Abstract
Background
Esophageal squamous cell carcinomas (ESCC) is the fourth most lethal cancer in China. Previous studies reveal several highly conserved mutational processes in ESCC. However, it remains unclear what are the true regulators of the mutational processes.
Results
We analyzed the somatic mutational signatures in 302 paired whole-exome sequencing data of ESCC in a Chinese population for potential regulators of the mutational processes. We identified three conserved subtypes based on the mutational signatures with significantly different clinical outcomes. Our results show that patients of different subpopulations of Chinese differ significantly in the activity of the “NpCpG” signature (FDR = 0.00188). In addition, we report ZNF750 and CDC27, of which the somatic statuses and the genetic burdens consistently influence the activities of specific mutational signatures in ESCC: the somatic ZNF750 status is associated with the AID/APOBEC-related mutational process (FDR = 0.0637); the somatic CDC27 copy-number is associated with the “NpCpG” (FDR = 0.00615) and the AID/APOBEC-related mutational processes (FDR = 8.69 × 10− 4). The burdens of germline variants in the two genes also significantly influence the activities of the same somatic mutational signatures (FDR < 0.1).
Conclusions
We report multiple factors that influence the mutational processes in ESCC including: the subpopulations of Chinese; the germline and somatic statuses of ZNF750 and CDC27 and exposure to alcohol and tobacco. Our findings based on the evidences from both germline and somatic levels reveal potential genetic regulators of the somatic mutational processes and provide insights into the biology of esophageal carcinogenesis.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4906-4) contains supplementary material, which is available to authorized users.
Keywords: Esophageal squamous cell carcinomas, ZNF750, CDC27, Mutational signature, Genetic burden
Background
Esophageal cancer (EC) is the second most common gastrointestinal cancer worldwide. EC causes 400,000 deaths annually, which accounts for 4.9% of total deaths from cancer [1]. EC consists of two major histological types, esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC). Each disease is characterized by distinct pathological and genomic features [2]. The incidence rates of EAC and ESCC differ significantly among different regions. The burden of EAC incidence is higher in Northern and Western Europe [3], while the burden of ESCC incidence is higher in South-Eastern and Central Asia (79%). 53% of global cases of ESCC (210,000 cases per year) are diagnosed in China, which makes it the fourth most lethal cancer in the country. The difference in the incidence rates is attributed to diverse epidemiological and genetic factors [4].
Cancers accumulate various somatic alternations in the genome. The somatic landscape of the tumor genome is a result of different mutational processes that occur throughout the course of the disease [5–7]. The mutational processes in cancers are driven by various determinants, the microenvironment, risk factors and genetic variations. The biological background of the mutational processes in diverse cancer types have been revealed by recent studies, such as APOBEC-induced deamination, deamination of methylated cytosine, tobacco and alcohol exposure and so on [8–10]. The mutational processes are represented by distinct patterns of frequencies of trinucleotide sequences surrounding the base of substitution, which is also known as the somatic mutational signatures [11]. The relative activities of the mutational signatures surrogate for the rate of the corresponding mutational processes. Many studies derive algorithms to detect mutational signatures from genome or exome sequences of cancer [12–14]. These algorithms are usually based on decomposition of the mutational profiles. The accuracy and power of the derivation are subject to the complexity of the model as well as the sample size. Recent methods, such as the “probabilistic mutation signature”, improve the reliability of the discovery of the mutational signatures from small sample by reducing the number of parameters [13, 15].
Notably, more and more studies show evidences that the somatic mutational processes in cancers are associated with the somatic or germline statuses of certain genes [12, 16]. For instance, the overall somatic mutational burden in melanoma is associated with germline MC1R status [17]; and in breast cancer it is associated with the germline RAD51B status (rs2588809) [18]. At the somatic level, the activity of the signature 5 in the Catalogue Of Somatic Mutations In Cancer (COSMIC) is associated with the somatic ERCC2 status in urothelial tumors [19]; and the activity of the APOBEC-related mutational signature in ESCC is associated with the somatic PIK3CA and ZNF750 statuses [20, 21]; Plus, specific mutational signatures are used to predict BRCA1 and BRCA2 deficiency in breast cancer [22]. These studies suggest that both germline and somatic alterations can drive the mutational processes in cancers and the driver genes may play a decisive role in the development of the disease.
In this study, we used paired whole-exome sequencing (WES) data to identify the regulators of the somatic mutational processes in ESCC in a Chinese population by combining evidences from both germline and somatic levels [20, 23–25]. We assessed the association between the activities of the somatic mutational processes and various gene sets including the significantly mutated genes (SMGs), the genes with somatic copy-number alterations (SCNAs) as well as the GWAS risk loci of ESCC. We considered the effects from the germline and somatic levels to suggest several potential determinants of the somatic mutational processes; and provide insights into esophageal carcinogenesis and inform the future diagnosis and therapy.
Results
Somatic mutational signatures
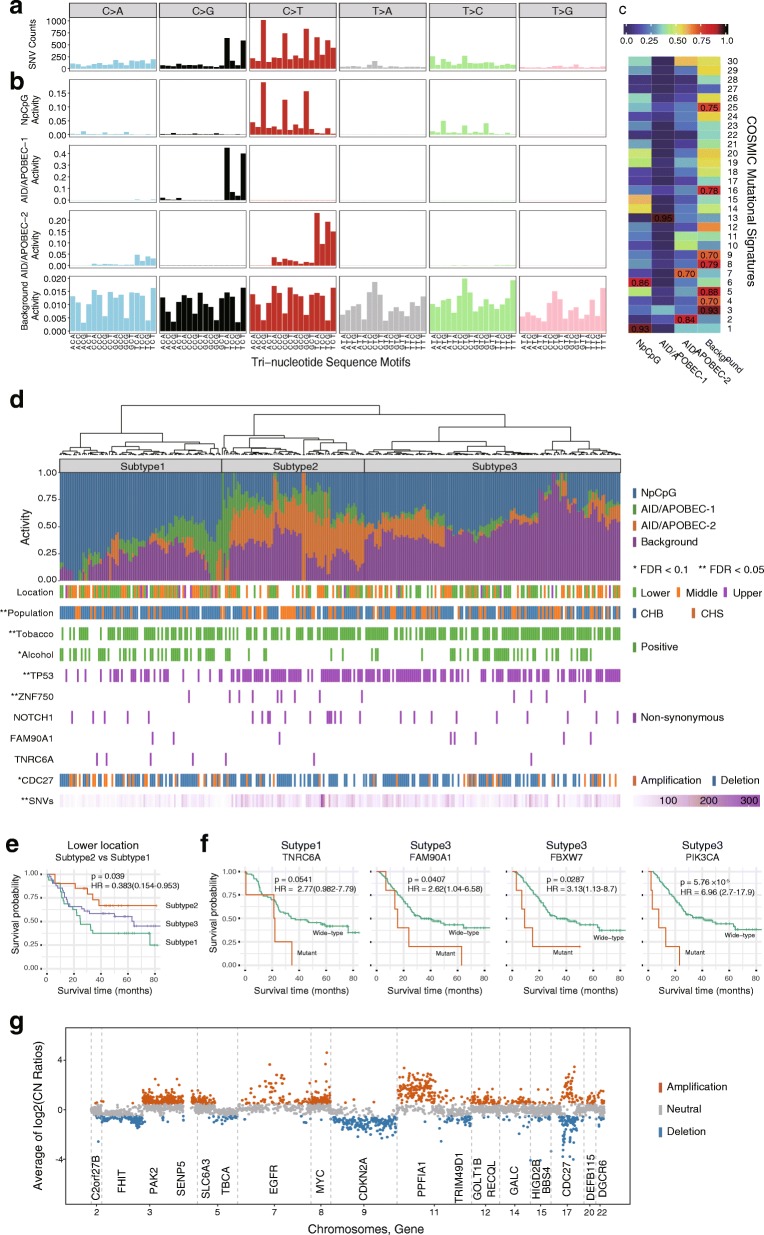
We identified 13,854 single nucleotide variants (SNVs) and 2274 insertions and deletions (InDels) from 302 paired exonic sequences of ESCC. The median rate of the mutations is 1.11 per megabase (range 0.03–9.21; Additional file 1: Table S1 and Additional file 2: Figure S1). Of all the SNVs and InDels, 73.5% (7331) are non-synonymous variants; 53.2% (8586) are annotated in the COSMIC v79 [26]. The six subtypes of base substitutions (C > A, C > G, C > T, T > A, T > C and T > G) are unevenly represented in the SNVs. C > T is the most common substitution in ESCC (6391, 39.6%), followed by C > G (2,234, 13.9%; Fig. 1a).
