Abstract
Data-driven respiratory gating techniques were developed to correct for respiratory motion in PET studies, without the help of external motion tracking systems. Due to the greatly increased image noise in gated reconstructions, it is desirable to develop a data-driven event-by-event respiratory motion correction method. In this study, using the Centroid-Of-Distribution (COD) algorithm, we established a data-driven event-by-event respiratory motion correction technique using TOF PET list-mode data, and investigated its performance by comparing with an external system-based correction method. Ten human scans with the pancreatic β-cell tracer 18F-FP-(+)-DTBZ were employed. Data-driven respiratory motions in Superior-Inferior (SI) and Anterior-Posterior (AP) directions were first determined by computing the centroid of all radioactive events during each short time frame with further processing. The Anzai belt system was employed to record respiratory motion in all studies. COD traces in both SI and AP directions were first compared with Anzai traces by computing the Pearson correlation coefficients. Then, respiratory gated reconstructions based on either COD or Anzai traces were performed to evaluate their relative performance in capturing respiratory motion. Finally, based on correlations of displacements of organ locations in all directions and COD information, continuous 3D internal organ motion in SI and AP directions was calculated based on COD traces to guide event-by-event respiratory motion correction in the MOLAR reconstruction framework. Continuous respiratory correction results based on COD were compared with that based on Anzai, and without motion correction. Data-driven COD traces showed a good correlation with Anzai in both SI and AP directions for the majority of studies, with correlation coefficients ranging from 63% to 89%. Based on the determined respiratory displacements of pancreas between end-expiration and end-inspiration from gated reconstructions, there was no significant difference between COD-based and Anzai-based methods. Finally, data-driven COD-based event-by-event respiratory motion correction yielded comparable results to that based on Anzai respiratory traces, in terms of contrast recovery and reduced motion-induced blur. Data-driven event-by-event respiratory motion correction using COD showed significant image quality improvement compared with reconstructions with no motion correction, and gave comparable results to the Anzai-based method.
Keywords: Respiratory Motion, Data-driven, Event-by-event Motion Correction
1. Introduction
Respiratory motion is a major cause of degradation of PET image quality (Nehmeh and Erdi, 2008). Since the PET acquisition duration is in the range of minutes to hours, image blurring and artifacts due to respiratory motion are unavoidable. Respiratory motion correction methods such as respiratory gating, motion compensation incorporated into image reconstruction, or post-reconstruction motion correction can be performed to reduce the effect of respiratory motion. These methods require motion information, typically from external motion tracking systems. Because of increased complexity and expense due to the use of external systems, various groups have investigated data-driven motion estimation methods. Based on reconstructed images, computing the center-of-mass (COM) of a manually defined region-of-interest (ROI) over a lesion on each reconstructed dynamic image was proposed to estimate respiratory motion (Bundschuh et al., 2007). Another image-based method which used total counts in a ROI placed over boundaries of organs also showed good performance in respiratory motion estimation (Visvikis et al., 2003). However, image-based methods are time-consuming because they require the reconstruction of all dynamic images in advance. Alternatively, many authors have studied data-driven methods based on raw count data such as variations of activity or evaluating the axial component of the COM (Buther et al., 2009; Buther et al., 2010; Kesner and Kuntner, 2010; Schleyer et al., 2009; Schleyer et al., 2011; Thielemans et al., 2011; Schleyer et al., 2014). These methods have included processing such as spectral analysis, and principal component analysis based on short time frame sinograms. All these studies showed promising results, although estimated motion traces can be noisy.
For most of these studies, the aim was to develop data-driven gating methods with respiratory motion signals estimated from raw PET data, without external tracking systems. Respiratory gating reduces the effect of respiratory motion by dividing PET data into gates based on either temporal phase or respiratory displacement amplitude, but with the cost of increased noise due to the use of a fraction of the PET data (Dawood et al., 2007; Nehmeh et al., 2004; Bai and Brady, 2011). To use all the acquired events in motion correction reconstruction and eliminate intragate motions, it is desirable to apply event-by-event (EBE) correction to achieve minimal motion-induced blurring (Victor et al., 2009). We have previously developed an EBE respiratory motion correction with 3D internal-1D external motion correlation (INTEX3D) (Chan et al., 2013). A 3D internal organ respiratory motion transformation matrix is estimated based on a linear correlation between the external respiratory trace and internal organ movement. The time-varying matrix is then incorporated into the Motion compensation OSEM List-Mode Algorithm for Resolution recovery reconstruction (MOLAR) with time-of-flight (TOF) capability for EBE motion correction (Carson et al., 2003; Jin et al., 2013). This method can correct for both inter- and intra-gate respiratory motion blur, and can uniquely be applied to dynamic data (Yu et al., 2016).
