Significance
Recent studies have established that metabolic restrains, such as glucose restriction, impair the activities of effector T cells in the tumor microenvironment. In the same context, a huge expansion of activated Treg cells in tumor tissues has been described in mice and humans, contributing to the suppression of protective antitumor immunity. Our data demonstrate that Tregs are committed to survive and proliferate in such a hostile milieu thanks to a metabolic advantage based on the combination of glycolysis and fatty acid synthesis and oxidation. This allows Tregs to prevail over conventional T cells that rely primarily on the glycolytic pathway for their metabolic demands. Awareness of the metabolic dynamics of Tregs in tumor could provide a means for cancer immunotherapy.
Keywords: Treg, tumor microenvironment, glycolysis, fatty acid synthesis, ox40
Abstract
The tumor microenvironment restrains conventional T cell (Tconv) activation while facilitating the expansion of Tregs. Here we showed that Tregs’ advantage in the tumor milieu relies on supplemental energetic routes involving lipid metabolism. In murine models, tumor-infiltrating Tregs displayed intracellular lipid accumulation, which was attributable to an increased rate of fatty acid (FA) synthesis. Since the relative advantage in glucose uptake may fuel FA synthesis in intratumoral Tregs, we demonstrated that both glycolytic and oxidative metabolism contribute to Tregs’ expansion. We corroborated our data in human tumors showing that Tregs displayed a gene signature oriented toward glycolysis and lipid synthesis. Our data support a model in which signals from the tumor microenvironment induce a circuitry of glycolysis, FA synthesis, and oxidation that confers a preferential proliferative advantage to Tregs, whose targeting might represent a strategy for cancer treatment.
Regulatory T cells (hereafter, “Tregs”), i.e., Foxp3-expressing CD4 T lymphocytes, are widely recognized as crucial controllers of immune responses, and the balance between Tregs and effector T cells determines the success of immune surveillance over cancer progression. Therefore, understanding the dynamics that dictate the relative Treg advantage over conventional T cells (Tconvs) is of utmost importance in the design of cancer immunotherapies (1). Both thymus-derived Treg (tTreg) and peripherally induced Treg (pTreg) subsets populate the tumor microenvironment (TME) (2): While the differentiation of pTregs as well as in vitro-induced Tregs (iTregs) is favored in conditions of poor T cell proliferation, both pTregs and tTregs can proliferate actively in response to adequate signals (3, 4) and independently contribute to generate the Treg pool in tumor-bearing hosts (5).
The T cell switch between quiescence and activation relies on a metabolic shift from oxidative to glycolytic pathways, respectively ensuring long-term survival and fueling fast energy supply, biosynthesis, and replication (6). Distinct cellular subsets display different metabolic signatures. Many pieces of evidence suggest that Tconvs require amplifying oxidative pathways, while blunting glycolysis, to undergo the terminal/restrained activation that ultimately favors their differentiation into iTregs/pTregs (7–9). First, mouse iTregs have been shown to preferentially use fatty acid oxidation (FAO) (7, 8, 10); however glycolysis is essential for human iTreg development upon suboptimal stimulation, since the “moonlighting” enzyme enolase-1 suppresses the transcription of the exon 2-containing FOXP3-splicing variant (FOXP3-E2, a crucial determinant of the regulatory function) unless engaged in glycolysis (11).
The proliferation of preestablished Tregs may have completely distinct metabolic requirements with respect to the conversion of Tconvs into iTregs/pTregs. Ex vivo, highly proliferative human Tregs show active mTOR and high levels of the glucose transporter Glut1; both glycolytic and FAO pathways contribute to their expansion in vitro (12–14), and mTOR drives a lipogenic and cholesterogenic signature in Tregs which crucially sustains their expansion in vivo (15). Overall, these data indicate that Tregs may arrange both glycolytic and oxidative routes and, similar to memory CD8 T cells (16), may also perform fatty acid synthesis (FAS). This may be especially true in contexts of limited nutrient availability such as the TME, where tumor cells glucose-restrict effector T cells, thus blocking their glycolytic shift and antitumor activity (17, 18).
Here we hypothesized that Tregs may overwhelm Tconvs in the hostile tumor context because of their peculiar ability to exploit supplemental metabolic routes and thus optimize nutrient usage. We first documented in a mouse tumor model that tumor-infiltrating Tregs preferentially capture and utilize glucose not only to fuel glycolysis but also to synthesize and accumulate fatty acids (FAs) and that Tregs relied on FAS and FAO to a higher extent than Tconvs in vitro. Genes involved in glycolysis and lipid biosynthesis are enriched in the signature of activated Tregs sorted from human liver cancer. Thus, exploiting a combination of different metabolic routes may be an immune escape mechanism that confers a selective advantage to Tregs in the TME.
Results
Proliferating Tregs in the TME Accumulate Intracellular Lipids.
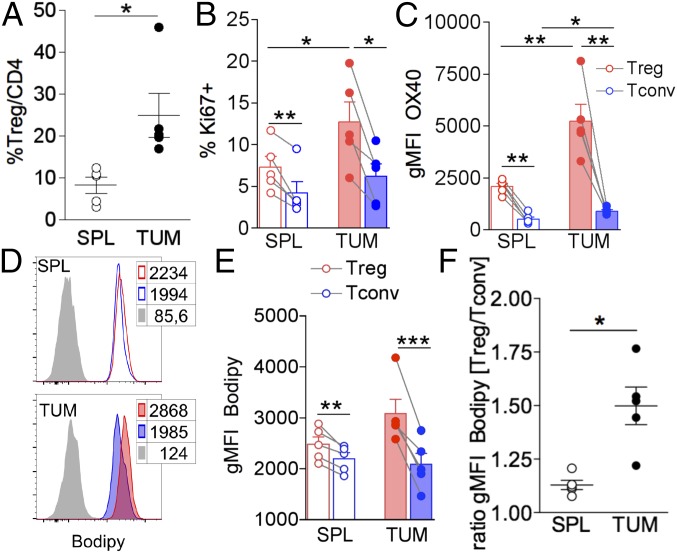
In the visceral adipose tissue (VAT), expanded Tregs accumulate intracellular lipids under the cell-intrinsic control of the adipogenic factor peroxisome proliferator-activated receptor gamma (PPARγ) (19). Therefore, we first checked whether lipid accumulation also occurred in Treg expansion in another context, i.e., in the TME. To this aim, CD4+Foxp3+ Tregs were analyzed by flow cytometry, in comparison with CD4+Foxp3− Tconvs, in cells extracted from tumor beds or, as control, from spleens of C57BL/6 wild-type mice bearing nodules of the s.c.-implanted colon carcinoma cell line MCA38 (the gating strategy is shown in SI Appendix, Fig. S1). Confirming previous studies in a variety of mouse tumor models and in human cancers (1), Treg frequency was significantly increased at the tumor site (Fig. 1A). Compared with spleen cells and Tconvs, Tregs extracted from the tumor bed (TUM-Tregs) were more proliferative, as pointed out by Ki67+ frequency (Fig. 1B), and expressed higher levels of the receptor OX40 (Fig. 1C), which we previously demonstrated sustains Treg fitness in mouse models of homeostatic proliferation and colitis (20) and promotes the expansion of stable and suppressive Tregs in human cancers (21, 22).
Fig. 1.
Expanded and activated tumor-infiltrating Tregs display high intracellular lipid content. MCA38 cells (5 × 105) were injected s.c. into C57BL/6 mice, and flow cytometry analysis was performed on lymphocytes extracted from spleen (SPL) and tumor (TUM) after 14 d. (A) Frequency of CD25+Foxp3+ Tregs in gated CD4+ lymphocytes. (B and C) Frequency of cells expressing Ki67 (B) and geometric mean fluorescent intensity (gMFI) of OX40 in gated CD25+Foxp3+ Tregs (red bars) and CD25−Foxp3− Tconvs (blue bars) (C) in spleens (empty bars) and tumors (filled bars) from tumor-bearing mice. (D and E) Representative histograms (D) and gMFI analysis (E) of Bodipy incorporation in gated CD25+Foxp3+ Tregs (red peaks and bars) and CD25−Foxp3− Tconvs (blue peaks and bars) in spleen (empty peaks and bars) and tumor (solid peaks and bars) from tumor-bearing mice. Peaks in gray represent the fluorescence-minus-one (FMO) controls. (F) Ratio of Bodipy gMFI between Tregs and Tconvs from spleen and tumor. Error bars show mean ± SEM; each dot corresponds to a single mouse. Data shown are from one representative of two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.005 by paired Student t test between Tregs and Tconvs in the same samples and by unpaired Student t test between populations of different samples.
When we stained the cells with Bodipy, a cell membrane-permeable fluorophore specific for neutral lipid stores, we could detect a higher intracellular lipid content in Tregs than in Tconvs from both spleen and tumor bed (Fig. 1 D and E and SI Appendix, Fig. S1B); however, this phenomenon especially affected TUM-Tregs, as evidenced by a significantly increased ratio of Bodipy levels between Tregs and Tconvs in the tumor bed compared with the spleen (Fig. 1F). Similar results were obtained in a distinct tumor model, i.e., the s.c.-injected melanoma B16F10 (SI Appendix, Fig. S2).
To ascertain whether the preferential lipid accumulation in Tregs was an exclusive feature of tumor or was shared with other conditions characterized by Treg expansion, we analyzed Treg frequency and intracellular lipid content in two distinct models of liver disease, namely the Mdr2−/− mouse in which an early-age inflammatory cholangitis progresses to cirrhosis and cancer (23) and the hepatitis C virus (HCV)-transgenic (HCVTg) mouse which spontaneously develops steatosis at advanced age with no evidence of inflammation (24). In young (3-wk-old) Mdr2−/− mice, hepatic Tregs displayed a higher frequency than in wild-type mice, and this was again associated with greater intracellular lipid content than in Tconvs (SI Appendix, Fig. S3 A–C). Conversely, hepatic Tregs were not expanded in aged (>10-mo-old) HCVTg compared with wild-type mice, and no preferential lipid accumulation was detected in Tregs (SI Appendix, Fig. S3 D–F). Overall these data indicate that Tregs enlarge their intracellular lipid pool when their expansion is triggered by either inflammation- or tumor-associated stimuli.
