Abstract
Background
Public biomedical data repositories often provide web-based interfaces to collect experimental metadata. However, these interfaces typically reflect the ad hoc metadata specification practices of the associated repositories, leading to a lack of standardization in the collected metadata. This lack of standardization limits the ability of the source datasets to be broadly discovered, reused, and integrated with other datasets. To increase reuse, discoverability, and reproducibility of the described experiments, datasets should be appropriately annotated by using agreed-upon terms, ideally from ontologies or other controlled term sources.
Results
This work presents “CEDAR OnDemand”, a browser extension powered by the NCBO (National Center for Biomedical Ontology) BioPortal that enables users to seamlessly enter ontology-based metadata through existing web forms native to individual repositories. CEDAR OnDemand analyzes the web page contents to identify the text input fields and associate them with relevant ontologies which are recommended automatically based upon input fields’ labels (using the NCBO ontology recommender) and a pre-defined list of ontologies. These field-specific ontologies are used for controlling metadata entry. CEDAR OnDemand works for any web form designed in the HTML format. We demonstrate how CEDAR OnDemand works through the NCBI (National Center for Biotechnology Information) BioSample web-based metadata entry.
Conclusion
CEDAR OnDemand helps lower the barrier of incorporating ontologies into standardized metadata entry for public data repositories. CEDAR OnDemand is available freely on the Google Chrome store https://chrome.google.com/webstore/search/CEDAROnDemand
Keywords: Ontology, Metadata, CEDAR, FAIR, BioPortal, NCBI, NCBO
Background
Biomedical data are increasingly being deposited in public repositories accompanied by descriptive metadata. These metadata are crucial for facilitating the discovery of the associated datasets and for reproducing the corresponding experiments. Many public data repositories provide web-based forms for researchers to enter metadata describing their datasets as part of the submission process. However, most repositories make limited use of controlled vocabularies in the metadata entry process and, as a result, metadata are often described using inconsistent terminologies [1]. This lack of standardization makes it difficult to access, find, interoperate, and reuse the datasets, and—crucially—to understand how the associated experiments were performed. Improvements are needed to make these datasets more FAIR (Findable, Accessible, Interoperable, and Reusable) [2]. The use of terms from controlled terminologies and ontologies can provide an important first step for creating FAIR metadata descriptions [3].
A wide array of ontology-based services have been developed in order to promote scientific data interoperability and reusability in biomedicine through the use of standard terminologies. These include BioPortal [4], the Ontology Lookup Service (OLS) [5], EBI Zooma [6], and NCBO Annotator and Recommender [7, 8]. In addition, data (metadata) standardization efforts have been established by different communities to ensure sufficient amount of information (metadata) be provided for reporting results in a way that facilitates reproducibility such as MIAIME (Minimum Information for Reporting Microarray Experiment) [9], MiAIRR (Minimal Information about Adaptive Immune Receptor Repertoire) [10, 11] and MIBBI (Minimum Information for Biological and Biomedical Investigations) [12]. The Center for Expanded Data Annotation and Retrieval (CEDAR) [13] has leveraged existing data standards and the ontologies available at BioPortal to develop the CEDAR Workbench with the goal of creating semantically rich metadata. A user either can create a new template (web form) or can use existing ontology-controlled templates to author standardized metadata within CEDAR Workbench. An example employing CEDAR Workbench for customized data submission is [14]. Expanding CEDAR’s approach of metadata creation outside of its environment, we have incorporated BioPortal ontologies and web services to develop a decentralized metadata authoring tool called “CEDAR OnDemand”. CEDAR OnDemand is a platform-independent program running as a web browser extension designed to help creating standardized metadata in repository-native web forms. The key advantage of this approach is that it enables users to seamlessly enter ontology-based metadata into existing web forms without requiring the individual repositories to provide these services.