Fig. 1.
Somatic mutational signatures in 302 ESCC. a Frequencies of 96 subtypes of base substitutions of SNV identified from the exomes of 302 ESCC. b Four somatic mutational signatures, “NpCpG”, “AID/APOBEC-1”, “AID/APOBEC-2” and “Background” retrieved from the mutational profiles of SNVs. c Comparison of the four somatic mutational signatures from ESCC with the known mutational signatures (No.1 to 30) in the COSMIC. The similarity measures labeled in the grids are based on “Cosine similarity”. d Subtypes of ESCC based on the clustering of the activities of the mutational signatures of the ESCC samples. The activities of each mutational signatures in each sample are denoted by the colored-bars below the dendrogram. The clinical and molecular features that are significantly associated with the subtypes are labeled beneath the bar plot. e The outcome of the three subtypes of ESCC differ in tumors in lower esophagus. The Kaplan-Meier curves are based on the fraction of overall survival in lower esophagus. The P values and Hazard Ratio (95% confidence interval) are estimated using Cox-regression. f The SMGs are predictive in subtype 1 and 3. Kaplan Meier curves are based on the somatic TNRC6A, FAM90A1, FBXW7 and PIK3CA mutational statuses. The P values and Hazard Ratio (95% confidence interval) are estimated using Cox-regression. g Log2 copy-number ratio of the genomic segments in ESCC. The segments of significant copy number alterations are shown in color (orange: amplification, blue: deletion) with the affected genes labeled beneath. A copy-number amplification was defined if the log2 copy number ratio is above 0.5; and the deletion if the value is below − 0.5
We retrieved four highly conserved somatic mutational signatures from the mutational profiles (Fig. 1b). The signatures are highly comparable to the known ones in the COSMIC database based on the cosine similarity (CS; Fig. 1c) [16, 26]. The “NpCpG” signature (characterized by C > T at NpCpG trinucleotide), which has been previously reported in ESCC, is highly similar to the COSMIC signature 1 (deamination of 5-methycytosine; CS = 0.93) and signature 6 (defective DNA mismatch repair; CS = 0.86) [20]. The “AID/APOBEC-1” signature (C > G at TpCpN trinucleotide) and “AID/APOBEC-2” signature (C > T at TpCpN trinucleotide) match the COSMIC signature 13 (CS = 0.95) and signature 2 (CS = 0.84), respectively. Both signatures are driven by AID/APOBEC family of cytidine deaminases activity [16, 26]. Moreover, “AID/APOBEC-2” signature is associated with COSMIC signature 7 (CS = 0.70), which presents in multiple squamous cancers. Finally, the “Background” signature represents the base level mutational processes in ESCC which correlates strongly to COSMIC signature 3 (the failure of double-strand-break repair by homologous recombination; CS = 0.93), signature 4 (tobacco exposure; CS = 0.70), signature 9 (the activity of AID during somatic hypermutation; CS = 0.70) and several other signatures of unknown background (COMIC signature 5, 8, 16, 25) [13].
We identified three conserved subtypes of ESCC based on the relative activities of the mutational signatures. Subtype 1 (n = 85) is characterized by the “NpCpG” signature activity and the relatively low burden of somatic SNVs (FDR = 3.85 × 10− 9; Fig. 1d). CDC27 somatic amplification is significantly enriched in this subtype (n = 22, FDR = 0.0682). Subtype 2 (n = 75) is characterized by the “AID/APOBEC-2” signature activity and patients who do not drink alcohol. The non-synonymous mutations of ZNF750 (n = 8, FDR = 0.0471; Fig. 1d) are overrepresented in this subtype. Finally, subtype 3 (n = 134) is characterized by the “Background” signature and smoking patients (n = 103, FDR = 0.0169; Fig. 1d). Besides, subtype 3 also significantly enriches TP53 mutations (n = 89, FDR = 7.38 × 10− 7; Fig. 1d).
The clinical outcomes of the three subtypes are also different. In the lower esophagus, subtype 2 show significantly higher overall survival rate than subtype 1 (HR = 0.383, 95% CI = 0.154–0.953, P = 0.039; Fig. 1e).
Significantly altered genes in ESCC
We identified 12 SMGs (FDR < 0.1; Additional file 2: Figure S1). We compared the SMGs to known cancer genes annotated in the COSMIC as well as other published studies, most of the SMGs we identified (TP53, NOTCH1, FAT1, ZNF750, RB1, PTCH1, PIK3CA, FBXW7, NFE2L2 and CDKN2A) are consistent to the ones reported previously in ESCC [20, 23–25]. Our results suggest two new candidate genes: FAM90A1 (family with sequence similarity 90, member A1, MIM: 613041, FDR < 1 × 10− 10) and TNRC6A (trinucleotide repeat containing 6A, MIM: 610739, FDR = 9 × 10− 10). For FAM90A1, we report an in-frame insertion (c.1031_1032insCGT [p.T344_S345insV]), which presents in 8 patients. And for TNRC6A, we report an in-frame deletion (c.333_344del12 [p.P115_Q118delPQPQ]) which presents in seven patients (Additional file 3: Table S2). It’s also noteworthy that both InDels are previously annotated in other TCGA cancer cohorts but not in ESCC [27–29].
In addition, the SMGs are selectively predictive in the subtypes of ESCC defined by the mutational signatures. For example, TNRC6A mutation is suggestively associated with poor overall survival in subtype 1 (HR = 2.77, 95% CI = 0.982–7.79, P = 0.0541); whereas the somatic statuses of FAM90A1 (HR = 2.62, 95% CI = 1.04–5.58, P = 0.0407), FBXW7 (HR = 3.13, 95% CI = 1.13–8.7, P = 0.0281) and PIK3CA (HR = 6.96, 95% CI = 2.7–17.9, P = 5.76 × 10− 5) are significantly predictive in subtype 3 (Fig. 1f).
On a different note, we identified 76 regions of significant somatic copy number alterations (SCNAs, FDR < 0.01; Additional file 4: Table S3) in ESCC, of which 39 are amplifications; 37 are deletions/losses. These regions include not only known SCNA events in ESCC (7 amplifications and 23 deletions) and ECA (6 amplifications and 6 deletions) [20, 24, 25, 30–33], but also 40 new events such as deletions in 5q14.1 (57.6%) and 17q21.32 (50.0%), amplifications in 10q11.21 (43.0%) and 17p11.1 (40.4%). We retrieved 20 genes located within the regions of the SCNAs, such as CDKN2A (9p21.3; FDR = 3.76 × 10− 92), MYC (8q24.21; FDR = 9.34 × 10− 54) and FHIT (3p14.2; FDR = 5.28 × 10− 28). We noticed that CDC27 (17q21.32) is both amplified in 15.90% (n = 48; FDR = 2.68 × 10− 10) and deleted in 51.6% (n = 156; FDR = 9.05 × 10− 22) of the patients (Fig. 1g; Additional file 5: Table S4).
We retrieved the protein-protein interaction (PPI) networks for both the SMGs (MutSigCV p < 0.05) and the SCNA-related genes, from which we identified 4 subnetworks undergoing significant somatic modification (p < 0.05; Additional files 2 and 6: Figures S1 and S2). The subnetworks significantly enrich for the KEGG pathways of cell cycle (FDR = 2.03 × 10− 9), P53 signaling (FDR = 4.10 × 10− 5), NOTCH signaling (FDR = 0.00163) and many other tumor related pathways (Additional file 7: Table S5a). Plus, the genes in the subnetworks significantly overrepresent DNA motifs of known transcription factor binding sites such as CEBPD (FDR = 0.00991), CEBPA (FDR = 0.00991) and many others (Additional file 7: Table S5b).
