OsPM1, encoded by an AWPM-19 family gene, mediates abscisic acid influx and is regulated by the transcription factor OsbZIP46 in the rice drought signaling pathway.
Abstract
Abscisic acid (ABA) regulates plant stress responses and development. However, how the ABA signal is transmitted in response to stresses remains largely unclear, especially in monocots. In this study, we found that rice (Oryza sativa) OsPM1 (PLASMA MEMBRANE PROTEIN1), encoded by a gene of AWPM-19 like family, mediates ABA influx through the plasma membrane. OsPM1 is predominantly expressed in vascular tissues, guard cells, and mature embryos. Phenotypic analysis of overexpression, RNA interference (RNAi), and knockout (KO) lines showed that OsPM1 is involved in drought responses and seed germination regulation. 3H-(±)ABA transport activity and fluorescence resonance energy transfer assays both demonstrated that OsPM1 facilitates ABA uptake into cells. The physiological isomer of ABA, (+)-ABA, is the preferred substrate of OsPM1. Higher ABA accumulation and faster stomatal closure in response to ABA treatment were observed in the overexpression lines compared with the wild-type control. Many ABA-responsive genes were upregulated more in the OsPM1-overexpression lines but less in the RNAi lines compared with wild-type plants. Further investigation revealed that OsPM1 expression is regulated by the AREB/ABF family transcription factor OsbZIP46. Our results thus revealed that OsPM1 is an ABA influx carrier that plays an important role in drought responses.
INTRODUCTION
Phytohormones are synthesized in certain cell types but regulate physiological responses throughout the plant (Weyers and Paterson, 2001; Boursiac et al., 2013). Abscisic acid (ABA) is an important hormone regulating stress responses, developmental processes, and seed dormancy. It is mainly synthesized in the vasculature, guard cells, and seeds, where the ABA biosynthetic genes, such as NCEDs, ABA2, and AAO3, are highly expressed (Ye et al., 2012; Bauer et al., 2013; Finkelstein, 2013; Merilo et al., 2015). ABA can be detected in both xylem and phloem bulk flow, while the closest sources producing ABA are phloem companion cells and xylem parenchyma cells, adjacent to the phloem sieve cells and xylem vessels, respectively (Koiwai et al., 2004; Kuromori et al., 2014). Therefore, exporting ABA from ABA-biosynthetic cells and importing ABA into target cells appear to be important for ABA signaling throughout plant.
ABA is a weak acid (pKa = 4.7) and exists as anionic (ABA−) and protonated (ABAH) forms. The uncharged ABAH diffuses passively through the plasma membrane, whereas ABA− cannot. Under normal conditions, 4 to 50% ABA is diffusible (Boursiac et al., 2013; Jones et al., 2014). Apoplastic ABA can be carried by water flow into the xylem by transpiration and then transported to distant tissues (Hartung et al., 2002). Under stress conditions, due to the alkalization of xylem sap and apoplastic fluid (Wilkinson and Davies, 1997; Hartung et al., 2002), the proton is dissociated from ABAH and the membrane permeability of ABA is dramatically reduced. Under these conditions, ABA relies on transporters to move into cells. Once ABA is intracellular, it binds to the receptors PYR (PYRABACTIN RESISTANCE)/PYL (PYR1-LIKE)/RCAR (REGULATORY COMPONENTS OF ABA RECEPTORS), inhibits the type 2C protein phosphatases (PP2Cs), and activates the SNF1-related type 2 protein kinases (SnRK2s). SnRK2s then phosphorylate downstream transcription factors, ion channels, and other mediators to regulate drought responses (Finkelstein, 2013; Hedrich and Geiger, 2017). Among the known downstream transcription factors, members of the AREB (ABA RESPONSIVE ELEMENT BINDING PROTEIN)/ABF (ABRE BINDING FACTOR) subfamily of the bZIP family play important roles in the ABA signaling pathway (Xiong et al., 2002; Finkelstein, 2013).
In recent years, several ATP binding cassette family G (ABCG)-type members, including AtABCG25, AtABCG30, AtABCG31, and AtABCG40; some NPF (NRT1/PTR FAMILY) proteins, such as NPF4.6/AIT1/NRT1.2 (ABA-IMPORTING TRANSPORTER/NITRATE TRANSPORTER1.2); and a DTX/MATE (DETOXIFICATION EFFLUX CARRIER/MULTIDRUG AND TOXIC COMPOUND EXTRUSION) family protein, AtDTX50, have been identified as ABA transporters in Arabidopsis thaliana (Kang et al., 2010, 2015; Kuromori et al., 2010; Kanno et al., 2012; Léran et al., 2014; Zhang et al., 2014; Corratgé-Faillie and Lacombe, 2017). They are expressed in different organs and participate in drought responses and/or the regulation of seed germination. Among them, AtABCG25 and AtDTX50 mediate ABA efflux (Kuromori et al., 2010; Zhang et al., 2014), whereas AtABCG40 and NPF4.6/AIT1/NRT1.2 mediate ABA influx (Kang et al., 2010; Kanno et al., 2012). In addition, four ABC transporters coordinate to control Arabidopsis seed germination, with AtABCG25 and AtABCG31 exporting ABA from the endosperm and AtABCG30 and AtABCG40 taking up ABA into the embryo (Kang et al., 2015).
No authentic ABA transporter has been reported in rice (Oryza sativa). Rice is a drought-sensitive and widely grown crop in many countries. Understanding the ABA signal transduction in rice is especially important to agricultural production. In this study, we identified an ABA importer in rice by both genetic and biochemistry approaches. This protein, OsPM1, belongs to a protein family named AWPM-19 (ABA-induced Wheat Plasma Membrane Polypeptide-19), whose functions were previously unknown. We further found that OsPM1 expression is regulated by the AREB/ABF family transcription factor OsbZIP46. Our work reveals both the function of OsPM1 and its regulatory mechanism.
RESULTS
OsPM1 Expression Is Regulated by ABA Signaling Pathways
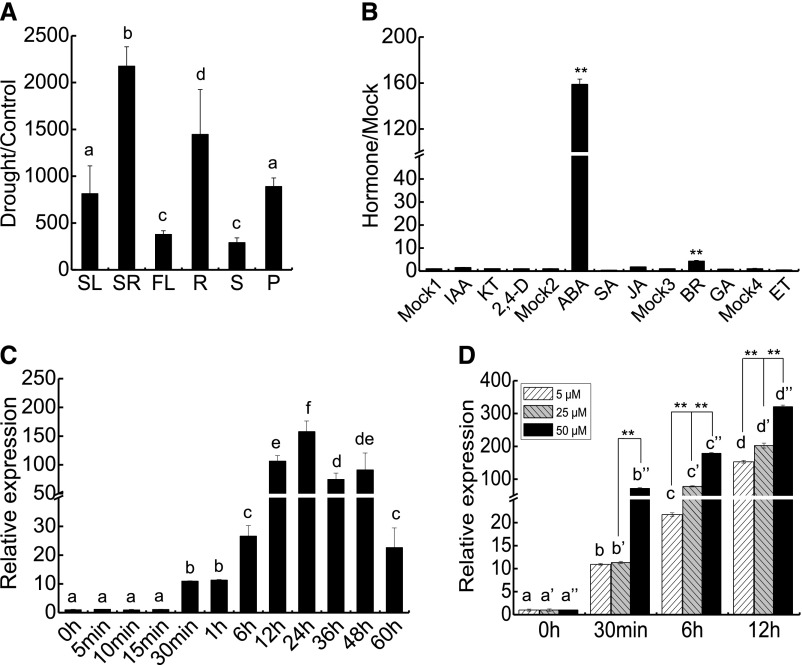
By comparative microarray analysis of florets from rice plants grown under normal growth conditions and water-deficit conditions (Jin et al., 2013), drought-responsive genes were identified. Among them, a gene named OsPM1 (ORYZA SATIVA PLASMA MEMBRANE PROTEIN1) showed strong responsiveness to drought in all developmental stages of florets. OsPM1 was first reported as a NAC045 upregulated gene in rice (Zheng et al., 2009). Sequence analysis showed that this gene (LOC_Os05g31670) belongs to the AWPM-19 gene family, members of which are found in monocots mainly as abiotic stress-inducible genes (Koike et al., 1997; Ranford et al., 2002; Rerksiri et al., 2013; Barrero et al., 2015; Chen et al., 2015), although their molecular functions remain unknown. The OsPM1 gene has also been designated as AWPM-19, OsPM19, or OsPM19L1 based on its similarity to wheat AWPM-19 gene (Chen et al., 2015) and the information given in the Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp). Results from RT-qPCR showed that OsPM1 was strongly induced by drought stress in various tissues at different developmental stages, including leaves and roots from young seedlings, and leaves, roots, stems, and panicles from reproductive stage plants (Figure 1A). Under normal growth conditions, OsPM1 was expressed weakly in leaves and roots, but more strongly in seeds (Supplemental Figure 1A). The temporal expression pattern of OsPM1 in developing seeds (Supplemental Figure 1B) is consistent with the temporal curve of ABA accumulation in developing rice seeds reported previously (Still et al., 1994).
Figure 1.
Expression Patterns of OsPM1 Show Relationship to ABA Signaling Pathways.
(A) OsPM1 was induced by drought in various organs at different developmental stages. Plants were drought stressed as described in Methods, and the indicated tissues were analyzed by RT-qPCR. SL and SR, leaves and roots from 14-d-old seedlings; FL, flag leaves; R, roots; S, stems; P, panicles. Flag leaves, roots, stems, and panicles were all from 2-month-old plants at reproductive stage.
(B) OsPM1 was highly induced by ABA treatment. Six-day-old rice seedlings grown in Kimura B liquid media were subjected to various phytohormone treatments; 5 μM ABA, IAA, kinetin (KT), 2,4-D, salicylic acid (SA), jasmonic acid (JA), and gibberellic acid (GA), and 10 nM BR were added to the media for treatments, and the whole seedlings 12 h after treatments were harvested for RT-qPCR. For ethylene treatment, 1 mM ethephon solution was used for producing ethylene in a sealed container. The solvent for each phytohormone was used in mock treatments. Mock1 (water), control for indole-3-acetic acid, kinetin, and 2,4-D treatments; Mock2 (ethanol), control for ABA, salicylic acid, and jasmonic acid treatments; Mock3 (DMSO), control for BR and gibberellic acid treatments; Mock4 (water), control for ethylene treatment.
(C) Temporal expression pattern of OsPM1 under ABA treatment. Fourteen-day-old seedlings grown in Kimura B liquid media were treated by ABA at a final concentration of 5 μM in the media. Leaves were harvested at the designated time points for RT-qPCR analysis. The expression at the time point 0 h was set as 1 and the relative expression levels of other time points are shown.
(D) The induced expression level of OsPM1 was positively correlated with the applied ABA concentration. Leaves were harvested from the 14-d-old seedlings treated by 5, 25, or 50 μM ABA in the media for different time periods and then subjected to RT-qPCR analysis. The expression level at time point “0 h” was set as 1 and the relative expression levels of other time points are shown.
In (A) to (D), data are shown as means ± sd, and three independent samples each pooled from three individual plants were used for assay. One-way ANOVA was performed for (A) and (C). Different letters indicate significant differences (P < 0.05). Student’s t test was performed for the significance of difference between treatment and the corresponding mock experiment in (B). Two-way ANOVA followed by Bonferroni’s post-hoc test was performed for (D). Different letters with the same superscript indicate significant differences (P < 0.05) between different time points treated by the same concentration of ABA. **P < 0.01 indicates significant differences between samples treated by different concentrations of ABA at the same time points (Supplemental Data Set 2).
