Significance
To avoid the mucosal barrier and attach to the intestinal epithelium, enteric pathogens have evolved a unique proteinaceous fiber called type IVb pilus (T4bP). Despite its importance for bacterial pathogenesis, little is known about the adhesion mechanisms of T4bP, especially regarding the role of the minor pilin subunit located at its tip. Here, we show that the type IVb minor pilin CofB of CFA/III from enterotoxigenic Escherichia coli (ETEC) plays a role not only in T4bP assembly by forming a trimeric initiator complex, but also in bacterial adhesion by anchoring a secreted protein, CofJ, at the trimerization interface of H-type lectin domain. These findings expand our knowledge of T4P biology and provide important insights for developing therapeutics against ETEC infection.
Keywords: ETEC, type IV pili, minor pilin, pilus assembly, protein crystallography
Abstract
Initial attachment and subsequent colonization of the intestinal epithelium comprise critical events allowing enteric pathogens to survive and express their pathogenesis. In enterotoxigenic Escherichia coli (ETEC), these are mediated by a long proteinaceous fiber termed type IVb pilus (T4bP). We have reported that the colonization factor antigen/III (CFA/III), an operon-encoded T4bP of ETEC, possesses a minor pilin, CofB, that carries an H-type lectin domain at its tip. Although CofB is critical for pilus assembly by forming a trimeric initiator complex, its importance for bacterial attachment remains undefined. Here, we show that T4bP is not sufficient for bacterial attachment, which also requires a secreted protein CofJ, encoded within the same CFA/III operon. The crystal structure of CofB complexed with a peptide encompassing the binding region of CofJ showed that CofJ interacts with CofB by anchoring its flexible N-terminal extension to be embedded deeply into the expected carbohydrate recognition site of the CofB H-type lectin domain. By combining this structure and physicochemical data in solution, we built a plausible model of the CofJ–CFA/III pilus complex, which suggested that CofJ acts as a molecular bridge by binding both T4bP and the host cell membrane. The Fab fragments of a polyclonal antibody against CofJ significantly inhibited bacterial attachment by preventing the adherence of secreted CofJ proteins. These findings signify the interplay between T4bP and a secreted protein for attaching to and colonizing the host cell surface, potentially constituting a therapeutic target against ETEC infection.
To attach to and colonize the host cell surface, bacterial pathogens have evolved myriad surface organelles, the majority of which form filamentous protein polymers termed pili or fimbriae (1–3). Comprehensive understanding of their structures and adhesion mechanisms is crucial to develop novel vaccines and/or antiadhesive therapies, but is still lacking, especially for enterotoxigenic Escherichia coli (ETEC), a major cause of diarrhea in travelers and children in developing countries (4, 5). Although their complexity and repertoire are increasing (5–7), at least 22 types of pilus-related colonization factors (CFs) of ETEC have been identified and categorized as either CF antigens (CFAs) or coli surface antigens (CSs) (8, 9). Of these 22 CFs, 17 are assembled by the chaperone–usher (CU) pathway through polymerization of major and minor pilus subunits (termed pilins) via the extensively studied “donor-strand exchange” mechanism (1, 2). Polymerization is initiated with a minor pilin that is located at the distal end, where it also functions as a tip adhesin (10). Most tip adhesins of ETEC CU pili fold into two Ig-like domains with an N-terminal receptor-binding lectin domain that recognizes glycoconjugates or glycosphingolipids, indicating glycan-mediated host–pathogen interaction as a common theme for ETEC infection (11–14).
ETEC also has two CFs, CS8 and CS21, alternatively known as CFA/III and Longus, respectively, that are processed by the type IV pilus (T4P) assembly pathway, which is substantially more complex than the CU pathway (1, 15–17). T4Ps, which are found in a wide variety of Gram-negative bacteria, are implicated in multiple biological functions, including surface motility, biofilm formation, cell adhesion, autoaggregation, host-cell invasion, and DNA uptake (16). Generally, they are further classified according to their type IV pilin signal sequence and molecular size as type IVa and IVb (18). Recently, an alternative classification method utilizing other T4P components (e.g., platform protein) has been proposed, which phylogenetically groups the tight adherence (Tad)-type pili as type IVc (19). Although the number of pili categorized as T4P is still limited and can be expected to increase upon future genomics studies of bacteria, current knowledge suggests that the type IVa pilus is a relatively homogeneous subclass and is distributed in many Gram-negative bacterial species, whereas type IVb pilus (T4bP), often encoded by an operon, is a more heterogeneous class that is found primarily in enteric bacteria (20). Notably, CFA/III and Longus are categorized as type IVb subclass and are structurally and functionally similar to other T4bPs from enteric pathogens, such as toxin-coregulated pilus (TCP) of Vibrio cholerae and CF Citrobacter of Citrobacter rodentium (20, 21).
Although little is known regarding T4bP, we and other researchers recently proposed a filament model of CFA/III, in which each major pilin (CofA) globular head is arranged in a three-start left-handed helix (or, equivalently, a one-start right-handed helix with helically arranged N-terminal α-helices) (22, 23). We also revealed that minor pilin, CofB, has a three-domain architecture and is located at the pilus tip (24). The N-terminal domain of CofB adopts a typical type IVb pilin α/β-roll fold, whereas the C-terminal region is uniquely composed of two β-strand-rich domains that homotrimerize to form a pilus assembly initiator complex that efficiently promotes T4bP assembly (24). Trimeric initiator complex-mediated pilus assembly is likely conserved in a wide variety of pilus or pseudopilus systems, such as T4P assembly and its evolutionarily related type II secretion systems (18, 24), and may represent a bacterial tactic evolved to efficiently assemble stable filamentous appendages for enhancing pathogenesis. Moreover, at the distal pilus end, the CofB C-terminal domain adopts an H-type lectin fold that bears substantial similarity with the discoidin I trimeric structure (25). As H-type lectins typically bind N-acetylgalactosamine (GalNAc) molecules at the conserved binding pocket of their association interfaces, this suggested that CofB might function as a lectin targeting the small intestinal mucosal glycome.
