Significance
Recently discovered biosynthetic gene clusters in plants are a striking example of the nonrandom complex structure of eukaryotic genomes. The mechanisms underpinning the formation of these clustered pathways are not understood. Here we carry out a systematic analysis of transposable elements associated with clustered terpene biosynthetic genes in plant genomes, and find evidence to suggest a role for miniature inverted-repeat transposable elements in cluster formation in eudicots. Our analyses provide insights into potential mechanisms of cluster assembly. They also shed light on the emergence of a “block” mechanism for the foundation of new terpene clusters in the eudicots in which microsyntenic blocks of terpene synthase and cytochrome P450 gene pairs duplicate, providing templates for the evolution of new pathways.
Keywords: biosynthetic gene clusters, genome evolution, gene relocation, transposons, terpenes
Abstract
Plants produce a plethora of natural products, including many drugs. It has recently emerged that the genes encoding different natural product pathways may be organized as biosynthetic gene clusters in plant genomes, with >30 examples reported so far. Despite superficial similarities with microbes, these clusters have not arisen by horizontal gene transfer, but rather by gene duplication, neofunctionalization, and relocation via unknown mechanisms. Previously we reported that two Arabidopsis thaliana biosynthetic gene clusters are located in regions of the genome that are significantly enriched in transposable elements (TEs). Other plant biosynthetic gene clusters also harbor abundant TEs. TEs can mediate genomic rearrangement by providing homologous sequences that enable illegitimate recombination and gene relocation. Thus, TE-mediated recombination may contribute to plant biosynthetic gene cluster formation. TEs may also facilitate establishment of regulons. However, a systematic analysis of the TEs associated with plant biosynthetic gene clusters has not been carried out. Here we investigate the TEs associated with clustered terpene biosynthetic genes in multiple plant genomes and find evidence to suggest a role for miniature inverted-repeat transposable elements in cluster formation in eudicots. Through investigation of the newly sequenced Amborella trichopoda, Aquilegia coerulea, and Kalanchoe fedtschenkoi genomes, we further show that the “block” mechanism of founding of biosynthetic gene clusters through duplication and diversification of pairs of terpene synthase and cytochrome P450 genes that is prevalent in the eudicots arose around 90–130 million years ago, after the appearance of the basal eudicots and before the emergence of the superrosid clade.
A growing number of biosynthetic gene clusters for the production of different types of natural products have recently been reported in plants, including for the synthesis of terpenes, hydroxamic acids, steroidal and benzylisoquinoline alkaloids, cyanogenic glucosides, and polyketides (1–23). These clusters have not arisen by horizontal gene transfer from microbes, but rather by recruitment of genes from elsewhere in the genome through duplication and neofunctionalization by as yet unknown mechanisms. There is also evidence of partial clustering of different types of plant pathway genes, and of duplication of functionally related gene pairs and modules (1, 17, 21, 24–27). Thus, some plant natural product pathways may have superficial similarities with microbial pathways in terms of genomic organization, yet their origins are distinct.
The terpenes are one of the largest families of natural products, with over 80,000 reported so far (28). In a recent genomics-based study, our investigations of multiple sequenced plant species revealed many known and potential gene clusters for terpene biosynthesis (17). We further found evidence of different evolutionary paths for cluster formation in monocots and eudicots. In eudicots, pairs of “signature” genes, encoding terpenoid synthases (TSs) and cytochrome P450-dependent monooxygenases (CYPs), appear to serve as templates for the formation of new clusters, while in the monocots the clusters had assembled de novo in each genome studied (17).
Transposable elements (TEs) can mediate genomic rearrangements by providing homologous sequences that enable illegitimate recombination and gene relocation (29, 30). This process has been extensively studied in the human genome, where the resultant duplications, deletions, inversions, and translocations are often associated with genetic disorders (31). Analysis of two Arabidopsis thaliana biosynthetic gene clusters (the thalianol and marneral clusters) has revealed that both are located in regions of the genome that are significantly enriched in TEs, and that have arisen since the last whole-genome duplication event (5, 8). Other plant biosynthetic gene clusters also contain TEs that may have played a role in gene rearrangements during cluster formation (2–4, 9, 10, 13, 17). Thus, TE-mediated recombination may contribute to plant biosynthetic gene cluster formation, and potentially also to the establishment of coregulation of these gene clusters. However, a systematic analysis of the TEs involved has not been carried out.
TSs generate terpene scaffold diversity, while CYPs modify and further diversify these scaffolds. These enzymes are the primary drivers of terpene diversification and together are responsible for the generation of a vast array of terpene structures. Our previous comprehensive analysis of 17 sequenced plant genomes yielded an inventory of >100 clustered TS/CYP gene pairs from monocot and eudicot genomes (17), paving the way for investigation of the evolutionary mechanisms responsible for cluster formation. Here we investigate the TEs associated with clustered and nonclustered TS and CYP genes across multiple plant species and find evidence for a role for miniature inverted-repeat transposable elements (MITEs) in biosynthetic gene cluster formation in eudicots. Our results suggest that common mechanisms are likely to underlie the assembly of eudicot biosynthetic gene clusters. They further suggest that the “block” mechanism of founding of biosynthetic gene clusters through duplication and diversification of TS/CYP gene-pair templates arose in a 10–30 My period sometime between 90 and 130 Mya, after the appearance of the basal eudicots and before the emergence of the superrosid clade.
Results and Discussion
Distribution of Terpene Biosynthetic Gene Clusters on Chromosomes.
In filamentous fungi, gene clusters for natural product pathways are often found close to the ends of chromosomes (32). The plant biosynthetic gene clusters that have been reported so far are in some cases also located in the subtelomeric regions. Examples include the maize 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA) cluster and the oat avenacin cluster (1, 33). In contrast, in A. thaliana the thalianol and marneral triterpene clusters are not subtelomeric, but instead are located in dynamic chromosomal regions that have formed since the last whole-genome duplication event (8). Our previous analysis of multiple plant genomes revealed an overall terminal cluster distribution profile for monocots, while in the eudicots the collective pattern was not explicitly terminal (17). In the present study, in-depth investigation of cluster-distribution profiles within individual eudicot species revealed that the eudicot clusters for the most part also follow the terminal pattern, with the exception of the two Brassicaceae species included in the analysis, A. thaliana and Brassica rapa. The majority of the A. thaliana TS/CYP gene clusters were located pericentrically, while the B. rapa cluster distribution profile appeared bimodal, TS/CYP pairs being located both pericentrically and in the terminal region (Fig. 1, Left). By mining three new genomes—the basal eudicot Aquilegia coerulea (completely assembled), the magnoliophyta Amborella trichopoda (scaffolds), and the superrosid Kalanchoe fedtschenkoi (scaffolds)—and augmenting our previous set of clustered TS/CYP genes, we identified an additional 38 clustered TS/CYP gene pairs (Dataset S1) that were also distributed terminally (included in the eudicot panels in Fig. 1).
Fig. 1.