Based on our previous work on EBE respiratory motion correction, which employed respiratory motion acquired from an external motion tracking device, in this study, we propose a data-driven EBE respiratory motion correction method based on motion information extracted from the listmode TOF data using the centroid of distribution (COD) algorithm (Jin, 2013). In this method, respiratory motion in three directions was estimated by computing the COD of all events in the field of view (FOV) with fine temporal resolution from the list-mode data. Then, EBE motion correction was performed using data-driven respiratory motion derived from the correlation of inter-gate displacement of internal organ motion and COD information. The performance of data-driven continuous respiratory motion correction method was compared with an external device-based method. Preliminary results from this work have been previously reported (Ren et al., 2014).
2. Methods
2.1. Patient Data
This study included abdominal PET data from an IRB approved protocol with 10 subjects injected with 9-fluoropropyl-(+)-dihydrotetrabenazine (18F-FP-(+)-DTBZ) (Normandin et al., 2012), which binds to the vesicular monoamine transporter-2 (VMAT2) in pancreatic β-cells. The averaged injected dose was 237±61 MBq. Listmode data were acquired for at least 120 min with the Siemens Biograph mCT PET/CT scanner (Siemens Medical Solutions, Knoxville, USA) with TOF information (Jakoby et al., 2011). During data acquisition, pancreas, liver, spleen, and kidneys were located in the scanner FOV. In all cases, an Anzai system (Anzai Medical, Tokyo, Japan) was used to acquire external respiratory traces at 40 Hz (25ms), which was rebinned into 100 ms per frame for comparison with the COD trace
2.2. Centroid-Of-Distribution (COD) Algorithm
To extract respiratory motion information from the PET data, the centroid of distribution (COD) was determined with fine temporal resolution. For each listmode event i, given the detector pair information, the line-of-response (LOR) can be determined, and specified as the (X,Y,Z) spatial coordinates of the two detectors, expressed in mm from the center of the FOV. With the TOF bin number available, the (Xi, Yi, Zi) coordinates of each event was determined as the center of its TOF bin along the LOR. For systems without TOF, the event could be backprojected to the center of its LOR. Coordinates were expressed in mm from the center of the FOV. These coordinates were averaged over a short time interval (100 ms), to produce raw COD traces in three directions: Cx: Left-Right (LR), Cy: Anterior-Posterior (AP), and Cz: Superior-Inferior (SI) directions, with the respiratory signal detectible in the AP and SI directions. Cz and Cy for each short time interval can be determined as:
| (1) |
with a total number of events of N during that interval.
In these dynamic scans, kinetic drift, i.e., COD change due to tracer redistribution, was detected, especially in the early data, as also seen in a recent study (Schleyer et al., 2014). Using the method proposed in (Schleyer et al., 2014), kinetic drift was extracted by applying a sliding average window, empirically set to 60 s, then, subtracting this signal. To further extract respiratory motion, a low-pass filter with a cut-off frequency that was determined by spectral analysis was applied. Specifically, the Fourier transform was applied to the Cz traces, after correction for kinetic drift, and the frequency associated with peak magnitude, fp, was determined. The cut-off frequency for the low-pass filter was set to fp +0.2Hz. These cut-off frequencies were in the range of 0.3 to 0.5Hz.
2.3. Data Analysis
COD traces were calculated for the first 90 minutes of each pancreas study. No detectible respiratory information was found in the X direction, so the motion correction was applied only to Y and Z directions. To evaluate the traces, Pearson correlation coefficients between COD and Anzai traces were calculated. Because of very low activity and rapid kinetic changes immediately following the injection, data-driven COD traces of the first few minutes of the acquisition may not show clear respiratory motion. For Y and Z directions, correlation coefficients were calculated over 1-min intervals from 10 to 90 minutes.
2.4. Data-Driven Respiratory Gated Reconstruction
Before implementing data-driven EBE respiratory motion correction, data-driven gated reconstructions were first performed. To perform gated reconstructions, the inspiration peak of each respiratory cycle must be determined. This can be done by finding local maxima in the time-course data. Two restrictions were applied to minimize noise effects on the detection of inspiration peaks. First, the time window between successive peaks was set to the range of 2–8s, to exclude noise spikes happening shortly after each inspiration peak. Second, noise spikes with much smaller magnitude than real inspiration peaks were excluded by setting a relative peak magnitude threshold, such as 0.3; in this manner, a new peak would only be accepted if its maximum was greater than this threshold multiplied by the average of all the previously detected inspiration peaks. Gating information was extracted from both Anzai and Cz traces. Phase-gated reconstructions were performed for all ten subjects. A ten-minute portion (10–20 min) in each of those studies was selected for phase-gated reconstruction with 8 gates with MOLAR (Carson et al., 2003), based on either COD-generated or Anzai-generated gating information. In order to exclude irregular breathing cycles, data from respiratory cycles were included in the reconstruction if their periods were within one standard deviation of the mean period duration of each 10-min frame between 10–90min post injection. For all subjects, over all time periods, the vast majority of data rejection rates were in the range of 5–15%, with a mean of 8% over all subjects (minimum: 1%, 25th percentile: 6, median: 7%, 75th percentile: 11%, maximum: 16%).
