Significance
Sex-chromosome dosage (SCD) effects on human gene expression are central to the biology of sex differences and sex-chromosome aneuploidy syndromes but are challenging to study given the cosegregation of SCD and gonadal status. We address this obstacle by systematically modeling SCD effects on genome-wide expression data from a large and rare cohort of individuals with diverse SCDs (XO, XX, XXX, XXXX, XY, XXY, XYY, XXYY, and XXXXY). Our findings update current models of sex-chromosome biology by (i) pinpointing a core set of X- and Y-linked genes with obligate SCD sensitivity, (ii) discovering several noncanonical modes of X-chromosome dosage compensation, and (iii) dissecting complex regulatory effects of X-chromosome dosage on large autosomal gene networks with key roles in cellular functioning.
Keywords: sex chromosomes, X-inactivation, sex differences, Turner syndrome, Klinefelter syndrome
Abstract
A fundamental question in the biology of sex differences has eluded direct study in humans: How does sex-chromosome dosage (SCD) shape genome function? To address this, we developed a systematic map of SCD effects on gene function by analyzing genome-wide expression data in humans with diverse sex-chromosome aneuploidies (XO, XXX, XXY, XYY, and XXYY). For sex chromosomes, we demonstrate a pattern of obligate dosage sensitivity among evolutionarily preserved X-Y homologs and update prevailing theoretical models for SCD compensation by detecting X-linked genes that increase expression with decreasing X- and/or Y-chromosome dosage. We further show that SCD-sensitive sex-chromosome genes regulate specific coexpression networks of SCD-sensitive autosomal genes with critical cellular functions and a demonstrable potential to mediate previously documented SCD effects on disease. These gene coexpression results converge with analysis of transcription factor binding site enrichment and measures of gene expression in murine knockout models to spotlight the dosage-sensitive X-linked transcription factor ZFX as a key mediator of SCD effects on wider genome expression. Our findings characterize the effects of SCD broadly across the genome, with potential implications for human phenotypic variation.
Disparity in sex-chromosome dosage (SCD) is fundamental to the biological definition of sex in almost all eutherian mammals: Females carry two X chromosomes, while males carry an X and a Y chromosome. The presence of the Y-linked SRY gene determines a testicular gonadal phenotype, while its absence allows development of ovaries (1, 2). Sexual differentiation of the gonads leads to hormonal sex differences that have traditionally been considered the major proximal cause for extragonadal phenotypic sex differences. However, diverse studies, including recent work in transgenic mice that uncouple Y chromosome and gonadal status, have revealed direct SCD effects on several sex-biased metabolic, immune, and neurological phenotypes (3).
These findings together with reports of widespread transcriptomic differences between preimplantation XY and XX embryos (4) suggest that SCD has gene-regulatory effects independently of gonadal status. However, the genome-wide consequences of SCD remain poorly understood, especially in humans, where experimental dissociation of SCD and gonadal status is not possible. Understanding these regulatory effects is critical for clarifying the biological underpinnings of phenotypic sex differences and the clinical features of sex-chromosome aneuploidy (SCA) [e.g., Turner (XO) and Klinefelter (XXY) syndrome (5)], which both involve altered risk for several common autoimmune disorders (ADs) and neurodevelopmental disorders (e.g., systemic lupus erythematosus and autism spectrum disorders) (6, 7). Here, we explore the genome-wide consequences of SCD through comparative transcriptomic analyses among humans across a range of dosages including typical XX and XY karyotypes as well as several rare SCA syndromes associated with 1, 3, 4, or 5 copies of the sex chromosomes. We harness these diverse karyotypes to dissect the architecture of dosage compensation among sex-chromosome genes and to systematically map the regulatory effects of SCD on autosomal gene expression in humans. These research goals also inform more general questions regarding the effects of aneuploidy on genome function. In particular, the wide range of chromosome-dosage variation in SCAs can help determine if previously reported inverse effects of aneuploidy on gene expression in maize and Drosophila (i.e., negative cis and trans effects of supernumerary chromosomes) (8, 9) also operate in humans.
We model SCD effects using gene-expression profiles in a total of 471 lymphoblastoid cell lines (LCLs) from (i) a core sample of 68 participants (12 XO, 10 XX, 9 XXX, 10 XY, 8 XXY, 10 XYY, and 9 XXYY) yielding for each sample genome-wide expression data for 19,984 autosomal and 894 sex-chromosome genes using the Illumina oligonucleotide BeadArray platform (Methods, SI Appendix, Text S1, and Dataset S1), and (ii) an independent set of validation/replication samples from 403 participants (6 XO, 146 XX, 22 XXX, 145 XY, 33 XXY, 16 XYY, 17 XXYY, 8 XXXY, and 10 XXXXY) with qRT-PCR measures of expression for genes of interest identified in our core sample (Methods, SI Appendix, Text S2, and Dataset S1). All SCA samples were from individuals with nonmosaic aneuploidy.
Results
Extreme Dosage Sensitivity of Evolutionarily Preserved X-Y Gametologs.
