The level of the cytokinin trans-zeatin in shoots controls the molecular, physiological, and growth responses of nitrate-supplied roots in a root system facing heterogeneous nitrate availability.
Abstract
Plants face temporal and spatial variation in nitrogen (N) availability. This includes heterogeneity in soil nitrate (NO3−) content. To overcome these constraints, plants modify their gene expression and physiological processes to optimize N acquisition. This plasticity relies on a complex long-distance root-shoot-root signaling network that remains poorly understood. We previously showed that cytokinin (CK) biosynthesis is required to trigger systemic N signaling. Here, we performed split-root experiments and used a combination of CK-related mutant analyses, hormone profiling, transcriptomic analysis, NO3− uptake assays, and root growth measurements to gain insight into systemic N signaling in Arabidopsis thaliana. By comparing wild-type plants and mutants affected in CK biosynthesis and ABCG14-dependent root-to-shoot translocation of CK, we revealed an important role for active trans-zeatin (tZ) in systemic N signaling. Both rapid sentinel gene regulation and long-term functional acclimation to heterogeneous NO3− supply, including NO3− transport and root growth regulation, are likely mediated by the integration of tZ content in shoots. Furthermore, shoot transcriptome profiling revealed that glutamate/glutamine metabolism is likely a target of tZ root-to-shoot translocation, prompting an interesting hypothesis regarding shoot-to-root communication. Finally, this study highlights tZ-independent pathways regulating gene expression in shoots as well as NO3− uptake activity in response to total N deprivation.
Continuous functional and morphological organ plasticity is one of the most fascinating differences between plants and animals. Indeed, plants employ a range of strategies to tolerate fluctuating resource availability, with plant roots particularly malleable, since they may be exposed to either a scarcity or excess of nutrients and/or water. The signaling networks controlling root adaptation are an important focus for a new green revolution aimed at optimizing root function for the benefit of aerial plant tissue development (Den Herder et al., 2010; Kong et al., 2014; Bishopp and Lynch, 2015).
Nitrogen (N) is a basic building block in the biosynthesis of proteins, nucleic acids, and essential pigments such as chlorophyll. Thus, plants need to deploy strategies to avoid N limitation and to optimize N use during their life cycle. The strong responsiveness of roots to N availability reflects the crucial capacity of plants to maintain N homeostasis. This is reflected, for example, by the plasticity of root system architecture to respond to N deficiency conditions (Gruber et al., 2013) and differentially to N availability based on the presence of other nutrients (Kellermeier et al., 2014).
Root responsiveness relies on the plant’s capacity to sense N availability. This N sensing is commonly divided into two major branches, namely, local perception of N in the vicinity of the root (particularly in the form of nitrate [NO3−]) and systemic perception of internal N/NO3− availability at the whole organism level. The latter relies on root-shoot-root signaling and the integration of information from different parts of the plant (Gansel et al., 2001; Walch-Liu et al., 2005; Alvarez et al., 2012; Li et al., 2014). This dual N sensing is integrated through an intricate signaling network that permits the reciprocal control of root N acquisition and plant growth to ensure N homeostasis (Krouk et al., 2011; Krouk, 2016).
In Arabidopsis thaliana, the perception and propagation of local NO3− signaling has been the focus of considerable research. The molecular players involved include the NO3− transceptor NPF6.3/NRT1.1/CHL1 (Ho et al., 2009; Krouk et al., 2010b), a number of kinases (CIPK8, CIPK23, CPK10, CPK30, and CPK32), the phosphatase ABI2 (Ho et al., 2009; Hu et al., 2009; Léran et al., 2015; Liu et al., 2017), the cleavage and polyadenylation factor CPSF30-L (Li et al., 2017), and several transcription factors (NLP6/7, TGA1/4, NRG2, and SPL9) (Castaings et al., 2009; Krouk et al., 2010a; Konishi and Yanagisawa, 2013; Marchive et al., 2013; Alvarez et al., 2014; Xu et al., 2016). Upon NO3− treatment, these transcription factors trigger the expression of genes involved in NO3− transport and assimilation, a molecular response also known as the primary nitrate response (PNR) (Medici and Krouk, 2014). Additionally, Ca2+ is a crucial secondary messenger in the establishment of this response (Riveras et al., 2015; Krouk, 2017; Liu et al., 2017). Plant responses to N provision involve additional molecular actors. For example, transcription factors such as ANR1, ARF8, and NAC4, as well as CLE peptides, are implicated in N-dependent root development (Zhang and Forde, 1998; Gifford et al., 2008; Vidal et al., 2013; Araya et al., 2014). The transcription factors bZIP1, LBD37/38/39, BT2, and HRS1/HHOs control N use (Baena-González et al., 2007; Rubin et al., 2009; Araus et al., 2016; Safi et al., 2018). The transcription factor TCP20, which is not involved in the PNR per se, physically interacts with NLP6/7 and regulates the expression of NO3−-responsive genes and a cell cycle marker gene (Li et al., 2005; Guan et al., 2014, 2017). Interestingly, TCP20 also regulates root foraging under heterogeneous NO3− supply conditions (Guan et al., 2014) and thus may serve as a gateway to understanding how local and systemic N regulation is integrated (Guan et al., 2017).
Unlike local N signaling, the mechanistic details of the systemic signaling cascade are still poorly understood. This knowledge gap may be attributed to the necessity for complex experimental approaches, such as split-root systems or grafting, to specifically address research questions about long-distance signaling. Indeed, split-root experiments provide a relevant framework to uncouple local and systemic signaling via comparisons of roots experiencing similar local hydro-mineral conditions but different distant media (i.e., in another part of the root) (Li et al., 2014). In this system, any differences in the properties of roots in the same local environment should be due to the effects of long-distance signals. Such an approach has helped to define the landscape of the systemic N-related signaling response and has revealed at least two coexisting systemic signaling pathways (Gansel et al., 2001; Ruffel et al., 2011; Li et al., 2014). The N-demand long-distance signal conveys the message that the whole plant is experiencing a distal N deprivation, whereas the N-supply long-distance signal conveys the message that some N has been found by the plant (Ruffel et al., 2011). Two major aspects of these signaling pathways have been elucidated, namely, the involvement of C-terminally encoded peptides (CEPs) (Tabata et al., 2014) and cytokinin (CK) biosynthesis (Ruffel et al., 2011, 2016).
The role of CEPs in systemic N-demand signaling was recently demonstrated. Upon N deprivation, CEPs are translocated to the shoots, where they are recognized by two leucine-rich repeat receptor kinases known as CEP Receptor 1 and 2 (Tabata et al., 2014). Within the shoot vascular system, this recognition leads to the expression of glutaredoxin-like small polypeptides that translocate toward the roots and, in combination with local NO3− sensing, participate in the specific control of the expression of NRT2.1, encoding the main high-affinity NO3− transporter in roots (Ohkubo et al., 2017; Ruffel and Gojon, 2017). However, in response to heterogeneous NO3− supply, roots display a wide range of responses, including the transcriptional regulation of hundred of genes, enhanced lateral root development, and enhanced NO3− acquisition (Gansel et al., 2001; Remans et al., 2006; Ruffel et al., 2008, 2011; Mounier et al., 2014). The roles for CEP-derived long-distance signals in these processes remain to be demonstrated.
In response to NO3− supply, CKs are synthesized in roots and translocated to the shoots, leading to the control of shoot growth (Takei et al., 2001, 2004; Sakakibara et al., 2006; Osugi et al., 2017). CKs also play crucial roles in root responses to long-distance signals (Ruffel et al., 2011). However, an important question remains concerning the role of CK in shoots in triggering root responses in a systemic context. In other words, are active CKs in shoots a component of the long-distance signaling that controls molecular and/or physiological responses to NO3− heterogeneity in roots? And which CK form in particular is involved?
Isoprenoid CKs, including N6-(Δ2-isopentenyl)adenine (iP), trans-zeatin (tZ), dihydrozeatin (DHZ), and cis-zeatin (cZ), are more abundant in plants than aromatic CKs, with the first two being the major forms in Arabidopsis. Briefly, the prenylation of adenosine mono-, di-, or tri-phosphate by the adenosine phosphate-isopentenyltransferase (IPT) enzymes leads first to the formation of iP-ribotides using the dimethylallyl diphosphate substrate predominantly from the methylerythritol phosphate pathway. These iP-ribotides are subsequently hydroxylated to synthetize tZ-ribotides. On the other hand, the prenylation of adenine on tRNA leads to the formation of cZ-ribotides, using the dimethylallyl diphosphate substrate from the mevalonate pathway. Therefore, tZ-type and cZ-type CKs have different metabolic origins, albeit they can be enzymatically interconverted by zeatin cis-trans isomerase. CK ribotides can be dephosphorylated to synthetize riboside-type CK precursors, which are considered to be the major translocation forms (Sakakibara, 2006). This does not exclude the possibility that free-base forms of CK, which are the actual active forms, are also translocated long-distance. Indeed, systemic transport of tZ was recently shown to regulate leaf size, whereas tZR controls meristem activity (Osugi et al., 2017). Ultimately, CK homeostasis is also modulated by sugar conjugation through O- or N-glycosylation, deactivating CK in a reversible (inactive storage) and irreversible manner, respectively (Sakakibara, 2006).
In this study, we further investigated the role of CK in systemic N signaling. We used genetic approaches to manipulate CK content and translocation combined with detailed measurements of CK forms and shoot transcriptome analysis. We show that tZ in shoots is required for the appropriate responses of roots to heterogeneous NO3− conditions. Specifically, the integration of tZ content in shoots likely plays an essential role in long-distance signaling that controls (1) transcriptional reprogramming of roots and shoots, (2) root growth, and (3) NO3− transport activity.