Finally, we report eight rarely mutated genes (mutational frequency < 2%), including CUL3, PTEN, RBPJ, EIF2S2, WAC, ANAPC10, CD7 and S100A2 (Additional files 2 and 6: Figures S1 and S2).
Risk factors associated with somatic mutational processes in ESCC
Our results show that the activity of “NpCpG” signature is significantly higher in the subpopulation of “Han Chinese in Beijing, China” (CHB) than those from the “Han Chinese South” (CHS; FDR = 0.00188); the activity of the “Background” signature is significantly higher in CHS than in CHB (FDR = 0.0108; Fig. 2a and b, Additional file 8: Figure S3). As for specific subtypes of substitutions, the frequency of C > A is higher in CHS (FDR = 0.00119) whereas the T > C frequency is higher in CHB (FDR = 0.00154; Fig. 2c and d). These results suggest that the population-genetical background strongly influences somatic mutational processes in ESCC.
Fig. 2.
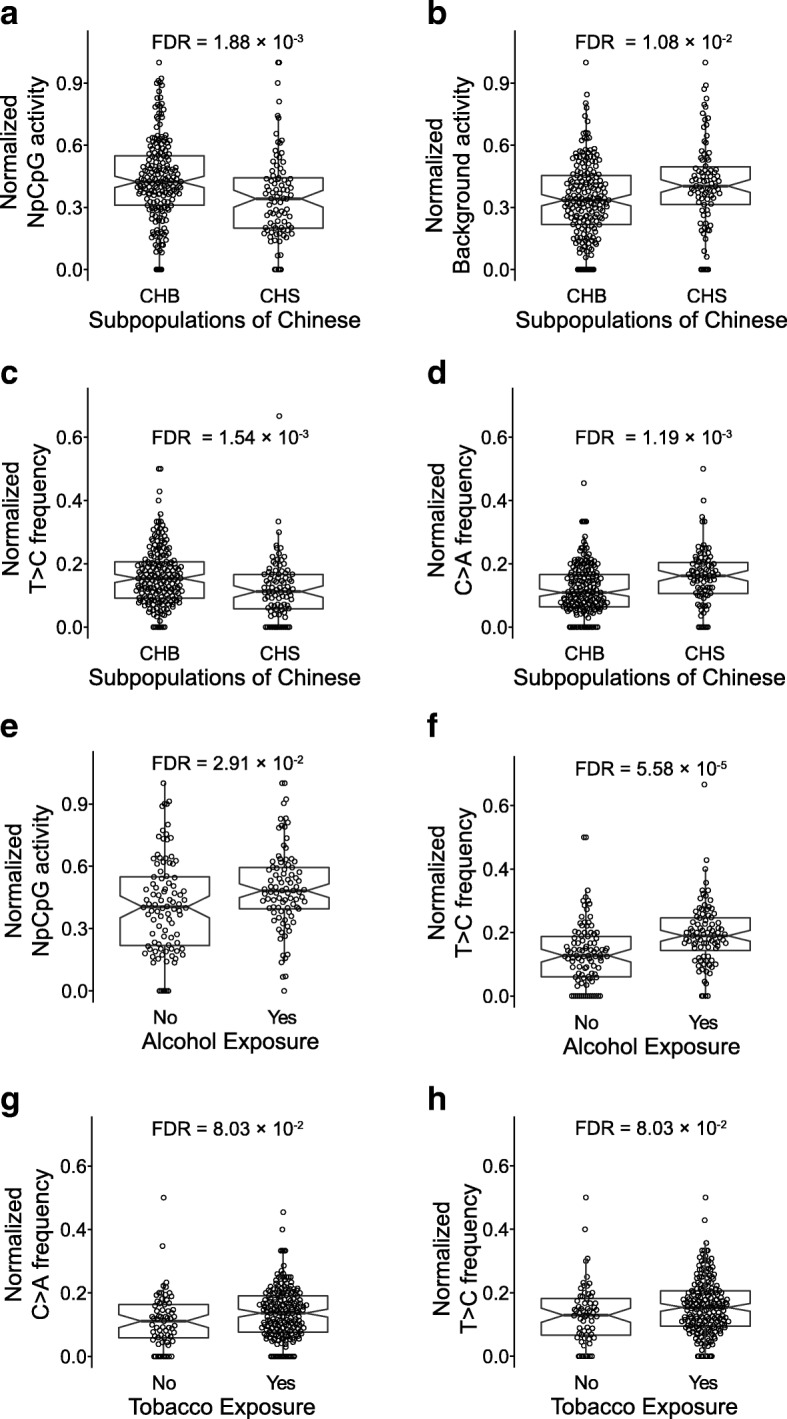
The mutational processes in ESCC are associated with subpopulations of Chinese, tobacco and alcohol exposure. Comparison of the activities of the “NpCpG” signature (a), the “Background” signature (b) and the frequency of C > A (c) and T > C (d) substitutions in CHBs and CHSs. The subpopulations are determined based on the reference populations of CHS and CHB from the TGP. Comparison of the activities of the “NpCpG” mutational signatures (e) and the frequencies of T > C substitutions (f) against the alcohol exposure. And comparison of the frequencies of C > A substitutions (g) and the frequencies of T > C substitutions (h) with tobacco exposure. The FDR is based on the adjusted Wilcoxon rank-sum test P values
We further assessed the association between other risk factors and the activity of the mutational signatures. As results, exposure to alcohol is strongly associated with higher frequency of T > C substitution (FDR = 5.58 × 10− 5) as well as the activity of the “NpCpG” signature (FDR = 0.0291; Fig. 2e and f). On the other hand, exposure to tobacco moderately associates with higher frequency of C > A (FDR = 0.0803) and T > C substitution (FDR = 0.0803; Fig. 2g and h).
Genetic regulators of the mutational signatures in ESCC
The identification of the driver genes is key to understand the mutational processes in cancers. Here we evaluated the gene sets aforementioned in ESCC for the effects on the mutational processes.
Somatic level
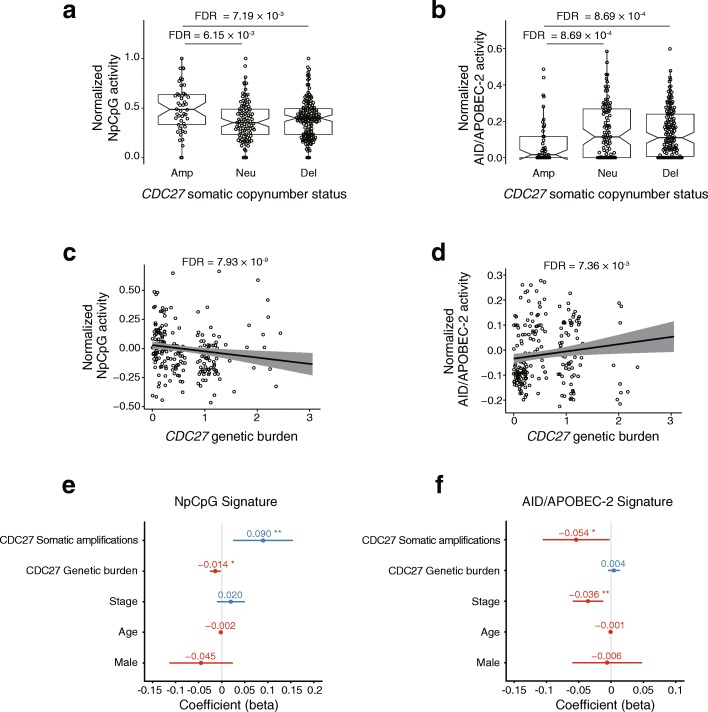
We find that the activity of the “NpCpG” signature increases in tumors carrying non-synonymous somatic PTCH1 mutations (FDR = 0.0895; Fig. 3a); but decreases in tumors with non-synonymous somatic mutations in TP53 (FDR = 1.67 × 10− 4; Fig. 3c) and ZNF750 (FDR = 0.00557; Fig. 4a). Moreover, the activity of the “AID/APOBEC-2” signature activity significantly increases with the somatic mutations of FBXW7 (FDR = 0.0283; Fig. 3b), PIK3CA (FDR = 0.0637; Fig. 3b), TP53 (FDR = 2.28 × 10− 9; Fig. 3d) and ZNF750 (FDR = 0.0637; Fig. 4b). When it comes to specific subtypes of substitutions, the mutational statuses of 5 SMGs (ZNF750, TP53, FAT1, FBXW7 and PIK3CA) are significantly associated with the increased overall burden of SNVs (FDR < 0.1; Additional file 9: Figure S4a). In particular, the somatic TP53 status is significantly correlated with high frequency of C > A, C > T, T > A and low frequency of T > C substitutions (FDR < 0.1; Additional file 9: Figure S4b).