Implementation
The CEDAR OnDemand script has been developed as a Google chrome browser extension [15] (a browser extension is essentially a small software program that can access contents of a web page, modify it and can enhance the functionality of a web browser). It is powered by the NCBO Annotator [7] and Recommender [8] Web services and facilitates users to suggest entry-time ontology controlled metadata to fill up web forms. After installation, the extension will appear as an icon on the chrome extension bar (upper right side of the browser). It is designed to be manually toggled on upon entry of a web form (it can be toggled off later if needed). Although CEDAR OnDemand can be programmed to be auto-activated, we used the manual activation method to minimize the system memory usage and to protect users from browser-based security attacks [16]. The extension operates in three phases (described below) that are initiated when a user visits a new web-based (metadata) entry form.
Identification of data entry fields
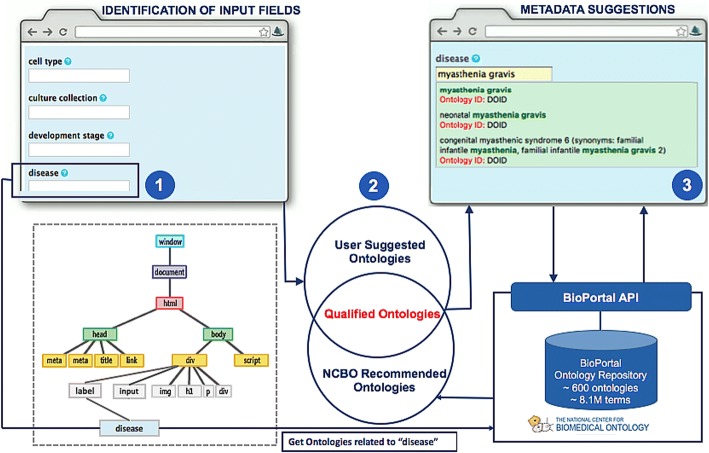
To detect data entry fields, the web page is analyzed to identify text input fields and the associated field labels (Fig. 1, left side). CEDAR OnDemand parses the content of a web page into the document object model (DOM) [17], which defines the content, structure and style of an HTML document (Fig. 1, left panel treeview). The current implementation of CEDAR OnDemand recognizes the standard INPUT fields (HTML5 and previous versions) and their associated labels (HTML5 element). The recognized fields are highlighted with light yellow color. The metadata entry of the detected input fields will be controlled by the list of ontologies chosen by the qualified ontologies.
Fig. 1.
CEDAR OnDemand Workflow. (1) CEDAR OnDemand is installed as a Google Chrome browser extension. When activated by the user (by toggling the icon), the web page in the browser will be analyzed. Users can customize a list of suggested ontologies used by CEDAR OnDemand and any point through a dialogue box with dropdown list (2) An HTML tags detection script identifies the text fields by analyzing the HTML INPUT tags and fetches their labels. Fetched labels are then passed to the NCBO to get the related ontology recommendations. This BioPortal ontology recommendation list is compared to the user suggested ontology list to find the qualified ontologies (3) CEDAR OnDemand associates the qualified ontology list to the detected input fields. Subsequently, NCBO Annotator service is invoked to match field values with the qualified set of ontologies to suggest ontology-based metadata
Ontologies recommendation algorithm
The CEDAR OnDemand ontology recommendation algorithm is designed to recommend ontologies relevant to each input field listed in a webform from the BioPortal [4] ontologies. CEDAR OnDemand takes each field label as input (as shown in Label 2 in Fig. 1) to the NCBO Recommender 2.0 service [8] to get a list of BioPortal ontologies (containing terms matching the field label). Moreover, a user can also define ontologies through a dialogue box which appears by toggling the CEDAR OnDemand extension. The CEDAR OnDemand algorithm takes the intersection of the set of user-defined ontologies and that of ontologies recommended automatically (by the NCBO recommender) to produce the set of qualified ontologies for each field. These field-specific qualified ontologies are then linked to each input field in a web form. If the intersection is an empty set, then the full user-defined list is used for as the qualified ontologies for controlling the field entry. By default, the user-defined list includes six ontologies: ChEBI Ontology [18], Human Disease Ontology (DOID) [19], Gene Ontology (GO) [20], Ontology for Biomedical Investigations (OBI) [21], Phenotypic Quality Ontology [22], Protein Ontology (PR) [23] (Fig. 1, Label 2). Not only do these ontologies cover a broad range of biological domains, but they are also ranked among the top ten by OBO Foundry in terms of their compliance to ontology best practice [24]. The user may change the default ontology list by adding/removing ontologies anytime during the metadata entry process. In its default behavior CEDAR OnDemand works fully automatically and does not require an ontology input from the user. However, customizing the default ontology list may help the user to get domain-specific metadata suggestions.
