Significance
We identified a one-of-a-kind duplication mutation in human transthyretin (TTR) that causes unusually aggressive systemic amyloidosis. To understand the poor response to treatment with a drug that stabilizes the TTR tetramer, we explored the structure, stability, and drug binding of recombinant proteins. The results suggested that amyloid formation could stem from global destabilization of the monomeric and tetrameric protein as well as the local disordering near the mutation site. This disordering induced proteolysis with release of aggregation-prone fragments. Alternatively, local disordering could trigger misfolding of the full-length protein by exposing an adhesive segment. Drug binding at a dimer interface distant from the mutation site did not significantly influence these pathological processes, indicating the need for alternative therapeutic targets.
Keywords: protein misfolding disease, small-molecule drug binding, kinetic stability, protein structural disorder, proteolysis
Abstract
Transthyretin (TTR) is a globular tetrameric transport protein in plasma. Nearly 140 single amino acid substitutions in TTR cause life-threatening amyloid disease. We report a one-of-a-kind pathological variant featuring a Glu51, Ser52 duplication mutation (Glu51_Ser52dup). The proband, heterozygous for the mutation, exhibited an unusually aggressive amyloidosis that was refractory to treatment with the small-molecule drug diflunisal. To understand the poor treatment response and expand therapeutic options, we explored the structure and stability of recombinant Glu51_Ser52dup. The duplication did not alter the protein secondary or tertiary structure but decreased the stability of the TTR monomer and tetramer. Diflunisal, which bound with near-micromolar affinity, partially restored tetramer stability. The duplication had no significant effect on the free energy and enthalpy of diflunisal binding, and hence on the drug–protein interactions. However, the duplication induced tryptic digestion of TTR at near-physiological conditions, releasing a C-terminal fragment 49–129 that formed amyloid fibrils under conditions in which the full-length protein did not. Such C-terminal fragments, along with the full-length TTR, comprise amyloid deposits in vivo. Bioinformatics and structural analyses suggested that increased disorder in the surface loop, which contains the Glu51_Ser52dup duplication, not only helped generate amyloid-forming fragments but also decreased structural protection in the amyloidogenic residue segment 25–34, promoting misfolding of the full-length protein. Our studies of a unique duplication mutation explain its diflunisal-resistant nature, identify misfolding pathways for amyloidogenic TTR variants, and provide therapeutic targets to inhibit amyloid fibril formation by variant TTR.
Protein misfolding diseases, including neurodegenerative (Alzheimer’s disease and Parkinson’s disease) and systemic amyloidoses, are a major public health challenge affecting millions of patients and their caregivers worldwide. More than 30 soluble proteins and peptides are currently known to misfold and deposit as insoluble amyloid fibrillar aggregates in various organs, causing organ damage and death (1). Such protein misfolding diseases may be either sporadic, involving deposition of wild-type (WT) proteins often associated with aging, or hereditary, caused by gene mutations.
A major form of human systemic amyloidosis arises from aggregation and misfolding of transthyretin (TTR), a globular plasma protein that functions as a minor transporter of thyroxine and retinol through a ternary complex with holoretinol-binding protein (2, 3). Normally, TTR circulates in serum as a stable ∼55-kDa homotetramer at concentrations of 18–45 mg/dL (4). However, in transthyretin amyloidosis (ATTR), the protein misfolds and forms fibrillar deposits that infiltrate vital organs, most frequently the heart and nerves, causing organ dysfunction. Treatment options for systemic amyloidoses such as ATTR are limited to several protein-specific drugs, organ transplantation, and gene silencing agents, fueling the search for new therapeutic targets. Sporadic ATTR occurs with aging and involves deposition of WT protein, while hereditary TTR amyloidosis (ATTRm) is caused by TTR mutations. Both diseases feature deposition of the full-length protein and/or C-terminal fragments ranging from residues 46–127 to 55–127 (5–7). Aggregation-prone fragment 49–127 is the major component of ex vivo amyloid fibers (7).
Of nearly 140 ATTRm mutations reported to date (8) and listed in the ATTRm database (www.amyloidosismutations.com/mut-attr.php), all but one result in single amino acid substitutions distributed across the protein surface, with Val122del being the lone deletion mutation (9). These point mutations destabilize the structure of the otherwise highly thermostable TTR tetramer, promoting the release of monomeric precursors of amyloid (10–12). Therefore, stabilization of the TTR tetramer is considered an important therapeutic target. To this end, small-molecule thyroxin-mimetic drugs have been developed targeting the hormone-binding site located across the “weak” dimer–dimer interface in the TTR tetramer (13–17). One small molecule shown to be effective in clinical testing is diflunisal, a nonsteroidal antiinflammatory drug repurposed for ATTR treatment (18). Although diflunisal does not cure ATTRm, it slows TTR amyloid fibril formation and inhibits disease progression. This therapeutic effect stems, in part, from diflunisal binding across the subunit interface to stabilize the TTR tetramer.
In the current study, we report a Glu51, Ser52 duplication (Glu51_Ser52dup) in TTR, a one-of-a-kind duplication mutation in a patient with ATTRm. The mutation led to an unusually aggressive amyloidosis that progressed unabated despite diflunisal treatment. To understand why diflunisal was ineffective, we generated a recombinant Glu51_Ser52dup variant and analyzed its structural, stability, and drug-binding properties. The results helped reveal the molecular underpinnings for the clinical presentation of the duplication mutation and propose two parallel molecular pathways for misfolding in Glu51_Ser52dup and, potentially, other ATTRm variants. Our study highlights the complex interplay between the global destabilization of a protein assembly and its local disordering, which influences proteostasis in vivo.
Results
Clinical Presentation and Histological Detection of Amyloid Deposits.
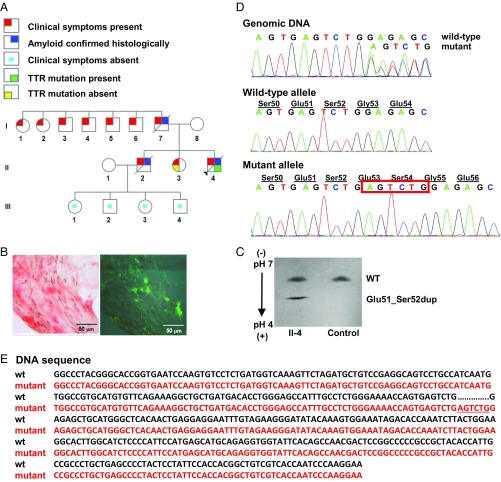
The proband (Fig. 1A, II-4), an African-American man, presented at the age of 37 y with length-dependent sensorimotor polyneuropathy in the lower extremities. At the age of 38 y (visit 1), he developed myopathy in the lower extremities with an elevated creatine kinase of 563 (39–259 U/L); cardiac biomarkers were normal. An abdominal fat pad aspirate identified amyloid deposits by Congo red staining (Fig. 1B). Tandem mass spectrometry proteomics of a right shin skin biopsy demonstrated TTR amyloid (SI Appendix, Fig. S1 A and B). The patient declined liver transplant evaluation, opting for diflunisal treatment (250 mg twice daily). After 9 mo of diflunisal therapy (visit 2), the patient exhibited progression of sensorimotor polyneuropathy to his upper extremities, bilateral carpal tunnel syndrome, and autonomic neuropathy. Despite 19 mo of diflunisal treatment (visit 3), polyneuropathy and myopathy progressed, rendering the patient unable to stand, feed himself, effectively swallow, or control bowel function. Cardiac biomarkers remained normal. Diflunisal was discontinued due to the lack of effect. He died at the age of 42 y of recurrent aspiration pneumonia and debilitating peripheral and autonomic neuropathy.
Fig. 1.
Identification of hereditary amyloidosis in proband II-4, which is linked to a TTR variant, Glu51_Ser52dup. (A) Family pedigree. Squares, males; circles, females; diagonal line, deceased members; black triangle, the proband. (B) Histological analyses. (Left) Congo red-positive amyloid deposits viewed by light microscopy in fat tissue. (Right) Polarized light view demonstrating apple-green birefringence of Congo red-positive amyloid deposits. (Magnification: Left and Right, 400×.) (C) Serum isoelectric focusing. Screening of serum from the proband (II-4) showed the presence of both WT and Glu51_Ser52dup variant proteins. Normal serum (Control) contained only the WT. (D) Genetic analyses. Partial sequence chromatograms of exon 3 of the TTR (Top), the subcloned WT allele (Middle), and the subcloned mutant allele (Bottom) are shown. A heterozygous duplication, c.212_217dupAGTCTG (red box), resulted in Glu51_Ser52dup protein variant. (E) Gene sequence alignment of the WT (wt; black) and mutant (red) TTR. The c.212_217dupAGTCTG duplication is underlined.