Fig. 3.
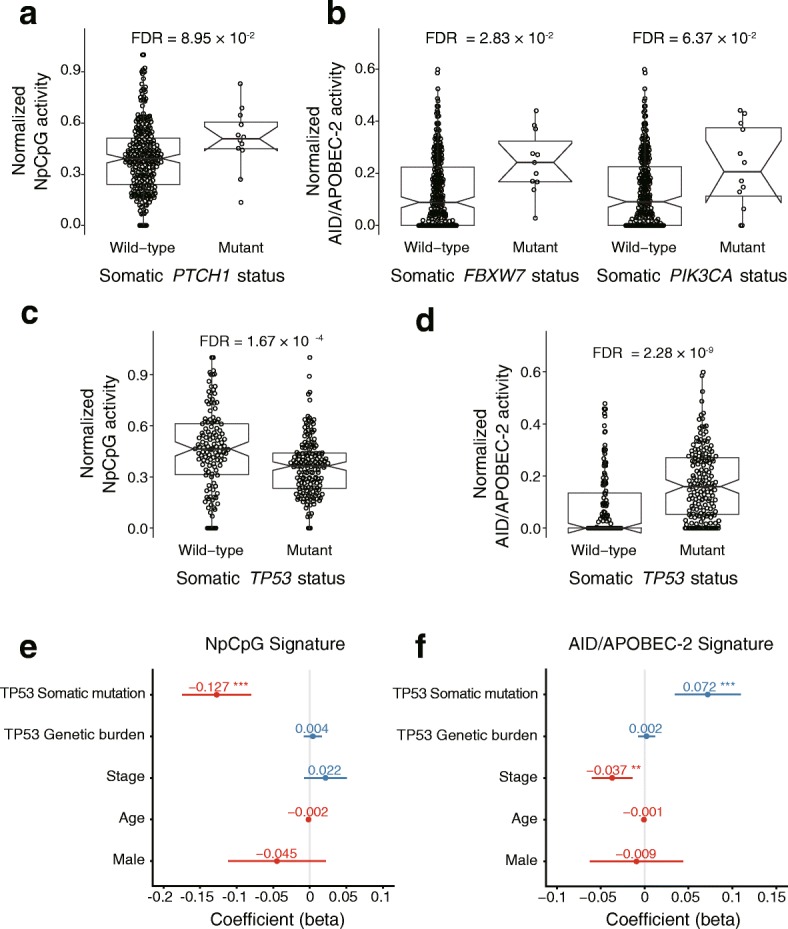
The mutational processes are associated with the somatic status of the SMGs. The activity of the “NpCpG” signature is associated with the somatic statuses of PTHC1 (a). The activity of the “AID/APOBEC-2” signature is associated with the somatic statuses of FBXW7 and PIK3CA (b). The somatic statuses of TP53 is associated with the activity of the “NpCpG” signature (c) and the “AID/APOBEC-2” signature (d). The FDR is based on the adjusted Wilcoxon rank-sum test P values. The effects for the somatic status and the genetic burden of TP53 on the activity of the “NpCpG” signature (e) and the “AID/APOBEC-2” signature (f) are shown with other clinical features. The P values are based on multivariate regression analysis: *P < 0.05, **P < 0.01, ***P < 0.001
Fig. 4.
The mutational processes are associated with the somatic status and the genetic burden of ZNF750. The activity of the “NpCpG” (a) and “AID/APOBEC-2” signature (b) is associated with the somatic ZNF750 statuses. The FDR is based on the adjusted Wilcoxon rank-sum test P values. The activity of the “NpCpG” (c) and “AID/APOBEC-2” signature (d) is associated with the genetic burden of ZNF750. The FDR is based on the adjusted SKAT P values, with the effects of the age, the clinical stage and ancestry being corrected. The effects of the somatic status and the genetic burden of ZNF750 on the activity of the “NpCpG” signature (e) and the “AID/APOBEC-2” signature (f) are shown with other clinical features. The P values are based on multivariate linear regression analysis: *P < 0.05, **P < 0.01, ***P < 0.001
Genes undergoing somatic copy number alterations can also influence the mutational processes in ESCC. For instance, MYC amplifications, CDC27 deletions and FHIT deletions are all significantly associated with the higher burden of SNVs (FDR < 0.1; Additional file 9: Figure S4c). And the CDC27 amplifications, alone, is associated with lower frequency of C > A substitution (FDR = 0.00216; Additional file 9: Figure S4d). As for the mutational signatures, the activity of the “NpCpG” signature increases significantly with somatic CDC27 amplifications (FDR = 0.00615; Fig. 5a) while the activity of the “AID/APOBEC-2” signature decreases with the same event (FDR = 8.69 × 10− 4; Fig. 5b).
Fig. 5.
The mutational processes are associated with somatic copy number status and genetic burden of CDC27. The activity of the “NpCpG” (a) and “AID/APOBEC-2” (b) signature is associated with the somatic copy number amplification of CDC27. The FDR is based on the adjusted Wilcoxon rank-sum test P values. The activity of the “NpCpG” (c) and “AID/APOBEC-2” (d) signature is associated with the genetic burden of CDC27. The FDR is based on adjusted SKAT P values, with the effects of the age, the clinical stage and ancestry being corrected. The effects of the somatic copy number amplification and the genetic burden of CDC27 on the activity of the “NpCpG” signature (e) and the “AID/APOBEC-2” signature (f) are shown with other clinical features. The P values are based on multivariate linear regression analysis: *P < 0.05, **P < 0.01, ***P < 0.001
Germline level
For the genes that influence the mutational processes somatically, we further interrogate if the burden of germline polymorphisms shows consistent effect. As results, the genetic burdens of ZNF750 (FDR = 0.0576; Fig. 4d) and PTCH1 (FDR = 0.00818; Additional file 10: Figure S5a) are significantly associated with increased activity of the “AID/APOBEC-2” signature. And the genetic burden of CDC27 is associated with both the activity of the “NpCpG” signature (FDR = 7.93 × 10− 9; Fig. 5c) and the “AID/APOBEC-2” signature (FDR = 0.00736; Fig. 5d). In addition, our data show that the genetic burdens of NOTCH1, TP53, PTCH1 and CDC27 are significantly associated with either the overall burden of SNVs or specific subtypes of substitutions (Additional files 10 and 11: Figures S5b, c and S6a). These findings suggest that ZNF750 and CDC27 are major candidate driver genes of the mutational processes in ESCC.
We investigated another 56 genes which are related to the known GWAS risk loci of ESCC in Chinese population. As results, four genes (DNAH11, CHEK2, HECTD4 and HEATR3) are associated with the activities of either mutation signatures or specific types of substitutions (FDR < 0.1, Additional file 12: Figure S7a and b). The activity of the “AID/APOBEC-1” signature is associated with the genetic burden CHEK2 (22q12.1; FDR = 0.0406), HEATR3 (16q12.1; FDR = 0.0406) and SMG6 (17p13.3; FDR = 0.0108; Additional file 12: Figure S7c). Whereas the activity of the “AID/APOBEC-2” signature is associated with the genetic burden of DNAH11 (7p15.3; FDR = 0.0941), HAP1 (17q21.2; FDR = 0.0941) and HECTD4 (12q24.13; FDR = 0.0941; Additional file 12: Figure S7d).