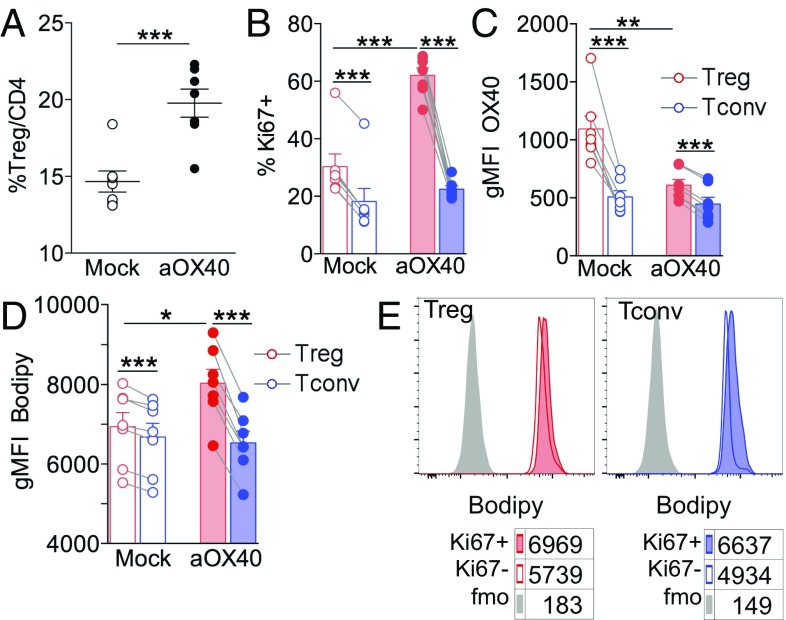
We have previously demonstrated that OX40 plays nonredundant roles in Treg expansion in a variety of contexts, including cancer (20, 21); therefore, we sought to test whether the OX40 signal could induce lipid accumulation in proliferating Tregs. To this aim, we injected an anti-OX40 agonist mAb, OX86, in naive mice. In agreement with published data (25), OX40 triggering induced a huge increase in Treg frequency (Fig. 2A). This expansion was likely due to the direct promotion of the selective proliferation of Tregs at the expense of Tconvs, as demonstrated by the increased percentage of Ki67+ Tregs but not Ki67+ Tconvs (Fig. 2B) and also corroborated by the decreased OX40 staining ex vivo (with the same OX86 clone) of Tregs but not Tconvs of αOX40-treated mice (Fig. 2C). Notably, αOX40-induced Treg expansion was associated with greater Bodipy staining of Tregs, which was even more pronounced in Ki67+ Tregs (Fig. 2 D and E). These results strengthen the link between Treg proliferation and FA accumulation and demonstrate that OX40 engagement alone can trigger the selective proliferation of lipid-laden Tregs.
Fig. 2.
OX40 triggering alone recapitulates Treg proliferation and FA accumulation in vivo. C57BL/6 naive mice were injected i.p. with 500 μg of anti-OX40 agonist mAb (OX86) or PBS as a mock control. After 6 d, flow cytometry analysis was performed in splenocytes. (A) Frequency of CD25+Foxp3+ Tregs in gated CD4+ lymphocytes. (B–D) Frequency of cells expressing Ki67 (B), gMFI analysis of OX40 (C), and gMFI analysis of Bodipy incorporation (D) in gated CD25+Foxp3+ Tregs (red bars) and CD25−Foxp3− Tconvs (blue bars) in αOX40-treated (solid bars) and mock-treated (open bars) mice. (E) Representative histograms showing Bodipy labeling in gated Ki67+ or Ki67− Tregs or Tconvs; numbers indicate Bodipy gMFI; peaks in gray represent the FMO controls. Error bars show mean ± SEM; each dot corresponds to a single mouse; six to seven mice were included in each experimental group. *P < 0.05, **P < 0.01, ***P < 0.005 by paired Student t test between Tregs and Tconvs in the same samples and by unpaired Student t test between populations of different samples.
FAS, Rather than FA Uptake, Shapes the TUM-Treg Lipid Pool and Promotes Treg Proliferation.
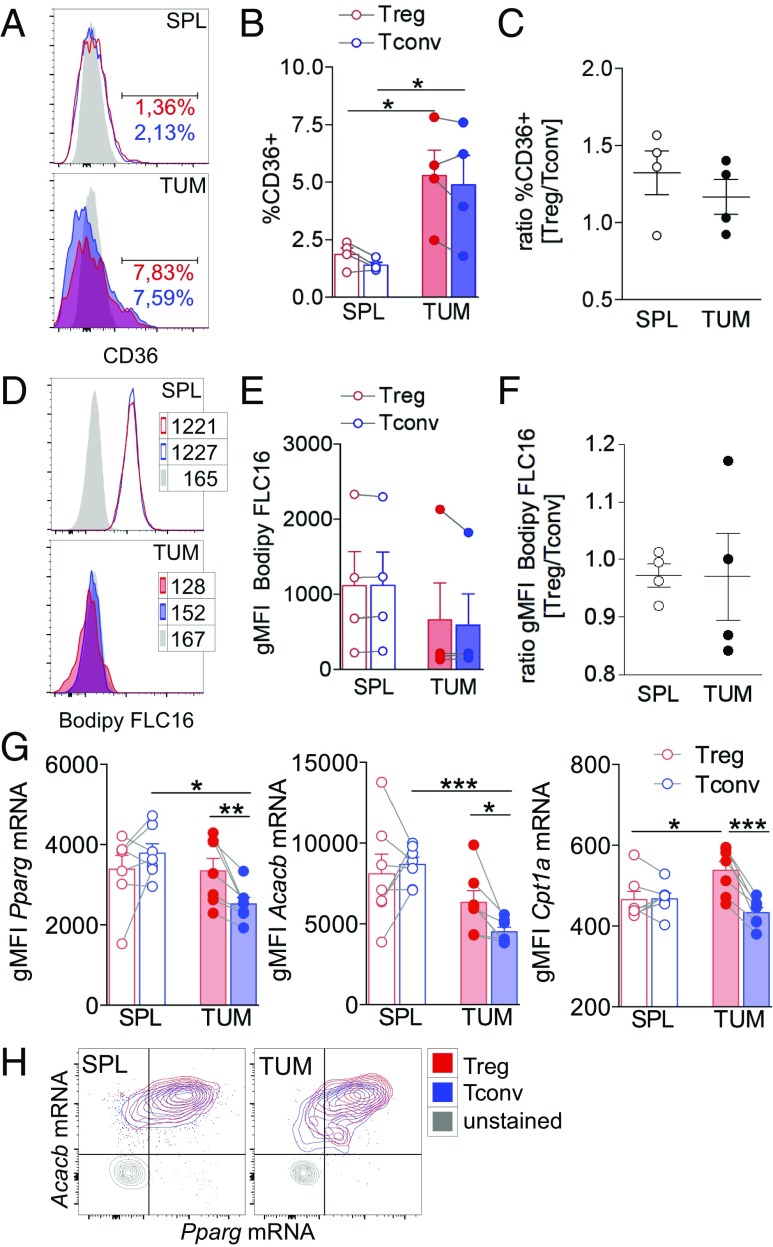
In the adipose tissue, lipid-laden Tregs also display higher levels of the FA translocator CD36 (19); therefore, we tested whether differential uptake of exogenous FAs might account for the higher lipid content in TUM-Tregs. We found that CD36 was modestly up-regulated at the tumor site in both Tregs and Tconvs but without any specific enrichment in TUM-Tregs (Fig. 3 A–C). Then we measured the absorption in vivo of a fluorescently labeled FA (Bodipy FL C16, a palmitic acid molecule conjugated to the Bodipy fluorophore), previously injected into tumor-bearing mice. Both Tregs and Tconvs tended to acquire less FA in the tumor site than in the spleen (despite the modest CD36 up-regulation) but without any major difference between the two subsets (Fig. 3 D–F). These results suggested a local defect in FA distribution and uptake shared by Tregs and Tconvs and unrelated to their CD36 levels, and excluded preferential FA scavenging by TUM-Tregs. Therefore we investigated whether, instead, intracellular FA metabolic routes were involved in lipid accumulation and Treg proliferation. First, in Tregs and Tconvs from tumor-bearing mice, we evaluated the expression levels of three selected genes, Pparg, Acacb, and Cpt1a, respectively required for FA storage, FAS, and FAO. To this aim, we adopted PrimeFlow RNA assay, allowing the simultaneous detection of transcripts and proteins by flow cytometry at single-cell resolution. With this approach, we could detect that TUM-Tregs expressed all three genes at significantly higher levels than TUM-Tconvs (Fig. 3G). Of note, Pparg and Acacb expression tended to cosegregate in all cell types and at higher levels in TUM-Tregs (Fig. 3H), suggesting a link between PPARγ expression and FAS programming specifically in these cells.
Fig. 3.
