This work provides an example of how parallel mutations in the same 5′ UTR region can shape phenotypic evolution in plants.
Abstract
Flowering time is an adaptive life history trait. Capsella rubella, a close relative of Arabidopsis thaliana and a young species, displays extensive variation for flowering time but low standing genetic variation due to an extreme bottleneck event, providing an excellent opportunity to understand how phenotypic diversity can occur with a limited initial gene pool. Here, we demonstrate that common allelic variation and parallel evolution at the FLC locus confer variation in flowering time in C. rubella. We show that two overlapping deletions in the 5′ untranslated region (UTR) of C. rubella FLC, which are associated with local changes in chromatin conformation and histone modifications, reduce its expression levels and promote flowering. We further show that these two pervasive variants originated independently in natural C. rubella populations after speciation and spread to an intermediate frequency, suggesting a role of this parallel cis-regulatory change in adaptive evolution. Our results provide an example of how parallel mutations in the same 5′ UTR region can shape phenotypic evolution in plants.
INTRODUCTION
Parallel evolution, which refers to different species or populations that share a common ancestor independently evolve the same trait by changing the same genes, is known to occur and can play a role in the evolution of adaptive traits (Wood et al., 2005). It has been studied in closely related Caenorhabditis species (McGrath et al., 2011) and in natural populations within the same species in sticklebacks (Colosimo et al., 2005; Chan et al., 2010). For example, parallel evolution at the melanocortin-1 receptor gene locus Mc1r confers adaptive variation in pigmentation in reptiles, birds, and mammals (Hoekstra, 2006; Arendt and Reznick, 2008; Protas and Patel, 2008; Elmer and Meyer, 2011). Mutations underlying parallel evolution have also been studied to understand the predictability of evolution, i.e., whether certain traits can be shaped by a specific gene(s) (Stern and Orgogozo, 2009; Martin and Orgogozo, 2013).
Flowering time is a crucial adaptive trait for plants, as flowering at the wrong time can mean failure to set seed and reproduce. Flowering time can vary tremendously both within and between species; therefore, analysis of flowering time offers an excellent opportunity to study parallel evolution. It is also a relatively well understood biological process at the molecular and genetic levels (Koornneef et al., 2004; Hepworth and Dean, 2015; Bloomer and Dean, 2017). Natural variation for flowering time within a species has attracted increasing attention owing to its likely correlation with adaptation to local environments (Salomé et al., 2011; Weigel, 2012). Although mutations in more than 100 genes are known to affect flowering time in Arabidopsis thaliana (Srikanth and Schmid, 2011; Lee et al., 2013; Posé et al., 2013; Wahl et al., 2013; Yan et al., 2014), only a few genes play important roles in regulating flowering time variation among natural accessions. These include FLOWERING LOCUS C (FLC), FLOWERING LOCUS M (FLM), FRIGIDA (FRI), PHYTOCHROME C, and SHORT VEGETATIVE PHASE (Johanson et al., 2000; Gazzani et al., 2003; Caicedo et al., 2004; Lempe et al., 2005; Shindo et al., 2005; Werner et al., 2005; Balasubramanian et al., 2006; Méndez-Vigo et al., 2013; Sureshkumar et al., 2016; Lutz et al., 2017; Zou et al., 2017).
Epistatic interactions between FRI and FLC regulate flowering time variation in natural populations of A. thaliana (Gazzani et al., 2003; Lempe et al., 2005; Shindo et al., 2005; Werner et al., 2005). Expression levels of FLC can explain up to 40% of the variation in flowering time between accessions of A. thaliana (Lempe et al., 2005; Shindo et al., 2005). Although natural variants of FLC have been described, they occur relatively rarely, and much of the variation in flowering time is explained by allelic variation in FRI and its epistatic interaction with FLC. Therefore, FRI appears to be the major determinant of flowering time variation in A. thaliana (Johanson et al., 2000; Lempe et al., 2005; Shindo et al., 2005).
In species other than A. thaliana, our molecular understanding of how variation for flowering time is achieved is still rudimentary. Flowering time variation has been studied in some relatives of A. thaliana, such as Arabidopsis lyrata (Leinonen et al., 2013), Arabis alpina (Wang et al., 2009), Brassica napus (Raman et al., 2016), Cardamine flexuosa (Zhou et al., 2013), and Capsella rubella (Guo et al., 2012). C. rubella is of particular interest for several reasons. During its speciation, C. rubella went through an extreme genetic bottleneck (a sharp reduction of population size due to environmental change or other factors) concurrent with the transition from an outcrossing to a selfing mating system, resulting in its current low genetic diversity (Foxe et al., 2009; Guo et al., 2009). Therefore, this relatively young species contains only limited genetic variation on which selection can operate. Nevertheless, there is considerable variation for flowering time among natural accessions of C. rubella (Guo et al., 2012). Investigation of the molecular basis of flowering time variation in C. rubella will therefore not only broaden our understanding of the genetic factors regulating this variation in nature but also address the fundamental question of how a young species can adapt to diverse environments, despite limited genetic variation. It is also likely to provide novel opportunities to study parallel evolution in plants with limited standing genetic variation.
Allelic variation at FLC and associated variation in gene expression has been shown to be associated with flowering time in A. thaliana relatives as well, including C. rubella (Slotte et al., 2009; Yuan et al., 2009; Hu et al., 2011; Guo et al., 2012; Raman et al., 2016). In a previous study, we reported an allele of FLC that causes early flowering in C. rubella (Guo et al., 2012), in which a single nucleotide polymorphism (SNP) deleted a splice acceptor site, that results in a frameshift and ultimately leading to a truncated, nonfunctional FLC product. However, this polymorphism is unique to a single accession and is therefore unable to explain variation in flowering time among other C. rubella accessions.