To compare COD-based and Anzai-based gating, for each respiratory-gated reconstruction, the pancreas was segmented using a level set (active contour) method (Chan et al., 2013), and the center of mass (COM) of the segmented pancreas was determined. The respiratory displacements between end- inspiration and end-expiration were identified as the maximum difference among the pancreas COM across the 8 gates in Y and Z directions. Gate 1 corresponded to the end-inspiration phase, and Gate 5 corresponded to the end-expiration phase. To quantitatively compare COD-based and Anzai-based gated reconstructions, for each subject, the difference between maximal respiratory displacements determined from COD-based and Anzai-based gated reconstructions were calculated in both Y and Z directions. Paired t test was used to statistically study if there was a significant difference between COD- and Anzai-based gated reconstruction results.
2.5. Data-driven Event-by-event Respiratory Motion Correction
Data-driven EBE respiratory motion correction was implemented using the INTEX3D approach (Chan et al., 2013). In this INTEX3D implementation, the external trace was replaced by the COD traces from the same 10-min frame of the pancreas studies used for respiratory-gated reconstructions. This analysis was performed analogously to that previously performed with Anzai data. Specifically, continuous motion information was generated by building the linear relationship between mean COD values for each gate, and internal organ motion calculated from the segmented pancreas from respiratory-gated images. For COD traces, we built the linear relationships in Z and Y directions separately, i.e., Cz was correlated with internal organ motion (COM) in the Z direction, and Cy with internal organ motion in the Y direction. Based on these correlations, continuous motions were estimated at 10 Hz by converting the Cz and Cy traces into organ displacements in Z and Y directions, respectively. These displacements were used by MOLAR for EBE motion correction, by relocating the LOR of each event to where it would have been if there was no motion. Anzai-based EBE respiratory motion correction was performed for comparison. For Anzai, the 1D Anzai signal was correlated with estimated internal organ motion in both Y and Z directions. Image reconstruction with no motion correction was also performed for comparison.
Reconstructions with COD-based, Anzai-based, and no motion correction were first compared visually by focusing on liver, pancreas, and kidney contours and fine structures, as well as the gallbladder area. Then, for quantitative comparison, intensity profiles were plotted in these regions.
3. Results
3.1. Raw COD Traces
In this work, ten subjects were employed for COD analysis. Figure 1 shows examples of raw COD signals in LR (Cx), AP (Cy), and SI (Cz) directions. Raw COD traces in Z and Y directions show respiratory motions to some extent but with noise. Cx traces do not show obvious breathing signals. The noise level of the COD signals can be estimated directly, see supplemental materials for this analysis.
Figure 1.
Raw COD signals from one minute of a pancreas study in three directions: Left-Right (LR, x), Anterior-Posterior (AP, y), and Superior-Inferior (SI, z).
3.2. COD and Anzai Traces Comparison
In Figure 2, COD traces in Y and Z directions were compared with the Anzai trace. For comparison, the mean was removed from both traces and the data were standardized to have zero mean and unit variance. Generally, COD traces of pancreas studies showed a strong correlation with Anzai data. An example of a 2-min period of data with a regular breathing pattern is shown in Figure 2(a) with COD and Anzai traces for SI and AP directions overlaid. COD traces in both directions show high correlation with Anzai traces, with Pearson correlation coefficients between COD and Anzai of 0.94 and 0.88 for Z and Y directions, respectively, capturing variation in respiratory amplitude and frequency. In addition, when irregular respiration patterns appeared, COD traces successfully captured the irregular pattern, as shown in Figure 2(b), giving correlation coefficients with Anzai of 0.88 and 0.82 for Z and Y directions, respectively. For deep breaths, the Anzai system did not record the portion of the traces that dropped below a certain threshold. In contrast, COD traces showed no saturation. Figure 2(c) shows an example with noisier COD traces than the previous two examples, with lower correlation coefficients of 0.79 and 0.73 for Z and Y direction. Although several peaks detected in the COD traces were not present in the Anzai data, the COD traces, particularly COD in the Z direction, captured the majority of respiratory cycles, and can thus be used for gated reconstruction and event-by-event motion correction.
Figure 2.
Cz and Cy traces (in red) overlaid on Anzai data (in black) from pancreas studies with (a) a 2-min period with a regular breathing pattern, (b) a 2-min period with an irregular breathing pattern. (c) a 2-min period with noisy COD traces in both Z and Y directions. Both COD and Anzai signals are standardized (0 mean and unit variance) for the convenience of visual comparison. Pearson correlation coefficients between Anzai and COD for both Z and Y directions were calculated as: a. 0.94 and 0.88, b. 0.88 and 0.82, c. 0.79 and 0.73.
