Significance
Double-stranded RNA (dsRNA) viruses infect hosts ranging from fungi to plants and to humans. Like most other dsRNA viruses, the aquareovirus RNA polymerase catalyzes the synthesis of RNA plus strands within the inner capsid, a process vital for the replication of virus progeny. Here we present a near-atomic resolution structure of the RNA polymerase in complex with its cofactor protein and genomic RNA within the aquareovirus. Some asymmetric structures within the elaborate viral machine that have never been previously determined have been resolved in our structure, and key interactions among the polymerase, cofactor protein, and dsRNA have been revealed. These findings provide insights into the mechanism underlying highly coordinated dsRNA virus transcription and assembly.
Keywords: dsRNA virus, transcription, reovirus, asymmetric reconstruction
Abstract
Most double-stranded RNA (dsRNA) viruses transcribe RNA plus strands within a common innermost capsid shell. This process requires coordinated efforts by RNA-dependent RNA polymerase (RdRp) together with other capsid proteins and genomic RNA. Here we report the near-atomic resolution structure of the RdRp protein VP2 in complex with its cofactor protein VP4 and genomic RNA within an aquareovirus capsid using 200-kV cryoelectron microscopy and symmetry-mismatch reconstruction. The structure of these capsid proteins enabled us to observe the elaborate nonicosahedral structure within the double-layered icosahedral capsid. Our structure shows that the RdRp complex is anchored at the inner surface of the capsid shell and interacts with genomic dsRNA and four of the five asymmetrically arranged N termini of the capsid shell proteins under the fivefold axis, implying roles for these N termini in virus assembly. The binding site of the RNA end at VP2 is different from the RNA cap binding site identified in the crystal structure of orthoreovirus RdRp λ3, although the structures of VP2 and λ3 are almost identical. A loop, which was thought to separate the RNA template and transcript, interacts with an apical domain of the capsid shell protein, suggesting a mechanism for regulating RdRp replication and transcription. A conserved nucleoside triphosphate binding site was localized in our RdRp cofactor protein VP4 structure, and interactions between the VP4 and the genomic RNA were identified.
Double-stranded RNA (dsRNA) viruses infect a wide range of hosts including vertebrates, invertebrates, fungi, bacteria, and plants. Most dsRNA viruses consist of a segmented dsRNA genome and a number of RNA-dependent RNA polymerases (RdRps) enclosed in a common architecture of a T = 1 icosahedral innermost capsid shell formed by 60 asymmetric dimers of a single capsid shell protein (1–7).
Aquareovirus belongs to the family of Reoviridae, which is one of the largest families of dsRNA viruses (8). The structures of aquareovirus capsid proteins are markedly similar to the counterparts of orthoreovirus (SI Appendix, Table S1), which is consistent with their high sequence identities (9–11). Aquareovirus/orthoreovirus has 11/10 dsRNA segments and approximately the same number of RdRps enclosed by an icosahedral inner capsid referred to as the “core” (sometimes as the “inner capsid particle”). The core of aquareovirus/orthoreovirus has the typical architecture of the dsRNA virus innermost capsid shell formed by two conformers of the innermost capsid shell protein, VP3A/λ1A and B. A distinct pentameric turret formed by five copies of the turret protein VP1/λ2 sits around the fivefold vertex on the shell and functions in the catalysis of mRNA 5′ cap synthesis (1, 12). The stability of the core is further reinforced by 120/150 copies of a clamp protein VP6/σ2 that sit on the innermost shell (1, 12) (orthoreovirus has additional 30 copies of the clamp protein on the twofold axis). A mature aquareovirus/orthoreovirus core is coated by 200 trimers of membrane penetration protein VP5/μ1 decorated with protection protein VP7/σ3, forming an incomplete icosahedral T = 13 outer capsid layer owing to the occupancy of the turrets on the fivefold vertices (12–14). The outer capsid layer is shed during membrane penetration, delivering the core into the cytoplasm (15).
The aquareovirus/orthoreovirus core, a multienzyme machine for RNA synthesis, remains intact in the cytoplasm. In the core, the RdRp protein VP2/λ3, probably assisted by its cofactor protein VP4/μ2, catalyzes the synthesis of RNA plus strands using minus strand in each genomic dsRNA segment as a template while avoiding the dsRNA-activated defense mechanisms of the host cell (8). Protein VP4/μ2 has been speculated to function as nucleoside triphosphatase (NTPase), RNA 5′-triphosphatase, and dsRNA helicase (16–18). The RNA plus strands, which are capped by the turret protein VP1/λ2, are released into the cytoplasm and serve as mRNA for viral protein translation and also as genomic plus strands for the core packaging. During assembly or in the newly assembled core, the RdRps switch mode to catalyze the synthesis of RNA minus strands forming genomic dsRNA segments using the RNA plus strands as the template (19).