RESULTS
Root-to-Shoot Translocation of Cytokinin Controls Transcriptional Responses to Systemic N-Demand Signaling
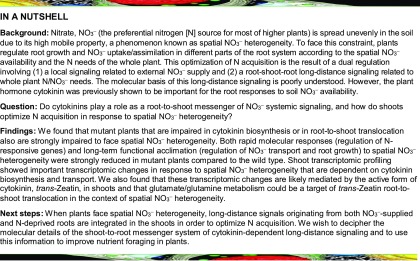
To investigate the role of root-to-shoot translocation of CK in N systemic signaling, we characterized an Arabidopsis mutant lacking the ATP Binding Cassette Transporter Subfamily G (ABCG14) transporter, which is impaired in delivering CK to the shoot (Ko et al., 2014; Zhang et al., 2014). Wild-type and mutant plants were grown in a hydroponic split-root system. Our experimental system consisted of three N supply conditions: (1) a heterogeneous split environment (Sp.KNO3/Sp.KCl: one compartment containing 1 mM KNO3 and the other containing 1 mM KCl), (2) a control homogeneous N-replete environment (C.KNO3: both compartments containing 1 mM KNO3), and (3) a control homogeneous N-deprived environment (C.KCl: both compartments containing 1 mM KCl), which thus provided four different root samples and three different shoot samples (Figure 1A). Any difference recorded between root C.KNO3 and Sp.KNO3 samples is indicative of a N-demand long-distance signal, whereas any difference recorded between root Sp.KCl and C.KCl samples is indicative of a N-supply long-distance signal. This logic was applied throughout the study (Figure 1A). This system was used to test the specific and rapid responses of systemic N-signaling sentinel genes that were identified previously in a root transcriptomic analysis following plant transfer to homogeneous or heterogeneous conditions (Ruffel et al., 2011). These sentinel genes have important functions, including roles in NO3− transport, i.e., NITRATE TRANSPORTER2.1 (NRT2.1; Filleur et al., 2001) and its functional partner NRT3.1/NAR2.1 (also known as WOUND-RESPONSIVE3; Yong et al., 2010), and NO3− assimilation, i.e., NITRITE REDUCTASE (NiR), GLUCOSE-6-PHOSPHATE DEHYDROGENASE3 (G6PD3), UROPHORPHYRIN METHYLASE1 (UPM1), and FERREDOXIN-NADP(+)-OXIDOREDUCTASE2 (FNR2).
Figure 1.
Perturbation of Root-to-Shoot Translocation of CK Impairs the Response of Sentinel Genes to Systemic N-Demand Signaling.
(A) Wild-type and mutant plants were grown under split-root conditions to decipher systemic N-demand and N-supply signaling by comparing root responses (molecular and integrated traits) in plants treated with the same local environment, as indicated by the equal signs. C.KNO3 and C.KCl indicate control conditions (homogeneous KNO3 or KCl supply). Sp.KNO3 and Sp.KCl indicate heterogeneous conditions with KNO3 supply for one-half of the root system and KCl supply for the other part.
(B) Relative mRNA accumulation of sentinel genes in wild-type (Col-0), ipt3,5,7, and abcg14 roots measured by qPCR 6.5 h after transfer to C.KNO3, Sp.KNO3/Sp.KCl, and C.KCl conditions. Bar graphs display the expression values of the six sentinel genes NRT2.1, NRT3.1, G6PD3, NiR, FNR2, and UPM1. Values are means (±se) of three independent experiments, each consisting of two biological replicates corresponding to a pool of three plants grown for 27 d under hydroponic conditions. Different letters indicate significant difference (two-way ANOVA, Tukey post-hoc test, P < 0.1).
In wild-type (Col-0) plants, the expression levels of these sentinel genes were globally higher in Sp.KNO3 roots compared with roots grown under the homogeneous control condition, C.KNO3. This result indicates that wild-type roots respond to heterogeneous NO3− availability through a systemic N-demand signal (Figure 1B) (Ruffel et al., 2011). Under these hydroponic split-root conditions, the isopentenyltransferase triple mutant ipt3 ipt5 ipt7 (ipt3,5,7), with altered CK biosynthesis (Miyawaki et al., 2006), displayed an altered response to systemic N-demand signaling (Figure 1B), as previously shown (Ruffel et al., 2011). In abcg14, the expression levels of sentinel genes in roots supplied with NO3− were lower compared with those in the wild type and ipt3,5,7, but more importantly, Sp.KNO3 roots did not display any significant increase in sentinel gene expression compared with C.KNO3 control roots. This demonstrates that abcg14 is also impaired in its response to systemic N-demand signaling (Figure 1B). Therefore, root-to-shoot translocation of CK may be essential for transcriptional plant responses to heterogeneous NO3− environments. Notably, for both mutants, some sentinel genes, such as G6PD3, were still responsive to local NO3− availability (C.KNO3 and Sp.KNO3 versus Sp.KCl and C.KCl; Figure 1B), indicating that these mutants have maintained their ability to detect NO3− per se.
CK Partitioning Is under the Control of the Combined Effect of NO3−, IPTs, and ABCG14
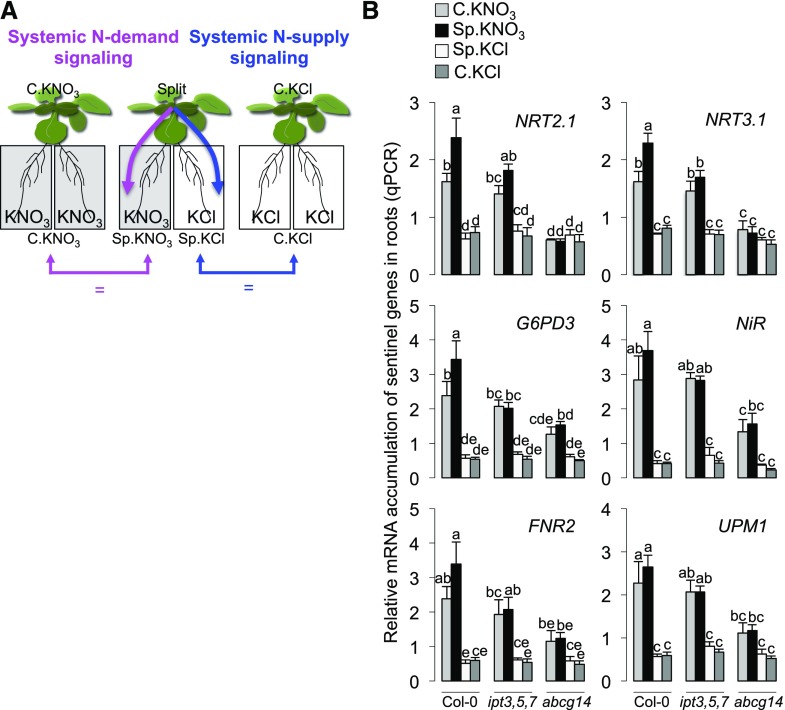
To determine how CK partitioning/homeostasis control root-specific responses to heterogeneous NO3− supply, we measured the levels of the four basic isoprenoid CK forms (iP, tZ, DHZ, and cZ) and their derivatives (ribotides, ribosides, O-glucosides, and N-glucosides) in roots and shoots. These measurements were performed at the same time point used to evaluate the responses of sentinel genes (Figure 1). To validate our experimental system, we first determined if the accumulation patterns of the four CK forms were indeed affected in ipt3,5,7 and abcg14 (Miyawaki et al., 2006; Ko et al., 2014) (Figure 2A). Moreover, we also explored the responses of CK accumulation to NO3− provision in the wild type and these mutants (C.KNO3 versus C.KCl; Figure 2A).
Figure 2.
Root and Shoot Concentrations of Various Types of CK Points to a Role for tZ Accumulation in Shoots.
(A) Stacked bar graphs showing total CK concentrations and distribution of tZ, iP, cZ, and DHZ types of CK in the roots (left graph) and shoots (right graph) of wild-type (Col-0) and mutant (ipt3,5,7 and abcg14) plants exposed to homogeneous C.KNO3 or C.KCl conditions for 6.5 h.
(B) Bar graphs showing the concentrations of active and transported (base and riboside) CK (tZ, iP, cZ, and DHZ) in the roots (left graphs) and shoots (right graphs) of wild-type (Col-0) and mutant (ipt3,5,7 and abcg14) plants exposed to homogeneous C.KNO3 and heterogeneous Sp.KNO3 or Sp.KCl conditions. The two insets in the two graphs at the bottom left are magnified versions of the corresponding graphs below.
In both panels, values are means (±se) of four to five biological replicates collected from four independent experiments, each consisting of a pool of three plants grown for 27 d under hydroponic conditions. Different letters indicate significant differences between treated plants (two-way ANOVA, Tukey post-hoc test, P < 0.05).
As expected, the triple mutation of IPT genes led to a drastic decrease in the levels of tZ and iP-type CKs in both roots and shoots (Figure 2A). In accordance with the predominant role of IPT3 and IPT5 in NO3−-dependent CK biosynthesis (Takei et al., 2004), tZ and iP-type CKs were still synthetized in ipt3,5,7 and were not regulated by NO3− provision (Figure 2A). Contrary to previous observations, the ipt3,5,7 mutations did not lead to a compensatory accumulation of cZ-type CK (Miyawaki et al., 2006) but rather led to a significant decrease in the accumulation of this CK-type in roots under our conditions (Figure 2A). Interestingly, an increase in global cZ-type accumulation was instead observed in abcg14 shoots in response to N deprivation (C.KCl) (Figure 2A). A more detailed analysis of cZ-type CK revealed that only the levels of O-glucosylated forms of cZ increased in ipt3,5,7 under all conditions (Supplemental Data Set 1), whereas in abcg14, the levels of all cZ-forms, except ribotides, increased in shoots as soon as N provision was limited (Supplemental Data Set 1). Altogether, these results demonstrate that cZ-type homeostasis is indeed modified when methylerythritol phosphate pathway-dependent CKs are perturbed. In addition, our experimental setup provides an interesting framework to investigate the role of cZ forms in maintaining the minimal CK activity in shoots required for abiotic stress responses (Schäfer et al., 2015).
The perturbations of CK partitioning and accumulation observed in the abcg14 mutant were consistent with previous data (Ko et al., 2014; Zhang et al., 2014). We indeed observed an increase in the accumulation of tZ-, iP-, and DHZ-type CKs in roots and a decrease in the accumulation of tZ-type CKs in shoots, in accordance with the role of ABCG14 in root-to-shoot translocation of CK (Figure 2A). Moreover, iP accumulation in the abcg14 mutant increased in response to NO3− provision (Supplemental Data Set 1). Interestingly, the shoot iP content of wild-type plants followed the level of root N provision, and this aspect was very strongly affected in the abcg14 mutant (Supplemental Data Set 1). In more detail, the abcg14 mutant exhibited reduced accumulation of active iP forms in C.KNO3 and higher accumulation of all iP-forms in C.KCl (Supplemental Data Set 1). Therefore, taken together, these results demonstrate that the dynamic accumulation of tZ in shoots is under the control of the ABCG14 protein and that this differential accumulation also controls the N-responsive accumulation of iP-type CKs.
