Significance
Antibiotics generally target one of five essential cellular functions in bacteria, but many of these targets are now compromised through rapidly spreading antibiotic resistance. Bedaquiline (BDQ), a new FDA-approved antitubercular drug, targets energy metabolism: defining cellular energetics as a new target space for antibiotics. This is a relatively unexplored area, as BDQ was only FDA approved in 2012. Several studies have recently found that BDQ stimulates mycobacterial respiration, in addition to inhibiting its molecular target, the F1Fo-ATP synthase. We show that BDQ is an ionophore, which shuttles H+ and K+ ions across membranes, and propose that this activity may contribute to killing of mycobacteria by BDQ. Combining ionophoric activity with high-affinity membrane protein inhibition may enhance the specificity and potency of antibiotics.
Keywords: bedaquiline, tuberculosis, respiration, uncoupler, ionophore
Abstract
Bedaquiline (BDQ), an inhibitor of the mycobacterial F1Fo-ATP synthase, has revolutionized the antitubercular drug discovery program by defining energy metabolism as a potent new target space. Several studies have recently suggested that BDQ ultimately causes mycobacterial cell death through a phenomenon known as uncoupling. The biochemical basis underlying this, in BDQ, is unresolved and may represent a new pathway to the development of effective therapeutics. In this communication, we demonstrate that BDQ can inhibit ATP synthesis in Escherichia coli by functioning as a H+/K+ ionophore, causing transmembrane pH and potassium gradients to be equilibrated. Despite the apparent lack of a BDQ-binding site, incorporating the E. coli Fo subunit into liposomes enhanced the ionophoric activity of BDQ. We discuss the possibility that localization of BDQ at F1Fo-ATP synthases enables BDQ to create an uncoupled microenvironment, by antiporting H+/K+. Ionophoric properties may be desirable in high-affinity antimicrobials targeting integral membrane proteins.
The paucity of new drug leads developed through target-based screening since 1999, compared with phenotypic screening, has largely been attributed to poorly resolved modes of action (1). Furthermore, compounds with new molecular effects are discovered through phenotypic screening methods, and the antitubercular medicine bedaquiline (BDQ, Sirturo), FDA approved in December 2012, is no exception (2, 3). An inhibitor of the mycobacterial F1Fo-ATP synthase (henceforth F1Fo), BDQ demonstrates that metabolism and energy generation is a promising new target space. However, despite only 5 y of clinical use, resistance in both laboratory and clinical settings has been reported (4–6), reinforcing the need to mine this new target space for second-generation compounds. However, this process will be slowed without thoroughly resolving the mode of action of first-generation inhibitors. Important aspects of BDQ’s mode of action are unresolved, including the time-dependent mechanism of killing and the molecular basis for selectivity between bacterial strains.
BDQ has been demonstrated to bind to the c-ring rotor of the Fo portion of the mycobacterial ATP synthase (7, 8); concomitantly the synthesis of ATP, an essential energy currency in biology, is inhibited and intracellular ATP levels drop (7, 9). BDQ is not reported to inhibit growth of nonmycobacterial strains (2) and in mammalian mitochondria the drug did not affect either ATP synthesis activity (10) or the membrane potential (11). Inhibition of mycobacterial growth by BDQ can be attributed to stereospecific inhibition of ATP synthase (7) leading to a decrease in intracellular ATP content (9, 12). The bactericidal activity and time-dependent killing of Mycobacterium tuberculosis by BDQ, on the other hand, is less well resolved. BDQ concentrations several orders of magnitude higher than that required for inhibition of growth are required for bactericidal activity (12, 13). It has also been demonstrated that BDQ stimulates oxygen consumption in Mycobacterium smegmatis (13) and M. tuberculosis (14). From these studies it has been proposed that BDQ is an uncoupler of respiration and ATP synthesis (11, 13), collapsing the transmembrane pH gradient component of the proton motive force (PMF) ultimately leading to cell death (13).
The PMF is an electrochemical gradient consisting of both a transmembrane pH gradient (acidoutside/alkalineinside) and the membrane potential (∆pH and ∆ψ, respectively), which is most well known for its utilization by the F1Fo synthase during ATP synthesis. Protonophores and ionophores are membrane diffusible chemicals that can bind and transport protons or other cations and can act to equilibrate/dissipate these gradients (15, 16). The cellular response to these chemicals is to increase respiration in an attempt to maintain the PMF, resulting in futile cycling of ions that is uncoupled from ATP synthesis, also known as “uncoupling.”
Protonophores generally are lipophilic weak acids, such as carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) or carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (17), which carry both protons and charge by directly binding and shuttling protons across the cell membrane. Extensive delocalization of the negative charge allows the deprotonated form to cross the lipid bilayer. Although less well characterized, cationic protonophores have been reported (18–20). These molecules are lipophilic weak bases, as opposed to weak acids, and delocalize a positive charge by similar mechanisms. Carrying protons without simultaneously moving a compensatory ion collapses both the ∆pH and ∆ψ (15). Ionophores are instead capable of binding and shuttling larger ions, sometimes in addition to protons. Nigericin is an example of a molecule that carries both cations and protons (15), by binding said ions through its carboxylate moiety. Nigericin antiports K+ and H+, an electroneutral exchange, to collapse only the ∆pH. Valinomycin instead carries only larger cations, not protons, and so collapses the ∆ψ while maintaining the ∆pH (15). BDQ has been proposed to function as a cationic protonophore (11). However, this result does not explain the observation that BDQ collapses only the ∆pH, but not the ∆ψ in M. smegmatis membrane vesicles and the dependence on ATP synthase binding (13). The counter ion, and the mechanism by which the counter ion is moved to maintain electroneutrality, is unresolved. Whether BDQ is a protono-/ionophore in its own right, requires the presence of an ATP synthase for its activity, or both, is unknown.