In this study, we report that FLC is an important determinant of flowering time variation in C. rubella. We studied three accessions, 86IT1, 762, and MTE, which represent three distinct points (early, intermediate, and late, respectively) on the flowering time gradient in C. rubella, to map the loci responsible for flowering time variation. Using quantitative trait locus analysis and fine mapping, we identified two overlapping deletions in the 5′ untranslated region (UTR) of CrFLC as causal mutations. These genetic variants were associated with the open chromatin state and histone modification levels at the FLC locus, and they affected FLC expression levels and flowering time. We also showed that allelic variation at FLC is pervasive and that the two causal 5′ UTR deletions originated independently in natural C. rubella populations post speciation and subsequently spread to intermediate frequencies. Overall, our results suggest that sequence variation in the 5′ UTR of FLC has a profound effect on C. rubella flowering time variation. They also support that parallel evolution of early flowering was regulated via two overlapping recurrent deletions in CrFLC.
RESULTS
Genetic Mapping Identifies CrFLC as the Cause of Flowering Time Variation in Two Distinct F2 Populations
To identify genetic variants controlling flowering time variation in C. rubella, we analyzed three accessions, MTE, 762, and 86IT1, which differ considerably in their flowering time. The reference accession MTE is one of the latest flowering accessions, 762 is a intermediate flowering accession, and 86IT1 is an early flowering accession (Supplemental Table 1). To dissect the genetic basis underlying the dramatic differences in flowering time among these accessions, we crossed MTE with 86IT1 and 762 (using 86IT1 and 762 as the female parents) and generated two F2 populations. F1 progeny from both crosses showed the late flowering phenotype, suggesting that late flowering is a potentially dominant phenotype.
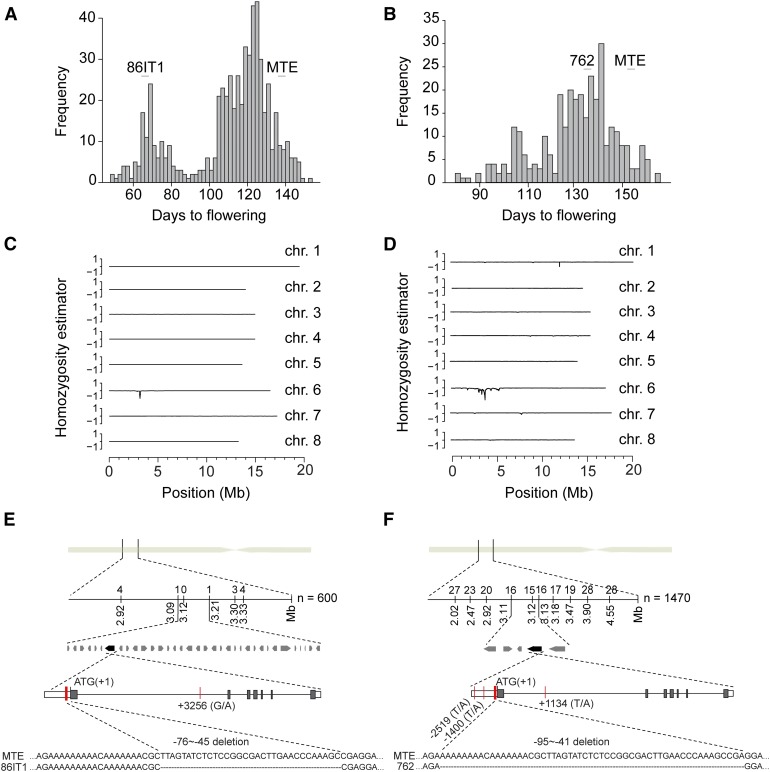
The 86IT1 × MTE F2 population (600 plants) showed a distinct bimodal distribution for flowering time (Figure 1A), suggesting the presence of a major causal locus regulating flowering time variation in this cross. A bimodal distribution for flowering time was also observed in the 762 × MTE F2 population (320 plants), indicating the presence of a large effect locus (Figure 1B). However, it was less noticeable than in 86IT1 × MTE population, suggesting that additional loci likely contribute to the flowering time variation we observed in the cross between MTE and 762.
Figure 1.
Mapping the Genetic Basis of the Flowering Time Variation in 86IT1 × MTE and 762 × MTE.
(A) and (B) Flowering time in 600 F2 individuals from the 86IT1 × MTE cross and 320 F2 individuals from the 762 × MTE cross. Plants were grown at 20°C in long days (16 h light/8 h dark) in 2012 and 2013, respectively. The parent population means are indicated above each graph.
(C) and (D) SHOREmap interval analysis of flowering time QTL in two combinations. The homozygosity estimator is 0 for equal allele frequencies in both parents. Progeny from the 86IT1 × MTE cross score 1 if homozygous for the late-flowering accession MTE and −1 if homozygous for the early-flowering accession 86IT1. Progeny from the 762 × MTE cross score 1 if homozygous for the late-flowering accession MTE and −1 if homozygous for the intermediate-flowering accession 762. Sliding windows of 150 and 200 kb with a step size of 10 kb were used for 86IT1 × MTE and 762 × MTE plants, respectively. The eight largest scaffolds of a preliminary C. rubella genome assembly are shown, corresponding to the majority of the eight chromosomes.
(E) and (F) Fine mapping by a genetic-linkage analysis and sequence variation of the candidate gene C. rubella FLC. The number of homozygous MTE and heterozygous individuals at the markers is indicated above the linkage map. Red vertical lines indicate Carubv10003343m.g (C. rubella FLC) gene sequence polymorphisms between MTE and 86IT1, or between MTE and 762, and indel variation in the 5′ UTR is shown as well.
To identify the causal locus (or loci), we used a mapping-by-sequencing approach, which integrates traditional genetic mapping with next-generation whole genome sequencing by analyzing allele frequencies in a pooled sample (Schneeberger et al., 2009). Tissues of 115 individuals from the 86IT1 × MTE F2 population and 46 individuals from the 762 × MTE F2 population, representing the early flowering fractions, were pooled to generate two distinct samples for resequencing. Short reads of each pooled DNA sample were aligned to the reference MTE genome to calculate allele frequencies using SNP data. We reasoned that regions enriched for 86IT1 or 762 SNPs would be linked to genes underlying early flowering.
