Significance
Copy-number variants are ubiquitous in nature, yet their immediate functional consequences are obscure. We conducted a spontaneous mutation accumulation experiment at varying sizes in Caenorhabditis elegans, thereby enabling the simultaneous investigation of the mutational input and strength of selection on the evolution of copy-number changes. Whole-genome sequencing reveals the highest genome-wide rate of gene duplication for any species thus far. Our transcriptome analysis further demonstrates that gene duplication frequently results in a greater than two-fold change in transcription. Despite the adaptive role of duplication as the primary source of novel genes, we find duplications and deletions of highly transcribed genes to be more detrimental to fitness and evidence for selection against increase in transcript abundance.
Keywords: gene duplication, experimental evolution, transcription, selection, mutation accumulation
Abstract
Gene duplication and deletion are pivotal processes shaping the structural and functional repertoire of genomes, with implications for disease, adaptation, and evolution. We employed a mutation accumulation (MA) framework partnered with high-throughput genomics to assess the molecular and transcriptional characteristics of newly arisen gene copy-number variants (CNVs) in Caenorhabditis elegans populations subjected to varying intensity of selection. Here, we report a direct spontaneous genome-wide rate of gene duplication of 2.9 × 10−5/gene per generation in C. elegans, the highest for any species to date. The rate of gene deletion is sixfold lower (5 × 10−6/gene per generation). Deletions of highly expressed genes are particularly deleterious, given their paucity in even the N = 1 lines with minimal efficacy of selection. The increase in average transcript abundance of new duplicates arising under minimal selection is significantly greater than twofold compared with single copies of the same gene, suggesting that genes in segmental duplications are frequently overactive at inception. The average increase in transcriptional activity of gene duplicates is greater in the N = 1 MA lines than in MA lines with larger population bottlenecks. There is an inverse relationship between the ancestral transcription levels of new gene duplicates and population size, with duplicate copies of highly expressed genes less likely to accumulate in larger populations. Our results demonstrate a fitness cost of increased transcription following duplication, which results in purifying selection against new gene duplicates. However, on average, duplications also provide a significant increase in gene expression that can facilitate adaptation to novel environmental challenges.
The fundamental role of gene acquisition and loss in the evolution of biodiversity has long been recognized (1), although other mutational classes (base substitutions and small indels) have been presumed to be the major contributors to genetic variation. The advent of genomics afforded the first glimpse into the widespread copy-number polymorphism existing within and among populations and their ubiquity across all domains of life (2). Gene acquisition and loss, both fundamental processes in genome evolution, embark on their evolutionary trajectories as copy-number variants (CNVs) in populations. CNVs have additional important consequences for evolutionary processes including genetic load and speciation, as well as implications for human disease given that somatic CNVs contribute to cancer origin and progression. Our current understanding of the evolutionary dynamics of CNVs largely comes from studies of their distribution and frequencies in natural populations and comparative genomics of extant organisms (3–6). However, while these observations provide insights into the long-term retention of CNVs, less is known about their initial establishment and fixation process within populations. Duplications and deletions in the wild have already been through the sieve of natural selection and therefore provide an incomplete understanding of the early evolutionary dynamics of CNVs where most of selection takes place. Furthermore, CNVs identified in natural populations are of variable or uncertain age and may have undergone additional modifications of their expression patterns and biological functions that no longer reflect their original consequences at conception. Lastly, the immediate phenotypic, average fitness, and transcriptional consequences of this class of mutations upon conception remain largely obscure.
The fate of mutations in populations depends on the rate at which they arise and the combined action of the evolutionary forces of genetic drift and natural selection (7–10). In general, population dynamics of new mutations are dominated by drift when the effective population size, Ne, is less than 1/s, where s is the fitness effect. In very small populations, the fate of most mutations is determined by drift rather than selection. In larger populations, purifying selection becomes more effective in purging deleterious mutations. In mutation accumulation (MA) experiments, multiple replicate lines descended from an ancestral inbred genotype are subjected to consecutive bottlenecks at a minimum population size that greatly impairs the efficacy of selection, facilitating the accumulation and study of the majority of newly originating mutations that would have been typically purged in natural populations (11).
We employed a spontaneous MA experiment design comprising three population size treatments in Caenorhabditis elegans (12–14). MA lines, all descended from a single N2 hermaphrodite ancestor, were bottlenecked each generation at N = 1, 10, or 100 hermaphrodites (SI Appendix, Fig. S1A) for >400 generations, allowing us to jointly assess the molecular and transcriptional consequences of segmental duplications and deletions under conditions of neutrality and with increasing intensity of selection. The predominantly self-fertilization mode of reproduction in hermaphroditic species such as C. elegans additionally results in a further reduction of Ne relative to the census population size (N) (9, 15). Hence, the genetic effective population sizes of our three population size treatments correspond to Ne = 1, 5, and 50 individuals wherein mutations with selection coefficients less than 1/2Ne are expected to accumulate at the neutral rate and contribute to mutational degradation, respectively (7, 16) (SI Appendix, Fig. S1B). These are approximations but the general relationship between population size and efficiency of selection still holds, namely that selection is more efficient in larger populations. A comparison of the accumulated mutations in experimental lines of different Ne subsequently facilitate inferences about the fitness consequences of different classes of mutations. Leveraging this experimental framework with high-throughput sequencing enabled us to identify de novo mutational and transcriptional variants within each line at a genome-wide scale since their divergence from a common ancestor. Another advantage of our experimental approach is the ability to identify the genomic and transcriptional properties of CNVs in their evolutionary infancy without the confounding effects of uncertain demography and evolutionary age.