The proband’s father (I-7) died at the age of 46 y of amyloidosis with symptoms of peripheral neuropathy, heart failure, and liver and kidney involvement. Two paternal aunts (I-1 and I-2) and four paternal uncles (I-3, I-4, I-5, and I-6) reportedly had symptoms and signs of peripheral neuropathy. The proband’s mother (I-8) was 57 y of age and healthy at the time of visit 1. The proband’s 40-y-old sister (II-3) had peripheral neuropathy with negative TTR mutation testing, while an older brother (II-2), diagnosed with amyloidosis by sural nerve biopsy, died at the age of 34 y with symptoms of cardiac involvement and peripheral and autonomic neuropathy. The proband’s brother had two daughters, aged 19 and 24 y (III-3 and III-1), and two sons, aged 13 and 22 y (III-4 and III-2), who were all symptom-free at the time of visit 1.
The proband’s serum TTR level was well below normal at visit 1, which is characteristic of patients who have ATTRm (19–21). While diflunisal treatment increased serum TTR levels at visits 2 and 3, the levels remained below the normal range (SI Appendix, Fig. S1C).
Identification of TTR Protein Glu51_Ser52dup.
Screening of serum from the proband by isoelectric focusing showed two distinct TTR bands (Fig. 1C). The higher band corresponded to WT TTR, while the lower, more anodal band corresponded to a more acidic mutant. Unexpectedly, direct DNA sequencing demonstrated a 6-nt duplication in exon 3 of the TTR, c.212_217dupAGTCTG (Fig. 1D, Top). Subcloning of genomic DNA showed WT and mutant alleles (Fig. 1D, Middle and Bottom), confirming the heterozygous nature of the mutation. This mutation resulted in the TTR duplication of Glu51, Ser52 in the mature protein, Glu51_Ser52dup. A duplication mutation has not been reported in the SNP database (https://www.ncbi.nlm.nih.gov/projects/SNP/) or Exome Aggregation Consortium (exac.broadinstitute.org). Moreover, no insertion mutations have been reported previously for human ATTRm. Protein Variation Effect Analyzer (provean.jcvi.org/) predicted the mutation to be deleterious with a score of −10.2 (a default threshold is −2.5).
The Duplication Mutation Greatly Destabilizes the TTR Structure.
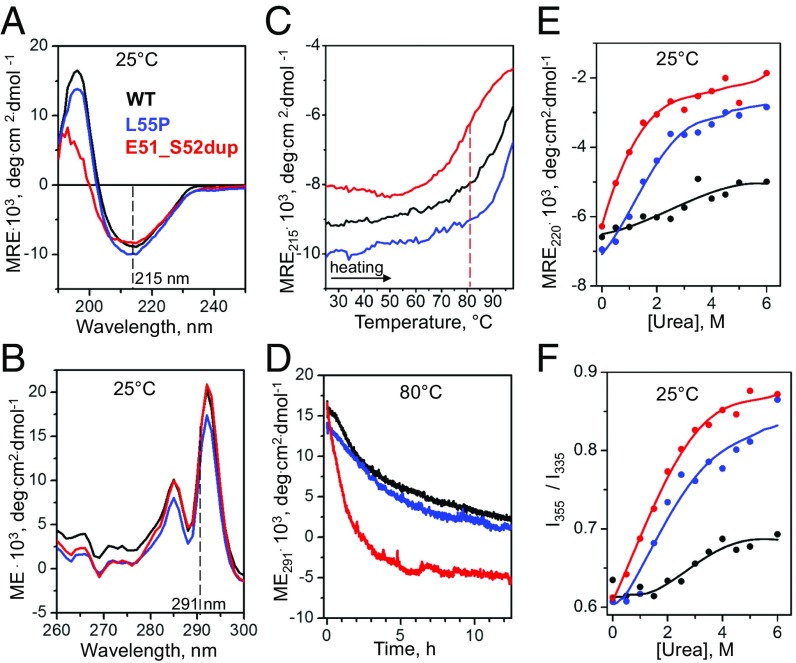
Ultrapure recombinant Glu51_Ser52dup TTR was generated and used to characterize the structure and stability of the protein by circular dichroism (CD) and intrinsic Trp fluorescence as described in Materials and Methods. For comparison, recombinant forms of WT and Leu55Pro, a particularly aggressive and destabilizing amyloidogenic variant (22), were studied. Far-UV CD spectra of all proteins showed a similar negative peak at 215 nm, and Glu51_Ser52dup showed a decreased positive peak at 193 nm (Fig. 2A), indicating similar β-sheet–rich secondary structure in these proteins with increased disorder in the duplication mutant. Near-UV CD spectra of all proteins showed a similar double peak (Fig. 2B), which was dominated by Trp79 buried in the core of the TTR monomer (10, 23, 24), suggesting a similar tertiary structure.
Fig. 2.
Effects of the duplication mutation on the TTR secondary and tertiary structure and stability. Recombinant proteins were analyzed by CD and fluorescence spectroscopy as described in Materials and Methods. WT (black) and Leu55Pro (blue) were used as controls for Glu51_Ser52dup (red). (A) Far-UV CD spectra (0.2 mg/mL protein in standard buffer at 25 °C) characterize protein secondary structure. (B) Near-UV CD spectra (0.2 mg/mL protein in standard buffer at 25 °C) show a double peak characteristic of Trp79 buried in the core of the native TTR monomer (2). (C) Melting data recorded by CD at 215 nm to monitor β-sheet unfolding during heating from 25 to 98 °C at a rate of 6 °C⋅h−1. Sample conditions are as in A. The vertical dashed line indicates the apparent melting temperature for Glu51_Ser52dup, Tm, app = 81 °C, determined from the first-derivative maximum in the melting data. (D) Time course of protein unfolding in a temperature jump to 80 °C. At t = 0, the sample temperature was rapidly increased from 25 to 80 °C, and changes in the Trp solvent exposure upon protein unfolding were monitored by CD at 291 nm. Sample conditions are as in B. (E) Equilibrium denaturation by urea monitored by CD at 220 nm for secondary structure unfolding after incubation in urea for 96 h. Protein concentrations are 0.2 mg/mL. (F) Equilibrium denaturation by urea tracked by the ratio of fluorescence intensity at 355 nm and 335 nm, I355/I335, to monitor solvent exposure of Trp. Sample conditions are 0.2 mg/mL protein. deg, degrees; ME, molar ellipticity; MRE, molar residue ellipticity.
Protein thermal stability was assessed by far- and near-UV CD in melting experiments and in temperature jumps, respectively (Fig. 2 C and D). Compared with WT and Leu55Pro, the melting data of Glu51_Ser52dup, recorded at 215 nm to track the β-sheet unfolding, shifted to lower temperatures by ∼10 °C (Fig. 2C, red) indicating greatly decreased stability. The temperature-jump data at 80 °C, recorded at 291 nm to track the Trp79 packing (23), showed a much faster unfolding of Glu51_Ser52dup, indicating greatly reduced kinetic stability compared with Leu55Pro or WT (Fig. 2D, red). Moreover, Glu51_Ser52dup showed equilibrium unfolding at lower urea concentrations compared with Leu55Pro or WT, as evidenced by chemical denaturation after 96 h of incubation in urea, which was monitored by far-UV CD and Trp fluorescence at 25 °C (Fig. 2 E and F). Therefore, consistent with thermal denaturation, chemical denaturation indicated that the duplication mutation decreases the thermodynamic stability of TTR (Fig. 2 C, E, and F), while the temperature-jump data (Fig. 2D) indicated that the mutation decreases the kinetic stability (25). Since the rate-limiting step in TTR unfolding is tetramer dissociation (23, 24, 26, 27), faster unfolding of Glu51_Ser52dup reflects a lower free-energy barrier for dissociation of the variant tetramer.
Together, the results in Fig. 2 revealed that the duplication mutation has a slight disordering effect on the secondary structure but does not affect the Trp79 packing, and hence the tertiary structure of the TTR monomer. However, the duplication decreases both the thermodynamic stability of the TTR monomer and the kinetic stability of the tetramer to a much greater extent than observed even for Leu55Pro, a particularly destabilizing ATTRm variant.
The Duplication Mutation Destabilizes TTR Heterotetramers.
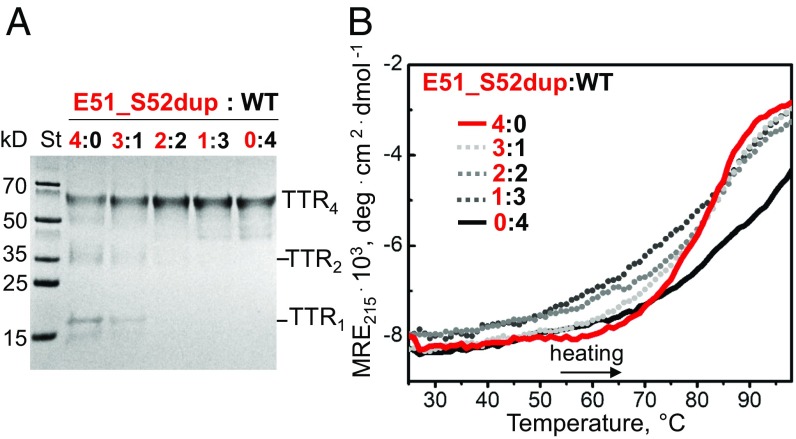
As ATTRm is an autosomal dominant disease with translationally active alleles, heterozygotes express WT and mutant TTR proteins in vivo, which circulate as heterotetramers (28, 29). To assess the effect of the duplication mutation on the structural integrity of such heterotetramers, Glu51_Ser52dup and WT were incubated at mutant/WT molar ratios from 4:0 (Glu51_Ser52dup alone) to 0:4 (WT alone) following published protocols to allow for the subunit exchange (30). SDS/PAGE of glutaraldehyde cross-linked proteins showed that the WT alone was entirely tetrameric (Fig. 3A, right lane), whereas Glu51_Ser52dup alone contained a large population of monomers and dimers (Fig. 3A, left lane) in addition to tetramers. Decreasing the mutant/WT ratio shifted the distribution from monomers and dimers to tetramers, and at a molar ratio of 2:2 or lower, only tetramers were detected (Fig. 3A, left to right).