Mapping analysis identified a region on chromosome 6 (Figures 1C and 1D) that is associated with early flowering in both populations. This raised the possibility that both 86IT1 and 762 might harbor common allelic variation resulting in early flowering. We genotyped early flowering individuals from the two F2 populations (94 early flowering plants out of 600 individuals from the 86IT1 × MTE F2 population and 188 early flowering plants out of 1470 individuals from the 762 × MTE F2 population) with additional genetic markers to further narrow down the mapping interval (Supplemental Tables 2 and 3). Using this approach, we fine-mapped the causal locus to a 120-kb interval (between markers nearby 3.09 and 3.21 Mb, containing 35 open reading frames) in the 86IT1 × MTE population and to a 13-kb interval (between markers nearby 3.113 and 3.126 M, containing five open reading frames) for the 762 × MTE population (Figures 1E and 1F). Interestingly, the mapping interval overlapped between the two populations, further supporting the possibility that there might be common allelic variation. Notably, the ortholog of A. thaliana FLC, a critical integrator of floral transition, was located within the overlap between the mapped region in the two populations (Figures 1E and 1F; Supplemental Tables 4 and 5). Given both that no other flowering time-associated genes are present in this interval and that FLC is known to be important for flowering time regulation, we investigated whether CrFLC could be the gene causing the phenotypic variation that we observed in both F2 populations.
Sequence and Expression Variation of FLC Affects Flowering Time in C. rubella
To analyze sequence variation at the FLC locus, we sequenced a 9554-bp genomic DNA fragment from the three accessions that included the ∼2.7-kb upstream region including the putative promoter, the entire coding region (∼5.8 kb), and ∼1 kb of downstream sequence. We detected two polymorphisms in the FLC locus of 86IT1 compared with MTE; a G-to-A SNP in intron 1 (SNP_3256) and a 32-bp deletion (Del-32) in the 5′ UTR that was located 44 bp upstream of the start codon (Figure 1E; Supplemental Figure 1). Four polymorphisms distinguished the FLC locus of 762 from the MTE allele: two T-to-A SNPs in the promoter (SNP_-2519 and SNP_-1400), a T-to-A SNP in intron 1 (SNP_1134), and a 54-bp deletion (Del-54) located 41 bp upstream of the start codon in the 5′ UTR (Figure 1F; Supplemental Figure 1). The 5′ UTR deletions overlapped, and the 54-bp deletion in accession 762 completely encompassed the 32-bp deletion observed in accession 861T1 (Figures 1E and 1F). We did not find any nonsynonymous polymorphisms, suggesting that the phenotypic differences were caused by regulatory polymorphisms.
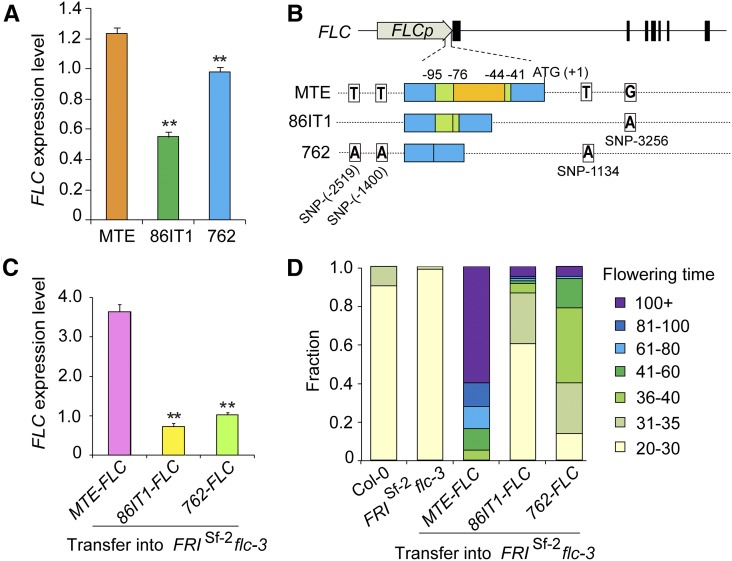
To test whether the observed polymorphisms affected gene expression, we measured FLC expression level in 14-d-old seedlings of three accessions. The expression levels of FLC differed significantly among the accessions (P < 0.01, Student’s t test) (Figure 2A), with levels being lowest in 861T1, followed by 762 and MTE. This correlated well with their flowering time phenotypes. To assess whether these different FLC alleles vary in their ability to delay flowering, we performed transgenic complementation experiments in A. thaliana. The entire FLC genomic fragment (∼9.5 kb) from all three different accessions (MTE-FLC, 86IT1-FLC, and 762-FLC; Figure 2B) was cloned and transformed into A. thaliana FRIsf2 flc-3 plants, which harbor a nonfunctional FLC allele and a functional FRI allele in the reference Col-0 background. The flowering phenotypes of the transgenic plants mimicked the parental strains harboring the respective alleles (Figure 2D; Supplemental Figure 2). Transgenic plants expressing the MTE-FLC allele showed strongly delayed flowering, while transgenic plants expressing the 86IT1-FLC and 762-FLC alleles flowered significantly earlier (P < 0.01, Student’s t test; Figure 2D; Supplemental Figure 2). Expression analysis revealed that their flowering phenotypes correlated with the expression levels of the transgene (Figures 2C and 2D). Consistent with these findings, the expression levels of FT and SOC1, downstream targets of FLC, were inversely correlated with the level of FLC expression (Supplemental Figure 3). Taken together, our results demonstrated that allelic variation at FLC is a major determinant of the variation in flowering time among the tested populations of C. rubella.
Figure 2.
Functional Analysis of C. rubella FLC Alleles in A. thaliana.
(A) RT-qPCR analyses of C. rubella FLC expression levels in the three C. rubella accessions. Expression data are standardized to the C. rubella TUBULIN (Carubv10017235m) transcript levels. Values shown are the means ± sd (n = 3 biological replicates, with 15 seedlings/replicate). **P < 0.01 for the comparison between two early-flowering accessions and MTE (Student’s t test).
(B) Schematic illustration of constructs in the MTE, 86IT1, and 762 alleles of FLC. The structure of C. rubella FLC is illustrated at the top, and the position of ATG is defined as +1. Below the structure are illustrations of C. rubella FLC sequence variation and indel variation in the 5′ UTR region among MTE, 86IT1, and 762.