Structures of the icosahedral capsids of Reoviridae viruses have been studied extensively (1, 9, 12, 13, 15, 20–30). By contrast, very few studies have been conducted to analyze the structures of the RdRps and cofactor proteins within the capsid shell because they lack icosahedral symmetry, and the intrinsic icosahedral average is imposed during structural determination. Efforts to understand the molecular mechanism of viral transcription and assembly have been hampered by the lack of the asymmetric structural information. Although the crystal structures of the recombinant RdRp proteins of orthoreovirus λ3 and rotavirus VP1 have been determined and fitted into their capsid structures (31–35), the structures of the viral particle-associated RdRps and cofactor proteins are still unknown. Recently, we reported the structure of the RdRp and cofactor protein within a single-layered cypovirus (36, 37). However, the structural differences between the RdRps of the double-layered aquareovirus/orthoreovirus and the single-layered cypovirus are relatively large. Therefore, we have previously been unable to analyze the structural changes between recombinant RdRp and particle-associated RdRp, in which the capsid shell protein is important for RdRp activation (38, 39). In addition, the difference between the complexes of the RdRp and cofactor protein within double-layered and single-layered Reoviridae viruses is expected to be notable because the outer capsid proteins play important roles in transcriptional regulation (40). The lack of asymmetric structural information of the RdRp VP2/λ3, cofactor protein VP4/μ2, and VP3A/λ1A N termini within the double-layered Reoviridae viruses prevents us from understanding the mechanism behind the roles of these proteins in the highly coordinated mRNA transcription process.
In this study, we determined the structure of the RdRp protein VP2 in complex with its cofactor protein VP4 and genomic RNA within the double-layered aquareovirus capsid using cryoelectron microscopy (cryo-EM) at 200 kV and our symmetry-mismatch reconstruction method (36, 41). Our structure shows that the VP2–VP4 complex is anchored at the capsid shell and interacts with genomic dsRNA and four of the five asymmetrically arranged N termini of capsid shell protein VP3A under the fivefold axis, implying roles for these N termini in virus assembly. Compared with the crystal structure of orthoreovirus λ3 elongation complex, the structure of our aquareovirus VP2 is almost identical to that of orthoreovirus λ3 (31). However, the binding site of a genomic RNA, which is located at the entrance of the RdRp VP2 channel for template entry, is different to the RNA cap binding site identified in the λ3 structure (31). In addition, some elements in the VP2, which is supposed to interact with template RNA and priming nucleoside triphosphate (NTP) during transcription, are flexible. A loop, which is thought to separate RNA template and transcript (31), was observed to interact with an apical domain of the shell protein VP3A, suggesting that the conformational change of the apical domain upon virus transcription activation (34, 42) could regulate RdRp replication and transcription. A conserved NTP binding site was localized in the VP4 structure, and both VP4 N-terminal and C-terminal domains are interacting with the genomic RNA.
Results and Discussion
Structure Determination of the Capsid, RdRp Complex, and Genome.
We obtained a 3D structure of an aquareovirus icosahedral capsid, cultured and purified from Ctenopharyngodon idellus kidney (CIK) cells, using cryo-EM and single-particle reconstruction. All images of aquareovirus were recorded using a 200-kV FEI Tecnai Arctica electron microscope equipped with a Falcon II camera. The aquareovirus, which consists of more than 1,500 molecules and is ∼820 Å in diameter, is, to our knowledge, the largest biological complex structure that has ever been determined at near-atomic resolution using cryo-EM at 200 kV. A total of 5,102 cryo-EM images were collected. Structure refinement and reconstruction were performed using our software package, as described in ref. 37. Approximately 41,000 particle images were selected based on our phase residue criteria (37) for the final reconstruction. The estimated resolution of the whole capsid structure according to the gold standard Fourier shell correlation is 3.3 Å (SI Appendix, Fig. S1). The estimated resolution of the inner capsid structure is 3.1 Å (SI Appendix, Fig. S1). In accordance with previous studies (33), fitting model of the orthoreovirus λ3 into the icosahedrally averaged density of VP2 under the fivefold vertex revealed that most α-helices match well with λ3 (SI Appendix, Fig. S2). However, the overall structural density is blurred by the icosahedral average during the reconstruction, and the structure and precise location of the RdRp cofactor protein VP4/μ2 are still unknown. To analyze the nonicosahedral structure of RdRp and associated proteins in detail, we further reconstructed the RdRp protein VP2 in complex with its cofactor protein VP4 and genomic RNA within the capsid using our symmetry-mismatch reconstruction method (36, 41). The estimated resolution of the capsid structure including VP2 and VP4 was 3.8 Å (SI Appendix, Fig. S1), allowing us to build full atomic models for VP2 and VP4.
Structural Comparison Between the Capsids of Our Aquareovirus and the Aquareovirus Infectious Subviral Particle.
Our aquareovirus capsid structure, which was reconstructed from cryo-EM images of the aquareovirus particles directly purified from the infected CIK cells, was essentially identical to the previously reported aquareovirus metastable, primed infectious subviral particle (ISVP) structure, which was reconstructed from cryo-EM images of the chymotrypsin-digested aquareovirus particles (13). The protection protein VP7 on the membrane penetration protein VP5 was partially resolved, allowing us to build an atomic model of the VP7 residues 3–88 (SI Appendix, Fig. S3). The density of the resolved VP7 region is slightly weaker than that of VP5, suggesting that these VP7 molecules are somewhat flexible. In fact, we observed that the identical VP7 structures are also present in the aquareovirus ISVP structure (13), indicating that the VP7 molecules in the ISVP were not fully removed by the treatment of α-chymotrypsin (SI Appendix, Fig. S3). The VP5 trimers in our structure are also identical to those in the aquareovirus ISVP structure (13). The cleavage site between Asn42 and Pro43 of VP5 and the myristoyl group linked to the N terminus of each VP5 molecule, which have been previously observed in the aquareovirus ISVP, are also present in all 10 VP5 subunits in the asymmetric unit of our aquareovirus structure (SI Appendix, Fig. S4). In addition, the structures of all three proteins VP1, VP3, and VP6 in the inner capsid of our aquareovirus are also identical to those in the aquareovirus ISVP.
