Significance
High-risk neuroblastomas (NBs) show undifferentiated/poorly differentiated morphology as a distinctive feature. We have identified the transcription factor ZNF281 as a factor that can counteract the neuronal differentiation of primary neurons in culture and NB cells. The expression of ZNF281 is inhibited by TAp73 and promoted by MYCN. In turn, ZNF281 inhibits the expression of GDNF and NRP2, two proteins associated with neuronal differentiation. In patients with NB, the expression of ZNF281 is higher in high-risk patients and is associated with worse prognosis. Understanding the molecular mechanisms that regulate neuronal differentiation is relevant for the identification of defects in this process that underlie the development of tumors such as NB, in which an aberrant differentiation arrest has occurred.
Keywords: ZNF281, MYCN, p73, miR34a, neuroblastoma
Abstract
Derangement of cellular differentiation because of mutation or inappropriate expression of specific genes is a common feature in tumors. Here, we show that the expression of ZNF281, a zinc finger factor involved in several cellular processes, decreases during terminal differentiation of murine cortical neurons and in retinoic acid-induced differentiation of neuroblastoma (NB) cells. The ectopic expression of ZNF281 inhibits the neuronal differentiation of murine cortical neurons and NB cells, whereas its silencing causes the opposite effect. Furthermore, TAp73 inhibits the expression of ZNF281 through miR34a. Conversely, MYCN promotes the expression of ZNF281 at least in part by inhibiting miR34a. These findings imply a functional network that includes p73, MYCN, and ZNF281 in NB cells, where ZNF281 acts by negatively affecting neuronal differentiation. Array analysis of NB cells silenced for ZNF281 expression identified GDNF and NRP2 as two transcriptional targets inhibited by ZNF281. Binding of ZNF281 to the promoters of these genes suggests a direct mechanism of repression. Bioinformatic analysis of NB datasets indicates that ZNF281 expression is higher in aggressive, undifferentiated stage 4 than in localized stage 1 tumors supporting a central role of ZNF281 in affecting the differentiation of NB. Furthermore, patients with NB with high expression of ZNF281 have a poor clinical outcome compared with low-expressors. These observations suggest that ZNF281 is a controller of neuronal differentiation that should be evaluated as a prognostic marker in NB.
Neuronal differentiation occurs through a complex array of epigenetic changes, posttranslational modifications, and the expression of specific genes. An example of the interplay between proteins and microRNAs is TAp73, a member of the p53 family (1, 2) that transcriptionally regulates the expression of miR34a in an axis necessary for the in vitro differentiation of cortical neurons and neuroblastoma (NB) cells (3, 4). Accordingly, the cortex and the hippocampus of TAp73 KO mice display developmental defects (3) and metabolic changes (5).
The zinc finger transcription factor Zfp281 is involved in the control of cell stemness by inhibiting Nanog expression in mice (6) through recruitment of the inhibitory complex NuRD on the Nanog promoter (7). Importantly, knock-out of Zfp281 in mice is embryonically lethal, suggesting a key role for this gene in development (6). However, this does not exclude other important functions performed by Zfp281 in adult cells. Indeed, ZNF281 (the human homolog of Zfp281) induces epithelial mesenchymal transition in colon cancer cells by controlling expression of SNAI1 and other key genes implicated in epithelial mesenchymal transition (8). In addition, ZNF281 knock-down promotes osteogenesis of multipotent stem cells (9). In line with this broader role of ZNF281 in somatic cells, we have recently demonstrated that ZNF281 is also involved in the regulation of the DNA damage response (10).
Here, using several experimental approaches, we show that ZNF281 is down-regulated during neuronal differentiation and negatively affects the differentiation process of murine cortical neurons and NB cells. High expression of ZNF281 defines a subset of patients with NB with poor clinical outcome. We also demonstrate that the expression of ZNF281 is induced by MYCN and inhibited by TAp73 acting through miR34a. In accordance with these data, we have identified neuronal differentiation-associated GDNF and NRP2 as two targets inhibited by ZNF281.
Results
ZNF281 Expression Is Down-Regulated During Neuronal Differentiation.