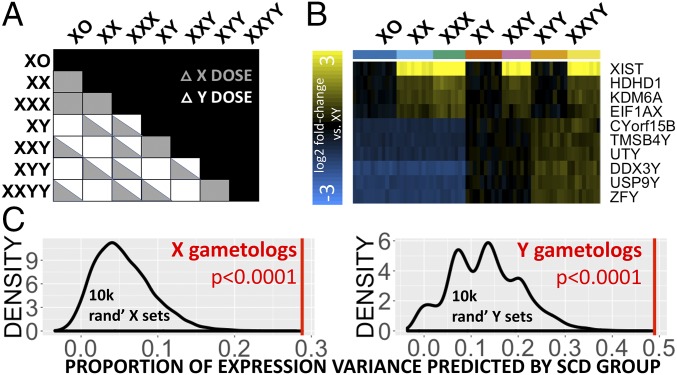
First, to verify our study design as a tool for probing SCD effects on gene expression and to identify core SCD-sensitive genes, we screened all 20,878 genes in our microarray dataset to define which, if any, genes showed a persistent pattern of significant differential expression (DE) (Methods and SI Appendix, Text S1) across all unique pairwise group contrasts involving a disparity in either X- or Y-chromosome dosage (n = 15 and n = 16 contrasts, respectively) (Fig. 1A). Disparities in X-chromosome dosage were always accompanied by statistically significant DE in four genes, which were all X-linked: XIST (the orchestrator of X inactivation) and three other genes known to escape X-chromosome inactivation (HDHD1, KDM6A, and EIF1AX) (10). Similarly, disparities in Y-chromosome dosage always led to statistically significant DE in six Y-linked genes: CYorf15B, DDX3Y, TMSB4Y, USP9Y, UTY, and ZFY. Observed expression profiles for these 10 genes perfectly segregated all microarray samples by karyotype group (Fig. 1B) and could be robustly replicated and extended using available qRT-PCR data for of these genes in the independent sample of 403 LCLs from participants with varying SCD (Methods and SI Appendix, Text S1 and Fig. S1).
Fig. 1.
Consistent gene-expression changes with altered sex-chromosome dosage. (A) Cross-table showing all pairwise SCD group contrasts within the microarray dataset shaded by X- and/or Y-chromosome disparity. (B) Expression heatmap for the 10 genes that show DE across all contrasts involving disparity in X- or Y-chromosome count. The column color bar encodes SCD group membership for each sample. (C) Density plots showing the observed mean proportion of expression variance explained by SCD for 14 gametolog genes (red line) vs. null distribution (black line) of this variable for 10,000 randomly sampled sets of nongametolog sex-linked genes of equal size. Results are provided separately for X and Y chromosomes.
Strikingly, 8 of the 10 genes showing obligatory SCD sensitivity (excepting XIST and HDHD1) are members of a class of 16 sex-linked genes with homologs on both the X and Y chromosomes (i.e., 16 X-Y gene pairs, henceforth referred to as “gametologs”) (11) that are distinguished from other sex-linked genes by (i) their selective preservation in multiple species across ∼300 My of sex-chromosome evolution to prevent male–female dosage disparity, (ii) the breadth of their tissue expression from both sex chromosomes, and (iii) their key regulatory roles in transcription and translation (Dataset S2) (11, 12). Broadening our analysis to all 14 X-Y gametolog pairs present in our microarray dataset, we found that these genes as a group exhibit a heightened degree of SCD sensitivity that distinguishes them from other sex-linked genes (Fig. 1C, empirical P < 0.0001). These findings provide direct evidence that the evolutionary maintenance, broad tissue expressivity, and enriched regulatory functions of X-Y gametologs (12) are indeed accompanied by a distinctive pattern of dosage sensitivity, which firmly establishes these genes as candidate regulators of SCD effects on wider genome function.
Observed Sex-Chromosome Dosage Effects on X- and Y-Chromosome Genes Modify Current Models of Dosage Compensation.
We next harnessed our study design to test the canonical (four-class) model for SCD compensation, which defines four mutually exclusive classes of sex-chromosome genes that would be predicted to have differing responses to changing SCD (13): (i) pseudoautosomal region (PAR) genes, (ii) Y-linked genes, (iii) X-linked genes that undergo X-chromosome inactivation (XCI), and (iv) X-linked genes that escape XCI (XCIE). Under the four-class model, PAR genes would be predicted to increase their expression with increases in X- or Y chromosome count, whereas expression of Y-linked genes would increase linearly with mounting Y-chromosome count. Due to the nonbinary nature of gene silencing with XCI (14), theorized SCD effects on the expression of XCI and XCIE genes represent the extreme ends of an X-chromosome dosage-sensitivity continuum: an X-linked gene that undergoes full silencing with XCI would show no expression change with changes in X-chromosome dosage, whereas an X-linked gene that undergoes complete escape from X-chromosome inactivation would show a linear increase in expression with increasing X-chromosome count.