Fig. 3.
Duplication mutation destabilizes the TTR tetramer. (A) SDS/PAGE of heterotetramers prepared at indicated molar ratios of mutant (red) to WT (black), from 4:0 (Glu51_Ser52dup alone) to 0:4 (WT alone), as shown on the lanes. The samples containing 0.2 mg/mol total protein were incubated at 4 °C for 48 h to allow for subunit exchange (31) and then cross-linked with glutaraldehyde. The gels were developed with Gel Code Blue stain reagent. TTR monomer (TTR1), dimer (TTR2), and tetramer (TTR4) are indicated. (B) Melting data were recorded by CD at 215 nm to monitor secondary structural unfolding in heterotetramers. Samples containing 0.2 mg/mL total protein, which were prepared as described in A, were heated from 25 to 98 °C at a rate of 6 °C⋅h−1. Mutant-to-WT molar ratios are shown by numbers and color-coded. deg, degrees; MRE, molar residue ellipticity; St, molecular weight standards.
To determine the effects of the duplication mutation on heterotetramer stability, far-UV CD melting data were recorded for the binary mixtures of the variant and WT proteins, and the results were compared with Glu51_Ser52dup or WT alone. The variant protein alone featured a narrow sigmoidal unfolding transition centered at an apparent melting temperature, Tm,app = 81 ± 1.5 °C, while WT protein unfolding occurred at much higher temperatures, was incomplete at 98 °C, and was much less cooperative (Fig. 3B, solid lines). Melting curves recorded from the mixtures containing 1:3–3:1 (mol/mol) Glu51_Ser52dup/WT (Fig. 3B, dashed lines) shifted to lower temperatures compared with the WT alone and partially overlapped the melting curve of the Glu51_Ser52dup alone. Moreover, increasing the mutant fraction in the mixture led to a progressive increase in the transition steepness, indicating increased unfolding cooperativity. This finding suggests that the unfolding of the relatively unstable Glu51_Ser52dup monomer is tightly coupled to its dissociation from the tetramer, and thus is highly cooperative. In contrast, WT monomer is relatively stable (24) and unfolds as one subunit with lower cooperativity. The melting data were nonadditive; instead of two distinct transitions representing WT (a broad transition centered above 90 °C) and variant (a sharp transition centered at 81 °C) homotetramers, the mixtures showed a single transition with intermediate width representing heterotetramers. Together, these findings demonstrate that even one Glu51_Ser52dup subunit is sufficient to destabilize a heterotetramer. Since the range of the mutant/WT ratios used in our experiments encompasses that found in circulation, the results predict that TTR heterotetramers are destabilized by the Glu51_Ser52dup mutation in vivo.
Diflunisal Stabilizes the Mutant Tetramer.
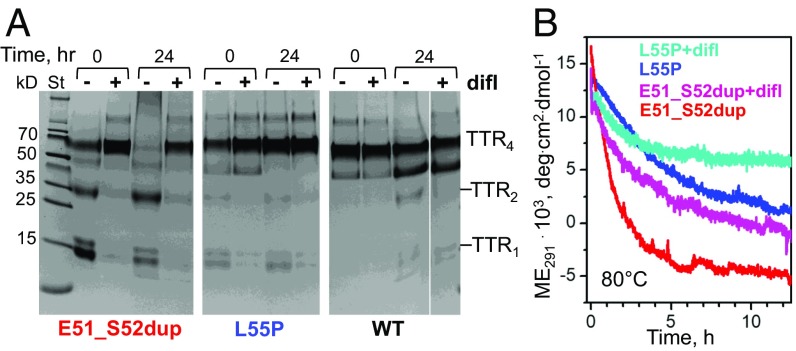
To understand why diflunisal was ineffective for the proband, we investigated the effects of the drug on the thermal stability of the duplication mutant. Glu51_Ser52dup, WT, and Leu55Pro proteins were incubated at 80 °C for 24 h alone or with diflunisal at the drug/TTR monomer ratio of 25:1 (mol/mol). The studied drug concentration (360 μM) was based on a maximal therapeutic serum level of ∼400 μM measured in patients who received 250 mg of diflunisal twice daily (32, 33), although the bioavailable levels of the drug are probably significantly lower due to albumin binding. The protein concentration of 14.4 μM mimicked that in the patient’s serum (SI Appendix, Fig. S1C). Hence, the drug/TTR molar ratio of 25:1 is representative of the maximal therapeutic levels of diflunisal. Proteins were cross-linked either before or after incubation (Fig. 4A, 0 or 24 h).
Fig. 4.
Diflunisal increases thermal stability of the mutant TTR. (A) SDS/PAGE of proteins (0.2 mg/mL) before (0) or after 24 h of incubation at 80 °C (10) either without (−) or with (+) diflunisal at 360 μM, 25:1 (mol/mol) drug to TTR monomers. The proteins were cross-linked by glutaraldehyde, and the gels were developed with Gel Code Blue stain reagent. TTR monomer (TTR1), dimer (TTR2), and tetramer (TTR4) are indicated. (B) Kinetics of protein unfolding during incubation at 80 °C in the absence or in the presence of diflunisal at 5:1 (mol/mol) drug to TTR monomers monitored by CD at 291 nm for Trp solvent exposure. The data for two ATTR mutants, Glu51_Ser52dup and Leu55Pro, are shown; WT remains fully folded under these conditions. deg, degrees; difl, diflunisal; ME, molar ellipticity.
Glu51_Ser52dup alone migrated as monomers, dimers, and tetramers under ambient conditions (Fig. 4A); preincubation with diflunisal shifted the equilibrium from the monomer and dimer to the tetramer (Fig. 4A, 0 h). After 24 h at 80 °C, Glu51_Ser52dup alone showed tetramer dissociation and formation of high-molecular-weight aggregates; diflunisal blocked this aggregation and stabilized the tetramers (Fig. 4A, Left). Compared with Glu51_Ser52dup, a clinically aggressive mutation (Leu55Pro) showed a much lower initial population of monomers and dimers; again, diflunisal shifted the distribution toward tetramers (Fig. 4A, Middle). WT migrated almost entirely as tetramers, with only trace amounts of monomers and dimers detected after 24 h at 80 °C in the absence of diflunisal, and only tetramers in the presence of the drug (Fig. 4A, Right). We conclude that diflunisal stabilizes TTR tetramers in vitro at ambient and elevated temperatures.
Temperature-jump data also revealed the effects of diflunisal on the protein unfolding kinetics. Changes in the tertiary structure at 80 °C were monitored by CD at 291 nm (23). To decrease the noise level resulting from UV absorption by diflunisal, a reduced drug/protein monomer molar ratio of 5:1 was used. Glu51_Ser52dup alone showed the fastest unfolding, which decelerated in the presence of diflunisal (Fig. 4B, red and purple). Compared with Glu51_Ser52dup either alone or with diflunisal, Leu55Pro alone exhibited slower unfolding that diflunisal further decelerated (Fig. 4B, blue and teal). WT showed little unfolding under these conditions. Together, the results in Fig. 4 revealed that the Glu51_Ser52dup mutation greatly destabilizes the TTR tetramer, and that diflunisal partially restores the structural integrity of the variant tetramer and increases its thermal stability.