Results
Extraordinarily High Rates of Spontaneous Duplication and Deletion.
We identified 161 simple and complex independent copy-number changes across 33 MA lines (SI Appendix, Tables S1–S4). The N = 1 populations should have a minimal influence of selection to provide the spontaneous mutation rate and the expected rate of neutral evolution. We detected 48 independent duplication events in the N = 1 populations, yielding a spontaneous rate of 6.5 × 10−3 duplication events/genome per generation. A substantial proportion of these duplications (∼31%) occurred within preexisting CNVs in the genome of the immediate ancestor of the MA lines. If we only consider duplications of single-copy sequences, this rate is 4.7 × 10−3 duplication events/genome per generation. These duplication tracts completely or partially duplicated 3,316 loci, yielding a direct, genome-wide spontaneous gene duplication rate of 2.88 × 10−5/protein-coding gene per generation (Table 1). The deletion rate is 5.0 × 10−6/protein-coding gene per generation (Table 1). However, 85% of the deletions are in fact the loss of duplicate copies in preexisting CNVs, and the spontaneous rate of loss of single-copy genes is only 7.3 × 10−7/protein-coding gene per generation.
Table 1.
Summary of gene duplication and deletion under three population size treatments
| Population size | Duplications* | Genes duplicated† | μduplication‡ | Deletions* | Genes deleted§ | μdeletion¶ |
| N = 1 | 48 | 3,316 | 2.88 × 10−5 | 44 | 588 | 0.50 × 10−5 |
| N = 10 | 30 | 97 | 0.10 × 10−5 | 15 | 27 | 0.03 × 10−5 |
| N = 100 | 16 | 42 | 0.08 × 10−5 | 15 | 45 | 0.06 × 10−5 |
Mutation rate estimates for the N = 1 populations represents the spontaneous rate of origin of gene duplications and deletions with minimal influence of selection. Rate estimates for the N = 10 and 100 populations denote copy-number changes with increasing intensity of natural selection.
Number of events.
Total number of genes duplicated.
Rate of duplication (/gene per generation).
Total number genes deleted.
Rate of deletion (/gene per generation).
One MA line in particular, 1T, contributed 93.6% and 75% of all duplications and deletions, respectively, to the pool of copy-number changes detected in the N = 1 lines. The genome of 1T contained six duplications and seven deletions across four chromosomes. Two extremely large duplications on chromosomes I and V by themselves contribute to 91.1% of all of the duplicated genes in the N = 1 lines (SI Appendix, Fig. S2). Some of the copy-number changes in 1T were duplication-inversion and duplication-deletion combinations reminiscent of complex genomic rearrangements (CGR) or chromothripsis detected in cancer cells (17, 18) and in mutagenized C. elegans (19). This MA line went extinct after 309 generations, one of three to do so over the course of the experiment. If we exclude 1T from the calculations of the spontaneous duplication and deletion rates in the N = 1 MA lines, we obtain rates of 2.64 × 10−6 and 1.19 × 10−6/protein-coding gene per generation, respectively, which are an order of magnitude greater than our preceding estimates for C. elegans (20).
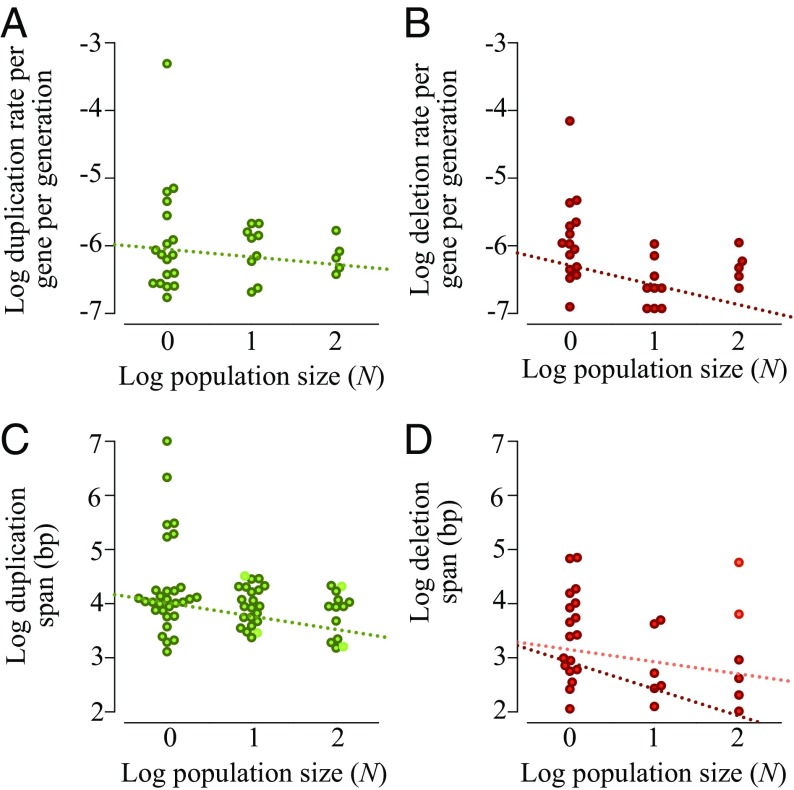
MA lines bottlenecked at population sizes of N = 10 and 100 individuals per generation enable insight into the dual influence of mutation and selection on standing genetic variation created by copy-number changes. We identified 97 and 42 complete or partial gene duplicates in the N = 10 and 100 lines, respectively, yielding accumulation rates of 1.03 × 10−6 and 8.02 × 10−7 duplications/gene per generation (Table 1). While the larger population sizes in our experiment have, on average, lower duplication accumulation rates than the N = 1 lines, the relationship between population size and duplication rate is not significant (Fig. 1A). In contrast, the deletion rates are significantly correlated with population size, providing evidence of their strongly deleterious effects on fitness and a potent role of selection in their eradication even at small population sizes (Fig. 1B).