Here, we examined the ability of CFA/III for bacterial attachment, and we unexpectedly found that it requires additional interaction with a secreted protein, CofJ, at the expected GalNAc-binding interfaces of the CofB trimer located at its tip, providing a clue for understanding the mechanism of ETEC infection.
Results
ETEC Adherence to Intestinal Epithelial Cells Requires CFA/III and Secreted Protein CofJ.
Based on its structural similarity with H-type lectin family proteins (24), we initially considered CofB as a tip-localized adhesin recognizing carbohydrate molecules. However, we found no indication of binding between CofB and GalNAc molecules in isothermal titration calorimetry (ITC) experiments (SI Appendix, Fig. S1), which is in part consistent with the inability of CFA/III-expressing ETEC to hemaglutinate erythrocytes (26).
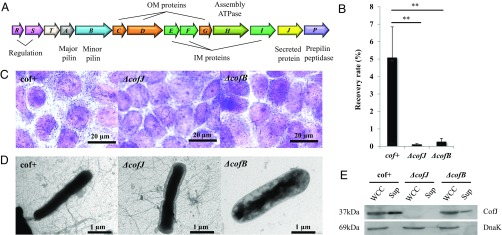
Although CFA/III is apparently required for bacterial attachment, it was also recently proposed to be involved in transporting CofJ (27), a secreted protein of unknown function encoded in the 14-gene CFA/III operon (cof operon) (Fig. 1A) (28). To clarify the relationship between CofJ and CFA/III, we performed cell adherence assays with different E. coli strains, including the CFA/III-positive HB101 strain carrying the recombinant plasmid pTT240 that harbors all cof operon genes (cof+ strain) (28) and the HB101 derivatives, CofJ- or CofB-deficient strains (ΔcofJ and ΔcofB strains, respectively), using human colon adenocarcinoma-derived Caco-2 cells (Fig. 1 B and C). The cof+ recovery rate calculated by dividing the number of colony-forming units (cfus) by that of the inoculum was 5.1%, whereas recovery rates of ΔcofJ and ΔcofB strains were markedly decreased to 0.01% and 0.3%, respectively (Fig. 1B). Because transmission electron microscopy (TEM) observation showed that cof+ and ΔcofJ strains expressed peritrichous CFA/III pili having lengths and structures similar to those of the wild-type ETEC 260-1 and 31-10 strains (Fig. 1D) (26, 27), these results suggested that ETEC adherence to intestinal cells requires not only the formation of CFA/III, but also the secretion of CofJ.
Fig. 1.
CFA/III-mediated E. coli adherence to Caco-2 cells. (A) Genetic organization of the cof gene cluster. Predicted functions for each gene product are indicated at the Top and Bottom of the figure. (B) Adherence values correspond to the recovery rate after incubation for 3 h. cof+ is an E. coli strain HB101 harboring all cof operon genes on the plasmid pTT240, ΔcofJ is a cofJ deletion mutant, and ΔcofB is a cofB deletion mutant. The experiments were performed five times. cof+, ΔcofJ, and ΔcofB recovery rates were 5.08 ± 1.76%, 0.12 ± 0.04%, and 0.27 ± 0.19%, respectively. **P < 0.005 vs. cof+. (C) Micrographs showing adhesion of cof+, ΔcofJ, and ΔcofB strains to Caco-2 cells. (D) TEM images of cof+, ΔcofJ, and ΔcofB strains grown on CFA agar plates. (E) Western blot analysis of whole cell culture (WCC, Left) and supernatant (Sup, Right) of cof+, ΔcofJ, and ΔcofB strains using anti-CofJ antibody. DnaK was detected with anti-DnaK antibody as a loading control.
Remarkably, no pili were observed for the ΔcofB strain by TEM, consistent with the previously proposed role of CofB in pilus assembly initiation (Fig. 1D) (23, 24), and Western blotting analysis clearly demonstrated equivalent amounts of CofJ were expressed by cof+ and ΔcofB strains in whole cell culture (Fig. 1E). However, CofJ secretion into the culture supernatant was substantially reduced in the ΔcofB strain (Fig. 1E). A similar, but even more significant reduction in CofJ secretion was confirmed in culture supernatants of CofA- or CofD-deficient strains (ΔcofA and ΔcofD strains, respectively), in which the gene encoding major pilin or outer membrane secretin, respectively, was disrupted (SI Appendix, Fig. S2). While CofJ secretion by the ETEC 31-10P-pcofJ strain that lacks all of the cof operon genes but harbors a cofJ gene-harboring plasmid reportedly was negligible (27), we detected residual amounts of secreted CofJ in the culture supernatants of the ΔcofA and ΔcofD strains, which we ascribed to cell fragility or protein leakage in these heterologous expression systems, as suggested previously (27). Nevertheless, the observed marked reduction in CofJ secretion from these deficient strains strikingly supports the previously proposed function of CFA/III in CofJ secretion, although the mechanism has not yet been studied.
CofJ Binds to the Minor Pilin CofB.
We hypothesized that the interaction between CofJ and CFA/III is a key step in establishing CofJ secretion and adhesion of ETEC to the extracellular milieu. To examine the interaction between CofJ and CFA/III and determine which of the two CFA/III subunits (CofA or CofB) interacts with CofJ, we performed pull-down assays using each pilin fused with a thioredoxin-His tag (Fig. 2). Because of the insolubility attributed to the pilin N-terminal characteristic hydrophobic segment (15), throughout the experiments, we used truncated CofA and CofB constructs for recombinantly expressed proteins, in which the N-terminal 28 residues are removed. Our experimental results showed that CofJ was detected only in the CofB coeluted fraction, indicating that CofJ specifically bound with the minor pilin CofB (Fig. 2). During the purification process, we also noted that a certain part (∼2 kDa) of CofJ was susceptible to degradation over time as demonstrated by the close doublet bands on SDS/PAGE; notably, these degraded CofJ proteins did not bind with CofB (SI Appendix, Fig. S3). We then solved the crystal structure of CofJ with extreme care to avoid protease digestion. The CofJ crystal structure solved by Dy derivative was determined at a resolution of 1.76 Å, with the crystal belonging to the space group of P212121 and a total of eight molecules in an asymmetric unit (SI Appendix, Fig. S4A). Comparison of these eight structures showed substantial structural similarity, with a root mean square deviation (rmsd) value of <0.237 Å (SI Appendix, Fig. S4 B and C). Owing to the intrinsic flexibility, however, the N-terminal 22 residues (2,214.3 Da) of all eight CofJ molecules were not included in the final models. The N terminus would be solvent exposed and hence susceptible to protease cleavage. Therefore, we suspected that the N-terminal region corresponds to the degraded 2-kDa fragment of CofJ.
