three additional sites, generating MCPC1 (res. 517–567), MCPC2 (res. 568–580), MCPC3 (res. 581–592) and MCPC4 (res. 593–606). Three of four cleavage sites in mavirus MCP are preceded by a diglycine motif (GG | X, Fig. 1B), the other cleavage occurred within GY | G. A sequence alignment of diverse virophage MCPs revealed at least four GG | X or GX | G motifs in 10 of 12 MCPs in the C terminus (SI Appendix, Fig. S2), suggesting that these cleavage motifs are highly conserved. Since the mavirus MCP is cleaved at C-terminal sites that are preserved in diverse virophage MCPs, we propose that processing by the cysteine protease has a key function in the virophage life cycle.
Fig. 1.
Mavirus MCP is processed by MVP in vitro and in native mavirus particles. (A) SDS/PAGE of flMCP turnover by MVP in vitro, performed in 50 mM SPG-NaOH pH 6.0, 1 mM TCEP, 5% (vol/vol) glycerol at 4 °C using 30 µM of each protein. flMCP, full-length MCP; L, protein ladder; MCPΔC, truncated MCP. Three C-terminal MCP fragments (MCPC1–C4) are indicated, but allocation of intermediate cleavage fragments was not possible based on the available data. (B) Scheme of MCP in vitro processing sites and fragments. N and C terminus and cleavage site residues refer to wild-type flMCP. (C) Native mavirus virion proteins. (C, Left) SDS/PAGE of mavirus particles. Indicated proteins were confirmed by MS PMF (i.e., significant Mascot score with P < 0.05). *No significant Mascot score for MV15 but 12 peptide mass matches. MV13N and MV13C refer to N- and C-terminal MV13 fragments, respectively. (C, Right) Immunoblot of MVP in mavirus virions. MVP, recombinant, purified protein (positive control). E. coli, cell lysate (negative control).
MCP and MV13 Are Processed in Native Mavirus Virions.
We analyzed native mavirus particles by MS peptide-mass fingerprinting (PMF) on excised SDS/PAGE bands to investigate MCP processing. Indeed, native MCP was detected as truncated ∼55-kDa fragment, and the presence of MCPΔC was confirmed by MS PMF (Fig. 1C, Left). Notably, no MCP C-terminal fragments could be identified by MS PMF in native mavirus virions, potentially due to both the specific MS method used and the low concentration of available native mavirus virions, but we can also not exclude that MCP C-terminal fragments are absent in these virions. In addition, five further native mavirus gene products were identified: MV02 (integrase), two cleavage fragments of MV13 (predicted α/β hydrolase), MV14 (unknown function), MV15 (FtsK/HerA-type ATPase), and MV17 (penton protein) (9). The SDS/PAGE fragments at ∼20 kDa and ∼60 kDa correspond to the N- and C-terminal part, respectively, of the 80.6-kDa MV13. Interestingly, the MV13 sequence between MV13N and MV13C that was not covered by tryptic peptide masses (res. 476–499) contains a potential MVP cleavage site, GK | G. We also identified MVP in native mavirus virions by immunoblotting (Fig. 1C, Right), supporting the hypothesis that it is the relevant protease for processing not only MCP but also MV13 in mavirus virions.
MCP Processing Results in Jelly-Roll Core Compaction and Tower Expansion.
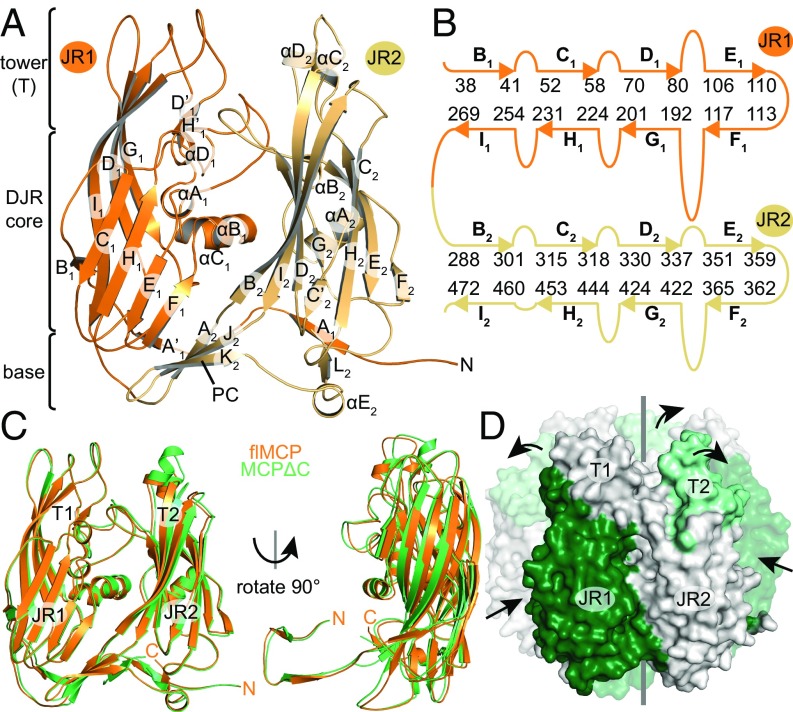
To elucidate the effect of processing on mavirus MCP regarding structure and stability, we recombinantly generated MCPΔC and compared its properties with flMCP. Both proteins form trimers in solution (SI Appendix, Fig. S1 A and D), but MCPΔC showed higher melting temperatures (Tm) than flMCP (SI Appendix, Fig. S3A), indicating higher thermal stability of MCPΔC. To answer whether processing alters the MCP conformation, we determined crystal structures of both MCP states (SI Appendix, Table S2). Although the flMCP electron density map at 2.5-Å resolution was unambiguous only for res. 2–504, the presence of the entire protein was confirmed by MS on washed and dissolved crystals. Thus, the C-terminal domain is likely flexible. The flMCP dataset contains a trimeric capsomer in the asymmetric unit that shows a central pore along the quasi-sixfold axis (SI Appendix, Fig. S3C). Protomers exhibit the DJR fold (Fig. 2 A and B and SI Appendix, Fig. S3B) and are highly symmetric with the highest root-mean-square deviation on Cα positions (Cα rmsd) of 0.135 Å. The capsomer includes a base and a tower, which would face the capsid interior and exterior, respectively, (Fig. 2A) by analogy to the Sputnik capsid structure (16). The base contains six three-stranded β-sheets, three of which are “pedestal connectors” (PCs) as described for the adenoviral hexon (21), which link JR1 and JR2 within each protomer, and the other are “basal interprotomer sheets” (BISs), which stabilize the trimer (SI Appendix, Fig. S3 C and D).