Ontology association and auto-completion of metadata
To associate ontology terms (e.g., “myasthenia gravis” from DOID) with the field entry (e.g., disease), CEDAR OnDemand matches the term(s) entered by the user with the terms defined in the qualified ontologies (Fig. 1, Label 3). This is done by invoking the NCBO Annotator web service [8] through AJAX (asynchronous JavaScript and XML) call [25]. AJAX communicates with NCBO BioPortal server [26] asynchronously (in the background) through XMLHttpRequest Object to send and retrieve the data. This asynchronous communication model of CEDAR OnDemand enables entry-time suggestion for ontology controlled metadata entry. The NCBO Annotator returns a ranked list of ontology term matches for the user to choose.
Results
We tested CEDAR OnDemand by entering metadata using the NCBI human BioSample web form1 [27]. In this use case, we first extended the user defined ontology list by adding several field-specific ontologies identified through NCBO recommender: Cell ontology (CL) [28], Cell Line Ontology (CLO) [29], NCI thesaurus NCIT [30], NCBI Taxonomy ontology NCBITAXON [31], and Uber Anatomy Ontology (UBERON) [32]. The NCBI human BioSample web form contains twenty-one text input fields. CEDAR OnDemand suggested eight ontologies based on the input fields in the NCBI human BioSample web form. After intersection with the user defined ontologies (extended list), the final ontology list recommended by the CEDAR OnDemand includes: NCI thesaurus NCIT [30], Cell Ontology [28], Cell Line Ontology [33], (UBERON) [32], Human disease Ontology [19], Gene Ontology (GO) [20] and OBI [21] (See Table 1). Controlled vocabularies do not make sense for some text fields, such as “Sample Name”, “Age” and “isolate”. Therefore, CEDAR OnDemand allows the user to override ontology suggestions for all fields with the user-defined entries. CEDAR OnDemand provides the field's specific metadata suggestion controlled by ontologies. Thus, users are no longer entering free text but they are instead using standardized ontology terms. An auto-completion feature is provided at runtime through a drop-down list. As an example (Fig. 1, Label 3), CEDAR OnDemand suggests “myasthenia gravis” as controlled term (defined in DOID) for the disease field.
Table 1.
CEDAR OnDemand Qualified Ontologies for each NCBI BioSample Field
| Field names | Qualified ontologies |
|---|---|
| Sample Name | Ontology for Biomedical Investigations (OBI), National Cancer Institute Thesaurus (NCIT) |
| Organism | National Cancer Institute Thesaurus (NCIT) |
| Isolate | National Cancer Institute Thesaurus (NCIT) |
| Age | National Cancer Institute Thesaurus (NCIT) |
| Biomaterial Provider | National Cancer Institute Thesaurus (NCIT) |
| Tissue | Uber Anatomy Ontology (UBERON), Ontology for Biomedical Investigations (OBI), National Cancer Institute Thesaurus (NCIT), Cell Ontology (CL), Cell Line Ontology (CLO) |
| Cell line | Cell Line Ontology (CLO), Ontology for Biomedical Investigations (OBI), National Cancer Institute Thesaurus (NCIT) |
| Cell subtype | Cell Ontology (CL), Gene Ontology (GO), National Cancer Institute Thesaurus (NCIT) |
| Cell type | Cell Ontology (CL), Cell Line Ontology (CLO), National Cancer Institute Thesaurus (NCIT) |
| Culture Collection | National Cancer Institute Thesaurus (NCIT) |
| Development Stage | Gene Ontology (GO), National Cancer Institute Thesaurus (NCIT) |
| Disease | Human Disease Ontology (DOID), Cell Line Ontology (CLO), Ontology for Biomedical Investigations (OBI), National Cancer Institute Thesaurus (NCIT) |
| Disease Stage | Human Disease Ontology (DOID), Cell Line Ontology (CLO), Ontology for Biomedical Investigations (OBI), National Cancer Institute Thesaurus (NCIT) |
| Ethnicity | National Cancer Institute Thesaurus (NCIT) |
| Health state | National Cancer Institute Thesaurus (NCIT) |
| Karyotype | National Cancer Institute Thesaurus (NCIT) |
| Phenotype | Ontology for Biomedical Investigations (OBI), National Cancer Institute Thesaurus (NCIT) |
| Population | Ontology for Biomedical Investigations (OBI) |
| Race | National Cancer Institute Thesaurus (NCIT) |
| Sample type | National Cancer Institute Thesaurus (NCIT) |
| Treatment | Ontology for Biomedical Investigations (OBI), National Cancer Institute Thesaurus (NCIT) |