Overall Organization of Genome and RdRp in Complex With Its Cofactor Protein Within the Capsid.
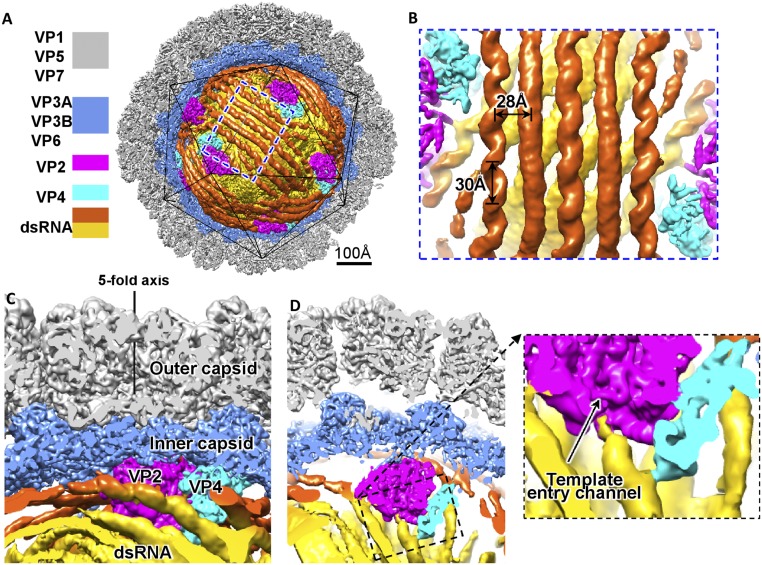
The overall organization of our aquareovirus genome and RdRp VP2 in complex with cofactor protein VP4 (SI Appendix, Fig. S5) is similar to that of cypovirus (36). The genome exhibits as discontinuous dsRNA fragments running in parallel (Fig. 1 and SI Appendix, Fig. S5). The discontinuities represent flexible regions of the genomic dsRNA. The distances between two adjacent parallel dsRNA fragments within the same layer are ∼28 Å, whereas two adjacent layers are ∼28 Å apart (Fig. 1A). The double helices of the dsRNA fragments located close to the inner capsid surface or interacting with the RdRp complexes are visible, and the helix pitch is ∼30 Å (Fig. 1B). Each RdRp complex, which consists of a copy of VP2 and a copy of VP4, is anchored at the inner surface, adjacent to the fivefold axis of the icosahedral capsid and surrounded by multiple layers of genomic dsRNA (Fig. 1 and SI Appendix, Fig. S5). The structures of the RdRp complexes and RNA genome are organized in a pseudo-D3 symmetric organization (SI Appendix, Fig. S5). There are two RdRp complexes in the D3 asymmetric unit—threefold RdRp complex and twofold RdRp complex (we followed the nomenclature used in ref. 36) (SI Appendix, Fig. S5). The two RdRp complexes are essentially identical except that the average density value of the twofold RdRp complexes amounts to ∼85% of the average density value of the threefold RdRp complexes. We therefore suggest that the six structures of the twofold RdRp complexes are the average of 5 RdRp complexes occupying the six positions, and the total number of RdRp complexes within the capsid is 11; this is consistent with the 11 RNA segments in aquareovirus.
Fig. 1.
Structures of the aquareovirus genome and RdRp complex within the capsid (filtered to 8-Å resolution). (A) Half of the icosahedral capsid is removed to show the structures of the genomic dsRNA (first layer in orange and second layer in yellow), RdRp VP2 (magenta), and cofactor protein VP4 (cyan). (B) Zoom-in view of the genome in A. (C and D) Cut open views show the interactions between the dsRNA, VP2, and VP4.
RdRp Protein VP2.
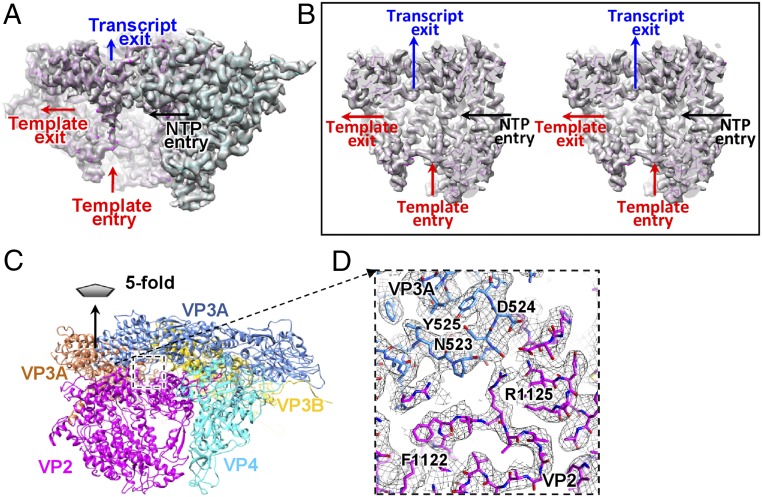
The structure of the aquareovirus RdRp protein VP2 was well resolved, which allowed us to build a full atomic model (SI Appendix, Fig. S6). The VP2 structure is almost identical to the crystal structure of the recombinant orthoreovirus λ3 (31) (SI Appendix, Fig. S6A), consistent with their high amino acid sequence identity of 43%. The Cα atoms of these two structures superimpose on each other with an RMSD of 1.8 Å. According to the domain nomenclature of λ3 (31), VP2 can also be divided into three domains (SI Appendix, Fig. S6E): an N-terminal domain (residues 1–386), a central polymerase domain (residues 387–897), and a C-terminal “bracelet” domain (residues 898–1,273). The four channels that are used for RNA template entry, NTP entry, template exit, and transcript exit are also present in the VP2 structure (Fig. 2 A and B and SI Appendix, Fig. S6).