Distribution of clustered TS/CYP pairs along chromosomes in monocot and eudicot genomes. The horizontal axis designates intervals of chromosome length expressed as a fraction of 1.0. The vertical axis of the panels on the Left designates the percentage of clustered TS/CYP gene pairs (black bars) or gene density at the whole genome scale (open bars) located in the intervals. The monocot profiles are derived from previously reported TS/CYP gene pairs from four genomes (B. distachyon, Oryza sativa, Sorghum bicolor, and Zea mays). The eudicot profiles are derived from previously reported TS/CYP gene pairs from six eudicot genomes (V. vinifera, P. trichocarpa, G. max, M. truncatula, S. lycopersicum, and S. tuberosum), along with an additional 19 pairs from the recently available genome sequence of the basal eudicot A. coerulea and an additional 7 pairs from M. truncatula, S. lycopersicum, and S. tuberosum. The A. thaliana and B. rapa profiles are shown separately because they have different profiles to the overall eudicot trend. The Center shows the clustered/global ratios of the two profiles depicted in the Left. The Right represents the pericentric ratios (see text) of the profiles from the Left. The results of the χ2 test (P values) for the significance of the difference between two distributions are shown for each on the Left and Right.
A potential drawback in interpreting the cluster profiles of species with the pericentric pattern of cluster distribution could be in that each individual chromosome has a specific centromere position, which results in metacentric, submetacentric, acrocentric, subtelocentric, or telocentric chromosomes. These different types of chromosomes are shown in SI Appendix, Fig. S1. Because of subtelocentric or telocentric centromere positions, a cluster profile may be terminally shifted from its center in cases when clusters are predominantly incorporated in the centromere region. However, investigation of karyotype data available for 11 of the 15 species included in our analysis indicated that the chromosomes of these species are primarily either metacentric (75.9%) or submetacentric (20.6%), with acrocentric and subtelocentric chromosomes constituting 2.5% and 0.9%, respectively, and no telocentric chromosomes (across a total of 320 chromosomes) (SI Appendix, Table S1). Our cluster distribution profiles are therefore unlikely to be skewed because of the species karyotypes. The seven most terminal B. rapa clusters are located on one metacentric chromosome and three submetacentric chromosomes, which suggests no consistency with the terminal centromere positions. The reason why the two Brassicaceae species have patterns that differ from the overall eudicot pattern is unclear. Forthcoming new genomes of species belonging to this family may in future shed light on this anomaly.
Because clustered genes could potentially follow global gene distribution along chromosomes, we analyzed the global profiles of gene density in the genomes studied to exclude possible effects on the cluster profiles. The global profiles for the eudicots and monocots were averaged and compared with the corresponding cluster profiles. Because numbers of clusters varied significantly in different genomes, from two in Glycine max to 19 in A. coerulea, the total cluster profiles for the eudicots and monocots were obtained by summarizing the corresponding values in the intervals of chromosomes instead of by finding the average and then calculating the percentage of each profile value. The Sorghum bicolor global gene-density profile significantly differed from the others and was omitted from the analysis. These results showed that across all genomes examined, genes were in general more prevalent toward the termini of chromosomes (Fig. 1, Left). This immediately suggests that there is no association between the distribution of coding sequences and the Brassicaceae pericentric clusters (Fig. 1, Left and Center, bottom two rows). At the same time, clustered TS/CYP genes were overrepresented at chromosomal termini in both the eudicot and monocot genomes compared with the corresponding global gene-density profiles (Fig. 1, Left and Center, top two rows). Further support in favor of the clustered TS/CYP terminal shift in these genomes is provided by the pericentric ratio profiles, which represent the ratios of each value in the profiles shown in the Left panels of Fig. 1 to the corresponding values of the pericentric intervals of 0.5 (Fig. 1, Right, top two panels). The pericentric ratios in this study characterize the degree to which the distributions of clusters or genes along chromosomes are shifted toward the ends. As can be seen from Fig. 1, the terminal pericentric ratios of the cluster-distribution profiles exceed the corresponding 0.5 interval values by around four- and sixfold for the monocots and eudicots, respectively. In contrast, those of the global gene distributions are only around twofold greater. The pericentric ratio profiles of the Brassicaceae genomes also suggest that their cluster profiles are independent of the global gene distribution (Fig. 1, Right, bottom two panels). The χ2 test for significance of difference between the corresponding cluster profiles and the global gene-density profiles represented in Fig. 1, Left and Right indicated highly significant differences in the eudicot, monocot species and Brassicaceae species (corresponding P values are shown in Fig. 1). Although the terminal cluster overrepresentation in the eudicots and monocots may at first glance appear low (Fig. 1, Center, top two panels), the strong statistical support makes it convincing (Fig. 1, Left and Right).
Previously, by random simulation, we have shown nonstochastic occurrence of clustered TS/CYP gene pairs in plant genomes (17). In this study, we used a similar approach (Materials and Methods) to show that the location of these clusters does not completely follow the global gene distribution. As it might be expected, no significant difference between the stochastic cluster profiles and the corresponding global gene profiles were found (P = 0.98 and 0.99 for eudicot and monocot species, respectively) (see also SI Appendix, Fig. S2). Conversely, the former were significantly different from the real cluster profiles (P = 3.5e-4 and 3.6e-5 for eudicots and monocots, respectively).
Global TE Profiling.
Several reports indicate that plant biosynthetic gene clusters are located in regions of high TE abundance (8, 10, 17). To carry out a systematic analysis of the types of TEs involved, we created a stand-alone BLAST database of 13 completely assembled plant genomes. Sequences of 2,460 retrotransposons, 528 DNA transposons, and 656 MITEs (TE class I, class II, and class III, respectively) were downloaded from the Plant Repeat Database (34). The DNA sequences of all three TE classes were separately piped through the plant genome database (Materials and Methods). The customized computer program then parsed the BLAST output file and searched for predicted TEs according to the coordinates of the TE alignments. Next, the same program used the predicted TEs of each class to build the corresponding profiles of the chromosomal TE distributions for each plant genome on the global scale (Table 1 and SI Appendix, Table S2).
Table 1.
The patterns of global TE distribution along chromosomes in the plant genomes
| TE class | ||||||
| Clade | Order | Family/subfamily | Species | I | II | III |
| Monocots | ||||||
| Commelinids | Poales | Pooideae | B. distachyon | P | P | T |
| Ehrhartoideae | O. sativa | P | T | T | ||
| Panicoideae | S. bicolor | P | T | T | ||
| Z. mays | P | T | T | |||
| Eudicots | ||||||
| Basal eudicots | Ranunculales | Ranunculaceae | A. coerulea | P | P | T |
| Rosids | Vitales | Vitaceae | V. vinifera | P | P | T |
| Rosid I (Fabidae) | Malpighiales | Salicaceae | P. trichocarpa | P | P | T |
| Fabales | Fabaceae | G. max | P | P | T | |
| M. truncatula | P | P | Eq | |||
| Rosid II (Malvidae) | Brassicales | Brassicaceae | A. thaliana | P | P | P |
| B. rapa | P | P | P | |||
| Asterid I | Solanales | Solanaceae | S. lycopersicum | P | T | T |
| S. tuberosum | P | T | Eq | |||
The represented genomes are in the status of “complete chromosomes.” Distribution patterns: Eq, equal; Ins, interspersed; P, pericentric; T, terminal; (for more details on the distribution patterns see text). TE classes: I, retrotransposons; II, DNA transposons; III, MITEs. The three monocot subfamilies (Pooideae, Ehrhartoideae and Panicoideae) represent the Poaceae family.