In this communication we report that BDQ inhibits ATP synthesis in Escherichia coli, an organism reported to resist BDQ growth inhibition, by dissipating the PMF. E. coli is a useful model organism due to the ease and high yield of F1Fo purification, the bidirectional nature of the enzyme’s activity (in contrast to the mycobacterial variant) (21), and the ability to separate the enzyme into its F1 and Fo subcomplexes for focused analysis. The E. coli F1Fo is not essential, unlike in mycobacteria (22), and so gene deletions are readily available (23). Further analysis in lipid vesicles demonstrates that BDQ can function as a cationic protonophore; but the addition of opposing salt gradients enhances this activity, suggesting BDQ is in fact a bona fide H+/K+ ionophore. The E. coli ATP synthase Fo subunit enhanced this activity, although dispensable, suggesting BDQ accumulates at an unresolved binding site. These activities occur at BDQ concentrations comparable to that required for bactericidal activity (cell killing) (12, 13), and therefore we invoke our model to provide a potential explanation for preliminary observations linking the stimulation of oxygen consumption to mycobacterial cell death (11, 13, 14). Combining an ionophoric moiety with a potent membrane protein-binding moiety may therefore be desirable in future antibiotic development.
Results
BDQ Inhibits ATP Synthesis in E. coli by Ionophoric Uncoupling.
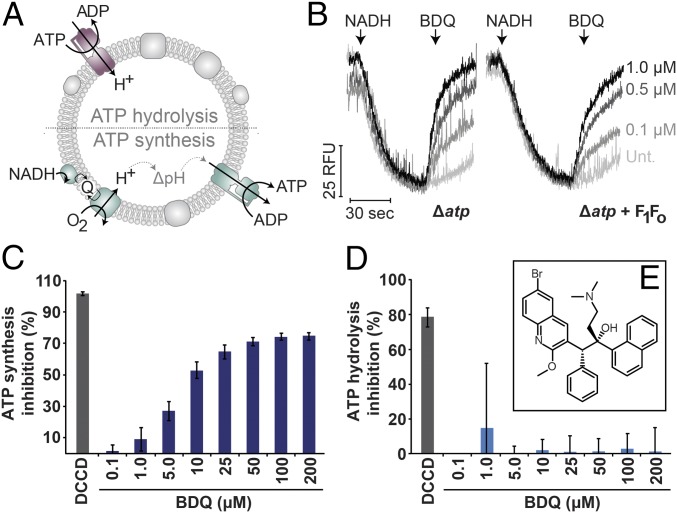
The cause of mycobacterial cell death upon bedaquiline addition is unclear, although several studies have implicated respiratory uncoupling (11, 13, 14). A correlation between uncoupling in E. coli and M. smegmatis membranes was previously observed (11), but the molecular mechanism is poorly resolved and hence the focus of our current study. The minimum inhibitory concentration (MIC) of BDQ against E. coli is reported to be >32 μg⋅mL−1 (58 μM) (2). In our own experiments we similarly found no growth inhibition for E. coli MG1655 (wild-type), testing up to 100 μM BDQ. In contrast to its lack of growth inhibition and consistent with previous reports (11), we found that BDQ could dissipate a ΔpH in inverted membrane vesicles (IMVs) (Fig. 1A) of E. coli that were energized by either NADH oxidation or ATP hydrolysis (Fig. 1B and SI Appendix, Fig. S2). Extending this finding, we found that BDQ was able to dissipate the ΔpH in IMVs of either E. coli with a deletion in the F1Fo operon (Fig. 1B) or the same strain overexpressing F1Fo (Fig. 1B). Expression was confirmed by activity and Western blots (SI Appendix, Fig. S3).
Fig. 1.
Uncoupling of E. coli IMVs by BDQ inhibits ATP synthesis. (A) Schematic for reactions performed by IMVs: ATP hydrolysis establishes a proton gradient, while ATP synthesis is energized by the proton gradient established by NADH oxidation and subsequent electron transport chain activity. (B) IMVs of E. coli C41 harboring an unc operon deletion (Δatp), or the same strain overexpressing F1Fo (Δatp + F1Fo), were assessed for PMF establishment using 250 nM acridine orange. Proton pumping was elicited by 200 μM NADH and the proton gradient then dissipated by the indicated amounts of BDQ. (C and D) IMVs of E. coli DK8 Δatp pBWU13 (F1Fo) were prepared and measured as endpoint assays for (C) inhibition of ATP synthesis or (D) inhibition of ATP hydrolysis. (E) The structure of BDQ. DCCD was used at 100 μM. Error bars represent SD from three independent experiments. B and C are kinetic traces representative of triplicate experiments.
The PMF is obligatory for ATP synthesis, but ATP hydrolysis is not a PMF-consuming process and can proceed in its absence (24). Consistently, BDQ was able to inhibit ATP synthesis in E. coli IMVs at concentrations similar to that causing ΔpH dissipation (Fig. 1C), with an inhibitory concentration for 50% of the response (IC50) of ∼5 μM. ATP hydrolysis was unaffected by the addition of BDQ, but strongly inhibited by the Fo inhibitor N,N′-dicyclohexylcarbodiimide (DCCD) (Fig. 1D). This suggests that BDQ is causing uncoupling by directly binding and shuttling protons (protonophore or ionophore) to collapse the ΔpH gradient. Nigericin was sufficient to inhibit ATP synthesis in our membrane preparations (SI Appendix, Fig. S4), suggesting our preparations produced a PMF composed mainly of a ΔpH. Acridine orange (ΔpH probe) and oxonol quenching (Δψ probe) profiles (SI Appendix, Figs. S5 and S6) suggest that valinomycin and nigericin are working as intended in our assay conditions, only uncoupling their respective component of the PMF, while the pore-forming gramicidin can completely equilibrate the entire PMF (SI Appendix, Fig. S6 A and B). Therefore, ATP synthesis results from this assay system may not inform on the role of the membrane potential. To address this, we performed oxonol quenching assays and found that BDQ does not collapse the Δψ in IMVs (SI Appendix, Fig. S6 E and F). This is similar to previous observations in M. smegmatis (13). The residual ATP synthesis activity (∼30%) in BDQ-treated IMVs (Fig. 1C) may represent Δψ-driven ATP synthesis.