Fig. 2.
Structures of VP2, VP4, and VP3A. (A) Transparent view of the density map of VP2 and VP4 superimposed with their atomic model (only the backbone is shown). The VP2 and VP4 are in magenta and cyan, respectively. The four channels in VP2 are indicated. (B) Cut open view showing the four channels of VP2 (shown in cross-eye stereo). (C) Interactions among two copies of VP3A, a copy of VP3B, VP2, and VP4. (D) Zoom-in view of the interaction between the VP3A apical domain and the switch loop. The conformational change in the apical domains is likely to trigger the conformational change in the switch loop and, consequently, switch the RdRp from the replicative to the transcriptional mode. The VP4 may play roles in dsRNA unwinding and in translocation of the template RNA to the VP2 catalytic site during transcription. The transcript exits through the transcript exit channel into a peripentonal channel formed by two copies of VP3A and one copy of VP3B and then moves to the turret for RNA capping reactions before finally exiting the particle (33, 42).
Our structure shows two major interactions between VP2 and dsRNA. A dsRNA fragment runs roughly parallel to the capsid shell and interacts closely with the RdRp bracelet domain and adjacent region of the polymerase domain (Fig. 1C and SI Appendix, Fig. S7), which is similar to the RdRp–dsRNA interaction in the nontranscribing cypovirus (36, 37). This dsRNA fragment traverses across and blocks the exit of channel for template exit. Another close interaction occurs where a dsRNA fragment end approaches the template entry channel binding at the entrance of the channel for template entry (Fig. 1D). This interaction resembles the RdRp–RNA interaction in the transcribing cypovirus (36); however, no RNA template or product was observed in the catalytic cavity. Notably, this RNA binding site is different from the cap binding site in the λ3 structure (SI Appendix, Fig. S6A), which was thought to recognize capped viral mRNA in the virus-packaging step and bind to the cap site of the 5′ end of the positive strand to facilitate insertion of the free 3′ end of the negative strand into the template channel in the transcriptional step (31).
Comparison between the structures of VP2 and λ3 elongation complex reveals that a number of elements are absent in the VP2 structure (SI Appendix, Fig. S8), resulting in more hollow central catalytic cavity. These absent elements, which can be ascribed to structural flexibility, can be classified into three categories according to their locations and the components they interact with. The first category of flexible elements, many of whose residues are positively charged, is involved in interactions with dsRNA (residues 488–492, 963–979, and 1,055–1,058) (SI Appendix, Fig. S8). The second category is involved in the interactions with priming NTP (residues 560–567, referred to as “priming loop” in ref. 31) and with the RNA template (residues 461–537 and 688–693) (SI Appendix, Fig. S8). The last flexible element (residues 1,112–1,120) consists of an α-helix (SI Appendix, Fig. S8), which is located at the exit of the channel for transcript exit and connects a loop (residues 1,121–1,132) located in the template exit channel.
We propose that all these flexible elements are subjected to movement in the dynamic process of RNA transcription. First, upon RdRp initiation, the genomic dsRNA, which is roughly parallel to the capsid shell, is released from VP2 to unblock the channel for template exit and ensure the sliding of the RNA template. Second, in orthoreovirus RdRp λ3, elongation of RNA synthesis requires shift of the priming loop, and any failure of this loop to shift would block elongation and would lead to abortive initiation (31). Therefore, the elongation of RNA synthesis may be facilitated by the flexibility of the priming loop. Third, some flexibility of the elements that interact with the template might be required for translocation of the template to the RdRp central catalytic site during transcription.
The loop (residues 1,121–1,132), which is located in the template exit channel and connects the aforementioned flexible α-helix, was suggested to separate RNA template and transcript and guide the nascent transcript into the transcript exit channel during transcription (31). Residues 1,127–1,132 of the loop in VP2 were fully conserved with those in λ3. We referred to this loop as the switch loop. An interaction between the switch loop and a loop (residues 521–524) in the apical domain of a copy of the innermost shell protein VP3A was observed (Fig. 2 C and D). We have tentatively designated it as the apical loop. The apical loops of the five copies of VP3A around the fivefold axis exhibit different conformations in our aquareovirus structure due to the asymmetric interactions, which explains why the apical loop in the icosahedrally reconstructed structure of aquareovirus VP3 (13) and the corresponding loop in crystal structure of orthoreovirus λ1 (1) were not resolved. The negatively charged residues Asp524 in the apical loop of VP3 are conserved in λ1, and the adjacent positively charged Arg1125 in the switch loop of VP2 is mutated to Lys1114 in λ3.
The apical domains of the innermost shell protein VP1A in a transcribing cypovirus (23, 42) and VP2A in a transcriptionally active rotavirus (34) have been observed to undergo a conformational change whereby they tilt outward. This conformational change of the apical domains appears to be required for the transcriptional activation of Reoviridae members, and it is likely to trigger the conformational change of the switch loop and, consequently, switch the RdRp from replicative to transcriptional mode. The similar interactions between the apical domain and the switch loop or adjacent residues have also been observed in cypovirus (37) and rotavirus (34). These findings may explain why polymerases from Reoviridae viruses are not fully active in the absence of their innermost shell proteins (38, 39).