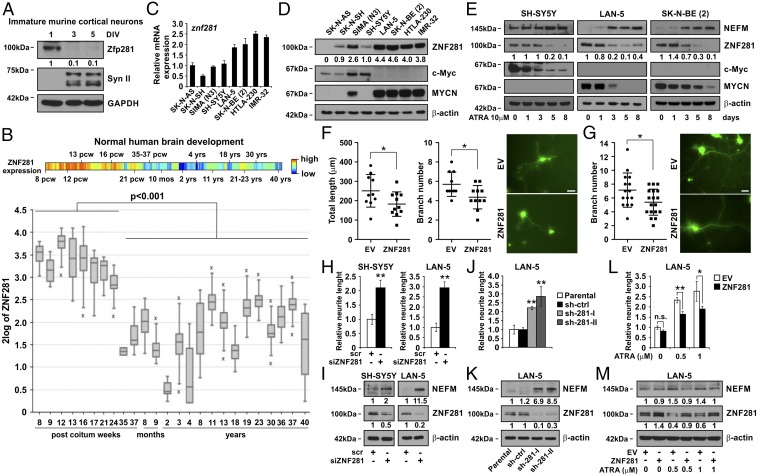
Immature cortical neurons from embryos at day 17.5 post coitum (p.c.) were allowed to complete differentiation in vitro for 5 d (3, 4). Western blot analysis showed a sharp decrease in ZNF281 expression during the 5-d culture (Fig. 1A). To further explore the behavior of ZNF281 during neural differentiation in man, we interrogated the R2 dataset that contains array-derived data on gene expression in human brains at different stages of embryonic and postnatal development. We demonstrated a significant decrease of ZNF281 expression comparing its expression levels before and after birth (Fig. 1B). To understand whether modulation of ZNF281 expression during the process of differentiation is also a common feature in transformed cells of neuro-ectodermal origin, we used NB cells that undergo neuronal differentiation on treatment with retinoids and/or neurotrophins (11). Under basal conditions, ZNF281 was expressed at higher levels in the MYCN-amplified cell lines LAN-5, SK-N-BE (2), HTLA-230, and IMR32 compared with the MYCN nonamplified lines SK-N-AS, SK-N-SH, SIMA, and SH-SY5Y (12) (Fig. 1 C and D). When we induced neuronal differentiation in NB cell lines SH-SY5Y, LAN-5, SK-N-BE (2) by treatment with all-trans-retinoic acid (ATRA; 10 μM) for 8 d, we detected neurite outgrowth (SI Appendix, Fig. S1A) and up-regulation of Neurofilament Medium Peptide (NEFM) (Fig. 1E) in ATRA-treated cells, as expected. Conversely, the expression of ZNF281 decreased throughout the differentiation process at both the protein (Fig. 1E) and transcript levels (SI Appendix, Fig. S1B). Decrease of ZNF281 expression during neuronal differentiation was also confirmed in other human NB cell lines (SK-N-AS, SK-N-SH, and IMR-32) (SI Appendix, Fig. S1C) and in the murine NB cell line N1E115 (SI Appendix, Fig. S1 D and E). Together, these results demonstrate a negative correlation between the expression of ZNF281 and the degree of neural differentiation in normal and transformed somatic cells.
Fig. 1.
ZNF281 expression during neuronal differentiation and in NB cell lines. (A) Immature murine cortical neurons were cultured in vitro, as described in Materials and Methods. Western blot (WB) analysis was carried out with antibodies to ZNF281 and Synapsin II (Syn II). Equal loading was checked by GAPDH detection. DIV, days in vitro. Numbers below the corresponding blot represent densitometric analysis normalized to the housekeeping gene and relative to DIV1. (B) Heat map (Upper) and box plot (Lower) analyses of ZNF281 expression during normal brain development. mos, months; pcw, postcoitum weeks; yrs, years. Data were retrieved from the R2 dataset (r2.amc.nl) Normal Brain Development (524 samples)–BrainSpan atlas of the developing human brain. Statistical significance of the difference between ZNF281 was calculated by unpaired two-tailed t test. (C and D) Reverse transcriptase real-time PCR (qPCR) (C) and Western blot (D) analyses of ZNF281 expression in human NB cell lines. qPCR carried out was repeated twice in triplicate. Western blot analysis was carried out with antibodies to ZNF281, c-Myc, and MYCN. β-actin was used for normalization. (E) WB analyses of ZNF281 expression during neuronal differentiation of human NB cell lines induced by ATRA 10 μM for the indicated times. Antibodies used were as in D. Blots are representative of two to four biological replicates. Numbers below the corresponding blot in D and E represent densitometric analysis normalized to the housekeeping gene. (F) ZNF281 negatively affects dendritic outgrowth of cortical neurons. DIV 1 murine cortical neurons were transfected with expression vectors encoding ZNF281 and a GFP-Spectrin fusion protein (pGFP-Spectrin) at a 15:1 ratio. After 48 h, neurons were analyzed by fluorescence microscopy. Total neurite length and quantification of branch number after transfection of ZNF281 or empty vector (EV) were performed as described in Methods. In each experiment, 10–11 cells were analyzed. Data represent mean ± SD of two different experiments (*P < 0.05, unpaired two-tailed Student’s t test). (Right) Representative image. (Scale bar, 25 μm.) (G) Quantification of branch number in DIV 3 murine cortical neurons was carried out the day after transfection. In each experiment, 15–18 cells were analyzed. Data represent mean ± SD of two different experiments (*P < 0.05, unpaired two-tailed Student’s t test). (Right) Representative image. (Scale bar: 25 μm.) (H) Neurite length of human NB cell lines transfected with a pool of siRNA against ZNF281 (siZNF281) or with scr was calculated by counting at least 60 cells. (I) WB analysis of ZNF281 and NEFM expression in the same cell lines transfected with siZNF281 or with scr. Numbers below the corresponding blot represent densitometric analysis performed by taking the expression of scr-transfected cells or controls as 1. (J) Neurite length of LAN-5 cells infected with the lentiviral vector pLVTHM-sh-scrambled, pLVTHM-sh281-I, and pLVTHM-sh281-II. Two independent clones infected with sh-ZNF281 (sh-281-I and sh-281-II) and a mixed population infected with sh-scrambled (sh-ctrl) were analyzed. Neurite counting was carried out as in H. (K) WB analysis of ZNF281 and NEFM expression in parental, sh-ctrl, sh-281-I and sh-281-II LAN-5 cells. Numbers below the corresponding blot represent densitometric analysis normalized to the housekeeping gene and relative to sh-ctrl cells. (L) Neurite length of LAN-5 cells treated with ATRA at the indicated concentrations and transfected with an expression vector for human ZNF281 was calculated by counting as in H. Differences in neurite length were evaluated for statistical significance taking the neurite length of the untreated cells as 1. Asterisks in H, J, and L indicate statistical significance (*P < 0.05; **P < 0.01) of the difference between scr and siZNF281 calculated with unpaired two-tailed Student’s t test. (M) WB analysis of LAN-5 cells treated with ATRA and transfected with an expression vector for ZNF281. Numbers below the corresponding blot represent densitometric analysis normalized to the housekeeping gene and relative to EV ctrl cells. Blots are representative of two to four biological replicates.