TEs can either be integrated in eukaryotic genomes at specific target sequences or inserted nonspecifically on a genome wide basis (interspersed TEs) (30, 35, 36). Retrotransposons (class I) have been shown to be preferentially integrated in centromeric heterochromatin regions in plants (37–39) and also to be specifically associated with telomeres in diverse eukaryotes (40), while interspersed genomic distribution (nonspecific TE integration) is characteristic of the L1 retrotransposon and nonautonomous elements in the human genome (41). A large proportion of maize and rice DNA transposons (class II), as well as MITEs (class III), are found in euchromatic regions, which suggests interspersed distribution (42, 43). In this study, it was important to use TE sequences without introducing bias for certain genomic targets, substantially telomeric or centromeric sequences, to allow the global distributions of each TE class within each genome to be determined regardless of the effects of specific TE integration. For this reason, the Plant Repeat Database was advantageous in that it provides a comprehensive resource of annotated TE sequences with no bias for mode of integration (34).
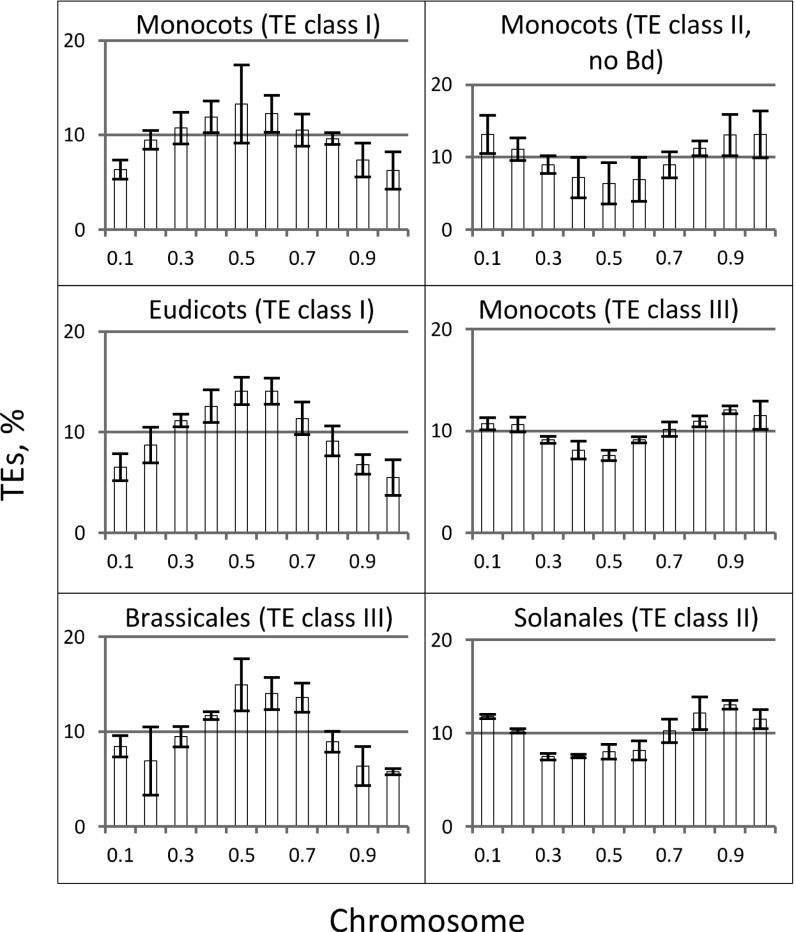
Our investigations revealed two principle patterns of global TE distribution against the general interspersed TE background: (i) pericentric distribution and (ii) terminal distribution (Table 1). The pericentric distribution pattern was characterized by a greater percentage of TEs within the centromeric region (shifted to the chromosomal center). Conversely, in the case of the terminal distribution, TEs were predominantly localized in the vicinity of chromosomal termini. Examples of both types of profile are shown in Fig. 2. The other less-significant pattern was the equal distribution pattern, which showed approximately equal TE distribution along chromosomes. The distribution patterns observed within different plant lineages are shown in Table 1. For example, class II DNA transposons are found predominantly in the terminal regions of chromosomes of the monocots with the exception of the grass, Brachypodium distachyon (pericentric pattern). In contrast, class II DNA transposons are predominantly pericentric in the eudicot species A. coerulea (basal eudicots), grapevine (Vitis vinifera), poplar (Populus trichocarpa), soybean (G. max), barrelclover (Medicago truncatula), A. thaliana and B. rapa (rosids), while in the Solanaceous species tomato (Solanum lycopersicum) and potato (Solanum tuberosum) (asterid I clade) have the terminal distribution pattern.
Fig. 2.
Examples of different patterns of global TE distribution in monocot and eudicot genomes. The vertical axis indicates the percentage of TEs with SDs located in each chromosome interval. The horizontal axis values are the fractions of the chromosome length. The B. distachyon (Bd) values for class II TEs were excluded from the monocot profile (Top Right) because the pattern was pericentric and differed from those of the other monocots (Table 1). The profiles are for TEs with ≥60% identity to the corresponding genomic sequences.
The MITE distribution profiles for the majority of the genomes were predominantly terminal, with the exception of the two Brassicaceae species, where they were pericentric (Table 1). The distributions of MITEs in M. truncatula and S. tuberosum differed from the pericentric or terminal patterns in that they were distributed approximately equally along the chromosomes (Table 1). The similarity in the MITE and the clustered TS/CYP distributions, both of which are terminal for the majority of the genomes and pericentric in the two Brassicaceae species, suggests the predominant participation of this TE class in cluster formation. MITEs are known to be frequently associated with coding sequences. Although the MITE profiles with the terminal distribution have similar profiles to their global gene-density counterparts (no significant difference was found between the average profiles with P = 0.97 and 0.98 for eudicots and monocots, respectively), this does not mean that the distributions of the clustered TS/CYP genes simply follow the gene distributions at the whole-genome scale due to the general gene/MITE association. This is inconsistent with the predominant pericentric location of both MITEs and TS/CYP pairs in Brassicaceae, which disagrees with the terminal global gene-density profiles, as well as with the uncorrelated cluster and global gene-density profiles in the eudicots and monocots, as noted.
The terminal profile pattern of the class II transposons, which is characteristic for the majority of the monocots and the Solanaceae species, suggests that these TEs may also contribute to the process in some lineages. In contrast, the class I retrotransposons are unlikely to contribute to genome reorganization events leading to cluster formation because these elements are located primarily in the pericentric regions, which contrasts with the terminal pattern of the cluster distributions in the majority of the genomes studied. Interestingly, in the case of the two Brassicaceae species, the profiles of all three TE classes are pericentric. Because the assemblies and annotations of many of the genomes studied are in a state of constant improvement, the possibility of pericentric or terminal patterns instead of the equal distribution found in some genomes should not be excluded.
Local TE Density.