To quantitatively compare COD and Anzai traces, the correlation coefficients of COD and Anzai traces were calculated over 1-min intervals from 10 to 90 minutes. Table 1 shows the mean correlation coefficients between COD and Anzai for each subject. Figure 3 shows the box plot for correlation coefficients of COD and Anzai in Z and Y directions for ten subjects, as shown in Table 1. The correlation coefficients range from 0.63 to 0.89, showing good correlation between COD and Anzai, both in Z and Y directions. The averaged correlation coefficients between COD and Anzai over all subjects were 79% and 76% for Z and Y directions, respectively. In most cases, Cz correlated with Anzai better than Cy, which is due to smaller respiratory motions and poorer signal-to-noise ratio in the Y direction. In the few cases with lower correlation coefficients and larger standard deviations, subjects displayed irregular breathing patterns, for example, multiple shallow and fast breaths in a row. In addition, shallow breaths with smaller motion amplitudes resulted in lower signal-to-noise ratios for COD traces, leading to lower correlation with the Anzai data.
Table 1.
Mean and SD of correlation coefficients of COD and Anzai traces calculated over 1-min intervals from 10 to 90 minutes in Z and Y directions.
| Subject No. | Correlation coefficients between Cz and Anzai |
Correlation coefficients between Cy and Anzai |
|---|---|---|
| 1 | 0.89 ± 0.05 | 0.78 ± 0.19 |
| 2 | 0.88 ± 0.02 | 0.86 ± 0.03 |
| 3 | 0.83 ± 0.05 | 0.72 ± 0.14 |
| 4 | 0.86 ± 0.05 | 0.81 ± 0.07 |
| 5 | 0.83 ± 0.06 | 0.80 ± 0.06 |
| 6 | 0.80 ± 0.08 | 0.76 ± 0.08 |
| 7 | 0.63 ± 0.13 | 0.78 ± 0.07 |
| 8 | 0.59 ± 0.14 | 0.70 ± 0.12 |
| 9 | 0.73 ± 0.09 | 0.69 ± 0.09 |
| 10 | 0.87 ± 0.04 | 0.75 ± 0.09 |
| Mean ± Std | 0.79 ± 0.11 | 0.76 ± 0.05 |
Figure 3.
Box plot of correlation coefficients of COD and Anzai traces in Z and Y directions for 10 subjects, showing the maximum, the 75th percentile, the median, the 25th percentile, and the minimum for each direction, respectively, as shown in Table 1. ”2D
Data-driven Respiratory Gated Reconstruction Comparison
A ten-minute portion (10–20 min) of each pancreas studies was selected for phase-gated reconstruction with 8 gates. Sample COD-gated maximum intensity projection (MIP) images of the end-inspiration phase (Gate 1) and the end-expiration phase (Gate 5) in coronal and sagittal views are shown in Figure 4, with the centroid of the segmented pancreas marked with a red dot. Comparing the pancreas location between phases, there is a noticeable displacement due to respiration. Displacements were calculated from the centroid of the segmented pancreas between phases. For COD, these displacements were 22.3 mm and 7.4 mm in Z and Y directions, respectively. Displacements in Anzai-gated images (data not shown) were comparable to the COD-gated images, with respiratory displacements of 20.6 mm and 7.3 mm in Z and Y directions, respectively.
Figure 4.
Maximum intensity projection (MIP) images of end-inspiration phase (Gate 1) and end-expiration phase (Gate 5) in coronal and sagittal views of a sample subject, with the centroid of the segmented pancreas denoted in a red dot. Respiratory displacements can be visualized by comparing the pancreas centroid locations between the phases.
Table 2 shows respiratory displacements (mm) of pancreas between maximum expiration and maximum inspiration, in Z and Y directions. These were calculated from the COM of the segmented pancreas from respiratory-gated images based on data-driven COD data and Anzai traces. The difference in the Z direction ranged from −0.8 to 1.9 mm with no significant difference between COD-based and Anzai-based respiratory gated reconstruction. Differences in the Y direction ranged from −0.8 to 2.3 mm, with all subjects but one below 1.0 mm. With smaller respiratory motion in the Y direction than in the Z direction, and with PET image resolution and possible segmentation errors taken into consideration, the slightly larger difference between the respiratory displacements in the Y direction is considered acceptable. Paired t tests comparing the respiratory displacements determined by COD and Anzai gave P values of 0.80 and 0.53 for Z and Y direction, respectively. Thus, overall, there was no significant difference in maximum respiratory displacement between COD-based and Anzai-based respiratory-gated images.
Table 2.