ZNF281 Affects Neuronal Differentiation of Murine Cortical Neurons and NB Cells.
To understand whether ZNF281 has a role in neuronal differentiation, we cultured murine cortical neurons at day 17.5 p.c. Cells were cotransfected with expression vectors encoding ZNF281 and a GFP-Spectrin fusion protein (pGFP-Spectrin) at a 15:1 ratio after 1 or 3 d in vitro (DIV1 and DIV3) to measure neurite length and number of dendrites. We detected a significant reduction in neurite length and branching in cells transfected at DIV1 with ZNF281/GFP-Spectrin compared with controls transfected with the empty vector/GFP-Spectrin after 48 h from transfection (Fig. 1F). Furthermore, cells transfected with ZNF281/GFP-Spectrin at DIV3 displayed reduction of neurite branching after 24 h from transfection (Fig. 1G). To analyze whether the effects of ZNF281 on neuronal differentiation were also detectable in transformed cells, we transfected SH-SY5Y and LAN-5 NB cell lines with a pool of siRNAs that specifically knock-down the expression of ZNF281. We detected the onset of neuronal differentiation in cells transfected with siRNA against ZNF281 compared with control siRNA, as evaluated by neurite extension and increase of NEFM expression (Fig. 1 H and I and SI Appendix, Fig. S2A). Furthermore, we labeled ZNF281-silenced SH-SY5Y cells with EdU to monitor their proliferation rate. FACS analysis of EdU-labeled cells did not detect a significant reduction in the percentage of cells in S phase between the ZNF281-silenced and the control cells, suggesting that the onset of differentiation driven by ZNF281 silencing is not a result of inhibition of proliferation (SI Appendix, Fig. S2 D and E). We confirmed the prodifferentiation effect of ZNF281 silencing by detecting an increase of NEFM expression and neurite outgrowth in the NB cell line LAN-5 infected with two lentiviral vectors encoding shRNAs directed against ZNF281 (Fig. 1 J and K and SI Appendix, Fig. S2B). To further explore the role of ZNF281 in NB differentiation, we ectopically expressed ZNF281 in LAN-5 cells that were subsequently treated with ATRA (0.5 or 1 μM). At both concentrations, neurite extension and NEFM expression decreased in ZNF281 overexpressing, ATRA-treated cells compared with control cells (Fig. 1 L and M and SI Appendix, Fig. S2C), suggesting that ZNF281 expression hinders ATRA-induced neuronal differentiation. Altogether, these data imply a role of ZNF281 in maintaining an undifferentiated phenotype in murine cortical neurons and in NB cells.
TAp73 Inhibits ZNF281 Expression Through miR34a.
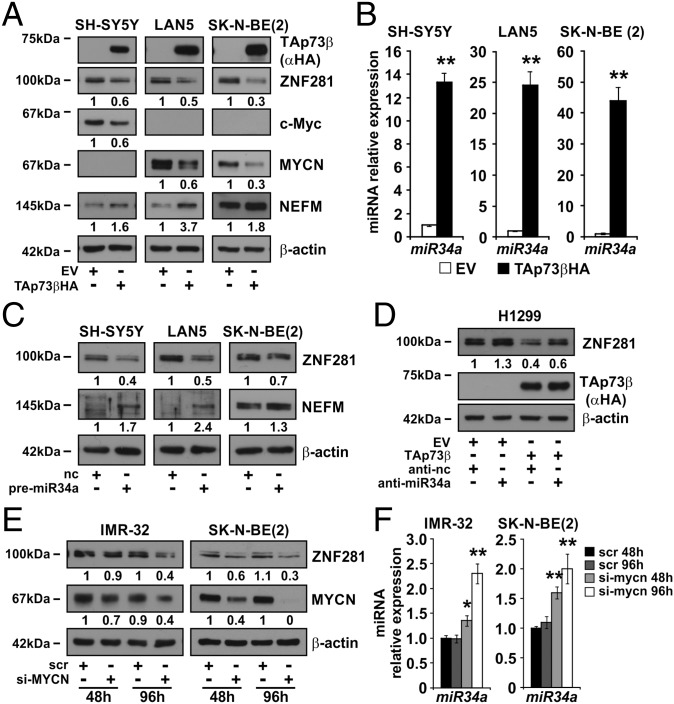
Previous work has highlighted a role for TAp73 in inducing neuronal differentiation through the activation of miR34a (3, 4). Ectopic expression of TAp73 in the NB cell lines SH-SY5Y, LAN-5, and SK-N-BE (2) induced a decrease of ZNF281 (Fig. 2A and SI Appendix, Fig. S3A) and MYCN expression (Fig. 2A) and a parallel increase of miR34a expression (Fig. 2B). In colon cancer cells, miR34a posttranscriptionally inhibits the expression of ZNF281 (8). To test whether miR34a had a similar role in NB cells, we cotransfected the NB cell line BE2 (M17) with a luciferase reporter vector containing the 3′-untranslated region of the human ZNF281 gene and the premiR34a. Luciferase activity was significantly decreased in cells transfected with the premiR34a compared with those transfected with the control premiR (SI Appendix, Fig. S3B). Cotransfection of premiR34a and a reporter vector containing the 3′-untranslated region of ZNF281 in which the miR34a binding site was mutated did not cause any significant variation in luciferase activity compared with controls, demonstrating that miR34a posttranscriptional inhibition of ZNF281 in NB cells depends on the presence of a miR34a binding site in the 3′-untranslated region of ZNF281 (SI Appendix, Fig. S3B). In keeping with these results, transfection of premiR34a in SH-SY5Y, LAN-5, and SK-N-BE (2) cells caused a decrease in ZNF281 expression, as opposed to an increase in NEFM (Fig. 2C), which further demonstrates the ability of miR34a to inhibit the expression of ZNF281 in NB cells. To evaluate whether there is a direct link among TAp73, miR34a, and ZNF281, we used the highly transfectable p53-null H1299 cell line (13) in which ZNF281 expression was reduced after transfection of TAp73β (SI Appendix, Fig. S3 C–E). We transfected H1299 cells with an anti-miR specific for miR34a (hereafter anti-miR34a) and, subsequently, with a plasmid encoding TAp73β. After 24 h, we detected a decrease of ZNF281 expression in cells transfected with TAp73β that was partially rescued by treatment with anti-miR34a (Fig. 2D). Of note, H1299 cells transfected with anti-miR34a (but not with TAp73β) showed an increase of ZNF281 expression as a result of the inhibition of the endogenous levels of miR34a (Fig. 2D). These data suggest a link among TAp73, miR34a, and ZNF281 that could be important in the control of neuronal differentiation.