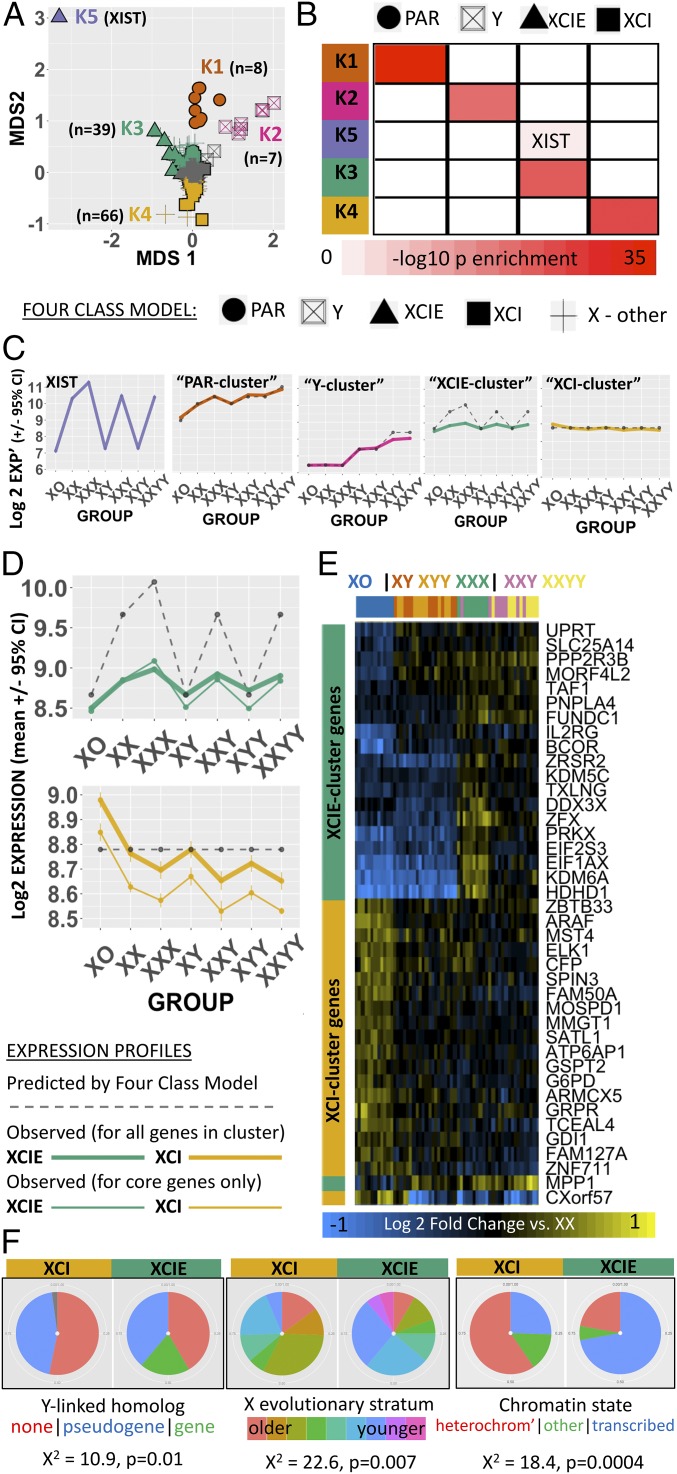
To test this canonical four-class model, we performed unsupervised k-means clustering of all sex-chromosome genes by their average expression in each of the seven karyotype groups represented in our microarray dataset (after normalization by average expression across all karyotype groups). We then compared this empirically defined grouping with that given a priori by the four-class model (Methods and SI Appendix, Text S3). k-Means clustering distinguished six gene clusters (Fig. 2A and SI Appendix, Fig. S2A) which were reproducible across 1,000 bootstrap draws from the LCL microarray dataset sample pool (SI Appendix, Text S3 and S4 and Fig. S2B). These clusters consisted of a single large set of 773 genes with low or undetectable expression levels in most samples (median detection rate of 4/68 samples) and no significant SCD sensitivity, and five smaller groups of genes (k1–k5) with high SCD sensitivity (Fig. 2A, SI Appendix, Fig. S2 A–C, and Datasets S3 and S4). The set of 773 genes with low expression by microarray also showed low/undetectable expression by RNA-sequencing (RNA-seq) in an independent study of 343 LCL lines from the Genotype-Tissue Expression (GTEx) project (SI Appendix, Text S4) (15) and is not considered further in this report.
Fig. 2.
Data-driven partitioning of sex-chromosome genes by dosage sensitivity. (A) 2D multidimensional scaling (MDS) plot of sex-chromosome genes by their mean expression profiles across all seven SCD groups. Genes are coded by both the four-class model (shape) and k-means cluster grouping (color). MDS2 arranges X-linked genes along the established gradient of X-linked dosage sensitivity that ranges from extreme XCIE (XIST) to full XCI. A cluster of low/nonexpressed genes that lack SCD sensitivity is colored gray (SI Appendix, Text S4 and Fig. S2). (B) Cross-table showing enrichment of k-means clusters for four-class model gene groups. (C) Dot and line plots showing observed (solid colored) and predicted (dashed gray) mean expression for each k-means gene cluster across karyotype groups. (D) Close-up of observed (solid colored) vs. predicted (dashed gray) mean expression profiles of XCIE and XCI gene clusters. Observed expression profiles still counter predictions when analysis is restricted to core genes in each cluster with XCIE/XCI statuses that have been confirmed across three independent reports (thick colored line). (E) Heatmap showing normalized (vs. XX mean) expression of dosage-sensitive genes in the XCIE and XCI clusters (rows) for each sample (columns; the color bar encodes the SCD group). (F) Pie-charts showing that XCIE and XCI gene clusters from k-means display mirrored over/underrepresentation for three genomic features that have been linked to XCIE in prior research: (i) persistence of a surviving Y-linked homolog; (ii) location within younger evolutionary strata of the X chromosome; and (iii) presence of euchromatic vs. heterochromatic epigenetic markers.
Each of the five SCD-sensitive clusters of sex-chromosome genes detected by k-means (k1–k5) was specifically enriched for a distinct set of genes within the a priori four-class model (Fig. 2B), defining a PAR cluster (k1), Y cluster (k2, preferential overrepresentation of Y gametologs: odds ratio = 5,213, P = 1.3 × 10−15), XCIE cluster (k3, preferential overrepresentation of X gametologs: odds ratio = 335, P = 3.4 × 10−11), XCI cluster (k4), and a separate cluster for XIST (k5) (Fig. 2B). For three of these clusters—Y-linked, XCIE, and XCI—we observed profiles of dosage sensitivity across karyotype groups that deviated from those predicted by the four-class model (Fig. 2 C and D).