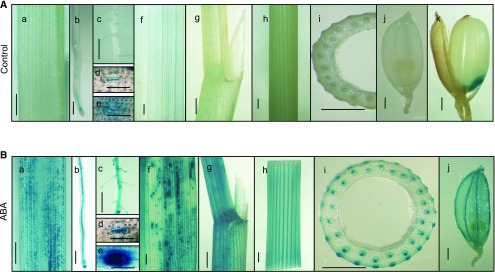
The expression pattern suggested that OsPM1 is closely related to the ABA signaling pathway. To determine whether OsPM1 is specifically regulated by ABA, we treated rice seedlings with various phytohormones and found that OsPM1 was dramatically induced by ABA, weakly induced by brassinosteroid, but not induced by other phytohormones (Figure 1B). OsPM1 responded to ABA application very quickly (within 1 to ∼2 min) in roots and leaves (Supplemental Figures 1C and 1D). When the roots were treated with 5 μM ABA, induction of OsPM1 in leaves could be detected at 30 min and the transcript level continued to increase for over a day and then declined (Figure 1C). The induction amplitude was higher when the concentration of exogenously applied ABA was increased (Figure 1D). GUS staining results of the rice plants transformed with an OsPM1pro:GUS reporter also showed that the gene was expressed weakly in the vasculature, guard cells, and root tip under normal conditions, but strongly induced by ABA at these sites (Figures 2A and 2B). The expression pattern of OsPM1 was generally consistent with the known ABA distribution pattern in tissues.
Figure 2.
Tissue-Specific Expression Pattern of OsPM1 before and after ABA Treatment.
(A) GUS staining of various tissues of the OsPM1pro:GUS transgenic reporter lines under normal growth conditions.
(B) GUS staining of various tissues of the OsPM1pro:GUS reporter lines after being treated by 50 μM ABA for 12 h.
a, leaf; b, root tip; c, root maturation zone; d and e, magnified image of guard cells in (A), representing the guard cells with weak signal (d) and strong signal (e). a to e were all from 14-d-old seedlings; f, g, and h, flag leaf, leaf auricle, and stem from rice plants at reproductive stage; i, the cross section of the stem in h; j, spikelet; k, mature seed. Bars = 1 mm in a to c and f to k, and 50 μm in d and e.
OsPM1 Regulates Rice Drought Tolerance
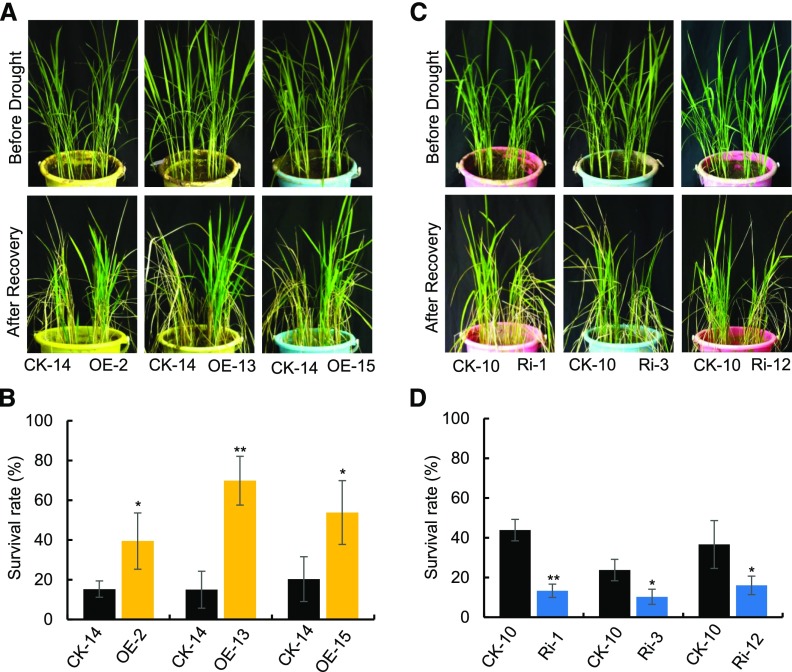
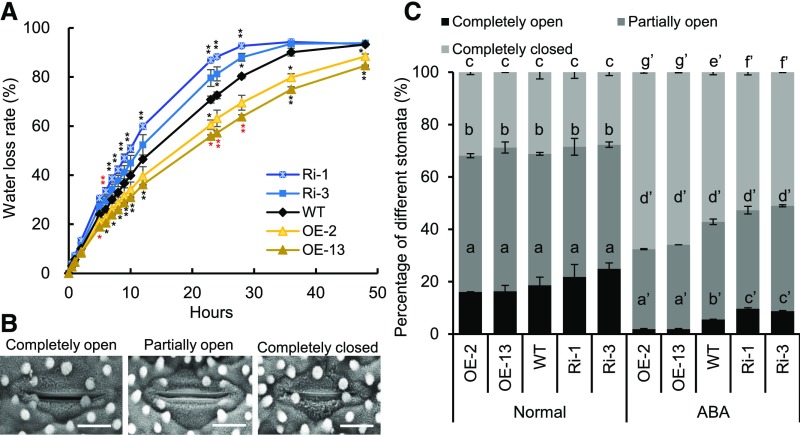
To investigate the physiological role of OsPM1 in rice, overexpression and RNA interference (RNAi) lines of OsPM1 were generated. After examination of the mRNA level of OsPM1 in these transgenic lines (Supplemental Figure 2), three overexpression lines (OE-2, OE-13, and OE-15) and three RNAi lines (Ri-1, Ri-3, and Ri-12) were selected for further studies. To understand the function of OsPM1 in drought response, the overexpression and RNAi lines were all subjected to drought stress, with wild-type plants grown side by side as a control. The overexpression lines showed an obvious drought resistance phenotype and the survival rates of the overexpression lines were much higher than that of the wild type under the same water stress conditions (24.2 to ∼54.9% increase) (Figures 3A and 3B), while those of the RNAi lines were lower than the wild type (13.5 to ∼30.6% decrease) (Figures 3C and 3D). In addition, if the drought stress was imposed during reproductive stage, the seed setting rate of the overexpression lines was significantly higher than that of the wild type (21.1 to ∼26.1% increase) (Supplemental Figures 3A and 3B), although 1000-grain weight showed no big differences (Supplemental Figure 3C), indicating that OsPM1 can improve rice yield by increasing seed setting rate instead of enhancing grain filling under drought stress.
Figure 3.
OsPM1 Regulates Drought Response.
(A) and (B) Phenotypes of OsPM1 overexpression (OE) lines before and after drought.
(C) and (D) Phenotypes of OsPM1-RNAi (Ri) lines before and after drought.
One-month-old plants were drought-stressed as described in Methods. OE plants in (A) were stressed for 14 d along with wild-type control plants (CK-14). Ri plants in (C) were stressed for 10 d along with wild-type control plants (CK-10) since they were more drought sensitive and exhibited phenotype in a shorter stress period. After a recovery period, the plants were photographed. The treatment was performed with at least 40 plants in total for each line. CK, control. OE-2, -13, and -15, three different overexpression lines of OsPM1; Ri-1, -3, and -12, three RNAi lines of OsPM1. In (C) and (D), data are shown as means ± sd for 40 plants from three replicates. Significance of differences between transgenic lines and the corresponding control plants was determined using Student’s t test. *P < 0.05 and **P <0.01 (Supplemental Data Set 2).
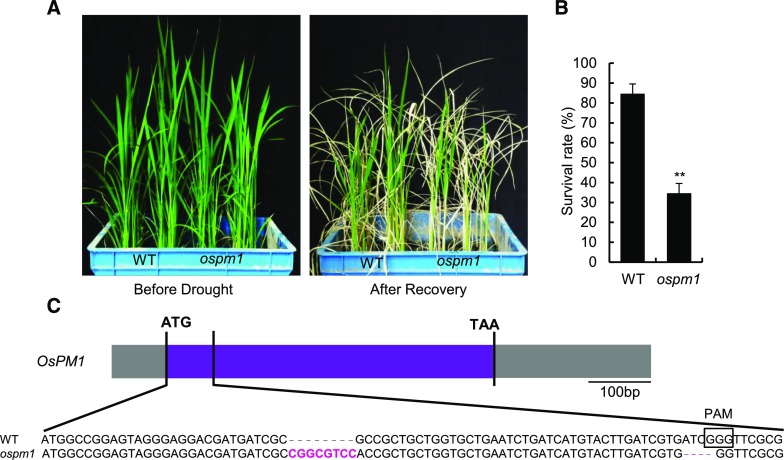
In recent years, the CRISPR/Cas9 gene editing technique has been developed for plants, and we also used this technique to construct a knockout (KO) mutant line of OsPM1. The ospm1 mutant also showed a drought-sensitive phenotype (Figures 4A and 4B), consistent with the phenotype of RNAi lines.
Figure 4.
The ospm1 Knockout Mutant Created by CRISPR-Cas9 Also Shows a Drought-Sensitive Phenotype.
(A) The phenotype of the ospm1 mutant under drought stress. Thirty-five-day-old plants were drought-stressed for 7 d along with wild-type control plants. After being rewatered for 7 d, the plants were photographed.
(B) The survival rates of the ospm1 mutant and wild-type plants under drought stress. Results are means ± sd from three repeats, with 12 plants in each repeat. **P < 0.01 compared with wild-type control (Student’s t test; Supplemental Data Set 2).
(C) The gene editing sites of OsPM1 in the ospm1 mutant. The ORF of OsPM1 is shown in a purple frame. The sequence of the mutated OsPM1 is aligned with that of the wild-type OsPM1 gene. Magenta letters and dashes indicate the InDels introduced by gene editing.
OsPM1 Is Involved in ABA-induced Stomatal Closure
To explore possible cellular processes affected by OsPM1 in improving rice drought tolerance, we compared the water loss rates and ABA-induced stomatal movement in OsPM1 overexpression lines and RNAi lines with that of the wild type. The water loss of detached leaves from RNAi plants was significantly faster than that of wild type, whereas the water loss of the overexpression lines was slower (Figure 5A). Under daylight conditions, rice stomata can be classified into three typical status categories: completely open, partially open, and completely closed (Cui et al., 2015; Figure 5B). There was little difference between transgenic and wild-type plants in the proportions of the three categories of stomata under normal growth conditions. However, the proportions of the completely open stomata in OE-2 and OE-13 plants after ABA treatment were significantly lower than those in the wild type, while the proportions of the completely closed stomata were remarkably higher. In Ri-1 or Ri-3 plants, the stomata showed the opposite changes compared with that of the overexpression lines (Figure 5C). Hence, we conclude that OsPM1 is involved in ABA-induced stomatal closure.
Figure 5.
Water Loss Rates and Stomatal Movement of the Transgenic Lines.
(A) Water loss rates of detached leaves from 70-d-old wild-type, OsPM1 overexpression, and RNAi plants. Results represent means ± sd (n = 5). Significance of difference between wild-type and the transgenic lines at the same time point was determined using Student’s t test. *P < 0.05 and **P < 0.01 (Supplemental Data Set 2).
(B) Scanning electron micrographs of the completely open, partially open, and completely closed stomata. Bars = 10 μm.
(C) Percentages of completely open, partially open, and completely closed stomata in wild-type, OsPM1 overexpression, and RNAi plants under normal and ABA treatment conditions. ABA (5 μM) was sprayed onto leaves and the stomata were observed 30 min afterwards. Results represent means ± sd (n = 500). Two-way ANOVA was performed, followed by Bonferroni’s post-hoc test. Different letters with the same superscript mark indicate significant differences (P < 0.05) between plants under the same conditions.
It has been reported that reduced atmospheric relative humidity (rh) near the leaf surface can trigger stomatal closure rapidly (Grantz, 1990; Assmann et al., 2000; Bauer et al., 2013). To determine whether OsPM1 participates in this process, we measured the changes of leaf conductance in response to an abrupt air rh drop from 70 to 25%. The stomata responded very quickly to low rh in all plants (Supplemental Figure 4). No significant differences could be observed among the wild-type, KO, and RNAi plants. These results suggest that OsPM1 is not required for the quick stomatal response to low air humidity stress.
Seed Germination Is Altered in the OsPM1 Transgenic Lines
Since OsPM1 is highly expressed in seeds under normal conditions, we further observed the seed germination processes of OsPM1 overexpression lines and RNAi lines (Supplemental Figure 5). Under normal conditions, the germination processes of the different lines exhibited only slight differences (Supplemental Figure 5B), with the RNAi lines germinating somewhat faster and the overexpression lines slightly slower than the wild type. When seeds were germinated in the presence of ABA, their germination rates differed more obviously. The overexpression line seeds showed higher sensitivity to ABA compared with wild-type seeds. By contrast, OsPM1-RNAi seeds were less sensitive to ABA (Supplemental Figure 5C). These observations strongly suggest that OsPM1 is involved in ABA-regulated seed germination.