Fig. 2.
The expression of ZNF281 is controlled by p73 and MYCN. (A) WB analysis of NB cell lines transfected with an expression vector encoding TAp73β-HA. Numbers below the corresponding blot represent densitometric analysis normalized to the housekeeping gene and relative to EV. (B) qPCR to measure the expression of miR34a. Analysis was performed twice in triplicate. Error bars represent SD. Asterisks indicate statistical significance (**P < 0.01) of the difference between EV and TAp73β-HA-transfected cells calculated with unpaired two-tailed Student’s t test. (C) WB analysis of the indicated NB cell lines transfected with premiR34a. Numbers below the corresponding blot represent densitometric analysis normalized to the housekeeping gene. (D) WB analysis of H1299 cells transfected with a backbone-modified anti miR34a or with anti-nc control and subsequently retransfected with an expression vector encoding for TAp73β-HA or with the EV. Numbers below the corresponding blot represent densitometric analysis normalized to the housekeeping gene and relative to the EV. (E) WB analysis of the indicated human NB cell lines transfected with a pool of anti-MYCN siRNA or with scrambled siRNAs control (src) for 48 and 96 h. Numbers below the corresponding blot represent densitometric analysis normalized to the housekeeping gene. (F) qPCR to measure the expression of miR34a in the cells of E. Asterisks indicate statistical significance (*P < 0.05; **P < 0.01) of the difference between scr (at 48 h) and other samples calculated with unpaired two-tailed Student’s t test. Error bars represent SD of triplicate biological replicates.
Silencing of MYCN Inhibits ZNF281 Expression.
To understand whether the oncogene MYCN could affect the expression of ZNF281 in NB cells, we transfected MYCN-amplified IMR32 and SK-N-BE (2) cells with siRNA against MYCN. The expression of ZNF281 was substantially inhibited in MYCN-silenced cells after 48 and 96 h (Fig. 2E) in parallel with an increase in miR34a expression (Fig. 2F). This result suggests that MYCN acts on ZNF281 through down-regulation of miR34a, similar to the action of c-Myc in colon carcinoma cells (8). In support of a role of c-Myc in controlling the expression of ZNF281 in NB cells, silencing of c-Myc in the MYCN nonamplified, c-Myc-expressing NB cell lines SH-SY5Y, SK-N-SH caused a decrease of ZNF281 (SI Appendix, Fig. S3F). Conversely, ectopic expression of c-Myc or MYCN in SK-N-AS cells increased the expression of ZNF281 (SI Appendix, Fig. S3G)
GDNF and NRP2 Are Targets of ZNF281.
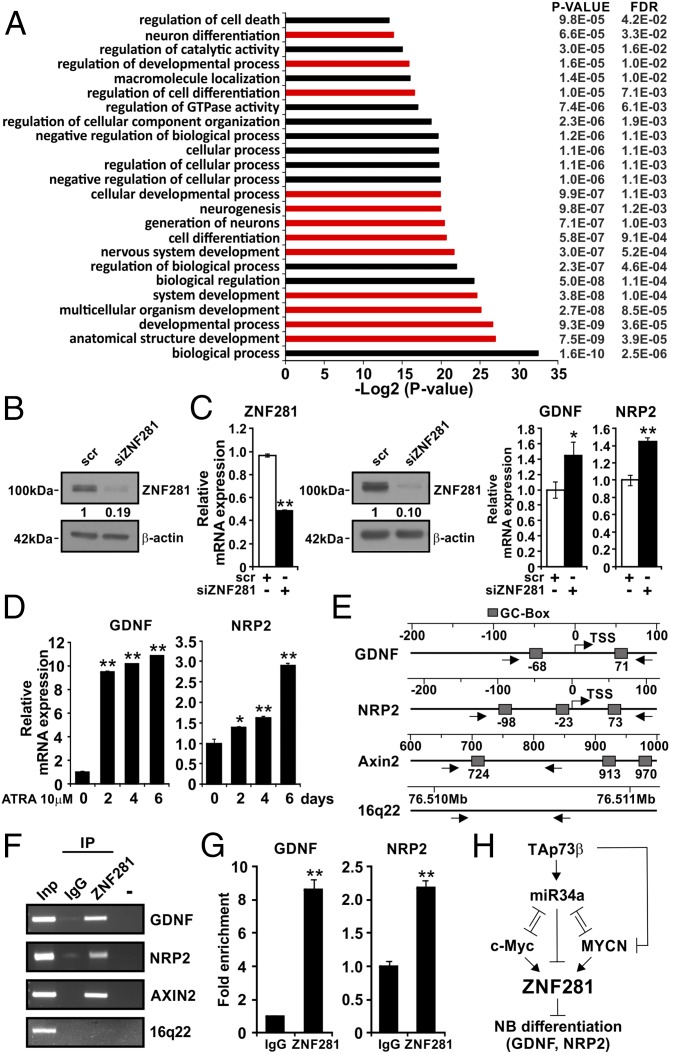
To identify targets of ZNF281 associated with neuronal differentiation, we carried out a microarray analysis comparing gene expression of BE2 (M17) cells [a transfectable subclone derived from SK-N-BE (2) cells] (14) silenced with specific siRNA against ZNF281 with that of the same cells transfected with scrambled (scr) siRNA. Four biological replicates of each treatment were used for the microarray analysis. Technical details are given in Materials and Methods. We used a threshold of 1.2-fold up- or down-regulation to select RNAs differentially expressed in cells treated with siRNA against ZNF281 compared with siRNA scr controls. We detected 960 genes that are associated with several cellular processes (Fig. 3A). The efficacy of ZNF281 silencing was checked by Western blot analysis to detect the levels of ZNF281 protein in ZNF281-silenced cells and in scr controls (Fig. 3B). Among the modulated genes, 587 were up-regulated and 373 were down-regulated on ZNF281 silencing. Gene ontology classification highlighted 116 genes that were involved in nervous system development. We focused our attention on two of these genes for their direct involvement in neuronal differentiation: Glia-Derived Neurotrophic Factor (GDNF) and Neuropilin 2 (NRP2) play a role in inducing neuronal differentiation and in axon guidance, respectively (15, 16). We confirmed that GDNF and NRP2 expression are indeed up-regulated by ZNF281 silencing in independent experiments in BE2-M17 cells (Fig. 3C). Of interest, the expression of both genes is also up-regulated during ATRA-induced differentiation of BE2-M17 cells (Fig. 3D). Thus, ZNF281 is able to suppress the expression of genes that promote neuronal differentiation.