To further investigate the potential role of the three different TE classes in cluster formation and function, we studied the abundance of TEs of each class in the vicinity of two cluster markers, the TS and CYP genes, and compared these values with nonclustered TS and CYP genes (global TS/CYP complement) of the corresponding plant genomes. For this purpose, we used the clustered TS and CYP genes from the complete genomes studied in our previous analysis (17), together with their global complements downloaded from the Phytozome database. These data were augmented with the 19 clustered TS/CYP gene pairs from the new complete A. coerulea genome, as well as a total of 11 TS/CYP pairs from two other genomes, A. trichopoda and K. fedtschenkoi (available as scaffold assemblies and scaffolds, respectively) (Dataset S1).
In the previous study, we found that the TE density (TE/kb) in 50-kb flanking regions of clustered and nonclustered TS/CYP genes was significantly different, although this difference in the case of the eudicot species studied was small (17). For this reason, to make statistical support more robust, 100-kb flanking regions of the TS and CYP genes for both the clustered and nonclustered complements from each genome were used to compute the TE local density for each of the three classes of TEs. The density values of each TE class for both clustered and nonclustered complements thus constituted separate distributions. Preliminary analysis showed that the empirical distributions were best fitted to the negative binomial distribution. The mean of this distribution is known as the μ-parameter (Materials and Methods).
We then compared the abundance of each TE class around the TS and CYP genes in each genome by determining the clustered/global ratio of the μ-values (i.e., the ratio of the TE distribution means for the flanks of the clustered and nonclustered TS/CYP genes). The results revealed a striking difference between the magnoliophyta A. trichopoda and the monocots, on the one hand, and the eudicots on the other hand (Table 2). Class I (retrotransposons) and class II (DNA transposons) TEs were either not overrepresented or slightly more abundant in the vicinity of clusters in all plant species studied. In contrast, the class III TEs (MITEs) were significantly overrepresented around the clustered TS and CYP genes in the eudicot genomes.
Table 2.
Local TE density in the vicinities of the clustered TS/CYP genes
| Clade | Order | Family/subfamily | Species | TE class | ||
| I | II | III | ||||
| Magnoliophyta | Amborellales | Amborellaceae | A. trichopoda | 1.1 ± 0.4 | 1.4 ± 0.2 | 1.6 ± 0.2 |
| Monocots | ||||||
| Commelinids | Poales | Pooideae | B. distachyon | 1.0 ± 0.1 | 1.5 ± 0.1 | 1.5 ± 0.2 |
| Ehrhartoideae | O. sativa | 1.3 ± 0.1 | 1.2 ± 0.1 | 1.0 ± 0.1 | ||
| Panicoideae | S. bicolor | 1.3 ± 0.2 | 1.0 ± 0.1 | 0.9 ± 0.1 | ||
| Pooideae | Z. mays | 1.4 ± 0.1 | 1.2 ± 0.1 | 1.6 ± 0.2 | ||
| Eudicots | ||||||
| Basal eudicots | Ranunculales | Ranunculaceae | A. coerulea | 1.3 ± 0.2 | 1.1 ± 0.2 | 4.2 ± 1.8 |
| Super-Rosids | Saxifragales | Crassulaceae | K. fedtschenkoi | 0.9 ± 0.3 | 1.2 ± 0.3 | 5.3 ± 1.5 |
| Rosids | Vitales | Vitaceae | V. vinifera | 1.4 ± 0.2 | 1.2 ± 0.2 | 2.5 ± 0.5 |
| Rosid I (Fabidae) | Malpighiales | Salicaceae | P. trichocarpa | 0.9 ± 0.2 | 1.5 ± 0.2 | 4.9 ± 0.8 |
| Fabales | Fabaceae | G. max | 1.0 ± 0.1 | 1.7 ± 0.3 | 4.7 ± 0.6 | |
| M. truncatula | 1.6 ± 0.4 | 2.5 ± 0.3 | 1.5 ± 0.2 | |||
| Rosid II (Malvidae) | Brassicales | Brassicaceae | A. thaliana | 1.7 ± 0.4 | 1.4 ± 0.3 | 5.6 ± 1.6 |
| B. rapa | 1.2 ± 0.1 | 1.9 ± 0.2 | 3.6 ± 0.5 | |||
| Asterid I | Solanales | Solanaceae | S. lycopersicum | 1.2 ± 0.1 | 1.6 ± 0.3 | 3.7 ± 0.6 |
| S. tuberosum | 0.9 ± 0.2 | 1.3 ± 0.1 | 3.9 ± 1.3 | |||
The ratios of the two means (clustered/global) of the TE numbers found in the flanking regions of TS and CYP genes are shown. The means are the μ-parameter values of the negative binomial distribution (Materials and Methods). TE classes I, II, and III are retrotransposons, DNA transposons, and MITEs, respectively.
Concerning the role of MITEs in cluster evolution, there are three possible scenarios. MITEs may insert into the flanking regions of TS and CYP genes, and then through duplication/recombination events independently bring different TS and CYP genes together to form various TS/CYP clusters. These may in turn provide the basis for the formation of larger biosynthetic gene clusters that also include genes encoding other types of enzymes. In this case, MITEs may have played a role in cluster formation.
In a second scenario MITEs, although overrepresented, may not have contributed to cluster formation. In this case, a few ancestral TS/CYP clusters may have formed through unknown mechanisms and by chance some MITEs inserted in the flanking regions of these ancestral TS/CYP clusters. The TS/CYP clusters and associated MITEs may then have amplified together in the genome through “block duplication.” In this scenario the MITEs did not play any role in cluster formation. In a third scenario, TS/CYP clusters may have formed independently of MITEs in different plant species/families. However, during the amplification of MITEs within plant genomes some of these elements may have inserted in the flanking regions of TS/CYP clusters and been maintained because they may have contributed favorably to coordinated expression of the gene clusters. In this scenario MITEs may have been important for establishment of cluster regulation but not for cluster formation per se.
To address the second possibility, we performed global interspecific pairwise alignments of 20-kb flanking regions of clustered TS/CYP pairs, as well as nonclustered TS and CYP genes. In the case of the monocot species, the average nucleotide identities were 43.18 ± 1.38% and 43.34 ± 0.68% for the flanking sequences of the clustered and nonclustered TS/CYP genes, respectively, while those for the eudicots were 43.31 ± 5.06% and 43.56 ± 3.57%, respectively. Flanking sequences of 20 kb were selected to reduce the likelihood of rearrangements. However, these 20-kb regions could include homologous coding regions that may affect identity values. To exclude this possibility, we then used the same approach to find average identities based on 200-bp flanking regions. These values constituted 43.81 ± 1.52% (clustered) and 43.53 ± 1.62% (nonclustered) for the monocots, and 45.59 ± 4.25% (clustered) and 45.36 ± 5.26% (nonclustered) for the eudicots. Thus, no significant difference was found either between the clustered and nonclustered TS/CYP complements or between two computations based on the 20-kb or 200-bp TS/CYP flanks. This lack of similarity of the flanking sequences of the TS/CYP clustered complement and nonclustered global complement among the species studied suggests that the second scenario is unlikely.
A third scenario may be that the MITEs may have inserted into TS/CYP regions postcluster formation, and that they may have functions in the establishment of coordinate expression of terpene biosynthetic gene clusters. Indeed, mapping of MITEs to 1,000-bp upstream regions of these genes revealed that MITEs were clearly more abundant in the 100-bp regions upstream of the eudicot clustered TS/CYP genes, compared with the eudicot global and both monocot complements (Fig. 3).