Fig. 2.
Crystal structures of mavirus flMCP and MCPΔC. (A) flMCP domains. JR1 and JR2 are orange and light orange, respectively. PC, pedestal connector. Secondary structure elements are labeled. (B) Scheme of flMCP JR strands and connecting loops. Starting and ending residues of JR strands are indicated. (C) The superposition of flMCP and MCPΔC structures was performed on trimeric capsomers but one protomer is illustrated here. (D) Core compaction and tower expansion after MCP processing. Side view of MCPΔC capsomer with the front protomer shown as solid surface and rear protomers transparently. DJR core compaction (dark green) and tower expansion (light green) are illustrated by arrows. Gray line, quasi-sixfold symmetry axis. Also see Movie S1.
Similar to flMCP, the crystal structure of MCPΔC at 1.5-Å resolution had a trimeric capsomer located in the asymmetric unit. Superposed flMCP and MCPΔC structures are highly similar (Cα rmsd = 1.21 Å), and both show intact DJR core, tower, and base (Fig. 2C). Both datasets allowed model building up to T504 close to the MVP cleavage site. In contrast to flMCP, the C-terminal region (res. 509–516) could be modeled for MCPΔC, which forms an additional β-strand (M2) at each BIS (SI Appendix, Fig. S3E). The C-terminal carboxylate group of MCPΔC is buried inside the capsomer pore and tightly bound by hydrogen bonds, suggesting that the observed conformation at the BIS was adopted after cleavage. Noteworthy, poor and ambiguous electron density for an additional BIS strand was also observed for flMCP but could not be traced (SI Appendix, Fig. S3D). Therefore, this strand could be formed by a different part in flMCP than in MCPΔC, suggesting that the MCP C terminus undergoes conformational changes at the BIS upon processing.
Although the largest local differences between flMCP and MCPΔC were observed in loops closing the central capsomer pore (SI Appendix, Fig. S3F), global changes were observed within capsomer domains (Fig. 2 C and D). The JR1 core is 0.9 Å closer at the capsomer center of mass (COM) in MCPΔC than in flMCP. In contrast, the JR2 tower (T2) is 1.5 Å further away from the capsomer COM in MCPΔC than in flMCP. Thus, the DJR core contracted, whereas T2 expanded, in MCPΔC compared with flMCP (Movie S1). These differences are not due to crystal artifacts since residues involved in crystal contacts (identified by the PISA server; ref. 22) are almost identical for flMCP and MCPΔC. These results suggest that DJR core contraction and tower expansion in MCP derive from processing by MVP.
High Structural Conservation of Virophage MCPs Extends to Processed C Termini.
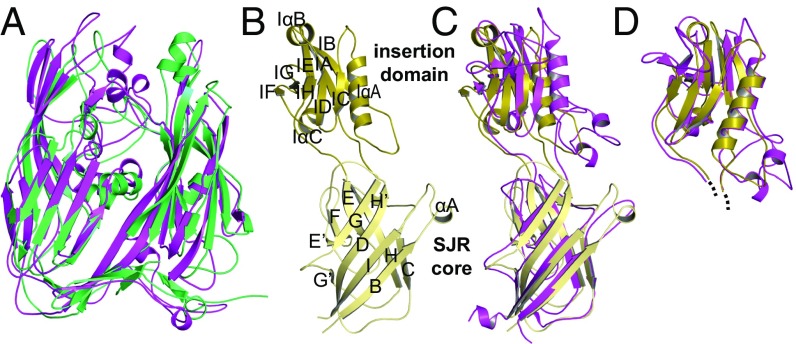
Using the DALI server (23), the highest structural similarity to mavirus flMCP was detected for the Sputnik virophage MCP, followed by the DJR proteins of members of the proposed order Megavirales (24), whereas the virophage MCP strongly diverged from both the adenoviral hexon and the MCP of bacteriophage PRD1 (SI Appendix, Table S3), as already proposed previously (25). Similar to mavirus MCP, Sputnik MCP was described as C-terminally truncated in mature capsids (16) and is called Sputnik MCPΔC in the following. The overall architecture including tower, DJR core, and base is conserved between mavirus and Sputnik MCPΔC (Fig. 3A, Cα rmsd: 4.36 Å). Interestingly, the MCPΔC C terminus is buried in the capsomer pore in both virophages (SI Appendix, Fig. S3G). The high structural similarity of mavirus and Sputnik MCPΔC including the position of processed C termini suggests that the implications of MCP processing on capsomer and capsid are conserved in virophages.