Fig. 2.
Interaction analysis of pilin subunits and secreted protein CofJ. Pull-down assay was carried out for CofJ alone and the following pairs: CofJ and Trx-His-CofA and CofJ and Trx-His-CofB. The mixed samples were loaded onto Ni Sepharose beads that were then washed with buffer containing a low concentration of imidazole. Bound proteins were eluted with buffer containing a high concentration of imidazole and detected by SDS/PAGE. The molecular weight markers are indicated on the Left, and the position of each protein is indicated on the Right.
To confirm this hypothesis, we prepared an N-terminal 24 residue-truncated mutant, ΔN24-CofJ, with the Arg24 C terminus being a possible CofJ digestion site. Strikingly, pull-down assay of ΔN24-CofJ showed that it did not bind to CofB (SI Appendix, Fig. S3). A subsequent ITC experiment to measure the interaction between a synthesized peptide consisting of CofJ residues 1–24 [CofJ (1–24) peptide] and CofB revealed that the peptide bound to CofB with an affinity of Kd = 8.8 ± 1.6 μM, whereas the intact CofJ molecule bound to CofB with higher affinity of Kd = 0.14 ± 0.01 μM, and ΔN24-CofJ showed no binding (SI Appendix, Fig. S5). These results clearly indicated that the CofJ N-terminal region mediates the binding with CofB.
Crystal Structure of CofJ (1–24)–CofB.
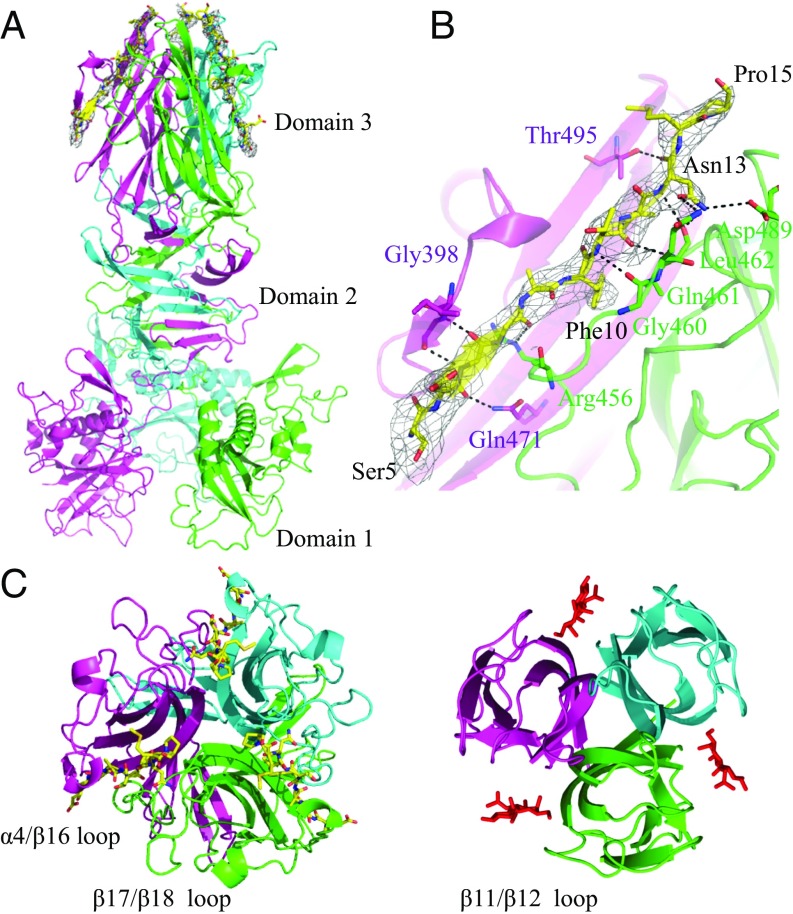
Although we were unable to obtain the cocrystal of CofB in complex with CofJ, the crystal of the CofJ (1–24)–CofB complex was successfully obtained. The crystal structure of the complex was determined by molecular replacement using the homotrimeric structure of CofB (24) as a search model that produced interpretable electron-density maps including well-defined densities responsible for three CofJ (1–24) peptides in the trimeric interfaces between each of two CofB molecule pairs (Fig. 3 A and B). The bulky Phe10 electron density of the CofJ (1–24) peptide was observed in each trimeric interface. The three CofJ (1–24) peptides were modeled using this Phe10 residue as a starting point. In contrast to the well-defined electron density observed for the central 5–15 residues of CofJ (1–24) peptide (Fig. 3B), the electron density map indicated that no structural model could be built for residues 1–4 and 16–24 owing to the scarce electron densities. After refinement, we determined the crystal structure of a CofJ (1–24)–CofB heterohexamer complex, including three CofB molecules and three CofJ (1–24) peptides, at 3.52-Å resolution (Fig. 3A).
Fig. 3.
Crystal structure of the CofJ (1–24)–CofB complex. (A) Side view of the structure with the CofB trimer (cyan, magenta, and green) shown in ribbon representation and bound CofJ (1–24) peptides shown as yellow sticks. A 2mFo−DFc omit map contoured at 1.0 σ corresponding to the region of peptide-binding grooves is shown. (B) Peptide-binding interface of CofJ (1–24)–CofB complex. Close-up view of the Ser5–Pro15 fragment of one of the three CofJ (1–24) peptides. CofJ (1–24) peptide is depicted as yellow sticks. The residues of CofB participating in hydrogen-bonding interactions are shown in stick representation. Hydrogen bonds are shown as black dotted lines. (C) Structural comparison of domain 3 of CofB with CofJ (1–24) peptides (Left) and the H-type lectin domain of discoidin I with GalNAc molecules (Right, PDB code 2WN3). In the CofJ (1–24)–CofB structure, the individual monomers are colored in cyan, magenta, and green. In the discoidin I structure, the three bound GalNAc molecules are represented as sticks.