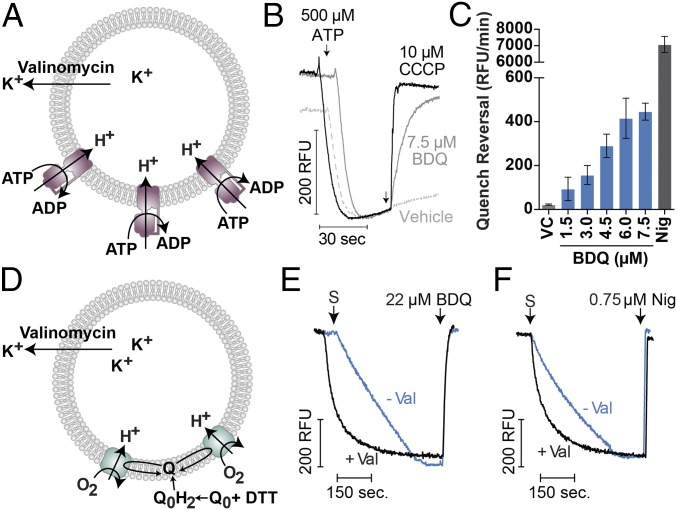
To confirm that some unspecified membrane protein (for example, H+-driven antiporters or efflux pumps) does not move ions in response to BDQ, we reconstituted the purified E. coli F1Fo (SI Appendix, Fig. S7) into proteoliposomes (Fig. 2A) and assessed the effects of BDQ in this system. BDQ could collapse a ΔpH gradient generated by ATP hydrolysis (Fig. 2 B and C), suggesting that uncoupling is indeed driven by a protonophoric or ionophoric mode of action. Similarly, a ΔpH gradient established by the activity of cytochrome bo3, when reconstituted into proteoliposomes (Fig. 2D), could be dissipated by BDQ (Fig. 2E). This is consistent with the lack of F1Fo-dependent effects in IMVs (Fig. 1 B and C). Compared with the positive control nigericin (Fig. 2F), 28-fold more BDQ was needed to achieve the same degree of dissipation. In the F1Fo system, the rate of requenching was maximal at 7.5 μM BDQ (Fig. 2C) and was 16-fold lower than that of 10 μM nigericin. The presence of a Δψ did not affect ATP hydrolysis inhibition (SI Appendix, Fig. S8) or ΔpH dissipation in cytochrome bo3-containing proteoliposomes (Fig. 2E). The lack of valinomycin dependency suggests an opposing Δψ was not a limiting factor. Although not necessarily as potent as nigericin, it is clear that BDQ at micromolar concentrations can collapse the ΔpH component of the PMF faster than any E. coli proton-pumping enzyme can establish it. The ability of BDQ to inhibit ATP synthesis in IMVs (Fig. 1) suggests this activity is kinetically faster than physiological rates of combined proton pumping by the respiratory chain.
Fig. 2.
Uncoupling of proton-pumping proteoliposome systems by BDQ. Schematics showing how proton pumping in proteoliposomal E. coli F1Fo (A) or E. coli cytochrome bo3 (D) is achieved by either ATP hydrolysis or reduced ubiquinone (QH2) addition, respectively. Unless otherwise indicated, 1 μM valinomycin is added to counteract inhibitory membrane potentials. (B) F1Fo proteoliposomes were incubated with ATP to establish a steady-state pH gradient and then the indicated compounds were added to reverse acridine orange quenching. (C) The initial rate of quenching reversal from B is quantified as relative fluorescence units (RFU) min−1, error bars represent SD from three independent experiments. Nig, 10 μM nigercin; VC, vehicle control. (E and F) Proton pumping in cytochrome bo3 proteoliposomes was initiated by the addition of 2.5 μM UQ0 (Q0 in figure) to establish a steady-state pH gradient, as determined by ACMA fluorescence quenching, in either the presence or absence of 1 μM valinomycin. Either (E) BDQ or (F) nigericin was added when indicated. Experiments are derived from kinetic traces.
BDQ Accumulates at Lipid Membranes to Collapse pH Gradients.
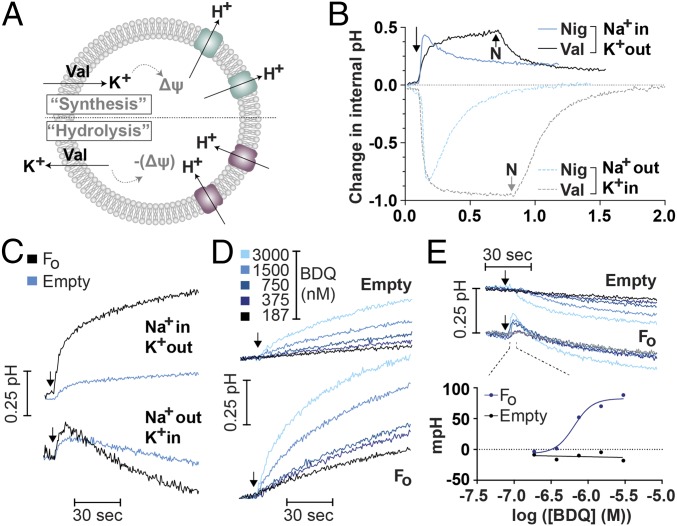
We prepared pyranine-containing phosphatidylcholine vesicles (liposomes) to examine these effects in a more controlled system. This technique quantifies the change in internal pH and is advantageous due to the ability to artificially manipulate pH and cation gradients. This method has previously been used to measure proton transport in isolated E. coli Fo complexes (25) and internal pH changes in protein-free liposomes (empty liposomes) (20). Empty liposomes are advantageous, as we found they can maintain artificially established gradients for far longer than Fo proteoliposomes (SI Appendix, Fig. S9). We quantified the ability of BDQ to equilibrate an artificially imposed ΔpH in the absence of any protein. Unlike the prior model systems, this pH gradient is finite.