Mean expression of the Y cluster increased in a sublinear stepwise fashion with greater Y-chromosome dosage, indicating that its constituent Y-linked genes may be subject to active dosage compensation. Groupwise fold changes observed by microarray for three out of three of these genes were highly correlated with groupwise fold changes observed by qRT-PCR in an independent sample of 403 participants with varying SCD (SI Appendix, Text S2 and Fig. S2E). Mean expression of the XCIE cluster increased with X-chromosome dosage (as predicted by the four-class model) but did so in a sublinear fashion (Fig. 2 C and D). For seven XCIE-cluster genes with recently published allelic expression imbalance data from LCLs in a female with skewed X-inactivation (16), we found that observed sublinear effects of X-chromosome dosage on expression by microarray were largely consistent with gene-level estimates of incomplete XCIE by RNA-seq (SI Appendix, Text S5 and Fig. S2D). Mean expression of the XCI cluster deviated from predictions of the four-class model by showing a statistically significant inverse relationship with X-chromosome dosage (coefficient for linear effect of X-chromosome count on expression = −0.12, P = 3.8 × 10−15). This phenomenon was reproducible for 60/66 (91%) of individual genes within the cluster (P < 0.05 for negative linear effect of X-chromosome count on expression). Furthermore, noting the relatively small log2 fold change (log2FC) values for reduced XCI-cluster expression with increasing X-chromosome count (Fig. 2D and Dataset S3), we also verified that for most (89%) XCI-cluster genes, observed log2FC differences in expression between XXX and XO groups were statistically significant relative to null fold-change distributions estimated within the same microarray dataset (SI Appendix, Text S6). Thus, observed patterns of gene expression for X-linked genes within the XCI-enriched cluster suggest that increasing X copy number may not involve only the silencing of these genes from the additional inactive X chromosome but also a further repression of their expression from the single active X chromosome. Remarkably, expression of the XCI cluster was also significantly decreased by the presence of a Y chromosome, at the level of both mean cluster expression (coefficient for linear effect of Y-chromosome count on expression = −0.09, P = 1.4 × 10−14) and expression profiles of 48/66 individual cluster genes (P < 0.05 for negative linear effect of Y count on expression). The XCIE cluster manifested an inverted version of this effect whereby increases in Y-chromosome dosage were associated with increased gene expression (P < 6.2 × 10−11 for mean cluster expression and P < 0.05 for 23/39 cluster genes). These findings provide evidence that Y-chromosome status in humans may influence the expression level of X-linked genes independently of circulating gonadal factors.
We took several steps to further probe the unexpected modes of dosage sensitivity observed for the XCI cluster and XCIE cluster. First, we established that observed patterns of dosage sensitivity for these clusters held when analysis was restricted to X-linked genes with only high-confidence annotations for XCIE and XCI status, respectively (Fig. 2D), suggesting that the observed expression profiles were unlikely to be explained by misclassification of X-linked genes by XCI status. Second, we confirmed that the distinct expression profiles for these two gene clusters were reproducible at the level of individual genes and samples. Indeed, unsupervised clustering of microarray samples based on expression of XCI- and XCIE-cluster genes distinguished three broad karyotype groups: females with one X chromosome (XO), males with one X chromosome (XY and XYY), and individuals with an extra X chromosome (XXX, XXY, and XXYY) (Fig. 2E). Third, we validated our data-driven discovery of XCI and XCIE clusters against independently generated X-chromosome annotations (Fig. 2F) which detail three distinct genomic predictors of inactivation status for X-linked genes. Specifically, XCI-cluster genes were relatively enriched (and XCIE cluster genes relatively impoverished) for (i) having lost a Y-chromosome homolog during evolution (χ2 = 10.9, P = 0.01) (17), (ii) being located in older evolutionary strata of the X chromosome (χ2 = 22.6, P = 0.007) (18), and (iii) bearing heterochromatic markers (χ2 = 18.4, P = 0.0004) (19).
Finally, qRT-PCR assays in LCLs from an independent sample of 403 participants with varying SCD validated the fold changes observed in microarray data for five of six of the most SCD-sensitive XCIE- and XCI-cluster genes (Methods and SI Appendix, Text S2 and Fig. S2F). To independently extend these observations, we measured gene expression by qRT-PCR in karyotype groups not represented in our microarray dataset (XXXY and XXXXY) (Methods and SI Appendix, Text S1) and were able to confirm reduction in expression with greater X-chromosome dosage for two of three XCI-cluster genes, NGFRAP1 and CXorf57 (SI Appendix, Fig. S2G,) and Y-chromosome dosage effects upon expression for four of six X-linked genes from the XCI and XCIE clusters (down-regulation: NGFRAP1 and CXorf57; up-regulation: PIM2, and PRKX) (SI Appendix, Fig. S2H). Taken together, these findings update the canonical four-class model of SCD compensation for specific Y-linked and X-linked genes and expand the list of X-linked genes capable of mediating wider phenotypic consequences of SCD variation. The existence of X-linked genes that decrease expression with increasing X-chromosome count also indicates that previously documented inverse effects of aneuploidy on gene expression in maize and Drosophila (8, 9) are also seen in humans.
Context-Specific Disruption of Autosomal Expression by Sex-Chromosome Aneuploidy.
We next leveraged the diverse SCAs represented in our study to assess how SCD variation shapes expression on a genome-wide scale. By counting the total number of differentially expressed genes (DEGs) (Methods) in each SCA group relative to its respective euploid control (i.e., XO and XXX compared with XX; XXY, XYY, and XXYY compared with XY), we detected order-of-magnitude differences in DEG count among SCAs across a range of log2FC cutoffs (SI Appendix, Fig. S3 A and B). We observed an order-of-magnitude increase in DEG count with X-chromosome supernumeracy in males vs. females, which, although previously undescribed, is congruent with the more severe phenotypic consequences of X-supernumeracy in males vs. females (20). Overall, increasing the dosage of the sex chromosome associated with the sex of an individual (i.e., X in females and Y in males) had a far smaller effect than other types of SCD change. Moreover, the ∼20 DEGs seen in XXX contrasted with >1,200 DEGs in XO, revealing a profoundly asymmetric impact of X-chromosome loss vs. gain on the transcriptome of female LCLs, which echoes the asymmetric phenotypic severity of X-chromosome loss (Turner syndrome) vs. gain (XXX) syndromes in females (6). The large number of DEGS in XO included a similar proportion with increases vs. decreases in expression (e.g., 782 XO > XX, 605 XO < XX). Autosomal genes accounted for >75% of all DEGs in females with X-monosomy (XO) and males with X-supernumeracy (XXY and XXYY) but <30% DEGs in all other SCD groups (Methods and SI Appendix, Fig. S3C). These results reveal that SCD changes vary widely in their capacity to disrupt genome function and indicate that differential involvement of autosomal genes is central to this variation. Moreover, SCA differences in LCL DEG count broadly recapitulate SCA differences in phenotypic severity.