(C) RT-qPCR analyses of the expression levels of FLC using pools of 40 independent T1 transgenic lines. Data are standardized relative to the abundance of A. thaliana TUBULIN (AT5G62690) transcripts. Values shown are the means ± sd (n = 3 biological replicates; collected from 40 independent T1 transgenic lines per replicate). **P < 0.01 between two early-flowering accessions and MTE allele transgenic lines (Student’s t test).
(D) Flowering time of transgenic plants in 20°C long-day conditions (16 h light/8 h dark). Values for two control lines (Col-0 and FRISf-2 flc-3, without hygromycin selection) are shown on the left. Values for T1 transgenic lines with the FRISf-2 flc-3 background expressing the indicated FLC allele driven from its native promoter are shown on the right (n = 80 T1 transgenic lines). The experiments shown in (A), (C), and (D) were repeated on three separate occasions (three experimental replicates) from screening the transgenic plants through to RNA extraction and RT-qPCR and phenotyping, with similar results. The results shown are from one of the three experimental replicates.
Deletions in the 5′ UTR of CrFLC Reduce Its Expression and Promotes Early Flowering
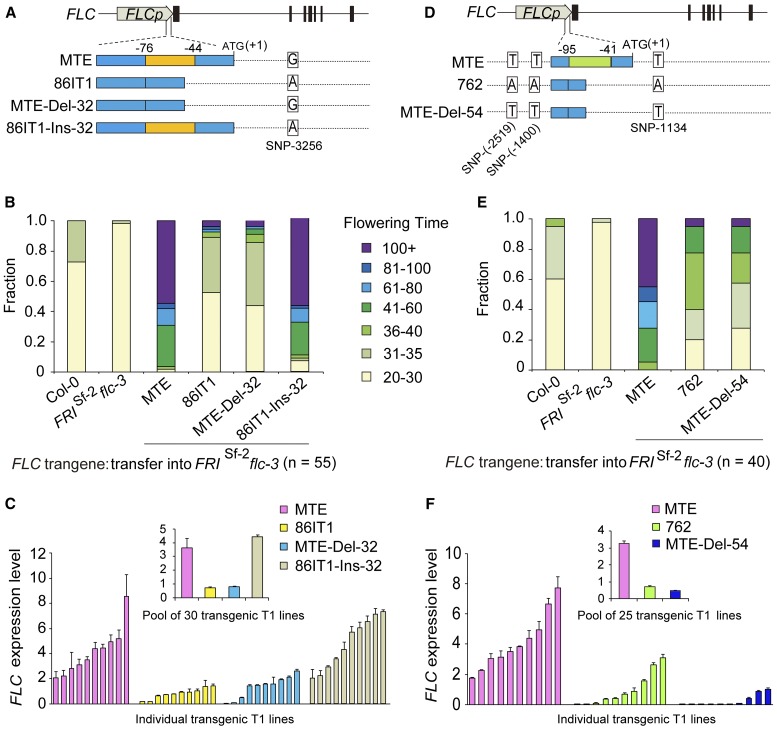
Among the polymorphisms that differentiated the three accessions (Figures 1E and 1F), only the deletion in the 5′ UTR was shared between the two earlier flowering accessions (86IT1 and 762) comparing to the later flowering accession MTE, although the length of the deletion varied between the accessions. Therefore, we asked whether these deletions were sufficient to explain the observed phenotypes. Deleting 32 bp from the 5′ UTR of the MTE allele MTE-FLC (MTE-Del-32) suppressed late flowering in transgenic A. thaliana plants, but reintroducing the deleted region in the 86IT1 allele 86IT1-FLC (86IT1-Ins-32) resulted in late flowering plants (Figures 3A and 3B; Supplemental Figure 4A). Consistent with these findings, the expression levels of FLC were changed, with the lines harboring the deletions showing reduced FLC expression (P < 0.01, Student’s t test; Figure 3C).
Figure 3.
Deletions in the C. rubella FLC 5′ UTR Result in Reduced FLC Expression and Early Flowering.
(A) and (D) Schematic illustration of the DNA regions introduced or removed to create the Del-32 and Del-54 constructs in the MTE-FLC, 86IT1-FLC and 762-FLC alleles. The 32- and 54-bp deletions in the FLC 5′ UTR are indicated with orange and green boxes, respectively. FLC is illustrated at the top, and the position of ATG is defined as +1.
(B) and (E) Flowering time for transgenic plants in the 20°C long-day condition. Values for two control lines (Col-0 and FRISf-2 flc-3, without hygromycin selection) are shown on the left of each graph. Values for transgenic lines with the FRISf-2 flc-3 background expressing different C. rubella FLC alleles with or without Del-32 or Del-54 are shown to the right of the control lines.
(C) and (F) RT-qPCR analyses of FLC expression levels using both a pool of independent T1 transgenic lines (inset) and 10 randomly selected independent T1 transgenic lines. Data are standardized relative to the abundance of A. thaliana TUBULIN (AT5G62690) transcripts and shown as means ± sd (n = 3 biological replicates; for individual lines, each replicate was collected from 3 individual plants; for insets, each replicate was collected from 30 or 25 T1 transgenic lines in [C] and [F], respectively). The experiments shown were repeated on three separate occasions (three experimental replicates), with similar results. The results shown are from one of the three experimental replicates.
A similar pattern was also observed in transgenic lines harboring the MTE allele with the 54-bp deletion MTE-FLC (MTE-Del-54), with early flowering and reduced C. rubella FLC expression (Figures 3D to 3F; Supplemental Figure 4B). Moreover, consistently, in all these transgenic lines, the expression levels of downstream target genes SOC1 and FT were inversely correlated with the FLC expression level (Supplemental Figures 5 and 6). Taken together, these experiments confirmed that a deletion in the 5′ UTR is sufficient to explain the observed differences in FLC expression and its associated phenotype.