Fig. 1.
Linear regression showing the rate of accumulation and span of copy-number changes as a function of population size. (A) No significant effect of population size on the accumulation of gene duplications (Kendall’s τ = −0.02, P = 0.87; Kruskal–Wallis H = 0.11, P = 0.95). (B) Significant negative association between population size and the accumulation of gene deletions (Kendall’s τ = −0.35, P = 0.02; Kruskal–Wallis H = 9.95, P = 0.01). (C) Negative correlation between duplication span and population size. The Pearson correlation for all unique duplications (including polymorphic) is −0.28 (P = 0.02; permutation 95% CI: −0.23, 0.25, P = 0.007). (D) No significant correlation between deletion spans of single-copy DNA and population size (r = −0.21, P = 0.25; permutation CI: −0.35, 0.36, P = 0.12, hatched orange line). There is a significant correlation between the span of fixed deletions and population size [r = −0.46, P = 0.01; permutation CI: (−0.37, 0.38), P = 0.005, hatched maroon line]. Fixed and polymorphic copy-number changes in C and D are indicated by darker and lighter filled circles, respectively.
Deleterious Fitness Consequences of Large Copy-Number Changes.
Under minimal selection in the N = 1 lines, the median duplication and deletion span was 10.6 kbp (range of 936 bp–10.1 Mbp) and 7.3 kbp (range of 114 bp–1.2 Mbp), respectively. The distribution of the duplication and deletion spans were not significantly different from previous estimates generated for a different set of C. elegans N = 1 MA lines using oligonucleotide array comparative genome hybridization (oaCGH) methods (Kruskal–Wallis H = 3.8, P = 0.05, and H = 0.94, P = 0.33, respectively) (20). In contrast, the spontaneous duplication and deletion spans in this study are significantly smaller than those observed in experimental C. elegans populations undergoing adaptation under strong selection (21) with a median duplication and deletion span of ∼191 kbp (Kruskal–Wallis H = 14.6, P = 0.00013) and ∼12.5 kbp (H = 4.7, P = 0.03), respectively. The difference in the duplication and deletion spans between MA and adapting populations may reflect selection on gene dosage in the adapting populations as larger duplication and deletion tracts have a greater likelihood of encapsulating genes under selection for altered gene dosage (20, 21).
How does purifying selection impinge on the size of copy-number changes? Larger duplication and deletion tracts spanning many loci may have a greater propensity for disturbing dosage balance and have the potential to impose a substantial fitness cost (2). However, smaller duplication tracts may fail to encapsulate the full repertoire of cis-regulatory elements and/or coding regions, thereby engendering some fitness cost (22) associated with superfluous expression of nonfunctional partial mRNAs. We compared the length distribution of duplication and deletion spans across the three population sizes to determine the role of selection, if any, in dictating the size of copy-number variants. Duplication span is negatively correlated with population size (Fig. 1C), primarily due to large (>100 kb) duplications that appear in the N = 1 lines but are absent in the larger populations. Deletion span is not significantly correlated with population size when all deletions are considered, but negatively correlated with population size when only fixed deletions (present in all individuals sampled from a population) are considered (Fig. 1D). The discrepancy in the results between fixed and polymorphic deletions is due to large deletions that were only found in single individuals in the larger populations sizes (N = 10 and 100). In fact, fixed deletions are significantly smaller than polymorphic deletions in these larger populations (Kruskal–Wallis H = 4.6, P = 0.03). These results provide evidence for significant deleterious fitness consequences of large copy-number changes and their eradication by selection even at small to moderate population sizes.
Greater than Twofold Increase in Transcription Following Gene Duplication Under Minimal Selection.