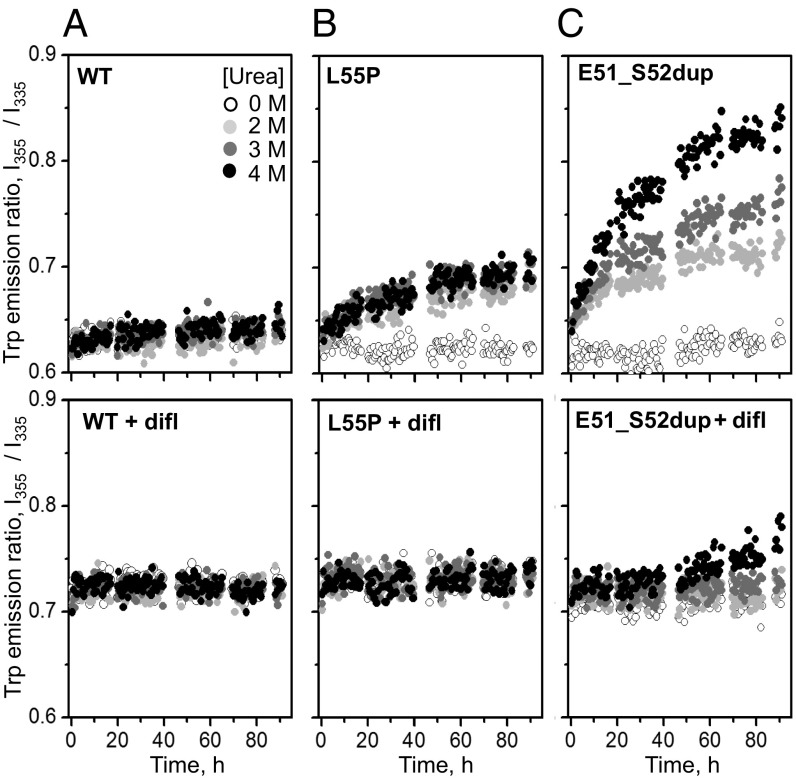
The effects of ATTRm mutations and diflunisal on the kinetics of TTR unfolding at ambient temperatures were tested by chemical denaturation. The proteins were incubated in 0–4 M urea at 25 °C for up to 96 h. The ratio of the fluorescence emission intensity at 355 nm and 335 nm, I355/I335, which reports on the solvent exposure of Trp upon protein unfolding (34), was measured with and without diflunisal at a drug/TTR monomer molar ratio of 25:1. As expected, WT showed no unfolding under these conditions, Leu55Pro showed some unfolding that was completely blocked by diflunisal, and Glu51_Ser52dup showed much greater unfolding that was only partially inhibited by diflunisal (Fig. 5). Compared with WT and Leu55Pro, Glu51_Ser52dup has much lower structural stability that clinically relevant diflunisal concentrations only partially ameliorated.
Fig. 5.
Diflunisal increases TTR stability to chemical denaturation. WT (A), Leu55Pro (B), and Glu51_Ser52dup (C) in the absence (Top) or presence (Bottom) of diflunisal were incubated in 0–4 M urea (color-coded light to dark) at 37 °C for up to 92 h, during which intrinsic Trp fluorescence was measured. The ratio of the emission intensity at 355 nm and 335 nm, I355/I335, versus time depicts the kinetics of protein unfolding. In diflunisal-containing samples, 360 μM diflunisal and a 25:1 drug/protein molar ratio were used.
Diflunisal Binds to WT and Mutant TTR with Comparable Affinity.
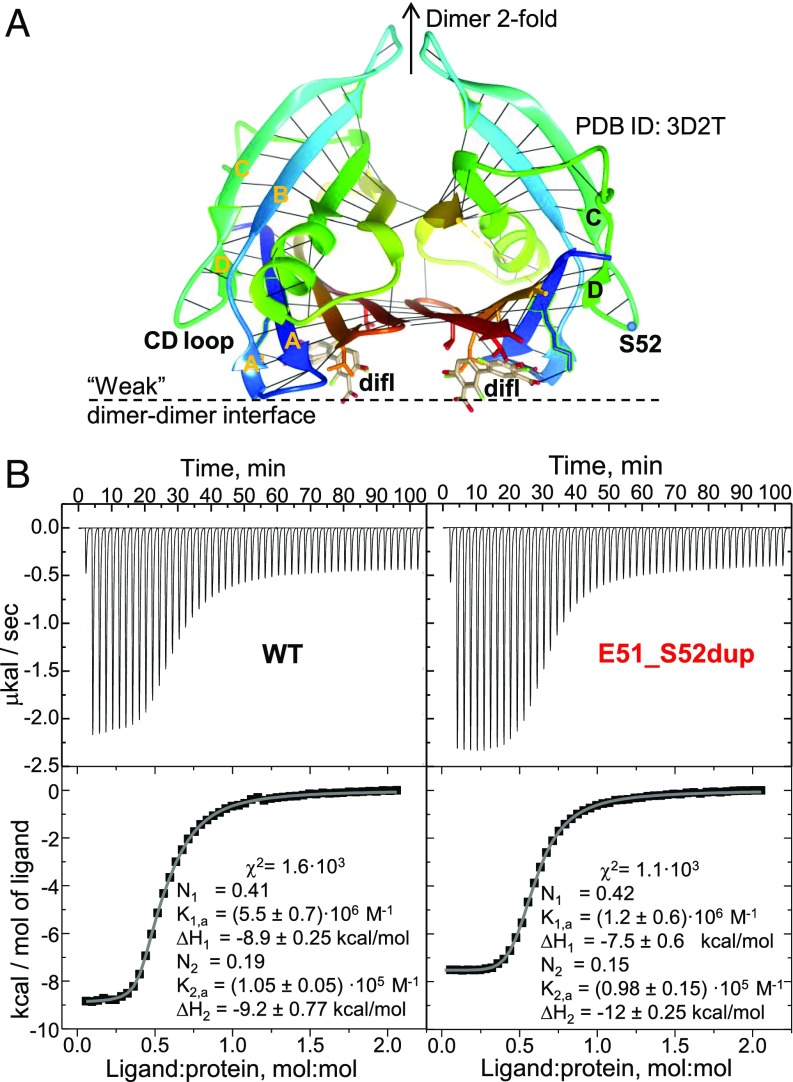
Isothermal titration calorimetry (ITC) was used to quantify diflunisal binding to Glu51_Ser52dup. WT tetramer has two diflunisal binding sites overlapping the thyroxin-binding sites that are located across the weak dimer–dimer interface, and are related via the dimer twofold symmetry (35) (Fig. 6A). To solubilize the hydrophobic drug at a 1.4 mM concentration necessary for ITC, we first used ethanol following published protocols (35); however, a low signal-to-baseline ratio in the ITC data, which resulted from the high dissolution heat of ethanol in water, precluded accurate data analysis (SI Appendix, Fig. S2). To diminish the baseline, diflunisal was dissolved in dimethyl sulfoxide as described in Materials and Methods, leading to useful ITC data (Fig. 6B).
Fig. 6.
Diflunisal binding to WT and Glu51_Ser52dup TTR. (A) TTR dimer binds diflunisal (difl) at two sites across the weak dimer–dimer interface. A ribbon representation of the 1.85 Å resolution crystal structure of WT TTR with bound diflusinal [Protein Data Bank (PDB) ID code 3D2T] reveals two alternative orientations of the drug molecule (in stick representation). Each protein molecule is rainbow-colored from the N terminus to the C terminus (blue to red); black lines indicate main chain H-bonds. (B) ITC of WT and Glu51_Ser52dup TTR. Raw data (Top) and binding isotherms (Bottom) are shown. Parameters determined by fitting these isotherms using a model with two independent binding sites are listed. TTR concentrations are ∼100 μM. K, dissociation constant; N, the site occupancy.
The isotherms for diflunisal binding to WT and Glu51_Ser52dup were similar, yielding similar binding parameters (Fig. 6B). Previous ITC studies used a two-site model to approximate diflunisal binding to WT (35). Although the binding isotherms in Fig. 6B showed only one observable binding event, a two-site binding model yielded much better data fitting (χ2 ∼ 103; Fig. 6B) compared with a single-site model (χ2 ∼ 105; SI Appendix, Fig. S2). For the one-site model, both proteins showed similar binding affinity with Kd ∼ 10 μM. For the two-site model, the dissociation constants were Kd,1 ∼ 10 μM and Kd,2 ∼ 600–900 ηM for Glu51S-Ser52dup and Kd,1 ∼ 10 μM and Kd,2 ∼ 200–300 ηM for the WT, which are comparable to the published values for the WT (35). The binding enthalpies for the two sites, ΔH1 ∼ ΔH2 = 9.5 ± 0.7 kcal/mol, were similar for both proteins, suggesting similar drug–protein interactions at these sites. Since diflunisal binding shifted the Glu51_Ser52dup population from monomers and dimers to tetramers (Fig. 4A), a decrease in the cratic (association) entropy was expected to provide an unfavorable contribution to the binding free energy of the mutant, but not the WT, TTR. However, this entropic effect was not detected (Fig. 6B), perhaps because the duplication mutant was largely tetrameric at the high protein concentrations used in ITC. In summary, ITC data showed that diflunisal binds weakly to both WT and Glu51_Ser52dup with comparable near-micromolar affinity (i.e., comparable free energy of binding), as well as similar enthalpy of binding, ΔH ∼ 10 kcal/mol, suggesting that the duplication mutation in the surface loop has little effect on the drug–protein interactions across the subunit interface.
We conclude that the Glu51_Ser52dup mutation carrier did not respond to diflunisal treatment because of reduced structural stability of this variant and insufficient drug binding at physiologically relevant concentrations (Figs. 4 and 5), and not because of decreased variant TTR affinity for the drug (Fig. 6B).
Duplication Mutation Helps Generate Proteolytic Fragments That Form Amyloid.