Respiratory displacement (mm) of pancreas between maximum expiration and maximum inspiration, in both Z and Y directions, in COD and Anzai-gated images
| Subject No. | Z Direction | Y Direction | ||||
|---|---|---|---|---|---|---|
| Respiratory displacement (mm) |
Difference of respiratory displacement (mm) |
Respiratory displacement (mm) |
Difference of respiratory displacement (mm) |
|||
| COD- gated |
Anzai- gated |
COD- gated |
Anzai- gated |
|||
| 1 | 11.2 | 11.1 | 0.1 | 1.5 | 1.4 | 0.1 |
| 2 | 11.0 | 10.1 | 0.9 | 3.8 | 3.5 | 0.3 |
| 3 | 10.5 | 10.4 | 0.1 | 3.7 | 1.4 | 2.3 |
| 4 | 22.3 | 20.6 | 1.7 | 7.4 | 7.3 | 0.1 |
| 5 | 7.6 | 8.1 | −0.5 | 2.6 | 3.1 | −0.5 |
| 6 | 7.2 | 8.4 | −1.2 | 2.6 | 3.2 | −0.6 |
| 7 | 6.9 | 7.9 | −1.0 | 4.0 | 3.2 | 0.8 |
| 8 | 9.8 | 10.6 | −0.8 | 3.0 | 2.9 | 0.1 |
| 9 | 7.9 | 6.0 | 1.9 | 3.0 | 3.8 | −0.8 |
| 10 | 14.7 | 15.0 | −0.3 | 2.9 | 2.9 | 0.0 |
| P value for Paired t-test | 0.80 | 0.53 | ||||
3.3. Data-driven Event-by-event Respiratory Motion Correction
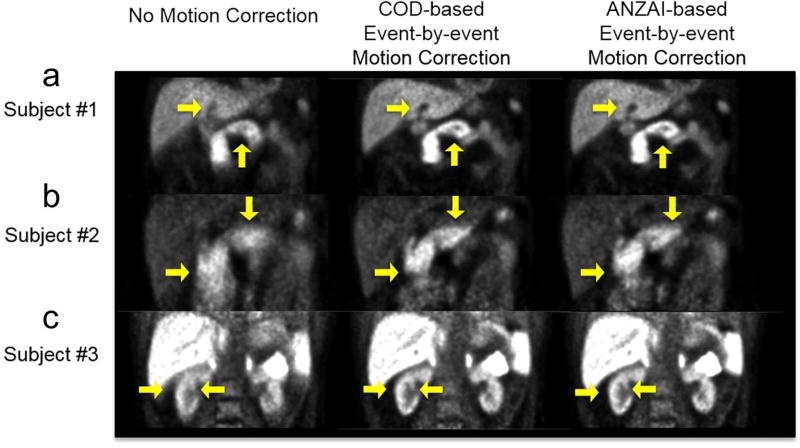
Figure 5 shows sample data-driven COD-based EBE respiratory motion correction results in three pancreas studies, with Anzai-based correction and reconstructions with no motion correction for comparison. For each study, one coronal slice was chosen to show liver and pancreas in A, pancreas in B, and kidneys in C, respectively. COD-based motion correction shows comparable visual results to the Anzai-based method. All show improvements in the gallbladder “cold” area, the pancreas region, and the kidney region with sharper contours of the fine structures, and with improved image quality compared with the no-correction results.
Figure 5.
Reconstructed images without motion correction, with COD-based data-driven event-by-event motion correction, and with Anzai-based correction. For each pancreas study, one coronal slice is displayed to show gallbladder and pancreas in (a), pancreas in (b), and kidneys in (c). Image intensity scale in (c) was saturated to show kidney structures.
Based on the reconstruction results shown in Figure 5, taking Subject#1 as an example, intensity profiles were plotted over the gallbladder and liver region, and the pancreas region to quantitatively compare data-driven COD-based and Anzai-based EBE motion correction performance. Figure 6(b) shows intensity profiles over gallbladder and liver at the level indicated by the horizontal line in Figure 6(a). Figure 6(c) shows intensity profiles over a section of the pancreas at the level indicated by the vertical line. Both COD and Anzai curves show decreased intensity values in the “cold” region (gallbladder) compared to results with no motion correction. In the pancreas, both COD and Anzai profiles show very similar radioactivity concentrations. The maximum value along the COD-based profile showed a 1% difference from the Anzai-derived value, while the maximum value of the no motion correction result differed by −25%. To quantitatively assess COD-based EBE motion correction over all subjects, the intensity profile across this region of the pancreas was plotted for each subject. The difference of maximum values of these profiles between COD and Anzai were −1±2%; the no motion correction results showed differences of −15±6% from the Anzai data. Thus, the COD-based method gives comparable intensity profiles to the Anzai-based method, indicating that data-driven COD-based EBE respiratory motion correction can achieve comparable performance to that achieved with an external tracking system.
Figure 6.
Intensity profiles of the reconstructions of Subject #1 in Figure. 4 (a), with no motion correction, COD-based and Anzai-based event-by-event motion correction. (b) Intensity profiles over gallbladder and liver at the level indicated by the horizontal bar in (a); (c) Intensity profiles over pancreas at the level indicated by the vertical bar in (a).
Discussion
In this study, we established methodology for data-driven EBE respiratory motion correction using raw PET data, based on respiratory traces determined by the centroid of distribution (COD) algorithm. The COD traces from the pancreas studies, visually and quantitatively, showed excellent agreement with the respiratory traces determined from the Anzai tracking system. Further, data-driven EBE respiratory motion correction using COD traces provided comparable results to the Anzai-based correction.