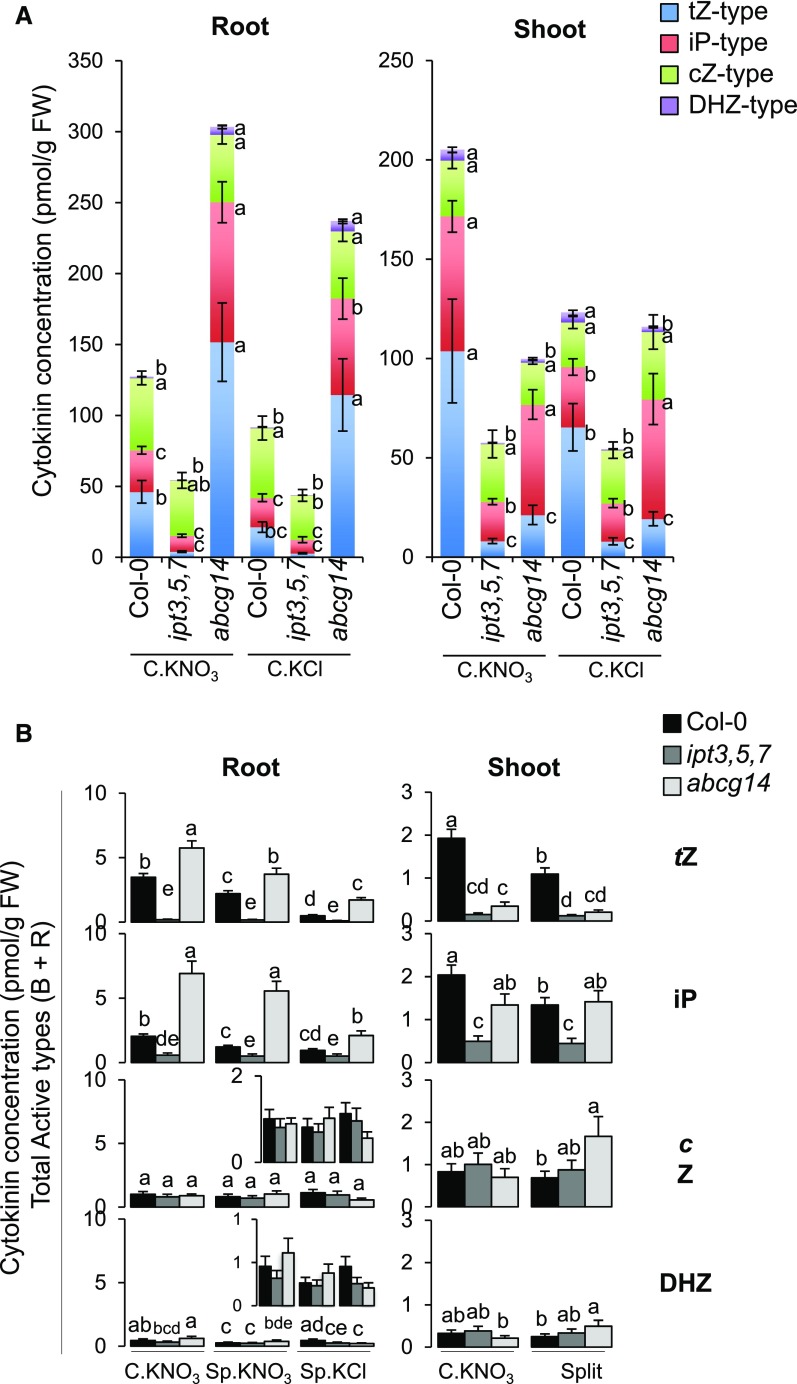
The Levels of the Active Forms of tZ in Shoots Control Root Responses to Systemic N-Demand Signaling
To investigate the early responses of sentinel genes to systemic N-demand signaling and their perturbations in the mutant backgrounds (Figure 1B), we analyzed the accumulation of active and transported forms of CK (Base and Riboside) in roots and shoots in the split-root system (Figure 1A). No obvious correlation was detected between gene expression (Figure 1B) and the accumulation of active forms of CK in roots (Figure 2B, left panel). Indeed, ipt3,5,7 and abcg14 plants displayed opposite phenotypes concerning tZ and iP accumulation, whereas under the same conditions, both genotypes displayed the same gene expression profile (i.e., the loss of N-demand signaling). We thus conclude that CK accumulation in roots cannot explain sentinel gene expression. However, tZ accumulation in shoots could explain the results of root transcriptomic profiling. Indeed, tZ accumulation in shoots was reduced in both ipt3,5,7 and abcg14 (Figure 2B, right panel). Moreover, iP concentrations in shoots could not explain sentinel gene expression patterns, since ipt3,5,7 but not abcg14 displayed a significant decrease in the expression of these genes compared with the wild type. Similarly, cZ and DHZ concentrations could not explain root responses, since abcg14 but not ipt3,5,7 showed a significant increase in these concentrations compared with the wild type. In conclusion, our analysis of ipt3,5,7 and abcg14 indicated that tZ accumulation in shoots likely mediates gene expression in roots in response to long-distance N-signaling.
Moreover, it is noteworthy that differential accumulation of tZ in roots between C.KNO3 and Sp.KNO3 conditions was observed in both the wild type and abcg14 (Figure 2B, left panel). This indicates that CK accumulation in roots is controlled by systemic N-demand signaling that is independent of CK itself. This resembles what we observed previously for lateral root elongation, which includes both CK-dependent and -independent branches (Ruffel et al., 2016).
Genetic Reprogramming in Shoots in Response to Heterogeneous NO3− Supply Is Perturbed in Cytokinin Biosynthesis and Translocation Mutants
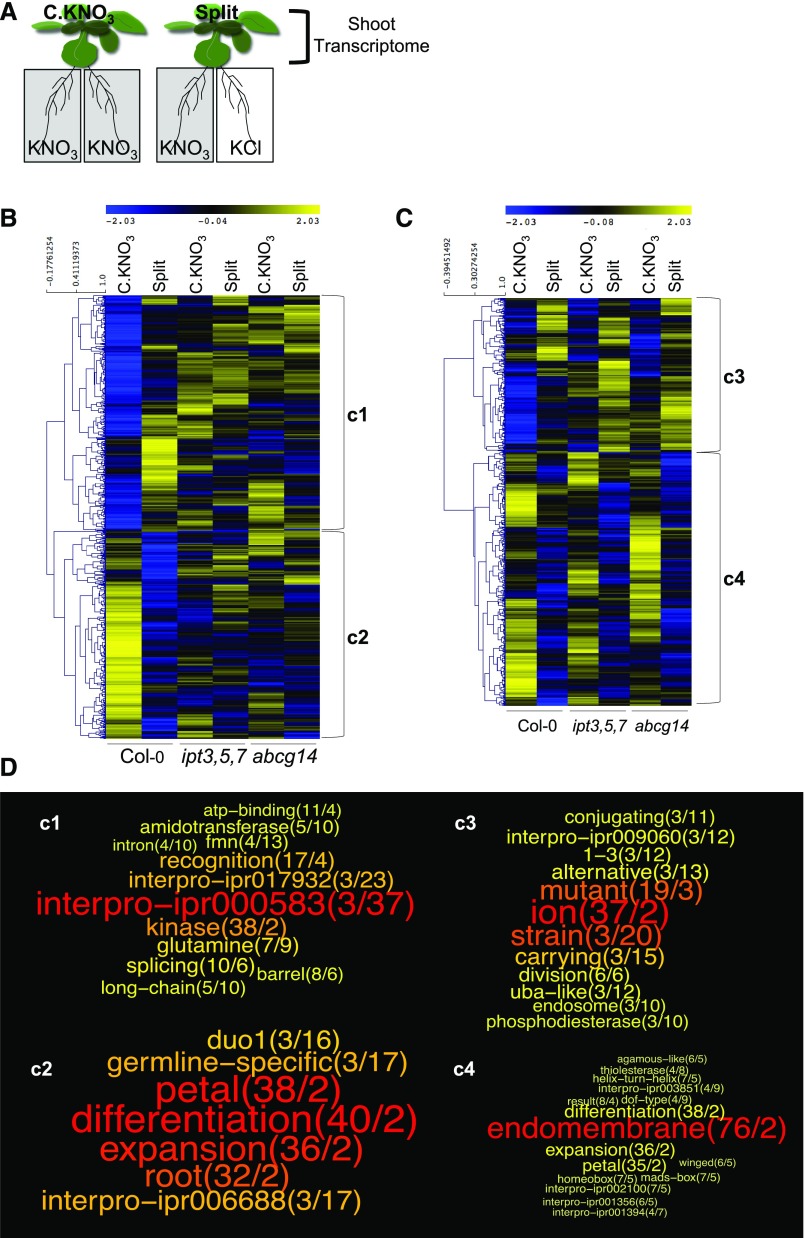
Since we demonstrated that tZ accumulation in shoots likely mediates molecular responses in roots to long-distance N-demand signaling, we decided to evaluate how the shoot transcriptome response in plants under homogeneous or heterogeneous NO3− supply is affected in ipt3,5,7 and abcg14 (Figure 3A), using Arabidopsis Gene1.1 ST Affymetrix array strips (see Methods for details on samples, arrays, and data analysis). By specifically analyzing gene expression data in the wild type, we found 745 nonambiguous significant differentially expressed genes between C.KNO3 and Split conditions in shoots (Supplemental Data Set 2). Among these, we found 397 upregulated genes under Split conditions compared with the control C.KNO3 and 348 downregulated genes under Split conditions compared with the control C.KNO3. Notably, we based our gene selection only on the results of the statistical test because at this stage of the analysis, we did not make any assumption about the level of gene expression differences that would functionally affect the shoot response. Hierarchical clustering of their expression level in the two treatments (homogeneous and heterogeneous NO3− supply) and the three genotypes revealed that the regulation of gene expression by heterogeneous NO3− provision detected in the wild type was strongly affected in the two mutants (Figure 3B). Therefore, in addition to being impaired in tZ accumulation in response to NO3− supply, the two CK mutants are affected in their capacity to reprogram gene expression in response to NO3− supply, which likely disrupts systemic N-signaling controlling root responses. Interestingly, the annotation of these genes revealed the enrichment of some functional terms, revealing biological functions likely under the control of CK accumulation in shoots. Among the genes upregulated under heterogeneous compared with homogeneous NO3− conditions, we found a significant enrichment of two interpro domains, ipr000583 and ipr017932, which both correspond to a glutamine amidotransferase class II domain found in three genes involved in glutamate biosynthesis (i.e., AT2G41220, AT3G24090, and AT5G04140) (Figure 3B, gene cluster c1, and Figure 3D, word cloud c1). Moreover, overrepresentation of the glutamine term was also found for four other genes annotated as glutamine amidotransferase class I and glutamate-ammonia ligase (i.e., AT1G53280, AT3G53180, AT4G26900, and AT4G30550) (Figure 3B, gene cluster c1, and Figure 3D, word cloud c1). These results suggest that, even if NO3− is the genuine signal that triggers systemic N-demand signaling (Ruffel et al., 2011), its heterogeneous supply triggers the modification of a N assimilation pathway in shoots in a CK-dependent manner. Similarly, we observed functional term enrichment among the genes downregulated under heterogeneous compared with homogeneous NO3− conditions that corresponded to the interpro domain ipr006688 found in three ADP-ribosylation factors (i.e., AT3G49860, AT5G14670, and AT1G02440) and duo1 (or germline-specific) found in two genes annotated as encoding C2H2 zinc-finger proteins and the HAPLESS2 gene (i.e., AT4G35280, AT4G35700, and AT4G11720) (Figure 3B, gene cluster c2, and Figure 3D word cloud c2).