BDQ was able to equilibrate the intraliposomal (internal) pH with the external (buffer) pH (Fig. 3B), regardless of whether the external pH was acidic or alkaline. The internal volume of liposomes containing the Fo subunit has previously been found to be 1.5–1.8 μL/mg lipid (25). The external buffer volume is therefore likely to be at least 100-fold in excess for all experiments, so we consider the external pH to be constant. Given sufficient time and/or concentration of BDQ, it was possible to fully equilibrate the internal pH with the external pH (SI Appendix, Fig. S9B). The effective concentration for 50% of the equilibration response (EC50) was 146 nM BDQ (Fig. 3C). In addition to equilibrating pH gradients, BDQ could additionally alkalize the liposome interior by ∼0.5 pH units in the absence of a ΔpH (Fig. 3D). This was also observed as an initial alkalization at external pH 6.53 (Fig. 3B). We attribute this to intraliposomal accumulation of BDQ and subsequent alkalization. Since BDQ is a weakly basic (pKa = 8.9) (11) and highly lipophilic compound (logP = 7.13, logD = 5.42), it is expected to partition into hydrophobic membranes and this result is an experimental confirmation of this expectation. Aside from this alkaline bias, BDQ mimics the pH equilibration profile of the protonophore CCCP (Fig. 3D). These results show that BDQ has the capacity to act as a cationic protonophore, consistent with the suggestion of Feng et al. (11). However, this is inconsistent with the lack of effects on the membrane potential in E. coli IMVs (SI Appendix, Fig. S6) or M. smegmatis IMVs (13).
Fig. 3.
BDQ accumulates in pyranine-containing liposomes and collapses pH gradients. (A) Schematic showing how the protonophore CCCP or the ionophore nigericin can manipulate the internal pH in empty liposome systems, depending on the type of imposed artificial gradient. (B) Suspensions of liposomes (internal pH ∼ 7.1) were incubated in buffers of the indicated pH and treated with BDQ, with stirring in a fluorimeter. The experiment is a kinetic trace representative of a technical triplicate. Subsequent experiments are treated analogously to B, but as endpoint assays performed in a plate reader (without stirring). (C) An initial pH gradient of ∼0.3 units (inside acidic) was established and the indicated amounts of BDQ added. The EC50 is indicated. (D) A total of 1 μM CCCP or BDQ was used as indicated and the internal pH after 30-min treatment was measured. Experiments used a 2-mM Mes-Mops-Tris buffer system (pH indicated). In C and D error bars indicate SD from triplicate measurements, although they are not visible in D.
E. coli Fo Subunits Enhance BDQ-Elicited Proton Transport.
We compared Fo-containing and empty pyranine liposomes, initially as a control to confirm the lack of F1Fo-dependent effects observed previously (Figs. 1 and 2). In this system, membrane potentials are manipulated to initiate proton transport through the Fo subunit (Fig. 4 A and B) (25). Unexpectedly, BDQ appeared to alleviate the requirement of valinomycin for inducing Fo-dependent proton transport, when using a K+ diffusion potential (Fig. 4C, K+out). This suggests that BDQ is able to shuttle K+ ions to create a Δψ using the starting gradient of KCl. Notably, BDQ does not show the same biphasic kinetics as nigericin (Fig. 4 B and C), although we cannot rule out that the timescale of the experiment is too small to observe a second phase of BDQ activity. Incorporation of Fo subunits enhanced the activity of BDQ, alkalizing the interior by 0.44 pH units more than empty liposomes after 90 s (Fig. 4C). A similar effect was observed when an inside acidic ΔpH was used (Fig. 4D), but this could not be observed when the salt gradients were reversed (Fig. 4C, K+in). Instead, BDQ appeared to show a bias for alkalization, similar to the empty liposome system (compare pH 6.53 in Fig. 3B). When an inside alkaline ΔpH was used (Fig. 4E), BDQ caused an initial alkalization of Fo-containing liposomes. This is despite the fact that the gradient used favors intraliposomal acidification (compare pH 6.02 in Fig. 3B). The EC50 for this effect was 647 nM (Fig. 4E). This suggests that the E. coli Fo subunit, despite the lack of mycobacterial BDQ binding site (8), has enhanced the ionophoric activity of BDQ. We were unable to outcompete this effect with DCCD, suggesting the binding site is not necessarily at the c-ring’s proton-binding site.
Fig. 4.
The E. coli Fo subunit enhances the activity of BDQ in proteoliposomes. (A) Schematic showing how proton transport is routinely initiated in Fo proteoliposomes, by either accumulating or depleting K+ to manipulate the membrane potential in these preparations. (B) Salt gradients were established by diluting 5 μL of Fo-containing liposomes with either 50 mM K2SO4 or 50 mM Na2SO4 (K+in, Na+in, respectively) into a 1-mL buffer containing 50 mM Na2SO4 or 50 mM K2SO4 (Na+out, K+out, respectively). The change in internal pH was measured. K+ is moved to generate a membrane potential as indicated in A. Valinomycin or nigericin (100 nM each) was added when indicated. Nigericin was additionally added to valinomycin experiments where indicated. (C) The same experiment as B was performed, with the salt compositions as indicated, except 1.5 μM BDQ was added at the arrow to either Fo-containing or empty liposomes as indicated. (D and E) pH gradients were established in either (D) ATP synthesis (inside acidic) or (E) ATP hydrolysis (inside alkaline) directions by diluting 5 μL of the indicated liposomes (∼pH 7.1 inside) into 1 mL of buffer either 1 unit more acidic or alkaline. The indicated amount of BDQ was added at the arrow. In E, the initial rapid alkalization is quantified below the trace. Experiments are representative of a technical triplicate and derived from kinetic traces.
BDQ Functions as a Proton/Monovalent-Cation Ionophore.