Fig. 3.
MITE landscape near 5′-ends of TS/CYP genes. The distributions of MITEs upstream of clustered and nonclustered genes are shown in blue and brown, respectively. MITEs prevail in the 100-bp upstream gene region in clustered TS/CYP pairs in eudicots (density peak) and are absent in eudicot nonclustered and monocot clustered complements, or are found as a single occurrence in the monocot nonclustered complement.
Because MITEs are overrepresented in the 100-bp regions upstream of the clustered genes, we then repeated the computational analysis of the local MITE density in the TS/CYP gene flanking regions of the eudicots with the following modification. In this case, to exclude possible effects of MITEs located in the immediate upstream 5′ regions and in gene introns, we used gene spacers within 100-kb TS/CYP flanks that lacked 100-bp sequences at both spacer ends (Materials and Methods). The lack of 100-bp sequences eliminated possible overrepresentation of MITEs in close proximity to the genes. The results were very similar to those obtained for the full-size flanking sequences consisting of gene-coding sequences, introns, and full-size spacers (Table 2 and SI Appendix, Table S3). Thus, although MITEs are overrepresented in the 100-bp 5′ regions of clustered TS/CYP genes in eudicots, they are also overrepresented along the spacer sequences within the continuous genomic region (100 kb in this study) adjacent to the TS/CYP pairs. This analysis provides further support for a structural role for MITEs in cluster formation by MITE-mediated recombination. Because MITEs have also previously been reported to influence the expression of nearby genes, either by acting as enhancers or through epigenetic repression (44–47), they may also have roles in regulation of expression of clustered genes.
Further analysis of these MITEs showed that different families of these TEs are overrepresented, underrepresented, or absent in the vicinities of the TS/CYP genes of the clustered and global complements. Overall, eight eudicot and four monocot genomes studied previously were analyzed, with the exception of the three new genomes of A. trichopoda, A. coerulea, and K. fedtschenkoi (Table 2). In total, among the eudicot species, the numbers of MITEs associated with the clustered and global TS/CYPs constituted 58 and 567, respectively. In the case of the monocot genomes, these numbers were 1,207 and 13,917, respectively. The MITE families that show twofold greater or lower abundance in the regions of clustered TS/CYP gene pairs compared with the counterpart complement (clustered or global) are shown in Table 3. Also shown are MITEs that are associated with only the clustered TS/CYP genes or only with the nonclustered TS/CYP genes on the whole-genome scale.
Table 3.
MITE families associated with the TS and CYP genes of the clustered and global complements of the eudicot and monocot plants
| Plant group | ||
| Presence | Eudicots | Monocots |
| Clustered, overrepresented | Explorer (PIF/Harbinger) | MITE-adh, type H (PIF/Harbinger) |
| Snabo (Mutator) | Helia (PIF/Harbinger) | |
| mPIF (PIF/Harbinger) | ||
| MITE-adh-2 (Tc1/Mariner) | ||
| MDM (Mutator) | ||
| Pop | ||
| Clustered, underrepresented | MITE-adh-11 (Tc1/Mariner) | MITE-adh-8 (PIF/Harbinger) |
| Buhui (PIF/Harbinger) | ||
| ID-4 (PIF/Harbinger) | ||
| Amy/LTP (Mutator) | ||
| p-SINE1 | ||
| Talisker | ||
| Only clustered | MITE-adh, type J (Tc1/Mariner) | Not found |
| Only global | MITE-adh, type K (PIF/Harbinger) | Stola (PIF/Harbinger) |
| mPIF (PIF/Harbinger) | MITE-adh-6 (PIF/Harbinger) | |
| MITE-adh-2 (Tc1/Mariner) | ID-2 (PIF/Harbinger) | |
| MITE-adh-12 (Tc1/Mariner) | Casin (PIF/Harbinger) | |
| Frequent Flyer (Tc1/Mariner) | MITE-adh-10 (Mutator) | |
| MITE-adh-7 | ECR (Mutator) | |
| MITE-adh-3 (Tc1/Mariner) | ||
| MITE-adh-4 (Tc1/Mariner) | ||
| Delay (hAT) | ||
MITEs with twofold greater or lower abundance compared with the counterpart complement are shown. Those associated only with the clustered TS/CYP genes or only with the nonclustered global complement of TS and CYP genes are also represented. See Dataset S5 for additional information.
Origin of Cluster Formation by TS/CYP Blocks in Eudicots.
Our previous work revealed a strong correlation between the TS/TS and CYP/CYP sequence identity values associated with different TS/CYP pairs in eudicots, providing evidence for a scenario in which terpene biosynthetic gene clusters may be established by a TS/CYP “block duplication” mechanism. Conversely, in monocots, where no such correlation was observed, clusters appear to have formed de novo by a mix-and-match mechanism (17).
The availability of the three recently annotated new genome sequences for A. trichopoda, A. coerulea, and K. fedtschenkoi opened up the opportunity to investigate the evolutionary time period within which this block mechanism of cluster formation arose. The new genomes are positioned at principle divergence points on the plant evolutionary tree: A. trichopoda (Amborellales order) represents the most ancient group of nonconifer higher plants (Magnoliophyta); A. coerulea (Ranunculales) is a member of the basal eudicots; and K. fedtschenkoi (Saxifragales order) belongs to the superrosids, the parent clade of the rosids (Fig. 4B).
Fig. 4.
(A) Correlation analysis of the relatedness of TS/CYP pairs from the three new genomes. A significant correlation of TPS/CYP71 clan pairs was found only in the case of K. fedtschenkoi (green circles; r = 0.42, P = 2⋅10−7). Analysis of A. trichopoda vs. eudicots (yellow circles) and A. coerulea vs. eudicots (brown circles) revealed no correlation between pairs of this combination (r = 0.02 and r = 0.14, respectively). The pairs of the TTC/CYP71 clan combination were not correlated either in the eudicot species (blue rectangles; r = 0.28, P = 0.69), or in A. coerulea vs. eudicots (brown rectangles; r = 0.20) and K. fedtschenkoi vs. eudicots (green rectangles; r = 0.11) and appear to have assembled de novo in all clades studied. The gray background represents highly correlated TS/CYP pairs from the eudicot genomes studied previously (17). (B) Schematic illustrating the key time points in the evolution of the main clades for which representatives were involved in this study (the asterid clade is not shown). A. trichopoda, A. coerulea, and K. fedtschenkoi, for which genome sequences have recently become available, represent principle divergence points in plant evolution. The two mechanisms of TS/CYP cluster formation (mix-and-match or block duplication) are shown on the right. The block mechanism arose within a time period of 10–30 My in between the emergence of the basal eudicots and superrosids. The numbers on the right designate the evolutionary times (Mya) of the origin of the Ranunculales and Saxifragales orders that belong to basal eudicots and superrosids, respectively.