Field Names column lists the Human Sample attributes of NCBI BioSample. Qualified Ontologies are the ontologies which CEDAR OnDemand algorithm recommends
Discussion
Although many public repositories, such as those run by the NCBI, provide easy-to-use tools and interfaces for entering and querying metadata, scientists who upload their datasets are generally not constrained to use standard terminologies when they define the necessary metadata. As a result, metadata are often described using inconsistent terminologies, limiting scientists’ ability to access, find, interoperate and reuse the datasets and to understand how the experiments were performed. Scientific data analysis or mining [34] often requires multiple datasets to be integrated within a single repository or across multiple repositories. Such integration would be easier if the datasets and their metadata were identified globally, described using standardized terminologies, and available in a standardized machine readable format. A common semantic schema [35] among different studies and data sources can be achieved by associating relevant ontology classes with each study's metadata. Despite the free availability of ontology resources [26, 36], only few repositories (e.g., IEDB -The immune epitope database [37]) and frameworks (e.g., SEBI-Semantic enrichment of biomedical Images [38, 39]) have integrated ontologies or structured controlled lists within their framework to collect standardized metadata.
PubMed uses Medical Subject Headings [40] as a controlled vocabulary for indexing and searching biomedical literature. Meshable [41] highlights an important issue in PubMed literature searching. In PubMed, biologists can use MeSH terms as queries to get the precise results. However, these are rarely used, and there is no convenient way to author standardized MeSH terms as queries. Through CEDAR OnDemand, users can suggest MeSH Ontology [42] replacing the default user-defined list and can get entry-time query suggestions from the MeSH controlled vocabulary.
CEDAR OnDemand has the potential to improve the FAIRness and overall quality of metadata to the available repositories. However, the current infrastructure has some limitations. For instance, the diversity in the input field coding schemes (e.g., <div, <inputfield and < text) limits the HTML tags detection script when there are custom-build tags are used to define the input fields. Our script identifies the standard HTML5 tags, Label was introduced in HTML5. However, input tag was present at the very beginning (i.e., <input type = “text”) to represent an input field. Though CEDAR OnDemand works with web forms designed in HTML4 or with older versions, the ontology recommendation algorithm does not make use of the field associated (labels) information for ontology recommendation in these cases, relying instead on the users suggested ontology list.
A key component of CEDAR OnDemand is the ability to analyze context and suggest appropriate ontologies for each particular field. The current qualified ontology selection process relies on NCBO ontology recommender service [8] and the user’s suggested ontology list. We have proposed this scheme as the NCBO recommended ontology list can be very long, and may not always recommend ontologies that are specific to a user’s particular domain. Allowing users to customize a set of suggested ontologies helps to address both these issues. Ideally, using the field context along with NCBO recommender would be able to identify and rank all of the relevant ontologies. In practice, it can be difficult to get sufficient context just from the web page and text surrounding a field. Even if enough context is present, it may be technically difficult to extract. For example, the web interfaces for some repositories have been designed using older versions of HTML and some with custom HTML tags.
We have tested CEDAR OnDemand with the latest Chrome version (59.0.3054) on Mac and Windows. The core of CEDAR OnDemand is a based on Javascript and should work with any version of chrome browser with its default setup on Windows, Mac OS and Linux operating systems. We are exploring the possibility of supporting other types of browsers (e.g., Firefox and Microsoft Edge).