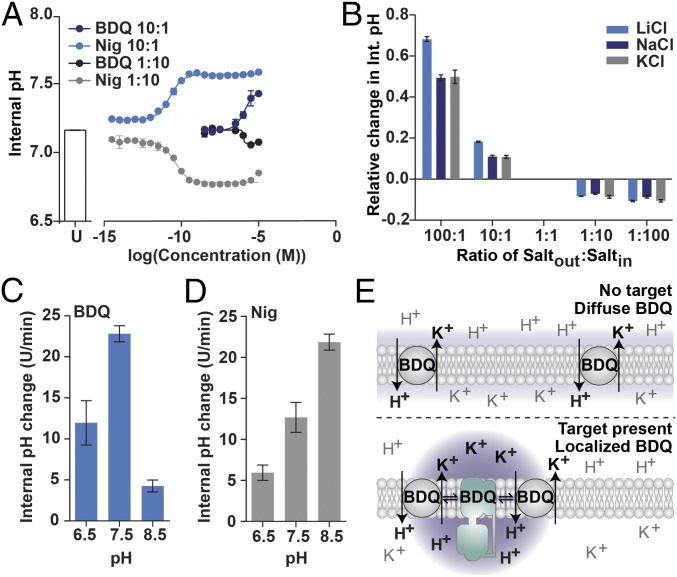
We observed that BDQ could alleviate the requirement of valinomycin in Fo proton transport assays (Fig. 4C), suggesting it could move K+ to generate a Δψ. Given that BDQ can also move H+ (Fig. 3), we hypothesized that BDQ functions as a H+/K+ exchanger. We used empty liposomes to test this hypothesis, to remove the contribution of Fo to intraliposomal pH change. Given the biphasic kinetics possible with multisalt systems (Fig. 4B), only a single type of salt was used for each experiment. Nigericin, a common H+/K+ antiporter, can convert a KCl gradient into a ΔpH gradient (15) and this was readily achievable in our experimental system (Fig. 5A). Nigericin caused either intraliposomal acidification or alkalization depending on whether a higher concentration of salt was inside the liposome or in the external buffer (Fig. 5A). BDQ could achieve a similar effect (Fig. 5A). A high-inside KCl gradient was sufficient for BDQ to cause intraliposomal acidification (Fig. 5A), despite BDQ’s alkaline bias, but BDQ could elicit a 4.3-fold greater change in liposomal pH for a high-outside KCl gradient. This agrees with the directional bias observed in the Fo-liposome system (Fig. 4). The response did not appear to be specific to K+, as LiCl and NaCl were able to achieve the same effect (Fig. 5B and SI Appendix, Fig. S10). It is possible that contaminating ions in soybean phosphatidylcholine (26) facilitates proton movement in the absence of added salt (i.e., in the conditions of Fig. 3). Changing the buffer used or the lipid used did not affect the result (SI Appendix, Fig. S11).
Fig. 5.
BDQ is a H+/K+ ionophore. (A) Liposomes were prepared in either 10 mM KCl or 100 mM KCl buffer and diluted in the opposite buffer to give a K+out:K+in ratio of 10:1 or 1:10, respectively. The ratio in the key refers to the K+out:K+in ratio. The indicated amount of BDQ was added and the 30-min endpoint was recorded. U, untreated control. (B) LiCl, NaCl, and KCl were compared for their ability to elicit proton movement upon BDQ addition. The 30-min endpoint was recorded. Saltout:saltin refers to the concentration of the indicated salt (where 1:1 is 10 mM inside and outside) Data are relative to a saltout:saltin ratio of 1:1. (C and D) In each experiment, a 10:1 K+out:K+in gradient is established, while the starting pH is the same across the liposome. (C) The rate of pH change caused by 10−5 M BDQ at different buffer pH values. (D) The rate of pH change caused by 10−9 M Nig at different buffer pH values. D and E are derived from kinetic traces from the multiwell-style experiment (Materials and Methods). Error bars indicate SD from triplicate experiments. (E) Model for how BDQ functions as an ionophore (Top) and how this might be localized at the site of a high-affinity binding partner. Purple shading represents intensity of uncoupling.
It is unlikely that Cl− ions are moved by BDQ, as this anion would preferentially move in the same (symport) direction of the H+ ion to prevent inhibitory counter potentials. In support of this, BDQ was able to collapse a ΔpH established by cytochrome bo3 when either potassium or sodium salts were used (SI Appendix, Fig. S12). This occurred with a slightly lower magnitude and a secondary slower rate when Na2SO4 was used, which is likely due to the stronger binding of Na+ to SO4− ion (the KD for dissociating Na+ from NaSO4− is less than Na2SO4) (27). Movement of SO42− would require dissociation of both Na+ ions first, a chemically unlikely phenomena under biological conditions, and this would not be consistent with a slower secondary rate. As K+ is biologically accumulated at the cytoplasmic face of the membrane, opposing the ΔpH, we continued to focus characterization on this particular cation.
BDQ Does Not Transport K+ as a Salt.
Nigericin transports K+ by forming a salt with the carboxylate group (15). The ionization state of nigericin therefore influences its K+ transport ability and so sufficient acidity should compete with the binding of K+. To test whether BDQ transports K+ similarly, we examined the ionization-state dependence of both BDQ and nigericin. Being a weak base (pKa ∼ 8.9), the unprotonated form of the drug only appreciably exists at alkaline pH (SI Appendix, Fig. S13A). If the amine groups coordinate K+, then increasing acidity should outcompete this binding. Instead, we find that the ability of BDQ to elicit H+ movement, using solely a KCl gradient, is best at pH 7.5 and worse at either alkaline or acidic pH (Fig. 5C and SI Appendix, Fig. S13B). In comparison, more acidic pH values inhibited the ability of nigericin to convert a KCl gradient into a ΔpH, consistent with the formation of carboxylate salts (Fig. 5D and SI Appendix, Fig. S13B). This suggests that, unlike nigericin, BDQ does not transport K+ as a salt. We propose that BDQ chelates K+ through a pH-sensitive mechanism, distinct to the amine protonation site. Overall, these data suggest that BDQ can function as a H+/K+ ionophore under the pH and salt conditions that emulate a standard neutrophilic bacterium, like E. coli or M. tuberculosis, and that this activity is enhanced at the location of a BDQ binding partner.
Discussion
Researchers place emphasis on characterizing the primary targets of lead therapeutics, yet this risks overlooking potentially meaningful and potentially bactericidal secondary effects. In this work we report that BDQ has the ability to act as a H+/K+ ionophore. This can result in inhibition of ATP synthesis in E. coli inverted membrane vesicles, despite its having no measurable sensitivity to BDQ at a whole cell level. Here, we will propose that target-dependent localization of BDQ enables specific and potent uncoupling, despite the ionophoric nature of its uncoupling mechanism.