Deletions in the 5′ UTR of CrFLC Correlate with a Reduced Response to Vernalization
FLC acts as a central control in the responses to prolonged cold treatment in A. thaliana and related species (Whittaker and Dean, 2017). If the 5′ UTR deletion affects vernalization, we would expect the lines MTE and 86IT1, which differed for the 5′ UTR deletion, to differ in their vernalization response. Consistent with this, a vernalization treatment reduced MTE flowering time by more than 60 d (from more than 132 to 56 d). By comparison, vernalization reduced the flowering time of 86IT1 by only 26 d (from 68 to 42 d; Supplemental Figure 7A). This suggested that the 5′ UTR deletion found in the 86IT1-FLC allele results in a considerably reduced sensitivity to vernalization.
FLC expression in A. thaliana is regulated by the antisense transcript COOLAIR long noncoding RNA, and increases during vernalization, accelerating the silencing of FLC (Csorba et al., 2014; Marquardt et al., 2014). Given that little is known about antisense transcripts in C. rubella, 5′ and 3′ RACE experiments were performed to identify putative CrCOOLAIR transcripts expressed in C. rubella during vernalization. Three CrCOOLAIR transcripts, all of which encompassed the FLC sense transcripts, were detected (Supplemental Figure 7B). However, in contrast to A. thaliana (Csorba et al., 2014), we did not find any short antisense transcripts only located at the 3′end of FLC (Supplemental Figure 7B).
Further analysis of the sense transcript and antisense transcripts of FLC in C. rubella revealed that the 32-bp deletion of 86IT1-FLC was located either in the 5′ UTR of the sense transcript of FLC or in the 3′ region of the antisense transcript CrCOOLAIR. We monitored the expression of FLC and COOLAIR in MTE and 86IT1 plants at different time points during an 8-week cold treatment. While FLC expression decreased during the vernalization treatment in both MTE and 86IT1, the effect was much stronger in 86IT1, with FLC levels being consistently higher in MTE plants (Supplemental Figure 7C). In contrast, COOLAIR expression in 86IT1 was ∼50% higher than in MTE, reaching its peak after one week of cold treatment in both accessions. However, at later stages, the difference in COOLAIR expression between the accessions was minimal (Supplemental Figure 7D). Therefore, while the two accessions show partially different patterns of COOLAIR expression in response to cold treatments, it is currently unclear whether they are linked to the deletion or can explain the differences in flowering time and responses to vernalization that are observed between MTE and 86IT1.
Deletions in the 5′ UTR of CrFLC Are Associated with Chromatin Changes
To elucidate the mechanism by which the 5′ UTR deletions lead to a reduced expression, we first checked whether the position of 32-bp region that is deleted in 86IT1-FLC 5′ UTR was important for early flowering. A fragment containing the 32 nucleotides that are deleted in 86IT1 was reinserted at the beginning of the upstream region (as opposed to its original position in the 5′ UTR) of a Pro86IT1:gMTE-FLC construct (the FLC genomic sequence of MTE driven by the 86IT1-FLC promoter), generating a modified construct hereafter referred to as Pro(32+86IT1):gMTE-FLC (Supplemental Figure 8A). The Pro(32+86IT1):MTE-FLC construct was then transformed into FRIsf2 flc-3 A. thaliana plants. These transgenic plants had a similar early-flowering phenotype as the Pro86IT1:gMTE-FLC transgenic plants (Supplemental Figure 8B). This result implied that the sequence context or location of the 32-bp deletion in 86IT1 is important for its role in modulating FLC activity.
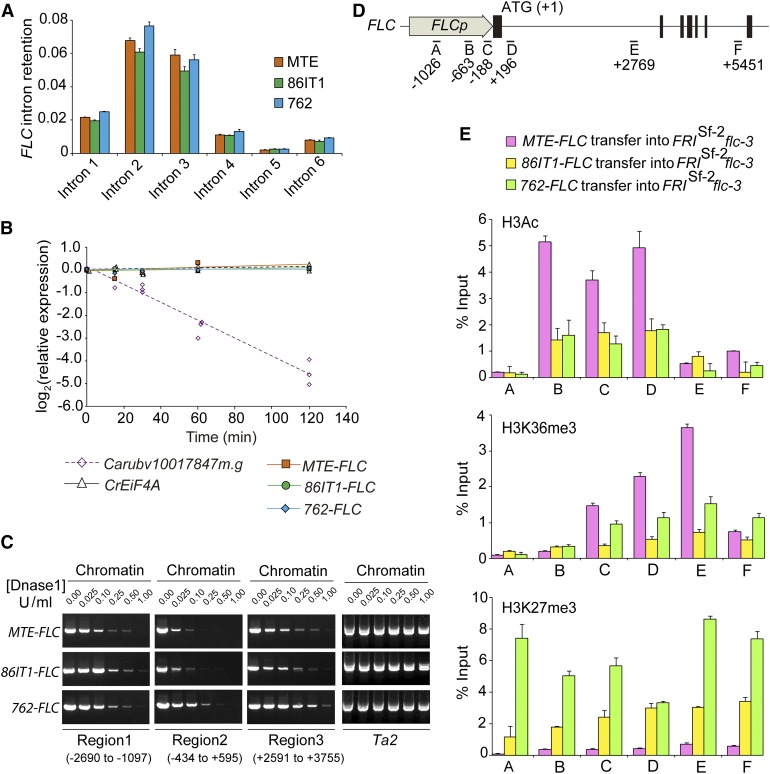
Subsequently, we considered whether FLC could be differentially spliced because this can lead to nonsense-mediated mRNA decay, as shown for the FLM locus in A. thaliana (Sureshkumar et al., 2016). To assess this, we verified intron retention in all three accessions, which would result in transcripts with premature stop codons. For all six tested FLC introns, there was no significant difference of intron retention among the three accessions, and the ratio of unspliced to total transcripts was generally low (∼2–8%; Figure 4A). Consistent with this, treatment with the transcription inhibitor cordycepin and subsequent expression analysis revealed no significant difference between the three accessions, which suggested that mRNA stability is unlikely to be the underlying cause for the reduction in FLC expression associated with the 5′ UTR deletion (Figure 4B).
Figure 4.
Sequence Deletion in the FLC 5′ UTR Is Correlated with Altered Chromatin Structure State and Histone Modifications.