Figure 3.
CK-Dependent and -Independent Changes in the Shoot Transcriptome in Response to Heterogeneous NO3− Supply to Roots.
(A) Schematic diagram of the experimental design. Transcriptomic analysis of gene expression was performed in shoots of wild-type (Col-0) and mutant (ipt3,5,7 and abcg14) plants following 24 h incubation under NO3− homogeneous (C.KNO3) or heterogeneous (split) conditions. Samples of four biological replicates, each consisting of a pool of three plants grown for 18 d in in vitro conditions, from four independent experiments were used to perform the microarray analysis using Arabidopsis Gene1.1 ST Array Strips (Affymetrix GeneAtlas).
(B) Hierarchical clustering of the 745 genes identified as differentially expressed under homogeneous versus heterogeneous conditions specifically in the wild type (Supplemental Data Set 2) was performed with MeV software (http://mev.tm4.org/). The scale on the left of the cluster indicates the distance threshold between gene expression levels based on Pearson correlation. The tree was drawn using the average linkage method. Each cluster is divided in two gene populations according to the most distant node, resulting in two subclusters named c1 and c2.
(C) Hierarchical clustering of the 669 genes identified as differentially expressed under homogeneous versus heterogeneous conditions in the three genotypes (Supplemental Data Set 3). Clustering was performed exactly as described in (B). The two subclusters are named c3 and c4.
(D) Word clouds based on functional term enrichment of the four subclusters examined via GeneCloud analysis (https://m2sb.org). The first number next to each term (in parentheses) corresponds to the number of genes containing the term and the second number indicates the fold enrichment. The words (or terms) are displayed in a range from red to yellow in order to highlight the different sizes of text, which are proportional to the product between the occurrence of the word and the enrichment ratio.
By integrating the expression levels of the whole genome data set (i.e., three genotypes and two treatments), we also identified 669 unique genes that responded similarly to NO3− treatment in the shoots of the three genotypes, without making any assumptions about gene expression ratios between the conditions (Supplemental Data Set 3). These genes are those whose regulation is likely not related to CK-dependent long-distance signaling. Hierarchical clustering displayed a first level of classification based on differential regulation under NO3− heterogeneous versus homogeneous conditions (Figure 3C). Functional term enrichment analysis of these genes revealed a variety of meaningful terms, including, for example, the term “uba-like” for three genes related to ubiquitination processes (i.e., AT2G17190, AT4G11740, and AT5G50870) (Figure 3D, word cloud c3).
Altogether, this shoot transcriptomic analysis revealed the massive and rapid reprogramming of gene expression accompanying the accumulation of distinct CKs in response to NO3− supply. Moreover, this allowed us to confirm the occurrence of CK-dependent and CK-independent branches of systemic N signaling (Ruffel et al., 2011, 2016) and shed light on the biological pathways associated with the respective signals.
ipt3,5,7 and abcg14 Plants Are Affected in Integrated Root Traits in Response to Heterogeneous NO3− Supply
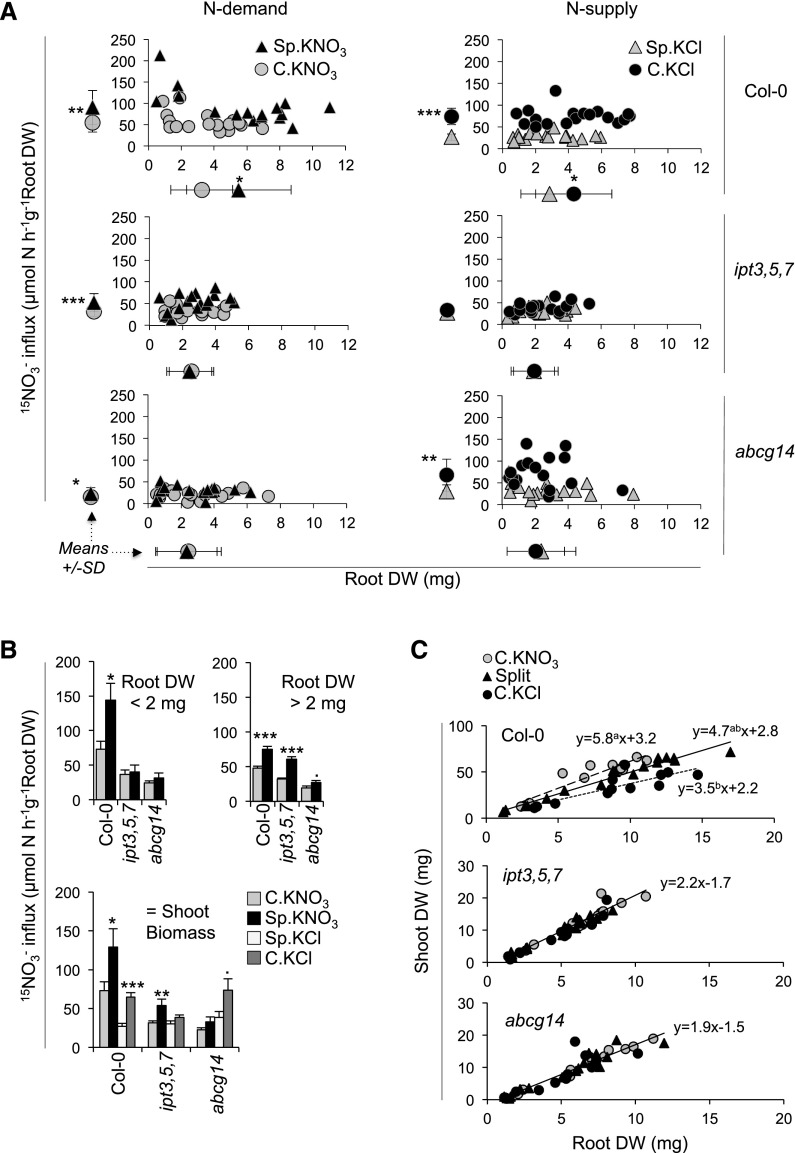
At the molecular level, sentinel genes that respond specifically and rapidly to a CK-dependent systemic N-demand signaling are largely involved in NO3− transport and assimilation (e.g., NRT2.1 and NiR) (Figure 1B). Therefore, we asked to what extent genetic perturbation of biosynthesis and root-to-shoot translocation of CK could affect the associated long-term acclimation of root physiology to variable N supply. We specifically assayed root NO3− influx capacity and biomass (e.g., root dry weight). We also investigated the relationship between these two components of N acquisition. To do so, we combined root traits on single graphs, which allowed us to take into account the variability of plant growth between independent biological replicates (independent plants and experiments) (Figure 4A).
Figure 4.
Responses of Integrated Traits to Systemic N Signaling Are Affected in CK Mutants.
(A) Individual 15NO3− influx in roots plotted against individual root biomass in wild-type (Col-0), ipt3,5,7, and abcg14 plants under C.KNO3, split, or C.KCl conditions for 4 d. Average (±se) root biomass and average (±se) root 15NO3− influx are shown below and to the left of each plot, respectively. In the left column plots, the effects of systemic N-demand signaling are shown via a comparison of C.KNO3 and Sp.KNO3 roots. In the right column plots, the effects of systemic N-supply signaling are shown via a comparison of C.KCl and Sp.KCl roots. Top, middle, and bottom row plots correspond to Col-0, ipt3,5,7, and abcg14, respectively. Data were obtained from five to six biological replicates of plants grown for 31 (27+4) d and collected from three independent experiments. Asterisks indicate significant differences (Student’s t test, *P < 0.05, **P < 0.01, and ***P < 0.001).
(B) Average (±se) root NO3− influx in wild-type (Col-0), ipt3,5,7, and abcg14 plants under NO3− homogeneous (C.KNO3), heterogeneous (Sp.KNO3/Sp.KCl) conditions or N-deprived homogeneous (C.KCl) conditions for a selected subset of plants presented in (A). To generate the two bar graphs on top, we used NO3− influx measurements from roots displaying dry weight less than (to the left) or greater than (to the right) 2 mg. This threshold was chosen according to the biomass distribution observed in Col-0 (graph on top left in [A]). For the third bar graph, we used root NO3− influx measurements from plants displaying an equivalent shoot biomass for the three genotypes for each condition. Asterisks indicate significant differences between C.KNO3 and Sp.KNO3 or between Sp.KCl and C.KCl (Student’s t test, ·P < 0.1, *P < 0.05, **P < 0.01, and ***P < 0.001).
(C) Shoot/root ratio in response to NO3− availability under split-root conditions. Scatterplots indicate the relationship between root and shoot dry biomass in wild-type (Col-0), ipt3,5,7, and abcg14 plants treated in C.KNO3, split, or C.KCl conditions using plants from (A). For each genotype and each condition, linear regression modeling was performed. Statistical differences between the slopes and the intercepts were tested by covariance analysis. For Col-0, the three equations are displayed beside each model. Different letters above the value of each slope indicate significant differences between the slopes (no differences between the intercepts). For the two CK mutants, no significant differences were observed between the linear models. Therefore, the equation corresponds to a linear regression including the three conditions.
In the wild type, both components (root dry weight and NO3− transport) responded significantly to systemic N-signaling (N-demand and N-supply), since the mean values were significantly higher under Sp.KNO3 (5.8 mg and 92 μmol N⋅h−1⋅g−1 root dry weight [DW]) compared with C.KNO3 (3.6 mg and 56 μmol N⋅h−1⋅g−1 root DW) conditions and lower under Sp.KCl (2.9 mg and 28 μmol N⋅h−1⋅g−1 root DW) compared with C.KCl (4.3 mg and 74 μmol N⋅h−1⋅g−1 root DW) conditions (Figure 4A, upper graphs). Interestingly, this analysis revealed the existence of a relationship between the two regulated processes measured in NO3− supplied roots (i.e., transport and biomass). Indeed, wild-type plants that had a strong developmental response displayed a lower NO3− transport response and vice versa, suggesting that the global level of NO3− acquisition by roots results from an interaction between development and transport (Figure 4A, top left).