Fig. 3.
High structural similarity of mavirus and Sputnik capsid proteins. The Sputnik structure (PDB ID code 3J26) used in A, C, and D was published in ref. 16. (A) Superposition of mavirus (green) and Sputnik (purple) MCPΔC. (B) Mavirus penton protein. SJR and ID are shown in light and dark yellow, respectively. Secondary structure elements are labeled. (C and D) Structural similarity between mavirus (colors as in B) and Sputnik (purple) penton protein shown by SJR superposition (C) and ID superposition (D).
Penton Protein Structures Are Preserved in Virophages, Especially the Variable Insertion Domain.
We explored whether structural and functional conservation can be extended from MCP to the second relevant virophage capsid protein, the penton protein, which could potentially be involved in host cell or giant virus recognition (12, 16). The crystal structure of mavirus penton protein at 2.7-Å resolution (SI Appendix, Table S2) comprises one pentamer in the asymmetric unit. Res. 2–302 could be modeled into the electron density map for all protomers, which contain a typical SJR domain and an insertion domain (ID). The 139-residue ID is located between SJR strands D and E (Fig. 3B and SI Appendix, Fig. S4 A and B), as observed for the Sputnik penton protein (16), and mainly consists of a β-sandwich attached to helix IαA. With ID β-strands labeled IA to IH, the β-sandwich is formed by a five- and a three-stranded antiparallel β-sheet, IB-IA-IE-IG-IF and IH-ID-IC, respectively. Although low in sequence identity (19%), mavirus and Sputnik penton proteins (PDB ID code 3J26; ref. 16) have high structural similarity (DALI Z score: 22.1; highest Cα rmsd of protomers: 5.07 Å). Superposing only the SJR of a mavirus and Sputnik penton protomer (highest Cα rmsd: 3.70 Å, Fig. 3C) revealed that the ID position relative to the SJR is the most distinctive feature, suggesting a certain degree of flexibility in the connecting loops. Notably, individual superposition of the mavirus and Sputnik IDs revealed a striking fold conservation (highest Cα rmsd: 2.39 Å, Fig. 3D). Such strong structural conservation combined with high sequence variability of the surface-exposed ID among virophages (12) supports the penton’s putative role in host cell or giant virus recognition.
MCP Spontaneously Assembles into Mavirus-Sized Virus-Like Particles in Escherichia coli.
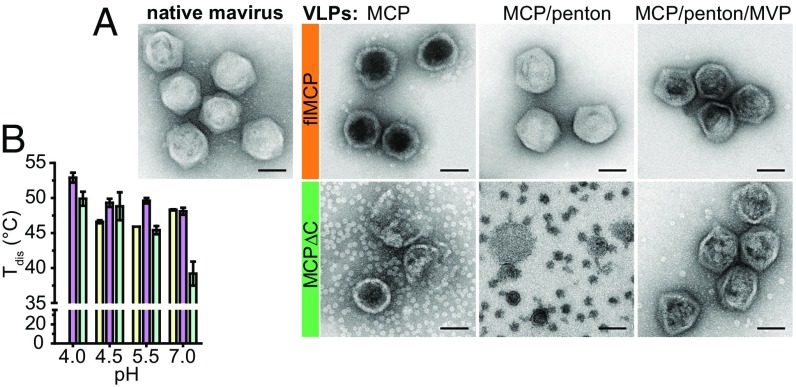
Based on the conservation of the cysteine protease (12) and the structural similarity of truncated mavirus and Sputnik MCP, we hypothesize that MCP processing is important for the formation of infectious virophage particles. To study mavirus capsid assembly, we expressed flMCP and MCPΔC in E. coli and analyzed lysates. Surprisingly, virus-like particles (VLPs) with diameters of 60–75 nm were observed independently of whether flMCP or MCPΔC was expressed (SI Appendix, Fig. S5B). These particles were comparable in size to native mavirus (Fig. 4A, d = 70–80 nm), indicating that exclusively MCP but no specific factors from mavirus, CroV, or host cell are required to form capsids. To compare the assembly efficiency of flMCP and MCPΔC, we used E. coli lysates containing the same amounts of expressed MCP and supplemented gold particles (GPs) as an internal VLP quantification standard for negative stain EM. For flMCP, 61 times more VLPs were present per micrograph than for MCPΔC (SI Appendix, Fig. S5A). In contrast to native mavirus virions, both flMCP- and MCPΔC-VLPs appeared more spherical (Fig. 4A), like “whiffle ball” capsids that lack vertex proteins (26). Notably, purified MCPΔC-VLPs showed a high number of isolated capsomers (Fig. 4A), although these should have been removed during purification, and most particles appeared damaged. These observations indicate that MCPΔC-VLPs assembled less efficiently than flMCP-VLPs and are less stable under harsh staining conditions for EM. We compared the protein melting temperature Tm of capsomers and VLPs by calculating a difference Tm (ΔTm = Tm(VLP) − Tm(capsomer)). We used ΔTm to assess how MCP stability is affected when capsomers assemble into VLPs. Interestingly, Tm1 and Tm2 of flMCP shifted to higher temperatures in assembled VLPs compared with capsomers by ΔTm of at least +4.7 ± 0.1 °C and +2.1 ± 0.1 °C, respectively (SI Appendix, Fig. S6A). This indicates that flMCP capsomers gained stability through VLP assembly. In contrast, MCPΔC stability marginally rose upon VLP formation with a maximum ΔTm1 of only +2.0 ± 0.1 °C (SI Appendix, Fig. S6A). Considering the low MCPΔC-VLP counts in E. coli lysates, damaged appearance and small gain in protein stability through capsid assembly compared with flMCP-VLPs, we propose that flMCP is the assembly relevant state for mavirus capsids.