OsPM1 Is a Plasma Membrane Protein
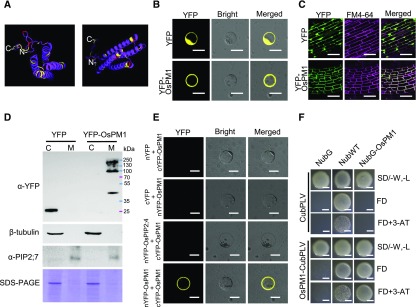
OsPM1 encodes a small protein of 173 amino acids with four typical α-helices (Figure 6A). The α-helices range between 20 and 30 amino acids in length and are abundant in hydrophobic amino acids, suggesting that OsPM1 might be a membrane-spanning protein. No typical signal peptide was identified in the protein using multiple signal peptide prediction programs such as Signal IP4.1 (http://www.cbs.dtu.dk/services/SignalP/). To explore the subcellular localization of rice OsPM1, an YFP-OsPM1 fusion protein was constructed and transiently expressed in rice protoplasts. The fluorescence signal was detected at the plasma membrane (Figure 6B). We also transformed 35Spro:YFP-OsPM1 into rice plants and observed the fluorescence signal in rice root cells. The YFP-OsPM1 signal colocalized with the fluorescence signal of the plasma membrane dye FM4-64 (Figure 6C), confirming its membrane localization. The 35Spro:YFP-OsPM1 transgenic rice had similar drought resistance and ABA-sensitive seed germination phenotypes as the 35Spro:OsPM1 transgenic lines, indicating that the YFP tag did not interfere with the function of the OsPM1 protein (Supplemental Figure 6). We further stably expressed YFP-OsPM1 in Arabidopsis plants and transiently expressed it in tobacco (Nicotiana tabacum) leaf epidermal cells (Supplemental Figure 7), and in all cases, fluorescence signals were localized to cell membrane (Supplemental Figures 7A and 7B). When the roots expressing YFP-OsPM1 were treated with sucrose for plasmolysis, the signal was clearly associated with the cell membrane instead of cell wall (Supplemental Figure 7C).
Figure 6.
OsPM1 Is Located at the Plasma Membrane.
(A) The secondary structure of OsPM1 predicted by PyMOL software (http://robetta.bakerlab.org/queue.jsp).
(B) Subcellular localization of OsPM1 in rice protoplasts. Bars = 20 μm.
(C) Subcellular localization of OsPM1 in transgenic rice plants. Young roots from 10-d-old rice seedlings expressing YFP-OsPM1 or YFP were observed. The fluorescence signal of FM4-64 indicates the plasma membrane. Bars = 50 μm.
(D) Immunoblot analysis of leaf cell lysates from Arabidopsis plants expressing YFP-OsPM1 and YFP. C, cytoplasm; M, plasma membrane. Antibody names are indicated on the left. Antitubulin and anti-PIP2;7 antibodies were used to verify the purity of cytoplasm and the plasma membrane portions, respectively. Coomassie blue staining of the SDS-PAGE gel was included as loading control.
(E) BiFC assay showing that OsPM1 interacts with itself in rice protoplasts. Coexpression of nYFP and cYFP-OsPM1, cYFP and nYFP-OsPM1, and an unrelated membrane protein control, nYFP-OsPIP2;4 and cYFP-OsPM1, were used as the negative controls for nYFP-OsPM1 and cYFP-OsPM1 coexpression in rice protoplasts. Bars = 20 μm.
(F) mbSUS assay showing that OsPM1 interacts with itself in yeast. Wild-type Nub (NubWT) and the mutant Nub (NubG) vectors served as the positive and negative controls of NubG-OsPM1, respectively. CubPLV acted as the negative control for OsPM1-CubPLV. FD indicates SD/-Ade, -His, -Trp, -Leu, -Met. Bars = 1 mm.
To further investigate the presence of OsPM1 at the plasma membrane, we performed cell fractionation to separate the plasma membrane and cytoplasm, and then detected YFP-OsPM1 using anti-YFP antibody. The results indicated clearly that YFP-OsPM1 appeared in the plasma membrane portion but not in the cytoplasm (Figure 6D). Notably, we detected the homodimeric (91.8 kD) and tetrameric (183.6 kD) forms of YFP-OsPM1 in addition to the monomeric form (45.9 kD), while only the monomeric form was seen for the tag protein YFP (27.8 kD). Experimental results from bimolecular fluorescence complementation (BiFC) assays with rice protoplasts and mating-based split ubiquitin system (mbSUS) assays in yeast cells confirmed that OsPM1 could interact with itself on the plasma membrane. Homodimerization of OsPM1 in rice protoplasts mediated the interaction of nYFP-OsPM1 with cYFP-OsPM1 to generate a reconstituted YFP fluorescence signal while the nYFP and cYFP split protein, and an unrelated membrane protein control OsPIP2;4 failed to do so (Figure 6E). In the mbSUS yeast two-hybrid assay, OsPM1-CubPLV interacted with NubG-OsPM1 to allow yeast growth on a stringent selection medium (SD/-Ade, -His, -Trp, -Leu, -Met) with 3-amino-1,2,4-triazol (Figure 6F).
OsPM1 Is an ABA Influx Carrier
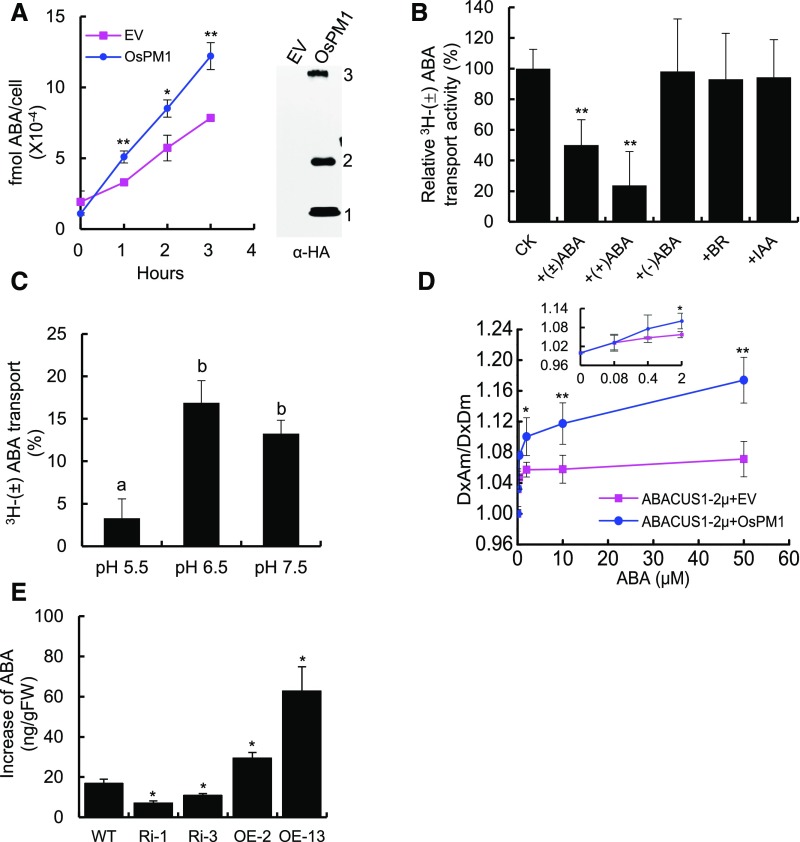
Because OsPM1 is localized at the plasma membrane and plays a role in ABA-induced stomatal movement and seed germination, and its expression pattern is highly correlated with ABA distribution in vivo, we hypothesized that OsPM1 may mediate ABA signal transduction and, more specifically, that it may be involved in ABA transport. To test this, we expressed OsPM1 in yeast cells and used 3H-(±)ABA as a substrate to investigate its transport activity. The yeast cells expressing OsPM1 accumulated more 3H-(±)ABA (Figure 7A). The physiologically active form of ABA, (+)ABA, not (−)ABA, competed with 3H-(±-ABA) in the transport assay, suggesting that (+)ABA is the preferred substrate of OsPM1 (Figure 7B). Although the expression level of OsPM1 is slightly induced by brassinosteroid (BR; Figure 1B), BR did not compete with 3H-(±)ABA in the assay, and neither did the hormone indole-3-acetic acid (IAA; Figure 7B), implying that OsPM1 specifically transported ABA. The transport activity at pH 6.5 and pH 7.5 was higher than that at pH 5.5 (Figure 7C), suggesting a more active role for OsPM1 under a nearly neutral pH condition. The pH values in the xylem and apoplast space increase from pH 5.0 to 6.1 to pH 7.2 to 7.4 under drought stress (Slovik and Hartung, 1992; Wilkinson and Davies, 1997; Hedrich et al., 2001; Boursiac et al., 2013), providing a possible physiological condition close to that for high OsPM1 activity.
Figure 7.
Transport Activity Assay of OsPM1.
(A) Time-dependent uptake of 3H-(±)ABA by yeast expressing OsPM1. The yeast transformed with empty vector (EV) was used as control. This assay was performed with 10 nM 3H-(±)ABA at pH 6.5. Results are means ± sd from three biological replicates. The experiments were repeated three times and results were similar. Immunoblots with anti-HA antibody were used to show the expression of HA-tagged OsPM1 protein in yeast.
(B) Relative 3H-(±)ABA transport activities of yeast expressing OsPM1 in the presence of 100-fold unlabeled ABA, BR, and IAA. 3H-(±)ABA radioactivity in yeast cells was determined at 2 h after the addition of radiolabeled ABA and the ABA transport value was calculated as described in Methods. 3H-(±)ABA transport activity without competitors was set as 100% (CK), and the relative activities in the presence of different potential 3H-(±)ABA competitors are shown. 3H-(±)ABA, 10 nM; (±)ABA, (+)ABA, (−)ABA, BR, and IAA, 1 μM; assay buffer, pH 6.5.
(C) 3H-(±)ABA uptake activities of yeast expressing OsPM1 under different pH conditions. The 3H-(±)ABA transport values were determined at 2 h after the addition of radiolabeled ABA. 3H-(±)ABA, 10 nM. One-way ANOVA with Bonferroni post-hoc test analysis was performed. Different letters indicate significant differences (P < 0.05).
(D) ABA concentration-dependent uptake activity of yeast expressing OsPM1 determined by ABACUS1 sensor. The NubG-OsPM1 construct and the ABA sensor plasmid ABACUS1-2μ were cotransformed into the protease-deficient yeast strain BJ5465 cells. The NubG empty vector cotransformed with ABACUS1-2μ plasmid was used as control. DxAm/DxDm indicates the ABA-induced FRET ratio. The inset shows the ABA transport activity in low ABA concentrations.
(E) The changes of ABA accumulation in wild-type, OsPM1 overexpression, and RNAi plants after exogenous ABA treatment. Twelve-day-old rice seedlings were treated with 5 μM ABA in the media for 1 h, then the leaf sheath ABA contents were determined. The values were obtained by subtracting the ABA level before treatment from the ABA level after treatment in each line. Data are represented as means ± sd (n = 80) from two replicates.
Significance of difference was determined using Student’s t test. *P < 0.05 and **P < 0.01 (Supplemental Data Set 2). OsPM1 compared with EV (A); competitors compared with control (B); ABACUS1-2μ + OsPM1 compared with ABACUS1-2μ + EV (C); transgenic lines compared with the wild type (D).
A fluorescence resonance energy transfer (FRET) method for monitoring ABA translocation and dynamics based on the ABA-PYR/PYL/RCAR-PP2C signaling cascade was reported by two groups recently (Jones et al., 2014; Waadt et al., 2014). The FRET sensor detects the ABA in cells and induces fluorescence changes. We adopted this method to examine the ABA transport activity of OsPM1 and found that the yeast cells expressing OsPM1 assimilated more ABA, to induce stronger fluorescence changes, than the cells transformed with the empty vector control (Figure 7D), consistent with the assay results using 3H-(±)ABA. In addition, to investigate the role of OsPM1 in ABA movement in planta, we monitored the ABA levels in rice leaf sheathes from wild-type, overexpression, and RNAi lines after exogenous ABA treatment. The in planta ABA level in overexpression lines was much higher than that of wild-type plants after ABA treatment (Figure 7E), supporting the role of OsPM1 in moving ABA into cells.