Fig. 3.
Array analysis of genes differentially expressed in BE2(M17) cells silenced for ZNF281 expression: ZNF281 binds to the promoters of GDNF and NRP2. (A) Gene ontology grouping of differentially expressed genes. The expression of ZNF281 was analyzed by Western blot in BE2(M17) cells silenced for ZNF281 compared with scr controls before microarray analysis. FDR, false discovery rate. (B) WB representative of four biological replicates used for the microarray analysis. Numbers below the blot represent densitometric analysis normalized to the housekeeping gene. (C) Validation analysis: the expression of ZNF281, GDNF, and NRP2 was analyzed by qPCR. Expression of ZNF281 was also analyzed in BE2(M17) cells silenced for ZNF281 by WB analysis (a representative result is shown). Graphs show the mean of 4 independent experiments (*P < 0.05; **P < 0.01). (D) The expression of GDNF and NRP2 was analyzed by qPCR in BE2(M17) cells treated with 10 μM ATRA for 6 d (*P < 0.05; **P < 0.01). (E) Scheme of the promoter regions of GDNF, NRP2, and Axin2 (positive control) and chromosome 16q22 region (negative control). TSS, transcription start site; arrows indicate the position of the primers used for the ChIP analysis; solid boxes represent GC-rich regions. (F) ChIP results analyzed by semiquantitative PCR. Inp, input; IgG immuno-precipitation carried out with IgG. (G) ChIP analyzed by real-time qPCR; results are expressed as fold enrichment respect to IgG (**P < 0.01). ChIP analyses were repeated twice with similar results. Error bars represent SD of triplicate biological replicates. Statistical analysis was performed using two-tailed Student’s t test. (H) Scheme of the controls operating on ZNF281 expression. Arrow-headed and bar-headed lines indicate activation and inhibition respectively.
ZNF281 Binds to the Promoters of GDNF and NRP2.
ZNF281 acts as a transcriptional repressor of Nanog by binding to its promoter (7). We hypothesized that ZNF281 could also bind to the promoters of GDNF and NRP2 to inhibit their expression. We analyzed the regulatory regions of both genes looking for GC-rich regions as potential binding sites for ZNF281 (8). We detected two CG-rich regions in the proximity of the transcription start site of GDNF and three near the transcription start site of NRP2 (Fig. 3E). We designed primers that included these regions to carry out a chromatin crosslinking immunoprecipitation (ChIP) analysis with a specific antibody for ZNF281. Results of these analyses clearly indicate binding of ZNF281 to the GDNF and NRP2 promoters (Fig. 3 F and G). We used Axin 2 as a positive control (10) and a region on chromosome 16 (16q22) as negative control (10) of ZNF281 binding (Fig. 3 F and G). These results suggest that ZNF281 inhibits the expression of GDNF and NRP2 through a direct mechanism.
Altogether our data indicate that ZNF281 in NB cells is positively controlled by MYCN and c-Myc, which in turn are repressed by TAp73, at least in part through miR34a; expression of ZNF281 inhibits NB differentiation by the negative regulation of differentiation-associated genes such as GDNF and NRP2 (Fig. 3H).
ZNF281 Expression Is a Prognostic Marker of NB.