Fig. 4.
Shape and stability of native mavirus virions and recombinantly generated VLPs. (A) Negative stain EM of purified native mavirus virions (Left) and VLPs (Right). For VLPs, the expressed MCP variant and additionally coexpressed proteins are indicated in the image. (Scale bars: 50 nm.) For the MCPΔC/penton coexpression, sucrose gradient fractions were imaged since no VLP light scattering band was detected in these gradients for extraction and further purification. (B) Thermal disassembly of mavirus and VLPs. Tdis, disassembly temperatures for flMCP/penton-VLPs (yellow), flMCP/penton/MVP-VLPs (purple), and native mavirus (turquoise). Average and SD (error bars) are shown (n = 3). No Tdis could be determined for flMCP/penton-VLPs at pH 4.0 although intact VLPs were observed under this condition by negative stain EM. Thus, it is assumed that disassembly and aggregation were not resolved by the measurement. Similarly, we detected disassembly neither for flMCP-VLPs nor MCPΔC-VLPs.
Penton Protein Confers Icosahedral Shape on VLPs.
To investigate whether mavirus MCP coassembles with penton proteins, we performed coexpression experiments in E. coli. We could observe and extract a VLP-containing light scattering band only for flMCP/penton but not for MCPΔC/penton coexpressions after sucrose gradient centrifugation (SI Appendix, Fig. S7A). Moreover, sucrose gradient fractions from MCPΔC/penton coexpressions did not show VLPs in negative stain EM (Fig. 4A), which is consistent with the low E. coli lysate n(VLP)/n(GP) count ratio of 0.016 ± 0.012 per micrograph (SI Appendix, Fig. S5A). These results imply that only flMCP but not MCPΔC is able to stably assemble into VLPs in the presence of penton protein. Purified flMCP/penton-VLPs indeed contained both capsid proteins (SI Appendix, Fig. S7B) and were icosahedral, resembling mavirus virions rather than spherical MCP-VLPs (Fig. 4A). Only VLPs that included both flMCP and penton protein appeared to be filled like native mavirus particles in negative stain EM images. Since the nucleic acid content, judged by the absorbance ratio A260nm/A280nm (SI Appendix, Table S4), was comparable for all VLPs and native mavirus particles, it is hypothesized that penton protein adds stiffness to flMCP-containing VLPs. We compared the thermal stability of flMCP-VLPs and flMCP/penton-VLPs and found only slightly higher thermal stability for flMCP/penton-VLPs with a maximum ΔTm1 of (1.3 ± 0.2) °C (SI Appendix, Fig. S6B). Thus, the incorporation of penton protein at the vertices only marginally increased VLP stability, suggesting that the gained free energy is used to induce strain or conformational changes.
MCP Processing Takes Place After Capsid Assembly.
We next aimed at generating mavirus-like VLPs containing both processed MCPΔC and penton protein by coexpressing flMCP with penton protein and MVP in E. coli. We indeed extracted mavirus-sized, icosahedral VLPs (Fig. 4A). They mainly contained MCPΔC but also residual flMCP, penton protein, and MVP, but no fragments of the flMCP CTD (SI Appendix, Fig. S7B). We termed these particles flMCP/penton/MVP-VLPs since they derive from flMCP expressions. Interestingly, coexpression of MCPΔC, penton protein, and MVP resulted in VLPs that resembled flMCP/penton/MVP-VLPs (Fig. 4A) and contained all three expressed proteins (MCPΔC/penton/MVP-VLPs, SI Appendix, Fig. S7B). These particles showed a migration behavior comparable to flMCP/penton/MVP-VLPs in rate zonal sucrose gradient centrifugation (SI Appendix, Fig. S7A) and similar thermal stability (SI Appendix, Fig. S6C), suggesting that these VLPs are highly similar.