In each trimeric interface, the CofJ (1–24) peptide fragment from Ser5 to Pro15 was bound to the hydrophobic groove sandwiched by two CofB C-terminal H-type lectin domains (domain 3s) (Fig. 3B and SI Appendix, Fig. S6). The structures of the three CofJ (1–24) peptides were essentially the same and well superimposed on each other, with rmsd values from 0.169 to 0.401 Å (SI Appendix, Fig. S7). Superposition of the complexed CofB trimer structure solved here on that determined previously of CofB alone showed that domains 2 and 3 of CofB do not undergo structural changes at the backbone level upon peptide binding, except the peptide-binding site regions and the arrangement of CofB molecule domain 1s in the CofJ (1–24)–CofB complex (SI Appendix, Figs. S8 and S9). CofB domain 3, which is composed of nine β-strands from β13 to β21 and one α-helix of α4, exhibits structural similarity with H-type lectins that form homotrimers recognizing GalNAc molecules (24, 25), but shows some structural differences with others. For example, the loop between CofB β17 and β18 is much longer than that of discoidin I (β11 and β12) and covers nearly the entire conserved sugar recognition site (Fig. 3C). Notably, by utilizing the long β17/β18 loop of one monomer and the α4/β16 loop of another, CofB recognizes the CofJ (1–24) peptide with slight local conformational changes in these loops (SI Appendix, Fig. S9). These unexpected features of CofB reasonably explain the fact that it precludes GalNAc binding in both the presence and absence of CofJ (SI Appendix, Fig. S1), suggesting functional “repurposing” of its H-type lectin domain.
In particular, the CofB trimer specifically recognizes only a limited part (Ser5–Pro15) of the CofJ (1–24) peptide by hydrophobic and hydrogen-bonding interactions, indicating that this region is a core fragment for CofB binding and that the central aromatic amino acid, Phe10, is critical for the interaction. Indeed, a synthetic 13-residue peptide comprising CofJ residues 4–16 [CofJ (4–16) peptide] that encompassed the above-mentioned recognizing sequence bound CofB trimer with an affinity (Kd = 4.8 ± 0.9 μM) similar to that of CofJ (1–24) peptide, and the substitution of Phe10 to Ala of CofJ (4–16) peptide completely abolished its binding ability (SI Appendix, Fig. S10). Sequence alignment showed that the corresponding aromatic residue is also found in operon-encoded secreted proteins of other T4bP assembly systems, along with meaningful N-terminal region sequence similarity (SI Appendix, Fig. S11) (21, 29–31). This highly conserved aromatic residue is thus likely key to binding with minor pilins at the region where the conserved sugar-binding site is located in H-type lectin family proteins. Although the mechanisms remain incompletely elucidated, it is noteworthy that the 20 flexible N-terminal residues, including the key aromatic residue at position 5, of the secreted protein TcpF of V. cholerae are reportedly critical for T4bP-mediated TcpF secretion, bacterial attachment, and microcolony formation (31), which is well explained by the interaction model provided here for CFA/III.
Structural Model of the CofJ–CFA/III Complex.
To determine the stoichiometry of CofB and intact CofJ molecules in solution, we subjected the purified CofJ–CofB complex to analytical ultracentrifugation. Sedimentation velocity experiments clearly demonstrated that the purified CofJ–CofB complex exists as a single species in solution, with a sedimentation coefficient of ∼8.7 S and an estimated molecular mass of 194 kDa (SI Appendix, Fig. S12). We previously reported that CofB in solution forms a trimer with a sedimentation coefficient of 7.3 S (158 kDa) (24). These results demonstrated that the CofJ–CofB complex in solution exists as a heterotetramer (∼198.1 kDa) with one intact CofJ molecule (∼37.5 kDa) and three CofB molecules (∼160.6 kDa).
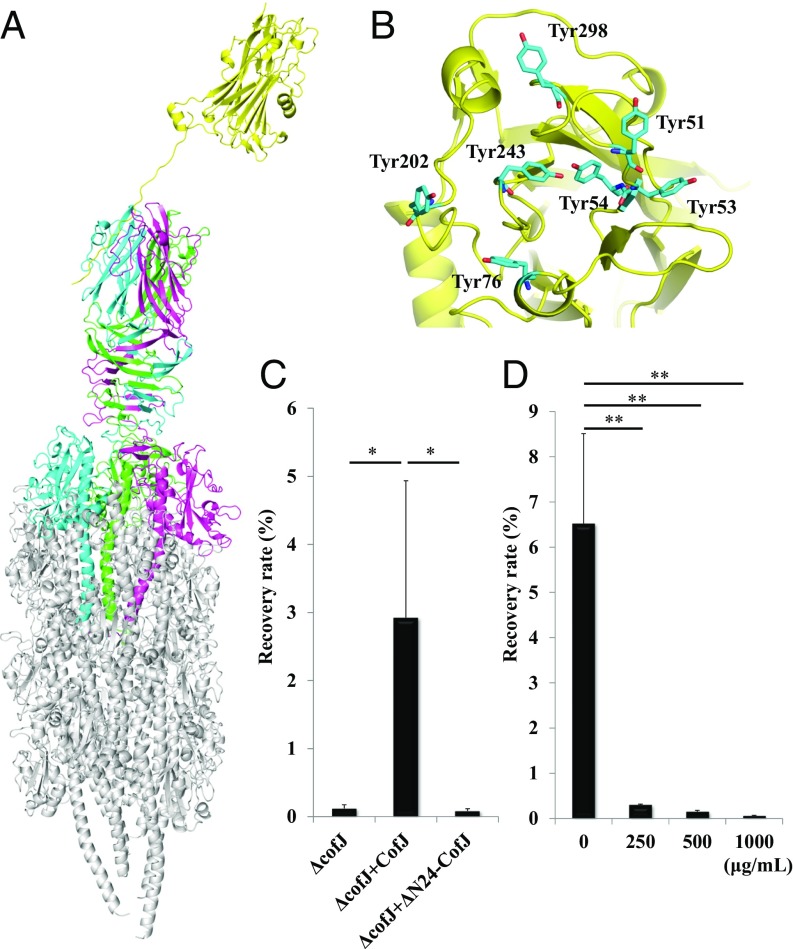
Based on these findings and the CFA/III pilus model reported previously (24), we generated a structural model of the CofJ–CFA/III complex in which the CofJ monomer was situated further above the minor pilin CofB at the tip of T4bP (Fig. 4A). The flexible gap (∼10 residues) between the globular CofJ domain and the N-terminal interaction site (Ser5–Pro15) of CofJ likely keeps the globular domain apart from the CofB trimer and sterically inhibits additional CofJ molecules from interacting with the top of the pilus tip, explaining the unique stoichiometry (1:3) of the CofJ–CofB complex. Notably, in the interaction model, the CofJ bottom surface is positively charged, whereas the CofB trimer top surface is negatively charged (SI Appendix, Fig. S13). This suggests that the globular domain of CofJ transiently interacts with CofB by electrostatic complementarity that may be responsible for the difference in affinity between intact CofJ and CofJ (1–24) peptide toward CofB as observed in the ITC experiments (SI Appendix, Fig. S5). The interaction model also revealed that the characteristic CofJ tyrosine cluster is located at the opposite side of the CofJ–CofB interface and is likely involved in target-cell recognition (Fig. 4B).