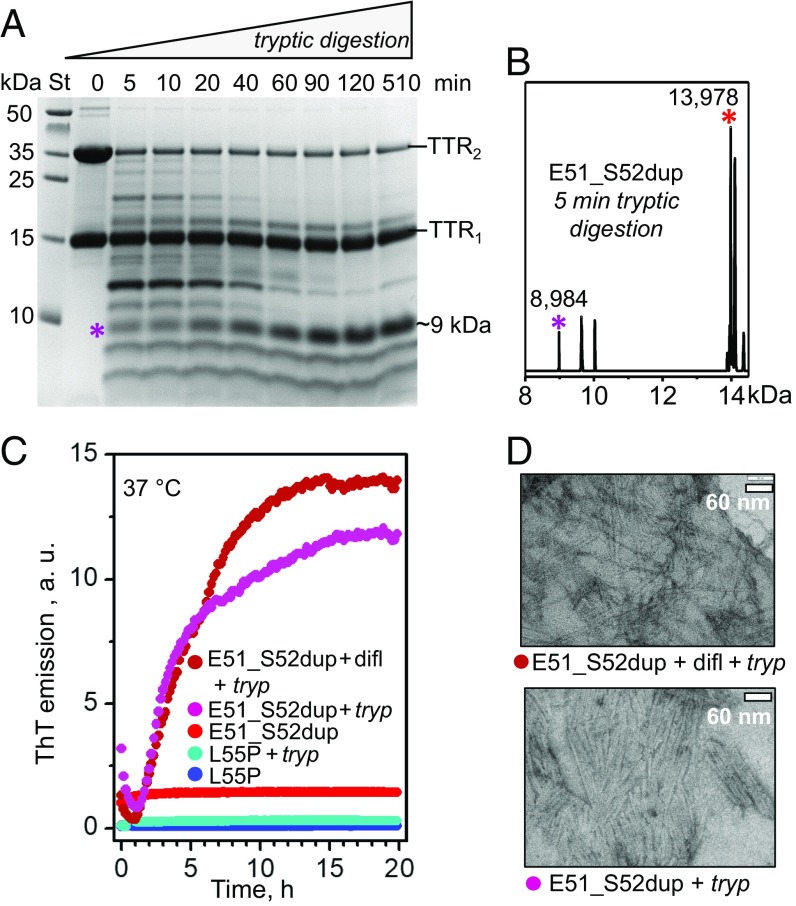
Protein fragmentation can importantly influence the development of amyloidosis, such as ATTRm. For example, trypsin can cleave TTR at the Thr48-Lys49 junction, releasing the C-terminal fragment, 49–127. This fragment is a major constituent of ATTRm amyloid deposits in vivo (7, 36) and also forms fibrils in vitro under ambient conditions, while the full-length protein does not (37). We used trypsin to mimic the yet-to-be-identified proteases that cleave TTR in vivo. To test whether the Glu51_Ser52dup mutation influences TTR cleavage and generation of a corresponding duplication-containing fragment, 49–129, the protein was subjected to limited tryptic digestion, and the products were analyzed by SDS/PAGE. All proteins showed two bands in SDS/PAGE corresponding to TTR monomers and dimers (Fig. 7A and SI Appendix, Fig. S3A), suggesting incomplete dimer unfolding in SDS. Unlike WT or Leu55Pro, which showed no fragmentation under these conditions (37) (SI Appendix, Fig. S3A), Glu51_Ser52dup was extensively proteolyzed after just 5 min of tryptic digestion, with formation of multiple fragments (Fig. 7A). The dimer population decreased much faster than the monomer population, suggesting that in the dimer, digestion of one subunit led to subunit dissociation, which protected the other subunit against proteolysis. Electrospray ionization mass spectrometry of the proteolytic products revealed a fragment of 8,984 Da corresponding to residues 49–129 (Fig. 7B).
Fig. 7.
Proteolysis of Glu51_Ser52dup by trypsin leads to amyloid formation. (A) SDS/PAGE of Glu51_Ser52dup at various stages of limited proteolysis. The protein (1 mg/mL) was incubated with trypsin at 37 °C, 200:1 substrate/enzyme weight ratio, for 5–510 min as indicated on the lanes; 0 indicates intact protein before incubation. An asterisk indicates the ∼9-kDa fragment that is formed after 5 min and gradually accumulates over time. St, molecular weight standards. (B) Electrospray ionization mass spectrometry of the incubation mixture at 5 min from A reveals a C-terminal fragment with molecular mass of 8,984 Da corresponding to residues 49–129 of Glu51_Ser52dup, along with the full-length duplication mutant (residues 1–129, 13,978 Da). (C) Amyloid formation upon limited proteolysis of mutant TTR monitored by ThT emission. Glu51_Ser52dup (warm colors) and Leu55Pro (cold colors) were incubated at 37 °C alone, with trypsin (1 mg/mL TTR, 200:1 weight ratio of TTR monomer to enzyme), or with trypsin and diflunisal [25:1 (mol/mol) drug/TTR monomer, 1.8 mM diflunisal]. Only samples containing E51_S52dup and trypsin, either with or without diflunisal, showed ThT binding to amyloid-like structures. (D) Transmission electron micrographs of negatively stained samples after incubation with trypsin for 5 d with double-orbital shaking at 168 rpm, 37 °C in a TECAN fluorescence microplate reader (Fisher Scientific). Samples containing Glu51_Ser52dup and trypsin, either with (Top, 25:1 drug/TTR monomer molar ratio) or without (Bottom) diflunisal, showed massive fibril formation, while other samples showed few fibrils. difl, diflunisal; tryp, trypsin.
To test whether the tryptic fragments of Glu51_Ser52dup formed amyloid, we monitored the time course of amyloid formation using thioflavin T (ThT), a fluorescent dye whose emission increases upon binding to an amyloid-like structure (38). The proteins were incubated with ThT in the presence or absence of trypsin, either with or without diflunisal (1.8 mM) at a 25:1 drug/protein molar ratio (Fig. 7C). As expected, Leu55Pro, which was not proteolyzed by trypsin under these conditions, showed no changes in the emission intensity, indicating that no amyloid was formed (37). In the absence of trypsin, Glu51_Ser52dup also showed no changes in ThT emission. In contrast, in the presence of trypsin, Glu51_Ser52dup showed a rapid increase in emission, which was observed either with or without diflunisal. This observation was consistent with the Glu51_Ser52dup fragmentation and accumulation of amyloidogenic fragments, particularly 49–129 (Fig. 7 A and B). In addition, this result was concordant with the location of the mutation and the cleavage sites on the exposed molecular surface away from the TTR subunit interface that binds diflunisal (Fig. 6A). Such a location explains why diflunisal binding at this interface did not interfere with the protein cleavage at the Thr48-Lys49 junction (SI Appendix, Fig. S3B), and hence had little effect on the amyloid formation (Fig. 7C).
To test whether the formation of the amyloid-like structure by tryptic fragments of Glu51_Ser52dup was on the pathway to formation of mature fibrils, the proteins at a concentration of about 1 mg/mL were incubated at 37 °C for 96 h with shaking and were visualized by negative-stain transmission electron microscopy. Massive clusters of nonbranched fibrils ∼10 nm in width, typical of mature amyloid fibrils, were observed in Glu51_Ser52dup proteolyzed by trypsin either in the absence or in the presence of diflunisal at a 25:1 drug/protein molar ratio, or 1.8 mM diflunisal (Fig. 7D).
We conclude that Glu51_Ser52dup mutation greatly enhances protein susceptibility to cleavage and the generation of amyloidogenic fragments, particularly 49–129, that readily form amyloid in a process not blocked by physiologically relevant diflunisal concentrations.
Effects of Mutation on the Local Protein Structure, Dynamics, and Amyloid Formation.