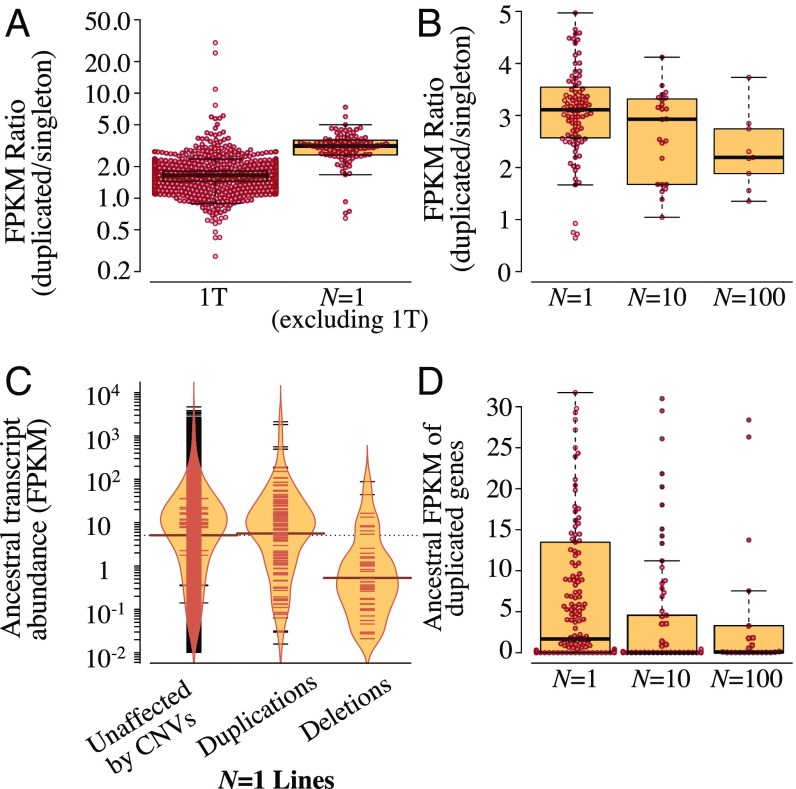
We conducted RNA-sequencing (RNA-sequencing) analysis of our MA lines to investigate the relative roles of divergent evolutionary forces such as drift and selection in shaping expression divergence following gene duplication. Surprisingly, there is a sizeable and significant difference in the change of mRNA abundance of duplicated genes between line 1T that bears several large duplications and copy-number changes relative to the remaining N = 1 lines (Fig. 2A). The average and median increase in transcript abundance of duplicated genes in line 1T relative to single copies of the same gene in other N = 1 MA lines is twofold and 1.7-fold, respectively. For the remaining N = 1 lines (n = 17), there was an average threefold (median 3.1-fold) increase in the transcript abundance of duplicated genes relative to other N = 1 MA lines bearing the same loci in single-copy form (Fig. 2A). The average increase in transcript abundance of gene duplicates significantly exceeds the twofold increase expected if transcriptional changes are additive and scale linearly with copy number (t = 10.64, P = 2.2 × 10−6).
Fig. 2.
Transcriptional consequences of duplications. (A) Lower increase in transcript abundance (FPKM) of duplicated genes in line 1T relative to all other N = 1 MA lines (t = 12.92, P < 2.2 × 10−16). (B) Lower increase in transcript abundance of duplicated genes in larger populations (N = 10 and 100 MA lines) relative to N = 1 MA lines (excluding 1T) (ANOVA, F = 9.4, P = 0.003; r = −0.27, P = 0.003). (C) The distribution of ancestral transcript abundance of duplicated genes in the N = 1 MA lines is not significantly different from that of all genes unaffected by copy-number changes (t = 0.73, P = 0.47). However, deleted genes in the N = 1 MA lines had significantly lower transcription in the ancestor relative to other genes in the genome (t = 13.32, P < 2.2 × 10−16). (D) The ancestral transcript abundance of newly duplicated genes differs significantly across MA lines of varying population size and is negatively correlated with population size (Kendall’s τ = −0.18, P = 0.0002; Kruskal–Wallis H = 16.98, P = 0.0002).
Lower Increase in Transcription of Gene Duplicates in Larger Populations.
MA lines maintained at larger population sizes (N = 10 and 100 individuals) engender a significantly smaller increase in transcript abundance due to duplicated genes relative to the N = 1 lines, presumably displaying the influence of greater intensity of purifying selection against duplications with a large transcriptional effect (Fig. 2B). Alternatively, there may be selection for chromatin-mediated gene silencing of duplicated loci at larger population sizes or selection for compensatory modifiers regulating duplicate gene expression.
Strong Selection Against the Duplication of Highly Expressed Genes in Larger Populations.
Does a gene’s transcriptional activity influence its propensity for retention following duplication or deletion? Transcriptome analysis of the ancestral control found no difference in the distribution of transcript abundance of genes that were subsequently duplicated in the N = 1 lines during the MA phase versus those that were not duplicated (Fig. 2C). Hence, the duplicated genes in the N = 1 lines constitute an unbiased sample of ancestral genes with respect to transcriptional level. In contrast, deleted genes in the N = 1 lines tend to have lower transcriptional levels in the ancestral control (Fig. 2C). Therefore, deletions of some highly transcribed genes are likely eradicated by selection even in the N = 1 lines with minimal efficacy of selection, suggesting their strongly detrimental average fitness effects. Additionally, there is a significant negative relationship between transcript abundance before duplication in the pre-MA ancestral control and population size (Fig. 2D). Duplications of genes with greater transcript abundance in the ancestral line are significantly less likely to accumulate in larger MA populations than in the N = 1 MA lines. Together these results demonstrate that the duplication of highly expressed genes is more deleterious and less tolerated at higher population sizes subject to greater intensity of selection. However, the results do not rule out that duplications of some highly transcribed genes were selected for in the larger population size treatments.
Structural and Genomic Features of Copy-Number Changes at Conception and Under Increasing Selection Intensity.