Sex-Chromosome Dosage Regulates Large-Scale Gene Coexpression Networks.
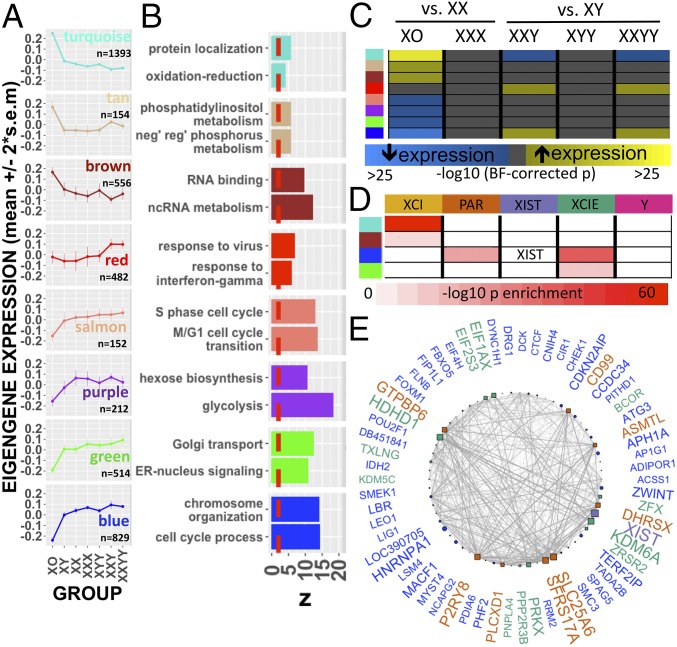
To provide a more comprehensive systems-level perspective on the impact of SCD on genome-wide expression patterns, we leveraged weighted gene coexpression network analysis (WGCNA) (Methods and SI Appendix, Text S7) (21). This analytic approach uses the correlational architecture of gene expression across a set of samples to detect sets (modules) of coexpressed genes. Using WGCNA, we identified 18 independent gene-coexpression modules in our dataset (Dataset S5). We established that these modules were not artifacts of the codifferential expression of genes between groups by demonstrating their robustness to removal of all group effects on gene expression by regression (SI Appendix, Fig. S4A) and after specific exclusion of XO samples (SI Appendix, Fig. S4B), given the extreme pattern of DE in this karyotype. We focused further analysis on modules meeting two independent statistical criteria after correction for multiple comparisons: (i) a significant omnibus effect of SCD group on expression and (ii) significant enrichment for one or more gene ontology (GO) process/function terms (Methods, Fig. 3 A and B, SI Appendix, Text S7, and Dataset S5). These steps defined eight functionally coherent and SCD-sensitive modules (Blue, Brown, Green, Purple, Red, Salmon, Tan, and Turquoise). Notably, the SCD effects we observed on genome-wide expression patterns appeared to be specific to shifts in sex-chromosome gene dosage, as application of our analytic workflow to publically available genome-wide Illumina BeadArray expression data from LCLs in patients with trisomy 21 (Down syndrome) revealed a profile of genome-wide expression change dissimilar to that observed in sex-chromosome trisomies (SI Appendix, Text S8 and Fig. S4C and Dataset S6).
Fig. 3.
WGCNA of sex-chromosome dosage effects. (A) Dot and line plots detailing mean expression (± 95% CI) by SCD group for eight SCD-sensitive and functionally coherent gene-coexpression modules. (B) Top two GO term enrichments for each module exceeding the threshold for statistical significance (dashed red line). (C) Heatmap showing statistically significant DE of gene-coexpression modules between karyotype groups. (D) Cross-tabulation showing enrichment of Turquoise, Brown, Blue, and Green modules for the dosage-sensitive clusters of sex-chromosome genes detected by k-means. (E) Gene-coexpression network for the Blue module showing the top decile of coexpression relationships (edges) between the top decile of SCD-sensitive genes (nodes). Nodes are positioned in a circle for ease of visualization. Node shape distinguishes autosomal (circle) from sex-chromosome (square) genes. Sex-chromosome genes within the Blue module are colored according to their k-means cluster designation. Larger node and gene name sizes reflect greater SCD sensitivity. Edge width indexes the strength of coexpression between gene pairs.
To specify SCA effects on module expression, we compared all aneuploidy groups with their respective gonadal controls (Fig. 3C). Statistically significant differences in modular eigengene expression were seen in the XO, XXY, and XXYY groups, as is consistent with these karyotypes causing larger total DEG counts than the other SCD variations (SI Appendix, Fig. S3A). The largest shifts in module expression were seen in XO and included robust up-regulation of the protein trafficking (Turquoise), metabolism of noncoding RNA (Brown), and phosphatidylinositol metabolism (Tan) modules, alongside down-regulation of the cell-cycle progression, DNA replication/chromatin organization (Blue and Salmon), glycolysis (Purple), and responses to endoplasmic reticular stress (Green) modules. Module DE in groups with supernumerary X chromosomes on an XY background (i.e., XXY and XXYY) involved an inversion of some XO effects, i.e., down-regulation of protein trafficking (Turquoise) and up-regulation of cell-cycle progression (Blue) modules, plus a more karyotype-specific up-regulation of the immune response pathways (Red) module.