OsPM1 Modulates the Expression of ABA-Responsive Genes
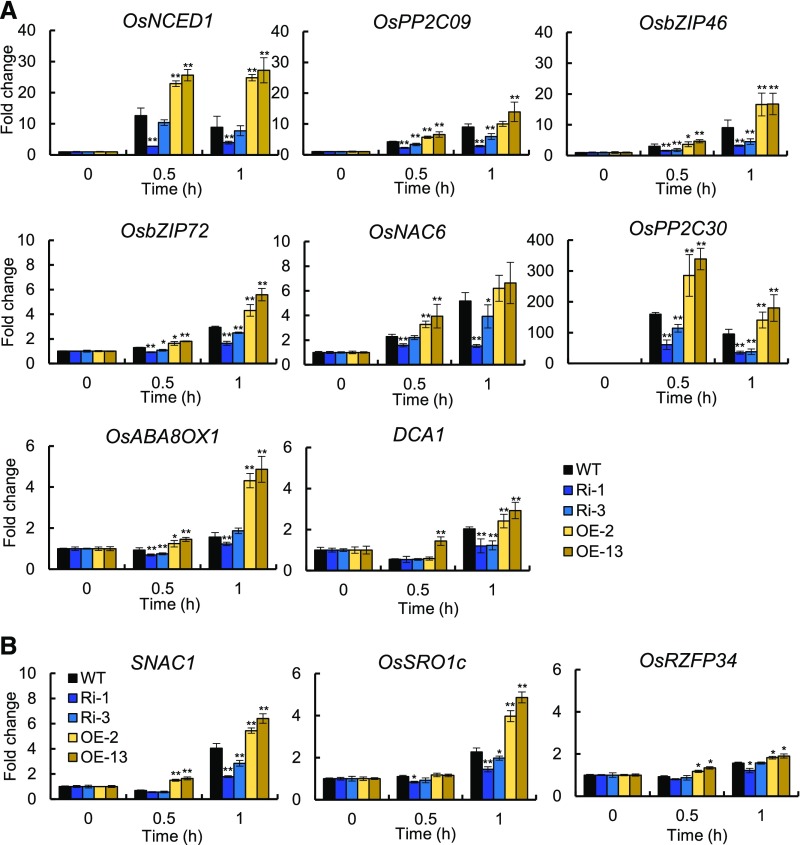
To gain more insights into the relationship between OsPM1 and ABA signaling pathway, we examined the expression levels of ABA-responsive genes in OsPM1 overexpression lines and RNAi lines after ABA treatment for 0.5 and 1 h, respectively (Figure 8). The ABA signaling pathway genes in roots including NCED1, ABA8OX1, PP2C, and the transcription factor-encoding bZIP46, bZIP72, NAC6, and DCA1 were all upregulated in the OsPM1 overexpression lines but downregulated in the RNAi lines compared with the wild type after ABA treatment (Figure 8A). SNAC1 (Hu et al., 2006) and OsSRO1c (You et al., 2013) are two ABA-responsive genes expressed in guard cells, while OsRZFP34 is a leaf ABA-responsive gene (Hsu et al., 2014). We also examined their expression levels for changes in leaves after ABA treatment (Figure 8B). Their transcriptional changes in response to ABA treatment were regulated by OsPM1 expression. These results support the conclusion that OsPM1 is an ABA influx carrier and can modulate the expression of genes functioning in the ABA signaling pathway.
Figure 8.
OsPM1 Modulates the Expression of ABA-Responsive Genes.
Fourteen-day-old seedlings grown in liquid media were treated with 50 μM ABA in the medium for 0.5 h and 1 h. The roots (A) and the leaves (B) were then harvested for RT-qPCR analysis of the indicated ABA-responsive genes. The expression level at the time point “0 h” was set as 1, and the change of expression relative to “0 h” was calculated as the fold change. Results are means ± sd from three replicates. Asterisks indicate the significant differences from the wild type: *P < 0.05 and **P < 0.01 (Student’s t test; Supplemental Data Set 2).
OsPM1 Is Regulated by the Transcription Factor OsbZIP46
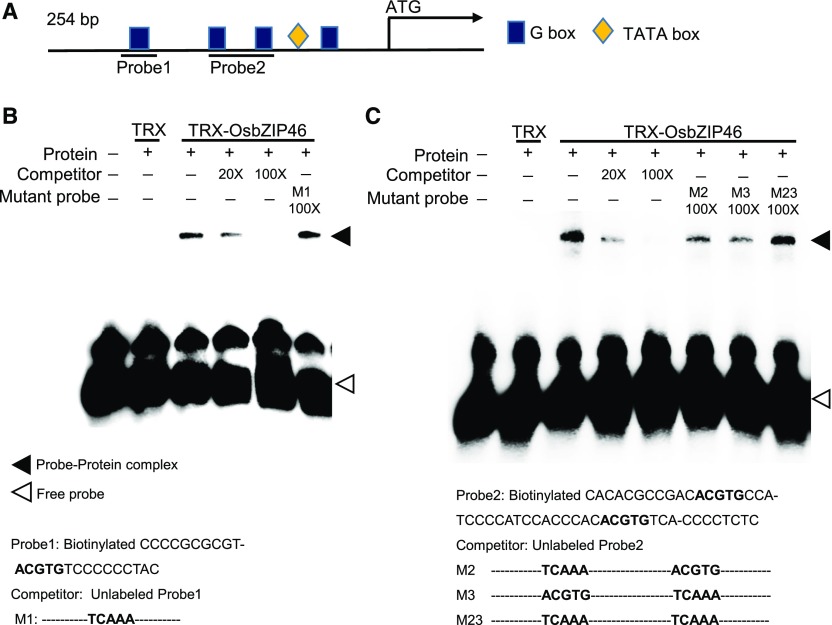
AREB/ABF transcription factors regulate the expression of stress-responsive genes in the ABA-dependent signaling pathway by binding to ABA-responsive elements (ABREs) with a core sequence of ACGT (G-box) (Lenka et al., 2009). In the proximal 254-bp region of the OsPM1 promoter, four ABREs were detected (Supplemental Figure 8A; Figure 9A). This region was sufficient for ABA induction of OsPM1 promoter activity in a dual-luciferase reporter assay with rice protoplasts (Supplemental Figures 8A and 8B), suggesting that OsPM1 is regulated by AREB/ABF transcription factors. Several bZIP transcription factors in rice, including OsbZIP12/ABF1 (Amir Hossain et al., 2010), OsbZIP16 (Chen et al., 2012), OsbZIP23 (Xiang et al., 2008; Zong et al., 2016), OsbZIP46 (Tang et al., 2012, 2016), OsbZIP72 (Lu et al., 2009), and OsbZIP66/TRAB1 (Hobo et al., 1999; Yoon et al., 2017), have been reported to bind to ABREs and act as positive regulators in drought responses. To find the regulator(s) of OsPM1, we performed electrophoretic mobility shift assays (EMSAs) using the OsPM1 promoter fragments containing different G-boxes as DNA probes. When OsbZIP46 was incubated with the probes, there was obvious gel retardation of the labeled probes (Figures 9B and 9C), indicating that OsbZIP46 can bind to the OsPM1 promoter sequence. The intensity of binding signal was reduced by adding excessive unlabeled probe in the reactions, but not by mutated unlabeled probes (Figures 9B and 9C), suggesting that the binding activity of OsbZIP46 to OsPM1 promoter was specific.
Figure 9.
OsbZIP46 Specifically Binds to OsPM1 Promoter in EMSA Analysis.
(A) The proximal promoter structure of OsPM1. Four G-boxes (ABREs) are identified in this region. The locations of the probes used for EMSA are also indicated.
(B) EMSA analysis using OsPM1 promoter Probe1. The thioredoxin-OsbZIP46 fusion protein (TRX-OsbZIP46) produced in Escherichia coli was used in the binding assay. Probe 1 contained a canonical G-box and was biotin-labeled at the N terminus. Unlabeled Probe 1 and the mutated Probe 1 (M1) that lacked the G-box sequence but retained the flanking sequence were used in the competition assay.
(C) EMSA analysis using OsPM1 promoter Probe 2. TRX-OsbZIP46 was used in the binding assays. Probe 2 contained two canonical G-boxes and was biotinylated. Unlabeled Probe 2, probes M2 and M3 each with a single mutated G-box, and M23 with two mutated G-boxes were used in the competition assays.
The probe sequences in (B) and (C) are shown below the EMSA images, with G-boxes and the mutated G-boxes highlighted in bold. Dashed lines in mutant probes indicate the same sequences as in the wild-type probe. The tag protein TRX was used as a control of TRX-OsbZIP46 in (B) and (C).
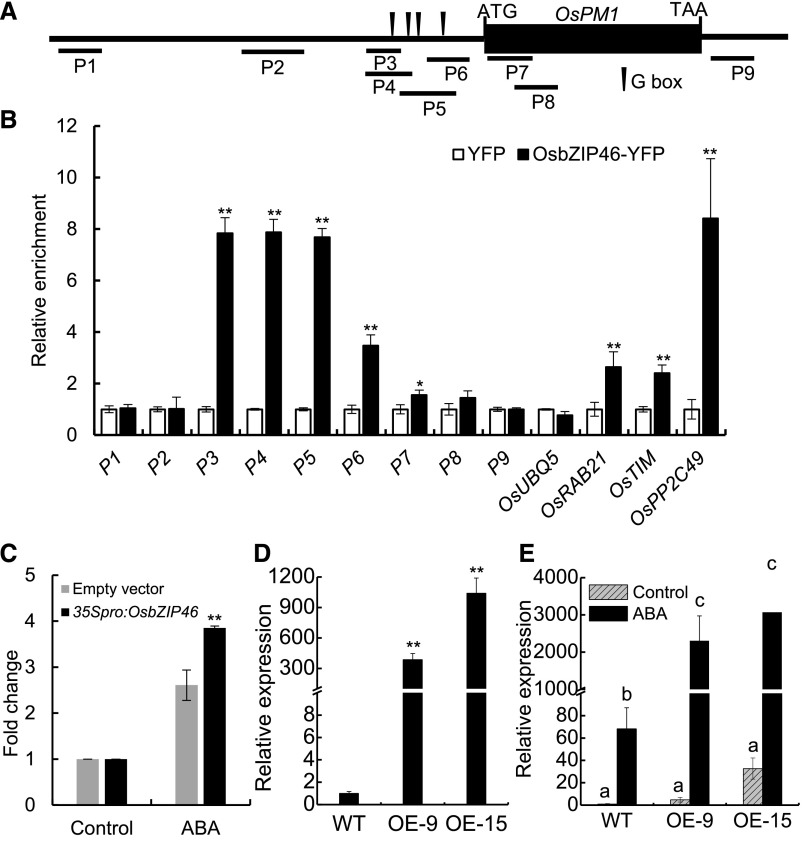
To further determine whether OsbZIP46 recognizes the OsPM1 promoter in vivo, we performed chromatin immunoprecipitation (ChIP)-qPCR assays. GFP-Trap beads binding with bZIP46-YFP were used for coimmunoprecipitation of the associated DNA fragments from the leaves of ABA-treated 35Spro:bZIP46-YFP transgenic plants. qPCR was used to quantify the fold enrichment of specific DNA regions of the OsPM1 promoter precipitated by the GFP antibody. Up to 8-fold enrichment was observed in the DNA regions of the OsPM1 promoter containing G-boxes, while no enrichment was found in other regions of the OsPM1 gene (Figures 10A and 10B). To further confirm the direct regulation of OsbZIP46 on OsPM1, we performed transient transcriptional activity assays in rice protoplasts using the dual-luciferase reporter assay system. OsbZIP46 significantly induced the expression of LUC driven by the OsPM1 promoter (Figure 10C). Furthermore, the expression of OsPM1 in the OsbZIP46 overexpression lines was substantially higher than that in the control plants after ABA treatment (Figures 10D and 10E). These data indicated clearly that OsbZIP46 regulates the expression of OsPM1 in vivo.
Figure 10.
OsbZIP46 Binds to the OsPM1 Promoter and Activates OsPM1 Expression in Vivo.
(A) Schematic diagram of OsPM1 gene showing the positions of P1-P9 fragments amplified by ChIP-qPCR.