BDQ is a lipophilic weak base (pKa = 8.9, logP = 7.13), so its ability to move protons is likely similar to the well-described weak acid CCCP and lipophilic weak bases such as ellipticine (15, 18), where the charge from its ionization is delocalized across π-orbitals. This would allow protonated BDQ to cross the plasma membrane and equilibrate pH gradients. In contrast, BDQ does not appear to bind K+ at the protonable amine groups. This is unlike nigericin, which binds K+ as a carboxylic salt (15), suggesting BDQ chelates K+ in a different manner. The apparent pH optimum of 7.5 for BDQ converting a KCl gradient into a pH gradient supports BDQ physiologically creating a futile cycle of K+ and H+ in a neutrophilic bacterial cell, like M. tuberculosis: BDQ acquires a proton from the acidic periplasm and moves to the neutral cytoplasm where the proton is displaced by K+, before returning to the periplasm and so on (Fig. 5E, Top). K+, being the predominant intracellular monovalent cation (28), is likely to be more physiologically relevant than Na+ and Li+.
Previously, a direct interaction of BDQ and the F1Fo of M. smegmatis was invoked, and subsequent disruption of the a–c subunit interface was proposed to allow uncontrolled proton influx (13). It has also been proposed that the basis of BDQ’s uncoupling is purely chemical (11). We invoke a revised mechanism to reconcile the combined data. It is appropriate to extrapolate our results to mycobacteria as the concentrations of BDQ required to kill M. tuberculosis are far in excess of its MIC and well within the effective concentrations used in this study (30× and 300× MIC or 1.6 μM and 16 μM, respectively; ref. 12). We note that purely chemical mechanisms are indeed possible in mycobacteria: the AtpED32V mutant in our previous study still had measurable pH gradient dissipation, albeit at a slower rate and requiring higher concentrations (14.4 μM and 7.2 μM) of BDQ (13). This strain is resistant to BDQ, so it is clear that this chemical effect alone is insufficient for cell killing. The recently published structure of the c-ring from Mycobacterium phlei with bound BDQ suggests that BDQ cannot bind to ATP synthase of nonmycobacterial species (8). However, BDQ appeared to accumulate with greater efficacy at liposome preparations containing the E. coli Fo subunit. The implications are twofold: (i) there may be a lower affinity, although not necessarily specific, BDQ-binding site in the E. coli Fo subunit; and (ii) binding BDQ may be necessary to localize its uncoupling activity to physiologically relevant locations in a cell.
To address the first implication, BDQ is an arginine mimetic (8) and may well have several lower affinity sites in the E. coli Fo subunit, for example at other glutamate or aspartate residues. Alternate binding sites are not without precedent, as Trp-16 of the M. tuberculosis epsilon subunit has been suggested to be a second BDQ-binding site (29, 30). To consider the second implication, we will use BDQ binding to the target mycobacterial Fo as an example. BDQ can bind and occupy all c subunits in the mycobacterial enzyme (8). However, binding interactions are inherently transient: the dissociation constants for BDQ binding to the mycobacterial F1Fo c subunit have been determined to be 1.5–19.7 μM, depending on the ionic strength of the buffer used (31). As one molecule is released, another may diffuse into the binding site. Continued on and off in this manner may localize BDQ at this binding site. Furthermore, the dependency of the dissociation constant on ionic strength (31) may be explained by BDQ binding cations. It is conceivable that K+ actively competes for BDQ, removing it from the a–c interface so that it can collapse the pH gradient. In this model, the microenvironment around the target protein would then be susceptible to uncoupling, while other areas in the membrane will be unaffected (Fig. 5E). A dependency on target-based localization allows for a stereospecific and target-specific uncoupling, even if the nature of the uncoupling is ionophoric and likely present in the other stereoisomers of BDQ.
The lack of apparent selectivity between Li+, Na+, and K+ suggests BDQ does not form a size-gated polar core like valinomycin (15). Ionophores with much broader ion specificities do exist, such as lasalocid A (32), but parallels are not readily drawn, owing to highly different chemical structures. Ellipticine, a cationic protonophore, has previously been reported to be most active around its pKa (18). In this work, BDQ was found to be most active at pH values around 1.0 units more acidic than its predicted pKa of 8.9 (11). It may be that the binding of salt and interactions with the lipid membrane result in a lower than predicted pKa. There is a possibility that several BDQ molecules may act to coordinate a single cation, which may explain the apparent lack of a singular cation-binding chemical motif and the ability of BDQ to act protonophorically: BDQ may transport protons as monomers and associate into multimers that complex K+, depending on the particular conditions.
While BDQ may well have weak uncoupling activity in other bacteria or mitochondria, our mechanism would suggest that it is not biologically relevant without a protein target. BDQ has no significant effect on oxygen consumption (10) and the membrane potential (11) of mammalian mitochondria. A small effect (∼35% inhibition at 200 μM) of BDQ on ATP synthesis has previously been observed in mitochondria (10) and may represent this weak uncoupling activity. However, BDQ has been found to have no effect on the oxygen consumption of intact HepG2 and RAW264.7 cell lines (14). The restricted antibacterial spectrum of BDQ is well known (2) and uncoupling may well be overcome by fermentation in other bacteria. BDQ may have arisen from a plethora of favorable conditions in mycobacteria: a high-affinity binding site for BDQ (8), a sensitivity to ionophores like nigericin and valinomycin (33), and its dependence on respiration due to the essentiality of F1Fo (22). Should uncouplers be targeted to high affinity protein-binding sites in other organisms, the result may well be a relevant therapeutic.
Oxidative phosphorylation is a very promising avenue for drug development and so it is important that there is sufficient knowledge of our current inhibitors, to allow well-informed decisions for future lead compounds. Our proposal is that bedaquiline is an atypical ionophore and that this ionophoric action explains killing of mycobacteria by bedaquiline (as suggested in ref. 13). This improves our understanding of the first-in-class antibiotic and highlights that ionophores and protonophores, typically associated with human toxicity (such as the case of dinitrophenol) (34), may well be rationally designed for potency and specificity. Designing high-affinity membrane protein inhibitors in this way may be a more effective strategy than tethering compounds to membrane-targeted compounds like TPP+ or plastoquinone (20, 35). These results also highlight the need to further understand the role of potassium ions in the mechanisms of new drug candidates. Finally, our work suggests new respiratory inhibitors must be considered in the context of entire respiratory chains and the PMF that intrinsically connects them.