The distinctive up-regulation of immune-system genes in samples of lymphoid tissue from males carrying a supernumerary X chromosome carries potential clinical relevance for one of the best-established clinical phenotypes in XXY and XXYY syndromes: a strongly (up to 18-fold) elevated risk for ADs such as systemic lupus erythematosus, Sjögren syndrome, and diabetes mellitus (7). In further support of this interpretation, we found the Red module to be significantly enriched (P = 0.01 by Fisher’s test and P = 0.01 by gene set permutation) for a set of known AD-risk genes compiled from multiple large-scale genome-wide association studies (GWAS) (Methods and SI Appendix, Text S7). The two GWAS-implicated AD-risk genes showing strongest connectivity within the Red module and up-regulation in males bearing an extra X chromosome were CLECL1 and ELF1, indicating that these two genes should be prioritized for further study in mechanisms of risk for heightened autoimmunity in XXY and XXYY males. Collectively, these results represent a systems-level characterization of SCD effects on genome function and provide convergent evidence that increased risk for AD risk in XXY and XXYY syndromes may be due to an up-regulation of immune pathways by supernumerary X chromosomes in male lymphoid cells.
To test for evidence of coordination between the expression changes in sex-chromosome genes imparted by SCD (Fig. 2) and the genome-wide transcriptomic variations detected through WGCNA (Fig. 3A), we asked if any SCD-sensitive gene-coexpression modules were enriched for one or more of the five SCD-sensitive clusters of sex-chromosome genes defined by k-means analysis (Fig. 2). Four WGCNA modules, all composed of >95% autosomal genes, showed such enrichment (Fig. 3D). XCI-cluster genes were enriched within Turquoise and Brown coexpression modules, indicating that the inverse cis effects of X-chromosome dosage on the expression of XCI-cluster genes (Fig. 2) are closely coordinated with inverse trans effects of X-chromosome dosage on autosomal expression (Fig. 3 A and C). Conversely, XCIE-cluster genes were enriched within the Green and Blue coexpression modules, with the Blue module being further distinguished by an additional enrichment in PAR-cluster genes and inclusion of XIST (Fig. 3D). We generated a network visualization to examine more closely SCD-sensitive genes and gene-coexpression relationships within the Blue module (Methods and Fig. 3E). This network highlights the high SCD sensitivity of XIST, select PAR genes (SLC25A6 and SFRS17A), and multiple X-linked genes from X-Y gametolog pairs [EIF1AX, KDM6A (UTX), ZFX, and PRKX] and shows that these genes are closely coexpressed with multiple SCD-sensitive autosomal genes including ZWINT, TERF2IP, and CDKN2AIP.
Our detection of highly organized coexpression relationships between SCD-sensitive sex-linked and autosomal genes hints at specific regulatory effects of dosage-sensitive sex-chromosome genes in mediating the genome-wide effects of SCD variation. To test this and to elucidate potential regulatory mechanisms, we performed an unbiased transcription factor binding site (TFBS) enrichment analysis of genes within the Blue, Green, Turquoise, and Brown WGCNA modules (Methods and SI Appendix, Text S9). This analysis converged on a single transcription factor—ZFX, encoded by the X-linked member of an X-Y gametolog pair—as the only SCD-sensitive transcription factor (see SI Appendix, Fig. S1 for qRT-PCR validation) showing significant TFBS enrichment in one or more modules. Remarkably, the gene ZFX was itself part of the Blue module, and ZFX-binding sites were enriched not only among Blue and Green module genes (increasing in expression with increasing X-chromosome dose) but also among Brown module genes that are down-regulated as X-chromosome dose increases (SI Appendix, Fig. S4D). To directly test if changes in ZFX expression are sufficient to modify the expression of Blue, Green, or Brown module genes in immortalized lymphocytes, we harnessed existing gene-expression data from murine T-lymphoblastic leukemia cells with and without ZFX knockout (Gene Expression Omnibus GSE43020) (22). These data revealed that genes down-regulated by ZFX knockout in mice have human homologs that are specifically and significantly overrepresented in Blue (e.g., PARP16 and TCEA1; odds ratio = 2.4, P = 0.0005, Fisher’s test) and Green (e.g., BAG3 and CCDC117; odds ratio = 2.4, P = 0.005, Fisher’s test) modules (P > 0.1 for each of the other six WGCNA modules), providing experimental validation of our hypothesized regulatory role for ZFX.
Discussion
In conclusion, our study, which systematically examined gene-expression data from 471 individuals representing nine different sex-chromosome karyotypes, yields several insights into sex-chromosome biology with consequences for basic and clinical science. First, our discovery and validation of X-linked genes that are up-regulated by reducing X-chromosome count (so that their expression is elevated in XO vs. XX, for example) runs counter to dominant models of sex-chromosome dosage compensation in mammals. This noncanonical mode of SCD sensitivity modifies current thinking regarding which subsets of X-chromosome genes could contribute to sex- and SCA-biased phenotypes (23). An inverse effect of X-chromosome dosage on X-chromosome gene expression could reflect repression of X-linked genes on the active X-chromosome by PAR or X-linked genes that are expressed from inactive X chromosomes or by stoichiometric imbalances (see below) between the products of PAR/X-linked and autosomal genes. Alternatively, inverse effects of X-chromosome dosage may reflect changes in nuclear heterochromatin dosage that would follow from varying numbers of inactivated X-chromosomes. The potential for heterochromatin-mediated consequences of SCD variation has been documented in mice, Drosophila, and humans (24, 25). Our findings also modify classic models of sex-chromosome biology by identifying X-linked genes that vary in their expression as a function of Y-chromosome dosage, indicating that the phenotypic effects of normative and aneuploid variations in Y-chromosome dose could theoretically be mediated by altered expression of X-linked genes. Moreover, the discovery of Y-chromosome dosage effects on X-linked gene expression provides routes for competition between maternally and paternally inherited genes beyond the previously described mechanisms of parental imprinting and genomic conflict, with consequences for our mechanistic understanding of sex-biased evolution and disease (26).