Our analyses of the local and global TE distributions in these new genomes revealed that MITE sequences were predominantly implicated in the formation of TS/CYP clusters in A. coerulea and K. fedtschenkoi, as well as in the rosid and asterid plants studied (Table 2). This suggests that, unlike the magnoliophyta and monocots, the principal involvement of MITEs in cluster formation in eudicot species may have occurred as early as in basal eudicot plants, of which A. coerulea is a representative. Nevertheless, the TS/CYP block duplication mechanism of building biosynthetic gene clusters is absent in this species and first appears in the evolutionarily younger superrosid clade, according to the correlation between the TS/TS and CYP/CYP identities in K. fedtschenkoi. This follows from Fig. 4A, which shows the dependencies between the clustered TPS/TPS and CYP/CYP (71 clan) sequence identities in A. trichopoda, A. coerulea, and K. fedtschenkoi (Fig. 4A, Top and Middle), compared with 51 such pairs from the 12 eudicot genomes studied previously (17) (gray background in Fig. 4A). In the present study, a correlation was observed in the case of K. fedtschenkoi (superrosids) but not in A. coerulea (basal eudicots), which suggests the emergence of the block duplication mechanism at some time between the origins of the basal eudicots and the superrosids.
TSs comprise the related superfamily of biosynthetic enzymes involved in the synthesis of the backbones of terpene natural products, that includes terpene synthases (TPSs) and triterpene cyclases (TTCs) (48). The CYPs similarly are a large superfamily of enzymes that can be classified into clans and their constituent families (49, 50). According to our previous results, three particularly abundant nonrandom TS/CYP combinations exist in eudicot species. They involve pairs of TPSs with CYPs belonging to the CYP71 clan (TPS/CYP71), and pairs of TTCs with CYPs belonging to either the CYP71 clan (TTC/CYP71 clan) or the CYP716 family (the latter belongs to the CYP85 clan) (TTC/CYP716) (17). The TTC/CYP71 clan combinations in the eudicot species in our earlier analysis included TTCs associated with CYP705 family CYPs in the Brassicaceae, and three other nonpseudogene CYPs from B. rapa, G. max, and S. lycopersicum (CYP76C9, CYP98A66, and CYP73A96, respectively). Two of these three pairs were considered to be random (no correlation) and were excluded from our previous analysis. With more representatives of the TTC/CYP71 clan combination from the A. coerulea and K. fedtschenkoi genomes, the present study revealed that there is no correlation between A. coerulea and eudicots, or between K. fedtschenkoi and eudicots. In addition, no correlation was found between the TTC/CYP71 clan pairs within the eudicot group. All these pairs are located aside of the correlated eudicot pairs (Fig. 4A, Bottom).
Thus, the block duplication mechanism in eudicot species is relevant for the TPS/CYP71 and TTC/CYP716 combinations, and also for the TTC/CYP705 pairs (the latter being specific to the Brassicaceae). In contrast, it appears that the TTC/CYP71 gene pairs have assembled de novo in all clades studied, including eudicots. Unlike the other combinations, a notable feature of this combination is that the TTC sequences in these pairs have significantly lower homology compared with their CYP71 clan counterparts.
A. coerulea and K. fedtschenkoi belong to the Ranunculales and Saxifragales orders, respectively. Therefore, phylogenetic data and paleobotanic findings concerning these systematic categories may shed light on the timing of emergence of the block mechanism for cluster assembly. Phylogenetic analysis indicates that the evolutionary age of the crown-group Ranunculales may be in the region of 113.2–132.5 Mya (51–54). A recently discovered fossil of the extinct eudicot Leefructus mirus, belonging to the Ranunculales order, has been dated at 122.6–125.8 Mya (55). Phylogenetic estimates of the date of origin of the Saxifragales order are in the region of 90–120 Mya (56, 57), while the fossil record suggests that radiation of the Saxifragales may have occurred between 89.5 and 110 Mya (58, 59). Collectively, these data indicate that the block mechanism arose in a 10- to 30-My period around 90–120 Mya (Fig. 4B).
Conclusion
In summary, TEs are likely to have played a key role in metabolic diversification in plants. They can mediate large changes in chromosomal architecture, causing deletions, inversions, translocations, and other rearrangements. They may also influence regulation of gene expression through cis-mediated and epigenetic effects. Our results suggest that MITEs may play a predominant role in cluster formation in eudicots, compared with the magnoliophyta and monocots, while the involvement of retrotransposons and DNA transposons in the process appears to have been less significant. The MITE global chromosomal profiles are consistent with the cluster distribution along the chromosomes in the majority of the monocot and eudicot species studied. Retrotransposons do not seem to have notable effect, with the possible exception of the Solanaceae. Overall, considering the global and local MITE distributions, local regions of MITE abundance seem to be more important for cluster formation.
Our understanding of the role of TEs in shaping plant genome architecture and in metabolic diversification will continue to expand as more sequenced plant genomes become available, and as we learn more about the diversity of TEs harbored within plant genomes. Our results further suggest that the block mechanism of operon-like cluster formation in eudicots arose within a 10- to 30-My period at some point between the emergence of the basal eudicots and the superrosids.
Materials and Methods
Databases and Genomic Sequences.
The genomic sequences, genomic and functional annotations, as well as genome versions of the eudicot and monocot species, with the exception of the three new genomes, are indicated in ref. 17. The new genomes of A. trichopoda (v.1.0), A. coerulea (v3.1), and K. fedtschenkoi (v.1.1) were obtained from the Phytozome database (www.phytozome.net).
The sequences of the TEs belonging to the three classes were downloaded from the Plant Repeat Database (34) (plantrepeats.plantbiology.msu.edu/index.html; for TE sequences see Datasets S2–S4). This database also provided MITE assignments. The PLACE database (pmite.hzau.edu.cn) (60) was used to assign the MITE sequences to corresponding superfamilies.
Mining of the genomes of A. trichopoda, A. coerulea, and K. fedtschenkoi for TS and CYP genes was carried out as described previously (17). Clustered TS/CYP gene pairs were initially detected using a 100-kb window size. However, to exclude stochastic pairs, 50 kb was used as a prerogative for inclusion of TS/CYP gene pairs in our analyses. TS genes were classified as either the TPS family or as TTCs (49). The CYP genes from the A. trichopoda and A. coerulea genomes were previously assigned by D. Nelson, University of Tennessee, Health Science Center, Memphis, TN (CYP database; drnelson.uthsc.edu/CytochromeP450.html). The K. fedtschenkoi CYP genes were assigned according to known sequences using the BLAST search engine (61) of the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov) and the Uniprot database (www.uniprot.org).
Software.