Materials and Methods
Bacterial strains, media and growth conditions, sample preparation (inverted membrane vesicles, F1Fo proteoliposomes, cytochrome bo3 proteoliposomes, Fo-containing and empty pyranine liposomes), determination of cell growth inhibition, and analytical methods are described in SI Appendix.
ATP Synthesis and Hydrolysis Assays.
For endpoint measurements in IMVs, ATP synthesis was measured using the hexokinase/glucose-6-phosphate dehydrogenase assay as previously described (10) and ATP hydrolysis was measured using the spectrophotometric Pi release assay as previously described (36). Real-time ATP synthesis measurments were made in an Oroboros O2k fluorespirometer, a clark-type oxygen electrode, modified to simultaneously measure ATP by the previously described luciferase assay (37). Further details are available in SI Appendix. F1Fo-proteoliposome samples were not preincubated with BDQ for ATP hydrolysis experiments and measured using the spectrophotometric ATP-regenerating assay as previously described (36). All assays were performed at 37 °C.
Fluorescence Quenching Dependent on ∆pH or ∆ψ.
Fluorescence quenching of the pH-responsive fluorophores 9-amino-6-chloro-2-methoxyacridine (ACMA) (excitation: 430 nm, emission: 470 nm) or acridine orange (excitation: 493 nm, emission: 530 nm) was performed essentially as previously described (13). The following modifications were made: 0.2 mg⋅mL−1 (final concentration) IMVs or 5 μL⋅mL−1 F1Fo proteoliposomes were added NADH or ATP were used to initiate quenching as indicated. Unless otherwise indicated the concentration of acridine orange was 5 μM. Assays were performed at 37 °C. For cytochrome bo3 (cbo3) proteoliposomes fluorescence quenching of the pH responsive fluorophore, ACMA, was measured and is described in SI Appendix. Fluorescence quenching of the ∆ψ responsive fluorophore oxonol VI was performed as previously described (13), except quenching was measured photometrically at 590–630 nm and NADH was simultaneously measured at 340 nm.
Internal pH Quantification by Pyranine Fluorescence.
The internal pH of pyranine-containing liposomes was determined as previously described (25), liposomes at a concentration of 60 mg (dry weight)/mL (lipid:dye ratio = 30 mg lipid/μmol pyranine) were 100-fold diluted in incorporation buffer with the salt and pH values indicated in text. A calibration curve of fluorescence ratio to pH was determined for each incorporation buffer, containing 20 nM pyranine, at known pH values (SI Appendix, Fig. S1A and Table S1). The contributions of trace external pyranine were removed according to the equations defined in ref. 25. Preparations of Fo-containing liposomes routinely had 50–60% of the liposomes with Fo inserted, as assessed by the ratio of proton transport observed from a K+/valinomycin diffusion potential vs. that of the protonophore CCCP (SI Appendix, Fig. S1B). We did not correct for this, to enable comparison with empty liposome controls. Our preparations were sensitive to DCCD (SI Appendix, Fig. S1C), confirming the fidelity (coupled activity) of our preparation. Kinetic traces were measured on a Varian Cary Eclipse fluorimeter with continuous stirring. Other experiments, presented as endpoint measurements, used a Varioskan Flash plate reader, although traces were routinely recorded to verify experimental integrity. Assays were performed at 37 °C.
Supplementary Material
Acknowledgments
We thank the anonymous reviewers for their insightful comments regarding the interpretation of these results. Bedaquiline was a kind gift of Koen Andries, Janssen Research and Development, Johnson and Johnson Pharmaceuticals. This research was funded by the Maurice Wilkins Centre for Molecular Biodiscovery and the Marsden Fund, Royal Society. K.H. was supported by a University of Otago doctoral scholarship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803723115/-/DCSupplemental.
References
- 1.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 2.Andries K, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 3.Jones D. Tuberculosis success. Nat Rev Drug Discov. 2013;12:175–176. doi: 10.1038/nrd3957. [DOI] [PubMed] [Google Scholar]
- 4.Somoskovi A, Bruderer V, Hömke R, Bloemberg GV, Böttger EC. A mutation associated with clofazimine and bedaquiline cross-resistance in MDR-TB following bedaquiline treatment. Eur Respir J. 2015;45:554–557. doi: 10.1183/09031936.00142914. [DOI] [PubMed] [Google Scholar]
- 5.Hartkoorn RC, Uplekar S, Cole ST. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014;58:2979–2981. doi: 10.1128/AAC.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andries K, et al. Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One. 2014;9:e102135. doi: 10.1371/journal.pone.0102135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koul A, et al. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat Chem Biol. 2007;3:323–324. doi: 10.1038/nchembio884. [DOI] [PubMed] [Google Scholar]
- 8.Preiss L, et al. Structure of the mycobacterial ATP synthase Fo rotor ring in complex with the anti-TB drug bedaquiline. Sci Adv. 2015;1:e1500106. doi: 10.1126/sciadv.1500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koul A, et al. Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis. J Biol Chem. 2008;283:25273–25280. doi: 10.1074/jbc.M803899200. [DOI] [PubMed] [Google Scholar]
- 10.Haagsma AC, et al. Selectivity of TMC207 towards mycobacterial ATP synthase compared with that towards the eukaryotic homologue. Antimicrob Agents Chemother. 2009;53:1290–1292. doi: 10.1128/AAC.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng X, et al. Antiinfectives targeting enzymes and the proton motive force. Proc Natl Acad Sci USA. 2015;112:E7073–E7082. doi: 10.1073/pnas.1521988112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koul A, et al. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat Commun. 2014;5:3369. doi: 10.1038/ncomms4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hards K, et al. Bactericidal mode of action of bedaquiline. J Antimicrob Chemother. 2015;70:2028–2037. doi: 10.1093/jac/dkv054. [DOI] [PubMed] [Google Scholar]
- 14.Lamprecht DA, et al. Turning the respiratory flexibility of Mycobacterium tuberculosis against itself. Nat Commun. 2016;7:12393. doi: 10.1038/ncomms12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholls DG, Ferguson SJ. Bioenergetics. 4th Ed. Elsevier; Amsterdam: 2013. Ion transport across energy-conserving membranes; pp. 13–25. [Google Scholar]
- 16.Cook GM, Greening C, Hards K, Berney M. 2014. Energetics of pathogenic bacteria and opportunities for drug development. Advances in Bacterial Pathogen Biology, ed Poole RK (Elsevier, Amsterdam), Vol 65, pp 1–62.