FAS, rather than FA uptake, shapes the lipid Treg pool and contributes to Treg proliferation. Mice bearing tumors 8 × 8 mm in size received an i.p. injection of the green fluorescent palmitate (Bodipy FL C16) 1 h before spleen and tumor collection. Then Bodipy FL C16 uptake and CD36 expression were quantified by flow cytometry in CD25+Foxp3+ Tregs and CD25−Foxp3− Tconvs. (A and B) Representative histograms (A) and frequency of cells expressing CD36 (B) in gated Tregs (red peaks and bars) and Tconvs (blue peaks and bars) in spleen (empty peaks and bars) and tumor (solid peaks and bars). (C) Ratio of the frequency of CD36-expressing Tregs and Tconvs in spleen and tumor. (D and E) Representative histograms (D) and gMFI of Bodipy FL C16 acquisition (E) in gated Tregs (red peaks and bars) and Tconvs (blue peaks and bars) in spleen (empty peaks and bars) and tumor (solid peaks and bars). (F) Ratio between Bodipy FL C16 gMFI of Tregs and Tconvs in spleen and tumor tissue. Peaks in gray represent the FMO controls. (G and H) The expression level of Pparg, Acacb, and Cpt1a mRNA was assessed by the PrimeFlow RNA assay in gated CD4+CD25+ Tregs and CD4+CD25− Tconvs. (G) gMFI of each gene expression in Tregs (red bars) and Tconvs (blue bars) in spleen (empty bars) and tumor (filled bars). (H) Representative overlay contour plots of Acacb- and Pparg-coexpressing cells in the indicated samples. Each dot corresponds to a single mouse; data shown are from one representative of four independent experiments, each including three to seven mice per group. Error bars show mean ± SEM; *P < 0.05, **P < 0.01, by paired Student t test between Tregs and Tconvs in the same sample and by unpaired Student t test between populations of different samples.
Therefore, we tested whether 5-(tetradecycloxy)-2-furoic acid (TOFA), an inhibitor of acetyl-CoA carboxylase, a key enzyme in the FAS cascade, impacted Treg proliferation. First, we administered TOFA to tumor-bearing mice, performing repeated intratumor injections starting when nodules were palpable. This treatment significantly suppressed tumor growth; however, this effect was unlikely to be due to immune-mediated events, as we failed to detect any significant increase in CD8 T cell frequency or IFNγ production in TOFA-treated compared with mock-treated nodules (SI Appendix, Fig. S4 A and B). Rather, direct TOFA toxicity against tumor cells might account for the antitumor activity observed in vivo. Indeed, TOFA also reduced the viability of the MCA38 cell line in a dose-dependent fashion in vitro, as evidenced in an XTT cytotoxicity assay (SI Appendix, Fig. S4C).
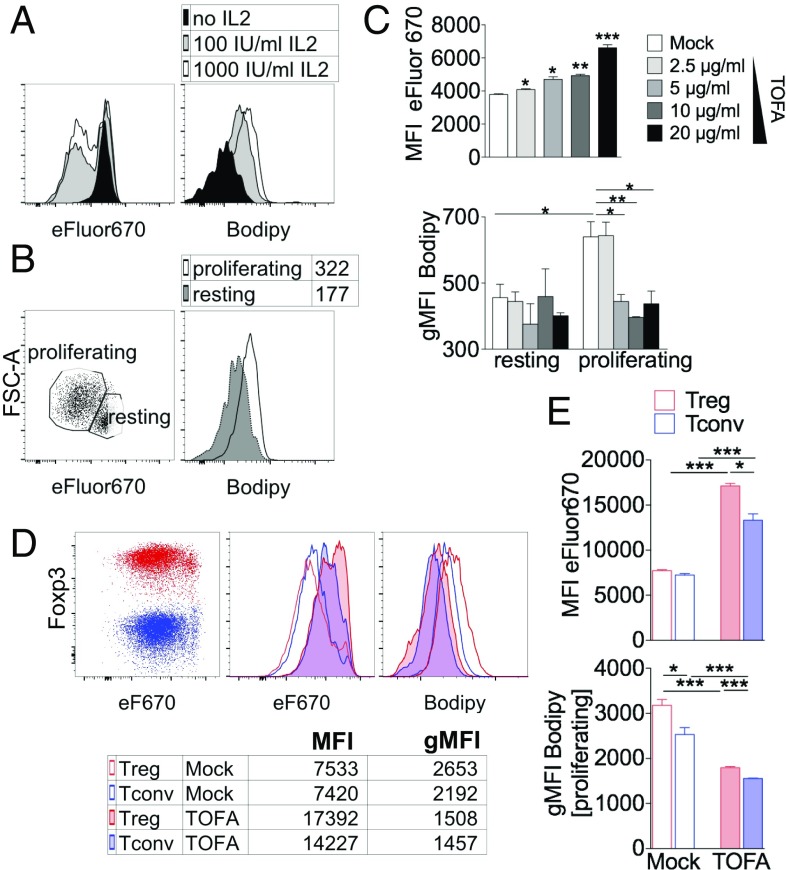
To assess the role of FAS directly on Tregs, we took advantage of a model of Treg proliferation in vitro which recapitulated the lipid accumulation occurring in TUM-Tregs in vivo. To this aim, Tregs were enriched from the spleens of naive mice and polyclonally stimulated in vitro in the presence of IL-2. In this context we could detect increased incorporation in Tregs of the Bodipy lipophilic fluorophore in proportion to the IL-2 dosage and to the extent of proliferation (Fig. 4A). Notably, Bodipy levels were higher in proliferating than in resting cells, indicating active lipid accumulation during Treg cell division (Fig. 4B). In this setting, TOFA treatment significantly inhibited the proliferation of Tregs in a dose-dependent fashion and completely abolished FA accumulation selectively in proliferating cells, demonstrating a key role for FAS in both the expansion and lipid pool generation of Tregs (Fig. 4C). Of note, unlike tumor cells, TOFA exposure did not exert a direct cytotoxic effect on Tconvs or Tregs but rather increased their survival (SI Appendix, Fig. S5).
Fig. 4.
Inhibition of FAS blocks proliferation and lipid accumulation preferentially in Tregs in vitro. (A–C) CD4+CD25+ Tregs, magnetically purified from splenocytes of naive C57BL/6 mice, were labeled with eFluor670 and polyclonally stimulated for 4 d and then were stained with Bodipy and analyzed by flow cytometry. (A) Representative histograms of eFluor670 dilution (Left) and Bodipy incorporation (Right) in Tregs cultured with different IL-2 concentrations. (B) Representative plot of the proliferating and resting Treg gating strategy (Left) and a representative histogram of Bodipy incorporation within each gate (Right). (C) Mean fluorescence intensity (MFI) of eFluor670 in gated Tregs (Upper) and gMFI of Bodipy incorporation in gated resting and proliferating Tregs (Lower) cultured with different TOFA concentrations. (D and E) Magnetically purified Tregs and Tconvs were cocultured at a 1:1 ratio with feeder cells, anti-CD3, and IL-2, with or without TOFA (5 μg/mL). After 4 d, cells were stained with Bodipy and Foxp3, and flow cytometry analysis was performed in gated Tregs (Foxp3+, red) or Tconvs (Foxp3−, blue). (D) Representative overlay of Tregs and Tconvs in the indicated conditions; numbers indicate MFI of eFluor670 or gMFI of Bodipy. (E) Analysis of eFluor670 MFI or Bodipy gMFI (gated on proliferating cells) of Tregs (red bars) or Tconvs (blue bars), either mock-treated (empty bars) or TOFA-treated (filled bars). Each condition was tested in triplicate; data shown are from one representative of two independent experiments. Error bars show mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.005 by unpaired Student t test.
To ascertain whether lipid accumulation and FAS reliance were preferential features of Tregs, we purified Tregs and Tconvs from spleens of naive mice, labeled them with a proliferation tracer, and cocultured them in the presence of IL-2. According to the literature (26), Tregs and Tconvs proliferate to a similar extent in the presence of IL-2 (Fig. 4D). Here, in basal conditions, we found that Tregs showed significantly higher Bodipy content but the same proliferation levels (as suggested by eFluor670 mean fluorescence intensity levels) as Tconvs, indicating that Tregs in vitro were also more prone to accumulate lipids during cell division. Notably, the addition of TOFA significantly reduced proliferation and Bodipy staining in both Tregs and Tconvs; however, Tregs were significantly more sensitive to such inhibition (Fig. 4 D and E).
Finally, we sought to test whether, when adoptively transferred in vivo, TOFA-treated Tregs were less able to suppress the immunity elicited by primary growing tumors against a secondary challenge, i.e., “concomitant immunity” (27). To this aim, mice bearing palpable tumors received a second injection of tumor cells alone or tumor cells admixed with TOFA- or mock-pretreated Tregs. The analysis of CD8 T cells infiltrating growing secondary tumors revealed a trend for decreased IFNγ and TNFα production in mice receiving tumor cells plus Tregs compared with mice receiving tumor cells alone. Notably, this suppressive effect was rescued when TOFA-treated Tregs were cotransferred with tumor cells (SI Appendix, Fig. S6). Together with in vitro data, these results point to a key role of FAS in Treg proliferation and possibly also in Tregs’ suppressive function in vivo.
Glycolysis and FAO both Contribute to Tregs’ Advantage in Cancer.