RdRp Cofactor Protein VP4.
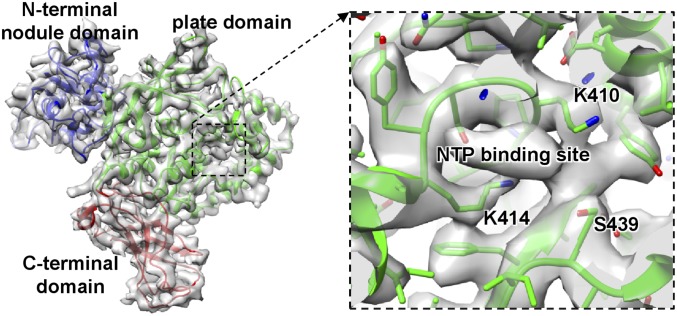
The VP4 in aquareovirus, which shares 22% of its amino acid sequence identity with the μ2 protein in orthoreovirus, interacts closely with VP2. We built a full atomic model for VP4 ab initio. The structure of VP4 can be divided into three domains: an N-terminal nodule domain (residues 1–265), a plate domain (266–599), and a C-terminal domain (600–715) (Fig. 3). The nodule domain resides in the corner between a copy of the shell protein VP3A and the RdRp protein VP2. A flexible density, which belongs to a part of the nodule domain, was also observed to interact with the dsRNA fragment roughly parallel to the capsid shell (Fig. 1C). The flexible part of the nodule domain (residues 83–190) was not resolved in our atomic model. The plate domain is roughly rectangular in shape and resides in the corner between the VP3A and the VP2 polymerase domain and interacts with a surface region of the VP2 polymerase domain between the channels for template entry and NTP entry. Structural comparison between the homology proteins of our aquareovirus VP4 and cypovirus VP4 (37) revealed that the VP4 N-terminal nodule domain exhibits almost no similarity to its counterpart in cypovirus VP4; by contrast, their central plate domains have a markedly similar structural topology (SI Appendix, Fig. S9), suggesting they must serve an identical critical function. Indeed, our VP4 plate domain contains fully conserved motifs (KxxxK and SDxxG, where uppercase letters indicate wholly conserved residues) for NTP binding in the μ2-homology proteins of the turreted Reoviridae viruses (16–18). An additional density feature, which cannot be assigned to any main chain or side chain, was observed to be surrounded by three of the five wholly conserved residues (Lys410, Lys414, and Ser439) in the plate domain of our VP4 (Fig. 3). Therefore, we attribute the density to an NTP located in the plate domain.
Fig. 3.
(Left) Transparent view of the density map of VP4 superimposed with their atomic model (ribbon). (Right) Atomic model VP4 superimposed on the density map (mesh). The additional density feature, which cannot be assigned to any main chain or side chain, is attributable an NTP.
Structural comparison also revealed an additional C-terminal domain in our aquareovirus VP4. Consistent with the fact that the C-terminal sequence of aquareovirus VP4 is longer than that of cypovirus VP4, this domain is absent from its cypovirus homology protein VP4 (SI Appendix, Fig. S9). The C-terminal domain is located at the entrance of the template entry channel and interacts with the dsRNA fragment approaching the RdRp template entry channel (Fig. 1D). The interaction is ∼30 Å away from the dsRNA end. The interactions noted between the aquareovirus VP4 and dsRNA agree with the RNA-binding activity of its homology protein μ2 (43). By contrast, no interaction between cypovirus VP4 and genomic RNA has been observed (36, 37).
The NTPase, RNA 5′-triphosphatase, and helicase activities have previously been assigned to the μ2 protein of orthoreovirus (16). Our structure indicates that the μ2-homology protein VP4 is not an RNA 5′-triphosphatase, because the positioning of the μ2-homology protein VP4 determines that the RNA transcript does not have access to it. The NTPase activity of VP4/μ2 may couple NTP hydrolysis to conformational change of the VP4 C-terminal domain, which interacts with the dsRNA adjacent to the entrance of the RdRp template entry channel. Therefore, the aquareovirus VP4 or orthoreovirus μ2 may be involved in dsRNA unwinding at the entrance of the template entry channel. In addition, the VP4 plate domain also interacts with the flexible template-interaction element of VP2, which interacts with the template RNA in the template entry channel. Therefore, the conformational change of the VP4 C-terminal domain may drive the back-and-forth movement of the VP2 template-interaction element and facilitate translocation of the template RNA to the VP2 catalytic site during transcription.
Interaction Between the N Termini of VP3A and the VP2–VP4 Complex.