To investigate whether capsid processing is regulated, we incubated flMCP-containing capsomers and VLPs with MVP and analyzed processing by SDS/PAGE (SI Appendix, Fig. S8). Notably, flMCP-VLPs were processed with a half-life (t1/2) of 24 ± 3 min (average ± SD, n = 3 for all given t1/2), which was sixfold faster than for flMCP capsomers (t1/2 = 136 ± 17 min) and 650-fold faster than the processing of flMCP/penton-VLPs (t1/2 = 15,600 ± 9,300 min). These results suggest two points: First, MVP can enter flMCP-VLPs to process MCP within the capsid. Second, MVP processes capsid-associated flMCP faster than free flMCP capsomers, providing a mechanism for timing capsid maturation after capsid assembly. The slow turnover of flMCP/penton-VLPs implies that penton-containing VLPs present a closed, protease-resistant shell.
MCP Processing Increases Capsid Stability at Acidic pH.
To study how processing affects VLP stability, we measured thermal disassembly of VLPs and native mavirus particles through light scattering. flMCP/penton-VLPs and processed flMCP/penton/MVP-VLPs showed similar disassembly temperatures (Tdis) at pH 7.0, indicating that processing does not alter VLP stability at neutral pH (Fig. 4B). However, VLP exposure to acidic environments revealed that capsids of processed flMCP/penton/MVP-VLPs have a higher thermal stability than flMCP/penton-VLPs. Interestingly, the stability of mavirus virions increased with decreasing pH and showed lower thermal stability (except for pH 4.5) than processed flMCP/penton/MVP-VLPs (Fig. 4B). Thus, despite the presence of genomic DNA and the complete set of virion proteins, native mavirus particles were less stable than artificially generated VLPs. VLP and mavirus capsid stability data suggest that MCP processing results in virion stabilization specifically in acidic environments, with potential implications for host cell infection.
Discussion
Virion Morphogenesis Is Conserved Among Virophages.
We found that mavirus MCP is processed at four C-terminal sites by MVP. These sites are conserved in diverse virophages, which is particularly striking since the MCP CTD shows high variability regarding sequence length and composition (SI Appendix, Fig. S2). MVP recognized two motifs, GG | X and GX | G, which are similar to cleavage motifs of the related adenoviral protease, (M/I/L/N/Q)XGG | X and (M/I/L/N/Q)XGX | G (27). Most clan CE proteases, such as African swine fever virus (ASFV) pS273R (28), recognize the GG | X motif or slight variations thereof. MVP recognition motifs could be more complex since the CTD of mavirus flMCP contains two more GXG sites for which we could not identify cleavage products in vitro.
We show that native mavirus virions contain processed MCPΔC (Fig. 1C), and it was shown previously that Sputnik MCP is processed at the same site in mature virions (16). The presence of MCPΔC in two virophages and detection of MVP in mavirus virions indicate that the cysteine protease has a conserved function among virophages. This hypothesis is further supported by the mavirus and Sputnik MCPΔC structures, which despite low sequence identity are highly similar, including the arrangement of N and processed C termini (Fig. 3 and SI Appendix, Fig. S3G). We also solved the crystal structure of the mavirus penton protein and found a striking similarity to the Sputnik equivalent (16) not only regarding the SJR domain but notably also the ID (Fig. 3). Interestingly, the ID fold has so far only been detected in the penton of virophages and adenoviruses, suggesting an evolutionary and potentially functional connection between these nonenveloped DJR viruses. In contrast, the virophage MCP appears to be more closely related to enveloped viruses of the proposed order Megavirales than to the adenoviral hexon (SI Appendix, Table S3). Therefore, virophages could provide an evolutionary link between these groups of eukaryotic DJR viruses as proposed by Krupovic and Bamford (18).
Considering the strong homology of virophage capsid proteins, we suggest that the capsid architectures of mavirus and Sputnik are similar. Based on the high structural conservation of the MCP, its processing sites and protease activity, we propose that our findings on mavirus capsid assembly and maturation processing are relevant for virophages in general.
Capsid Assembly Is Enabled by Strain Through the flMCP C-Terminal Domain.
We found that mavirus MCP, both full-length and truncated, assembled into mavirus-sized VLPs in E. coli, implying that no specific virus or host protein is required to initiate assembly or determine size. This finding is in contrast to other DJR viruses, e.g., PRD1 (29) and adenovirus (30), for which tape measure proteins are proposed to determine virion size. Thus, mavirus shows an assembly mechanism yet undescribed for DJR viruses. Although mavirus VLPs readily formed in E. coli, in vitro capsid assembly (including crystallization batches) was not pronounced under our experimental conditions. We assume that the cellular conditions with high local protein concentrations and the presence of molecular chaperones to avoid aggregation, among other factors, assist capsid formation in E. coli.
Our crystal structures of flMCP and MCPΔC show that MCP processing results in DJR core compaction and tower expansion (Fig. 2 and Movie S1). In addition, our VLP assembly (Fig. 4) and thermal stability data (SI Appendix, Fig. S6) suggest that flMCP rather than MCPΔC is relevant for capsid assembly (Fig. 5). We can only speculate how structural and stability changes in capsomers affect mavirus capsid assembly. The seemingly flexible flMCP CTD could transfer its high free energy to the DJR core via PCs and BISs, producing tension and causing core expansion, which could affect the ability to form capsids. Similarly, the tailed dsDNA bacteriophage HK97 uses the Δ-domain of capsid protein gp5 as scaffolding domain by generating strain on capsomers, rendering them assembly competent (31). Thus, we propose that the CTD of flMCP serves as scaffolding domain during mavirus capsid assembly. We cannot rule out that the flMCP CTD directly supports capsid assembly, e.g., by contacting neighboring capsomers, as proposed for PRD1 (29).