Fig. 4.
Structural model of the CofJ–CFA/III pilus complex. (A) Side view of the CofJ–CFA/III pilus depicted as a ribbon model, built by superimposing the CofJ (1–24)–CofB crystal structure onto our previously reported CFA/III pilus model with PyMOL. The CofJ N-terminal disordered residues (Ser1 to Ser4 and Lys16 to Asp22) were modeled and connected with the CofJ globular domain situated above the CofB homotrimer by using the program Coot (52). The CofJ monomer (yellow) is situated further above the minor pilin CofB homotrimer (cyan, magenta, and green) at the major pilin CofA (gray) pilus tip. (B) Ribbon representation of CofJ shown in the Top region with seven tyrosine residues (cyan stick representation). (C) Adherence values correspond to recovery rates after 3-h incubation. ΔcofJ is the cofJ deletion mutant. Experiments were performed five times. ΔcofJ alone, ΔcofJ plus CofJ, and ΔcofJ plus ΔN24-CofJ recovery rates were 0.12 ± 0.05%, 2.92 ± 2.01%, and 0.08 ± 0.03%, respectively. *P < 0.05 vs. ΔcofJ mixed with CofJ. (D) Results of cof+ adherence inhibition assay using anti-CofJ polyclonal antibody Fab fragments. Adherence values are as in C. Bottom values are anti-CofJ IgG antibody Fab fragment concentration. Experiments were performed five times. cof+ alone, cof+ plus 250, 500, and 1,000 µg/mL Fab fragment recovery rates were 6.53 ± 1.98%, 0.30 ± 0.01%, 0.15 ± 0.03%, and 0.06 ± 0.01%. **P < 0.005 vs. cof+ alone.
Adherence assays of the ΔcofJ strain to Caco-2 cells with addition of intact CofJ or ΔN24-CofJ yielded recovery rates of 2.9% and 0.1%, respectively, again confirming the importance of interaction between CofJ and CFA/III (Fig. 4C). In addition, adherence inhibition assays using Fab fragments of an anti-CofJ IgG antibody demonstrated that the Fab fragments could prevent the attachment of cof+ strain to Caco-2 cells (Fig. 4D). These results further supported the hypothesis that CofJ serves in an anchor role, bridging both the host-cell surface and the CofB trimer at the tip of CFA/III in the process of ETEC adherence.
Discussion
To attach to the host intestinal epithelium at a safe distance from the mucosal barrier, ETEC express long polymeric fibers termed “type IVb pili.” The most distal end of the pilus is likely involved in the initial contact with the epithelium, and we have shown that the minor pilin with an H-type lectin domain is located at that position (Fig. 4). However, although a CFA/III-positive E. coli strain lacking the cofJ gene that encodes the secreted protein successfully produced T4bP, it exhibited negligible adherence to Caco-2 cells. These results show the importance of the interplay of T4bP with a secreted protein for bacterial attachment during ETEC infection. Subsequent ITC experiments clearly demonstrated that CofJ interacts with the CofB trimer with submicromolar affinity. Clues regarding interaction details were provided by the serendipitous observation that lack of the ∼24-residue N-terminal flexible region completely diminished CofJ binding activity. The crystal structure of CofB in complex with a CofJ N-terminal 24-residue synthetic peptide revealed an unprecedented binding mode, by which the peptide is deeply embedded into the expected GalNAc-binding pocket typically conserved at the trimeric interface of the H-type lectin family (25). This finding provides a conceptual advance in T4P biology, wherein T4bP anchors the secreted protein at the pilus tip for pathogenesis, as well as a plausible answer for the currently debated association state—either monomeric or trimeric—of the type IVb minor pilin in action (24, 32–34), based on the evidence that the trimeric association of CofB is prerequisite for its stable interaction with CofJ (32–35).
Together with their gene synteny (10), the sequence similarities among minor pilins and among secreted-protein N-terminal regions suggest that this interplay is likely conserved in T4bP-expressing enteropathogens, including at least ETEC, V. cholerae, and C. rodentium (SI Appendix, Figs. S11 and S14). A database search using the CofB H-type lectin domain suggested that a wide variety of other enterobacteria potentially utilize similar T4bP assembly systems for attaching to and colonizing host intestinal epithelium, possibly through interaction with its cognate secreted protein (SI Appendix, Fig. S14 and Table S1).