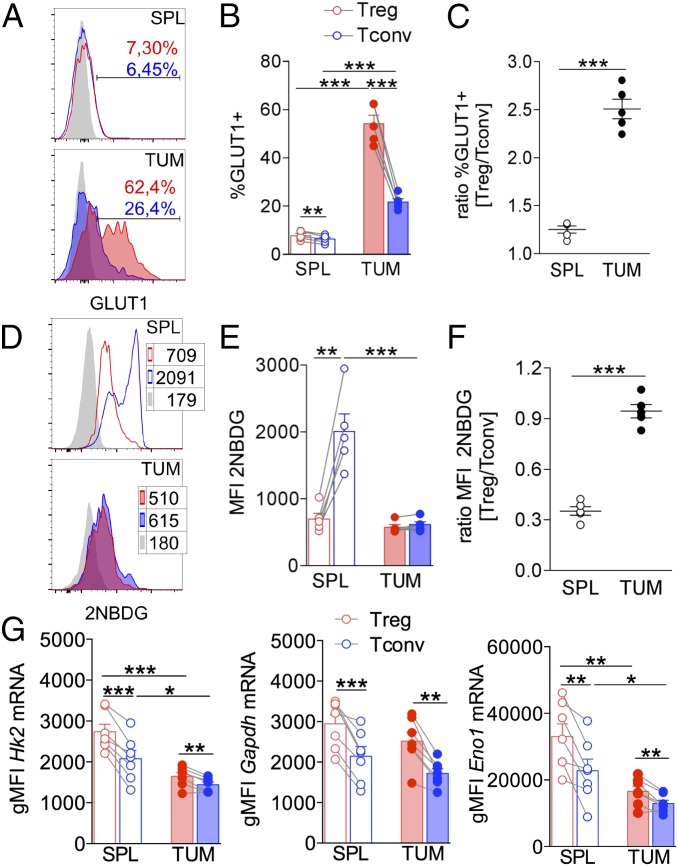
Memory CD8 T cells were shown to synthesize FAs from glucose (16); therefore we hypothesized that Tregs also might fuel FAS through a greater absorption of glucose relative to effector T cells, which instead are damaged by glucose deprivation in the TME (17, 18). First, in Tregs and Tconvs from tumors and spleens, we analyzed the expression of Glut1, a prominent glucose transporter in CD4 T cell activation (28) that is also relevant for Treg proliferation (12). We found that Glut1 was up-regulated in both Tconvs and Tregs at the tumor site compared with the spleen; however, such induction was significantly higher in Tregs, resulting in an increased relative ratio of Glut1 expression in Tregs versus Tconvs in tumors (Fig. 5 A–C). To track glucose uptake in vivo directly, we injected the fluorescent glucose analog 2-NBDG into tumor-bearing mice. In line with previous data (17, 18), Tconvs showed a massive defect in glucose uptake in tumor compared with spleen; conversely, Tregs displayed a relatively low glucose absorption in spleen that was maintained in tumor, resulting in a significant rescue of the Treg/Tconv relative ratio of glucose uptake at the tumor site (Fig. 5 D–F).
Fig. 5.
Glucose uptake and usage occur preferentially in Tregs in tumors. Mice bearing tumors 8 × 8 mm in size received an i.p. injection of the fluorescent glucose analog 2-NBDG, and spleens and tumors were collected 15 min after injection. 2-NBDG uptake and GLUT1 expression were quantified by flow cytometry in CD25+Foxp3+ Tregs and CD25−Foxp3− Tconvs. (A and B) Representative histograms (A) and frequency of cells expressing GLUT1 (B) in gated Tregs (red peaks and bars) and Tconvs (blue peaks and bars) in spleen (empty peaks and bars) and tumor (solid peaks and bars); gray peaks represent the FMO controls. (C) Ratio between frequency of GLUT1-expressing Tregs and Tconvs in spleen and tumor. (D and E) Representative histograms (D) and MFI of 2-NBDG acquisition (E) in gated Tregs (red peaks and bars) and Tconvs (blue peaks and bars) in spleen (empty peaks and bars) and tumor (solid peaks and bars). (F) Ratio between 2-NBDG MFI of Tregs versus Tconvs at different sites. (G) Expression levels of the glycolysis-related genes Hk2, Gapdh, and Eno1, evaluated through the PrimeFlow RNA assay in gated CD4+CD25+ Tregs (red bars) and CD4+CD25− Tconvs (blue bars) from spleen (empty bars) and tumor (filled bars). Each dot corresponds to a single mouse; data shown are from one representative of three independent experiments each including three to seven mice. Error bars show mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.005, by paired Student t test between Tregs and Tconvs in the same sample and by unpaired Student t test between populations of different samples.
Then, we analyzed the expression of selected glycolysis-related genes (Hk2, Gapdh, and Eno1) in Tregs and Tconvs through the PrimeFlow RNA assay. All three genes were significantly more represented in TUM-Tregs than in TUM-Tconvs (Fig. 5G). Interestingly, in both Tregs and Tconvs, Hk2 and Eno1 [the latter being a master regulator of human Treg metabolism and development (11)] were repressed in tumors compared with spleens, in line with a general contraction of glycolysis in the tumor context; however, their expression remained higher in Tregs in both compartments, pointing to a preferential preservation of glycolytic activity in Tregs in the TME (Fig. 5G, Right).
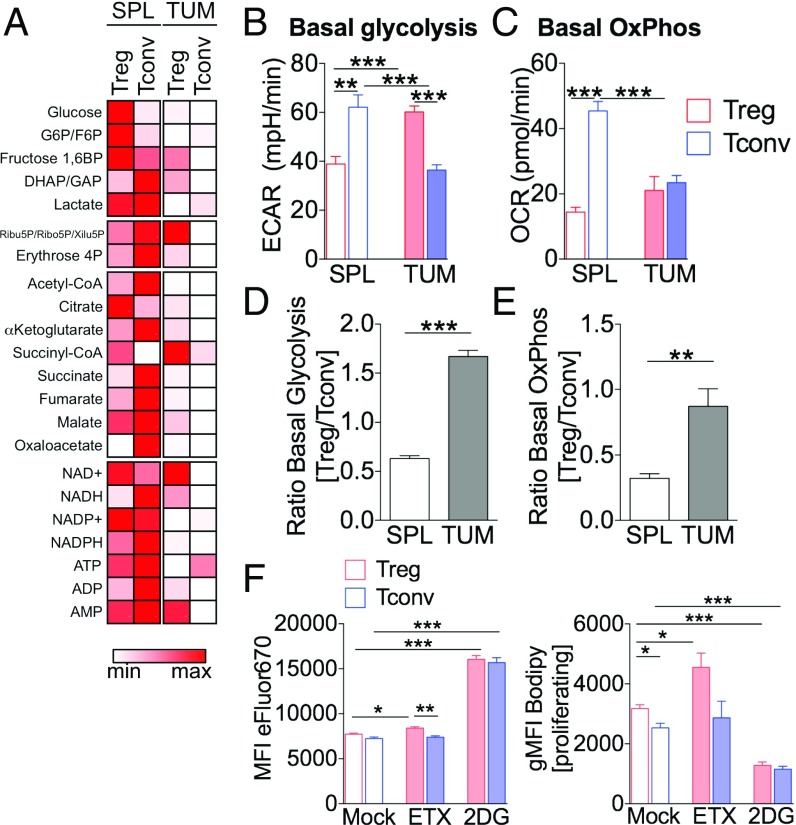
The results described above delineate a scenario in which the Treg advantage in tumor may be sustained by the concomitant engagement of glucose and lipid metabolic routes. To address this possibility, we performed a metabolomic analysis of Tregs and Tconvs isolated from tumors or spleens; profoundly different profiles emerged from these T cell subsets (Fig. 6A). In more detail, we could detect a series of metabolic changes in TUM-Tregs compared with Tregs extracted from the spleen (SPL-Tregs): TUM-Tregs displayed a preserved pentose phosphate pathway but higher glycolytic flux and higher consumption of acetyl-CoA and of the first intermediates of the tricarboxylic acid (TCA) cycle (citrate and α-ketoglutarate) (SI Appendix, Fig. S7A). Of note, this profile was paralleled by increased NADH/NAD+ and NADPH/NADP+ ratios and lower ATP/ADP ratios (SI Appendix, Fig. S7B), suggesting that in TUM-Tregs improved glucose usage may fuel the TCA cycle, whose intermediates are likely diverted to FA biosynthetic pathways rather than being fully oxidized to generate ATP.
Fig. 6.
TUM-Tregs display higher levels of both glycolytic and oxidative metabolism. (A) CD4+CD25+ Tregs and CD4+CD25− Tconvs were magnetically purified from pooled spleens and tumors of tumor-bearing mice, and metabolomic profiling was performed directly ex vivo by LC-MS/MS. The heatmap displays the levels of each metabolite (in femtograms per cell) in the indicated samples (minimum/maximum related to each metabolite). G6P/F6P, glucose 6P/fructose 6P; Ribu5P/Ribo5P/Xilu5P, Ribulose 5P/Ribose 5P/Xilulose 5P; DHAP/GAP, dihydroxyacetone phosphate/glyceraldehyde phosphate. (B–E) CD4+CD25+ Tregs and CD4+CD25− Tconvs were magnetically purified from pooled spleens and tumors of tumor-bearing mice, and quantification of the ECAR (B) and OCR (C) was performed by Seahorse analysis after a 12-h stimulation with anti-CD3/anti-CD28. (B) Basal glycolysis, (C) basal OxPhos, and (D and E) respective Treg/Tconv ratios for each index, in the indicated conditions. Data shown are from one representative of two independent experiments. Data are expressed as mean ± SEM of three measurements, each collected in two to eight replicates. **P < 0.01, ***P < 0.005, by unpaired Student t test. (F) Magnetically purified Tregs and Tconvs were cocultured at a 1:1 ratio for 4 d with or without Etx (100 μM) or 2-DG (1 mM), stained with Bodipy and Foxp3, and analyzed by flow cytometry. Analysis of eFluor670 MFI or Bodipy gMFI (gated on proliferating cells) in Tregs (red bars) or Tconvs (blue bars) is shown in the indicated conditions. Data are shown as mean ± SEM; each condition was tested in triplicate; data shown are from one representative of two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.005, by unpaired Student t test.
To functionally validate the peculiar TUM-Treg metabolic signature, we analyzed the bioenergetic profile of Tregs and Tconvs isolated from spleen or tumor and cultured in vitro for 12 h through Seahorse assays. Basal glycolysis, as measured by the extracellular acidification rate (ECAR) in tumor compared with spleen, was repressed in Tconvs, as expected (18), but was strikingly enhanced in Tregs (Fig. 6B). The rate of basal oxidative phosphorylation (OxPhos), as assessed by oxygen-consumption rate (OCR), was again inhibited in Tconvs at the tumor site but was maintained in Tregs (Fig. 6C). For all the metabolic indexes analyzed, Tregs showed a relative advantage over Tconvs specifically at the tumor site, as evidenced by higher Treg/Tconv ratios (Fig. 6 D and E).