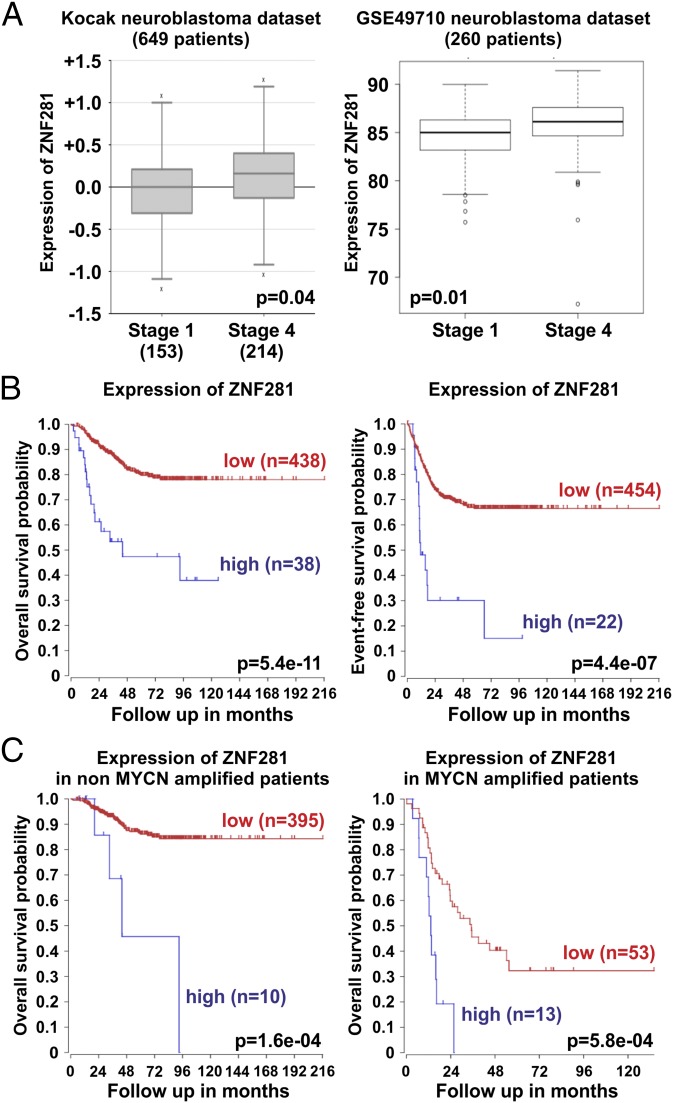
We used public datasets generated by microarray analyses of RNA from patients with NB to understand whether the expression of ZNF281 is compartmentalized in specific subsets of patients. We found that the expression of ZNF281 was increased in stage 4 (metastatic, high-risk tumors) (17) compared with less aggressive stage 1 (Fig. 4A). ZNF281 expression is also elevated in high-risk (stage 4) compared with low- to intermediate-risk patients (stages 1, 2, 3, and 4S; SI Appendix, Fig. S4A). Kaplan Meier analysis revealed that patients expressing high levels of ZNF281 have a significantly lower overall and event-free survival probability compared with low-expressing patients (Fig. 4B). Because the expression of ZNF281 is associated with MYCN amplification in NB cell lines (Fig. 1D) and MYCN amplification defines a subset of high-risk NBs, we sought to understand whether the expression of ZNF281 could have a prognostic value in subgroups of patients with or without MYCN amplification. Kaplan Meier analysis of patients without MYCN amplification indicated that high levels of ZNF281 expression predicts significantly lower overall (Fig. 4C) and event-free (SI Appendix, Fig. S4B) survival probability compared with low-expressors. Even in MYCN amplified patients, high expression of ZNF281 identified a subgroup of patients with worse overall and event-free survival probability (Fig. 4C and SI Appendix, Fig. S4B, respectively). In NB, clinical outcome is worse in patients >18 mo compared with those <18 mo of age (17). Kaplan Meier analyses in subgroups >18 mo and <18 mo of age highlighted a worse overall and event-free (SI Appendix, Fig. S4 C and D) survival probability for those patients expressing high levels of ZNF281 in both subgroups. We also evaluated whether the expression of ZNF281 could be prognostic in subgroups of NB patients >18 and <18 mo of age with or without MYCN amplification. Overall survival in all MYCN-amplified subgroups and event-free survival in MYCN-amplified >18 mo, in MYCN-amplified <18 mo, and in non-MYCN amplified >18 mo was worse in patients highly expressing ZNF281 (SI Appendix, Fig. S4 E and F).
Fig. 4.
Expression of ZNF281 as a prognostic marker in NB datasets. (A) Expression of ZNF281 in NB patients at stage 1 versus stage 4. All data used for the analysis are from the GSE 45547 Tumor Neuroblastoma–Kocak dataset (649 patients) in R2 genomics analysis and visualization platform (r2.amc.nl), and from the GSE49710 NB dataset (260 patients). Boxes indicate the median (horizontal line); whiskers indicate distances from the highest and lowest value to each end of the box that are within 1.5× box length; outliers are represented as dots. Mean values were compared with the two-tailed unpaired t test. (B) Kaplan-Meier (KM) overall and event-free survival analysis of the total Kocak cohort. (C) KM analyses in the MYCN nonamp and in the MYCN-amp subcohorts according to ZNF281 expression. The R2 system gives the cutoff value of ZNF281 expression levels. The difference between the curves for ZNF281 high and ZNF281 low groups are compared by χ2 test.
Discussion
Here, we show that ZNF281 expression is associated with the undifferentiated state and decreases during terminal differentiation of murine cortical neurons and of NB cells induced by ATRA. The inverse association between ZNF281 expression and neural differentiation is further revealed by the decrease in ZNF281 levels that occurs in human brains from embryonic to adult life. In line with these observations, our data indicate that ZNF281 acts as a brake for the differentiation potential of murine cortical neurons and human NB cells, whereas its down-regulation triggers the onset of differentiation.