Although the requirement of CofJ in ETEC adhesion is proposed here, the mechanistic details remain obscure. The crystal structure of CofJ, determined here and reported previously (27), demonstrates that CofJ shows marked structural homology with pore-forming toxins (PFTs), such as α-hemolysin from Staphylococcus aureus (36). Spatial and directional location of the CofJ N-terminal region is moreover also similar to that of the characteristic “amino-latch” N-terminal segment of α-hemolysin (SI Appendix, Fig. S15). In prepore-to-pore transition on the target-cell surface, the amino latch (typically 15–20 amino acids in length), initially flexible and highly exposed, plays an important role in stabilizing oligomeric association by anchoring neighboring subunits of PFTs (37, 38), whereas its truncation severely affects the ability to form functional pores (38). Notably, the CofB trimer specifically interacts with the N-terminal CofJ extension, which potentially hampers pore formation by this secreted protein. Although no experimental evidence indicating pore formation of CofJ is currently available, the architectural plan encompassing the prepore state of PFTs may serve as a molecular bridge that binds to both the cell surface and other molecules (Fig. 5).
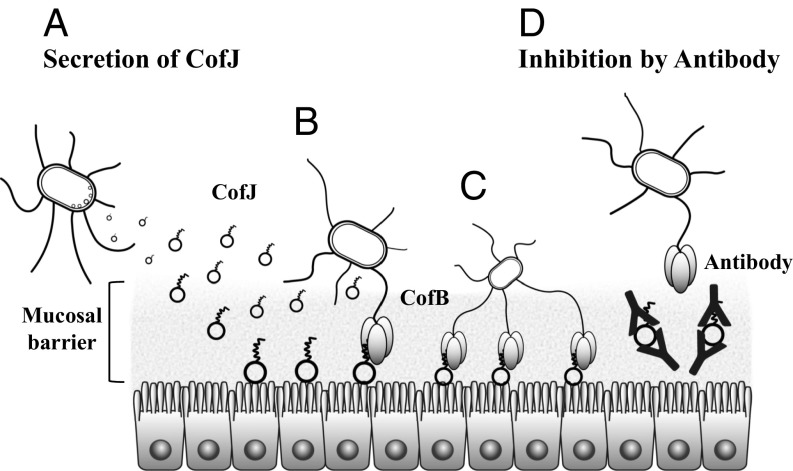
Fig. 5.
Schematic representation of initial attachment of ETEC and its inhibition by anti-CofJ antibody. (A) ETEC entering the small intestine first secretes CofJ molecules. These secreted CofJ molecules bind to the intestinal epithelial cell surface. (B) ETEC achieves adhesion to the intestinal cells by interaction between the N-terminal fragment of CofJ and the CofB trimer present at the tip of type IVb pilus. (C) A number of CofJ molecules on the cell surface are utilized for attachment of another ETEC. Multivalent binding is also possible using several pili to tightly adhere to cells. (D) Polyclonal anti-CofJ antibody inhibits the attachment of ETEC by blocking the interaction between CofJ and the CofB trimer.
In this context, the cluster of tyrosine residues forming an aromatic patch at the side opposite to the CofJ N-terminal interaction site is particularly interesting (Fig. 4 A and B). The presence of an aromatic patch is commonly observed in PFTs and might act as a multivalent lipid-binding site that recognizes lipid head groups of the cell membrane (39–41), and CofJ reportedly can bind lipid vesicles as well as epithelial cells, such as HeLa and Caco-2 (34). The lipid-binding nature of CofJ provides advantages for the bacteria by reserving the preferential adhesion spot for initial and/or future attachment to the epithelium as well as in competing with other bacterial species, including commensal species, for subsequent microcolony formation (Fig. 5).
To date, no broadly applicable vaccine for ETEC has been developed (42). This is partly explained by the high complexity of virulence factors, including more than at least 25 types of CFs, as well as several other secreted non-CF proteins, each of which functions at different stages of infection (43). Before intimate association with the target-cell surface, possibly via CFs of relatively short (1–5 μm) CU pili (44, 45), another specific mechanism is apparently required for initial attachment while avoiding the mucosal barrier consisting of an inner layer of ∼15–30 μm (46). One such mechanism was recently proposed for the non-CF ETEC virulence factor EtpA, which is secreted and interacts as a molecular bridge with target cells and the tip of flagellum, a common bacterial appendage with typical length of 10–15 μm (47, 48). Notably, the wild-type CFA/III strain is nonflagellated and nonmotile, and some ETEC strains that possess Longus, one of the most prevalent CFs among ETEC clinical isolates that has substantial structural homology with CFA/III, are also nonflagellated (49, 50). Our functional model of CofJ indicated it acts as a molecular bridge, in striking resemblance with EtpA; therefore, it is tempting to speculate that the interplay between the secreted protein and relatively long (5–15 μm or more) proteinaceous fibers, such as flagellum and T4bP (48), is a common strategy for initial attachment of ETEC, avoiding the mucosal barrier. Recent reports demonstrate that EtpA-targeted vaccination is effective (51), and our results showed that Fab fragments of an antibody against CofJ significantly inhibited adhesion of a CFA/III-expressing E. coli strain (Figs. 4D and 5). Given that the initial attachment of bacteria is a critical step common to all enteropathogens, the interplay between the secreted protein and T4bP shown here may constitute an attractive therapeutic target for vaccination and/or antiadhesive treatment against ETEC infection.
Materials and Methods
Details of experimental procedures are available in SI Appendix, SI Materials and Methods. The E. coli K12 derivative carrying recombinant plasmid harboring the entire cof gene cluster (cof+ strain) and its gene-deficient mutants (ΔcofA, ΔcofB, ΔcofD, and ΔcofJ) were used for CFA/III functional analyses. Recombinant CofA and CofB proteins were overexpressed and purified according to previously reported methods (22, 24). For recombinant CofJ and ΔN24-CofJ proteins, the E. coli expression strain SHuffle T7 Express LysY (New England Biolabs) was used for overexpression. Purification was carried out using standard techniques with affinity, ion-exchange, and size-exclusion chromatography. The Fab fragment of the anti-CofJ IgG antibody purified from rabbit serum and digested by papain was used for adherence inhibition assay. For evaluating the recovery rate, bacterial adherence assays were performed using Caco-2 cells with E. coli strains cof+, ΔcofB, and ΔcofJ grown on CFA agar (28) for efficient CFA/III expression. For each strain, CFA/III formation was checked by TEM. Western blot analyses were conducted using the strains cof+, ΔcofA, ΔcofB, ΔcofD, and ΔcofJ. CofJ in the culture supernatant was detected by using anti-CofJ IgG. Pull-down assay and ITC experiments were conducted using recombinant proteins, synthetic peptides, and GalNAc to analyze their potential interactions. CofJ was crystallized by the hanging-drop vapor diffusion method and the structure was determined by the SAD technique using the dysprosium heavy atom derivative. CofB in complex with CofJ (1–24) peptide was cocrystallized by the sitting-drop vapor diffusion method and the structure was solved by molecular replacement using the previously reported CofB structure (24). To determine the stoichiometry of the CofJ–CofB complex, sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation techniques were used.