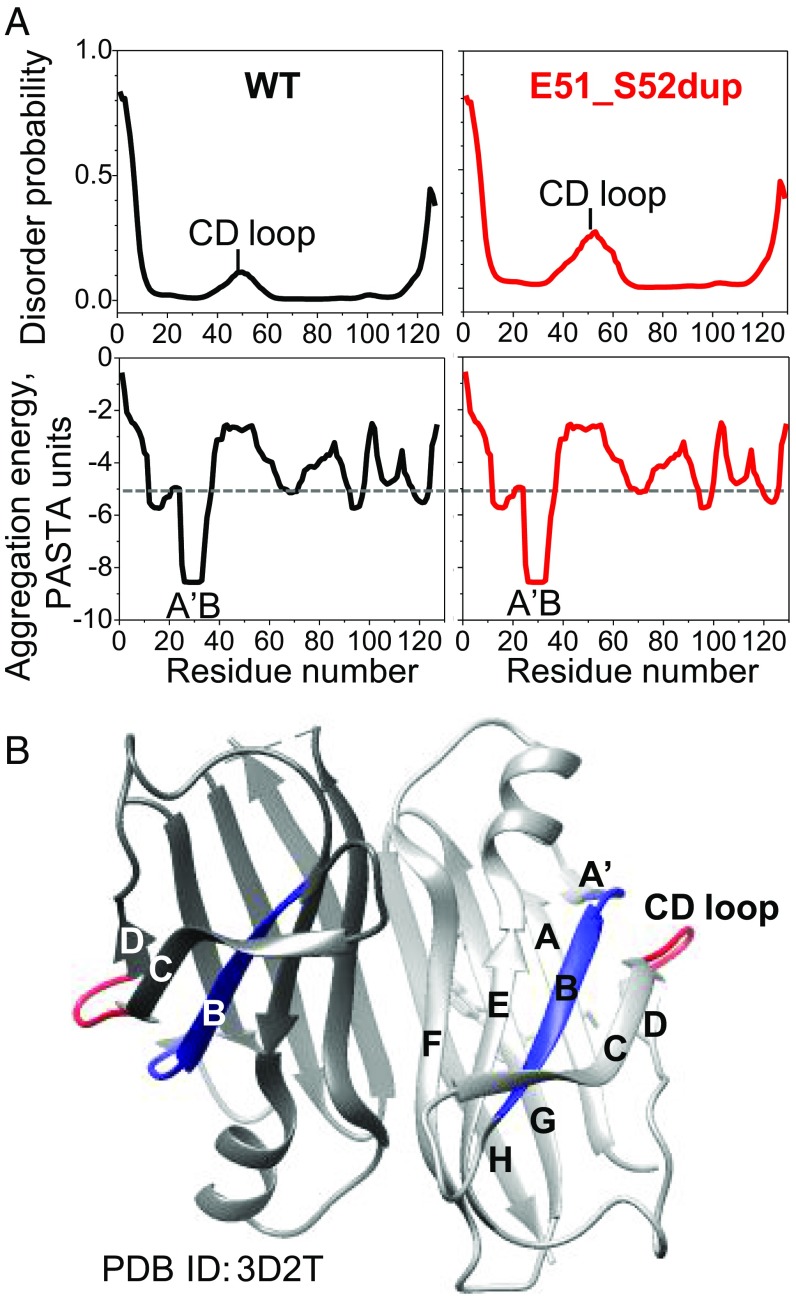
To probe how the Glu51_Ser52dup mutation affects protein dynamics and aggregation propensity, we performed amino acid sequence analysis of TTR using the bioinformatics server PASTA 2.0 (39), and mapped the results on the crystal structure of WT with bound diflunisal (Fig. 8). The duplication site is located in the CD loop (WT residues 49–53, TSESG) linking β-strands C and D (Figs. 6A and 8A). In the WT protein, this loop was predicted to be relatively disordered (Fig. 8B, Top). This prediction was substantiated by previous hydrogen-deuterium exchange NMR studies of WT showing low structural protection in this surface loop compared with other regions of the TTR molecule (40–42) (Fig. 8A, red). The Glu51_Ser52dup mutation was predicted to further increase the disorder in residue segment 40–60 containing the CD loop (Fig. 8B, Top). This prediction was substantiated by a rapid release of the 49–129 fragment upon tryptic digestion of the duplication mutant (Fig. 7 A and B). Hence, both experimental and bioinformatics studies showed that the duplication mutation increases local structural disorder in the CD loop. This result is likely relevant to the disease since TTR cleavage at the 48–49 junction generates the aggregation-prone C-terminal fragments found in ATTRm deposits (7).
Fig. 8.
Effects of Glu51_Ser52dup mutation on the local protein structure predicted by bioinformatics methods. (A) Amino acid sequence analysis of WT and mutant TTR using the PASTA 2.0 server. (Top) In WT, increased disorder is predicted in residues 40–60 containing the CD loop, and is confirmed by hydrogen-deuterium exchange NMR (40, 42). The Glu51_Ser52dup mutation in the CD loop is predicted to further increase the disorder in this region. (Bottom) However, the mutation does not affect the amyloid-forming sequence propensity of TTR (shown as aggregation-free energy). Residues 25–34 from the A′B region are predicted to have the strongest amyloid-forming propensity in both WT and mutant TTR, with favorable (negative) aggregation-free energy exceeding the threshold of −5 PASTA units (dotted line; 1 unit = 1.92 kcal/mol). (B) X-ray crystal structure of WT TTR with bound diflunisal shows packing of the amyloidogenic segment (residues 25–34, in blue) from the A′B loop/strand, which is buried in the native structure, against the CD surface loop (residues 49–53, in red). Hence, the CD loop screens the amyloidogenic segment 25–34 from the solvent. Two dimer-forming molecules in ribbon representation are shown in different shades of gray. PDB, Protein Data Bank.
The duplication did not alter the protein’s amyloid-forming sequence propensity, as evident from the aggregation-free energy plots (Fig. 8B, Bottom). These plots showed that residue segment 25–34 (AINVAVHVFR) from the A′B region has the strongest amyloidogenic sequence propensity, with a favorable (negative) free energy of aggregation approaching −9 kcal/mol. Other prediction methods (e.g., the consensus software AmylPred2 or zipperDB) also identified these residues as amyloidogenic, which was confirmed experimentally in synthetic peptide studies (43). Hence, bioinformatics and experimental studies consistently showed that residue segment 25–34 is strongly amyloidogenic.
In globular proteins, adhesive segments are normally protected from aggregation by their native packing. One example is WT TTR segment 25–34, which forms a part of the A′B loop and the B strand, shows high structural protection by hydrogen-deuterium exchange NMR (40, 42), and is well-ordered in the crystal structure (Fig. 8A). The protection of the loop residues 25–28 (AINV) is conferred mainly via tertiary interactions with adjacent segments such as the C-strand and CD loop, including the formation of two main chain hydrogen bonds (H-bonds): Asn27 to Ser50 and Ala29 to Lys48. The extension of the CD loop in the Glu51_Ser52dup variant must perturb these native interactions.
We propose that increased local disorder caused by the duplication mutation in the CD loop (Fig. 8B) decreases the structural protection of the nearby adhesive segment A′B (Fig. 8A), thereby promoting the aggregation and misfolding of the full-length Glu51_Ser52dup mutant. Moreover, such an increased disorder promotes cleavage with a release of the aggregation-prone C-terminal residue fragment 49–129 (Fig. 7).
Discussion
Molecular Origins of Unusual Clinical Features of Glu51_Ser52dup ATTRm.
This study reports ATTRm linked to a unique duplication mutation with an unusually severe phenotype. Diflunisal, a proven TTR tetramer stabilizer, failed to slow neurological disease progression. The duplication mutation decreased the apparent melting temperature of TTR by about 10 °C (Fig. 2C) and destabilized the protein structure even more than Leu55Pro, a particularly destabilizing and clinically aggressive ATTRm variant (22). Since the secondary and tertiary structures of Glu51_Ser52dup (Fig. 2 A and B) and other ATTRm mutants are very similar to those of the WT (37), the deleterious effects of the duplication mutation probably stem from the altered quaternary structure and/or protein dynamics. Several factors could potentially contribute to this aggressive drug-resistant form of ATTRm: (i) decreased thermodynamic stability of the variant TTR monomer, which promotes protein misfolding; (ii) decreased kinetic stability of the variant tetramer, which induces the release of aggregation-prone monomers; (iii) decreased affinity of the variant tetramer for diflunisal; and (iv) increased local disorder in the variant protein at sites that promote amyloid formation but are not affected by diflunisal binding. Our biophysical studies of Glu51_Ser52dup, Leu55Pro, and WT TTR proteins, together with bioinformatics and structural studies, revealed that factors i, ii, and iv pertained to the duplication mutant.
Structural Basis for TTR Destabilization by the Duplication Mutation.
In the TTR crystal structures, such as the WT with bound diflunisal (35) (Fig. 6A; Protein Data Bank ID code 3D2T, 1.85 Å resolution), the CD loop is well-ordered (main chain B-factors ∼ 35 Å2) and forms a tight β-turn at Ser52 stabilized by a 2.8 Å main chain H-bond between O of Ser50 and N of Gly53. The polypeptide chain is bent at ∼120° at Gly53, which helps to position the short strand D (Gly54-Leu55) antiparallel to strand A (residues 11–18) to form main chain H-bonds linking Val14 to Leu55 and Lys15 to Asp54. The importance of this H-bonded network is suggested by very similar H-bonding observed in various TTR crystal structures and by the high degree of conservation of the A and D strands in vertebrates (44).
This intramolecular H-bonding is perturbed by mutations in the CD loop. For example, an aggressive amyloidogenic mutation, Ser52Pro, perturbs the side-chain H-bonding in this region, reducing the protein thermodynamic stability by 2.3 kcal/mol (37). Notably, this conservative mutation places a proline in a preferred position of the β-turn without perturbing the main chain interactions. In contrast, Glu51_Ser52dup, which extends the CD loop, is expected to perturb both the side- and main-chain H-bonding, which helps explain the greatly reduced thermodynamic stability of the Glu51_Ser52dup monomer detected by equilibrium unfolding (Fig. 2 E and F).
Furthermore, in the WT tetramer, the β-turn at Ser52 is only several angstroms away from the C terminus of another subunit located across the weak dimer–dimer interface (Fig. 6A). In the duplication mutant, the extension of the CD loop toward this interface may interfere with the tetramer packing, perhaps by generating an uncompensated charge and/or creating steric clashes with adjacent subunits, and thereby favoring the release of the TTR dimers and monomers. This idea helps explain why the equilibrium population in Glu51_Ser52dup was shifted from the tetramer to the monomer and dimer (Figs. 3A and 4A) and the kinetic stability of the Glu51_Ser52dup homotetramer was greatly decreased (Fig. 2D). Moreover, the duplication mutation destabilized WT/mutant heterotetramers, wherein the presence of just one mutant subunit was sufficient for tetramer destabilization (Fig. 3). This observation is particularly relevant, as mixed heterotetramers are hallmarks of heterozygous ATTRm mutants in vivo (28, 29). In summary, the duplication mutation probably perturbs intramolecular H-bonding in the TTR monomer and intermolecular packing in the tetramer, leading to decreased structural stability of the monomer and tetramer, which is proamyloidogenic.