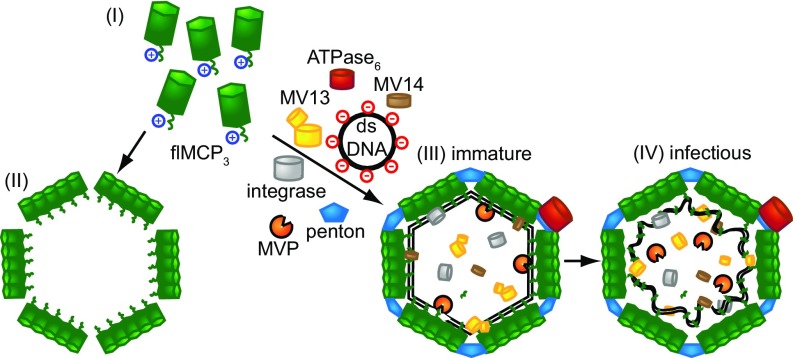
Fig. 5.
Model for virophage assembly and function of MCP processing. (I) flMCP is the assembly-relevant state and can self-assemble into whiffle balls in the absence of other virophage proteins (II). (III) All mavirus genes are preceded by the same late CroV promoter motif (9), suggesting parallel expression and coassembly. The negatively charged dsDNA genome interacts with the positively charged flMCP CTD. (IV) Protease-mediated processing of flMCP detaches the DNA–CTD complex from the capsid.
The penton protein confers icosahedral shape on mavirus VLPs and renders them nearly inaccessible for external MVP (SI Appendix, Fig. S8). Thus, MVP must be packaged during assembly, and we indeed detected MVP in mavirus virions (Fig. 1C). Since all mavirus genes are preceded by the same late CroV promoter motif (9), mavirus MCP and penton protein likely coassemble in virion factories (Fig. 5), which we propose to be mediated through strain on capsomers induced by the flMCP CTD.
Virophage Processing Prepares Virions for Infection.
We hypothesize that virophage processing is a prerequisite to generate infectious virions, as described for, e.g., adenoviruses (27, 32) and poxviruses (33). We observed that flMCP-VLPs are processed faster by MVP than flMCP capsomers (SI Appendix, Fig. S8). Such regulation of MCP processing ensures efficient capsid assembly before maturation cleavage. We propose that a conformational change in flMCP CTD improves cleavage site accessibility upon assembly, but we cannot rule out different scenarios. Regulation of virion processing has also been described for other DJR viruses encoding clan CE cysteine proteases, such as smallpox (34) and adenovirus (35). Whether MVP can be stimulated by cofactors or is affected by oligomerization remains open.
We found that processing of mavirus MCP stabilized VLPs at acidic pH, and stability of mature mavirus particles increased with decreasing pH (Fig. 4B). These observations are in contrast to adenovirus, for which maturation and acidic environments destabilize the particle (36), but are reminiscent of bacteriophages HK97 and P22 (31, 37). Moreover, increased HK97 capsid stability was observed when an inactive gp4 protease was retained in the capsid, and gp4 was proposed to be not only relevant for capsid processing but also capsid assembly (38). Similarly, MVP could cause higher stability of processed flMCP/penton/MVP-VLPs compared with flMCP/penton-VLPs. Capsid stabilization by MVP might explain why we observed VLPs from coexpression of MCPΔC/penton/MVP, but not MCPΔC/penton. The fact that VLPs were not detectable for MCPΔC/penton coexpressions hints at a labile interaction between processed MCP and penton at the vertices. Such vertex instability is reported for mature adenovirus (36) and Sputnik (16), which release pentons under stress conditions, e.g., acidification. Penton release is important during adenoviral infection (39) and could be relevant for virophages as well.
Beside penton destabilization, we propose that MCP processing prepares the mavirus genome for infection. Due to the strong positive charge of the flMCP CTD [theoretical isoelectric point (pI): 10.15], especially in fragments MCPC2, MCPC3, and MCPC4, it could cause an interaction between flMCP and packaged dsDNA, tightly attaching the genome to the capsid interior (Fig. 5). flMCP processing would detach the genome–CTD complex from the capsid interior, preparing it for cell entry. Using PSIPRED v3.3 (40) and WHAT 2.0 (41), we identified a potential amphipathic α helix in fragment MCPC1 that could be involved in endosomal membrane rupture to enable entering the cytoplasm. We could not detect CTD fragments of flMCP in native mavirus virions, either due to low concentration or intense fragmentation by trypsin, which was used for MS PMF. Thus, we cannot rule out that CTD fragments exit mavirus particles after processing.