(B) ChIP-qPCR analysis of the gene fragments enriched by OsbZIP46 in rice plants. Seventy-day-old rice plants expressing YFP-OsbZIP46 were sprayed with 100 μM ABA and used for ChIP using an anti-YFP antibody. Plants expressing YFP alone were also treated and used as a control. The relative amounts of coprecipitation of each DNA fragment by YFP-OsbZIP46 were quantified by qPCR and the level by YFP control was set as 1. OsUBQ5 was used as a negative control. OsRAB21, OsTIM ,and OsPP2C49 are positive controls known to be regulated by OsbZIP46.
(C) Dual-luciferase reporter assay in rice protoplasts. The pGreenII 0800-LUC reporter plasmid carrying the OsPM1 promoter was introduced together with 35Spro:OsbZIP46 construct or with the empty vector control into rice protoplasts. Firefly and Renilla luciferase activities in the protoplasts with or without 50 μM ABA treatment were quantified after 12 h. The OsPM1 promoter-driven firefly luciferase activity was normalized by the internal Renilla luciferase activity.
(D) Relative expression of OsbZIP46 in OsbZIP46 overexpression lines. The expression level of OsbZIP46 in the wild type was set as 1. Zhonghua 11 was used as wild-type control. OE-9 and OE-15 are two overexpression transgenic lines generated with Zhonghua 11 background.
(E) OsPM1 expression in OsbZIP46-overexpression lines. Fourteen-day-old rice seedlings were treated with or without 50 μM ABA for 12 h. The leaves were sampled for RT-qPCR analysis. The OsbZIP46 expression level in the wild type without treatment (control) was set as 1.
In (B) to (E), results are represented as means ± sd from three replicates. For (B) to (D), Student’s t test was performed (Supplemental Data Set 2): *P < 0.05 and **P < 0.01. For (E), two-way ANOVA was used (Supplemental Data Set 2). Different letters indicate significant differences (P < 0.05).
DISCUSSION
Among plant hormones, auxin is the one with the most clearly understood transport mechanism to date. Auxin transporters including AUX1/LAX family, PIN family, and ABC family proteins have been identified and extensively studied (Petrásek and Friml, 2009; Zazimalova et al., 2010). These transporters coordinate auxin flow, although there is no sequence similarity among these protein families. To date, the majority of the identified ABA transporters come from G-type subfamily of ABC transporters, which transport a large array of molecules in plants (Kuromori and Shinozaki, 2010). Some members of the B-type ABC family proteins are also able to transport auxin, such as ABCB1 and ABCB19 (Noh et al., 2001; Verrier et al., 2008). NPF4.6/AIT1/NRT1.2 and DTX50 are two ABA transporters distinct from ABC family proteins. NPF4.6/AIT1/NRT1.2 was originally considered to transport nitrate (Kanno et al., 2012, 2013), and DTX/MATE family proteins can extrude toxic compounds and organic acids from cells (Omote et al., 2006). No sequence similarities are found among these known ABA transporters of different protein families. Therefore, it seems common that proteins with different origins play a similar role in hormone transport.
AWPM-19 homologs have been found in different species (Koike et al., 1997; Ranford et al., 2002; Rerksiri et al., 2013; Barrero et al., 2015; Chen et al., 2015). AWPM-19 protein was first isolated from plasma membranes of ABA-treated wheat suspension cells. After treatment with ABA, the wheat suspension cells accumulate abundant AWPM-19 protein and acquire freezing tolerance (Koike et al., 1997). An AWPM-19 like protein was also identified in the oil bodies of Arabidopsis (Vermachova et al., 2011). In wheat (Triticum aestivum), two AWPM-19 family members, PM19-A1 and -A2, were identified from near-isogenic lines as major quantitative trait loci controlling seed dormancy (Barrero et al., 2015). They had much higher expression levels in the maturing grain in dormant genotypes than nondormant ones, but the underlying molecular functions are unknown.
To understand the evolutionary relationships among AWPM-19 family members, we searched for homologous sequences of OsPM1 in public databases and constructed a phylogenetic tree (Supplemental Figure 9). Seven AWPM-19 like proteins (OsPM1 to OsPM7) were identified in rice. The amino acid sequence identities among them are 30.5 to 78.6%, with the C-terminal region being most variable (Supplemental Data Set 1). In Arabidopsis, four AWPM-19 like proteins were found, and the member closest to OsPM1 is At1g04560 (identity 60.1%). This gene is also highly expressed in the embryo at the later developmental stages, but almost undetectable in other tissues. It is moderately induced by mannitol and ABA but only weakly by salt (http://bbc.botany.utoronto.ca/). The relative gene expression level compared with UBIQUITIN after induction is far lower than that in rice when determined by RT-qPCR (Supplemental Figure 10). It remains to be investigated if it plays the same role as rice OsPM1.
Among the rice AWPM-19 like genes, OsPM1 shows the strongest response to ABA, drought, and osmotic stress (Supplemental Figure 11). OsPM2, OsPM3, and OsPM7 were also responsive but the expression levels were much lower. Other family members did not show obvious ABA-inducible expression (Supplemental Figure 11). We detected the dimeric and tetrameric forms of OsPM1 in both plant and yeast cells, suggesting that OsPM1 may exist at the plasma membrane as homomultimers. However, different rice AWPM-19 members may also form heteromultimers. In wheat suspension cells, the coisolated 19-kD polypeptides can be further separated as a major band (AWPM-19) and several minor components (Koike et al., 1997), suggesting that the family members may exist as heteromers on the plasma membrane in wheat.
OsPM1 is a basic protein with a molecular mass of 18.1 kD and pI of 9.5. No ATP binding domain or other known structural domains have been found in this protein. Four highly hydrophobic regions were predicted to form membrane-spanning α-helices. Both N-terminal (11 amino acids) and C-terminal (32 amino acids) regions are short and basic and predicted to be cytosol-oriented according to the positive-inside rule (von Heijne, 1992). The extracellular loop regions are also rich in basic amino acids. It remains unknown whether OsPM1 cotransports other ions. A similar channel with four α-helices, formed by the M2 integral membrane protein found in influenza virus, facilitates proton entry into cells (Schnell and Chou, 2008). Structural determination will be necessary to uncover the mechanism of its action.
Under normal conditions, OsPM1 was expressed weakly in most tissues except in developing seeds. The expression pattern of OsPM1 was consistent with the ABA accumulation pattern in different tissues (Supplemental Figures 1A and 1B). Disruption of OsPM1 did not cause a striking growth phenotype under nonstressed growth conditions, suggesting that OsPM1 is not critical for normal plant growth except seed development. However, the adaptive stress responses were altered in the KO and RNAi plants, similar to the phenotypes observed for mutants defective in previously identified ABA transporters. Under drought stress, ABA is actively synthesized, and the signal is transduced to the whole plant to coordinate cellular responses in various tissues. In the meantime, apoplastic space and xylem sap undergo physiological alkalization (Wilkinson and Davies, 1997; Boursiac et al., 2013), resulting in more apoplastic ABA− retention due to the deprotonation of ABA (Davies and Zhang, 1991). Thus, ABA transporters are important for facilitating the passage of the nondiffusible ABA− across the membrane. The stronger transport activity of OsPM1 at near neutral pH conditions could be due to the fact that more ABA is converted into ABA− at a neutral pH condition than that at acidic conditions. In addition, the negatively charged form, ABA−, would be more attracted to the OsPM1 protein, which is positively charged in the extracellular domains according to the secondary structure prediction. An optimal activity between pH 6.0 and pH 7.0 is also observed for another ABA transporter AtDTX50 (Zhang et al., 2014) and for the auxin permease AUX1 (Yang et al., 2006).
Although our studies have shown that OsPM1 was involved in ABA uptake under drought stress, it did not affect dry air-induced stomatal closure. Recent studies indicated that rapid stomatal response to dry air is guard cell autonomous and does not need exogenous ABA import (Bauer et al., 2013; Merilo et al., 2015). This may explain why the stomatal response to low air humidity was not significantly changed in OsPM1 OE, KO, and RNAi plants.
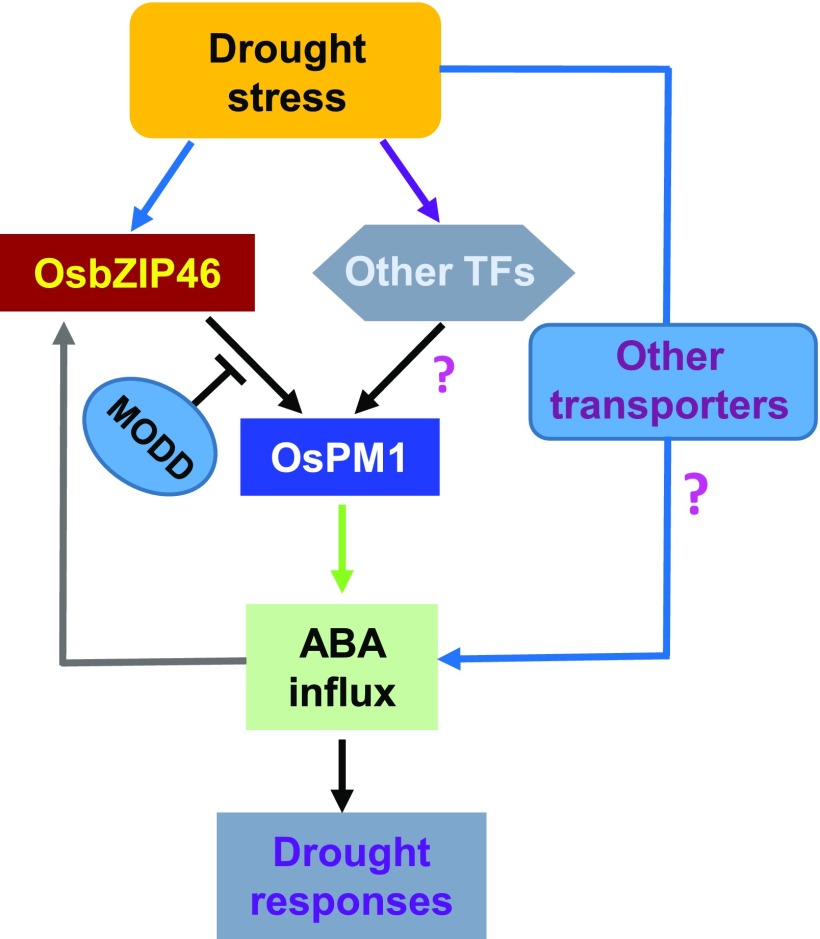
A working model for OsPM1 in the ABA signaling pathway under drought stress is shown in Figure 11. OsPM1 is regulated by OsbZIP46 and plays a role in facilitating ABA influx into cells to induce drought responses. OsbZIP46 is a major transcription factor mediating drought response in rice and can regulate the expression of many ABA-responsive genes (Tang et al., 2012, 2016). The transcriptional activity of OsbZIP46 is activated due to phosphorylation by SAPK kinases (Tang et al., 2012), and overexpression of a constitutively active form of OsbZIP46 enhances rice drought tolerance (Tang et al., 2012). Our results showed that OsPM1 was induced by ABA within a very short time (1–2 min), while the transcription upregulation of OsbZIP46 could only be observed from 0.5 to 1 h for roots, and at 6 h for leaves after ABA treatment (Supplemental Figure 12), indicating that the rapid ABA-induced OsPM1 expression is not due to the slow OsbZIP46 transcriptional change. However, it is possible that preexisting inactive OsbZIP46 protein is activated by phosphorylation quickly following ABA treatment and then activates OsPM1 expression. Furthermore, the increased OsPM1 level can also promote the expression of OsbZIP46, providing a potential positive feedback loop, allowing rapid enhancement of ABA responses regulated by OsbZIP46. To prevent overresponse, a braking system would be necessary to downregulate ABA signaling. It was reported that MODD, a nuclear protein homologous to the rice ABSCISIC ACID-INSENSITIVE5 binding protein AFP, acts as a negative regulator of ABA signaling by mediating deactivation and degradation of OsbZIP46 (Tang et al., 2016). We also observed that at 24 h after ABA treatment, the expression of OsPM1 was downregulated (Figure 1C), consistent with an attenuation mechanism of ABA signaling.
Figure 11.
A Working Model for OsPM1 Function under Drought Stress.