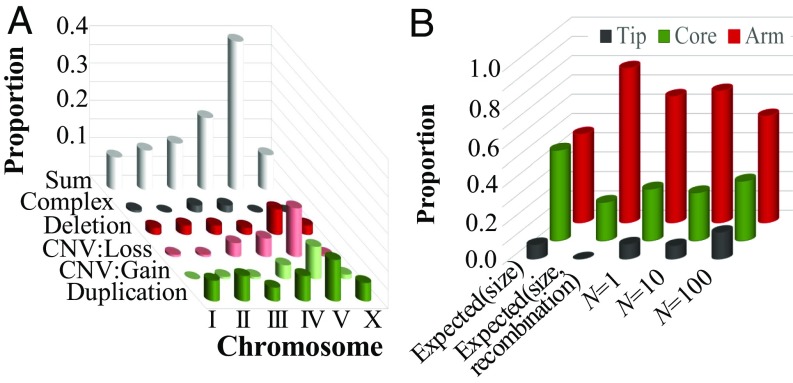
The distribution of copy-number changes is nonrandom across the C. elegans chromosomes (Fig. 3A and SI Appendix, Fig. S3). A disproportionate number of simple independent duplications and deletions occurred on chromosome V (SI Appendix, Table S5). Chromosome V, which comprises 21% of the genome, incurred 42% of simple duplications of single-copy genes, 52% of all deletion events, and 58% of all copy-number changes to preexisting duplications. The majority of copy-number gains or losses in preexisting duplications occur only once in any particular MA line. However, three preexisting duplications in chromosome V have unusually high numbers of copy-number gains or losses. These three CNVs, all located within 2 Mbp of one another on the far end of the right arm of chromosome V, are the sites of 12 independent copy gains and 13 copy losses across all of the MA lines. The significant elevation in spontaneous copy-number changes in chromosome V relative to the other chromosomes predicts that natural populations should also exhibit greater variation in gene copy number on chromosome V relative to other chromosomes. Indeed, the right arm of chromosome V is a region of deletion enrichment in natural isolates of C. elegans (4, 23, 24).
Fig. 3.
Genomic distribution of copy-number changes. CNV gains and losses refer to copy-number changes in preexisting duplications; duplications and deletions refer to copy-number changes of single-copy genes only. (A) The chromosomal distribution of spontaneous copy-number changes across all MA lines is significantly different from the expected distribution based on chromosomal length (G = 55.23, P = 5.01 × 10−7). Chromosome V harbors more spontaneous copy-number changes than expected by chance. (B) The expected and observed intrachromosomal distribution of spontaneous copy-number changes on chromosomal tips, cores, and arms in MA lines across different population sizes. The expected quantities are calculated from the proportion of the genome that is located in tips, cores, and arms. In addition, expected quantities were calculated from the product of the size and the recombination frequencies in different genomic regions. The observed distribution is significantly different from that expected based on size alone (G = 48.8; P = 2.55 × 10−11) and when recombination frequency is taken into account (G = 420.5, P < 2.2 × 10−16).
The breakpoints of copy-number changes are also nonrandomly distributed within chromosomes (Fig. 3B and SI Appendix, Table S6). Breakpoints are more frequent in the arms and less frequent in the cores than expected by chance. This difference between the arms and the cores may be partly explained by differences in recombination frequency observed within the C. elegans genome with low recombination rates in gene-rich cores and gene-depauperate tips in contrast to gene-poor arms with high recombination frequencies (25). However, the frequency of copy-number breakpoints in the tips is similar to expectations based on size alone and greater than expected when the recombination frequency is taken into account (Fig. 3B). Hence, the intrachromosomal distribution of spontaneous copy-number changes in our C. elegans study cannot be explained by the size of different chromosomal regions alone or by a combination of size and recombination frequency.
Individual gene duplication events in the N = 1 lines comprised one to 3,102 protein-coding genes, with a median of four protein-coding genes per duplication. We determined the degree of structural homology between the derived and ancestral paralogs to categorize them as complete, partial, or chimeric duplicates (Fig. 4A) using previously described methods (22, 26). The vast majority (98.6%) of gene duplicates arising in the N = 1 lines are complete, in that the derived copy contains all ancestral exonic, intronic, and untranslated regions in their entirety (Fig. 4B). The high fraction of complete duplicates is a function of duplication span because larger duplications are more likely to contain completely duplicated genes, and the largest duplications arose in the N = 1 lines. Copy-number breakpoints in internal exonic regions result in either partial gene duplicates, which capture only part of the ancestral ORF, or chimeric gene duplicates, which fuse coding sequences from different genes. Seventy protein-coding genes were intersected by such breakpoints across all duplications and complex rearrangements in the N = 1 lines, generating 18 chimeric (0.5%) and 27 partial (0.8%) gene duplicates, whereas the structure of six CNV events could not be delineated due to the lack of quality split reads. The relative proportions of complete, partial, and chimeric gene duplicates varied significantly with the intensity of selection in the experimental lines, with higher frequencies of structurally heterogeneous duplicates (partial and chimeric) observed at larger population sizes (N = 10 and 100 individuals) (Fig. 4B). The reduction in the fraction of complete duplicates likely stems from the eradication of very large segmental duplications (which harbor many complete duplicates) via purifying selection.
Fig. 4.
Degree of structural homology between gene paralogs and copy-number change in rRNA genes. (A) Duplication events can yield gene copies with varying degrees of structural resemblance to the ancestral locus depending on the span of duplication and the location of duplication breakpoints. G1 and G2 represent ancestral loci; G1′ and/or G2′ represent duplications of G1 and G2 genes, respectively. Complete gene duplicates were identified as those resulting from a duplication event that spanned, at a minimum, the entire ORF and untranslated regions of one ancestral locus (in this case, G1′ is a complete duplicate of G1). Partial gene duplicates originate when only a portion of the ORF of the ancestral locus is duplicated due to the presence of at least one duplication breakpoint falling within the ancestral ORF (G1′ is a partial duplicate of G1). Chimeric gene duplicates comprise duplicated segments of two protein-coding genes resulting in a single ORF. (B) The frequencies of three structural categories of gene duplicates vary significantly with population size (G = 155.2, P < 2.2 × 10−16). No significant difference is found between the distribution of N = 10 vs. N = 100 MA lines (G = 0.2, P = 0.9). (C) Greater than fourfold variation in rRNA gene copy number in the C. elegans MA lines. There is no relationship between population size and rDNA copy number (F = 1.57, P = 0.22). However, the average copy number of rRNA genes across the MA lines differed significantly from that of the ancestral control (t = 4.96, P = 2.23 × 10−5; red dashed line).