Multivariate analysis
To control for the confounding effects in the univariate analysis, we verified the associations aforementioned using multivariate regression, which accounts for the known clinical features of ESCC such as age, gender, stage. As results, we verified the associations between the somatic TP53 mutations and the activities of the “NpCpG” signature (P = 2.21 × 10− 7; Fig. 3e), the “AID/APOBEC-2” signature, respectively (P = 2.00 × 10− 4; Fig. 3f). For ZNF750, we verified the associations with the activity of the “NpCpG” signature (P = 6.43 × 10− 4 for somatic status; Fig. 4e) and the “AID/APOBEC-2” signature (P = 3.47 × 10− 2 for somatic status and P = 1.26 × 10− 6 for genetic burden; Fig. 4f).And for CDC27, we verified its significant association with the activities of the “NpCpG” signature (P = 1.77 × 10− 2 for genetic burden and P = 6.71 × 10− 3 for somatic amplification, Fig. 5e) and the “AID/APOBEC-2” signature (P = 4.09 × 10− 2 for somatic amplification; Fig. 5f). It is also worth noting that the effects of these genes on the mutational signatures are highly consistent as revealed by both univariate and multivariate analyses.
We further looked into the genes which are significantly associated with somatic mutational signatures within each subtype. As results, we show that the associations between each gene and the activities of the mutational signatures varies considerably among the three subtypes of ESCC (Additional file 13: Figure S8).
Discussion
The somatic landscape of the cancers are results of diverse mutational processes driven by both germline and somatic alterations in certain genes. Identification of the determinants of the mutational processes can inform the search for driver mutations and genes and thus enable a better understanding of esophagus carcinogenesis.
In this study, we report two new SMGs from a Chinese cohort of ESCC. FAM90A1 is an unfavorable prognostic marker in endometrial cancer [34]. TNRC6A is an efficient biomarker for diagnosis and prognosis of non-small-cell lung cancer; and target of miR-30a [35]. All of the novel genes are functionally annotated in other tumor types. We also report some rare mutants with proved functions in ESCC. For instance, CD7, “a cell surface glycoprotein member of the immunoglobulin superfamily”, is up-regulated in Chinese ESCC patients and plays an essential role in T-cell and T-cell/B-cell interactions during early lymphoid development [36]. And S100A2, “a member of the S100 family of calcium-binding proteins”, is downregulated in a cohort of Chinese ESCC patients and related to the progression of ESCC [37].
Recent studies claim that both germline and somatic variants can influence the mutational processes in cancers [17–21]. In our analysis, we identified potential regulators of the mutational processes based on the evidences of associations at both germline and somatic levels. Our results highlight two genes, ZNF750 and CDC27, of which the non-synonymous somatic mutations and germline genetic burden show consistent, significant effects on the activities of the mutational signatures. ZNF750 functions as a regulator of epidermal cell differentiation by promoting differentiation genes while inhibiting progenitor factors. The tumor suppressing activity of ZNF750 with significant prognostic power in multiple squamous cell carcinomas is confirmed by independent reports [38–42]. As for the other gene, Cell Division Cycle 27 is an activator of cAMP-dependent phosphorylation kinase activity [43]. The somatic copy-number amplification of CDC27, as an event of gain-of-function, is significantly associated with higher activity of the “NpCpG” signature whereas the high genetic burden in CDC27 attenuates the activity of the “NpCpG” signature.
The driver mutations in cancer is often confounded by outnumbered passenger mutations, which can only be distinguished by systematic functional validation. We resort to the evidences from the germline genetics to prioritize the candidate somatic driver genes, and suggested determinants of the mutational processes in ESCC. While the clinical implication of ZNF750 has been empirically proved by independent studies, CDC27 is newly discovered for its effects on the mutational processes of ESCC. The method we described can be applied to many other cancer types to suggest candidate driver genes.
On the other hand, the discovery is based on a relatively small population, which means other subtle, independent mutational signatures in ESCC are yet to be discovered. The sample size can also hinder the discovery of other novel candidate genes due to the limitation in the statistical power. Plus, the current in silico analysis still does not fully reveal the underlying biology of the associations due to the lack of matched information such as mRNA transcription levels and complete pathological reports. The functional impacts of the candidate genes can be better addressed by further empirical validation in cell lines or mouse models.
Finally, our results suggest that the mutational processes in ESCC differ significantly between subpopulations of Chinese. Similar associations between the population-genetical background and the somatic mutations is previously reported in prostate cancer [44]. We also show that genes related to the GWAS risk loci can also influence the mutational processes in ESCC. Together these facts strongly suggest that germline variants help to better identify the driver genes of the somatic mutational processes in cancer.
Conclusions
As conclusions, we report multiple factors that influence specific mutational processes in ESCC in a Chinese population, including the subpopulations, the SMGs, the genes related to the GWAS risk loci as well as exposure to alcohol and tobacco. We highlight ZNF750 and CDC27 as potential regulators of the mutational processes in ESCC by combining evidences at both germline and somatic levels. These findings inform the esophageal carcinogenesis and provide testable candidates for future functional studies.
Methods
Exome-sequencing data of ESCC
We obtained 9 ESCC samples from Zhongshan Hospital Xiamen University (ZHXU) in Xiamen, China. Tumor and matched normal tissues were collected after diagnosis and were stored at − 80 °C for subsequent DNA extraction. No patients were treated by chemotherapy or radiotherapy before the operation. All tumor tissues contained at least 80% malignant cells. We extracted DNA using QIAamp DNA Mini Kit (Qiagen) and prepared the whole-exome library using Agilent “SureSelect Human All Exon V5” following the standard protocol and perform 100 bp paired-end sequencing using Illumina HiSeq2000. All methods were performed in accordance with the relevant guidelines and regulations. This study is approved by the ethics committee/institutional review board (IRB) of ZHXU (approval no. 2017028) with written consent from all participants. The study complied with Declaration of Helsinki principles. The rest of 293 pairs of WES data sets were collected from published studies [20, 23–25]. All the sequencing data sets were based on paired ESCC and normal tissues from Chinese patients. The clinical information of the patients is either obtained by collected by ZHXU or previous studies (Additional file 14: Table S6).
Somatic mutation analysis and genotyping
For the 302 WES data, we aligned the paired-end reads to human reference genome (hg19) using BWA (v0.7.12) [45]. The duplicated reads were removed using Picard Tools (v1.119; http://broadinstitute.github.io/picard/). We called somatic and germline variants using VarScan2 (v2.3.9) by controlling for mapping quality (≥ 30) and coverage (≥ 10) [46, 47]. The resulting SNVs and InDels were further filtered for alternate allele depth ≥ 10. We used GATK (v3.4.0) HaplotypeCaller for germline variants calling as well [48]. Then we chose the intersection of variants calls from GATK and VarScan2 for further analysis.
To determine the genotypes of the germline variants, we filtered the loci based on mapping quality (≥ 30), coverage of alternate allele (≥ 5) and the total coverage (≥ 20). We then determined the genotype based on alternate allele coverage rate (ACR) as the followings: (1) ACR ≤ 0.1, homozygote of the reference allele; (2) 0.2 ≤ ACR ≤ 0.8, heterozygote; (3) ACR ≥ 0.9, homozygote of the alternate allele. We intersected the resulting germline variants with the variants from the 1000 Genomes Project (TGP) phase 3 databases [49] then performed principal component analysis in a combined cohort of 103 CHB and 105 CHS from TGP and 302 ESCC samples.
Somatic mutational signatures in ESCC
We retrieved highly conserved somatic mutational signatures using “pmsignature” [13]; and compared the results to the signatures in the COSMIC (http://cancer.sanger.ac.uk) [26] using CS measure.
Somatic copy-number alterations
We used VarScan2 (v.2.3.9) to identify SCNAs with filtering conditions on the coverage (≥ 20), the base quality (≥ 20) and the mapping quality (≥ 20). The breakpoints were defined based on significant change of the ratio-of-depth between the tumor and the matched normal (p < 0.05 with Fisher’s Exact Test). We obtained the autosomal segments of copy-number changes using circular binary segmentation (CBS) and determined the somatic copy-number status of each segment by the log2-adjusted ratio-of-depth. A copy-number gain was defined if the corresponding value is above 0.5; and loss if the value is below − 0.5. JISTIC was used to find the significant regions and genes of SCNAs [50].
Significantly mutated genes and pathways in ESCC
The somatic mutations were annotated using Oncotator (v1.5.1.0) [51]. Then we evaluated the enrichment of the non-synonymous somatic mutations (SMGs are FDR < 0.1) in each gene using MutSigCV (v1.4) [52]. We analyzed the pathways of genes that are either significantly frequently mutated (MutSigCV p < 0.05) and strongly associated with SCNAs using HotNet2 [53].