Under drought stress, the transcription factor OsbZIP46 is activated by phosphorylation, which then activates the expression of OsPM1. OsPM1 protein allows ABA influx, which amplifies and enhances ABA signaling in a very short time, including the transcriptional upregulation of OsbZIP46 and many other ABA signaling pathway genes. MODD protein may be necessary to interrupt the feedback loop between OsbZIP46 and OsPM1. In addition, the initial ABA influx is mediated by low level of OsPM1 or other unknown ABA transporters at the plasma membrane, and other transcription factors besides OsbZIP46 may also regulate the expression of OsPM1 under drought stress. Arrowheads indicate positive regulation. Question marks represent presumed pathways.
In addition to OsbZIP46, OsPM1 may also be regulated by other transcription factors (Figure 11), considering its quick induction and dramatic expression increase under drought stress. Possible candidates include the transcription factors binding to ABREs or other cis-elements found in the promoter region, such as potential MYB, MYC, and WRKY binding sites (Supplemental Figure 8A). It has been reported that a transcription factor in the ABA-independent signaling pathway, NAC045, is a positive regulator of OsPM1 (Zheng et al., 2009). Therefore, OsPM1 may be regulated by transcription factors from both ABA-dependent and -independent pathways. Moreover, we also propose that other unidentified ABA transporters exist in rice (Figure 11), based on the findings of ABA transporters in Arabidopsis. The most likely candidates may come from the ABC transporter family. It was reported that the half-size ABCG protein RCN1/OsABCG5 is involved in ABA accumulation in guard cells, although its direct ABA transport activity has not been demonstrated (Matsuda et al., 2016).
Taken together, our study identified an ABA transporter in rice and revealed that this transporter is positively regulated by the transcription factor OsbZIP46. Our results also showed that overexpression of OsPM1 confers drought tolerance without a growth penalty under normal conditions, indicating that it is an ideal candidate for breeding rice cultivars with improved drought tolerance, considering that rice growth area is frequently confronted with arid weather.
METHODS
Plant Materials and Growth Conditions
All rice materials used in this study are of Oryza sativa japonica cv Nipponbare background. They were grown under a 12-h-light (30°C)/12-h-dark (22°C) photoperiod with 50% humidity unless otherwise specified. Full spectrum natural light was used, with 500-W sodium lamps and halogen lamps arranged at an interval of one meter for supplemental lighting when the natural light intensity is less than 375 μmol m−2 s−1.
Generation of Transgenic Lines
For generating OsPM1 (LOC_Os05g31670.1) overexpression lines, the open reading frame (ORF) of OsPM1 was amplified from Nipponbare rice by RT-PCR and then cloned into the binary expression vector pCAMBIA1301U using BamHI/SacI sites with the maize ubiquitin promoter replacing the cauliflower 35S promoter (Guo et al., 2016). The construct was transformed into rice by Agrobacterium tumefaciens-mediated transformation method (Hiei and Komari, 2008). For generating OsPM1 RNAi lines, the OsPM1 cDNA-specific sequence was amplified and cloned into an intermediate vector pGEM-RNAi to generate a hairpin fragment, which was then cloned into the pCAMBIA1301U vector and used for further rice transformation. The primers used in clone construction for overexpression and RNAi lines are listed in Supplemental Table 1. The homozygous transgenic lines were verified at the genomic DNA and RNA levels before use for further experiments (for primers, see Supplemental Table 1).
Mutation of OsPM1 Gene by the CRISPR-Cas9 Technique
The vectors and methods for editing rice genes by the CRISPR-Cas9 technique were described previously (Miao et al., 2013). Spacer sequence of OsPM1 was designed by CRISPR Primer Designer (http://ship.plantsignal.cn/CRISPR/crispr_primer_designer.html). The primers for cloning spacer sequence into pOs-sgRNA vector are given in Supplemental Table 1. The fragment containing U3 promoter, OsPM1 spacer, and sgRNA sequence was cloned into the vector pH-Ubi-cas9-7 by LR reaction (Miao et al., 2013). The construct was then transformed into Nipponbare rice by Agrobacterium-mediated transformation method. To identify the mutant, the OsPM1 gene fragment was amplified (for primers, see Supplemental Table 1) with PrimeSTAR GXL DNA Polymerase (TaKaRa) and sequenced.
Phenotype Observation and Expressional Studies
Drought Stress at Vegetative Stage
Transgenic and wild-type plants were grown in the same pot for 30 d. For drought treatment of the OsPM1-overexpression lines and wild-type control, water was withheld until the leaves of wild-type plants became completely wilted, then the soil water content was monitored and maintained at 15% by replenishing a small amount of water every day for 14 d. For drought treatment of OsPM1-RNAi lines and wild-type control, water was withheld until the leaves of OsPM1-RNAi lines wilted completely, then the soil water content was maintained at 15% as described above for 10 d. After drought treatment, all plants were allowed to recover for an additional 7 d under well-watered conditions, then photographed and analyzed for the survival rates.
Drought Stress at Reproductive Stage
Rice plants were grown for about two months in greenhouse until the plants switched into reproductive growth. The initiation of reproductive growth was confirmed by checking for the appearance of panicle primordium. The reproductive-stage plants were deprived of water supply until the leaves wilted completely. A soil water content of 15% was then maintained for 10 d by supplementing the water lost every day. After rewatering, all plants were grown under standard growth conditions until ripening. Panicles were photographed after harvest. Seed setting rates as well as the 1000-grain weights were calculated as previously described (Jin et al., 2013).
For expressional studies under drought stress, rice plants grown in soil for 2 weeks (seedlings) or about 2 months (reproductive-stage plants) in greenhouse were used for drought treatment as described above. Once the leaves start to wilt, different tissues were sampled for detection of the expression level of OsPM1 by RT-qPCR. Tissues harvested from the plants at the same stage without treatment were used as control.
To check the transcriptional changes of OsPM1 induced by different phytohormones, rice plants were grown in a growth chamber for 6 d under standard conditions, then a final concentration of 5 μM ABA, IAA, kinetin, 2,4-D, salicylic acid, jasmonic acid, and gibberellic acid, or 10 nM BR were added into the culture media to treat the plants for 12 h. The solvent for each hormone was used as control to perform the same treatment. For ethylene treatment, the seedlings were put in a sealed container and treated for 12 h by ethylene released from 200 mL 1 mM ethephon solution in the container. The same amount of water was used instead of ethephon solution as the mock experiment. The whole seedlings were sampled for detecting the expression level of OsPM1.
RNA Isolation and RT-qPCR
Total RNA was extracted using the TRIzol reagent (TaKaRa) and reverse-transcribed with PrimeScript RT reagent (TaKaRa). qPCR was performed with the SYBR Premix Ex Taq kit (TaKaRa) using the CFX96 real-time PCR detection system (Bio-Rad). Rice UBQ5 gene was used as an internal control to normalize different samples. The RT-qPCR primers are given in Supplemental Table 1.
GUS Staining
OsPM1pro:GUS reporter vector was constructed by cloning 2152-bp promoter fragment upstream of the start codon of OsPM1 into pCAMBIA1300 vector using HindIII/BamHI sites (for primers, see Supplemental Table 1), then used for agrobacteria-mediated rice transformation. GUS activity was analyzed by staining the transgenic T2 plants. Tissues were treated by 90% cold acetone for 5 min and then washed with staining buffer without X-Gluc. The staining solution [0.0216 M NaH2PO4, 0.029 M Na2HPO4, 2 mM K3Fe(CN)6, 2 mM K4Fe(CN)6, 0.1% Triton X-100, and 1 mg/mL X-Gluc] was then added into the samples, which were treated under a vacuum for 10 min. After staining at 37°C overnight, the tissues were cleared in ethanol and photographed using the S8APO stereomicroscope (Leica).
Rice Stomata Imaging and Analysis
Plants (21 d old) were sprayed with 5 μM ABA until the solution ran off the surface. The plants without ABA treatment were used as control. After 30 min, the leaves were detached and immediately fixed in FAA solution (2% formaldehyde, 5% acetic acid, 63% alcohol, and 5% glycerol) at 4°C for 24 h. After dehydration in an ethanol series (60–100%, with 5% as a gradient interval), the leaves were submerged in a graded series of tert-butyl alcohol (TBA) solutions (ethanol:TBA [v/v], 3:1, 2:2, 1:3) for 15 min each and twice in 100% TBA. Then, the materials were transferred into 100% t-butanol. After incubation in t-butanol at 4°C for 30 min, they were freeze-dried using a VFD-21S t-BuOH freeze-dryer (SHINKKU VD). The materials were stuck onto the sample stage and sprayed with gold by a MSP-1S magnetron sputter (SHINKKU VD). Stomata were imaged using a scanning electron microscopy (TM3000) according to the manufacturer’s instructions. Five hundred stomata of each line were observed and the completely open, partially open, and completely closed stomata were analyzed as described previously (Zhang et al., 2011a; Cui et al., 2015).
Water Loss Rate Measurement
Flag leaves of similar size from 70-d-old plants were detached and placed in Petri dishes at room temperature, and the fresh weights were monitored at indicated time points. The water loss rate (WLR) was calculated based on the following formula WLR (%) = (fresh weight – desiccated weight)/fresh weight × 100% (Wei et al., 2014).
Seed Germination Assay
Fifty seeds were sterilized and placed on wet filter papers in a Petri dish and then germinated in a growth chamber under standard conditions. The filter papers were wetted thoroughly with water or 2.5 or 5 μM ABA for control or ABA treatments, respectively.
Subcellular Localization
35Spro:YFP-OsPM1 was obtained by cloning the OsPM1 ORF fragment into pCAM-N-EYFP vector using SacI/KpnI sites. Rice protoplasts was prepared from sheaths of 10-d-old rice seedlings based on the methods described before (Zhang et al., 2011b). 35Spro:YFP and 35Spro:YFP-OsPM1 plasmids were transformed into rice protoplasts by the polyethylene glycol (PEG)-mediated transformation method (Yoo et al., 2007; Zhang et al., 2011b). YFP florescence was detected using a confocal laser scanning microscope (Leica).
35Spro:YFP and 35Spro:YFP-OsPM1 were transformed into rice and Arabidopsis thaliana for stable expression (Clough and Bent, 1998; Hiei and Komari, 2008). Rice roots from 10-d-old transgenic plants were subjected to YFP signal observation. FM4-64 was used to stain the plasma membrane as described previously (Lee et al., 2010; Eom et al., 2011). One-month-old Arabidopsis leaves were used for plasma membrane and cytoplasm fractionation based on the published method (Alexandersson et al., 2004). The anti-YFP (Abmart; M20004), anti-β-tubulin (Beyotime; AT819), and anti-PIP2;7 (Agrisera; AS12 2110) antibodies were used for detecting the proteins in the immunoblot. For transient expression in tobacco (Nicotiana tabacum) leaves, the vectors were transformed into Agrobacterium GV3101 and then used for infiltration into tobacco leaves by syringe injection. Three days later, the fluorescence signal was observed.
mbSUS Assay
OsPM1 ORF fragment was cloned into pMetYCgate vector and pNXgate32-3HA vector based on the instructions (Grefen et al., 2007) to generate Met:OsPM1-CubPLV and ADH1:NubG-OsPM1 plasmids, respectively (for primers, see Supplemental Table 1). The plasmids were then cotransformed into the yeast strain THY.AP4. Diploid yeasts were selected on SC medium lacking leucine and tryptophan. The successfully mated yeasts were replica-stamped onto FD plates (SD/-Ade, -His, -Trp, -Leu, -Met) or FD plates including 5 mM 3-amino-1,2,4-triazol as a competitive inhibitor of the HIS3 reporter gene product for more stringent selection.
BiFC Assay
OsPM1 ORF fragment was cloned into the pXY105 and pXY106 vectors using BamHI/XbaI sites (Yu et al., 2008), to generate 35Spro:cYFP-OsPM1 and 35Spro:nYFP-OsPM1 plasmids (primers are given in Supplemental Table 1). The nYFP and cYFP empty vectors and an unrelated membrane protein OsPIP2;4 (Kumar et al., 2014) construct 35Spro:nYFP-OsPIP2;4 were used as the negative controls for the assay. The nYFP and cYFP construct pairs were cotransformed into rice protoplasts by PEG-mediated transformation method. Fluorescence signals were visualized with a confocal laser scanning microscope (Leica).