Conclusions
CEDAR OnDemand is a chrome browser extension that enables users to seamlessly enter ontology-controlled metadata using existing web-based submission forms provided by metadata repositories. The use of controlled vocabularies for entering metadata can help improve the quality of metadata submitted to repositories and ultimately contributes to the creation of FAIR data.
Availability and requirements
Availability: https://chrome.google.com/webstore/search/CEDAROnDemand
Code Availability: https://github.com/ahmadchan/CEDAROnDemand
Project name: CEDAR OnDemand.
Operating system(s): Operating system independent works within web browser.
Programming language: Javascript.
License: GPL.
Any restrictions to use by non-academics: none.
Acknowledgements
We acknowledge the BioPortal and CEDAR team for their valuable suggestions during this research work.
Funding
This work was supported by grant U54 AI117925 awarded by the National Institute of Allergy and Infectious Diseases through funds provided by the trans-NIH Big Data to Knowledge (BD2K) initiative (https://commonfund.nih.gov/bd2k).
Authors’ contributions
Study conception and design: SACB, KHC, SHK, JB, MAM. Code Implementation: SACB. Validated and interpreted the results: SACB, JB, MOC, DB, ALE. Drafting of manuscript: SACB, SHK, KHC. Critical revision: SACB, MMR. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Contributor Information
Syed Ahmad Chan Bukhari, Email: ahmad.chan@yale.edu.
Marcos Martínez-Romero, Email: marcosmr@stanford.edu.
Martin J. O’ Connor, Email: sunid@stanford.edu
Attila L. Egyedi, Email: attila.egyedi@stanford.edu
Debra Willrett, Email: willrett@stanford.edu.
John Graybeal, Email: jgraybeal@stanford.edu.
Mark A. Musen, Email: musen@stanford.edu
Kei-Hoi Cheung, Email: kei.cheung@yale.edu.
Steven H. Kleinstein, Email: steven.kleinstein@yale.edu
References
- 1.Gonçalves RS, O’Connor MJ, Martínez-Romero M, Graybeal J, Musen MA: Metadata in the BioSample Online Repository are Impaired by Numerous Anomalies. arXiv [cs.DB] 2017.
- 2.Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJG, Groth P, Goble C, Grethe JS, Heringa J, t Hoen PAC, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone S-A, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shadbolt N, Berners-Lee T, Hall W. The semantic web revisited. IEEE Intell Syst. 2006;21:96–101. doi: 10.1109/MIS.2006.62. [DOI] [Google Scholar]
- 4.Whetzel PL, NCBO Team NCBO Technology: Powering semantically aware applications. J Biomed Semantics. 2013;4(Suppl 1):S8. doi: 10.1186/2041-1480-4-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ison J, Kalas M, Jonassen I, Bolser D, Uludag M, McWilliam H, Malone J, Lopez R, Pettifer S, Rice P. EDAM: an ontology of bioinformatics operations, types of data and identifiers, topics and formats. Bioinformatics. 2013;29:1325–1332. doi: 10.1093/bioinformatics/btt113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ZOOMA text annotations tool. http://www.ebi.ac.uk/spot/zooma/.
- 7.Jonquet C, Shah NH, Youn CH, Callendar C, Storey M-A, Musen MA. NCBO annotator: semantic annotation of biomedical data. International Semantic Web Conference, Poster and Demo session. 2009. https://pdfs.semanticscholar.org/9956/898d4012bb87374931085a643eb06b18ac9f.pdf.