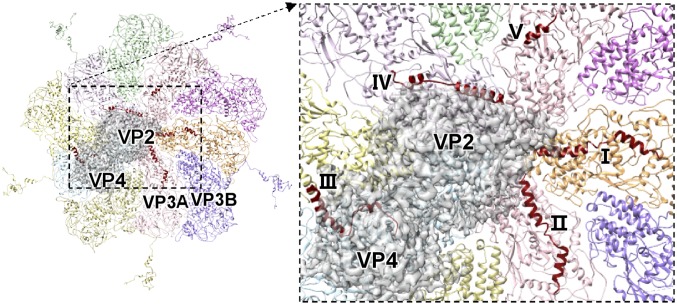
The N-terminal regions of five copies of VP3A, which have previously been resolved only from residue 188 through icosahedral reconstruction, have now been extended to residue 149 or 177 in our structure (Fig. 4 and SI Appendix, Fig. S10). These five N-terminal regions exhibit four different conformations to adapt to the occupation of the VP2–VP4 complex under the fivefold vertex. Four of these five newly resolved N-terminal regions consist of two α-helices connected by a loop, and the other contains one α-helix (Fig. 4 and SI Appendix, Fig. S10). Two of the N-terminal regions, in which the distal ends of the resolved N termini project toward the fivefold axis and interact with outside surface of the RdRp bracelet domain, share the same conformation (Fig. 4 and SI Appendix, Fig. S10). The third resides between the VP2–VP4 complex and the VP3A to mediate the interaction between the innermost shell and the complex. The fourth is located on the surface region of the VP2 bracelet domain near the exit of the template exit channel (Fig. 4 and SI Appendix, Fig. S10). The last, which is the shortest one (residues 177–187), is ∼25 Å away from the nearest surface of the RdRp and does not interact with the VP2–VP4 complex (Fig. 4 and SI Appendix, Fig. S10), suggesting the unresolved part of this N terminus is flexible. This conformation of the shortest N terminus implies that all of the fixed structures of the other four longer N termini is mediated by the presence of the VP2–VP4 complex, rather than the VP3A itself, and that these N termini should hang randomly before the VP2–VP4 complex are anchored to the capsid shell. The third N terminus, which resides between the VP2–VP4 complex and the VP3A inner surface, suggests that the individual VP3A N termini bind to the VP2–VP4 complex first, and then the complexes are anchored to the innermost capsid shell. Therefore, we concluded that the VP3A N termini are critical in recruiting the VP2 and VP4 during the virus assembly.
Fig. 4.
Interactions between the N-terminal structures of five copies of VP3A around the fivefold axis and the VP2–VP4 complex. (Left) Five copies of VP3A and five copies of VP3B around the fivefold axis are shown in ribbon. (Right) Five N termini marked I–V in the zoom-in view. The N-terminal structures of the five copies of VP3A are in red. The VP2–VP4 complex is shown in transparent gray.
An unresolved long sequence remains at each of the five VP3A N termini (residues 1–148). The resolved N-terminal structure of the other VP3 conformer VP3B (9) suggests that the unresolved N-terminal structure of VP3A contains a zinc finger structural motif. Biochemical studies have indicated that the orthoreovirus λ1 protein, homolog of VP3, possesses NTPase, RNA 5′-triphosphatase, helicase, and dsRNA-binding activities (44, 45). These unresolved VP3A N termini, which are located adjacent to the RdRp template exit channel, may be involved in guiding the plus-strand side of the transcription bubble and reannealing the RNA.
Materials and Methods
Cell Culture and Viral Growth.
A CIK cell line was used in this study for the propagation of grass carp reovirus (GCRV), a pathogenic aquareovirus isolate. The proliferation and purification of GCRV were performed as described elsewhere (12, 46). The cells were grown at 28 °C in Eagle’s minimum essential medium (MEM; Invitrogen) containing 2 mM l-glutamine. The media was supplemented with 10% FBS (Gibco BRL). The GCRV was propagated in CIK cell cultures in MEM supplemented with 2% FBS and incubated for 3 d at 28 °C until mature virions were released from the infected cells. The collected GCRV culture suspension was pelleted and further purified through sucrose density gradient centrifugation. The purified virions were suspended in PBS to create a concentration of ∼1.5 mg/mL for cryo-EM.
Cryo-EM Imaging, Image Processing, and Structural Analysis.
An aliquot of 3.5 μL purified GCRV sample was applied to a quantifoil grid, blotted for 2–4 s, and plunged into liquid ethane using an FEI Vitrobot. The viruses were imaged with an FEI Tecnai Arctica 200-kV electron microscope equipped with a Falcon II camera at a nominal magnification of 110,000× corresponding to a pixel size of 0.932 Å. A dose rate of ∼21 e−/Å2/s was used with a total exposure time of 1.2 s. The full electron dose of ∼25 e−/A2 was fractionated into 19 movie frames. The 19 frames were aligned and averaged to a single image (47). The defocus values of the images, which were set to ∼1.2–3.2 μm, were determined by CTFFIND3 (48). Determination of orientations and centers for all particle images and 3D reconstruction of the icosahedral capsid were carried out using software (37) we developed based on the common-line algorithm (49, 50). The genome and RdRp complex structure within the capsid was reconstructed using our symmetry-mismatch reconstruction method as described elsewhere (36, 41). Each cryo-EM image of a virus particle can be considered the sum of a genome image and a capsid image. We obtained the genome images by subtracting the capsid images, which were obtained by projecting the 3D density map of the capsid on the capsid orientation and applying CTF modulation on the projection, from the cryo-EM images of virus particles. Because the 60 equivalent orientations of the icosahedral capsid have been determined by icosahedral reconstruction and the asymmetric genome structure has a fixed orientation related to the symmetric capsid, the correct orientation of each genome image is one of the 60 equivalent orientations. The correct orientation of each genome image was obtained by searching the 60 equivalent orientations of the icosahedral capsid. The genome structure was determined by iterative orientation refinement and 3D reconstruction. Protein subunit densities were segmented from the maps and visualized using Chimera (51).
Atomic Model Building and Refinement.