Customized software was used to identify TEs and calculate TE density (TE/kb) as described previously (17). To find predicted TEs in plant genomes, we took advantage of the BLASTn search engine (62). A stand-alone BLASTv2.2.28+ (61) database of the plant genomes was created, and sequences of each of the three TE classes were piped through it to give a BLAST output file with DNA/DNA alignments to corresponding sequences in each of the plant genomes. The BLAST command line included the following parameters: word_size 11, reward 1, penalty -1, gapopen 3, gapextend 1, evalue 0.001. The program then parsed the output file and extracted all necessary information, essentially alignment coordinates and identities. The identified genomic sequences with coverage of 50% or greater and nucleotide sequence identity of 60% or greater were automatically selected. These stringent parameters were chosen to select recent TEs. Next, the program joined overlapping coordinates and computed coordinates of predicted TE sequences. To find the TE density in the clustered TS/CYP, a separate BLAST database with the TS and CYP flank sequences was built. Because the genomic coordinates of the clustered TS/CYP genes used in this study are known, their 100-kb flanks were readily extracted from the genomic sequences (see below). The same procedure was used for the global TS and CYP complements of each genome. TE sequences were piped through this BLAST database in the same way. Based on the suggested TE coordinates, and the known chromosome sizes and the size of the TS/CYP flanks, the same customized software was used to build the global TE distribution profiles and find the TE density values in the TS/CYP flanks. The latter were used to compute the μ-parameters for the clustered and global TS/CYP complements, their ratios, and SDs. This approach was also used to study MITE local density in the reduced TS/CYP flanks consisting of only the spacer sequences, which lacked introns and upstream 100 bp of the 5′-gene regions (see also below).
Because chromosomal location of clustered TS/CYP pairs corresponds to a coordinate of the principle cluster member, which is TS, it is relevant to consider coordinates of clustered TSs as chromosomal cluster locations. Based on this assumption, a customized program was created to simulate random cluster distribution along chromosomes. For each genome, the program randomly shuffled chromosomal gene coordinates (gene start), selected a number of random coordinates according to a number of clustered TS/CYP pairs located on the chromosome in question, placed each random coordinate in a relevant chromosomal interval from 0.1 to 1.0, and computed frequencies of coordinate occurrence in each interval. Finally, the frequencies were expressed as their percentage values. This process was cycled 100 times for each genome.
The customized PERL script based on the Bioperl modules was used to extract the TS/CYP flanking sequences (200 bp, 20 kb, and 100 kb; see Results and Discussion) from each genome studied according to the known gene coordinates. The same script was used to extract the gene spacer sequences at the whole-genome scale, select those spacers that were located within the 100 kb TS/CYP flanks, and join them for each nonclustered TS or CYP, or for each clustered TS/CYP pair. The spacer coordinates were reduced by 100 bp at the both spacer ends to eliminate upstream 100 bp of the 5′-gene regions.
Another PERL script was used to find identities between the TS/CYP flanking sequences (200-bp, full-size 20 kb, or reduced 100-kb flanks consisted of spacers) across the genomes studied. The CLUSTALW program (63) incorporated in this script was used for the alignments of the TS/CYP flanking sequences.
Statistics.
The “R”-functions fitdistr() and goodfit() were used to find the best fitting of the distributions of the TE density values for each TE class in each genome. The empirical data were best fitted to the negative binomial distribution with the mean represented by the μ-parameter. A bootstrap approach was used to find SDs for each ratio of two μ-parameters (clustered/global). The MITE frequencies in 1,000-bp upstream gene regions were found using the Excel program. The same program was used to find gene frequencies (global gene density profiling) in the chromosomal intervals of each genome studied. The χ2 test was used to compare different profiles.
Supplementary Material
Acknowledgments
A.E.O.’s laboratory is supported by the United Kingdom Biotechnological and Biological Sciences Research Council Institute Strategic Programme Grant ‘Molecules from Nature’ (BB/P012523/1) and the John Innes Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721318115/-/DCSupplemental.
References
- 1.Frey M, et al. Analysis of a chemical plant defense mechanism in grasses. Science. 1997;277:696–699. doi: 10.1126/science.277.5326.696. [DOI] [PubMed] [Google Scholar]
- 2.Qi X, et al. A gene cluster for secondary metabolism in oat: Implications for the evolution of metabolic diversity in plants. Proc Natl Acad Sci USA. 2004;101:8233–8238. doi: 10.1073/pnas.0401301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ. Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 2004;135:2098–2105. doi: 10.1104/pp.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimura K, et al. Identification of a biosynthetic gene cluster in rice for momilactones. J Biol Chem. 2007;282:34013–34018. doi: 10.1074/jbc.M703344200. [DOI] [PubMed] [Google Scholar]
- 5.Field B, Osbourn AE. Metabolic diversification—Independent assembly of operon-like gene clusters in different plants. Science. 2008;320:543–547. doi: 10.1126/science.1154990. [DOI] [PubMed] [Google Scholar]
- 6.Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ. CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell. 2009;21:3315–3325. doi: 10.1105/tpc.108.063677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falara V, et al. The tomato terpene synthase gene family. Plant Physiol. 2011;157:770–789. doi: 10.1104/pp.111.179648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Field B, et al. Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc Natl Acad Sci USA. 2011;108:16116–16121. doi: 10.1073/pnas.1109273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takos AM, Rook F. Why biosynthetic genes for chemical defense compounds cluster. Trends Plant Sci. 2012;17:383–388. doi: 10.1016/j.tplants.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Winzer T, et al. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science. 2012;336:1704–1708. doi: 10.1126/science.1220757. [DOI] [PubMed] [Google Scholar]
- 11.Castillo DA, Kolesnikova MD, Matsuda SP. An effective strategy for exploring unknown metabolic pathways by genome mining. J Am Chem Soc. 2013;135:5885–5894. doi: 10.1021/ja401535g. [DOI] [PubMed] [Google Scholar]
- 12.Itkin M, et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science. 2013;341:175–179. doi: 10.1126/science.1240230. [DOI] [PubMed] [Google Scholar]
- 13.Krokida A, et al. A metabolic gene cluster in Lotus japonicus discloses novel enzyme functions and products in triterpene biosynthesis. New Phytol. 2013;200:675–690. doi: 10.1111/nph.12414. [DOI] [PubMed] [Google Scholar]
- 14.Matsuba Y, et al. Evolution of a complex locus for terpene biosynthesis in solanum. Plant Cell. 2013;25:2022–2036. doi: 10.1105/tpc.113.111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King AJ, Brown GD, Gilday AD, Larson TR, Graham IA. Production of bioactive diterpenoids in the euphorbiaceae depends on evolutionarily conserved gene clusters. Plant Cell. 2014;26:3286–3298. doi: 10.1105/tpc.114.129668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang Y, et al. Plant science. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science. 2014;346:1084–1088. doi: 10.1126/science.1259215. [DOI] [PubMed] [Google Scholar]
- 17.Boutanaev AM, et al. Investigation of terpene diversification across multiple sequenced plant genomes. Proc Natl Acad Sci USA. 2015;112:E81–E88. doi: 10.1073/pnas.1419547112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuba Y, Zi J, Jones AD, Peters RJ, Pichersky E. Biosynthesis of the diterpenoid lycosantalonol via nerylnerly diphosphate in Solanum lycopersicum. PLoS One. 2015;10:e0119302. doi: 10.1371/journal.pone.0119302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sohrabi R, et al. In planta variation of volatile biosynthesis: An alternative biosynthetic route to the formation of the pathogen-induced volatile homoterpene DMNT via triterpene degradation in Arabidopsis roots. Plant Cell. 2015;27:874–890. doi: 10.1105/tpc.114.132209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hen-Avivi S, et al. A metabolic gene cluster in the wheat W1 and the barley Cer-cqu loci determines β-diketone biosynthesis and glaucousness. Plant Cell. 2016;28:1440–1460. doi: 10.1105/tpc.16.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nützmann H-W, Huang A, Osbourn A. Plant metabolic clusters—From genetics to genomics. New Phytol. 2016;211:771–789. doi: 10.1111/nph.13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider LM, et al. 2017. The Cer-cqu gene cluster determines three key players in a β-diketone synthase polyketide pathway synthesizing aliphatics in epicuticular waxes. J Exp Bot, 10.1093/jxb/erw105; erratum in J Exp Bot (2017) 68:5009.