Increase in rDNA Copy Number.
Ribosomal RNA (rRNA) genes (18s, 28s, and 5.8s) in C. elegans occur in long tandem arrays at one terminal end of chromosome I (27), a region that exhibits wide copy-number variation within C. elegans wild isolates (24) and among different nematode species (28). This repeat region exhibited a threefold variation in ribosomal DNA (rDNA) copy number across our 33 MA lines (SI Appendix, Fig. S3). These changes in rDNA copy number based on read depth were strongly supported by oaCGH results (r = 0.9, P = 2 × 10−12). rDNA copy number per genome did not differ significantly as a function of population size (Fig. 4C). However, the average rDNA copy number of 128 in the MA lines is significantly greater than that of the ancestor (98 copies) (Fig. 4C). The results support the hypothesis of an intrinsic drive toward copy-number increase in rDNA genes in C. elegans, at least under standard laboratory conditions (28).
Discussion
The genome-wide, spontaneous gene duplication rate measured in our MA lines is the highest for any organism to date and two orders of magnitude greater than previously reported for C. elegans (20). Gene duplication rates from spontaneous MA experiments in Saccharomyces cerevisiae (29), Drosophila melanogaster (30), and Daphnia pulex (31) were estimated to be 3.4 × 10−6, 3.7 × 10−7, and 2.3 × 10−5/gene per generation, respectively, representing an approximately two orders of magnitude difference. A combination of technical differences and sample size contribute to the differences between the estimates from this study and our previous analysis of gene duplication and deletion rates in C. elegans (20). The preceding results were based on oaCGH arrays with unique probes only. In contrast, this study employed whole genome sequencing in combination with oaCGH arrays containing probes to unique and duplicated sequences, further enabling the detection of copy-number changes in preexisting duplications. Furthermore, the larger sample size in this study increases the probability of detecting rare large duplications. These astoundingly high rates of duplication and their inherent plasticity with respect to expansion and contraction suggest an important role for this form of genetic variation in adaptation to changing or fluctuating environments.
Several recent studies aiming to characterize the influence of segmental gene duplications in shaping gene expression patterns via a transcriptome approach have reached conflicting conclusions, arguing for (i) limited or no change in expression associated with duplication (6, 32–34) versus (ii) a significant increase in transcript abundance with increasing gene copy number (35, 36). Conclusions garnered from the study of gene expression patterns in natural populations or laboratory strains that are subject to intense natural (usually purifying) or artificial selection are not ideal for making inferences about the average transcriptional consequences of any class of mutation at origin, including gene duplications. Elucidating how gene copy-number changes are targeted by selection first requires an understanding of how nascent gene duplicates engender the evolution of gene expression, conditions that are fulfilled in MA experiments where the efficacy of selection is minimized. The transcriptional activity of duplicated loci in the N = 1 MA lines barring line 1T are congruent with those observed in a preceding study in D. melanogaster, which found greater than twofold expression of synthetically constructed tandem duplications of the Adh gene (35). Indeed, the average transcript abundance of a duplicated gene in the N = 1 MA lines was increased by threefold, suggesting that genes in segmental duplications are frequently overactive at inception. The exact mechanism(s) contributing to this transcriptional overexpression remain obscure and require further investigation but may be owing to positional effects of adjacent paralogs (35) or some form of dosage imbalance between duplicated genes and their regulatory elements.
Although gene duplications are frequently a source of adaptive genetic variation, some evidence suggests that they are, on average, deleterious (2, 5, 20, 30, 37–39). The detrimental effects of duplications for fitness may be owing to (i) the cost of superfluous gene expression, (ii) dosage imbalance between duplicated genes and other genes in the genome that remain in single copy, and (iii) inappropriate expression of gene duplicates that are under the control of a different regulatory system (40–43). Our results demonstrate that an increased transcript abundance of gene duplicates contributes to their fitness cost. The average increase in transcript abundance following the duplication of a gene is negatively associated with population size, a consequence of increased efficiency of selection in our larger MA lines. Large copy-number changes appear to have greater fitness costs and are eradicated by selection even at small population sizes (10–100 individuals). Moreover, highly transcribed single-copy genes in the ancestor were more likely to be observed in duplicated form in the N = 1 lines, relative to the larger MA populations (N = 10 and 100 lines). This observed difference is expected if duplications of highly expressed genes have a greater fitness cost and are selected against in larger populations. Therefore, the cost of overexpression of duplicated genes also appears to be related to their normal transcriptional activity. Our RNA-Seq analysis of long-term C. elegans MA lines maintained at differing population sizes under both minimal and increasing intensity of selection can reconcile the discrepancy in conclusions about the influence of gene duplicates on gene expression from preceding studies that have argued for (36, 37) and against (6, 32–34) a change in gene expression following gene duplication. The presence of an extra copy of a gene engenders a greater than twofold increase in gene expression in the absence of selection, as is observed in Drosophila (35) and the N = 1 MA lines in this study. If the presence of a duplicate gene copy is deleterious to fitness and perturbs the ancestral gene dosage level, populations with greater efficacy of selection can be expected to eradicate the extra gene copy or silence the ensuing higher expression such that subsequent expression levels are rendered closer to the ancestral optimum. This will manifest as an overall lack of change in gene expression following gene duplication, as is observed in natural populations with extreme intensity of selection (6, 32–34). The fact that the fitness cost is associated with increase in transcript abundance can explain why segmental duplications in natural populations often do not show transcriptional activity proportionate to their copy number.