The atomic models of VP3 (Protein Data Bank ID 3IYL) (13) were docked into the cryo-EM map within the UCSF Chimera, and the atomic models for the VP2, VP4, and VP7 were built ab initio using an automatic model building program, EMBuilder, developed specifically for cryo-EM maps (52). Then, the missing residues in the five N termini of VP3A were added into the model according to our cryo-EM map, and the whole model was manually adjusted in the COOT software (53). Thereafter, the models were refined using real-space refinement including stereochemistry and secondary structure restraints as implemented in the Phenix.real_space_refine program (54). Refinement and validation statistics are shown in SI Appendix, Table S2.
Supplementary Material
Acknowledgments
We thank the Computing and Cryo-EM Platforms of Tsinghua University, Branch of the National Center for Protein Sciences (Beijing), for providing facilities; Anbao Jia for help with computer setup; and Jianwei Zeng for assistance with model refinement. This research was supported by the National Research and Development Program of China (2016YFA0501100 and 2015CB910104), the National Natural Science Foundation of China (91530321, 31672693, 31570727, 31372565, and 31570742), and Natural Science Foundation of Hunan Province (2017RS3033).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.R. is a guest editor invited by the Editorial Board.
Data deposition: The electron density maps and atomic models have been deposited in the Electron Microscopy Data Bank and Protein Data Bank under accession codes EMD-6969 (icosahedral reconstruction), EMD-6968 (nonicosahedral reconstruction), 5ZVT (icosahedral asymmetric unit), and 5ZVS (VP2, VP4, and a VP3 decamer).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803885115/-/DCSupplemental.
References
- 1.Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 A resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 2.Duquerroy S, et al. The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. EMBO J. 2009;28:1655–1665. doi: 10.1038/emboj.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El Omari K, et al. Plate tectonics of virus shell assembly and reorganization in phage φ8, a distant relative of mammalian reoviruses. Structure. 2013;21:1384–1395. doi: 10.1016/j.str.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naitow H, Tang J, Canady M, Wickner RB, Johnson JE. L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism. Nat Struct Biol. 2002;9:725–728. doi: 10.1038/nsb844. [DOI] [PubMed] [Google Scholar]
- 5.Janssen ME, et al. Three-dimensional structure of a protozoal double-stranded RNA virus that infects the enteric pathogen Giardia lamblia. J Virol. 2015;89:1182–1194. doi: 10.1128/JVI.02745-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan J, et al. Atomic structure reveals the unique capsid organization of a dsRNA virus. Proc Natl Acad Sci USA. 2009;106:4225–4230. doi: 10.1073/pnas.0812071106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ochoa WF, et al. Partitivirus structure reveals a 120-subunit, helix-rich capsid with distinctive surface arches formed by quasisymmetric coat-protein dimers. Structure. 2008;16:776–786. doi: 10.1016/j.str.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mertens P. The dsRNA viruses. Virus Res. 2004;101:3–13. doi: 10.1016/j.virusres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Cheng L, et al. Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics. J Mol Biol. 2010;397:852–863. doi: 10.1016/j.jmb.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Tao Y, Reinisch KM, Harrison SC, Nibert ML. Orthoreovirus and aquareovirus core proteins: Conserved enzymatic surfaces, but not protein-protein interfaces. Virus Res. 2004;101:15–28. doi: 10.1016/j.virusres.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Attoui H, et al. Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of golden shiner reovirus, grass carp reovirus, striped bass reovirus and golden ide reovirus (genus Aquareovirus, family Reoviridae) J Gen Virol. 2002;83:1941–1951. doi: 10.1099/0022-1317-83-8-1941. [DOI] [PubMed] [Google Scholar]
- 12.Cheng L, Fang Q, Shah S, Atanasov IC, Zhou ZH. Subnanometer-resolution structures of the grass carp reovirus core and virion. J Mol Biol. 2008;382:213–222. doi: 10.1016/j.jmb.2008.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liemann S, Chandran K, Baker TS, Nibert ML, Harrison SC. Structure of the reovirus membrane-penetration protein, Mu1, in a complex with is protector protein, Sigma3. Cell. 2002;108:283–295. doi: 10.1016/s0092-8674(02)00612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dryden KA, et al. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: Analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Parker JS, Murray KE, Nibert ML. Nucleoside and RNA triphosphatase activities of orthoreovirus transcriptase cofactor mu2. J Biol Chem. 2004;279:4394–4403. doi: 10.1074/jbc.M308637200. [DOI] [PubMed] [Google Scholar]
- 17.Nibert ML, Kim J. Conserved sequence motifs for nucleoside triphosphate binding unique to turreted reoviridae members and coltiviruses. J Virol. 2004;78:5528–5530. doi: 10.1128/JVI.78.10.5528-5530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noble S, Nibert ML. Core protein mu2 is a second determinant of nucleoside triphosphatase activities by reovirus cores. J Virol. 1997;71:7728–7735. doi: 10.1128/jvi.71.10.7728-7735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knipe DM, Howley PM. Fields Virology. 6th Ed Lippincott Williams & Wilkins; Philadelphia: 2013. [Google Scholar]