Apart from MCPΔC, penton protein, and MVP, four further mavirus gene products were identified in mature virions: MV02, two cleavage fragments of MV13, MV14, and MV15 (Fig. 1C). The FtsK/HerA-type ATPase MV15 (9) could be associated with the capsid exterior to package the mavirus genome during virion morphogenesis as observed for Vaccinia virus (42). Apart from MV15, we presume that the detected virion proteins play a role during mavirus infection. The MV02 retroviral integrase likely interacts with the mavirus genome to allow host genome integration as observed recently (14). Besides C-terminal MCP fragments, MV13C and MV14 could also interact with the mavirus genome due to their high theoretical pIs (≥9.6). Interestingly, two nuclear localization signals (NLSs) were predicted for MV13 and one for the flMCP CTD stretching across MCPC2-C4 (43). One of these NLSs could enable nuclear import of the proposed mavirus genome/integrase complex during C. roenbergensis infection. The cleavage into MCPC2, MCPC3, and MCPC4 could act in a regulatory way on the proposed genome-interacting and NLS functions of MCPC2-C4, similar to the processing-based regulation of nuclear import for the adenoviral protein VI (44).
In conclusion, this study provides mechanistic insights into virion assembly, stability, and processing in virophages. Our findings have direct implications for virophage maturation and infection, enriching our understanding of these unique parasites of giant viruses. Parallels could be identified not only to other virophages, but also to more distantly related DJR viruses such as adenovirus and poxvirus, which share morphogenetic proteins with virophages. These homologies extend to the even larger Mimiviridae, the actual host of virophages. Virophages thus appear to be the smallest and least complex member of the PRD1-adenovirus lineage described yet, which makes them particularly exciting for further characterization with implications for the entire viral lineage and potential biotechnological applications as nanocontainers.
Materials and Methods
Detailed materials and methods are described in SI Appendix.
Cloning.
Mavirus genes (GenBank: HQ712116.1) were cloned into plasmids pETM-11 (provided by Gunter Stier) or pASK-IBA3C (IBA). For VLP preparation, capsid protein genes were cloned into pRSFDuet-1.
Protein Expression and Purification.
Mavirus genes were expressed in E. coli BL21-CodonPlus(DE3)-RIL or BL21(DE3). Hexahistidine-tagged penton protein, flMCP, and MCPΔC were mainly purified by Ni2+ affinity chromatography. Size exclusion chromatography (SEC) was the final step for all proteins. MVP was purified by ion exchange chromatography and SEC. Mavirus was prepared as described previously (14). Mavirus and VLPs were purified by sucrose gradient centrifugation.
Crystal Structures.
Experimental phases of MCPΔC and penton protein from selenomethionine single-wavelength anomalous dispersion data were used for molecular replacement (MR) to phase native datasets. The flMCP dataset was phased by MR on the native MCPΔC model. Protein Data Bank ID codes are 6G41–6G45 (SI Appendix, Table S2).
Negative Stain EM.
Samples on glow discharged Formvar/carbon-coated 100 mesh Cu grids were stained by 0.5% uranyl acetate for 90 s and imaged by a Tecnai T20 electron microscope.
Supplementary Material
Acknowledgments
We thank the Heidelberg and Villigen X-ray crystallography teams for measurements at beamline X10SA (Swiss Light Source, Villigen, Switzerland); Ilme Schlichting and Miroslaw Tarnawski for support with crystallization; Sabine Zimmermann and Susanne Eisel for technical support; Martine Haehsler for help with VLP preparations; Robert L. Shoeman and Sebastian Fabritz for help with mass spectrometry; and Dennis H. Bamford, Modesto Redrejo-Rodríguez, and David Stuart for information in the initial stages of the project. We acknowledge financial support by the Heidelberg Biosciences International Graduate School (D.B.) and by the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6G41–6G45).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805376115/-/DCSupplemental.
References
- 1.Raoult D, Forterre P. Redefining viruses: Lessons from mimivirus. Nat Rev Microbiol. 2008;6:315–319. doi: 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- 2.Claverie JM. Viruses take center stage in cellular evolution. Genome Biol. 2006;7:110. doi: 10.1186/gb-2006-7-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.La Scola B, et al. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 4.Abrahão JS, et al. Acanthamoeba polyphaga mimivirus and other giant viruses: An open field to outstanding discoveries. Virol J. 2014;11:120. doi: 10.1186/1743-422X-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krupovic M, Kuhn JH, Fischer MG. A classification system for virophages and satellite viruses. Arch Virol. 2015;161:233–247. doi: 10.1007/s00705-015-2622-9. [DOI] [PubMed] [Google Scholar]
- 6.Desnues C, et al. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc Natl Acad Sci USA. 2012;109:18078–18083. doi: 10.1073/pnas.1208835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaia M, et al. Broad spectrum of mimiviridae virophage allows its isolation using a mimivirus reporter. PLoS One. 2013;8:e61912. doi: 10.1371/journal.pone.0061912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaia M, et al. Zamilon, a novel virophage with Mimiviridae host specificity. PLoS One. 2014;9:e94923. doi: 10.1371/journal.pone.0094923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer MG, Suttle CA. A virophage at the origin of large DNA transposons. Science. 2011;332:231–234. doi: 10.1126/science.1199412. [DOI] [PubMed] [Google Scholar]