The Duplication Mutation Does Not Interfere with Diflunisal Binding.
Although diflunisal treatment was inefficient in vivo, the duplication mutant did bind diflunisal in vitro at physiologically relevant drug concentrations and drug/protein ratios. As expected, this binding stabilized the tetramer, although at 360 μM diflunisal, which mimics the maximal therapeutic serum levels (32, 33), it did not restore the variant protein stability to the levels seen in the WT (Fig. 4). Interestingly, free energy and enthalpy of diflunisal binding to the WT and Glu51_Ser52dup were similar within the error of their determination by ITC (Fig. 6B). This implies that the drug–protein interactions at the weak dimer–dimer interface were not significantly altered by the duplication mutation in the surface loop; conversely, diflunisal binding did not significantly influence local conformation near the mutation site (discussed in the next section). Therefore, the drug-resistant nature of the duplication mutation must result from factors other than the decreased TTR affinity for diflunisal. These factors could include destabilized structure of the variant tetramer and monomer, along with local disordering in sensitive regions, particularly those close to the mutation site.
Proamyloidogenic Cleavage in the CD Loop Is Insensitive to Diflunisal.
Besides structural destabilization of the Glu51_Ser52dup variant upon thermal or chemical stress (Fig. 2), reduced protein stability was also evident from rapid tryptic digestion at ambient conditions (Fig. 7). Of all ATTRm variants explored to date, only Ser52Pro underwent tryptic digestion at similar ambient conditions (37), while other variants became susceptible only upon additional perturbations, such as sheer stress (45, 46). Notably, both Glu51_Ser52dup and Ser52Pro mutation sites are located in the flexible CD loop that undergoes cleavage in vivo. In the duplication mutant, increased disorder in this loop was predicted by bioinformatics tools (Fig. 8A, Top) and was confirmed by limited proteolysis, with release of the amyloidogenic C-terminal residue fragment 49–129 and rapid formation of amyloid (Fig. 7). A similar fragment is the principal constituent of type A human amyloid deposits formed by WT or variant proteins (7, 47). We posit that enhanced cleavage at Lys48-Thre49 of the Glu51_Ser52dup variant with release of the amyloidogenic C-terminal fragment contributes to amyloid deposition in vivo. This process is representative of WT and other ATTRm variants (45, 46), but is particularly pronounced in the duplication variant.
Importantly, diflunisal did not inhibit this pathogenic cleavage in the Glu51_Ser52dup variant even at 360 μM diflunisal and a 25:1 molar excess of the drug (Fig. 7 C and D), which mimics the maximal therapeutic serum levels of diflunisal administered twice daily at 250 mg but exceeds its bioavailable levels. At such a drug concentration, both binding sites in the TTR tetramer are largely occupied. This is evident from the previous in vitro studies of other ATTRm variants using similar drug concentrations and 0.5–4.0 (mol/mol) diflunisal/TTR (45), as well as from the clinical studies of diflunisal occupancy in the TTR tetramer in vivo (33). Consequently, increasing the dose of diflunisal is not expected to stabilize the tetramer and block its pathological cleavage in vivo.
Similar to Glu51_Ser52dup, diflunisal did not inhibit the proamyloidogenic cleavage of Ser52Pro variant (37). These observations help explain the drug-resistant nature of the duplication mutation and the limited efficacy of diflunisal for ATTRm therapy in general. Other small molecules targeting the thyroxine-binding site, including bivalent ligands that bind both sites simultaneously, showed limited protection for other ATTRm variants against cleavage at Lys48 under mechanical stress in vitro (45, 46). However, these small molecules did not protect Ser52Pro variant against this pathogenic cleavage (45), and hence are not expected to protect Glu51_Ser52dup variant, necessitating the search for additional drug targets.
Besides formation of amyloidogenic proteolytic fragments, increased local disorder in the CD loop can promote amyloid formation by decreasing the structural protection of the nearby adhesive segment A′B (Fig. 8), which favors the misfolding of the full-length protein. In fact, mass spectral analysis demonstrated the presence of full-length Gly51_Ser52dup in amyloid deposits from the proband (SI Appendix, Fig. S1).
In summary, our results suggest that the aggressive character of the duplication mutation may stem from the global reduction in stability of the variant TTR monomer and the tetramer (Figs. 2, 3, and 7A) and local disordering near the mutation site. This disordering may either promote the generation of the amyloidogenic C-terminal fragments (Fig. 7) or favor the misfolding of the full-length protein by decreasing the structural protection in the adhesive residue segment 25–34 (Fig. 8). Notably, these two processes are insensitive to diflunisal binding at a subunit interface, which helps explain why diflunisal was ineffective in treating Glu51_Ser52dup ATTRm. The observation that diflunisal binds with comparable affinity to the WT and the mutant proteins (Fig. 8), but that this binding only partially restored the TTR tetramer stability even at a high drug concentration when both sites were occupied (Fig. 4), suggests that increasing the drug dose would not necessarily benefit the mutation carriers. Rather, new therapeutic strategies should be developed to target the mutation and the cleavage sites, and not only to stabilize the tetramer.
Materials and Methods
The study was conducted according to the Declaration of Helsinki. Informed consent was obtained from the patient with the permission of the Boston Medical Center Institutional Review Board, which approves all experiments involving human subjects. Recombinant full-length human TTR, including WT, Glu51_Ser52dup, and Leu55Pro, was expressed in Escherichia coli and purified to 95%+ purity following published protocols (48). All other materials, which were commercially obtained as described in SI Appendix, SI Materials and Methods, were of the highest available purity. Methods of sample preparation and analysis using biophysical and biochemical techniques are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank all members of the Amyloidosis Center clinical team who took care of the patient. We thank Donald L. Gantz for help with electron microscopy and Dr. Shobini Jayaraman for useful advice and help. This study was supported by NIH Grants GM067260, AG031804, and HL007969; the Wildflower Foundation; the Gruss Foundation; the Stewart Amyloidosis Endowment Fund; the Young Family Amyloidosis Endowment Fund; and the Amyloidosis Research Fund at Boston University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The gene sequence of the TTR variant has been deposited in the Clinvar database (accession no. SCV000778367).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802977115/-/DCSupplemental.