Beyond their theoretical implications, our data help pinpoint specific genes that are likely to play key roles in mediating SCD effects on wider genome function. Specifically, we establish that a distinctive group of sex-linked genes notable for their evolutionary preservation as X-Y gametolog pairs across multiple species and the breadth of their tissue expression in humans (12) are further distinguished from other sex-linked genes by their exquisite sensitivity to SCD and exceptionally close coexpression with SCD-sensitive autosomal genes. These results add critical evidence in support of the idea that X-Y gametologs play a key role in mediating SCD effects on wider genome function. In convergent support of this idea, we show that (i) multiple SCD-sensitive modules of coexpressed autosomal genes are enriched with TFBS for an X-linked transcription factor from the highly dosage-sensitive ZFX-ZFY gametolog pair and (ii) ZFX deletion causes targeted gene-expression changes in such modules. Inclusion of ZFX in a coexpression module (Blue) with enriched annotations for chromatin organization and cell-cycle pathways is especially striking given the rich bodies of experimental data which have independently identified ZFX as a key regulator of cellular renewal and maintenance (27).
Gene-coexpression analysis also reveals the diverse domains of cellular function that are sensitive to SCD, spanning cell-cycle regulation, protein trafficking, and energy metabolism. Many of these processes are known to be sensitive to shifts in cellular karyotype more generally (28), although we find several which appear to be specific to shifts in SCD, as they are not induced by trisomy of chromosome 21. Furthermore, gene-coexpression analysis of SCD effects dissects out specific immune activation pathways that are up-regulated by supernumerary X-chromosomes in males and enriched for genes known to confer risk for ADs that are overrepresented among males bearing an extra X chromosome. Thus, we report a coordinated genomic response to SCD that could potentially explain observed patterns of disease risk in SCA. Finally, coordinated genomic responses to SCA also feature extensive trans-acting inverse effects of SCD on autosomal expression. Such inverse trans effects of aneuploidy have been well described in model systems such as maize and Drosophila but remain relatively undocumented in humans (8, 9). Inverse trans effects in these model systems have been linked to specific regulatory genes on aneuploidy chromosomes and altered stoichiometry between the products of these genes and partner molecules encoded by nonaneuploid chromosomes (29), suggesting that similar processes may underlie our observations in humans. Collectively, our findings help refine current models of sex-chromosome biology and advance our understanding of genomic pathways through which sex chromosomes can shape phenotypic variation in health and sex-chromosome aneuploidy.
Materials and Methods
Acquisition and Preparation of Biosamples.
RNA was extracted by standard methods (Qiagen) from LCLs from 471 participants recruited through studies of SCA at the NIH Health Intramural Research Program and Thomas Jefferson University (Dataset S1) (30). Sixty-eight participants provided RNA samples for microarray analyses (12 XO, 10 XX, 9 XXX, 10 XY, 8 XXY, 10 XYY, and 9 XXYY), and 403 participants provided RNA samples for a separate qRT-PCR validation/extension study (6 XO, 146 XX, 22 XXX, 145 XY, 33 XXY, 16 XYY, 17 XXYY, 8 XXXY, and 10 XXXXY). The microarray and qRT-PCR samples were fully independent of each other (biological replicates), with the exception of four XO participants in the microarray study who each also provided a separate LCL sample for the qRT-PCR study. All participants with X/Y aneuploidy were nonmosaic based on visualization of at least 50 metaphase spreads in peripheral blood. Stability of karyotype across LCL derivation was confirmed by chromosome FISH in all members of a randomly selected subset of nine LCL samples representing each of the four supernumerary SCA groups included in our microarray analysis. The research protocol was approved by the institutional review board at the National Institute of Mental Health, and informed consent or assent was obtained from all children who participated in the study, as well as consent from their parents if the child was under the legal age of majority.
Microarray Data Preparation, Differential Expression Analysis, Annotation, and Probe Selection.
Gene expression was profiled using the Illumina HT-12 v4 Expression BeadChip Kit (Illumina, Inc.). Preprocessing and annotation of microarray data (SI Appendix, Text S1) resulted in high-quality measures of expression for 19,984 autosomal and 894 sex-chromosome genes in each of 68 independent samples from seven different karyotype groups. For each SCD group contrast, differentially expressed genes survived correction for multiple comparison across all probes, with q (the expected proportion of falsely rejected nulls) set at 0.05, and showed a log2FC >0.26. This log2 cutoff was selected empirically, by defining the log2 fold increment associated with the greatest drop in DEG count for each SCA group and averaging these thresholds all five groups (SI Appendix, Fig. S3).
Testing the Four-Class Model.
All 894 sex-chromosome genes with microarray expression data were uniquely assigned to PAR, Y-linked, XCIE, and XCI classes using known PAR boundaries and a consensus classification of X-linked genes by X-inactivation/escape status (SI Appendix, Text S3) (10). These a priori gene groupings were compared with groups defined by k-means clustering of genes by their profile of mean expression across SCD groups. k-Means clustering defined five clusters of expressed genes with SCD sensitivity and a remainder cluster of genes with low or undetectable expression levels across all samples (SI Appendix, Text S3 and Fig. S2A). k-Means cluster stability was assessed using bootstrap methods (SI Appendix, Text S4 and S5), and cluster overlaps with four-class model groupings were assessed by two-tailed Fisher’s tests. General linear models were used to estimate X- and Y-chromosome dosage effects on mean expression of each k-means gene cluster (SI Appendix, Text S3). χ2 tests were used to compare k-means clustering of X-linked genes with independently published X-chromosome annotations for (i) chromatin state (19), (ii) evolutionary strata (18), and (iii) presence vs. absence of a surviving Y-chromosome homolog (SI Appendix, Text S3) (10).
Comparison of DEG Count and Genomic Distribution Across SCA Groups.