Transport Assay Using 3H-(±)ABA
OsPM1 ORF fragment was cloned into pNXgate32-3HA vector (for primers, see Supplemental Table 1). The construct was transformed into the yeast strain w3031b. Yeast cells were cultured in SD/-Trp medium and harvested during the exponential growth phase. After washing twice with the reaction buffer (1 mM KCl, 1 mM MES, and 20 mM glucose, at pH 6.5), the cells were resuspended in the same buffer at a concentration of OD600 = 16. 3H-(±)ABA (50 μM, 0.74 TBq mmol−1; ARC) was then added into the cell suspensions at a final concentration of 10 nM. At the time points indicated, 200 μL of homogeneous cell suspensions was filtered with nitrocellulose membranes (Sartorius Stedim). Cells retained on the filter membrane were washed five times with 2 mL of ice-cold reaction buffer, and the radioactivity on the filter was determined by a liquid scintillation counter (LS6500; Beckman). The 3H-(±)ABA transport value was obtained by the formula (T-C)/T: The total radioactivity of the OsPM1-expressing yeast (T) minus the radioactivity of the yeast transformed with empty vector control (C) and then divided by T. For relative 3H-(±)ABA transport activity assay with different competitors, 4 mM ATP was added into the reaction buffer for all samples 1 h before the assay.
Transport Assay Using a FRET Sensor
The pNXgate32-3HA vector carrying OsPM1 and the ABA uptake sensor plasmid ABACUS1-2μ (Jones et al., 2014) were cotransformed into the yeast strain BJ5465. ABA uptake was detected as described previously (Jones et al., 2014) with minor modifications. Cells were cultured in SD/-Trp, -Ura medium and harvested during the exponential growth phase. After washing twice, the cells were resuspended in 20 mM MES reaction buffer (pH 4.7) to OD600 = 1.6. Different concentrations of ABA (in 20 mM MES buffer) were added and incubated with the yeast cells for 5 min. Then the fluorescence signals of the yeast culture were determined by the Cell Imaging Multi-mode Reader Cytation 3 System (BioTek). The donor fluorescence (eCFP) DxDm was determined at 428-nm excitation and 485-nm emission; acceptor fluorescence (eYFP) AxAm was determined at 500-nm excitation and 535-nm emission; and fluorescence transferred from donor to acceptor DxAm was determined at 428-nm excitation (eCFP) and 530-nm emission (eYFP). The ratio DxAm/DxDm was calculated as the FRET ratio.
In Planta ABA Level Measurement
Twelve-day-old rice seedlings were treated with 5 μM ABA added in the Kimura B nutrient medium (Tanoi et al., 2011; Anugoolprasert et al., 2012) for 1 h. The seedlings grown in the same medium without ABA were used as the control. Rice sheaths were collected and weighed, then frozen in liquid nitrogen and ground into powder. ABA in tissues was extracted by 80% methanol buffer with 200 mg/L sodium diethyldithiocarbamate trihydrate (Liu et al., 2014). The ABA content was measured according to the manufacturer’s instructions of Phytodtek ELISA kit (Agdia).
Protein Expression and EMSA
Recombinant OsbZIP46 protein was expressed in Escherichia coli and used for EMSA analysis. OsbZIP46 ORF fragment was cloned into pET-32a (+) vector using BamHI/XhoI sites (primers are included in Supplemental Table 1) and then transformed into Origami (DE3) based on the standard protocol (Novagen). The pET-32a (+) empty vector was also transformed for tag protein expression, and the tag protein Thioredoxin (TRX) served as a negative control in EMSA. Recombinant proteins were purified with a column filled with Nickle Sepharose 6 Fast Flow (GE Healthcare). EMSA analysis was performed according to the manufacturer’s instructions of the LightShift Chemiluminescent EMSA Kit (Thermo Scientific).
Dual-Luciferase Assay
The 254-bp promoter fragment upstream of the start codon of OsPM1 was cloned into pGreenII 0800-LUC reporter vector (Hellens et al., 2005). The coding region of OsbZIP46 was integrated into pCAMBIA1306-3XFlag vector using SacI/XbaI sites for OsbZIP46 expression (primers are included in Supplemental Table 1). The two recombinant plasmids were cotransformed into rice protoplasts by PEG-mediated transformation method (Yoo et al., 2007; Zhang et al., 2011b). The pGreenII 0800-LUC vector carrying OsPM1 promoter was cotransformed with the pCAMBIA1306-3XFlag empty vector to act as a negative control. Sixteen hours after incubation in dark at 28°C, the rice protoplasts were harvested. Firefly and Renilla luciferase activities in the protoplasts were quantified using a dual-luciferase assay kit based on the manual (Promega). The chemoluminescence was determined by the Synergy 2 Multi-Mode Microplate Reader (Bio-Tek).
ChIP-qPCR Analysis
The ChIP experiments were performed as described previously (Saleh et al., 2008) with some modifications. YFP and OsbZIP46-YFP overexpression plants were grown in the greenhouse with 12-h-light 30°C/12-h-dark 22°C cycle and 50% humidity for 70 d, then sprayed with 100 μM ABA. After 16 h, the second youngest leaf from five plants of each line was sampled and fixed in 1% formaldehyde by vacuum infiltration for 1.5 h in total. Nuclei were then isolated from these leaves and sonicated with a Bioruptor Plus sonication device UCD300 (Diagenode) to obtain DNA fragments around 0.5 kb. GFP-Trap agarose beads (ChromoTek) were used to pull down the OsbZIP46-YFP associated complex by incubation with the sonicated samples overnight at 4°C with rotation. The beads were then washed, and the DNA associated was separated. The coprecipitated level of each DNA fragment was quantified by qPCR. The primer sets of different genes for qPCR are listed in Supplemental Table 1. The input DNA level was normalized with UBIQUITIN as the internal control. The coprecipitated levels of the specific gene promoter regions from 35Spro:YFP transgenic plants was set as 1, and the relative enrichment of the gene fragments from 35Spro:ObZIP46-YFP plants compared with that from 35Spro:YFP plants was calculated as the enrichment fold.
Phylogenetic Analysis
Sequences of the land plant lineages except Ginkgo biloba were chosen according to the blast results online (https://phytozome.jgi.doe.gov/pz/portal.html) via the embedded BLASTP algorithm with an expectation threshold value of 0.00001. The sequences in G. biloba were obtained from the genome sequencing results (Guan et al., 2016). Multiple amino acid sequences were aligned using the MAFFT (Multiple Alignment using Fast Fourier Transform) v7.273 software (http://mafft.cbrc.jp/alignment/software/; Katoh and Standley, 2013), and the sequences of low quality were automatically removed using the trimAl v1.4 software with default values (Capella-Gutiérrez et al., 2009). The approximate maximum likelihood tree was generated by FastTree v2.1.7 software (http://www.microbesonline.org/fasttree/; Price et al., 2009). Support values were computed by Shimodaira-Hasegawa test with 1000 resamples (Guindon et al., 2010). The sequence IDs used for this analysis are given in Supplemental Table 2.
Accession Numbers
Sequence data for the genes described in this article can be found in the rice database RAP-DB (http://rapdb.dna.affrc.go.jp/) under the following accession numbers: OsPM1, LOC_Os05g31670; OsPM2, LOC_Os07g24000; OsPM3, LOC_Os02g49860; OsPM4, LOC_Os01g50440; OsPM5, LOC_Os01g14530; OsPM6, LOC_Os05g33220; OsPM7, LOC_Os10g32720; OsbZIP46; LOC_Os06g10880; OsNCED1, LOC_Os03g44380; OsPP2C09, LOC_Os01g62760; OsPP2C30, LOC_Os03g16170; OsbZIP72, LOC_Os09g28310; OsNAC6, LOC_Os01g66120; OsABA8OX1, LOC_Os02g47470; DCA1, LOC_Os10g31850; OsUBQ5, LOC_Os01g22490; OsTIM, LOC_Os03g19290; OsRAB21, LOC_Os11g26790; OsPP2C49, LOC_Os05g38290; SNAC1, LOC_Os03g60080; OsSRO1c, LOC_Os03g12820; OsRZFP34, LOC_Os01g52110; and OsPIP2;4, LOC_Os07g26630; or from the Arabidopsis database TAIR (https://www.arabidopsis.org/) under the following numbers: AtPM1, At1g04560; and AtUBQ5, At3G62250. Accession numbers of sequences used in the phylogenetic analysis can be found in Supplemental Table 2.
Supplemental Data
Supplemental Figure 1. Spatial and temporal expression pattern of OsPM1.
Supplemental Figure 2. The expression level of OsPM1 in transgenic plants.
Supplemental Figure 3. Overexpression of OsPM1 improves rice seed yield under drought stress.
Supplemental Figure 4. Stomatal responses of different OsPM1 transgenic lines to low relative humidity treatment.
Supplemental Figure 5. OsPM1 regulates rice seed germination.
Supplemental Figure 6. The 35Spro:YFP-OsPM1 transgenic plants show drought tolerance and ABA-sensitive seed germination phenotypes.
Supplemental Figure 7. OsPM1 is located at the plasma membrane in tobacco leaves and in Arabidopsis roots.
Supplemental Figure 8. Diagram of OsPM1 promoter and the luciferase reporter assay of the promoter activity.
Supplemental Figure 9. Phylogenetic analysis of the AWPM-19 family proteins.
Supplemental Figure 10. The relative expression level of AtPM1 in Arabidopsis seedlings is far less than that of OsPM1 in rice seedlings.
Supplemental Figure 11. Comparison of the transcriptional responses of rice AWPM-19 family genes to different treatments.
Supplemental Figure 12. Comparison of the temporal expression patterns of OsbZIP46 and OsPM1 after ABA treatment.
Supplemental Table 1. Primers used in this study.
Supplemental Table 2. AWPM-19 family members from different species used for phylogenetic analysis.
Supplemental Data Set 1. Amino acid sequences for phylogenetic analysis in Supplemental Figure 9.
Supplemental Data Set 2. Statistical analysis results.
Acknowledgments
We thank Lizhong Xiong (Huazhong Agricultural University) for providing the 35Spro:OsbZIP46 transgenic seeds, Alexander M. Jones and Wolf B. Frommer (Carnegie Institution for Science) for providing the ABA FRET assay system, and Xinguang Zhu and Weining Sun (Shanghai Institute of Plant Physiology and Ecology) for providing the Li-6400XT photosynthesis system and anti-PIP2;7 antibody, respectively. We also thank Yaoqian Liu for vector construction and abiotic stress treatments. This research was supported by grants from the National Natural Science Foundation of China (31770274), from the Chinese Genetically Modified Organisms Major Project (2014ZX0800914B), and by the Chinese Ministry of Science and Technology 973 program (2012CB114300).
AUTHOR CONTRIBUTIONS
X.G. and L.Y. designed research. L.Y., X.C., Z.G., W.H., and S.L. performed research. L.W., Y.-F.W., and H.M. contributed new reagents/analytic tools. L.Y. and X.G. analyzed data. L.Y., P.X., H.M., and X.G. wrote the article.