Supplementary Material
Acknowledgments
The synchrotron X-ray diffraction experiments were performed with the approval of the SPring-8 Proposal Review Committee (2009A1283, 2009B1417, 2015A1101, 2015B2101, 2016A2569, and 2016B2569) and the Photon Factory Advisory Committee (2008G184 and 2010G071). Electron microscopy was performed by using the Analysis and Development System for Advanced Materials at the Research Institute for Sustainable Humanosphere, Kyoto University. This study was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science & Technology in Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure-factor information for CofJ and CofJ (1–24)-CofB complex have been deposited in the Protein Data Bank (PDB), www.wwpdb.org (PDB code: 5YQ0 and 5YPZ, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805671115/-/DCSupplemental.
References
- 1.Kline KA, Fälker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Hospenthal MK, Costa TRD, Waksman G. A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat Rev Microbiol. 2017;15:365–379. doi: 10.1038/nrmicro.2017.40. [DOI] [PubMed] [Google Scholar]
- 3.Soto GE, Hultgren SJ. Bacterial adhesins: Common themes and variations in architecture and assembly. J Bacteriol. 1999;181:1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization New frontiers in the development of vaccines against enterotoxigenic (ETEC) and enterohaemorrhagic (EHEC) E. coli infections. Wkly Epidemiol Rec. 1999;13:98–100. [PubMed] [Google Scholar]
- 5.von Mentzer A, et al. Identification of enterotoxigenic Escherichia coli (ETEC) clades with long-term global distribution. Nat Genet. 2014;46:1321–1326. doi: 10.1038/ng.3145. [DOI] [PubMed] [Google Scholar]
- 6.Sheikh A, et al. Highly conserved type 1 pili promote enterotoxigenic E. coli pathogen-host interactions. PLoS Negl Trop Dis. 2017;11:e0005586. doi: 10.1371/journal.pntd.0005586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Canto F, et al. Chaperone-Usher pili loci of colonization factor-negative human enterotoxigenic Escherichia coli. Front Cell Infect Microbiol. 2017;6:200. doi: 10.3389/fcimb.2016.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaastra W, Svennerholm AM. Colonization factors of human enterotoxigenic Escherichia coli (ETEC) Trends Microbiol. 1996;4:444–452. doi: 10.1016/0966-842x(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 9.Qadri F, Svennerholm AM, Faruque AS, Sack RB. Enterotoxigenic Escherichia coli in developing countries: Epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev. 2005;18:465–483. doi: 10.1128/CMR.18.3.465-483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madhavan TP, Sakellaris H. Colonization factors of enterotoxigenic Escherichia coli. Adv Appl Microbiol. 2015;90:155–197. doi: 10.1016/bs.aambs.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Wennerås C, Holmgren J, Svennerholm AM. The binding of colonization factor antigens of enterotoxigenic Escherichia coli to intestinal cell membrane proteins. FEMS Microbiol Lett. 1990;54:107–112. doi: 10.1016/0378-1097(90)90266-s. [DOI] [PubMed] [Google Scholar]
- 12.Madhavan TPV, Riches JD, Scanlon MJ, Ulett GC, Sakellaris H. Binding of CFA/I pili of enterotoxigenic Escherichia coli to Asialo-GM1 is mediated by the minor pilin CfaE. Infect Immun. 2016;84:1642–1649. doi: 10.1128/IAI.01562-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Greve H, Wyns L, Bouckaert J. Combining sites of bacterial fimbriae. Curr Opin Struct Biol. 2007;17:506–512. doi: 10.1016/j.sbi.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Jansson L, et al. Sulfatide recognition by colonization factor antigen CS6 from enterotoxigenic Escherichia coli. PLoS One. 2009;4:e4487. doi: 10.1371/journal.pone.0004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 16.Chang YW, et al. Architecture of the type IVa pilus machine. Science. 2016;351:aad2001. doi: 10.1126/science.aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang YW, et al. Architecture of the Vibrio cholerae toxin-coregulated pilus machine revealed by electron cryotomography. Nat Microbiol. 2017;2:16269. doi: 10.1038/nmicrobiol.2016.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giltner CL, Nguyen Y, Burrows LL. Type IV pilin proteins: Versatile molecular modules. Microbiol Mol Biol Rev. 2012;76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellison CK, et al. Obstruction of pilus retraction stimulates bacterial surface sensing. Science. 2017;358:535–538. doi: 10.1126/science.aan5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roux N, Spagnolo J, de Bentzmann S. Neglected but amazingly diverse type IVb pili. Res Microbiol. 2012;163:659–673. doi: 10.1016/j.resmic.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 21.Mundy R, et al. Identification of a novel type IV pilus gene cluster required for gastrointestinal colonization of Citrobacter rodentium. Mol Microbiol. 2003;48:795–809. doi: 10.1046/j.1365-2958.2003.03470.x. [DOI] [PubMed] [Google Scholar]
- 22.Fukakusa S, et al. Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 Å resolution by sulfur-SAD phasing. Acta Crystallogr D Biol Crystallogr. 2012;68:1418–1429. doi: 10.1107/S0907444912034464. [DOI] [PubMed] [Google Scholar]
- 23.Kolappan S, Roos J, Yuen AS, Pierce OM, Craig L. Structural characterization of CFA/III and Longus type IVb pili from enterotoxigenic Escherichia coli. J Bacteriol. 2012;194:2725–2735. doi: 10.1128/JB.00282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawahara K, et al. Homo-trimeric structure of the type IVb minor pilin CofB suggests mechanism of CFA/III pilus assembly in human enterotoxigenic Escherichia coli. J Mol Biol. 2016;428:1209–1226. doi: 10.1016/j.jmb.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Mathieu SV, Aragão KS, Imberty A, Varrot A. Discoidin I from Dictyostelium discoideum and interactions with oligosaccharides: Specificity, affinity, crystal structures, and comparison with discoidin II. J Mol Biol. 2010;400:540–554. doi: 10.1016/j.jmb.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honda T, Arita M, Miwatani T. Characterization of new hydrophobic pili of human enterotoxigenic Escherichia coli: A possible new colonization factor. Infect Immun. 1984;43:959–965. doi: 10.1128/iai.43.3.959-965.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuen AS, Kolappan S, Ng D, Craig L. Structure and secretion of CofJ, a putative colonization factor of enterotoxigenic Escherichia coli. Mol Microbiol. 2013;90:898–918. doi: 10.1111/mmi.12407. [DOI] [PubMed] [Google Scholar]