In the CK mutants, root biomass and NO3− influx responses to systemic N-demand signaling were strongly perturbed (Figure 4A, left panel), suggesting that CK and especially root-to-shoot translocation of CK is a limiting factor in root responses to a heterogeneous NO3− environment. The stimulation of root biomass production observed in the wild type under Sp.KNO3 conditions compared with C.KNO3 conditions was abolished in the CK mutants (Figure 4A, left panel). According to the strategy of wild-type plants described above, one would expect that the smallest root systems would display a greater increase in NO3− influx as an alternative strategy to compensate for distal N deprivation. Thus, the CK mutants might compensate for their root biomass phenotype by increasing their NO3− transport activity. However, this effect was not observed. Indeed, if we consider only roots with a dry biomass under 2 mg (those with the greatest adaptations in NO3− transport in the wild type), wild-type roots under Sp.KNO3 conditions displayed a significant (2-fold) increase in NO3− influx. By contrast, the CK mutants did not display any response to this treatment (Figure 4B, top left graph). However, when the dry biomass was >2 mg, the ipt3,5,7 mutant displayed a NO3− influx response (Figure 4B, top right graph). This suggests that despite the dramatic alteration of CK contents (Figure 2), signaling pathways can still operate to some extent in the triple mutant to trigger an acclimated NO3− transport response to heterogeneous NO3− supply. Overall, CK appears to be central for the two intertwined long-term responses to heterogeneous NO3− supply, namely, NO3− transport and root development.
The two CK mutants did not behave similarly in response to systemic N-supply signaling (Figure 4A, right panel). Whereas CK biosynthesis was required to stimulate root proliferation and NO3− influx when N was completely absent from the medium (Figure 4A, graph on middle right), abcg14 still maintained a NO3− influx capacity similar to that in the wild type (i.e., 71 and 74 μmol N⋅h−1⋅g−1 root, respectively), albeit abcg14 displayed higher variability than the wild type (Figure 4A, graphs on top and bottom right).
Given that CK mutants displayed reduced shoot biomass (Figure 4C, y axis), we exploited the phenotypic variability in order to rule out the notion that their response defect was due to their small size. Specifically, we extracted biological repeats among the three genotypes displaying the same shoot biomass for each condition (i.e., the smallest in Col-0 and largest in the mutants). Using these samples, we confirmed the trend in NO3− influx responses described above (Figure 4A). Indeed, the ipt3,5,7 mutant displayed the systemic regulation of NO3− influx based on its increased capacity only in roots under Sp.KNO3 conditions, whereas abcg14 displayed an increased capacity only in roots under C.KCl conditions (Figure 4B, bottom graph). These results confirm that the two mutants are affected in their response to systemic N signaling, which cannot be explained by their shoot growth defects (Figure 4B, bottom graph).
Unambiguously, root growth responses to systemic N demand and N supply are impaired by the alteration of CK biosynthesis or partitioning. Therefore, we looked at the impact of CK mutation on the modulation of an even more integrative trait controlled by N provision, that is, the shoot/root ratio (Figure 4C). Compared with the wild type, whose shoot/root ratio varied according to the level of N supply, alterations in CK biosynthesis and partitioning resulted in an unchanged shoot/root ratio in the mutants. This result suggests that these mutants would not be able to direct their growth toward the shoots or roots to acclimate the growth of the whole plant to fluctuating environmental conditions, i.e., variations in NO3− availability in the soil.
We conclude that CKs are likely integral to the control of long-term plant acclimation to NO3− heterogeneity, but with some specificity according to the type of systemic N signaling. For instance, root-to-shoot tZ translocation or tZ levels in shoots are not likely involved in regulating NO3− uptake capacity in roots when plants are totally N-deprived (Figure 4A, graph bottom right).
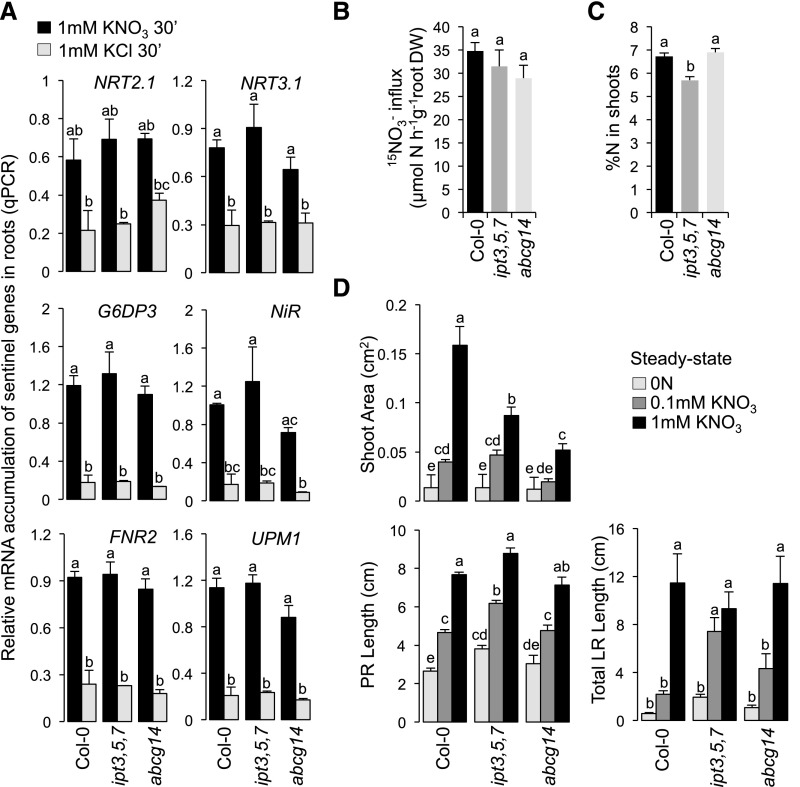
Local NO3− Response Is Not Impaired in Cytokinin Mutants
We previously showed that root responses to heterogeneous NO3− supply depend on a long-distance signaling network triggered by NO3− per se (Ruffel et al., 2011). Indeed, the regulation of the specific and early sentinel genes under heterogeneous conditions is similar between wild-type and NR-null (nitrate reductase-null) mutant plants, leading to the conclusion that the perception of NO3− is a prerequisite to trigger the response to systemic signaling (Ruffel et al., 2011). Thus, we performed further experiments to rule out the possibility that ipt3,5,7 and abcg14 plants are impaired in their molecular and physiological responses to heterogeneous NO3− conditions only because they cannot perceive local NO3− supply. We first tested the PNR (Medici and Krouk, 2014) in the two mutants by transferring plants from N-free solution to 1 mM KNO3 or 1 mM KCl for 30 min. In the two mutants, the activation of PNR was similar to that observed in wild-type plants (Figure 5A). The capacity of ipt3,5,7 and abcg14 plants to react to NO3− addition was also supported by the similar root NO3− influx level between the three genotypes at the same time point selected to evaluate PNR (Figure 5B).
Figure 5.
The Primary NO3− Response, NO3− Transport, Shoot N Status, and N-Dependent Root Development Are Not Impaired in CK Mutants.
(A) Relative mRNA accumulation of six sentinel genes of the primary NO3− response (NRT2.1, NRT3.1, G6PD3, NiR, FNR2, and UPM1) in roots of wild-type (Col-0), ipt3,5,7, and abcg14 plants following 30 min incubation in medium containing 1 mM KNO3 (dark bars) or 1 mM KCl (gray bars) as the control. Data are means (±se) obtained from two independent experiments, each including two pools of five to six plants grown for 27 d under hydroponic conditions.
(B) Root 15NO3− influx in 0.2 mM K15NO3 in wild-type (Col-0), ipt3,5,7, and abcg14 plants after being exposed to KNO3 1 mM for 30 min. Data are means (±se) obtained from two independent experiments, each including 12 individual plants grown for 27 d under hydroponic conditions.
(C) Percentage of N in shoots of the three genotypes from plants collected at a stage corresponding to time 0 of the split-root treatments. Data are means (±se) obtained from two independent experiments, each including 12 individual plants grown for 27 d under hydroponic conditions.
(D) Shoot area, lengths of primary and total lateral roots (PR and LR, respectively) in wild-type (Col-0) and mutant (ipt3,5,7 and abcg14) plants grown for 17 d in N-free (0 N) or 0.1 mM or 1 mM KNO3-containing medium. Data are means (±se) determined from 6 to 12 plants.
Different letters indicate a significant difference (two-way ANOVA, Tukey post-hoc test, P < 0.05 for [A] and [C] and one-way ANOVA for [B]).
Also, in order to rule out the possibility that ipt3,5,7 and abcg14 are impaired in their responses to heterogeneous NO3− conditions only because the N status of the shoots is different from the wild type, we determined the percentage of N in the shoots of the three genotypes. To do so, we used plants at the stage corresponding to the beginning of the split-root treatments. Interestingly, ipt3,5,7 and abcg14 displayed significantly different N% from each other (Figure 5C). Moreover, abcg14 and wild-type plants displayed the same N% (Figure 5C), although their responses to heterogeneous NO3− conditions were the most different (Figures 1B and 4). These results suggest that the loss of responses to systemic N signaling in ipt3,5,7 and abcg14 mutants cannot be explained by some differences of the N status of the mutant plants.
Similarly, the absence of changes in root biomass in response to systemic N signaling in the mutants (Figure 4A) prompted us to verify that they are not simply restrained in their capacity to grow, independent of N supply conditions. When wild-type and mutant plants were grown on N-free, 0.1 mM KNO3, or 1 mM KNO3 containing medium, the primary root length and total lateral root length increased with increasing N concentration for all three genotypes (Figure 5D, bottom graphs), with a longer primary root observed in ipt3,5,7, as previously reported (Miyawaki et al., 2006). Moreover, under steady state conditions of N supply, wild-type and abcg14 plants were similarly able to direct root growth matching the level of N provided to the plant (Figure 5D, bottom graphs), even though shoot growth was particularly impaired in the mutant background (Figure 5D, top graph). Thus, we confirmed that these mutants have the capacity for increased growth with increasing N supply.
Altogether, we conclude that CK biosynthesis and root-to-shoot translocation of CK do not affect NO3− perception, the potential to use N, or plant growth rates in accordance with N supply. Therefore, we confirmed that CK plays an important role in the fine-tuned functional root response to systemic N signaling that operates in a heterogeneous NO3− environment.