References
- Alexandersson E., Saalbach G., Larsson C., Kjellbom P. (2004). Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol. 45: 1543–1556. [DOI] [PubMed] [Google Scholar]
- Amir Hossain M., Lee Y., Cho J.I., Ahn C.H., Lee S.K., Jeon J.S., Kang H., Lee C.H., An G., Park P.B. (2010). The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 72: 557–566. [DOI] [PubMed] [Google Scholar]
- Anugoolprasert O., Kinoshita S., Naito H., Shimizu M., Ehara H. (2012). Effect of low pH on the growth, physiological characteristics and nutrient absorption of sago palm in a hydroponic system. Plant Prod. Sci. 15: 125–131. [Google Scholar]
- Assmann S.M., Snyder J.A., Lee Y.R.J. (2000). ABA-deficient (aba1) and ABA-insensitive (abi1-1, abi2-1) mutants of Arabidopsis have a wild-type stomatal response to humidity. Plant Cell Environ. 23: 387–395. [Google Scholar]
- Barrero J.M., et al. (2015). Transcriptomic analysis of wheat near-isogenic lines identifies PM19-A1 and A2 as candidates for a major dormancy QTL. Genome Biol. 16: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H., et al. (2013). The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr. Biol. 23: 53–57. [DOI] [PubMed] [Google Scholar]
- Boursiac Y., Léran S., Corratgé-Faillie C., Gojon A., Krouk G., Lacombe B. (2013). ABA transport and transporters. Trends Plant Sci. 18: 325–333. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Chen W., Zhou J., He H., Chen L., Chen H., Deng X.W. (2012). Basic leucine zipper transcription factor OsbZIP16 positively regulates drought resistance in rice. Plant Sci. 193-194: 8–17. [DOI] [PubMed] [Google Scholar]
- Chen H., Lan H., Huang P., Zhang Y., Yuan X., Huang X., Huang J., Zhang H. (2015). Characterization of OsPM19L1 encoding an AWPM-19-like family protein that is dramatically induced by osmotic stress in rice. Genet. Mol. Res. 14: 11994–12005. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Corratgé-Faillie C., Lacombe B. (2017). Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 68: 3107–3113. [DOI] [PubMed] [Google Scholar]
- Cui L.G., Shan J.X., Shi M., Gao J.P., Lin H.X. (2015). DCA1 acts as a transcriptional co-activator of DST and contributes to drought and salt tolerance in rice. PLoS Genet. 11: e1005617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies W.J., Zhang J. (1991). Root signals and the regulation of growth and development of plants in drying soil. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42: 55–76. [Google Scholar]
- Eom J.S., et al. (2011). Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol. 157: 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. (2013). Abscisic acid synthesis and response. Arabidopsis Book 11: e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantz D.A. (1990). Plant response to atmospheric humidity. Plant Cell Environ. 13: 667–679. [Google Scholar]
- Grefen C., Lalonde S., Obrdlik P. (2007). Split-ubiquitin system for identifying protein-protein interactions in membrane and full-length proteins. Curr. Protoc. Neurosci. 5: 27. [DOI] [PubMed] [Google Scholar]
- Guan R., et al. (2016). Draft genome of the living fossil Ginkgo biloba. Gigascience 5: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Guo C., Yao L., You C., Wang S., Cui J., Ge X., Ma H. (2016). MID1 plays an important role in response to drought stress during reproductive development. Plant J. 88: 280–293. [DOI] [PubMed] [Google Scholar]
- Hartung W., Sauter A., Hose E. (2002). Abscisic acid in the xylem: where does it come from, where does it go to? J. Exp. Bot. 53: 27–32. [PubMed] [Google Scholar]
- Hedrich R., Geiger D. (2017). Biology of SLAC1-type anion channels - from nutrient uptake to stomatal closure. New Phytol. 216: 46–61. [DOI] [PubMed] [Google Scholar]
- Hedrich R., Neimanis S., Savchenko G., Felle H.H., Kaiser W.M., Heber U. (2001). Changes in apoplastic pH and membrane potential in leaves in relation to stomatal responses to CO2, malate, abscisic acid or interruption of water supply. Planta 213: 594–601. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Komari T. (2008). Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 3: 824–834. [DOI] [PubMed] [Google Scholar]
- Hobo T., Kowyama Y., Hattori T. (1999). A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 96: 15348–15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K.H., Liu C.C., Wu S.J., Kuo Y.Y., Lu C.A., Wu C.R., Lian P.J., Hong C.Y., Ke Y.T., Huang J.H., Yeh C.H. (2014). Expression of a gene encoding a rice RING zinc-finger protein, OsRZFP34, enhances stomata opening. Plant Mol. Biol. 86: 125–137. [DOI] [PubMed] [Google Scholar]
- Hu H., Dai M., Yao J., Xiao B., Li X., Zhang Q., Xiong L. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 103: 12987–12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Yang H., Wei Z., Ma H., Ge X. (2013). Rice male development under drought stress: phenotypic changes and stage-dependent transcriptomic reprogramming. Mol. Plant 6: 1630–1645. [DOI] [PubMed] [Google Scholar]
- Jones A.M., Danielson J.A., Manojkumar S.N., Lanquar V., Grossmann G., Frommer W.B. (2014). Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife 3: e01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Hwang J.U., Lee M., Kim Y.Y., Assmann S.M., Martinoia E., Lee Y. (2010). PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA 107: 2355–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Yim S., Choi H., Kim A., Lee K.P., Lopez-Molina L., Martinoia E., Lee Y. (2015). Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 6: 8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y., Hanada A., Chiba Y., Ichikawa T., Nakazawa M., Matsui M., Koshiba T., Kamiya Y., Seo M. (2012). Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. USA 109: 9653–9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y., Kamiya Y., Seo M. (2013). Nitrate does not compete with abscisic acid as a substrate of AtNPF4.6/NRT1.2/AIT1 in Arabidopsis. Plant Signal. Behav. 8: e26624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M., Takezawa D., Arakawa K., Yoshida S. (1997). Accumulation of 19-kDa plasma membrane polypeptide during induction of freezing tolerance in wheat suspension-cultured cells by abscisic acid. Plant Cell Physiol. 38: 707–716. [DOI] [PubMed] [Google Scholar]
- Koiwai H., Nakaminami K., Seo M., Mitsuhashi W., Toyomasu T., Koshiba T. (2004). Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol. 134: 1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K., Mosa K.A., Chhikara S., Musante C., White J.C., Dhankher O.P. (2014). Two rice plasma membrane intrinsic proteins, OsPIP2;4 and OsPIP2;7, are involved in transport and providing tolerance to boron toxicity. Planta 239: 187–198. [DOI] [PubMed] [Google Scholar]
- Kuromori T., Shinozaki K. (2010). ABA transport factors found in Arabidopsis ABC transporters. Plant Signal. Behav. 5: 1124–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T., Miyaji T., Yabuuchi H., Shimizu H., Sugimoto E., Kamiya A., Moriyama Y., Shinozaki K. (2010). ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. USA 107: 2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T., Sugimoto E., Shinozaki K. (2014). Intertissue signal transfer of abscisic acid from vascular cells to guard cells. Plant Physiol. 164: 1587–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Jeong H.J., Kim S.A., Lee J., Guerinot M.L., An G. (2010). OsZIP5 is a plasma membrane zinc transporter in rice. Plant Mol. Biol. 73: 507–517. [DOI] [PubMed] [Google Scholar]
- Lenka S.K., Lohia B., Kumar A., Chinnusamy V., Bansal K.C. (2009). Genome-wide targeted prediction of ABA responsive genes in rice based on over-represented cis-motif in co-expressed genes. Plant Mol. Biol. 69: 261–271. [DOI] [PubMed] [Google Scholar]
- Léran S., et al. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19: 5–9. [DOI] [PubMed] [Google Scholar]
- Liu N., Ding Y., Fromm M., Avramova Z. (2014). Endogenous ABA extraction and measurement from Arabidopsis leaves. Biol. Protoc. 4: e1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Gao C., Zheng X., Han B. (2009). Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229: 605–615. [DOI] [PubMed] [Google Scholar]
- Matsuda S., Takano S., Sato M., Furukawa K., Nagasawa H., Yoshikawa S., Kasuga J., Tokuji Y., Yazaki K., Nakazono M., Takamure I., Kato K. (2016). Rice stomatal closure requires guard cell plasma membrane ATP-Binding Cassette transporter RCN1/OsABCG5. Mol. Plant 9: 417–427. [DOI] [PubMed] [Google Scholar]
- Merilo E., Jalakas P., Laanemets K., Mohammadi O., Hõrak H., Kollist H., Brosché M. (2015). Abscisic acid transport and homeostasis in the context of stomatal regulation. Mol. Plant 8: 1321–1333. [DOI] [PubMed] [Google Scholar]
- Miao J., Guo D., Zhang J., Huang Q., Qin G., Zhang X., Wan J., Gu H., Qu L.J. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23: 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B., Murphy A.S., Spalding E.P. (2001). Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omote H., Hiasa M., Matsumoto T., Otsuka M., Moriyama Y. (2006). The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 27: 587–593. [DOI] [PubMed] [Google Scholar]
- Petrásek J., Friml J. (2009). Auxin transport routes in plant development. Development 136: 2675–2688. [DOI] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranford J.C., Bryce J.H., Morris P.C. (2002). PM19, a barley (Hordeum vulgare L.) gene encoding a putative plasma membrane protein, is expressed during embryo development and dormancy. J. Exp. Bot. 53: 147–148. [PubMed] [Google Scholar]
- Rerksiri W., Zhang X., Xiong H., Chen X. (2013). Expression and promoter analysis of six heat stress-inducible genes in rice. Sci. World J. 2013: 397401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Schnell J.R., Chou J.J. (2008). Structure and mechanism of the M2 proton channel of influenza A virus. Nature 451: 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovik S., Hartung W. (1992). Compartmental distribution and redistribution of abscisic acid in intact leaves: II. Model analysis. Planta 187: 26–36. [DOI] [PubMed] [Google Scholar]
- Still D.W., Kovach D.A., Bradford K.J. (1994). Development of desiccation tolerance during embryogenesis in rice (Oryza sativa) and wild rice (Zizania palustris): Dehydrin expression, abscisic-acid content, and sucrose accumulation. Plant Physiol. 104: 431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N., et al. (2016). MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell 28: 2161–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N., Zhang H., Li X., Xiao J., Xiong L. (2012). Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 158: 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoi K., Saito T., Iwata N., Kobayashi N.I., Nakanishi T.M. (2011). The analysis of magnesium transport system from external solution to xylem in rice root. Soil Sci. Plant Nutr. 57: 265–271. [Google Scholar]
- Vermachova M., Purkrtova Z., Santrucek J., Jolivet P., Chardot T., Kodicek M. (2011). New protein isoforms identified within Arabidopsis thaliana seed oil bodies combining chymotrypsin/trypsin digestion and peptide fragmentation analysis. Proteomics 11: 3430–3434. [DOI] [PubMed] [Google Scholar]
- Verrier P.J., et al. (2008). Plant ABC proteins--a unified nomenclature and updated inventory. Trends Plant Sci. 13: 151–159. [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1992). Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225: 487–494. [DOI] [PubMed] [Google Scholar]
- Waadt R., Hitomi K., Nishimura N., Hitomi C., Adams S.R., Getzoff E.D., Schroeder J.I. (2014). FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife 3: e01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S., et al. (2014). A rice calcium-dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility. BMC Plant Biol. 14: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyers J.D.B., Paterson N.W. (2001). Plant hormones and the control of physiological processes. New Phytol. 152: 375–407. [DOI] [PubMed] [Google Scholar]
- Wilkinson S., Davies W.J. (1997). Xylem sap pH increase: A drought signal received at the apoplastic face of the guard cell that involves the suppression of saturable abscisic acid uptake by the epidermal symplast. Plant Physiol. 113: 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Tang N., Du H., Ye H., Xiong L. (2008). Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 148: 1938–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Schumaker K.S., Zhu J.K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 (suppl.): S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Hammes U.Z., Taylor C.G., Schachtman D.P., Nielsen E. (2006). High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 16: 1123–1127. [DOI] [PubMed] [Google Scholar]
- Ye N., Jia L., Zhang J. (2012). ABA signal in rice under stress conditions. Rice (N.Y.) 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoon S., Lee D.K., Yu I.J., Kim Y.S., Choi Y.D., Kim J.K. (2017). Overexpression of the OsbZIP66 transcription factor enhances drought tolerance of rice plants. Plant Biotechnol. Rep. 11: 53–62. [Google Scholar]
- You J., Zong W., Li X., Ning J., Hu H., Li X., Xiao J., Xiong L. (2013). The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J. Exp. Bot. 64: 569–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li L., Li L., Guo M., Chory J., Yin Y. (2008). Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 7618–7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazimalova E., Murphy A.S., Yang H.B., Hoyerova K., Hosek P. (2010). Auxin transporters: Why so many? Cold Spring Harb. Perspect. Biol. 2: a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhu H., Pan Y., Yu Y., Luan S., Li L. (2014). A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol. Plant 7: 1522–1532. [DOI] [PubMed] [Google Scholar]
- Zhang L., Xiao S., Li W., Feng W., Li J., Wu Z., Gao X., Liu F., Shao M. (2011a). Overexpression of a Harpin-encoding gene hrf1 in rice enhances drought tolerance. J. Exp. Bot. 62: 4229–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., Wang P., Li Y., Liu B., Feng D., Wang J., Wang H. (2011b). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Chen B., Lu G., Han B. (2009). Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem. Biophys. Res. Commun. 379: 985–989. [DOI] [PubMed] [Google Scholar]
- Zong W., Tang N., Yang J., Peng L., Ma S., Xu Y., Li G., Xiong L. (2016). Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol. 171: 2810–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]