These data demonstrated that tumor-associated Tregs engage both glycolysis and OxPhos for their expansion. To assess whether these metabolic routes are differentially required for Treg and Tconv proliferation, we performed the Treg/Tconv coculture assay described above in the presence of 2-deoxyglucose (2DG) or etomoxir (Etx), selective inhibitors of glycolysis or FAO, respectively. In line with previous results with human Tregs (14), 2DG profoundly restrained proliferation and lipid accumulation to a similar extent in both Tconvs and Tregs, suggesting that both cell types require glycolysis to support cell division and FAS. Conversely, Etx slightly, although significantly, affected Treg, but not Tconv, proliferation, and it promoted a further increase in Bodipy staining only in Tregs, indicating that FAO consumed the intracellular lipid pool and contributed to cell division specifically in Tregs (Fig. 6F). In analogy with the above-described effect of TOFA, we could not test whether Etx blocked Treg proliferation in tumors in vivo, due to its direct toxicity against tumor cells that overshadowed any immune-mediated effect (SI Appendix, Fig. S4 D–F). However, taken together, the greater sensitivity to FAO blockade in the coculture assay in vitro and the higher relative rate of OxPhos from tumors ex vivo highlight the reliance of Tregs on FA utilization and suggest that Tregs may display a relative resistance to glucose restriction in the TME due to their special ability to engage multiple metabolic routes, such as glycolysis and FAS/FAO.
Glycolytic and Lipid Biosynthetic Pathways Are Active in Human Tumor-Associated Tregs.
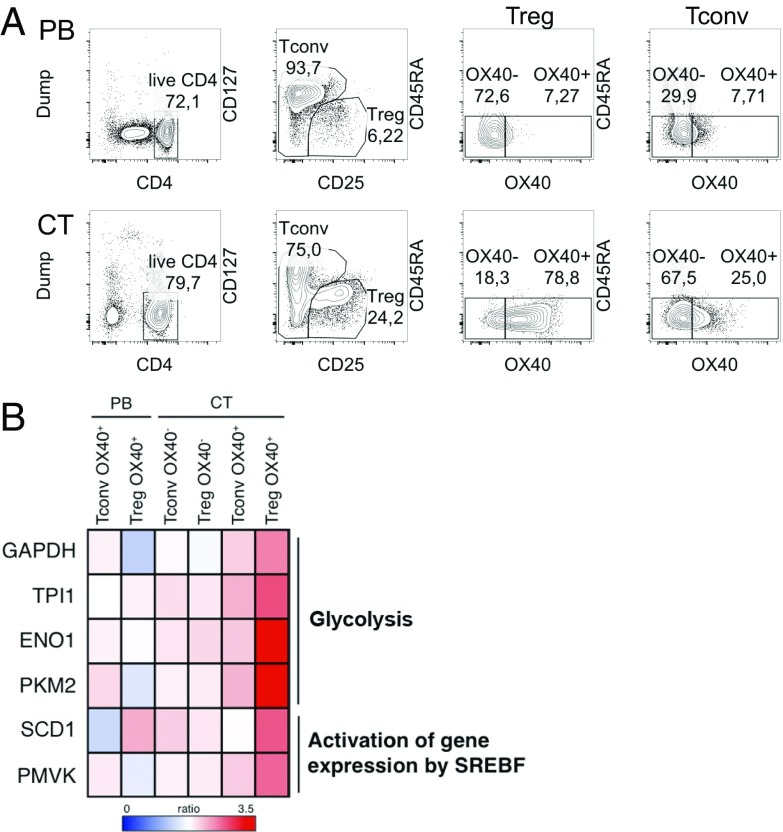
Compared with mouse tumors, Tregs infiltrating human cancers display greater complexity and heterogeneity, which are not completely understood especially in their immunometabolic aspects. Human cancer-infiltrating Tregs reveal a peculiar gene signature characterized by specific chemokine receptors, immune checkpoints, and costimulatory molecules such as OX40 (29, 30). Based on our recent work demonstrating that OX40-expressing Tregs were expanded in hepatic cirrhosis and liver and colon cancers and that OX40 was connected with tumor-associated Tregs’ high proliferation, stability, and suppression (21, 22), we explored the gene signature of OX40+ Tregs freshly extracted from specimens of liver cirrhosis and tumor (CT) or from the peripheral blood (PB) of patients with chronic HCV infection and hepatocellular carcinoma (HCC), in comparison with OX40− Tregs and the same subsets of Tconvs (Fig. 7A). We selected 216 genes showing a greater than twofold increase selectively in OX40+ CT-Tregs and performed a pathway enrichment analysis. Notably, among the significantly enriched pathways, we could reveal two metabolic routes, namely glycolysis (P = 0.00148) and activation of gene expression by SREBF (P = 0.0466) (Fig. 7B and SI Appendix, Tables S1 and S2). Glycolysis-related genes included GAPDH, TPI1, ENO1, and PKM2; all are involved in the glycolytic cascade, and PKM2, in particular, is involved in substrate phosphorylation. The up-regulated genes belonging to the SREBF-related pathway were SCD1 and PMVK, respectively involved in the synthesis of monounsaturated FAs and in the mevalonate pathway for cholesterol biosynthesis. In conclusion, these data demonstrate that human Tregs shift between different metabolic routes in specific tissue contexts.
Fig. 7.
Human Treg proliferation ex vivo from diseased liver is associated with glycolytic and FAS-related gene expression. OX40+/OX40− CD45RAlow Tregs and Tconvs were extracted and sorted from the PB and CT of five patients with chronic HCV infection, and gene expression analysis was performed. (A) Sorting strategy. (B) The fold-change was calculated for each gene over the respective control (OX40− PB Tconvs for all Tconv samples and OX40− PB Tregs for all Treg samples), and 216 genes up-regulated more than twofold only in Treg OX40+ CT were selected for pathway analysis (https://reactome.org/). The heatmap displays the fold-change over control in the genes accounting for two pathways of interest among those showing statistically significant enrichment (P < 0.05).
Discussion
The TME poses metabolic hurdles, such as hypoxia and glucose restriction, to the development of protective antitumor immunity. Nevertheless, this context favors the accumulation of proliferating and activated Tregs. Our study demonstrates that this advantage may rely on Tregs’ capacity to compete for glucose and perform FAS at higher rates than Tconvs.
Whether Tregs utilize glycolysis for their functions has been a matter of controversy in recent years. While it has been shown that murine iTregs (mostly polarized in the presence of TGFβ) show low glycolytic rates and that glycolysis inhibition promotes their differentiation (7–10), human iTregs (differentiated with suboptimal stimulation) rely strictly on glycolysis for development and function (11). Human Tregs are highly glycolytic when freshly analyzed from human blood and strongly require both FAO and glycolysis for their proliferation in vitro, while Tconvs rely only on glycolysis (14). Our present data confirm the same requirements in murine cells in vitro and demonstrate that Treg expansion in vivo in the tumor context is sustained by a strong increase in the glycolytic rate of Tregs compared with Tconvs.
Others have shown that Glut1 is higher in proliferating Tregs from the spleens of naive mice and that Glut1 overexpression increased both glycolytic and oxidative metabolism in Tregs and provoked Treg expansion in the spleen. However, these Tregs showed signs of reduced stability and suppressive function (12, 28). These results suggest that an artificial boost in glycolysis may uncouple proliferation from suppressive function in Tregs. However, increasing data indicate that these two events are tightly interconnected, especially in tissue contexts where Tregs are activated. Indeed, despite their refractoriness to conventional stimulation in vitro, Tregs are a highly proliferative population in vivo in both mice and humans, and many pieces of evidence demonstrate that activated Tregs, displaying a higher proliferation rate, retain or even increase their suppressive ability in several contexts (31). A few examples are the spontaneous development of lethal autoimmunity due to impaired expansion of suppressive Tregs in mTORC1 deficiency (15), the impaired ability of OX40-null Tregs to suppress colitis because of defective competitive fitness (20), the ability of OX40/OX40L to induce the proliferation of highly suppressive human Tregs (21), and the lower proliferative potential of Tregs in multiple sclerosis patients with more severe disease (32). In cancer mouse models and cancer patients, a huge amount of data demonstrates that highly proliferative and highly suppressive Tregs accumulate in tumors (2). Accordingly, our current results show higher frequency of Ki67+ and OX40+ Tregs at the tumor site, in association with an increased glycolytic rate, higher Glut1 expression, and improved competition for glucose. Combined with the well-established notion of the superior suppressive function of tumor-infiltrating Tregs, our data contribute in delineating a picture in which a glycolytic shift may boost the proliferation and, consequently, the suppressive function of Tregs in certain tissues and especially in cancer.
Foxp3 expression alone is sufficient to shift the metabolic program from glycolysis to oxidative phosphorylation and enables Tregs to resist low-glucose, high-lactate exposure (12, 33). Such a default program may help Tregs maintain their quiescent state in adverse conditions. However, our results indicate the presence of active Treg proliferation in the tumor context, possibly sustained by glycolysis, and associated with the expression of OX40 as a marker of Treg activation. Furthermore, OX40 engagement directly promotes the selective proliferation of lipid-laden Tregs in vivo. Overall, considering these results, we propose that, on the one hand, Foxp3 induces a default oxidative program to maintain Treg survival in a quiescent state; on the other, the signal elicited through OX40 and possibly other costimulatory receptors and other members of the TNF receptor superfamily may superimpose a glycolytic and anabolic boost over the oxidative program, allowing Tregs to exit from quiescence, undergo active proliferation, and perform an efficient suppressive function.