DISCUSSION
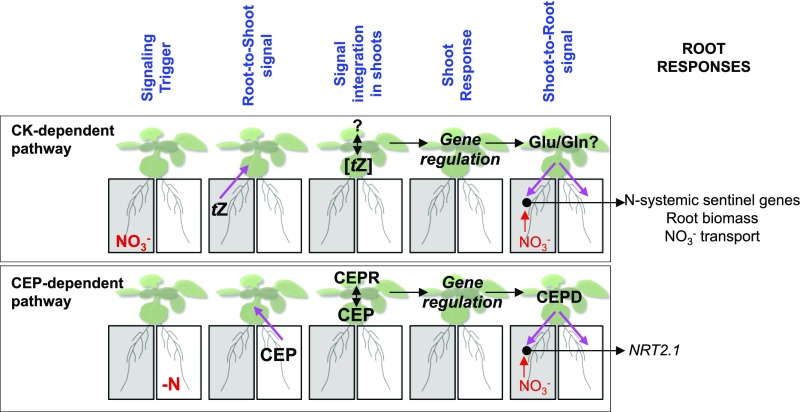
Functional root responses to NO3− availability are the result of a complex signaling network that integrates localized sensing of root NO3− availability with long-distance signaling aimed at coordinating the needs of different parts of the plant. Here, we showed that the integration of tZ content in shoots is likely an essential component of the long-distance N-signaling network controlling root responses. Our results, supported by previous works focusing on the functional characterization of genes involved in CK biosynthesis (Sakakibara et al., 2006; Kiba et al., 2013; Osugi et al., 2017), suggest that NO3− triggers tZ biosynthesis mainly in roots. This tZ would then be transported to the shoots via ABCG14, where it modifies gene expression and possibly associated metabolic processes. Therefore, our model proposes that NO3− provision leads to tZ accumulation in roots, which is subsequently transported to the shoots. Under this scenario, tZ translocation could even constitute part of the systemic signal itself. The accumulation of tZ, which differs between homogeneous and heterogeneous NO3− conditions, would then be interpreted in the shoots. This would lead to differential control of root responses according to NO3− supply to the roots through a shoot-to-root signal that still needs to be identified (Figure 6). Recently, major advances have been made regarding the role of CK in shoot growth and development, including some links with NO3− availability (Osugi et al., 2017; Landrein et al., 2018). These works shed light on the roles of tZ and tZR in signaling mechanisms controlling leaf expansion or shoot apical meristem development through the regulation of WUSCHEL (regulator of stem cell homeostasis) expression. Therefore, it would be interesting to understand the extent to which these pathways are connected to the transcriptional regulation in shoots that we described here and how they play a role in adjusting molecular responses in roots to N availability.
Figure 6.
Multiple Systemic Signaling Pathways Likely Coexist to Coordinate Plant Responses to a Heterogeneous NO3− Environment.
In the CK-dependent pathway, NO3− itself appears to be the triggering signal of systemic N-demand signaling, since NO3− supplied roots are the main providers of tZ. The integration of tZ concentrations in shoots would lead to the differential regulation of genes under heterogeneous compared with homogeneous NO3− conditions, including the regulation of genes involved in glutamate and glutamine biosynthesis. Our model hypothesizes that these amino acids correspond to a shoot-to-root signal that, in combination with local NO3− presence, would enhance the expression of N-systemic sentinel genes as well as root biomass and NO3− transport. This proposed model diverges from the CEP-dependent pathway because CEP synthesis is triggered in N-deprived roots. These peptides, moving from roots to shoots, are perceived by CEPR (CEP receptor) kinase receptors. This recognition in shoots triggers the synthesis of CEPD (CEP downstream) peptides (glutaredoxin-like) corresponding to a shoot-to-root signal that, in combination with local NO3− presence, enhances NRT2.1 expression (Tabata et al., 2014; Ohkubo et al., 2017; Ruffel and Gojon, 2017).
The regulatory pathways of physiological and developmental acclimation to systemic N signaling (N demand and N supply) in roots and the role of CK in this signaling appear to be complex. For instance, the plant will compensate for a small root system by increasing NO3− uptake capacity (Figure 4A, top left graph), suggesting tight and reciprocal control of the regulatory pathways downstream of systemic N-demand signaling. This control does not seem to occur for root acclimation to total N deprivation, since NO3− uptake was stable across various sizes of root systems under C.KCl conditions (Figure 4A, top right graph). This tends to confirm the genetic uncoupling of signaling pathways that control the same root responses in roots under Sp.KNO3 and C.KCl conditions (Ruffel et al., 2011). If on one hand, the role of CK in the control of root growth by systemic N signaling is clearly required, on the other hand, the role of CK in regulating NO3− transport capacity is more complex. Indeed, the ipt3,5,7 triple mutant displayed an altered root response to systemic N-demand signaling, except for NO3− transport capacity (Figure 4A). This could be the result of a compensatory mechanism in the ipt3,5,7 background. A more detailed analysis of CK concentrations in ipt3,5,7 across the duration of split-root treatment could allow us to determine if CK biosynthesis by other IPT members or IPT-independent CK biosynthesis could be responsible for the acclimation of the NO3− uptake response. Similarly, the two mutants displayed different NO3− uptake levels under total N-deprivation conditions (C.KCl roots). In this case, we would be tempted to speculate that the stimulation of NO3− influx capacity in abcg14 plants depends on the CK species, whose concentrations do not dramatically differ between the wild type and abcg14. Therefore, it would be interesting to assess the role of cZ level in roots (Figure 2A, left graph, C.KCl condition) to determine, specifically under this condition, the extent to which this particular CK-type participates to responses to growth limitation (Schäfer et al., 2015).
In shoots, glutamine biosynthesis is a functional term likely influenced by tZ accumulation (Figure 3D). Interestingly, amino acids are thought to serve as reporters of the N status of a plant (Cooper and Clarkson, 1989; Muller and Touraine, 1992). We therefore hypothesize that CK-dependent N-related signals might modify glutamine and/or glutamate metabolism in shoots and that this might be part of a branch of the shoot-to-root signal, as previously proposed by others (Imsande and Touraine, 1994; Girin et al., 2010; Gent and Forde, 2017). Of course, further studies are needed to validate this hypothesis, but our work provides experimental and genome-wide clues about the potential roles of amino acids in shoot-to-root N signaling.
We confirmed that CK accumulation is considerably affected in ipt3,5,7 and abcg14 plants (Figure 2) (Miyawaki et al., 2004; Ko et al., 2014; Zhang et al., 2014). Despite the strong repressive effect of local CK status on root development (Laplaze et al., 2007; Werner et al., 2010), we found that these mutants, particularly abcg14, still maintain a certain growth plasticity, particularly in response to homogenous NO3− supply (Figure 5C). This could be explained by recent findings regarding CK partitioning between the apoplasm and the cytosol. Indeed, an important aspect of CK signaling is that CKs are perceived in the apoplasm and that the PUP14 transporter is crucial for importing bioactive CKs into the cytosol and suppressing the CK response (Zürcher et al., 2016). In this regard, we believe that CK accumulation is important but active CK transport at the cellular level in roots can, to some extent, explain the responsiveness of ipt3,5,7 and abcg14 plants.
Finally, this work refines the model of the integration of different N-related long-distance signaling pathways, which displays some differences to the CEP-related long-distance pathway. Indeed, CEPs are synthesized in N-deprived roots and transported to the shoots where they are recognized by the related receptor kinase, CEPR (Tabata et al., 2014). This recognition activates the biosynthesis of CEPD polypeptides, which are transported to the root where they activate NRT2.1 expression (Ohkubo et al., 2017). Thus, signaling in the CEP-dependent pathway is triggered by N deprivation. In our model, we suggest that the long-distance signal is generated by NO3− itself. Indeed, we observed that tZ accumulation is correlated with NO3− supply (Figure 2B). We thus hypothesize that the two pathways likely rely on different signaling modules (Figure 6). In nature, the shoot of a plant that is experiencing heterogeneous NO3− conditions in roots likely receives a combination of different long-distance signals arising from the different parts of the plant, which include CEP from N-deprived roots and CK from N-supplied roots. Future investigations will aim to resolve how plants integrate these different signaling pathways in shoots to enact a coherent response in roots.
METHODS
Plant Materials
Arabidopsis thaliana in the Col-0 background was used as the wild type. The abcg14 (SK_15918) mutant line was kindly provided by Donghwi Ko (The Sainsbury Laboratory, Cambridge, UK). The ipt3,5,7 triple mutant line was kindly provided by Sabrina Sabatini (University La Sapienza, Rome).
Plant Growth Conditions
All plants were grown under a short-day photoperiod (8 h light 23°C/16 h dark 21°C) at 260 μmol m−2 s−1 light intensity using a mix of sodium-vapor and metal halide 400-W lamps (in growth chambers used for hydroponic culture) and Osram 18-W 840 Lumilux neon tubes (for in vitro plant growth). Split-root in vitro culture was performed as previously described (Ruffel et al., 2011). Briefly, plants were grown on solid (1% agar type A) N-free modified basal Murashige and Skoog (MS) medium supplemented with 0.5 mM NH4-succinate and 0.1 mM KNO3 as N sources. On day 10, the primary root was cut off below the second lateral root in order to obtain two new primary roots. On day 14, the plants were transferred to 1 mM NH4-succinate split medium in order to separate the root system into two isolated parts. On day 18, the plants were transferred to new split plates containing basal MS medium supplemented with (1) 1 mM KNO3 on one side (Sp.KNO3) and 1 mM KCl on the other side (Sp.KCl), (2) 1 mM KNO3 on both sides (C.KNO3), or (3) 1 mM KCl on both sides (C.KCl). For the split-root assay in the hydroponic system, seeds were sown on upside down Eppendorf caps with 1-mm holes filled with water-agar 0.7% solution and grown for 7 d on tap water. The seedlings were grown in nutrient solution containing KH2PO4 1 mM; MgSO4,7H2O 1 mM; K2SO4 0.25 mM; CaCl2,2H2O 2.5 mM; Na-Fe-EDTA 0.1 mM; KCl 50 μM; H3BO3 7.5 μM; MnSO4,H2O 1.25 μM; ZnSO4,7H2O 0.25 μM; CuSO4, 5H2O 0.25 μM; and (NH4)6 Mo7 O24, 4H2O 0.025 μM supplemented with 1 mM NH4Cl and 0.1 mM KNO3 as N sources, pH 5.8. Nutrient solution was renewed every 4 d. At 17 d after sowing, the primary root was cut off below the second lateral root in order to obtain two root systems. Four days later, the plants were transferred to a split root system with 1 mM NH4Cl as the sole N source for an additional 4 d to let the roots grow under split conditions. Twenty-four hours before treatment, the nutrient solution was replaced with N-free nutrient solution. Treatments were then applied by adding concentrated KNO3 or KCl solution to each compartment up to a final concentration of 1 mM. For PNR analysis, plants were grown exactly as described for the split-root experiments, except that the primary root was not cut and, thus, the root system was not split into two parts at the time of treatment. To analyze root developmental traits, seeds were sown and plants grown for 17 d in vitro in square plates containing modified N-free basal MS containing 0.3 mM sucrose, 0.5 g/L MES, and 1% agar type A, supplemented with either 0, 0.1, or 1 mM KNO3. Collection and analysis of plants tissues were performed as indicated in Results.