Compared to COD, the Anzai system has the disadvantage of patient set-up. Further, the respiratory trace depends on the external motion of the patient’s lower abdomen, with no direction of motion specified. Since COD signals are calculated based on each event’s backprojected coordinates, the values are provided in X, Y and Z directions, having millimeter units. As expected, the Cz and Cy signals showed respiratory motions of inner organs. In the COD method, respiratory motion in Z and Y directions are calculated separately, providing 2 measures of motion. For the Anzai, or for other data-driven methods, only one measure is produced for each time. For COD, Cz and Cy were highly correlated, having an average correlation coefficient of 0.82 over 10 subjects. Correlations less than 1 may be cause by noise in the COD data or may reflect that there is independent signal in the Cz and Cy data. Additionally, it was observed in some subjects that irregular breathing patterns, such as coughing, occurred. This led to sudden and large movements, and the Anzai data showed saturated values because of the threshold limit of the system. For COD, there is no such limit, as can be seen in Figure 2(b). It is worth noting that the Anzai system records motion at a frequency of 40Hz (recently improved to 50Hz). When calculating COD curves, we used a 10Hz sampling frequency, which was sufficient to capture continuous respiratory motion. COD data could be acquired at a higher frequency, although it would produce noisier signals.
We used the Anzai data as the gold standard in this study to evaluate the data-driven COD method and its performance in continuous respiratory motion correction. The first measured quantity was the Pearson correlation coefficient between COD and Anzai in Z and Y directions. Most studies showed excellent agreement between COD and Anzai. Some studies showed lower correlation coefficients; explanations include 1) subjects took shallow breaths, which leads to low signal-to-noise in COD traces and lower correlation with Anzai, and 2) subjects had some very deep breaths and Anzai data saturated. For shallow breaths, the Anzai system is likely to provide more accurate detection of respiratory motion, since the COD signals have higher noise levels. For irregular breathers, the same may be true, if the filtering used in the COD data eliminates some high-frequency components in the respiratory signal. However, in the cases of very deep breaths, the Anzai signals could be saturated, in which case, COD may provide more reliable motion estimation than Anzai. The second measured quantity was the respiratory displacement between end-inspiration and end-expiration, determined from gated reconstructions based on either COD or Anzai signals. Not surprisingly, COD traces that were in good agreement with Anzai generated reliable gating triggers for gated reconstructions, yielding matched respiratory displacements, with no significant differences in Z and Y directions, respectively. Finally, when evaluating the performance of data-driven EBE respiratory motion correction, in addition to the comparisons of image sharpness and organ details, intensity profiles were compared. Better correction should give higher intensity values in high-uptake regions and lower values in low-activity areas, such as gallbladder. As shown in Figure 6, COD- and Anzai-based corrections gave similar intensity and increased contrast values in the gallbladder and liver regions. In the pancreas region of the 10 subjects, COD and Anzai images had highly similar intensity profiles, with higher values than those of uncorrected images. Therefore, in these studies, COD-based EBE correction can correct respiratory motion with comparable performance to the Anzai-based method.
In our respiratory gated reconstructions, a 10-min interval was chosen to demonstrate the feasibility of data-driven respiratory gated reconstruction, and performance was compared with the Anzai-based method in terms of captured respiratory motion. Gated reconstruction can also be performed on a longer scan duration, however use of longer scan data will often require additional corrections for body motion. See supplemental materials for a comparison of gated reconstructions between 10–20min and 10–90min study.
Our data-driven EBE respiratory motion correction was implemented using INTEX3D. EBE respiratory motion correction can correct for intra-gate motion, which is not possible in respiratory-gated reconstruction, which only corrects for inter-gate motion. In our COD-based INTEX3D, internal motions in Z and Y directions were determined from the motion of the centroid of the segmented pancreas from reconstructed images of 8 gates. Thus the estimated internal motion is highly dependent on the segmentation of pancreas. If the internal organ motion estimated from organ-based segmentation is not accurate, the accuracy of the EBE respiratory motion correction is likely to decrease, leading to additional image blurring. Level-set segmentation was employed in this study, however, this approach may not provide reliable results in cases where the gated image quality is poor. More robust segmentation methods, or direct registration between gated images (Rohlfing et al., 2004), should be evaluated to improve the reliability of estimated internal motion.
In this implementation, rigid motion correction was performed, which relocated every LOR in each short time interval by the same motion displacement. Rigid correction clearly produced improved results, however, it is not completely accurate. It does not correct for nonrigid motion within and between organs; also, tissues such as the spine, that are not affected by respiratory motion, were over-corrected in rigid motion correction. Thus, it is desirable to develop data-driven EBE nonrigid respiratory motion correction, an approach which is feasible, using a nonrigid EBE correction framework, which is under development in our group (Chan, 2014). Better image quality and more accurate quantitative results are expected with nonrigid motion correction.