De novo synthesized intracellular lipids may be utilized not only for metabolic but also for functional purposes. Lipids are a preferential source of acetyl groups for histone acetylation and epigenetic reprogramming (34), and short-chain FA uptake has been shown to support colonic Treg homeostasis by regulating histone acetylation (35, 36). The stability of Foxp3 itself is increased by acetylation (37) and thus may be sensitive to the supply of lipid-derived acetyl groups. Lipids are also necessary for the palmitoylation of crucial signaling proteins, for building cell membranes and shaping their lipid composition and fluidity, and for the synthesis of pro- as well as antiinflammatory lipid mediators. Our results suggest that TUM-Tregs may perform FAS not only to fuel FAO but also for other purposes. Indeed, the high neutral lipid content and the metabolite signature observed in TUM-Tregs ex vivo indicates that FAS may prevail over FA consumption. The glycolytic rate was boosted in TUM-Tregs to a greater extent than oxidative indexes, and the coinduction of Pparg and Acacb was higher than the Cpt1a increase in TUM-Tregs. Finally, in vitro, Treg proliferation was more sensitive to the blockade of FAS than of FAO. Human OX40+ tumor-infiltrating Tregs displayed a gene signature oriented to glycolysis and lipid biosynthesis. Overall, these data indicate that lipid biosynthetic pathways may dominate in active Treg expansion in cancer and also in inflammatory conditions. Accordingly, impaired Treg homeostasis in mTORC1-deficient mice is associated with defective lipid biosynthesis (15).
According to our gene-expression analysis in OX40+ Tregs extracted from human tumor, an SREBP-driven program may support lipid biosynthesis in these cells. While no comprehensive data exist on the role of this family of transcription factors in Treg homeostasis and suppressive function, their function has been clearly elucidated in CD8 T cell activation: SREBP is essential for T cell transition from quiescence to activation through the active promotion of membrane synthesis (38). We speculate that, likewise, a program of lipid biosynthesis addressed to the construction of functional membranes might support the local proliferation of OX40+ Tregs in the TME. Indeed, the two SREBP-related genes up-regulated in tumor OX40+ Tregs, SCD1 and PMVK, may contribute to the biosynthesis of those lipids, namely monounsaturated FAs and especially cholesterol, that more profoundly impact on membrane fluidity, lipid-raft formation, and T cell receptor clustering and signaling, as recently demonstrated in CD8 T cells (39).
We report here that OX40 triggering in naive mice prompts the proliferation of lipid-laden Tregs. Therefore, the OX40/OX40L signal, occurring in the HCC microenvironment (21, 40), may directly sustain Treg lipogenic program. Indeed, OX40 ligation induces TRAF6 activation in CD4 T cells (41), and TRAF6 is a crucial driver of lipid metabolism in memory T cell development (42). Therefore, we suggest that the signal of OX40, possibly together with other members of the TNF receptor family, may sustain the transition of Tregs into the effector phase through the promotion of lipid biosynthetic pathways.
Our data show that Tregs accumulate intracellular lipids not only in cancer but also in the context of inflammation-driven expansion (early cholangitis in Mdr2−/− mice) and not in hepatic steatosis in the absence of inflammation (HCVTg mice at advanced age). Therefore, Treg lipid loading seems to occur as a result of Treg-intrinsic pathways (such as OX40 signaling) and irrespective of the lipid profile in the surrounding tissue. Intriguingly, we also could find high expression of PPARγ in TUM-Tregs. PPARγ is a nuclear factor that, in response to lipid ligands, controls a program of FA uptake, glucose uptake, and lipogenesis in the adipose tissue and also regulates a variety of other functions in other organs and in immune cells (43). In VAT-Tregs, PPARγ was shown to drive CD36 expression and FA incorporation; however, treatment of PPARγ transfectants with a synthetic agonist induced not only the Cd36 gene but also other genes involved in lipid metabolism, such as Scd1 and Cpt1a, among others (19). Therefore, PPARγ may be active in both VAT- and TUM-Tregs, driving a similar program of lipid rearrangement that is specifically instrumental to Treg expansion. Whether certain endogenous FAs generated by lipogenesis may bind (at relatively low affinity) and activate PPARγ is still controversial (44). It is reasonable to think that PPARγ may be activated as a consequence of increased FAS in TUM-Tregs; in line with this hypothesis, we found a preferential cosegregation of Pparg and Acacb expression in TUM-Tregs.
At the systemic level, starvation leads to a transient Treg expansion through the mediation of the leptin/mTOR axis (13); indeed, conditions of poor nutrient availability may shift the balance from growth and defense to maintenance of homeostasis and switch the immune status from inflammation to immune regulation. We propose that the tumor setting might configure as a context of nutrient starvation at the microenvironmental level, fostering immune-regulatory events. Whether leptin deprivation plays a role in the local tumor-associated immune dysfunction, and especially in Treg expansion, remains to be investigated. While the link between obesity, chronic inflammation, and cancer risk is well established, it is still unclear whether systemic/regional nutrient availability, adiposity, or metabolic inflammation affects immune regulation at the tumor site. However, we have recently elucidated that OX40+ Tregs accumulate in the VAT of obese human subjects unless affected by colorectal cancer, suggesting that the adipose tissue of obese individuals may represent a regional reservoir for Tregs to be recruited to the tumor site (45). Our present results suggest that perturbations in nutrient accessibility at the microenvironmental level may impact not only tumor cell but also immune cell functions. For instance, therapies targeting FAS, directly or indirectly, are under development for cancer treatment (46); based on our data, this approach may off-target Tregs, and especially activated tumor-associated Tregs, and it thus may act as a double-edged sword against both tumor cell proliferation and immune regulation.
Materials and Methods
Tumor Cell Lines.
MCA38 colon adenocarcinoma and B16F10 metastatic melanoma cell lines, kindly provided by M. P. Colombo (Fondazione Istituto di Ricovero e Cura a Carattere Scientifico Istituto Nazionale Tumori, Milan), were cultured in complete DMEM with high glucose (Gibco) supplemented with 10% FBS (Gibco), 2 mM l-glutamine (Sigma-Aldrich), penicillin/streptomycin, nonessential amino acids, sodium pyruvate (EuroClone), 50 μM 2-mercaptoethanol (Sigma-Aldrich), and 10 mM Hepes (Aurogene) at 37 °C in a humidified 5% CO2 atmosphere. To obtain transplanted tumors, tumor cells were s.c. injected into the right flank of C57BL/6NCrl wild-type male mice (Charles River Laboratories).
Mouse Models.
HCVTg mice of the FL-N/35 transgenic lineage, expressing the HCV polyprotein under the albumin promoter (24) were kindly provided by H. Lerat (Institut National de la Santé et de la Recherche Médicale, Unité U955, Université Paris-Est, Créteil, France) and were backcrossed for more than 10 generations to the C57BL/6NCrl background before analysis. HCVTg and C57BL/6NCrl mice were bred and maintained under conventional conditions at the animal facility of the Dipartimento di Scienze Anatomiche, Istologiche, Medico legali e dell’Apparato locomotore (SAIMLAL), in Sapienza Università di Roma, under protocols approved by the Italian Ministry of Health (authorization no. 481/2015-PR). Mdr2−/− (FVB.129P2-Abcb4 tm1Bor/J) mice were obtained from the Jackson Laboratory and were maintained under pathogen-free conditions at the Pasteur Institute animal facility (Paris). For all experiments, mice were killed by cervical dislocation. Tumor-bearing C57BL/6NCrl mice were killed 2 wk after s.c. tumor cell transplantation, and tumor volume (in cubic millimeters) was calculated using the formula: (smaller diameter)2 × larger diameter. For OX40 stimulation in vivo, C57BL/6 mice received an i.p. injection of 500 μg of anti-OX40 mAb (OX86; BioXcell); after 6 d, flow cytometry analysis was performed on splenocytes.
In Vivo Uptake Assays.
To test in vivo uptake of glucose and lipids, tumor-bearing mice received a single i.p. injection of 50 μg Bodipy-conjugated palmitate (Bodipy FL C16; Life Technologies) 1 h before analysis or a single i.p. injection of 100 μg the fluorescent glucose analog 2-NBDG [2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose; Life Technologies] 15 min before analysis.
TOFA and Etx Intratumor Treatments.
To test TOFA or Etx antitumor activity in vivo, tumor-bearing mice received repeated intratumor injections of 250 μg TOFA (Sigma) or 200 μg Etx (Sigma) at days 5, 8, 11, and 13 after s.c. tumor transplantation. As control, DMSO or water was administered, respectively; mice were killed at day 14 for analysis.
All in vivo experiments were authorized by the Italian Ministry of Health and were performed in accordance with the institutional animal care and use committee (Organismo preposto al Benessere Animale, SAIMLAL Department, Sapienza University of Rome) and the national law (Dlgs 26/2014).
Lymphocyte Extraction from Murine Tissues.
Murine tumor samples were mechanically dissociated using a gentleMACS Octo Dissociator according to the manufacturer’s instructions (Miltenyi Biotec), and mononuclear cells were enriched through a 40/80 Percoll (GE Healthcare) density gradient, collecting cells at the interface between the 40% and 80% Percoll solutions. The same protocol was applied for hepatic lymphocyte extraction. Splenocytes were obtained by spleen mechanical dissociation on 70-μm filter followed by incubation with ammonium-chloride-potassium (ACK) lysing buffer (Gibco) for 4 min at 4 °C. Lymph nodes were mechanically dissociated on a 70-μm filter. Cells were collected in complete RPMI-1640 Dutch-modified medium containing 10% FBS (Gibco), 2 mM l-glutamine (Sigma-Aldrich), penicillin/streptomycin, nonessential amino acids, sodium pyruvate (EuroClone), and 50 μM 2-mercaptoethanol (Sigma-Aldrich).