Association analyses
We performed hierarchical clustering of the samples based on the Euclidean distances of the relative activities of the mutational signatures using Ward’s linkage method. We evaluated the associations between the subtypes and various risk factors (tobacco, alcohol exposure and populations), the total number of SNVs and the somatic statuses of genes. For the association analysis, we used χ2 tests for categorical variables and analysis of variance (ANOVA) for quantitative features.
We also evaluated the associations between mutational processes in ESCC and a set of genes of interests. We chose the total number of SNVs, the activities of the mutational signatures, the frequencies of each of the six types of base-substitutions (C > A, C > G, C > T, T > A, T > C and T > G) as proxies to the mutational processes. We determined the somatic statuses based on whether any non-synonymous somatic mutations present in a gene. We then compared the proxies between subsets defined by the somatic statuses of the genes of interests using Wilcoxon’s rank sum test.
To address the genetic burdens, we used the SNP-set Kernel Association Test (SKAT) to evaluate the association between the genetic burden of a given gene and the proxies of the somatic mutational processes [54]. For each gene, we chose all the exonic germline SNPs with no more than 50% missing calls in the population as the SNP-set. To avoid any confounding effects, we used the patients’ age, clinical stage and the first two principal components of the germline genotype (as surrogates for population variation) as covariates of the SKAT model. Thus, we tested 12 SMGs, 20 SCNA-related genes and 56 genes that are associated with known ESCC germline risk loci.
To better understand the association, we performed multivariate linear regression in ESCC samples and subgroup of ESCC:
where εkli~N (0, σ2) is a Gaussian error term; Sigki corresponds to the kth signature of the ith sample; Gli corresponds to the genetic burden of the lth gene in the ith sample; Si corresponds to the somatic mutational status of the lth gene in the ith sample; genderi corresponds to the gender of the ith sample; agei corresponds to the age of the ith sample;
where Wj corresponds to the weight of the jth SNP of the lth gene, glij corresponds to the genotype of the jth SNP of the lth gene in the ith sample;
where MAFlj corresponds to the minimum allele frequency of the jth SNP of the lth gene;
where Nli corresponds to the number of the non-synonymous mutation of the lth gene in the ith sample.
Thus, we looked into the genes which were significantly association with somatic mutational signatures in the previously individually correlation analysis.
Additional files
Table S1. Somatic mutations in 302 ESCC patients. (XLSX 785 kb)
Figure S1. Significantly mutated genes in 302 ESCC. The samples are sorted by the counts of somatic mutations per megabase, with the synonymous and non-synonymous mutations shown in different colors. (top); the Significantly mutated genes (FDR < 0.1) are plotted for each patient carrying the mutations. Each row corresponds to a gene and each column a patient. The different types of mutations are color-coded (middle). (PDF 162 kb)
Table S2. The functional annotations of in-frame mutations of TNRC6A and FAM90A1. (XLSX 10 kb)
Table S3. The significant somatic copy-number alterations in 302 ESCC samples. The copy-number statuses, FDR and the mutated sample counts of peak regions are based on JISTIC. (XLSX 14 kb)
Table S4. Genes of significant somatic copy-number alterations in ESCC. The copy-number GSCORE and FDR, the somatic copy-number statuses of each gene are based on JSTIC. A: amplification, D: deletion and N: neutral. (XLSX 34 kb)
Figure S2. Significantly modified subnetworks in ESCC. The subnetworks are identified by HotNet2 from public protein-protein interactions databases of HINT+HI2012 (a), HPRD (b), iRefIndex (c) and MultiNet (d). The colored nodes represent the genes with different types of somatic alterations in ESCC, the sizes of the nodes correspond to the frequency of alteration in the population. All the subnetworks are identified with the minimum edge weight (δ), the minimum size of subnetwork (k) and the P less than 0.05. (PDF 158 kb)
Table S5. The results of Gene Set Enrichment Analysis. (a) The subnetworks significantly enrich for KEGG pathways. (b) The subnetworks significantly enrich for Transcription factor targets. (XLSX 11 kb)
Figure S3. Population stratification of 302 ESCC patients. Two hundred eight genotyped reference individuals are obtained from TGP including 103 CHB and 105 CHS. After filtering 20 outliers, the remaining 282 samples are classified into CHB and CHS at a threshold level of 0 for PC2. (PDF 165 kb)
Figure S4. Comparison of total SNV counts and the frequencies of specific base substitutions with the somatic statuses of certain genes. (a) The total SNV counts are compared to the somatic statuses of TP53, ZNF750, FAT1, FBXW7 and PIK3CA. (b) The frequencies of substitutions are compared to the somatic statuses of TP53. (c) The total SNV counts are compared to the somatic copy-number statuses of MYC, CDC27 and FHIT. (d) The frequency of C > A substitution is compared to the somatic copy-number statuses of CDC27. FDR is based on the adjusted Wilcoxon rank-sum test P values. (PDF 418 kb)
Figure S5. Correlation of the somatic events with the genetic burdens of the SMGs in ESCC. (a) The genetic burdens of PTCH1 are associated with the activity of the “AID/APOBEC-2” signature. (b) The genetic burdens of NOTCH1 are associated with the frequencies of C > G. (c) The genetic burdens of TP53 and PTCH1 are associated with the frequencies of T > C. FDR is based on the adjusted SKAT P values, in which the age, the clinical stage and ancestry are considered as covariates. (PDF 278 kb)
Figure S6. Correlation of the somatic events with the genetic burdens of the SCNA-related genes in ESCC. The genetic burdens of CDC27 are associated with the total number of SNVs (a) and the frequencies of C > G (b). The genetic burdens of SLC6A3 are associated with the frequencies of T > G (c). The genetic burdens of CDC27, RECQL, DGCR6 and EGFR are associated with the frequencies of T > C (d). FDR is based on the adjusted SKAT P values, in which the age, the clinical stage and ancestry are considered as covariates. (PDF 428 kb)
Figure S7 Correlation of the somatic events with the genetic burdens of the risk-associated genes in ESCC. The genetic burdens of CHEK2 and HECTD4 are associated with the frequencies of C > G substitution (a). The genetic burdens of HEATR3 are associated with the frequencies of C > T substitution (b). The genetic burdens of CHEK2, HEATR3 and SMG6 are associated with the “AID/APOBEC-1” signature (c). The genetic burdens of DNAH11, HAP1, HECTD4 and HLA-DQA1 are associated with the “AID/APOBEC-2” signature (d). FDR is based on the adjusted SKAT P values, in which the age, the clinical stage and ancestry are considered as covariates. (PDF 407 kb)
Figure S8. The effects of TP53, ZNF750, CDC27 and other clinical features on the activity of the “NpCpG” signature and the “AID/APOBEC-2” signature in the subtypes of ESCC. The somatic status and the genetic burden of TP53 influence the activities of the “NpCpG” signature (a) and the “AID/APOBEC-2” signature (b) in subtype 1 to 3, independent of other clinical features. And the somatic status and the genetic burden of ZNF750 influence the activities of the “NpCpG” signature (c) and the “AID/APOBEC-2” signature (d) in subtype 1 to 3, independent of other clinical features. The somatic copy number amplification and the genetic burden of CDC27 influence the activity of the “NpCpG” signature (e) and the “AID/APOBEC-2” signature (f) in subtype 1 to 3, independent of other clinical features. P values are based on multivariate linear regression analysis: *P < 0.05, **P < 0.01, ***P < 0.001. (PDF 892 kb)
Table S6. Summary of the clinical features of the 302 ESCC patients included in this study. (XLSX 26 kb)
Acknowledgments
This study makes use of data generated by Prof. Jie He of Cancer Institute and Hospital Chinese Academy of Medical Sciences; and data generated by Prof Qimin Zhan of the Molecular Oncology Laboratory and the Cancer Research Department of BGI.
Funding
This study is supported by grants from the National Natural Science Foundation of China (NSFC, 31371289 to JG and QL), the Major Disease Research Projects of Xiamen, China (3502Z20149030 to YLZ), Natural Science Foundation of Fujian Province, China (Grant No. 2018J01054 to YZ) and the Education and Research Foundation for Young Scholars of Education department of Fujian Province (Grant No. JAT170004 to YZ).