(A) FLC splicing efficiency in three C. rubella accessions. Intron retention was calculated as the ratio of unspliced to total (spliced + unspliced) transcripts for all six FLC introns. Results were normalized to C. rubella TUBULIN (Carubv10017235m) results and are shown as means ± sd (n = 3 biological replicates; with 15 seedlings per replicate).
(B) RNA stability assay in three C. rubella accessions. After the addition of cordycepin, the rate of mRNA degradation was measured by RT-qPCR to determine the level of remaining transcripts relative to that of an untreated sample. C. rubella EIF4A and Carubv10017847m.g were used as controls, representing a stable and an unstable transcript, respectively.
(C) Sensitivity of three regions (regions 1 to 3) of C. rubella FLC to increasing dosages of mononuclease in three transgenic plants. Chromatin with loose nucleosome occupation is more susceptible to mononuclease digestion and is inefficiently amplified by PCR. The A. thaliana Ta2 region in three transgenic plants was used as an insensitive control (Shu et al., 2013). The position of each region is labeled, and the A of ATG is referred to as +1.
(D) Regions of C. rubella FLC used for ChIP-qPCR assays.
(E) ChIP-qPCR analyses of six regions of C. rubella FLC. Data are from pooled T2 transgenic plants using antibodies against H3Ac (top), H3K36me3 (middle), and H3K27me3 (bottom), respectively. ChIP values were normalized to their respective DNA inputs. Data are shown are the means ± sd (n = 3 biological replicates; each replicate was obtained from at least 3 g of tissue corresponding to roughly 500 10-d-old seedlings from 10 independent T2 transgenic lines).
The gene expression level could also be affected by patterns of DNA methylation (He et al., 2011). Therefore, we considered whether the deletion could influence the methylation status of FLC, which would likely modulate its expression levels. Bisulphite sequencing of the locus revealed that the total methylation levels of 86IT1 and 762 were not significantly different from those of MTE for all tested regions. Also, the methylation levels of all tested regions were very low (Supplemental Figure 9). This suggested that the observed reduction in FLC expression was unlikely to be due to changes in methylation.
Another possible mechanism through which the 5′ UTR deletion could affect FLC expression is by modulating openness of the chromatin. One distinct characteristic of the genomic regions of open chromatin that are associated with active gene expression is a pronounced sensitivity to cleavage by the endonuclease DNase I (Wu, 1980; Zhang et al., 2012, 2017). To test whether the deletions altered the chromatin organization at the FLC locus, we evaluated nucleosome organization by a mononuclease digestion-PCR in A. thaliana plants expressing the three FLC alleles. We found that the region around the transcriptional start site and 5′ UTR (region 2; Figure 4C) is the most sensitive to mononuclease digestion and that the MTE-FLC allele was more sensitive to mononuclease digestion compared with the 86IT1-FLC or 762-FLC alleles in all the three tested regions. This is consistent with higher levels of FLC expression in MTE-FLC transgenic plants (Figure 2C).
Our results suggested nucleosome occupancy may be altered at the FLC locus. Because epigenetic regulation is a critical component of the regulation of FLC expression in A. thaliana and the chromatin structure state is closely associated with epigenetic modifications (He et al., 2003; Zhao et al., 2005; Jiang et al., 2007, 2008; Liu et al., 2007, 2010; Pien et al., 2008; Xu et al., 2008; He, 2009; Zhang et al., 2012; Lee et al., 2015), we tested whether the 5′ UTR deletions are associated with changes in histone modifications using transgenic A. thaliana plants expressing the MTE-FLC, 86IT1-FLC, and 762-FLC alleles. We examined histone modification levels in six regions (A–F; Figure 4D) covering the whole C. rubella FLC locus. H3K36 trimethylation and acetylated H3, which marks active transcription, displayed significant enrichment in the MTE-FLC allele compared with the 86IT1-FLC and 762-FLC alleles (Figure 4E; P < 0.01, Student’s t test). In contrast, the H3K27me3 mark, which is associated with repressive chromatin states, was significantly enriched in the 86IT1-FLC and 762-FLC alleles compared to MTE-FLC (Figure 4E; P < 0.01, Student’s t test). The differential enrichments were more pronounced in the 5′ end of the gene in region C, which includes the 32-bp/54-bp deletions found in 86IT1-FLC and 762-FLC, and in region D, which includes the nucleation region of FLC (Figure 4E) (Qüesta et al., 2016; Yuan et al., 2016). This is consistent with the mononuclease digestion-PCR results, indicating that MTE-FLC transgenic plants have a more open chromatin state than that of 86IT1-FLC or 762-FLC in these tested regions (Figures 4C to 4E). These results are also consistent with the dramatic upregulation of FLC expression in MTE-FLC transgenic plants compared with the expression levels for 86IT1-FLC and 762-FLC (Figure 2C). Taken together, these results suggest that the deletions in the FLC 5′ UTR is associated with changes in histone modification and chromatin structure state, which is likely to be the underlying mechanism for the observed transcriptional changes.