- 28.Taniguchi T, et al. Gene cluster for assembly of pilus colonization factor antigen III of enterotoxigenic Escherichia coli. Infect Immun. 2001;69:5864–5873. doi: 10.1128/IAI.69.9.5864-5873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez-Duarte OG, et al. Genetic diversity of the gene cluster encoding longus, a type IV pilus of enterotoxigenic Escherichia coli. J Bacteriol. 2007;189:9145–9149. doi: 10.1128/JB.00722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirn TJ, Bose N, Taylor RK. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol Microbiol. 2003;49:81–92. doi: 10.1046/j.1365-2958.2003.03546.x. [DOI] [PubMed] [Google Scholar]
- 31.Megli CJ, Taylor RK. Secretion of TcpF by the Vibrio cholerae toxin-coregulated pilus biogenesis apparatus requires an N-terminal determinant. J Bacteriol. 2013;195:2718–2727. doi: 10.1128/JB.01122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolappan S, Ng D, Yang G, Harn T, Craig L. Crystal structure of the minor pilin CofB, the initiator of CFA/III pilus assembly in enterotoxigenic Escherichia coli. J Biol Chem. 2015;290:25805–25818. doi: 10.1074/jbc.M115.676106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng D, et al. The Vibrio cholerae minor pilin TcpB initiates assembly and retraction of the toxin-coregulated pilus. PLoS Pathog. 2016;12:e1006109. doi: 10.1371/journal.ppat.1006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saldaña-Ahuactzi Z, et al. Effects of lng mutations on LngA expression, processing, and CS21 assembly in enterotoxigenic Escherichia coli E9034A. Front Microbiol. 2016;7:1201, and erratum (2017) 8:26. doi: 10.3389/fmicb.2016.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y, Hauke CA, Marles JM, Taylor RK. Effects of tcpB mutations on biogenesis and function of the toxin-coregulated pilus, the type IVb pilus of Vibrio cholerae. J Bacteriol. 2016;198:2818–2828. doi: 10.1128/JB.00309-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song L, et al. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 37.Dal Peraro M, van der Goot FG. Pore-forming toxins: Ancient, but never really out of fashion. Nat Rev Microbiol. 2016;14:77–92. doi: 10.1038/nrmicro.2015.3. [DOI] [PubMed] [Google Scholar]
- 38.Jayasinghe L, Miles G, Bayley H. Role of the amino latch of staphylococcal alpha-hemolysin in pore formation: A co-operative interaction between the N terminus and position 217. J Biol Chem. 2006;281:2195–2204. doi: 10.1074/jbc.M510841200. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka K, Caaveiro JMM, Morante K, González-Mañas JM, Tsumoto K. Structural basis for self-assembly of a cytolytic pore lined by protein and lipid. Nat Commun. 2015;6:6337. doi: 10.1038/ncomms7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huyet J, et al. Structural insights into Clostridium perfringens delta toxin pore formation. PLoS One. 2013;8:e66673. doi: 10.1371/journal.pone.0066673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson R, Nariya H, Yokota K, Kamio Y, Gouaux E. Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel. Nat Struct Biol. 1999;6:134–140. doi: 10.1038/5821. [DOI] [PubMed] [Google Scholar]
- 42.Zhang W, Sack DA. Current progress in developing subunit vaccines against enterotoxigenic Escherichia coli-associated diarrhea. Clin Vaccine Immunol. 2015;22:983–991. doi: 10.1128/CVI.00224-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fleckenstein JM, et al. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect. 2010;12:89–98. doi: 10.1016/j.micinf.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans DG, Silver RP, Evans DJ, Jr, Chase DG, Gorbach SL. Plasmid-controlled colonization factor associated with virulence in Esherichia coli enterotoxigenic for humans. Infect Immun. 1975;12:656–667. doi: 10.1128/iai.12.3.656-667.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Girón JA, Gómez-Duarte OG, Jarvis KG, Kaper JB. Longus pilus of enterotoxigenic Escherichia coli and its relatedness to other type-4 pili–A minireview. Gene. 1997;192:39–43. doi: 10.1016/s0378-1119(97)00039-5. [DOI] [PubMed] [Google Scholar]
- 46.McGuckin MA, Lindén SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 47.Roy K, et al. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature. 2009;457:594–598. doi: 10.1038/nature07568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar P, et al. Dynamic interactions of a conserved enterotoxigenic Escherichia coli adhesin with intestinal mucins govern epithelium engagement and toxin delivery. Infect Immun. 2016;84:3608–3617. doi: 10.1128/IAI.00692-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Girón JA, Levine MM, Kaper JB. Longus: A long pilus ultrastructure produced by human enterotoxigenic Escherichia coli. Mol Microbiol. 1994;12:71–82. doi: 10.1111/j.1365-2958.1994.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 50.Girón JA, et al. Prevalence and association of the longus pilus structural gene (lngA) with colonization factor antigens, enterotoxin types, and serotypes of enterotoxigenic Escherichia coli. Infect Immun. 1995;63:4195–4198. doi: 10.1128/iai.63.10.4195-4198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo Q, Vickers TJ, Fleckenstein JM. Immunogenicity and protective efficacy against enterotoxigenic Escherichia coli colonization following intradermal, sublingual, or oral vaccination with EtpA Adhesin. Clin Vaccine Immunol. 2016;23:628–637. doi: 10.1128/CVI.00248-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.