Availability of data and materials
Most of the data support the findings of this study are available in the European Genome-phenome Archive (https://ega-archive.org) with the identifiers EGAD00001000760 [24] and EGAD00001001006 [25]; and NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) with the identifiers SRP033394 [23] and SRA112617 [20]. The other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- ACR
Alternate-allele Coverage Rate
- ANOVA
Analysis of variance
- CBS
Circular Binary Segmentation
- CHB
Han Chinese in Beijing
- CHS
Southern Han Chinese
- COSMIC
Catalogue of Somatic Mutations in Cancer
- CS
Cosine Similarity
- EAC
Esophageal Adenocarcinoma
- EC
Esophageal Cancer
- ESCC
Esophageal Squamous Cell Carcinoma
- InDels
Insertions and Deletions
- PPI
Protein-protein Interaction
- SCNAs
Somatic Copy-number Alterations
- SKAT
SNP-set Kernel Association Test
- SMGs
Significantly Mutated Genes
- SNVs
Single Nucleotide Variants
- TGP
The 1000 Genomes Project
- WES
Whole-exome Sequencing
Authors’ contributions
QL conceived the study. BG provided tissue samples and the clinical information. QL, JG and YZ wrote, reviewed and revised the manuscript. BG reviewed and revised the manuscript. JH, YLZ, HL, LH, LZ, DG performed the experiments. JG, HL, LY, YZ and YYZ analyzed and interpreted the data. All authors read, approved and contributed to the final manuscript.
Ethics approval and consent to participate
This study was approved by the ethics committee/institutional review board (IRB) of ZHXU (approval no. 2017028). The study complied with Declaration of Helsinki principles. Written consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Jintao Guo, Jiankun Huang and Ying Zhou contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4906-4) contains supplementary material, which is available to authorized users.
Contributor Information
Jintao Guo, Email: guojt-4451@163.com.
Jiankun Huang, Email: huangjiankun@xmu.edu.cn.
Ying Zhou, Email: yingzhou@xmu.edu.cn.
Yulin Zhou, Email: zhou_yulin@126.com.
Liying Yu, Email: 513259453@qq.com.
Huili Li, Email: lhlbio@163.com.
Lingyun Hou, Email: xiaoyu2015xiaoyu@sina.com.
Liuwei Zhu, Email: zhuliuwei1125@163.com.
Dandan Ge, Email: gedandan0412@163.com.
Yuanyuan Zeng, Email: 15959225967@163.com.
Bayasi Guleng, Email: bayasi@xmu.edu.cn.
Qiyuan Li, Phone: +86 592 2185175, Email: qiyuan.li@xmu.edu.cn.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012: Globocan 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Napier KJ. Esophageal cancer: a review of epidemiology, pathogenesis, staging workup and treatment modalities. World J Gastrointest Oncol. 2014;6:112. doi: 10.4251/wjgo.v6.i5.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381–387. doi: 10.1136/gutjnl-2014-308124. [DOI] [PubMed] [Google Scholar]
- 4.Gao Y, Hu N, Han X, Giffen C, Ding T, Goldstein A, et al. Family history of cancer and risk for esophageal and gastric cancer in Shanxi, China. BMC Cancer. 2009;9:269. [DOI] [PMC free article] [PubMed]
- 5.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93. [DOI] [PMC free article] [PubMed]
- 6.Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polak P, Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, Tiao G, et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet. 2017; Available from: http://www.nature.com/doifinder/10.1038/ng.3934. [cited 28 Aug 2017] [DOI] [PMC free article] [PubMed]
- 10.Letouzé E, Shinde J, Renault V, Couchy G, Blanc J-F, Tubacher E, et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat Commun. 2017;8:1315. Available from: http://www.nature.com/articles/s41467-017-01358-x. [cited 27 Mar 2018]. [DOI] [PMC free article] [PubMed]
- 11.Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev. 2014;24:52–60. doi: 10.1016/j.gde.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human Cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiraishi Y, Tremmel G, Miyano S, Stephens M. A simple model-based approach to inferring and visualizing Cancer mutation signatures. PLOS Genet. 2015;11:1005657. Marchini J, editor [DOI] [PMC free article] [PubMed]
- 14.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milne RL, Kuchenbaecker KB, Michailidou K, Beesley J, Kar S, Lindström S, et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat Genet. 2017;49(12):1767–1778. doi: 10.1038/ng.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. [DOI] [PMC free article] [PubMed]
- 17.Robles-Espinoza CD, Roberts ND, Chen S, Leacy FP, Alexandrov LB, Pornputtapong N, et al. Germline MC1R status influences somatic mutation burden in melanoma. Nat Commun. 2016;7:12064. doi: 10.1038/ncomms12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu B, Mukherjee A, Machiela MJ, Song L, Hua X, Shi J, et al. An investigation of the association of genetic susceptibility risk with somatic mutation burden in breast cancer. Br J Cancer. 2016;115:752–60. [DOI] [PMC free article] [PubMed]
- 19.Kim J, Mouw KW, Polak P, Braunstein LZ, Kamburov A, Tiao G, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet. 2016;48:600–606. doi: 10.1038/ng.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Zhou Y, Cheng C, Cui H, Cheng L, Kong P, et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet. 2015;96:597–611. doi: 10.1016/j.ajhg.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawada G, Niida A, Uchi R, Hirata H, Shimamura T, Suzuki Y, et al. Genomic landscape of esophageal squamous cell carcinoma in a Japanese population. Gastroenterology. 2016;150:1171–1182. doi: 10.1053/j.gastro.2016.01.035. [DOI] [PubMed] [Google Scholar]
- 22.Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23:517–525. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin D-C, Hao J-J, Nagata Y, Xu L, Shang L, Meng X, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. 2014;46:467–473. doi: 10.1038/ng.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song Y, Li L, Ou Y, Gao Z, Li E, Li X, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–95. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 25.Gao Y-B, Chen Z-L, Li J-G, Hu X-D, Shi X-J, Sun Z-M, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46:1097–102. [DOI] [PubMed]
- 26.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Getz G, Gabriel SB, Cibulskis K, Lander E, Sivachenko A, Sougnez C, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patil V, Pal J, Somasundaram K. Elucidating the cancer-specific genetic alteration spectrum of glioblastoma derived cell lines from whole exome and RNA sequencing. Oncotarget. 2015;6:43452–43471. doi: 10.18632/oncotarget.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng C, Cui H, Zhang L, Jia Z, Song B, Wang F, et al. Genomic analyses reveal FAM84B and the NOTCH pathway are associated with the progression of esophageal squamous cell carcinoma. GigaScience. 2016;5:1. doi: 10.1186/s13742-015-0107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Bowlby R, Mungall AJ, Robertson AG, Odze RD, Cherniack AD, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/541464d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Li X, Cheng Y, Sun X, Sun X, Self S, et al. Copy number alterations detected by whole-exome and whole-genome sequencing of esophageal adenocarcinoma. Hum Genomics. 2015;9:22. doi: 10.1186/s40246-015-0044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 34.Tang R, Liang L, Luo D, Feng Z, Huang Q, He R, et al. Downregulation of MiR-30a is associated with poor prognosis in lung Cancer. Med Sci Monit. 2015;21:2514–2520. doi: 10.12659/MSM.894372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao Y, Chai D, Ma L, Zhang T, Feng Z, Cheng Z, et al. Identification of distinct gene expression profiles between esophageal squamous cell carcinoma and adjacent normal epithelial tissues. Tohoku J Exp Med. 2012;226:301–311. doi: 10.1620/tjem.226.301. [DOI] [PubMed] [Google Scholar]
- 36.Cao L-Y, Yin Y, Li H, Jiang Y, Zhang H-F. Expression and clinical significance of S100A2 and p63 in esophageal carcinoma. World J Gastroenterol. 2009;15:4183. doi: 10.3748/wjg.15.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao J-J, Lin D-C, Dinh HQ, Mayakonda A, Jiang Y-Y, Chang C, et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat Genet. 2016;48:1500–1507. doi: 10.1038/ng.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hazawa M, Lin D-C, Handral H, Xu L, Chen Y, Jiang Y-Y, et al. ZNF750 is a lineage-specific tumour suppressor in squamous cell carcinoma. Oncogene. 2017;36:2243–2254. doi: 10.1038/onc.2016.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otsuka R, Akutsu Y, Sakata H, Hanari N, Murakami K, Kano M, et al. ZNF750 expression as a novel candidate biomarker of Chemoradiosensitivity in esophageal squamous cell carcinoma. Oncology. 2017;93(3):197–203. doi: 10.1159/000476068. [DOI] [PubMed] [Google Scholar]