In this study, we tested the COD method with the tracer 18F-FPDTBZ in the abdomen. Organs in the abdomen tend to move together with respiration, which gives a strong COD signal. One limitation of COD, like other data-driven methods, is that it depends upon the uptake pattern in the FOV. The greater the contrast between moving organs and the background, the stronger the COD signals will be. To better extract COD signals, especially in studies with higher noise, more signal processing methods may be needed. For example, in the PCA data-driven respiratory motion detection method, principal component analysis was used to detect respiration motion from sinograms rebinned into 500ms per frame (Thielemans et al., 2011). Thus, it is worth investigating if methods such as PCA can improve our extracted respiratory signals. Additionally, in studies with lesions, methods that restrict the analysis to a portion of the FOV should be further investigated.
One limitation of data-driven methods such as COD is that respiratory information is unreliable in periods of very low activity and rapid tracer kinetics, e.g., in the first few minutes postinjection. This may affect quantitative modeling studies that use early data. Note that slow kinetic drifts can be easily removed.
In this study, we did not evaluate the effect of COD-based EBE respiratory motion correction on estimation of kinetic parameters. In a previous work by our group (Yu et al., 2016), the effect of Anzai-based EBE respiratory motion correction on kinetic parameter estimator was evaluated for this tracer, and found that more accurate parameters can be expected after correction. Based on the excellent agreement between COD and Anzai, we can expect that COD-based EBE respiratory correction can also improve kinetic parameter estimation.
Further investigation is needed to assess how the proposed data-driven method can be applied for other body regions and other tracers, including oncology scans using 18F-FDG. These methods could improve image quality greatly, and hence the diagnostic utility of abdominal PET.
Conclusion
Data-driven event-by-event respiratory motion correction using TOF PET listmode data based on Centroid-of-Distribution motion estimation was successfully implemented in dynamic pancreas studies using the tracer 18F-FP-(+)-DTBZ. Data-driven COD respiratory motion traces showed excellent agreement with the Anzai external tracking system, and data-driven respiratory gating had comparable performance to the Anzai-based method. Finally, COD’s performance in improving image quality with event-by-event motion correction was comparable to that of the Anzai data.
Supplementary Material
Acknowledgments
We thank the staff at the Yale PET Center for their support and the studies that formed the basis of this work. This work was also supported by NIH grants 1S10RR029245-01 and by CTSA Grant Number UL1 TR000142 from the National Center for Advancing Translational Science, a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH.
References
- Bai W, Brady M. Motion correction and attenuation correction for respiratory gated PET images. IEEE Trans Med Imaging. 2011;30:351–65. doi: 10.1109/TMI.2010.2078514. [DOI] [PubMed] [Google Scholar]
- Bundschuh RA, Martinez-Moeller A, Essler M, Martinez MJ, Nekolla SG, Ziegler SI, Schwaiger M. Postacquisition detection of tumor motion in the lung and upper abdomen using list-mode PET data: A feasibility study. Journal of Nuclear Medicine. 2007;48:758–63. doi: 10.2967/jnumed.106.035279. [DOI] [PubMed] [Google Scholar]
- Buther F, Dawood M, Stegger L, Wubbeling F, Schafers M, Schober O, Schafers KP. List Mode-Driven Cardiac and Respiratory Gating in PET. Journal of Nuclear Medicine. 2009;50:674–81. doi: 10.2967/jnumed.108.059204. [DOI] [PubMed] [Google Scholar]
- Buther F, Ernst I, Dawood M, Kraxner P, Schafers M, Schober O, Schafers KP. Detection of respiratory tumour motion using intrinsic list mode-driven gating in positron emission tomography. Eur. J. Nucl. Med. Mol. Imaging. 2010;37:2315–27. doi: 10.1007/s00259-010-1533-y. [DOI] [PubMed] [Google Scholar]
- Carson RE, Barker WC, Jeih-San L, Johnson CA. Design of a motion-compensation OSEM list-mode algorithm for resolution-recovery reconstruction for the HRRT. 2003 IEEE Nuclear Science Symposium. Conference Record (IEEE Cat. No.03CH37515) 2003;5:3281–5. [Google Scholar]
- Chan C. IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC) Seattle, USA: 2014. Non-Rigid Event-by-Event Motion Correction for PET with Motion-Dependent PSF. Presentation. [Google Scholar]