- 20.Cheng L, et al. Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping. Proc Natl Acad Sci USA. 2011;108:1373–1378. doi: 10.1073/pnas.1014995108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyazaki N, et al. Structural evolution of reoviridae revealed by oryzavirus in acquiring the second capsid shell. J Virol. 2008;82:11344–11353. doi: 10.1128/JVI.02375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan X, et al. Virion structure of baboon reovirus, a fusogenic orthoreovirus that lacks an adhesion fiber. J Virol. 2011;85:7483–7495. doi: 10.1128/JVI.00729-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu B, et al. Identification of the active sites in the methyltransferases of a transcribing dsRNA virus. J Mol Biol. 2014;426:2167–2174. doi: 10.1016/j.jmb.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy P. Bluetongue virus structure and assembly. Curr Opin Virol. 2017;24:115–123. doi: 10.1016/j.coviro.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Desselberger U. Rotaviruses. Virus Res. 2014;190:75–96. doi: 10.1016/j.virusres.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Nason EL, Samal SK, Venkataram Prasad BV. Trypsin-induced structural transformation in aquareovirus. J Virol. 2000;74:6546–6555. doi: 10.1128/jvi.74.14.6546-6555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill CL, et al. The structure of a cypovirus and the functional organization of dsRNA viruses. Nat Struct Biol. 1999;6:565–568. doi: 10.1038/9347. [DOI] [PubMed] [Google Scholar]
- 28.Lawton JA, Estes MK, Prasad BVV. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Baker ML, Jiang W, Estes MK, Prasad BVV. Rotavirus architecture at subnanometer resolution. J Virol. 2009;83:1754–1766. doi: 10.1128/JVI.01855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mertens PPC, Diprose J. The bluetongue virus core: A nano-scale transcription machine. Virus Res. 2004;101:29–43. doi: 10.1016/j.virusres.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Tao Y, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage–Structural studies of reovirus polymerase lambda3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 32.McClain B, Settembre E, Temple BRS, Bellamy AR, Harrison SC. X-ray crystal structure of the rotavirus inner capsid particle at 3.8 A resolution. J Mol Biol. 2010;397:587–599. doi: 10.1016/j.jmb.2010.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 A. Nat Struct Biol. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Estrozi LF, et al. Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles. J Mol Biol. 2013;425:124–132. doi: 10.1016/j.jmb.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu X, et al. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure. 2008;16:1678–1688. doi: 10.1016/j.str.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, Cheng L. Cryo-EM shows the polymerase structures and a nonspooled genome within a dsRNA virus. Science. 2015;349:1347–1350. doi: 10.1126/science.aaa4938. [DOI] [PubMed] [Google Scholar]
- 37.Li X, et al. Near-atomic resolution structure determination of a cypovirus capsid and polymerase complex using cryo-EM at 200 kV. J Mol Biol. 2017;429:79–87. doi: 10.1016/j.jmb.2016.11.025. [DOI] [PubMed] [Google Scholar]
- 38.Starnes MC, Joklik WK. Reovirus protein lambda 3 is a poly(C)-dependent poly(G) polymerase. Virology. 1993;193:356–366. doi: 10.1006/viro.1993.1132. [DOI] [PubMed] [Google Scholar]
- 39.Patton JT, Jones MT, Kalbach AN, He YW, Xiaobo J. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J Virol. 1997;71:9618–9626. doi: 10.1128/jvi.71.12.9618-9626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farsetta DL, Chandran K, Nibert ML. Transcriptional activities of reovirus RNA polymerase in recoated cores. Initiation and elongation are regulated by separate mechanisms. J Biol Chem. 2000;275:39693–39701. doi: 10.1074/jbc.M004562200. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Liu H, Cheng L. Symmetry-mismatch reconstruction of genomes and associated proteins within icosahedral viruses using cryo-EM. Biophys Rep. 2016;2:25–32. doi: 10.1007/s41048-016-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang C, et al. Cryo-EM structure of a transcribing cypovirus. Proc Natl Acad Sci USA. 2012;109:6118–6123. doi: 10.1073/pnas.1200206109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brentano L, Noah DL, Brown EG, Sherry B. The reovirus protein mu2, encoded by the M1 gene, is an RNA-binding protein. J Virol. 1998;72:8354–8357. doi: 10.1128/jvi.72.10.8354-8357.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bisaillon M, Lemay G. Characterization of the reovirus lambda1 protein RNA 5′-triphosphatase activity. J Biol Chem. 1997;272:29954–29957. doi: 10.1074/jbc.272.47.29954. [DOI] [PubMed] [Google Scholar]
- 45.Bisaillon M, Bergeron J, Lemay G. Characterization of the nucleoside triphosphate phosphohydrolase and helicase activities of the reovirus lambda1 protein. J Biol Chem. 1997;272:18298–18303. doi: 10.1074/jbc.272.29.18298. [DOI] [PubMed] [Google Scholar]
- 46.Fang Q, Shah S, Liang Y, Zhou ZH. 3D reconstruction and capsid protein characterization of grass carp reovirus. Sci China C Life Sci. 2005;48:593–600. doi: 10.1360/062004-105. [DOI] [PubMed] [Google Scholar]
- 47.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 49.Thuman-Commike PA, Chiu W. Improved common line-based icosahedral particle image orientation estimation algorithms. Ultramicroscopy. 1997;68:231–255. doi: 10.1016/s0304-3991(97)00033-8. [DOI] [PubMed] [Google Scholar]
- 50.Fuller SD, Butcher SJ, Cheng RH, Baker TS. Three-dimensional reconstruction of icosahedral particles–The uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 51.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 52.Zhou N, Wang H, Wang J. EMBuilder: A template matching-based automatic model-building program for high-resolution cryo-electron microscopy maps. Sci Rep. 2017;7:2664. doi: 10.1038/s41598-017-02725-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.