- 40.Dai W, Ko JMY, Choi SSA, Yu Z, Ning L, Zheng H, et al. Whole-exome sequencing reveals critical genes underlying metastasis in esophageal squamous cell carcinoma: genomic study for metastatic ESCC. J Pathol. 2017;242:500–10. [DOI] [PubMed]
- 41.Otsuka R, Akutsu Y, Sakata H, Hanari N, Murakami K, Kano M, et al. ZNF750 expression is a potential prognostic biomarker in esophageal squamous cell carcinoma. Oncology. 2018;94:142–8. [DOI] [PMC free article] [PubMed]
- 42.Sansregret L, Patterson JO, Dewhurst S, López-García C, Koch A, McGranahan N, et al. APC/C dysfunction limits excessive Cancer chromosomal instability. Cancer Discov. 2017;7:218–233. doi: 10.1158/2159-8290.CD-16-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khani F, Mosquera JM, Park K, Blattner M, O’Reilly C, MacDonald TY, et al. Evidence for molecular differences in prostate Cancer between African American and Caucasian men. Clin Cancer Res. 2014;20:4925–4934. doi: 10.1158/1078-0432.CCR-13-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koboldt DC, Larson DE, Wilson RK. Using VarScan 2 for Germline Variant Calling and Somatic Mutation Detection. In: Bateman A, Pearson WR, Stein LD, Stormo GD, Yates JR, editors. Curr Protoc Bioinforma. Hoboken: Wiley; 2013. pp. 15.4.1–15.4.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, et al. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;43:11.10.1–11.1033. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez-Garcia F, Akavia UD, Mozes E, Pe’er D. JISTIC: identification of significant targets in cancer. BMC Bioinformatics. 2010;11:189. doi: 10.1186/1471-2105-11-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, et al. Oncotator: Cancer variant annotation tool. Hum Mutat. 2015;36:E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leiserson MDM, Vandin F, Wu H-T, Dobson JR, Eldridge JV, Thomas JL, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet. 2014;47:106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence kernel association tests for the combined effect of rare and common variants. Am J Hum Genet. 2013;92:841–853. doi: 10.1016/j.ajhg.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Somatic mutations in 302 ESCC patients. (XLSX 785 kb)
Figure S1. Significantly mutated genes in 302 ESCC. The samples are sorted by the counts of somatic mutations per megabase, with the synonymous and non-synonymous mutations shown in different colors. (top); the Significantly mutated genes (FDR < 0.1) are plotted for each patient carrying the mutations. Each row corresponds to a gene and each column a patient. The different types of mutations are color-coded (middle). (PDF 162 kb)
Table S2. The functional annotations of in-frame mutations of TNRC6A and FAM90A1. (XLSX 10 kb)
Table S3. The significant somatic copy-number alterations in 302 ESCC samples. The copy-number statuses, FDR and the mutated sample counts of peak regions are based on JISTIC. (XLSX 14 kb)
Table S4. Genes of significant somatic copy-number alterations in ESCC. The copy-number GSCORE and FDR, the somatic copy-number statuses of each gene are based on JSTIC. A: amplification, D: deletion and N: neutral. (XLSX 34 kb)
Figure S2. Significantly modified subnetworks in ESCC. The subnetworks are identified by HotNet2 from public protein-protein interactions databases of HINT+HI2012 (a), HPRD (b), iRefIndex (c) and MultiNet (d). The colored nodes represent the genes with different types of somatic alterations in ESCC, the sizes of the nodes correspond to the frequency of alteration in the population. All the subnetworks are identified with the minimum edge weight (δ), the minimum size of subnetwork (k) and the P less than 0.05. (PDF 158 kb)
Table S5. The results of Gene Set Enrichment Analysis. (a) The subnetworks significantly enrich for KEGG pathways. (b) The subnetworks significantly enrich for Transcription factor targets. (XLSX 11 kb)
Figure S3. Population stratification of 302 ESCC patients. Two hundred eight genotyped reference individuals are obtained from TGP including 103 CHB and 105 CHS. After filtering 20 outliers, the remaining 282 samples are classified into CHB and CHS at a threshold level of 0 for PC2. (PDF 165 kb)
Figure S4. Comparison of total SNV counts and the frequencies of specific base substitutions with the somatic statuses of certain genes. (a) The total SNV counts are compared to the somatic statuses of TP53, ZNF750, FAT1, FBXW7 and PIK3CA. (b) The frequencies of substitutions are compared to the somatic statuses of TP53. (c) The total SNV counts are compared to the somatic copy-number statuses of MYC, CDC27 and FHIT. (d) The frequency of C > A substitution is compared to the somatic copy-number statuses of CDC27. FDR is based on the adjusted Wilcoxon rank-sum test P values. (PDF 418 kb)
Figure S5. Correlation of the somatic events with the genetic burdens of the SMGs in ESCC. (a) The genetic burdens of PTCH1 are associated with the activity of the “AID/APOBEC-2” signature. (b) The genetic burdens of NOTCH1 are associated with the frequencies of C > G. (c) The genetic burdens of TP53 and PTCH1 are associated with the frequencies of T > C. FDR is based on the adjusted SKAT P values, in which the age, the clinical stage and ancestry are considered as covariates. (PDF 278 kb)
Figure S6. Correlation of the somatic events with the genetic burdens of the SCNA-related genes in ESCC. The genetic burdens of CDC27 are associated with the total number of SNVs (a) and the frequencies of C > G (b). The genetic burdens of SLC6A3 are associated with the frequencies of T > G (c). The genetic burdens of CDC27, RECQL, DGCR6 and EGFR are associated with the frequencies of T > C (d). FDR is based on the adjusted SKAT P values, in which the age, the clinical stage and ancestry are considered as covariates. (PDF 428 kb)
Figure S7 Correlation of the somatic events with the genetic burdens of the risk-associated genes in ESCC. The genetic burdens of CHEK2 and HECTD4 are associated with the frequencies of C > G substitution (a). The genetic burdens of HEATR3 are associated with the frequencies of C > T substitution (b). The genetic burdens of CHEK2, HEATR3 and SMG6 are associated with the “AID/APOBEC-1” signature (c). The genetic burdens of DNAH11, HAP1, HECTD4 and HLA-DQA1 are associated with the “AID/APOBEC-2” signature (d). FDR is based on the adjusted SKAT P values, in which the age, the clinical stage and ancestry are considered as covariates. (PDF 407 kb)
Figure S8. The effects of TP53, ZNF750, CDC27 and other clinical features on the activity of the “NpCpG” signature and the “AID/APOBEC-2” signature in the subtypes of ESCC. The somatic status and the genetic burden of TP53 influence the activities of the “NpCpG” signature (a) and the “AID/APOBEC-2” signature (b) in subtype 1 to 3, independent of other clinical features. And the somatic status and the genetic burden of ZNF750 influence the activities of the “NpCpG” signature (c) and the “AID/APOBEC-2” signature (d) in subtype 1 to 3, independent of other clinical features. The somatic copy number amplification and the genetic burden of CDC27 influence the activity of the “NpCpG” signature (e) and the “AID/APOBEC-2” signature (f) in subtype 1 to 3, independent of other clinical features. P values are based on multivariate linear regression analysis: *P < 0.05, **P < 0.01, ***P < 0.001. (PDF 892 kb)
Table S6. Summary of the clinical features of the 302 ESCC patients included in this study. (XLSX 26 kb)
Data Availability Statement
Most of the data support the findings of this study are available in the European Genome-phenome Archive (https://ega-archive.org) with the identifiers EGAD00001000760 [24] and EGAD00001001006 [25]; and NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) with the identifiers SRP033394 [23] and SRA112617 [20]. The other data supporting the findings of this study are available from the corresponding author upon reasonable request.