- Chan C, Jin X, Fung EK, Naganawa M, Mulnix T, Carson RE, Liu C. Event-by-event respiratory motion correction for PET with 3D internal-1D external motion correlation. Medical physics. 2013;40:112507. doi: 10.1118/1.4826165. [DOI] [PubMed] [Google Scholar]
- Dawood M, Buther F, Lang N, Schober O, Schafers KP. Respiratory gating in positron emission tomography: a quantitative comparison of different gating schemes. Medical physics. 2007;34:3067–76. doi: 10.1118/1.2748104. [DOI] [PubMed] [Google Scholar]
- Jakoby BW, Bercier Y, Conti M, Casey ME, Bendriem B, Townsend DW. Physical and clinical performance of the mCT time-of-flight PET/CT scanner. Phys Med Biol. 2011;56:2375–89. doi: 10.1088/0031-9155/56/8/004. [DOI] [PubMed] [Google Scholar]
- Jin X. PhD dissertation, Yale University. ProQuest Dissertation & Theses Global. 2013. Event-by-Event Motion Correction in Positron Emission Tomography: Development, Evaluation and Applications. [Google Scholar]
- Jin X, Chan C, Mulnix T, Panin V, Casey ME, Liu C, Carson RE. List-mode reconstruction for the Biograph mCT with physics modeling and event-by-event motion correction. Phys. Med. Biol. 2013;58:5567–91. doi: 10.1088/0031-9155/58/16/5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesner AL, Kuntner C. A new fast and fully automated software based algorithm for extracting respiratory signal from raw PET data and its comparison to other methods. Medical physics. 2010;37:5550–9. doi: 10.1118/1.3483784. [DOI] [PubMed] [Google Scholar]
- Nehmeh SA, Erdi YE. Respiratory motion in positron emission tomography/computed tomography: A review. Semin. Nucl. Med. 2008;38:167–76. doi: 10.1053/j.semnuclmed.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Nehmeh SA, Erdi YE, Pan T, Yorke E, Mageras GS, Rosenzweig KE, Schoder H, Mostafavi H, Squire O, Pevsner A, Larson SM, Humm JL. Quantitation of respiratory motion during 4D-PET/CT acquisition. Medical physics. 2004;31:1333–8. doi: 10.1118/1.1739671. [DOI] [PubMed] [Google Scholar]
- Normandin MD, Petersen KF, Ding YS, Lin SF, Naik S, Fowles K, Skovronsky DM, Herold KC, McCarthy TJ, Calle RA, Carson RE, Treadway JL, Cline GW. In Vivo Imaging of Endogenous Pancreatic beta-Cell Mass in Healthy and Type 1 Diabetic Subjects Using F-18-Fluoropropyl-Dihydrotetrabenazine and PET. Journal of Nuclear Medicine. 2012;53:908–16. doi: 10.2967/jnumed.111.100545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Xiao J, Chung C, Jian Y, Mulnix T, Liu C, Carson R. Data-driven respiratory motion estimation and correction using TOF PET list-mode centroid of distribution; 2014 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC); 2014. pp. 1–5. [Google Scholar]
- Rohlfing T, Maurer CR, O’Dell WG, Zhong J. Modeling liver motion and deformation during the respiratory cycle using intensity-based nonrigid registration of gated MR images. Medical physics. 2004;31:427–32. doi: 10.1118/1.1644513. [DOI] [PubMed] [Google Scholar]
- Schleyer PJ, O'Doherty MJ, Barrington SF, Marsden PK. Retrospective data-driven respiratory gating for PET/CT. Phys. Med. Biol. 2009;54:1935–50. doi: 10.1088/0031-9155/54/7/005. [DOI] [PubMed] [Google Scholar]
- Schleyer PJ, O'Doherty MJ, Marsden PK. Extension of a data-driven gating technique to 3D, whole body PET studies. Phys. Med. Biol. 2011;56:3953–65. doi: 10.1088/0031-9155/56/13/013. [DOI] [PubMed] [Google Scholar]
- Schleyer PJ, Thielemans K, Marsden PK. Extracting a respiratory signal from raw dynamic PET data that contain tracer kinetics. Phys. Med. Biol. 2014;59:4345–56. doi: 10.1088/0031-9155/59/15/4345. [DOI] [PubMed] [Google Scholar]
- Thielemans K, Rathore S, Engbrant F, Razifar P. Device-less gating for PET/CT using PCA; 2011 IEEE Nuclear Science Symposium Conference Record; 2011. pp. 3904–10. [Google Scholar]
- Victor WZ, Andre ZK, Steven RM, Roger F. A scheme for PET data normalization in event-based motion correction. Phys. Med. Biol. 2009;54:5321. doi: 10.1088/0031-9155/54/17/016. [DOI] [PubMed] [Google Scholar]
- Visvikis D, Barret O, Fryer T, Turzo A, Lamare F, Rest CCL, Bizais Y. A posteriori respiratory motion gating of dynamic PET images. 2003 IEEE Nuclear Science Symposium. Conference Record (IEEE Cat. No.03CH37515) 2003;5:3276–80. [Google Scholar]
- Yu Y, Chan C, Ma T, Liu Y, Gallezot JD, Naganawa M, Kelada OJ, Germino M, Sinusas AJ, Carson RE, Liu C. Event-by-Event Continuous Respiratory Motion Correction for Dynamic PET Imaging. J Nucl Med. 2016;57:1084–90. doi: 10.2967/jnumed.115.167676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.