Gene Expression Analysis
Total RNA was extracted from frozen and ground root or shoot tissue using TRIzol reagent (15596026; Thermo Fisher Scientific) following the manufacturer’s instructions. RNA integrity and concentration were determined using a 2100 Bioanalyzer Instrument (Agilent) and Agilent RNA 6000 Nano kit (5067-1511). DNA contamination was removed by digestion with DNase I (AMPD1; Sigma-Aldrich). For RT-qPCR analysis, reverse transcription of mRNAs was done using ThermoScript RT-PCR (11146016; Thermo Fisher Scientific) according to the manufacturer’s protocol. Gene expression levels were determined using a LightCycler 480 Instrument (Roche) and SYBR Premix Ex Taq (RR420L; TaKaRa). Expression levels of the tested genes were normalized using the expression levels of ACTIN2/8 and CLATHRIN. All specific primers used in this study are listed in Supplemental Data Set 4. Genome-wide expression analysis in shoots was based on four biological replicates obtained from four independent experiments including the three treatments (i.e., C.KNO3, Split, and C.KCl) and the three genotypes (i.e., Col-0, ipt3,5,7, and abcg14). Gene expression measurements were performed using Arabidopsis Affymetrix Gene 1.1 ST array strips designed to measure whole transcript accumulation of 28,501 genes (or transcripts clusters), based on 600,941 probes designed based on TAIR10 genome annotation. Biotin labeled and fragmented cRNAs were obtained using a GeneChip WT PLUS Reagent kit (902280; Thermo Fisher Scientific) following the manufacturer’s instructions. Hybridization on array strips was performed for 16 h at 48°C. The arrays were washed, stained, and scanned using a GeneAtlas HWS Kit (901667; Thermo Fisher Scientific) on the GeneAtlas Fluidics and Imaging Station.
Statistical Analysis and Bioinformatics
All data analysis was performed using the program R and the ANOVA, TukeyHSD, or t.test functions (Supplemental Table 1). Microarrays raw data were processed with GCRMA available in the Expression Console Software package developed by Affymetrix. Genes differentially expressed specifically in the wild type were identified using Student’s t test (P value < 0.05). Genes responding to the treatment in the three genotypes were identified using a two-way ANOVA that was modeled as follows: where Y is the normalized expression signal of a gene, μ is the global mean, the α and β coefficients correspond to the effects of NO3− availability (homogeneous or heterogeneous) of the genotype and of the interaction between both factors, and ε represents unexplained variance. All genes for which at least the βtreatment was significant (P value<0.05) to explain variation of expression were selected. Hierarchical clustering of gene expression was performed using Multiple Experiment Viewer v4.8 (MeV) software (Saeed et al., 2003). Functional analysis of gene lists was performed using the GeneCloud platform, and functional term enrichment was displayed using word clouds (https://m2sb.org; Krouk et al., 2015).
where Y is the normalized expression signal of a gene, μ is the global mean, the α and β coefficients correspond to the effects of NO3− availability (homogeneous or heterogeneous) of the genotype and of the interaction between both factors, and ε represents unexplained variance. All genes for which at least the βtreatment was significant (P value<0.05) to explain variation of expression were selected. Hierarchical clustering of gene expression was performed using Multiple Experiment Viewer v4.8 (MeV) software (Saeed et al., 2003). Functional analysis of gene lists was performed using the GeneCloud platform, and functional term enrichment was displayed using word clouds (https://m2sb.org; Krouk et al., 2015).
Determination of Cytokinin Levels
CK purification was performed according to previously described methods (Svačinová et al., 2012) with some modifications (Šmehilová et al., 2016). Briefly, CKs were extracted from 30 mg of frozen powdered tissue in modified Bieleski buffer (methanol/water/formic acid, 15/4/1, v/v/v) containing a cocktail of stable isotope-labeled internal standards (0.25 pmol of CK bases, ribosides, and N-glucosides; 0.5 pmol of CK O-glucosides and nucleotides added per sample) and purified using two solid phase extraction columns. CK concentration was determined by UHPLC-MS/MS (ultra-high-performance liquid chromatography coupled to a triple quadrupole mass spectrometer equipped with an electrospray interface). Quantification was performed with Masslynx software (v4.1; Waters) using a standard isotope dilution method. The ratio of endogenous CK to the appropriate labeled standard was determined and used to quantify the level of endogenous compounds in the original extract according to the known quantity of the added internal standard.
Determination of Root Biomass and Nitrate Influx Capacity
Root 15NO3− influx was assayed as described previously (Muños et al., 2004). Root systems were rinsed with 0.1 mM CaSO4 solution for 1 min, transferred to nutrient solution containing 0.2 mM 15NO3− (99% atom excess 15N), pH 5.8, for 5 min, and washed with 0.1 mM CaSO4 solution for 1 min. Roots and shoots were harvested separately and dried in an oven at 70°C for 48 h. Dry weight was determined and the total N and atom % 15N were determined by continuous-flow isotope ratio mass spectrometry using a Euro-EA Euro Vector elemental analyzer coupled with an IsoPrime mass spectrometer (GV Instruments).
Measurement of Shoot and Root Developmental Traits
Scans of square plates containing plants were performed at 600 dpi in TIFF format using an HP scanner. Shoot area, primary and lateral root length, and the number of lateral roots were measured using ImageJ software (https://imagej.nih.gov/ij/).
Accession Numbers
Sequence data for the genes mentioned in this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: IPT3 (AT3G63110), IPT5 (AT5G19040), IPT7 (AT3G23630), ABCG14 (AT1G31770), NRT2.1 (AT1G08090), NRT3.1 (AT5G50200), G6PD3 (AT1G24280), NiR (AT2G15620), FNR2 (AT1G30510), and UPM1 (AT5G40850). The Affymetrix GeneChip data have been deposited in NCBI’s Gene Expression Omnibus in compliance with MIAME standards (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through Gene Expression Omnibus Series accession number GSE114162.
Supplemental Data
Supplemental Table 1. ANOVA, Tukey, and Student’s t test tables.
Supplemental Data Set 1. Detailed CK concentrations (pmol/gFW), organized by type and form, in shoots and roots of Col-0, ipt3,5,7, and abcg14.
Supplemental Data Set 2. 745 genes significantly regulated by heterogeneous NO3− provision (split) compared with homogeneous NO3− conditions (C.KNO3) specifically in Col-0 shoots.
Supplemental Data Set 3. 669 genes significantly regulated by heterogeneous NO3− provision (split) compared with homogeneous NO3− conditions (C.KNO3) in the shoots of the three genotypes.
Supplemental Data Set 4. RT-qPCR primers used in this study.
Acknowledgments
We thank Hugues Baudot and his team for taking care of the plant culture system, Franck Lecocq and Chantal Brachet for technical support, Pascal Tillard for 15N measurements, and Hana Martínková for her help with phytohormone analyses. We also thank Denis Vile for his help for statistical analysis of the data. This work was supported by the Institut National de La Recherche Agronomique (CJS Fellowship to A.P. and BAP project VARNET to S.R.), by the National Science Foundation (IOS 1339362-NutriNet with a fellowship to A.C.), and by Agence Nationale de la Recherche (IMANA ANR-14-CE19-0008). I.P and O.N. were supported by the Czech Science Foundation (Project GA17-06613S) and the Ministry of Education, Youth, and Sports of the Czech Republic (National Program for Sustainability I; Grant LO1204).
AUTHOR CONTRIBUTIONS
A.P., A.C., I.P., and O.N. performed the research. A.P. and S.R. analyzed the data. G.K., B.L., and S.R. designed the research and wrote the manuscript.