References
- 1.Sipe JD, et al. Amyloid fibril proteins and amyloidosis: Chemical identification and clinical classification International Society of Amyloidosis 2016 nomenclature guidelines. Amyloid. 2016;23:209–213. doi: 10.1080/13506129.2016.1257986. [DOI] [PubMed] [Google Scholar]
- 2.Branch WT, Robbins J, Edelhoch H. Thyroxine-binding prealbumin. Conformation in urea and guanidine. Arch Biochem Biophys. 1972;152:144–151. doi: 10.1016/0003-9861(72)90202-0. [DOI] [PubMed] [Google Scholar]
- 3.Monaco HL, Rizzi M, Coda A. Structure of a complex of two plasma proteins: Transthyretin and retinol-binding protein. Science. 1995;268:1039–1041. doi: 10.1126/science.7754382. [DOI] [PubMed] [Google Scholar]
- 4.Ritchie R, editor. Serum Proteins in Clinical Medicine. Vol 1. Foundation for Blood Research; Scarborough, ME: 1996. pp. 9.01–9.06. [Google Scholar]
- 5.Gustavsson A, et al. Amyloid fibril composition and transthyretin gene structure in senile systemic amyloidosis. Lab Invest. 1995;73:703–708. [PubMed] [Google Scholar]
- 6.Kingsbury JS, et al. Detailed structural analysis of amyloidogenic wild-type transthyretin using a novel purification strategy and mass spectrometry. Anal Chem. 2007;79:1990–1998. doi: 10.1021/ac061546s. [DOI] [PubMed] [Google Scholar]
- 7.Ihse E, et al. Amyloid fibrils containing fragmented ATTR may be the standard fibril composition in ATTR amyloidosis. Amyloid. 2013;20:142–150. doi: 10.3109/13506129.2013.797890. [DOI] [PubMed] [Google Scholar]
- 8.Rowczenio DM, et al. Online registry for mutations in hereditary amyloidosis including nomenclature recommendations. Hum Mutat. 2014;35:E2403–E2412. doi: 10.1002/humu.22619. [DOI] [PubMed] [Google Scholar]
- 9.Uemichi T, Liepnieks JJ, Benson MD. A trinucleotide deletion in the transthyretin gene (delta V 122) in a kindred with familial amyloidotic polyneuropathy. Neurology. 1997;48:1667–1670. doi: 10.1212/wnl.48.6.1667. [DOI] [PubMed] [Google Scholar]
- 10.Lashuel HA, Lai Z, Kelly JW. Characterization of the transthyretin acid denaturation pathways by analytical ultracentrifugation: Implications for wild-type, V30M, and L55P amyloid fibril formation. Biochemistry. 1998;37:17851–17864. doi: 10.1021/bi981876+. [DOI] [PubMed] [Google Scholar]
- 11.Quintas A, Saraiva MJM, Brito RM. The tetrameric protein transthyretin dissociates to a non-native monomer in solution. A novel model for amyloidogenesis. J Biol Chem. 1999;274:32943–32949. doi: 10.1074/jbc.274.46.32943. [DOI] [PubMed] [Google Scholar]
- 12.Jiang X, Buxbaum JN, Kelly JW. The V122I cardiomyopathy variant of transthyretin increases the velocity of rate-limiting tetramer dissociation, resulting in accelerated amyloidosis. Proc Natl Acad Sci USA. 2001;98:14943–14948. doi: 10.1073/pnas.261419998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolstoe SE, et al. Trapping of palindromic ligands within native transthyretin prevents amyloid formation. Proc Natl Acad Sci USA. 2010;107:20483–20488. doi: 10.1073/pnas.1008255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sant’Anna R, et al. Repositioning tolcapone as a potent inhibitor of transthyretin amyloidogenesis and associated cellular toxicity. Nat Commun. 2016;7:10787–10799. doi: 10.1038/ncomms10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulawa CE, et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc Natl Acad Sci USA. 2012;109:9629–9634. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coelho T, et al. Long-term effects of tafamidis for the treatment of transthyretin familial amyloid polyneuropathy. J Neurol. 2013;260:2802–2814. doi: 10.1007/s00415-013-7051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Planté-Bordeneuve V, et al. Long-term treatment of transthyretin familial amyloid polyneuropathy with tafamidis: A clinical and neurophysiological study. J Neurol. 2017;264:268–276. doi: 10.1007/s00415-016-8337-3. [DOI] [PubMed] [Google Scholar]
- 18.Berk JL, et al. Diflunisal Trial Consortium Repurposing diflunisal for familial amyloid polyneuropathy: A randomized clinical trial. JAMA. 2013;310:2658–2667. doi: 10.1001/jama.2013.283815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buxbaum J, Anan I, Suhr O. Serum transthyretin levels in Swedish TTR V30M carriers. Amyloid. 2010;17:83–85. doi: 10.3109/13506129.2010.483118. [DOI] [PubMed] [Google Scholar]
- 20.Skinner M, et al. Lowered prealbumin levels in patients with familial amyloid polyneuropathy (FAP) and their non-affected but at risk relatives. Am J Med Sci. 1985;289:17–21. doi: 10.1097/00000441-198501000-00003. [DOI] [PubMed] [Google Scholar]
- 21.Connors LH, Gertz MA, Skinner M, Cohen AS. Nephelometric measurement of human serum prealbumin and correlation with acute-phase proteins CRP and SAA: Results in familial amyloid polyneuropathy. J Lab Clin Med. 1984;104:538–545. [PubMed] [Google Scholar]
- 22.Jacobson DR, McFarlin DE, Kane I, Buxbaum JN. Transthyretin Pro55, a variant associated with early-onset, aggressive, diffuse amyloidosis with cardiac and neurologic involvement. Hum Genet. 1992;89:353–356. doi: 10.1007/BF00220559. [DOI] [PubMed] [Google Scholar]
- 23.Lai Z, Colón W, Kelly JW. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry. 1996;35:6470–6482. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 24.Jiang X, et al. An engineered transthyretin monomer that is nonamyloidogenic, unless it is partially denatured. Biochemistry. 2001;40:11442–11452. doi: 10.1021/bi011194d. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez-Ruiz JM. Protein kinetic stability. Biophys Chem. 2010;148:1–15. doi: 10.1016/j.bpc.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Colon W, Kelly JW. Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 27.Hurshman Babbes AR, Powers ET, Kelly JW. Quantification of the thermodynamically linked quaternary and tertiary structural stabilities of transthyretin and its disease-associated variants: The relationship between stability and amyloidosis. Biochemistry. 2008;47:6969–6984. doi: 10.1021/bi800636q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dwulet FE, Benson MD. Primary structure of an amyloid prealbumin and its plasma precursor in a heredofamilial polyneuropathy of Swedish origin. Proc Natl Acad Sci USA. 1984;81:694–698. doi: 10.1073/pnas.81.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekijima Y, et al. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121:73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 30.Robinson LZ, Reixach N. Quantification of quaternary structure stability in aggregation-prone proteins under physiological conditions: The transthyretin case. Biochemistry. 2014;53:6496–6510. doi: 10.1021/bi500739q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider F, Hammarström P, Kelly JW. Transthyretin slowly exchanges subunits under physiological conditions: A convenient chromatographic method to study subunit exchange in oligomeric proteins. Protein Sci. 2001;10:1606–1613. doi: 10.1110/ps.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller SR, Sekijima Y, Kelly JW. Native state stabilization by NSAIDs inhibits transthyretin amyloidogenesis from the most common familial disease variants. Lab Invest. 2004;84:545–552. doi: 10.1038/labinvest.3700059. [DOI] [PubMed] [Google Scholar]
- 33.Sekijima Y, Dendle MA, Kelly JW. Orally administered diflunisal stabilizes transthyretin against dissociation required for amyloidogenesis. Amyloid. 2006;13:236–249. doi: 10.1080/13506120600960882. [DOI] [PubMed] [Google Scholar]
- 34.Hammarström P, Jiang X, Deechongkit S, Kelly JW. Anion shielding of electrostatic repulsions in transthyretin modulates stability and amyloidosis: Insight into the chaotrope unfolding dichotomy. Biochemistry. 2001;40:11453–11459. doi: 10.1021/bi010673+. [DOI] [PubMed] [Google Scholar]
- 35.Adamski-Werner SL, Palaninathan SK, Sacchettini JC, Kelly JW. Diflunisal analogues stabilize the native state of transthyretin. Potent inhibition of amyloidogenesis. J Med Chem. 2004;47:355–374. doi: 10.1021/jm030347n. [DOI] [PubMed] [Google Scholar]
- 36.Bergström J, et al. Amyloid deposits in transthyretin-derived amyloidosis: Cleaved transthyretin is associated with distinct amyloid morphology. J Pathol. 2005;206:224–232. doi: 10.1002/path.1759. [DOI] [PubMed] [Google Scholar]
- 37.Mangione PP, et al. Proteolytic cleavage of Ser52Pro variant transthyretin triggers its amyloid fibrillogenesis. Proc Natl Acad Sci USA. 2014;111:1539–1544. doi: 10.1073/pnas.1317488111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naiki H, Higuchi K, Hosokawa M, Takeda T. Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal Biochem. 1989;177:244–249. doi: 10.1016/0003-2697(89)90046-8. [DOI] [PubMed] [Google Scholar]
- 39.Walsh I, Seno F, Tosatto SCE, Trovato A. PASTA 2.0: An improved server for protein aggregation prediction. Nucleic Acids Res. 2014;42:W301–W307. doi: 10.1093/nar/gku399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu K, et al. Deuterium-proton exchange on the native wild-type transthyretin tetramer identifies the stable core of the individual subunits and indicates mobility at the subunit interface. J Mol Biol. 2000;303:555–565. doi: 10.1006/jmbi.2000.4164. [DOI] [PubMed] [Google Scholar]
- 41.Liu K, Kelly JW, Wemmer DE. Native state hydrogen exchange study of suppressor and pathogenic variants of transthyretin. J Mol Biol. 2002;320:821–832. doi: 10.1016/s0022-2836(02)00471-0. [DOI] [PubMed] [Google Scholar]
- 42.Olofsson A, Ippel JH, Wijmenga SS, Lundgren E, Öhman A. Probing solvent accessibility of transthyretin amyloid by solution NMR spectroscopy. J Biol Chem. 2004;279:5699–5707. doi: 10.1074/jbc.M310605200. [DOI] [PubMed] [Google Scholar]
- 43.Saelices L, et al. Uncovering the mechanism of aggregation of human transthyretin. J Biol Chem. 2015;290:28932–28943. doi: 10.1074/jbc.M115.659912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manzon RG, Neuls TM, Manzon LA. Molecular cloning, tissue distribution, and developmental expression of lamprey transthyretins. Gen Comp Endocrinol. 2007;151:55–65. doi: 10.1016/j.ygcen.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 45.Marcoux J, et al. A novel mechano-enzymatic cleavage mechanism underlies transthyretin amyloidogenesis. EMBO Mol Med. 2015;7:1337–1349. doi: 10.15252/emmm.201505357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verona G, et al. Inhibition of the mechano-enzymatic amyloidogenesis of transthyretin: Role of ligand affinity, binding cooperativity and occupancy of the inner channel. Sci Rep. 2017;7:182. doi: 10.1038/s41598-017-00338-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suhr OB, Lundgren E, Westermark P. One mutation, two distinct disease variants: Unravelling the impact of transthyretin amyloid fibril composition. J Intern Med. 2017;281:337–347. doi: 10.1111/joim.12585. [DOI] [PubMed] [Google Scholar]
- 48.Kingsbury JS, Klimtchuk ES, Théberge R, Costello CE, Connors LH. Expression, purification, and in vitro cysteine-10 modification of native sequence recombinant human transthyretin. Protein Expr Purif. 2007;53:370–377. doi: 10.1016/j.pep.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.