- 17.McLaughlin SG, Dilger JP. Transport of protons across membranes by weak acids. Physiol Rev. 1980;60:825–863. doi: 10.1152/physrev.1980.60.3.825. [DOI] [PubMed] [Google Scholar]
- 18.Schwaller M-A, Allard B, Lescot E, Moreau F. Protonophoric activity of ellipticine and isomers across the energy-transducing membrane of mitochondria. J Biol Chem. 1995;270:22709–22713. doi: 10.1074/jbc.270.39.22709. [DOI] [PubMed] [Google Scholar]
- 19.Sun X, Garlid KD. On the mechanism by which bupivacaine conducts protons across the membranes of mitochondria and liposomes. J Biol Chem. 1992;267:19147–19154. [PubMed] [Google Scholar]
- 20.Antonenko YN, et al. Derivatives of rhodamine 19 as mild mitochondria-targeted cationic uncouplers. J Biol Chem. 2011;286:17831–17840. doi: 10.1074/jbc.M110.212837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haagsma AC, Driessen NN, Hahn M-M, Lill H, Bald D. ATP synthase in slow- and fast-growing mycobacteria is active in ATP synthesis and blocked in ATP hydrolysis direction. FEMS Microbiol Lett. 2010;313:68–74. doi: 10.1111/j.1574-6968.2010.02123.x. [DOI] [PubMed] [Google Scholar]
- 22.Tran SL, Cook GM. The F1Fo-ATP synthase of Mycobacterium smegmatis is essential for growth. J Bacteriol. 2005;187:5023–5028. doi: 10.1128/JB.187.14.5023-5028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferguson SA, Cook GM, Montgomery MG, Leslie AGW, Walker JE. Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum. Proc Natl Acad Sci USA. 2016;113:10860–10865. doi: 10.1073/pnas.1612035113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholls DG, Ferguson SJ. Bioenergetics. 4th Ed. Elsevier; Amsterdam: 2013. ATP synthases and bacterial flagella rotary motors; pp. 197–220. [Google Scholar]
- 25.Wiedenmann A, Dimroth P, von Ballmoos C. Deltapsi and DeltapH are equivalent driving forces for proton transport through isolated F(0) complexes of ATP synthases. Biochim Biophys Acta. 2008;1777:1301–1310. doi: 10.1016/j.bbabio.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Soga N, Kinosita K, Jr, Yoshida M, Suzuki T. Kinetic equivalence of transmembrane pH and electrical potential differences in ATP synthesis. J Biol Chem. 2012;287:9633–9639. doi: 10.1074/jbc.M111.335356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hnedkovsky L, Wood RH, Balashov VN. Electrical conductances of aqueous Na2SO4, H2SO4, and their mixtures: Limiting equivalent ion conductances, dissociation constants, and speciation to 673 K and 28 MPa. J Phys Chem B. 2005;109:9034–9046. doi: 10.1021/jp045707c. [DOI] [PubMed] [Google Scholar]
- 28.Epstein W. Ion Transport in Prokaryotes. Academic; New York: 2014. Potassium transport in bacteria; p. 85. [Google Scholar]
- 29.Kundu S, Biukovic G, Grüber G, Dick T. Bedaquiline targets the ε subunit of mycobacterial F-ATP synthase. Antimicrob Agents Chemother. 2016;60:6977–6979. doi: 10.1128/AAC.01291-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biukovic G, et al. Variations of subunit varepsilon of the Mycobacterium tuberculosis F1Fo ATP synthase and a novel model for mechanism of action of the tuberculosis drug TMC207. Antimicrob Agents Chemother. 2013;57:168–176. doi: 10.1128/AAC.01039-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haagsma AC, et al. Probing the interaction of the diarylquinoline TMC207 with its target mycobacterial ATP synthase. PLoS One. 2011;6:e23575. doi: 10.1371/journal.pone.0023575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeiffer DR, Taylor RW, Lardy HA. Ionophore A23187: Cation binding and transport properties. Ann N Y Acad Sci. 1978;307:402–423. [Google Scholar]
- 33.Rao SPS, Alonso S, Rand L, Dick T, Pethe K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2008;105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grundlingh J, Dargan PI, El-Zanfaly M, Wood DM. 2,4-dinitrophenol (DNP): A weight loss agent with significant acute toxicity and risk of death. J Med Toxicol. 2011;7:205–212. doi: 10.1007/s13181-011-0162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunn EA, et al. Incorporation of triphenylphosphonium functionality improves the inhibitory properties of phenothiazine derivatives in Mycobacterium tuberculosis. Bioorg Med Chem. 2014;22:5320–5328. doi: 10.1016/j.bmc.2014.07.050. [DOI] [PubMed] [Google Scholar]
- 36.Ferguson SA, Keis S, Cook GM. Biochemical and molecular characterization of a Na+-translocating F1Fo-ATPase from the thermoalkaliphilic bacterium Clostridium paradoxum. J Bacteriol. 2006;188:5045–5054. doi: 10.1128/JB.00128-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki T, Ozaki Y, Sone N, Feniouk BA, Yoshida M. The product of uncI gene in F1Fo-ATP synthase operon plays a chaperone-like role to assist c-ring assembly. Proc Natl Acad Sci USA. 2007;104:20776–20781. doi: 10.1073/pnas.0708075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.