- 8.Martínez-Romero M, Jonquet C, O’Connor MJ, Graybeal J, Pazos A, Musen MA. NCBO Ontology Recommender 2.0: an enhanced approach for biomedical ontology recommendation. J Biomed Semantics. 2017;8:21. doi: 10.1186/s13326-017-0128-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brazma A. Minimum information about a microarray experiment (MIAME)--successes, failures, challenges. Sci World J. 2009;9:420–423. doi: 10.1100/tsw.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubelt F, Busse CE, Bukhari SAC, Bürckert J-P, Mariotti-Ferrandiz E, Cowell LG, Watson CT, Marthandan N, Faison WJ, Hershberg U, Laserson U, Corrie BD, Davis MM, Peters B, Lefranc M-P, Scott JK, Breden F. AIRR community, Luning Prak ET, Kleinstein SH: adaptive immune receptor repertoire community recommendations for sharing immune-repertoire sequencing data. Nat Immunol. 2017;18:1274–1278. doi: 10.1038/ni.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breden F, Luning Prak ET, Peters B, Rubelt F, Schramm CA, Busse C, Vander Heiden JA, Christley S, Bukhari SAC, Thorogood A, Matsen F, Wine Y, Laserson U, Klatzmann D, Douek D, Lefranc M-P, Collins AM, Bubela T, Kleinstein S, Watson CT, Cowell LG, Scott JK, Kepler TB. Perspective: Reproducibility and Reuse of Adaptive Immune Receptor Repertoire Data. Front Immunol. 2017;8. [DOI] [PMC free article] [PubMed]
- 12.Kettner C, Field D, Sansone S-A, Taylor C, Aerts J, Binns N, Blake A, Britten CM, de Marco A, Fostel J, Gaudet P, González-Beltrán A, Hardy N, Hellemans J, Hermjakob H, Juty N, Leebens-Mack J, Maguire E, Neumann S, Orchard S, Parkinson H, Piel W, Ranganathan S, Rocca-Serra P, Santarsiero A, Shotton D, Sterk P, Untergasser A, Whetzel PL. Meeting report from the second “minimum information for biological and biomedical investigations” (MIBBI) workshop. Stand Genomic Sci. 2010;3:259–266. doi: 10.4056/sigs.147362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musen MA, Bean CA, Cheung K-H, Dumontier M, Durante KA, Gevaert O, Gonzalez-Beltran A, Khatri P, Kleinstein SH, O’Connor MJ, Pouliot Y, Rocca-Serra P, Sansone S-A, Wiser JA. CEDAR team: the center for expanded data annotation and retrieval. J Am Med Inform Assoc. 2015;22:1148–1152. doi: 10.1093/jamia/ocv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bukhari SAC, O'Connor MJ, Graybeal J, Musen MA, Cheung K-H, Kleinstein SH. Leveraging the CEDAR Workbench for Ontology-linked Submission of Adaptive Immune Receptor Repertoire Data to the Sequence Read Archive (SRA). 10.6084/m9.figshare.4244126.v3.
- 15.Mehta P. Introduction to Google Chrome Extensions. In: Creating Google Chrome Extensions: Apress. New Delhi: Spinger; 2016. p. 1–33. https://link.springer.com/content/pdf/10.1007/978-1-4842-1775-7.pdf.
- 16.Shital P. Web browser security: different attacks detection and prevention techniques. IJCAI. 2017;170:35–41. doi: 10.5120/ijca2017914938. [DOI] [Google Scholar]
- 17.Wood L, Nicol G, Robie J, Champion M, Byrne S. Document object model (DOM) level 3 core specification. MIT, INRIA, KEO: W3C; 2000.
- 18.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schriml LM, Arze C, Nadendla S, Chang Y-WW, Mazaitis M, Felix V, Feng G, Kibbe WA. Disease ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012;40:D940–D946. doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R. Gene ontology consortium: the gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bjoern P and OBI consortium. Ontology for Biomedical Investigations. Available from Nature Precedings; 2009.
- 22.Quality Control in Phenotypic Analysis by Flow Cytometry. In: Robinson JP, Darzynkiewicz Z, Dobrucki J, Hyun WC, Nolan JP, Orfao A, Rabinovitch PS, editors. Current Protocols in Cytometry. Hoboken: Wiley; 2001. p. 26:13.
- 23.Natale DA, Arighi CN, Barker WC, Blake J, Chang T-C, Hu Z, Liu H, Smith B, Wu CH. Framework for a protein ontology. BMC Bioinformatics. 2007;8(Suppl 9):S1. doi: 10.1186/1471-2105-8-S9-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Consortium OBI, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone S-A, Scheuermann RH, Shah N, Whetzel PL, Lewis S. The OBO foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulson LD. Building rich web applications with Ajax. Computer. 2005;38(10):14-7.