- 10.Yau S, et al. Virophage control of antarctic algal host-virus dynamics. Proc Natl Acad Sci USA. 2011;108:6163–6168. doi: 10.1073/pnas.1018221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, et al. Diversity of virophages in metagenomic data sets. J Virol. 2013;87:4225–4236. doi: 10.1128/JVI.03398-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roux S, et al. Ecogenomics of virophages and their giant virus hosts assessed through time series metagenomics. Nat Commun. 2017;8:858. doi: 10.1038/s41467-017-01086-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanc G, Gallot-Lavallée L, Maumus F. Provirophages in the Bigelowiella genome bear testimony to past encounters with giant viruses. Proc Natl Acad Sci USA. 2015;112:E5318–E5326. doi: 10.1073/pnas.1506469112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer MG, Hackl T. Host genome integration and giant virus-induced reactivation of the virophage mavirus. Nature. 2016;540:288–291. doi: 10.1038/nature20593. [DOI] [PubMed] [Google Scholar]
- 15.Krupovic M, Koonin EV. Polintons: A hotbed of eukaryotic virus, transposon and plasmid evolution. Nat Rev Microbiol. 2015;13:105–115. doi: 10.1038/nrmicro3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, et al. Structure of Sputnik, a virophage, at 3.5-Å resolution. Proc Natl Acad Sci USA. 2012;109:18431–18436. doi: 10.1073/pnas.1211702109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun S, et al. Structural studies of the Sputnik virophage. J Virol. 2010;84:894–897. doi: 10.1128/JVI.01957-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krupovic M, Bamford DH. Virus evolution: How far does the double beta-barrel viral lineage extend? Nat Rev Microbiol. 2008;6:941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- 19.Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 20.Yutin N, Raoult D, Koonin EV. Virophages, polintons, and transpovirons: A complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol J. 2013;10:158. doi: 10.1186/1743-422X-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Athappilly FK, Murali R, Rux JJ, Cai Z, Burnett RM. The refined crystal structure of hexon, the major coat protein of adenovirus type 2, at 2.9 A resolution. J Mol Biol. 1994;242:430–455. doi: 10.1006/jmbi.1994.1593. [DOI] [PubMed] [Google Scholar]
- 22.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colson P, et al. “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol. 2013;158:2517–2521. doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravantti J, Bamford D, Stuart DI. Automatic comparison and classification of protein structures. J Struct Biol. 2013;183:47–56. doi: 10.1016/j.jsb.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, et al. Control of virus assembly: HK97 “Whiffleball” mutant capsids without pentons. J Mol Biol. 2005;348:167–182. doi: 10.1016/j.jmb.2005.02.045. [DOI] [PubMed] [Google Scholar]
- 27.Mangel WF, San Martín C. Structure, function and dynamics in adenovirus maturation. Viruses. 2014;6:4536–4570. doi: 10.3390/v6114536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrés G, Alejo A, Simón-Mateo C, Salas ML. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J Biol Chem. 2001;276:780–787. doi: 10.1074/jbc.M006844200. [DOI] [PubMed] [Google Scholar]
- 29.Abrescia NG, et al. Insights into assembly from structural analysis of bacteriophage PRD1. Nature. 2004;432:68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- 30.Reddy VS, Nemerow GR. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc Natl Acad Sci USA. 2014;111:11715–11720. doi: 10.1073/pnas.1408462111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gertsman I, et al. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–650. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber J. Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J Virol. 1976;17:462–471. doi: 10.1128/jvi.17.2.462-471.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ansarah-Sobrinho C, Moss B. Role of the I7 protein in proteolytic processing of vaccinia virus membrane and core components. J Virol. 2004;78:6335–6343. doi: 10.1128/JVI.78.12.6335-6343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aleshin AE, et al. Activity, specificity, and probe design for the smallpox virus protease K7L. J Biol Chem. 2012;287:39470–39479. doi: 10.1074/jbc.M112.388678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangel WF, McGrath WJ, Toledo DL, Anderson CW. Viral DNA and a viral peptide can act as cofactors of adenovirus virion proteinase activity. Nature. 1993;361:274–275. doi: 10.1038/361274a0. [DOI] [PubMed] [Google Scholar]
- 36.Pérez-Berná AJ, et al. The role of capsid maturation on adenovirus priming for sequential uncoating. J Biol Chem. 2012;287:31582–31595. doi: 10.1074/jbc.M112.389957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galisteo ML, King J. Conformational transformations in the protein lattice of phage P22 procapsids. Biophys J. 1993;65:227–235. doi: 10.1016/S0006-3495(93)81073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang RK, et al. The Prohead-I structure of bacteriophage HK97: Implications for scaffold-mediated control of particle assembly and maturation. J Mol Biol. 2011;408:541–554. doi: 10.1016/j.jmb.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiethoff CM, Wodrich H, Gerace L, Nemerow GR. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J Virol. 2005;79:1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2013;41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhai Y, Saier MH., Jr A web-based program (WHAT) for the simultaneous prediction of hydropathy, amphipathicity, secondary structure and transmembrane topology for a single protein sequence. J Mol Microbiol Biotechnol. 2001;3:501–502. [PubMed] [Google Scholar]
- 42.Cassetti MC, Merchlinsky M, Wolffe EJ, Weisberg AS, Moss B. DNA packaging mutant: Repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J Virol. 1998;72:5769–5780. doi: 10.1128/jvi.72.7.5769-5780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: A simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wodrich H, et al. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 2003;22:6245–6255. doi: 10.1093/emboj/cdg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.