References
- Alvarez J.M., Vidal E.A., Gutiérrez R.A. (2012). Integration of local and systemic signaling pathways for plant N responses. Curr. Opin. Plant Biol. 15: 185–191. [DOI] [PubMed] [Google Scholar]
- Alvarez J.M., Riveras E., Vidal E.A., Gras D.E., Contreras-López O., Tamayo K.P., Aceituno F., Gómez I., Ruffel S., Lejay L., Jordana X., Gutiérrez R.A. (2014). Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 80: 1–13. [DOI] [PubMed] [Google Scholar]
- Araus V., Vidal E.A., Puelma T., Alamos S., Mieulet D., Guiderdoni E., Gutiérrez R.A. (2016). Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiol. 171: 1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya T., Miyamoto M., Wibowo J., Suzuki A., Kojima S., Tsuchiya Y.N., Sawa S., Fukuda H., von Wirén N., Takahashi H. (2014). CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. USA 111: 2029–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E., Rolland F., Thevelein J.M., Sheen J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942. [DOI] [PubMed] [Google Scholar]
- Bishopp A., Lynch J.P. (2015). The hidden half of crop yields. Nat. Plants 1: 15117. [DOI] [PubMed] [Google Scholar]
- Castaings L., Camargo A., Pocholle D., Gaudon V., Texier Y., Boutet-Mercey S., Taconnat L., Renou J.P., Daniel-Vedele F., Fernandez E., Meyer C., Krapp A. (2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57: 426–435. [DOI] [PubMed] [Google Scholar]
- Cooper H.D., Clarkson D.T. (1989). Cycling of amino-nitrogen and other nutrient between shoots and roots in cereals: a possible mechanism integrating shoot and root in the regulation of nutrient uptake. J. Exp. Bot. 40: 753–762. [Google Scholar]
- Den Herder G., Van Isterdael G., Beeckman T., De Smet I. (2010). The roots of a new green revolution. Trends Plant Sci. 15: 600–607. [DOI] [PubMed] [Google Scholar]
- Filleur S., Dorbe M.F., Cerezo M., Orsel M., Granier F., Gojon A., Daniel-Vedele F. (2001). An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 489: 220–224. [DOI] [PubMed] [Google Scholar]
- Gansel X., Muños S., Tillard P., Gojon A. (2001). Differential regulation of the NO3− and NH4+ transporter genes AtNrt2.1 and AtAmt1.1 in Arabidopsis: relation with long-distance and local controls by N status of the plant. Plant J. 26: 143–155. [DOI] [PubMed] [Google Scholar]
- Gent L., Forde B.G. (2017). How do plants sense their nitrogen status? J. Exp. Bot. 68: 2531–2539. [DOI] [PubMed] [Google Scholar]
- Gifford M.L., Dean A., Gutierrez R.A., Coruzzi G.M., Birnbaum K.D. (2008). Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 105: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girin T., El-Kafafi S., Widiez T., Erban A., Hubberten H.M., Kopka J., Hoefgen R., Gojon A., Lepetit M. (2010). Identification of Arabidopsis mutants impaired in the systemic regulation of root nitrate uptake by the nitrogen status of the plant. Plant Physiol. 153: 1250–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber B.D., Giehl R.F., Friedel S., von Wirén N. (2013). Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163: 161–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P., Wang R., Nacry P., Breton G., Kay S.A., Pruneda-Paz J.L., Davani A., Crawford N.M. (2014). Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. USA 111: 15267–15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P., Ripoll J.-J., Wang R., Vuong L., Bailey-Steinitz L.J., Ye D., Crawford N.M. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. USA 114: 2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.H., Lin S.H., Hu H.C., Tsay Y.F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194. [DOI] [PubMed] [Google Scholar]
- Hu H.C., Wang Y.Y., Tsay Y.F. (2009). AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57: 264–278. [DOI] [PubMed] [Google Scholar]
- Imsande J., Touraine B. (1994). N demand and the regulation of nitrate uptake. Plant Physiol. 105: 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermeier F., Armengaud P., Seditas T.J., Danku J., Salt D.E., Amtmann A. (2014). Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 26: 1480–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Takei K., Kojima M., Sakakibara H. (2013). Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev. Cell 27: 452–461. [DOI] [PubMed] [Google Scholar]
- Ko D., et al. (2014). Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proc. Natl. Acad. Sci. USA 111: 7150–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X., Zhang M., De Smet I., Ding Z. (2014). Designer crops: optimal root system architecture for nutrient acquisition. Trends Biotechnol. 32: 597–598. [DOI] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S. (2013). Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 4: 1617. [DOI] [PubMed] [Google Scholar]
- Krouk G., et al. (2010b). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18: 927–937. [DOI] [PubMed] [Google Scholar]
- Krouk G. (2016). Hormones and nitrate: a two-way connection. Plant Mol. Biol. 91: 599–606. [DOI] [PubMed] [Google Scholar]
- Krouk G. (2017). Nitrate signalling: Calcium bridges the nitrate gap. Nat. Plants 3: 17095. [DOI] [PubMed] [Google Scholar]
- Krouk G., Mirowski P., LeCun Y., Shasha D.E., Coruzzi G.M. (2010a). Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 11: R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G., Ruffel S., Gutiérrez R.A., Gojon A., Crawford N.M., Coruzzi G.M., Lacombe B. (2011). A framework integrating plant growth with hormones and nutrients. Trends Plant Sci. 16: 178–182. [DOI] [PubMed] [Google Scholar]
- Krouk G., Carré C., Fizames C., Gojon A., Ruffel S., Lacombe B. (2015). GeneCloud reveals semantic enrichment in lists of gene descriptions. Mol. Plant 8: 971–973. [DOI] [PubMed] [Google Scholar]
- Landrein B., Formosa-Jordan P., Malivert A., Schuster C., Melnyk C.W., Yang W., Turnbull C., Meyerowitz E.M., Locke J.C.W., Jönsson H. (2018). Nitrate modulates stem cell dynamics in Arabidopsis shoot meristems through cytokinins. Proc. Natl. Acad. Sci. USA 115: 1382–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léran S., Edel K.H., Pervent M., Hashimoto K., Corratgé-Faillie C., Offenborn J.N., Tillard P., Gojon A., Kudla J., Lacombe B. (2015). Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci. Signal. 8: ra43. [DOI] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colón-Carmona A., Gutiérrez R.A., Doerner P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 102: 12978–12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Krouk G., Coruzzi G.M., Ruffel S. (2014). Finding a nitrogen niche: a systems integration of local and systemic nitrogen signalling in plants. J. Exp. Bot. 65: 5601–5610. [DOI] [PubMed] [Google Scholar]
- Li Z., Wang R., Gao Y., Wang C., Zhao L., Xu N., Chen K.E., Qi S., Zhang M., Tsay Y.F., Crawford N.M., Wang Y. (2017). The Arabidopsis CPSF30-L gene plays an essential role in nitrate signaling and regulates the nitrate transceptor gene NRT1.1. New Phytol. 16: 1205–1222. [DOI] [PubMed] [Google Scholar]
- Liu K.H., et al. (2017). Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545: 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C., Roudier F., Castaings L., Bréhaut V., Blondet E., Colot V., Meyer C., Krapp A. (2013). Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4: 1713. [DOI] [PubMed] [Google Scholar]
- Medici A., Krouk G. (2014). The primary nitrate response: a multifaceted signalling pathway. J. Exp. Bot. 65: 5567–5576. [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Matsumoto-Kitano M., Kakimoto T. (2004). Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 37: 128–138. [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Tarkowski P., Matsumoto-Kitano M., Kato T., Sato S., Tarkowska D., Tabata S., Sandberg G., Kakimoto T. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 103: 16598–16603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier E., Pervent M., Ljung K., Gojon A., Nacry P. (2014). Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ. 37: 162–174. [DOI] [PubMed] [Google Scholar]
- Muller B., Touraine B. (1992). Inhibition of NO3− uptake by various phloem-translocated amino acids in soybean seedlings. J. Exp. Bot. 43: 617–623. [Google Scholar]
- Muños S., Cazettes C., Fizames C., Gaymard F., Tillard P., Lepetit M., Lejay L., Gojon A. (2004). Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16: 2433–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo Y., Tanaka M., Tabata R., Ogawa-Ohnishi M., Matsubayashi Y. (2017). Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 3: 17029. [DOI] [PubMed] [Google Scholar]
- Osugi A., Kojima M., Takebayashi Y., Ueda N., Kiba T., Sakakibara H. (2017). Systemic transport of trans-zeatin and its precursor have differing roles in Arabidopsis shoots. Nat. Plants 3: 17112. [DOI] [PubMed] [Google Scholar]
- Remans T., Nacry P., Pervent M., Filleur S., Diatloff E., Mounier E., Tillard P., Forde B.G., Gojon A. (2006). The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. USA 103: 19206–19211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveras E., Alvarez J.M., Vidal E.A., Oses C., Vega A., Gutiérrez R.A. (2015). The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiol. 169: 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G., Tohge T., Matsuda F., Saito K., Scheible W.R. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S., et al. (2008). Systemic signaling of the plant nitrogen status triggers specific transcriptome responses depending on the nitrogen source in Medicago truncatula. Plant Physiol. 146: 2020–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S., Gojon A. (2017). Systemic nutrient signalling: On the road for nitrate. Nat. Plants 3: 17040. [DOI] [PubMed] [Google Scholar]
- Ruffel S., Krouk G., Ristova D., Shasha D., Birnbaum K.D., Coruzzi G.M. (2011). Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc. Natl. Acad. Sci. USA 108: 18524–18529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S., Poitout A., Krouk G., Coruzzi G.M., Lacombe B. (2016). Long-distance nitrate signaling displays cytokinin dependent and independent branches. J. Integr. Plant Biol. 58: 226–229. [DOI] [PubMed] [Google Scholar]
- Saeed A.I., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378. [DOI] [PubMed] [Google Scholar]
- Safi A., Medici A., Szponarski W., Marshall-Colon A., Ruffel S., Gaymard F., Coruzzi G., Lacombe B., Krouk G. (2018). HRS1/HHOs GARP transcription factors and reactive oxygen species are regulators of Arabidopsis nitrogen starvation response. bioRxiv doi/10.1101/164277. [Google Scholar]
- Sakakibara H. (2006). Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57: 431–449. [DOI] [PubMed] [Google Scholar]
- Sakakibara H., Takei K., Hirose N. (2006). Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci. 11: 440–448. [DOI] [PubMed] [Google Scholar]
- Schäfer M., Brütting C., Meza-Canales I.D., Großkinsky D.K., Vankova R., Baldwin I.T., Meldau S. (2015). The role of cis-zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J. Exp. Bot. 66: 4873–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmehilová M., Dobrůšková J., Novák O., Takáč T., Galuszka P. (2016). Cytokinin-specific glycosyltransferases possess different roles in cytokinin homeostasis maintenance. Front. Plant Sci. 7: 1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svačinová J., Novák O., Plačková L., Lenobel R., Holík J., Strnad M., Doležal K. (2012). A new approach for cytokinin isolation from Arabidopsis tissues using miniaturized purification: pipette tip solid-phase extraction. Plant Methods 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R., Sumida K., Yoshii T., Ohyama K., Shinohara H., Matsubayashi Y. (2014). Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346: 343–346. [DOI] [PubMed] [Google Scholar]
- Takei K., Sakakibara H., Taniguchi M., Sugiyama T. (2001). Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol. 42: 85–93. [DOI] [PubMed] [Google Scholar]
- Takei K., Ueda N., Aoki K., Kuromori T., Hirayama T., Shinozaki K., Yamaya T., Sakakibara H. (2004). AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 45: 1053–1062. [DOI] [PubMed] [Google Scholar]
- Vidal E.A., Moyano T.C., Riveras E., Contreras-López O., Gutiérrez R.A. (2013). Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc. Natl. Acad. Sci. USA 110: 12840–12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P., Filleur S., Gan Y., Forde B.G. (2005). Signaling mechanisms integrating root and shoot responses to changes in the nitrogen supply. Photosynth. Res. 83: 239–250. [DOI] [PubMed] [Google Scholar]
- Werner T., Nehnevajova E., Köllmer I., Novák O., Strnad M., Krämer U., Schmülling T. (2010). Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22: 3905–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Wang R., Zhao L., Zhang C., Li Z., Lei Z., Liu F., Guan P., Chu Z., Crawford N.M., Wang Y. (2016). The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell 28: 485–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong Z., Kotur Z., Glass A.D. (2010). Characterization of an intact two-component high-affinity nitrate transporter from Arabidopsis roots. Plant J. 63: 739–748. [DOI] [PubMed] [Google Scholar]
- Zhang H., Forde B.G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409. [DOI] [PubMed] [Google Scholar]
- Zhang K., Novak O., Wei Z., Gou M., Zhang X., Yu Y., Yang H., Cai Y., Strnad M., Liu C.J. (2014). Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nat. Commun. 5: 3274. [DOI] [PubMed] [Google Scholar]
- Zürcher E., Liu J., di Donato M., Geisler M., Müller B. (2016). Plant development regulated by cytokinin sinks. Science 353: 1027–1030. [DOI] [PubMed] [Google Scholar]