Flow Cytometry and PrimeFlow RNA Assay.
Bodipy (BODIPY 505/515, 4,4-Difluoro-1,3,5,7-Tetramethyl-4-Bora-3a,4a-Diaza-s-Indacene; catalog number D3921; Thermo Fisher Scientific) is a cell membrane-permeant fluorophore specific for neutral lipid stores known to stain lipid droplets in CD8 T cells (16). For Treg analysis, cells were incubated 15 min at 37 °C with Fixable Viability Dye eFluor780 (eBioscience) plus Bodipy or Nile Red (Enzo Life Sciences) dyes for neutral lipid staining. Then staining with the following antibodies for surface antigens was performed for 20 min at 4 °C: CD4 BrilliantViolet605, CD25 BrilliantViolet510, and CD134 (OX-40) BrilliantViolet421 (all from BioLegend); CD36 APC (BD Biosciences); and Glut1 PE (Novus Biologicals). Finally, intracellular staining with Foxp3 PE-eFluor610 and Ki67 PE-Cyanine7 (both from eBioscience) was performed for 30 min at RT after cell fixation and permeabilization for 30 min at 4 °C, using the Foxp3/Transcription Factor Staining Buffer Set according to the manufacturer’s instructions (eBioscience). For CD8 analysis, cells were restimulated 4 h with Cell Stimulation Cocktail plus Protein Transport Inhibitor (eBioscience), stained with Viability Dye eFluor780, CD8a BrilliantViolet785 (BioLegend), and CD44 BrilliantViolet510 (BioLegend), fixed/permeabilized with Cytofix/Cytoperm solution according to the manufacturer’s instructions (BD Biosciences), and finally stained with IFNγ PE (eBioscience) and TNFα PECy7 (BioLegend).
The PrimeFlow RNA Assay (Affymetrix/eBioscience) was performed according to the manufacturer’s instructions. After surface staining and fixation/permeabilization, target probe hybridization was performed using type 1 (AlexaFluor647-conjugated) probes for Pparg and Eno1 mRNAs, type 4 (AlexaFluor488-conjugated) probes for Cpt1a and Hk2 mRNAs, and type 6 (AlexaFluor750-conjugated) probes for Acacb and Gapdh mRNAs. Mouse β-Actin (Actb), conjugated with each probe type, was used as a positive control. Cells were incubated for 2 h with the target probes at 40 °C in a thermal incubator for microtubes and then were incubated with the preamplification reagent for 2 h and with the amplification reagent for an additional 2 h at 40 °C. After signal amplification, cells were incubated with label probes at 40 °C for 1 h and then were washed and resuspended in staining buffer for acquisition.
Data were acquired on an LSR Fortessa cell analyzer (Becton Dickinson) and were analyzed with FlowJo software (version 10.1r5; Tree Star Inc.).
In Vitro Proliferation Assays.
CD4+CD25+ Tregs and CD4+CD25− Tconvs were magnetically purified from splenocytes of wild-type C57BL/6 mice using the CD4+CD25+ Regulatory T Cell Isolation Kit (Miltenyi Biotec), labeled with 10 μM of the cell proliferation dye eFluor670 (eBioscience) by incubation for 15 min at 37 °C in complete RPMI Dutch-modified medium, and cultured or cocultured at 1:1 ratios in the presence of equal numbers of irradiated (3,500 rad) autologous splenocytes plus soluble anti-CD3 (1 μg/mL; eBioscience) and IL-2 (100 IU/mL; Roche), with or without TOFA (Sigma), Etx (Sigma), or 2-DG (Sigma) at different concentrations. After 4 d of culture, analysis of eFluor670 dilution and Bodipy incorporation was performed in gated Foxp3+ Tregs or Foxp3− Tconvs by flow cytometry.
Adoptive Treg Transfer and Suppression of Concomitant Immunity in Vivo.
CD4+CD25+ Tregs were magnetically purified from splenocytes of wild-type C57BL/6 mice and were cultured for 40 h in complete RPMI medium with coated anti-CD3 (1 μg/mL) plus IL-2 (100 IU/mL) in the presence of TOFA (5 μg/mL) or DMSO as a control. Recipient mice first received a primary tumor inoculum of 105 MCA38 cells s.c. in the right flank. After 10 d, when primary tumors were palpable, mice received a challenge inoculum in the opposite flank, containing 105 MCA38 cells, either alone or admixed at a 1:1 ratio with Tregs prepared as described above. After further 13 d, flow cytometry was performed in lymphocytes extracted from draining lymph nodes and tumors.
Seahorse Analysis.
OCR and ECAR real-time measurements were performed using an XFe-96 Extracellular Flux Analyzer (Seahorse Bioscience) in basal condition in XF medium and also in response to 5 μM oligomycin, 1.5 μM carbonylcyanide-4- (trifluoromethoxy)-phenylhydrazone (FCCP), or 1 μM of antimycin and rotenone (all from Sigma-Aldrich) for the OCR profile and in response to 10 mM glucose, 5 μM oligomycin, and 100 mM 2-DG (all from Sigma-Aldrich) for ECAR analysis. Basal OxPhos (before oligomycin addition) and ATP-linked OxPhos (the difference between basal OCR and oligomycin-induced OCR) were calculated from the OCR profile as indices of mitochondrial respiratory function. Basal glycolysis (after the addition of glucose), maximal glycolysis (after the addition of oligomycin), and glycolytic capacity (the difference between oligomycin-induced ECAR and 2-DG–induced ECAR) were calculated from the ECAR profile as indices of glycolytic pathway activation.
Metabolomic Analysis.
CD4+CD25+ Tregs and CD4+CD25− Tconvs were magnetically purified from pooled splenocytes and tumors of C57BL/6 mice (n = 15) using the CD4+CD25+ Regulatory T Cell Isolation Kit (Miltenyi Biotec). Pellets were then resuspended in 250 µL methanol/acetonitrile 1:1 containing [U-13C6]-Glucose 1 ng/µL (internal standard; 389374; Sigma Aldrich) and lysed by TissueLyser (Qiagen) for 3 min at the highest frequency. Lysates were spun at 20,000 × g for 5 min at 4 °C. Supernatants were then passed through a regenerated cellulose filter, dried, and resuspended in 100 µL of MeOH for subsequent analysis. Metabolomic data were obtained on an API-4000 triple quadrupole mass spectrometer (AB Sciex) coupled with an HPLC system (Agilent) and a CTC PAL HTS autosampler (PAL System). The identity of all metabolites was confirmed using pure standards. Quantification of different metabolites was performed with an LC-MS/MS method using a cyano-phase LUNA column (50 mm × 4.6 mm × 5 μm; Phenomenex). Methanolic samples were analyzed by a 5-min run in negative ion mode with 30 multiple reaction monitoring (MRM) transitions. The mobile phase A was 5 mM ammonium acetate (pH 7.00) in MeOH. The gradient was 100% A for all the analysis with a flow rate of 500 μL/min. MultiQuant software (version 3.0.2; SCIEX) was used for data analysis and peak review of chromatograms. Quantitative evaluation of all metabolites was performed based on calibration curves with pure standards; then data were normalized on numbers of total purified cells.
XTT Cytotoxicity Assay.
To assess the cytotoxic effects of TOFA and Etx on tumor cells in vitro, MCA38 cells were seeded at a concentration of 5 × 103 cells per well into 200 μL of complete DMEM in a flat-bottomed 96-well plate in the presence of different concentrations of TOFA and Etx or DMSO and water as respective controls. Conditions without cells and cell treatment with 0.05% saponin (Sigma-Aldrich) were used as negative and positive controls, respectively. After 24 h of culture, the analysis of cell viability was performed with the XTT Cell Proliferation Assay Kit (Cayman Chemical), and absorbance was measured with a Multiskan FC microplate reader (Thermo Scientific).
Human Samples.
PB from healthy volunteers was obtained from the buffy coats of healthy blood donors, anonymously provided by the Immunohematology and Transfusion Center of Policlinico Umberto I. PB and liver specimens were obtained from five patients with chronic hepatitis C, cirrhosis, and HCC (four males and one female, age 53–80 y) undergoing surgery or liver transplantation at Istituto Nazionale dei Tumori Regina Elena or Sapienza Università di Roma-Policlinico Umberto I. Human studies were performed in accordance with the ethical guidelines of the 1975 Declaration of Helsinki and were approved by the Institutional Ethical Committee (Comitato etico dell’Azienda Policlinico Umberto I, Authorization: RIF.CE: 4259). Informed consent was obtained from all patients.
Human Treg/Tconv Sorting and Gene-Expression Analysis.
Liver fragments from patients were perfused and grossly mashed with HBSS (EuroClone) containing 0.5 mg/mL collagenase IV (Sigma-Aldrich) and 50 ng/mL DNase I (Worthington). Samples were incubated for 10 min at 37 °C and then were mechanically disaggregated into single-cell suspensions. Mononuclear cells from patient PB were also enriched by Lympholyte (Cedarlane Laboratories) density gradient. T cells were enriched from mononuclear cells using the Pan T cell Isolation kit II (Miltenyi Biotec) and, after treatment with Fixable Viability Dye eFluor780 (eBioscience) and with FcBlock (eBioscience) for 15 min at 4 °C, were stained for surface markers: dump channel (CD14 APCeFluor780, CD56 APCeFluor780, and CD8 APCeFluor780, all from eBioscience); CD4 AlexaFluor488 (eBioscience); OX40 PE (ACT35; BD); CD45RA PerCPCy5.5 (BioLegend); CD127 PECy7 (BioLegend); and CD25 APC (BioLegend). Then OX40+ and OX40− subsets in gated CD45RAlow Tregs and Tconvs were sorted using a FACSAria II cell sorter (Becton Dickinson).