Conclusion
Diverse models have been proposed for the maintenance and evolution of gene duplicates, and it is imperative that the creativity and proliferation in theory be matched with biological realism. Retrospective analysis of sequenced genomes has yielded a wealth of information about the importance of copy-number changes in evolution. A comprehensive understanding of the population dynamics and evolution of CNVs would be bolstered by information about their consequences for gene expression, phenotype, and fitness. Our experimental evolution study manipulating the strength of selection via differing population size bottlenecks establishes that copy-number changes upon conception can impose a significant fitness cost by perturbing ancestral transcriptional levels. However, gene duplications are the primary source of novel genes as well as provide the genetic fodder for adaptation to novel environmental regimes (44–47). Determining the context dependence of these genetic variants for organismal fitness hinges heavily on future investigations into their phenotypic, functional, and distribution of fitness effects using modern and emerging technologies of high-throughput genomics and genome editing.
Materials and Methods
C. elegans is a self-fertilizing nematode with several key characteristics that are particularly amenable to experimental evolution studies (SI Appendix, SI Experimental Procedures). The descendants of a single wild-type Bristol (N2) hermaphrodite were used to establish 35 MA lines that were subsequently maintained at three constant population sizes of N = 1, 10, and 100 individuals per generation (12, 13) (SI Appendix, Fig. S1). After the conclusion of the MA phase, the descendants of one, four, and five worm(s) from each N = 1, N = 10, and N = 100 MA line, respectively (SI Appendix, Fig. S4), were prepared for genome sequencing as previously described (13). Copy-number changes in the MA lines were additionally assessed by oaCGH using the ancestral control line as a common reference. Raw reads generated from WGS were aligned to the reference N2 Genome and putative variants were identified using five CNV/SV detection programs. Replicate RNA samples for sequencing were prepared for all 33 MA lines, as well as the pre-MA ancestral control. RNA-Seq was performed on an Illumina HiSeq 4000 platform and the relative transcript abundance for each protein-coding gene estimated. Gene duplications and deletions were annotated based on the N2 reference genome (version WS247). Direct estimates of the spontaneous rates of origin were estimated from the N = 1 lines under minimal selection. CNVs were analyzed with respect to their structural and genomic characteristics including the degree of structural homology, span, and chromosomal location. Differential transcription of duplicated and deleted genes was assessed by comparisons to ancestral transcript abundance profiles. Details of the design of the MA experiment, procedures for oaCGH, whole-genome sequencing and RNA-Seq, identification of CNVs, and analyses of transcript abundance can be found in the SI Appendix, SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank the editor and one anonymous referee for helpful suggestions in improving the manuscript, Lucille Packard for assistance in the creation of the MA lines, and Philip Green from the University of Washington for providing the program Phaster. This research was supported by National Science Foundation Grant MCB-1330245 (to V.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.L. is a guest editor invited by the Editorial Board.
Data deposition: oaCGH data for this study has been deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE112821). The genome sequence data have been submitted to the National Center for Biotechnology Information BioProject, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA448413 (accession no. SRP145329).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801930115/-/DCSupplemental.
References
- 1.Ohno S. Evolution by Gene Duplication. Springer; New York: 1970. [Google Scholar]
- 2.Katju V, Bergthorsson U. Copy-number changes in evolution: Rates, fitness effects and adaptive significance. Front Genet. 2013;4:273. doi: 10.3389/fgene.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad DF, Hurles ME. The population genetics of structural variation. Nat Genet. 2007;39(7) Suppl:S30–S36. doi: 10.1038/ng2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maydan JS, Lorch A, Edgley ML, Flibotte S, Moerman DG. Copy number variation in the genomes of twelve natural isolates of Caenorhabditis elegans. BMC Genomics. 2010;11:62. doi: 10.1186/1471-2164-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheeseman IH, et al. Population structure shapes copy-number variation in malaria parasites. Mol Biol Evol. 2016;33:603–620. doi: 10.1093/molbev/msv282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers RL, Shao L, Thornton KR. Tandem duplications lead to novel expression patterns through exon shuffling in Drosophila yakuba. PLoS Genet. 2017;13:e1006795. doi: 10.1371/journal.pgen.1006795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge Univ Press; Cambridge, UK: 1983. [Google Scholar]
- 8.Ohta T. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst. 1992;23:263–286. [Google Scholar]
- 9.Charlesworth B. Fundamental concepts in genetics: Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 10.Yampolsky LY, Stoltzfus A. Bias in the introduction of variation as an orienting factor in evolution. Evol Dev. 2001;3:73–83. doi: 10.1046/j.1525-142x.2001.003002073.x. [DOI] [PubMed] [Google Scholar]
- 11.Halligan DL, Keightley PD. Spontaneous mutation accumulation studies in evolutionary genetics. Annu Rev Ecol Evol Syst. 2009;40:151–172. [Google Scholar]
- 12.Katju V, Packard LB, Bu L, Keightley PD, Bergthorsson U. Fitness decline in spontaneous mutation accumulation lines of Caenorhabditis elegans with varying effective population sizes. Evolution. 2015;69:104–116. doi: 10.1111/evo.12554. [DOI] [PubMed] [Google Scholar]