- 23.Zhou Y, et al. Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nat Plants. 2016;2:16183. doi: 10.1038/nplants.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aubourg S, Lecharny A, Bohlmann J. Genomic analysis of the terpenoid synthase ( AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics. 2002;267:730–745. doi: 10.1007/s00438-002-0709-y. [DOI] [PubMed] [Google Scholar]
- 25.Dutartre L, Hilliou F, Feyereisen R. Phylogenomics of the benzoxazinoid biosynthetic pathway of Poaceae: Gene duplications and origin of the Bx cluster. BMC Evol Biol. 2012;12:64. doi: 10.1186/1471-2148-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medema MH, Osbourn A. Computational genomic identification and functional reconstitution of plant natural product biosynthetic pathways. Nat Prod Rep. 2016;33:951–962. doi: 10.1039/c6np00035e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schläpfer P, et al. Genome-wide prediction of metabolic enzymes, pathways and gene clusters in plants. Plant Physiol. 2017;173:2041–2059, and erratum (2018) 176:2583. doi: 10.1104/pp.16.01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christianson DW. Structural and chemical biology of terpenoid cyclases. Chem Rev. 2017;117:11570–11648. doi: 10.1021/acs.chemrev.7b00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang CRL, Burns KH, Boeke JD. Active transposition in genomes. Annu Rev Genet. 2012;46:651–675. doi: 10.1146/annurev-genet-110711-155616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennetzen JL, Wang H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol. 2014;65:505–530. doi: 10.1146/annurev-arplant-050213-035811. [DOI] [PubMed] [Google Scholar]
- 31.Konkel MK, Batzer MA. A mobile threat to genome stability: The impact of non-LTR retrotransposons upon the human genome. Semin Cancer Biol. 2010;20:211–221. doi: 10.1016/j.semcancer.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmeister D, Keller NP. Natural products of filamentous fungi: Enzymes, genes, and their regulation. Nat Prod Rep. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- 33.Chu HY, Wegel E, Osbourn A. From hormones to secondary metabolism: The emergence of metabolic gene clusters in plants. Plant J. 2011;66:66–79. doi: 10.1111/j.1365-313X.2011.04503.x. [DOI] [PubMed] [Google Scholar]
- 34.Ouyang S, Buell CR. The TIGR plant repeat databases: A collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004;32:D360–D363. doi: 10.1093/nar/gkh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kejnovsky E. Plant transposable elements: Biology and evolution. In: Wendel JF, et al., editors. Plant Genome Diversity. Vol 1. Springer; Vienna: 2012. pp. 17–33. [Google Scholar]
- 37.Neumann P, et al. Plant centromeric retrotransposons: A structural and cytogenetic perspective. Mob DNA. 2011;2:4. doi: 10.1186/1759-8753-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsukahara S, et al. Centromere-targeted de novo integrations of an LTR retrotransposon of Arabidopsis lyrata. Genes Dev. 2012;26:705–713. doi: 10.1101/gad.183871.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du J, et al. Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: Insights from genome-wide analysis and multi-specific comparison. Plant J. 2010;63:584–598. doi: 10.1111/j.1365-313X.2010.04263.x. [DOI] [PubMed] [Google Scholar]
- 40.Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci USA. 2007;104:9352–9357. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 42.Bureau TE, Wessler SR. Tourist: A large family of small inverted repeat elements frequently associated with maize genes. Plant Cell. 1992;4:1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 44.Wessler SR, Bureau TE, White SE. LTR-retrotransposons and MITEs: Important players in the evolution of plant genomes. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 45.Naito K, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461:1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- 46.Yang G, et al. A two-edged role for the transposable element Kiddo in the rice ubiquitin2 promoter. Plant Cell. 2005;17:1559–1568. doi: 10.1105/tpc.104.030528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei L, et al. Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice. Proc Natl Acad Sci USA. 2014;111:3877–3882. doi: 10.1073/pnas.1318131111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao Y, Honzatko RB, Peters RJ. Terpenoid synthase structures: A so far incomplete view of complex catalysis. Nat Prod Rep. 2012;29:1153–1175. doi: 10.1039/c2np20059g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson D, Werck-Reichhart D. A P450-centric view of plant evolution. Plant J. 2011;66:194–211. doi: 10.1111/j.1365-313X.2011.04529.x. [DOI] [PubMed] [Google Scholar]
- 50.Nelson DR. The cytochrome p450 homepage. Hum Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson CL, Bremer K, Friis EM. Dating phylogenetically basal eudicots using rbcL sequences and multiple fossil reference points. Am J Bot. 2005;92:1737–1748. doi: 10.3732/ajb.92.10.1737. [DOI] [PubMed] [Google Scholar]
- 52.Magallón S, Castillo A. Angiosperm diversification through time. Am J Bot. 2009;96:349–365. doi: 10.3732/ajb.0800060. [DOI] [PubMed] [Google Scholar]
- 53.Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 2015;207:437–453. doi: 10.1111/nph.13264. [DOI] [PubMed] [Google Scholar]
- 54.Tank DC, et al. Nested radiations and the pulse of angiosperm diversification: Increased diversification rates often follow whole genome duplications. New Phytol. 2015;207:454–467. doi: 10.1111/nph.13491. [DOI] [PubMed] [Google Scholar]
- 55.Sun G, Dilcher DL, Wang H, Chen Z. A eudicot from the Early Cretaceous of China. Nature. 2011;471:625–628. doi: 10.1038/nature09811. [DOI] [PubMed] [Google Scholar]
- 56.Jian S, et al. Resolving an ancient, rapid radiation in Saxifragales. Syst Biol. 2008;57:38–57. doi: 10.1080/10635150801888871. [DOI] [PubMed] [Google Scholar]
- 57.Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA. 2010;107:4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magallón S, Crane PR, Herendeen PS. Phylogenetic pattern, diversity and diversification of eudicots. Ann Mo Bot Gard. 1999;86:297–372. [Google Scholar]
- 59.Hermsen EJ, Gandolfo MA, Nixon KC, Crepet WL. The impact of extinct taxa on understanding the early evolution of angiosperm clades: An example incorporating fossil reproductive structures of Saxifragales. Plant Syst Evol. 2006;260:141–169. [Google Scholar]
- 60.Chen J, Hu Q, Zhang Y, Lu C, Kuang H. P-MITE: A database for plant miniature inverted-repeat transposable elements. Nucleic Acids Res. 2013;42:D1176–D1181. doi: 10.1093/nar/gkt1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tao T. 2008 BLAST Help (National Center for Biotechnology Information, Bethesda). Available at https://www.ncbi.nlm.nih.gov/books/NBK1762/. Accessed January 2, 2013.
- 62.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.