Total RNA was isolated using the mirVana miRNA Isolation Kit (Ambion) following the standard protocol. Briefly, the lysates were extracted once with acid-phenol chloroform and further purified to yield total RNA. Extracted RNA was quantified with the RiboGreen RNA Quantitation Kit (Molecular Probes) on an Infinite F200 plate reader (Tecan Trading AG). All extracted RNA samples were quality controlled for integrity with a 2100 Bioanalyzer system (Agilent Technologies).
Gene expression of whole transcriptomes was performed according to the standard protocol. Total RNA was isolated, quality controlled, and quantified as described above. For each sample 100 ng of total RNA were reverse transcribed according to the Illumina TotalPrep RNA Amplification kit (Ambion), and biotinylated cRNA was generated by in vitro transcription. Washing, staining, and hybridization were performed according to the standard Illumina protocol: Briefly, 750 ng of cRNA of each sample in a final volume of 15 μL were hybridized onto Illumina HumanHT-12 v4 Expression BeadChip arrays. Hybridization and scanning were performed according to the manufacturer’s indications on an Illumina iScan System, and data were processed with BeadStudio v.3 (Illumina). Arrays were quantile normalized, with no background subtraction, and average signals were calculated on gene-level data for genes whose detection P value was lower than 0.001 in at least one sample.
Statistical Analysis.
Statistical analysis was performed using Prism software (version 6.0; GraphPad). A two-tailed unpaired Student’s t test was used to analyze in vitro data and to compare ex vivo data from Tregs and Tconvs in different samples. The two-tailed paired Student’s t test was applied to compare ex vivo data from Tregs and Tconvs in the same sample. Every in vitro and ex vivo assay was performed in duplicate, triplicate, or quadruplicate when possible; for all experiments, the number of repetitions is indicated in the figure legend. In all graphs, bars show means ± SEM. In all tests, P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Massimo Rossi, Nicola Guglielmo, and Gian Luca Grazi for liver specimens; Mario P. Colombo for providing mouse tumor cell lines; Natalia Pediconi and Massimo Levrero for discussions and sharing reagents; Alfonso Grimaldi for collaboration in the revision process; Giovanni Bernardini for sharing reagents; and Hervé Lerat for providing HCVTg colony founders. This work was supported by Associazione Italiana per la Ricerca sul Cancro Grants IG-2014 15199 and IG-2017 19939 (to V.B.) and IG-2017 19784 (to S.P.); Ministry of Education, University and Research (MIUR) Grants RF-2010-2310438 and RF 2010-2318269; Fondazione Italiana Sclerosi Multipla Grants code 2015/R/04 (to V.B.), code 2013/R/9 (to E.M.C.), code 2016/R/18 (to G.M.), and code 2014/R/19 (to M.S.); MIUR Progetti di Ricerca di Interesse Nazionale Grant 2010-2011 protocol 2010LC747T_004; MIUR Fondo per gli Investimenti della Ricerca di Base 2011/13 Grant no. RBAP10TPXK; Istituto Pasteur Italia–Fondazione Cenci Bolognetti Grant 2014-2016; International Network Institut Pasteur Programmes Transversaux de Recherche Grant 20-16; Fondazione Roma Grant for Biomedical Research NCDS-2013-000000345; the European Union’s Seventh Framework Program (FP7) under Grant Agreement 259743 (MODHEP consortium) (to Y.W.); Fondazione Cariplo Grant 2016-0852 (to G.D.N.); Ministero della Salute Grants GR-2011-02346974 (to G.D.N.) and GR-2013-02355011 (to F.B.); and European Union IDEAS Programme European Research Council Starting Grant menTORingTregs 310496 and Telethon Grant GGP17086 (to G.M.). C.F. was supported by a 2015 Fellowship from Fondazione Veronesi.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray dataset has been deposited in the Gene Expression Omnibus databank (accession no. GSE103523).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720113115/-/DCSupplemental.
References
- 1.Roychoudhuri R, Eil RL, Restifo NP. The interplay of effector and regulatory T cells in cancer. Curr Opin Immunol. 2015;33:101–111. doi: 10.1016/j.coi.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Burocchi A, Colombo MP, Piconese S. Convergences and divergences of thymus- and peripherally derived regulatory T cells in cancer. Front Immunol. 2013;4:247. doi: 10.3389/fimmu.2013.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kretschmer K, et al. Inducing and expanding regulatory T cell populations by foreign antigen. Nat Immunol. 2005;6:1219–1227. doi: 10.1038/ni1265. [DOI] [PubMed] [Google Scholar]
- 4.Weiss JM, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med. 2012;209:1723–1742, S1. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou G, Levitsky HI. Natural regulatory T cells and de novo-induced regulatory T cells contribute independently to tumor-specific tolerance. J Immunol. 2007;178:2155–2162. doi: 10.4049/jimmunol.178.4.2155. [DOI] [PubMed] [Google Scholar]
- 6.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: Insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berod L, et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat Med. 2014;20:1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- 8.Michalek RD, et al. Cutting edge: Distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerriets VA, et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest. 2015;125:194–207. doi: 10.1172/JCI76012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Rosa V, et al. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nat Immunol. 2015;16:1174–1184. doi: 10.1038/ni.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerriets VA, et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat Immunol. 2016;17:1459–1466. doi: 10.1038/ni.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Procaccini C, et al. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. 2010;33:929–941. doi: 10.1016/j.immuni.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Procaccini C, et al. The proteomic landscape of human ex vivo regulatory and conventional T cells reveals specific metabolic requirements. Immunity. 2016;44:406–421. doi: 10.1016/j.immuni.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng H, et al. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Sullivan D, et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho PC, et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang CH, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cipolletta D, et al. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piconese S, et al. A non-redundant role for OX40 in the competitive fitness of Treg in response to IL-2. Eur J Immunol. 2010;40:2902–2913. doi: 10.1002/eji.201040505. [DOI] [PubMed] [Google Scholar]
- 21.Piconese S, et al. Human OX40 tunes the function of regulatory T cells in tumor and nontumor areas of hepatitis C virus-infected liver tissue. Hepatology. 2014;60:1494–1507. doi: 10.1002/hep.27188. [DOI] [PubMed] [Google Scholar]
- 22.Timperi E, et al. Regulatory T cells with multiple suppressive and potentially pro-tumor activities accumulate in human colorectal cancer. OncoImmunology. 2016;5:e1175800. doi: 10.1080/2162402X.2016.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mauad TH, et al. Mice with homozygous disruption of the mdr2 P-glycoprotein gene. A novel animal model for studies of nonsuppurative inflammatory cholangitis and hepatocarcinogenesis. Am J Pathol. 1994;145:1237–1245. [PMC free article] [PubMed] [Google Scholar]
- 24.Lerat H, et al. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology. 2002;122:352–365. doi: 10.1053/gast.2002.31001. [DOI] [PubMed] [Google Scholar]
- 25.Ruby CE, et al. Cutting edge: OX40 agonists can drive regulatory T cell expansion if the cytokine milieu is right. J Immunol. 2009;183:4853–4857. doi: 10.4049/jimmunol.0901112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thornton AM, Donovan EE, Piccirillo CA, Shevach EM. Cutting edge: IL-2 is critically required for the in vitro activation of CD4+CD25+ T cell suppressor function. J Immunol. 2004;172:6519–6523. doi: 10.4049/jimmunol.172.11.6519. [DOI] [PubMed] [Google Scholar]
- 27.Turk MJ, et al. Concomitant tumor immunity to a poorly immunogenic melanoma is prevented by regulatory T cells. J Exp Med. 2004;200:771–782. doi: 10.1084/jem.20041130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macintyre AN, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20:61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Simone M, et al. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity. 2016;45:1135–1147. doi: 10.1016/j.immuni.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plitas G, et al. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45:1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Attridge K, Walker LS. Homeostasis and function of regulatory T cells (Tregs) in vivo: Lessons from TCR-transgenic Tregs. Immunol Rev. 2014;259:23–39. doi: 10.1111/imr.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carbone F, et al. Regulatory T cell proliferative potential is impaired in human autoimmune disease. Nat Med. 2014;20:69–74. doi: 10.1038/nm.3411. [DOI] [PubMed] [Google Scholar]
- 33.Angelin A, et al. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 2017;25:1282–1293.e7. doi: 10.1016/j.cmet.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonnell E, et al. Lipids reprogram metabolism to become a major carbon source for histone acetylation. Cell Rep. 2016;17:1463–1472. doi: 10.1016/j.celrep.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 37.van Loosdregt J, Coffer PJ. Post-translational modification networks regulating FOXP3 function. Trends Immunol. 2014;35:368–378. doi: 10.1016/j.it.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Kidani Y, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang W, et al. Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature. 2016;531:651–655. doi: 10.1038/nature17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piconese S, Valzasina B, Colombo MP. OX40 triggering blocks suppression by regulatory T cells and facilitates tumor rejection. J Exp Med. 2008;205:825–839. doi: 10.1084/jem.20071341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao X, et al. OX40 signaling favors the induction of T(H)9 cells and airway inflammation. Nat Immunol. 2012;13:981–990. doi: 10.1038/ni.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmadian M, et al. PPARγ signaling and metabolism: The good, the bad and the future. Nat Med. 2013;19:557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bensinger SJ, Tontonoz P. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature. 2008;454:470–477. doi: 10.1038/nature07202. [DOI] [PubMed] [Google Scholar]
- 45.Donninelli G, et al. Distinct blood and visceral adipose tissue regulatory T cell and innate lymphocyte profiles characterize obesity and colorectal cancer. Front Immunol. 2017;8:643. doi: 10.3389/fimmu.2017.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Röhrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer. 2016;16:732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.