- 13.Konrad A, et al. Mitochondrial mutation rate, spectrum and heteroplasmy in Caenorhabditis elegans spontaneous mutation accumulation lines of differing population size. Mol Biol Evol. 2017;34:1319–1334. doi: 10.1093/molbev/msx051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katju V, Packard LB, Keightley PD. Fitness decline under osmotic stress in Caenorhabditis elegans populations subjected to spontaneous mutation accumulation at varying population sizes. Evolution. 2018;72:1000–1008. doi: 10.1111/evo.13463. [DOI] [PubMed] [Google Scholar]
- 15.Pollak E. On the theory of partially inbreeding finite populations. I. Partial selfing. Genetics. 1987;117:353–360. doi: 10.1093/genetics/117.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ota T. Fixation probability of a mutant influenced by random fluctuation of selection intensity. Genet Res. 1972;19:33–38. [PubMed] [Google Scholar]
- 17.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itani OA, Flibotte S, Dumas KJ, Moerman DG, Hu PJ. Chromoanasynthetic genomic rearrangement identified in a N-ethyl-N-nitrosourea (ENU) mutagenesis screen in Caenorhabditis elegans. G3 (Bethesda) 2015;6:351–356. doi: 10.1534/g3.115.024257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipinski KJ, et al. High spontaneous rate of gene duplication in Caenorhabditis elegans. Curr Biol. 2011;21:306–310. doi: 10.1016/j.cub.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farslow JC, et al. Rapid Increase in frequency of gene copy-number variants during experimental evolution in Caenorhabditis elegans. BMC Genomics. 2015;16:1044. doi: 10.1186/s12864-015-2253-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katju V. In with the old, in with the new: The promiscuity of the duplication process engenders diverse pathways for novel gene creation. Int J Evol Biol. 2012;2012:341932. doi: 10.1155/2012/341932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maydan JS, et al. Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative genomic hybridization. Genome Res. 2007;17:337–347. doi: 10.1101/gr.5690307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson O, et al. The million mutation project: A new approach to genetics in Caenorhabditis elegans. Genome Res. 2013;23:1749–1762. doi: 10.1101/gr.157651.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 2009;5:e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katju V, Lynch M. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics. 2003;165:1793–1803. doi: 10.1093/genetics/165.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 28.Bik HM, Fournier D, Sung W, Bergeron RD, Thomas WK. Intra-genomic variation in the ribosomal repeats of nematodes. PLoS One. 2013;8:e78230. doi: 10.1371/journal.pone.0078230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch M, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci USA. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schrider DR, Houle D, Lynch M, Hahn MW. Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster. Genetics. 2013;194:937–954. doi: 10.1534/genetics.113.151670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keith N, et al. High mutational rates of large-scale duplication and deletion in Daphnia pulex. Genome Res. 2016;26:60–69. doi: 10.1101/gr.191338.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henrichsen CN, et al. Segmental copy number variation shapes tissue transcriptomes. Nat Genet. 2009;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 33.Qian W, Liao BY, Chang AY, Zhang J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010;26:425–430. doi: 10.1016/j.tig.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guschanski K, Warnefors M, Kaessmann H. The evolution of duplicate gene expression in mammalian organs. Genome Res. 2017;27:1461–1474. doi: 10.1101/gr.215566.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loehlin DW, Carroll SB. Expression of tandem gene duplicates is often greater than twofold. Proc Natl Acad Sci USA. 2016;113:5988–5992. doi: 10.1073/pnas.1605886113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cardoso-Moreira M, et al. Evidence for the fixation of gene duplications by positive selection in Drosophila. Genome Res. 2016;26:787–798. doi: 10.1101/gr.199323.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320:1629–1631. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- 38.Langley CH, et al. Genomic variation in natural populations of Drosophila melanogaster. Genetics. 2012;192:533–598. doi: 10.1534/genetics.112.142018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adler M, Anjum M, Berg OG, Andersson DI, Sandegren L. High fitness costs and instability of gene duplications reduce rates of evolution of new genes by duplication-divergence mechanisms. Mol Biol Evol. 2014;31:1526–1535. doi: 10.1093/molbev/msu111. [DOI] [PubMed] [Google Scholar]
- 40.Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 41.Veitia RA. Gene dosage balance in cellular pathways: Implications for dominance and gene duplicability. Genetics. 2004;168:569–574. doi: 10.1534/genetics.104.029785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reams AB, Kofoid E, Savageau M, Roth JR. Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics. 2010;184:1077–1094. doi: 10.1534/genetics.109.111963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birchler JA, Veitia RA. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA. 2012;109:14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nair S, et al. Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance in malaria parasites. Mol Biol Evol. 2007;24:562–573. doi: 10.1093/molbev/msl185. [DOI] [PubMed] [Google Scholar]
- 45.Patrick WM, Quandt EM, Swartzlander DB, Matsumura I. Multicopy suppression underpins metabolic evolvability. Mol Biol Evol. 2007;24:2716–2722. doi: 10.1093/molbev/msm204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Axelsson E, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- 47.Assogba BS, et al. The ace-1 locus is amplified in all resistant Anopheles gambiae mosquitoes: Fitness consequences of homogeneous and heterogeneous duplications. PLoS Biol. 2016;14:e2000618. doi: 10.1371/journal.pbio.2000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.