- 26.Noy NF, Shah NH, Whetzel PL, Dai B, Dorf M, Griffith N, Jonquet C, Rubin DL, Storey M-A, Chute CG, Musen MA. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009;37:W170–W173. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, Karsch-Mizrachi I, Kimelman M, Pruitt KD, Resenchuk S, Tatusova T, Yaschenko E, Ostell J. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012;40:D57–D63. doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meehan TF, Masci AM, Abdulla A, Cowell LG, Blake JA, Mungall CJ, Diehl AD. Logical development of the cell ontology. BMC Bioinformatics. 2011;12:6. doi: 10.1186/1471-2105-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarntivijai S, Lin Y, Xiang Z, Meehan TF, Diehl AD, Vempati UD, Schürer SC, Pang C, Malone J, Parkinson H, Liu Y, Takatsuki T, Saijo K, Masuya H, Nakamura Y, Brush MH, Haendel MA, Zheng J, Stoeckert CJ, Peters B, Mungall CJ, Carey TE, States DJ, Athey BD, He Y. CLO: the cell line ontology. J Biomed Semantics. 2014;5:37. doi: 10.1186/2041-1480-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar A, Smith B. Artificial Intelligence in Medicine. Berlin, Heidelberg: Springer; 2005. Oncology ontology in the NCI thesaurus; pp. 213–220. [Google Scholar]
- 31.Federhen S. The NCBI taxonomy database. Nucleic Acids Res. 2012;40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mungall CJ, Torniai C, Gkoutos GV, Lewis SE, Haendel MA. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012;13:R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarntivijai S, Xiang Z, Meehan TF, Diehl AD, Vempati U, Schürer SC, Pang C, Malone J, Parkinson HE, Athey BD. Others: cell line ontology: redesigning the cell line knowledgebase to aid integrative translational informatics. ICBO. 2011;833:25–32. [Google Scholar]
- 34.Kamath C. Scientific data mining: a practical perspective. SIAM; 2009. https://epubs.siam.org/doi/book/10.1137/1.9780898717693.
- 35.Tandareanu N, Ghindeanu M. Properties of derivations in a semantic Schema. Annals of the University of Craiova-Mathematics and Computer Science Series. 2006;33:147–153. [Google Scholar]
- 36.Hartmann J, Palma R, Gómez-Pérez A. Handbook on Ontologies. Berlin, Heidelberg: Springer; 2009. Ontology repositories; pp. 551–571. [Google Scholar]
- 37.Vita R, Overton JA, Greenbaum JA, Sette A, Peters B. Query enhancement through the practical application of ontology: the IEDB and OBI. J Biomed Semantics. 2013;4(Suppl 1):S6. doi: 10.1186/2041-1480-4-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bukhari SAC, Krauthammer M, Baker CJO. SEBI: an architecture for biomedical image discovery, interoperability and reusability based on semantic enrichment. In: SWAT4LS: Citeseer. Berlin: 7th International Workshop on Semantic Web Applications and Tools for life sciences; 2014.
- 39.Bukhari SAC. Semantic enrichment and similarity approximation for biomedical sequence images. Canada: University of New Brunswick (Canada) and ProQuest Dissertations Publishing; 2017.
- 40.Lipscomb CE. Medical subject headings (MeSH) Bull Med Libr Assoc. 2000;88:265–266. [PMC free article] [PubMed] [Google Scholar]
- 41.Kim S, Yeganova L, Wilbur WJ. Meshable: searching PubMed abstracts by utilizing MeSH and MeSH-derived topical terms. Bioinformatics. 2016;32:3044–3046. doi: 10.1093/bioinformatics/btw331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beissinger TM, Morota G. Medical subject heading (MeSH) annotations illuminate maize genetics and evolution. Plant Methods. 2017;13:8. doi: 10.1186/s13007-017-0159-5. [DOI] [PMC free article] [PubMed] [Google Scholar]