In an attempt to define mechanisms that govern ZNF281 expression in neural cells, we investigated the possible involvement of p73 (18, 19). In NB, where it is often expressed (3, 20), the TAp73 isoform plays a prodifferentiating role through its target miR34a (3). Our results indicate that TAp73 inhibits the expression of ZNF281, at least in part, through the posttranscriptional repression exerted by miR34a. Furthermore, we looked for activators of ZNF281 in NB cells. Here, MYCN plays a central role in controlling proliferation and differentiation of NB cells and tumors (21, 22). During NB differentiation, MYCN expression decreases and elevated MYCN levels are associated with an aggressive and undifferentiated phenotype (23). MYCN expression is negatively regulated by TAp73 (24). We now show that inhibition of MYCN expression elicits a substantial reduction of ZNF281 expression. The latter observation suggests that MYCN promotes the expression of ZNF281 in NB cells. Thus, our data point to a control axis involving MYCN as a positive regulator of ZNF281 in contrast to TAp73, which negatively regulates its expression through its action on miR34a and through its inhibition of MYCN.
As a transcription factor, ZNF281 can affect the expression of several genes (6, 8) whose identity is most likely dependent on the cellular context. Indeed, recent data demonstrated that ZNF281 stimulates cardiac reprogramming by acting in association with GATA4 on cardiac enhancers and by inhibiting inflammation-associated genes through the recruitment of components of the NuRD complex on their regulatory regions (25). We envisaged that in neuroectodermally derived NB cells, ZNF281 could control genes directly involved in the neuronal differentiation process. To test this hypothesis, we silenced ZNF281 in NB cells to analyze the resulting modulation of expressed genes. We focused our attention on two genes, GDNF and NRP2, whose expression increased significantly in cells silenced for ZNF281. GDNF belongs to a family of neurotrophic factors that includes Artemin, Persephin, and Neurturin (26). The GDNF signaling pathway includes the receptor GFRα1 and the Ret and NCAM coreceptors. GDNF has been shown to induce neurite outgrowth and branching morphology in sympathetic and dopaminergic neurons (15). Furthermore, GDNF is capable of inducing neuronal differentiation in many NB cell lines (15). NRP2, together with NRP1, plays an important role in gangliogenesis, axon guidance, and innervation of targets in the sympathetic nervous system (16). Functionally, NRP2 is a receptor for which SEMA3F is a ligand (16). Because NB cells derive from the neural crest as well as the sympathetic nervous system, it is conceivable that NRP2 plays a role in neurite outgrowth and, more generally, in neuronal differentiation in NB. GDNF and NRP2 together represent two examples of how ZNF281 can exert its inhibitory action of differentiation of NB cells by inhibiting the drivers of this process. The most common way through which ZNF281 inhibits its targets is through direct binding to their regulatory regions and the recruitment of inhibitory complexes of the NuRD type (7, 25). Our data demonstrate ZNF281 binding to GDNF and NRP2 promoters. In both cases, binding occurs in GC-rich regions very close to the transcription start site. These observations are consistent with a mechanism of transcriptional repression involving chromatin remodeling proteins.
NB derives from embryonic neuroectodermal cells that stop their differentiation process and remain in a proliferative phase. The causes of this differentiation block are not entirely understood, although the genomic amplification and the resulting overexpression of the MYCN oncogene play an important role (23, 27). More recently, the GPC2 gene, whose expression is promoted by MYCN or by the amplification of the GPC2 locus, was demonstrated to be essential for NB proliferation (28). Accordingly, GPC2 is mostly expressed in high-risk NB, but is generally absent in normal childhood tissues (28). Interrogation of two NB datasets indicates that the expression of ZNF281 is increased in patients with stage 4 NB whose tumors present less histologically differentiated features with respect to stage 1. Accordingly, patients with NB with high expression of ZNF281 have a poor clinical outcome compared with low-expressors. These observations on patient-derived samples are in agreement with a primary role of ZNF281 on the differentiation control of NB that we demonstrated with our experimental results.
In brief, our data suggest that ZNF281 acts in maintaining the undifferentiated state of normal and transformed neural cells. In NB cells, the expression of ZNF281 is controlled through an axis involving TAp73, miR34a, and MYCN. The inhibitory activity of ZNF281 on NB differentiation is exerted by the negative control of the expression of differentiation-associated genes such as GDNF and NRP2. Finally, the enhanced expression of ZNF281 in advanced stages of NB should be further evaluated as a prognostic factor of this disease.
Materials and Methods
Primary cortical neurons cultures were prepared from embryonic day 17.5 murine embryos, as previously described (4). Cortical neurons were cotransfected at the indicated day of culture, with an expression vector containing the coding region of ZNF281 (pcDNA3-ZNF281HA) and a vector expressing a GFP-Spectrin fusion protein (pGFP-Spectrin) at a 15:1 ratio, using Lipofectamine 2000 (Invitrogen), as previously described (4). We analyzed the total dendritic length and branch number of each individual GFP-positive neuron, as previously described (4). Images were collected using a Leica DMI6000B digital inverted microscope (LEICA microsystem) and analyzed with LAS AF (Leica Application Suite Advanced Fluorescence) software (LEICA microsystem). The research on animals described in this paper was approved by the Ethical Committee of the University of Rome “Tor Vergata.”
Additional information on cell lines, protein analysis and antibodies, RNA extraction, cell transfection, siRNA silencing and real-time qPCR analyses, functional assays, lentiviral infections, cell proliferation, array analysis, chromatin crosslinking immunoprecipitation, NB dataset analysis, and immunofluorescence analysis is provided in SI Appendix, SI Materials and Methods.
Uncropped images from WB and ChIP analyses are shown in SI Appendix, Figs. S5 and S6.