Total DEG counts were compared across SCD groups across a range of log2FC cutoffs as described above and are reported in SI Appendix, Fig. S3 A and B. Observed DEG counts across four genomic regions—autosomal, PAR, Y-linked, and X-linked—were compared with the background distribution of total gene counts across these regions using the prop.test function in R.
qRT-PCR Validation of Differentially Expressed Genes in Microarray.
For selected genes showing significant DE between karyotype groups in our core sample, we used qRT-PCR measures of gene expression (Fluidigm) to validate and extent observed fold changes in an independent sample of 403 participants representing all the karyotypes in our core sample plus two additional SCAs: XXXY and XXXXY (SI Appendix, Text S2, Dataset S1).
WGCNA.
Gene-coexpression modules were generated and assessed for reproducibility using the R package WGCNA (SI Appendix, Text S7). Modular GO term enrichments were assessed using GO elite (31) and Gorilla (32). Fisher’s exact test and resampling methods were used to assess modular enrichments for AD-risk genes from a reference catalog of GWAS findings (https://www.ebi.ac.uk/gwas/). See SI Appendix, Text S8 for details of TFBS enrichment analysis and experimental validation for the hypothesized regulatory role of ZFX assessed using gene-expression data from lymphocytes in a murine ZFX-knockout model (22).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802889115/-/DCSupplemental.
References
- 1.Hughes JF, Rozen S. Genomics and genetics of human and primate y chromosomes. Annu Rev Genomics Hum Genet. 2012;13:83–108. doi: 10.1146/annurev-genom-090711-163855. [DOI] [PubMed] [Google Scholar]
- 2.Bachtrog D, et al. Tree of Sex Consortium Sex determination: Why so many ways of doing it? PLoS Biol. 2014;12:e1001899. doi: 10.1371/journal.pbio.1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold AP. The end of gonad-centric sex determination in mammals. Trends Genet. 2012;28:55–61. doi: 10.1016/j.tig.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A. Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc Natl Acad Sci USA. 2010;107:3394–3399. doi: 10.1073/pnas.0913843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belling K, et al. Klinefelter syndrome comorbidities linked to increased X chromosome gene dosage and altered protein interactome activity. Hum Mol Genet. 2017;26:1219–1229. doi: 10.1093/hmg/ddx014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong DS, Reiss AL. Cognitive and neurological aspects of sex chromosome aneuploidies. Lancet Neurol. 2014;13:306–318. doi: 10.1016/S1474-4422(13)70302-8. [DOI] [PubMed] [Google Scholar]
- 7.Seminog OO, Seminog AB, Yeates D, Goldacre MJ. Associations between Klinefelter’s syndrome and autoimmune diseases: English national record linkage studies. Autoimmunity. 2015;48:125–128. doi: 10.3109/08916934.2014.968918. [DOI] [PubMed] [Google Scholar]
- 8.Guo M, Birchler JA. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science. 1994;266:1999–2002. doi: 10.1126/science.266.5193.1999. [DOI] [PubMed] [Google Scholar]
- 9.Sun L, et al. Dosage compensation and inverse effects in triple X metafemales of Drosophila. Proc Natl Acad Sci USA. 2013;110:7383–7388. doi: 10.1073/pnas.1305638110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balaton BP, Cotton AM, Brown CJ. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol Sex Differ. 2015;6:35. doi: 10.1186/s13293-015-0053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skaletsky H, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 12.Bellott DW, et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499, and erratum (2014) 514:126. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng X, Berletch JB, Nguyen DK, Disteche CM. X chromosome regulation: Diverse patterns in development, tissues and disease. Nat Rev Genet. 2014;15:367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 15.GTEx Consortium Human genomics. The Genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tukiainen T, et al. GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson Sayres MA, Makova KD. Gene survival and death on the human Y chromosome. Mol Biol Evol. 2013;30:781–787. doi: 10.1093/molbev/mss267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey RS, Wilson Sayres MA, Azad RK. Detecting evolutionary strata on the human x chromosome in the absence of gametologous y-linked sequences. Genome Biol Evol. 2013;5:1863–1871. doi: 10.1093/gbe/evt139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aksglaede L, Juul A. Testicular function and fertility in men with Klinefelter syndrome: A review. Eur J Endocrinol. 2013;168:R67–R76. doi: 10.1530/EJE-12-0934. [DOI] [PubMed] [Google Scholar]
- 21.Parikshak NN, Gandal MJ, Geschwind DH. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet. 2015;16:441–458. doi: 10.1038/nrg3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weisberg SP, et al. ZFX controls propagation and prevents differentiation of acute T-lymphoblastic and myeloid leukemia. Cell Rep. 2014;6:528–540. doi: 10.1016/j.celrep.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Disteche CM. Dosage compensation of the sex chromosomes and autosomes. Semin Cell Dev Biol. 2016;56:9–18. doi: 10.1016/j.semcdb.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wijchers PJ, Festenstein RJ. Epigenetic regulation of autosomal gene expression by sex chromosomes. Trends Genet. 2011;27:132–140. doi: 10.1016/j.tig.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Trolle C, et al. Widespread DNA hypomethylation and differential gene expression in Turner syndrome. Sci Rep. 2016;6:34220. doi: 10.1038/srep34220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cocquet J, et al. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 2012;8:e1002900. doi: 10.1371/journal.pgen.1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galan-Caridad JM, et al. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129:345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheltzer JM, Torres EM, Dunham MJ, Amon A. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci USA. 2012;109:12644–12649. doi: 10.1073/pnas.1209227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birchler JA, Veitia RA. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA. 2012;109:14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zinn AR, et al. A Turner syndrome neurocognitive phenotype maps to Xp22.3. Behav Brain Funct. 2007;3:24. doi: 10.1186/1744-9081-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zambon AC, et al. GO-elite: A flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28:2209–2210. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.