Parallel Mutations in the Same Region of CrFLC Determine Flowering Time
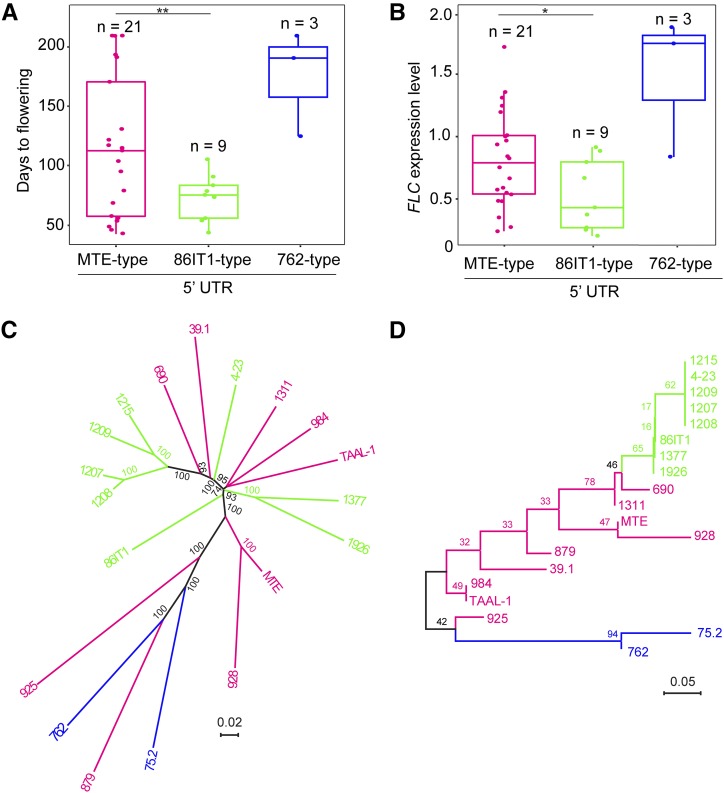
Having established that allelic variation at the 5′ UTR is the underlying cause for variation in flowering time of the accessions we tested, we then asked how common these variants are among C. rubella accessions. To assess this, we genotyped 34 C. rubella accessions for these deletions and identified all three haplotypes (MTE-type, 86IT1-type, and 762-type) of FLC based on the two deletions in the 5′ UTR (Supplemental Table 1). The MTE type was the most abundant haplotype with 22 accessions (65%). Nine other accessions belonged to the 86IT1 type (26%) and only three accessions belonged to the 762 type (9%) (Figure 5A; Supplemental Table 1). Accession 1408 carries a unique loss-of-function allele of FLC (Guo et al., 2012), which was included in the MTE-type haplotype.
Figure 5.
Parallel Mutations in the Same Region of C. rubella FLC Shape Flowering Time.
(A) and (B) Flowering time and FLC expression levels of C. rubella accessions. Accessions were classified into three different haplotypes based on the promoter sequence at FLC (∼2.7 kb, presence/absence of the 32-bp/54-bp deletion in 5′ UTR). The details about the C. rubella accessions used to provide data for this figure are listed in Supplemental Table 1, except accession 1408, whose early flowering was caused by the splice variation (Guo et al., 2012). Data are plotted for n individuals per haplotype; all of the data points are presented with box plots showing the median, upper, and lower quartiles, and minimum and maximum range values for each haplotype. **P < 0.01 and *P < 0.05 between the 86IT1 and MTE haplotypes (Student’s t test) for flowering time and C. rubella FLC expression level, respectively.
(C) Phylogenetic tree of 19 C. rubella accessions based on whole-genome sequences. Bootstrap values (100 replicates) are indicated at branches.
(D) Phylogeny of FLC alleles of 19 C. rubella accessions based on the FLC full-length fragment (∼9.5 kb). Bootstrap values (1000 replicates) are indicated at branches. Different FLC 5′ UTR haplotypes are indicated by different colors. Orange indicates MTE-type 5′ UTR accessions, green indicates 86IT1-type 5′ UTR accessions, and blue indicates 762-type 5′ UTR accessions. Note that the 86IT1-type and 762-type accessions formed a clade on the phylogenetic tree of FLC alleles but were separated on the phylogenetic tree based on the whole-genome sequences.
Having established the pervasive nature of allelic variation at the FLC locus, we assessed how much variation in flowering time could be explained by variation in FLC expression levels. Our analysis revealed that FLC expression could account for 60% of variation (P < 0.0001, linear regression analysis of log-transformed expression levels versus days to flowering [DTF] 20°C long-day condition) in flowering time among accessions of C. rubella, confirmed that FLC expression was a key determinant of flowering time variation, as has been reported in A. thaliana (Shindo et al., 2005). To test how much of this variation in FLC expression could be attributed to deletions of 5′ UTR at FLC locus, we analyzed the association between FLC expression and allelic variation at FLC. We detected a significant association between allelic variation at FLC’s 5′ UTR and the expression levels (P < 0.01). Allelic variation at FLC’s 5′ UTR could account for 32% of the variation in FLC expression. Given that 60% of the variation in flowering time could be attributed FLC expression variation, and 32% of variation in FLC expression could be explained by allelic variation at FLC’s 5′ UTR, we asked how much of the variation in flowering time could be attributed to allelic variation at FLC’s 5′ UTR. Regression analysis revealed that 23% of the variation in flowering time could be attributed to allelic variation at FLC’s 5′UTR in C. rubella (P < 0.05).
To assess the specific contributions of the different 5′ UTR deletions that we identified, we analyzed their individual effect on flowering time. We found that accessions carrying the 86IT1 haplotype displayed significantly early flowering than MTE-type accessions (Figure 5A; Supplemental Figure 10A; P < 0.05). However, 762-type accessions did not show a significant difference in flowering time when compared with MTE. This could be due to the limited sample size for 762-type accessions (only three accessions) and/or the presence of additional genetic factors affecting flowering time in these accessions, consistent with the more continuous distribution for flowering time we observed in the 762 × MTE F2 population. FLC expression levels were significantly (P < 0.05, Student’s t test) higher in MTE-type accessions than that in 86IT1-type accessions but lower than those in 762-type accessions (Figure 5B).
We then tested for a correlation between flowering time and climate variables (19 ecological factors downloaded from the WorldClim database; www.worldclim.org) using 13 accessions with the MTE-type 5′ UTR and nine accessions with the 86IT1-type 5′ UTR for which we have the accurate latitude and longitude collection information (Supplemental Figure 10B). Interestingly, we found that the values of the two environmental factors “mean temperature of driest quarter” and “precipitation of wettest quarter” are significantly (P < 0.05) different between MTE-type 5′ UTR accessions and 86IT1-type 5′ UTR accessions, even though there was no general correlation between flowering time and climate variables. This is suggestive that the deletion of 5′ UTR might be correlated to the adaptation of C. rubella populations to the local climates.