Supplementary Material
Acknowledgments
This work has been supported by the Medical Research Council, United Kingdom; grants from Associazione Italiana per la Ricerca contro il Cancro (2017 IG20473 to G.M.) and Fondazione Roma malattie Non trasmissibili Cronico-Degenerative (to G.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE112029).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801435115/-/DCSupplemental.
References
- 1.Kaghad M, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 2.Pozniak CD, et al. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289:304–306. doi: 10.1126/science.289.5477.304. [DOI] [PubMed] [Google Scholar]
- 3.Agostini M, et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc Natl Acad Sci USA. 2011;108:21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agostini M, et al. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci USA. 2011;108:21099–21104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agostini M, et al. Metabolic reprogramming during neuronal differentiation. Cell Death Differ. 2016;23:1502–1514. doi: 10.1038/cdd.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fidalgo M, et al. Zfp281 functions as a transcriptional repressor for pluripotency of mouse embryonic stem cells. Stem Cells. 2011;29:1705–1716. doi: 10.1002/stem.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fidalgo M, et al. Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proc Natl Acad Sci USA. 2012;109:16202–16207. doi: 10.1073/pnas.1208533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn S, Jackstadt R, Siemens H, Hünten S, Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013;32:3079–3095. doi: 10.1038/emboj.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo KW, et al. ZNF281 knockdown induced osteogenic differentiation of human multipotent stem cells in vivo and in vitro. Cell Transplant. 2013;22:29–40. doi: 10.3727/096368912X654948. [DOI] [PubMed] [Google Scholar]
- 10.Pieraccioli M, et al. ZNF281 contributes to the DNA damage response by controlling the expression of XRCC2 and XRCC4. Oncogene. 2016;35:2592–2601. doi: 10.1038/onc.2015.320. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan DR, Matsumoto K, Lucarelli E, Thiele CJ. Eukaryotic Signal Transduction Group Induction of TrkB by retinoic acid mediates biologic responsiveness to BDNF and differentiation of human neuroblastoma cells. Neuron. 1993;11:321–331. doi: 10.1016/0896-6273(93)90187-v. [DOI] [PubMed] [Google Scholar]
- 12.Thiele CJ. Neuroblastoma. In: Masters J, editor. Human Cell Culture. Vol 1. Kluwer Academic Publishers; Lancaster, UK: 1998. pp. 21–53. [Google Scholar]
- 13.Lin RK, et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 2010;70:5807–5817. doi: 10.1158/0008-5472.CAN-09-4161. [DOI] [PubMed] [Google Scholar]
- 14.Biedler JL, Roffler-Tarlov S, Schachner M, Freedman LS. Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res. 1978;38:3751–3757. [PubMed] [Google Scholar]
- 15.Yoong LF, Wan G, Too HP. GDNF-induced cell signaling and neurite outgrowths are differentially mediated by GFRalpha1 isoforms. Mol Cell Neurosci. 2009;41:464–473. doi: 10.1016/j.mcn.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Maden CH, et al. NRP1 and NRP2 cooperate to regulate gangliogenesis, axon guidance and target innervation in the sympathetic nervous system. Dev Biol. 2012;369:277–285. doi: 10.1016/j.ydbio.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohn SL, et al. INRG Task Force The International Neuroblastoma Risk Group (INRG) classification system: An INRG Task Force report. J Clin Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Killick R, et al. p73: A multifunctional protein in neurobiology. Mol Neurobiol. 2011;43:139–146. doi: 10.1007/s12035-011-8172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nemajerova A, et al. Non-oncogenic roles of TAp73: From multiciliogenesis to metabolism. Cell Death Differ. 2018;25:144–153. doi: 10.1038/cdd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Laurenzi V, et al. Induction of neuronal differentiation by p73 in a neuroblastoma cell line. J Biol Chem. 2000;275:15226–15231. doi: 10.1074/jbc.275.20.15226. [DOI] [PubMed] [Google Scholar]
- 21.Bosse KR, Maris JM. Advances in the translational genomics of neuroblastoma: From improving risk stratification and revealing novel biology to identifying actionable genomic alterations. Cancer. 2016;122:20–33. doi: 10.1002/cncr.29706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maris JM, Matthay KK. Molecular biology of neuroblastoma. J Clin Oncol. 1999;17:2264–2279. doi: 10.1200/JCO.1999.17.7.2264. [DOI] [PubMed] [Google Scholar]
- 23.Westermark UK, Wilhelm M, Frenzel A, Henriksson MA. The MYCN oncogene and differentiation in neuroblastoma. Semin Cancer Biol. 2011;21:256–266. doi: 10.1016/j.semcancer.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Horvilleur E, et al. p73alpha isoforms drive opposite transcriptional and post-transcriptional regulation of MYCN expression in neuroblastoma cells. Nucleic Acids Res. 2008;36:4222–4232. doi: 10.1093/nar/gkn394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou H, et al. ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression. Genes Dev. 2017;31:1770–1783. doi: 10.1101/gad.305482.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Airaksinen MS, Saarma M. The GDNF family: Signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3:383–394. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- 27.Thiele CJ, Reynolds CP, Israel MA. Decreased expression of N-myc precedes retinoic acid-induced morphological differentiation of human neuroblastoma. Nature. 1985;313:404–406. doi: 10.1038/313404a0. [DOI] [PubMed] [Google Scholar]
- 28.Bosse KR, et al. Identification of GPC2 as an oncoprotein and candidate immunotherapeutic target in high-risk neuroblastoma. Cancer Cell. 2017;32:295–309.e12. doi: 10.1016/j.ccell.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.