Our findings revealed that common allelic variation at the 5′ UTR of FLC is a major determinant of flowering time variation. By contrast, despite FLC being one of the most studied genes in A. thaliana, only one case of a natural 50-bp deletion at 5′ UTR affecting flowering time in the L1-0 accession was described. This deletion does not affect prevernalization levels of FLC expression, but enhances the response to vernalization (Sánchez-Bermejo et al., 2012). The deletion overlapped with the 5′ end of the deletions identified in this study (Supplemental Figure 11). Therefore, we tested whether these deletions are present in other species and found that the deletions were not detected in Capsella grandiflora, Capsella orientalis, A. lyrata, or Arabidopsis halleri (Supplemental Figure 11). Unlike the genetic variation in the first intron of FLC orthologs in the Arabis genus, which plays an important role on the flowering time variation (Kiefer et al., 2017), the 5′ UTR deletions exist only in C. rubella. To assess the origin of these deletions in C. rubella, we performed a phylogenetic analysis of 19 accessions based on whole-genome sequencing data (Agren et al., 2014) (Figure 5C) and FLC sequences (Figure 5D; Supplemental File 1). These analyses revealed that the two deletions originated independently in C. rubella.
DISCUSSION
Allelic Variation at FLC Contributes to Natural Variation in Flowering Time in C. rubella
We have demonstrated that allelic variation at FLC is a major determinant of flowering time variation in C. rubella. Flowering time is one of the adaptive traits for which variation has been reported and well studied in a variety of species (Salomé et al., 2011; Weigel, 2012). In the model species A. thaliana, the epistatic interaction between FRI/FLC accounts for much of the natural variation in flowering time, with changes in FLC expression levels resulting mostly from loss/reduction of function polymorphisms at the FRI locus (Sanchez-Bermejo and Balasubramanian, 2016). Interestingly, common allelic variations at FRI in A. thaliana occur primarily through deletions involving 5′ UTR (Johanson et al., 2000). Although allelic variation at FLC has been reported, it is not the major determinant of flowering time variation in A. thaliana.
FLC, however, plays a more important role in natural variation for flowering time in other species of the Brassicaceae family (Slotte et al., 2009; Wang et al., 2009; Yuan et al., 2009; Hu et al., 2011; Leinonen et al., 2013; Raman et al., 2016). Sequence analysis of the FRI locus in C. rubella did not reveal the same level of allelic variation that is observed in A. thaliana (Guo et al., 2012). Therefore, it appears that the C. rubella evolutionary strategy differs from that of A. thaliana. Whereas changes in FLC expression retain a central importance in the regulation of flowering time, in C. rubella these changes are caused by allelic variation at FLC itself rather than at the FRI locus.
This is not surprising, given that, being downstream of its activator FRI, FLC is likely to be more capable of generating phenotypic variation without affecting other functions. However, it is not clear why A. thaliana exhibits a high frequency of FRI mutations related to flowering time regulation. More case studies are needed to clarify the general mechanisms underlying these divergent evolutionary patterns.
Loss/Reduction-of-Function Alleles of FLC Have Evolved Independently Multiple Times in C. rubella
The study of parallel evolution provides insight into the predictability of evolution; whether different species or populations that share a common ancestor could independently evolve the same trait by changing the same genes. In numerous cases in diverse organisms, different mutations in the same gene across species or natural populations are responsible for parallel phenotypic evolution (Martin and Orgogozo, 2013; Stern, 2013; Storz, 2016). In plants, there are several cases of parallel evolution documented in flower pigmentation (Schwinn et al., 2006; Smith and Rausher, 2011). However, the extent to which mutations in the same region in independent lineages have shaped evolutionary history in plants is largely unknown.
Early flowering has evolved multiple times independently in A. thaliana associated with loss or reduction in function of alleles of FRI, FLC, and FLM (Michaels and Amasino, 1999; Lempe et al., 2005; Shindo et al., 2005; Salomé et al., 2011; Sánchez-Bermejo et al., 2012; Lutz et al., 2017). Given the recent origin and very low genetic variation of C. rubella, the study of the evolution of phenotypic differences in this species could, to some extent, be considered akin to the experimental evolution studies often performed in microorganisms to understand how phenotypic diversity arises. We have demonstrated that in C. rubella, FLC loss of function has evolved multiple times, conferring variation in flowering time (this study; Guo et al., 2012). Here, we have shown that the cause of the early-flowering 86IT1-FLC and 762-FLC alleles is two overlapping deletions in the 5′ UTR of FLC that have originated independently. In addition, in a previous study, we have demonstrated the existence of an independent a loss-of-function allele at the FLC locus (Guo et al., 2012). While the latter case appears to be restricted to a single accession, the deletions are pervasive in nature. Therefore, it is clear that loss/reduction-of-function alleles at FLC have repeatedly originated in C. rubella populations, similar to the situation in the Arabis genus (Albani et al., 2012; Kiefer et al., 2017).
Flowering Time Variation beyond FLC in C. rubella
Despite the association between flowering time variation and the 5′ UTR deletions in FLC, a large fraction of the variation observed among accessions of C. rubella cannot be explained by these polymorphisms. For example, the two accessions in the 762-type group, 1482 and 75.2, are late flowering despite carrying an early-flowering allele of FLC. It is likely that other loci, aside from FLC, are responsible for determining flowering time in these accessions. This is further supported by the distribution for flowering time in the 762 × MTE F2 population (Figure 1B), suggesting the presence of other loci affecting the timing of flowering in these accession (762 and 75.2 are closely related; Figure 5). The deletion in the FLC 5′ UTR in these two accessions promotes early flowering, but other loci may have the opposite effect. Additionally, transgenic introduction of either the 86IT1-FLC or the 762-FLC allele into A. thaliana results in similarly early-flowering phenotypes (Figure 2). However, significantly later flowering was observed in C. rubella 762 plants relative to 86IT1 plants (Supplemental Figure 10A and Supplemental Table 1). We cannot exclude the possibility that alleles of FLC behave differently when introduced into A. thaliana versus C. rubella owing to differences between the regulatory networks controlling flowering in the two species. However, it seems more plausible that loci other than FLC are responsible for the difference in flowering time between 86IT1 and 762.
In this study, we focused on the role of FLC as a key determinant of flowering time variation in C. rubella. Although we have conclusively shown both that the deletions are the underlying cause for early flowering and that they are associated with altered chromatin, exactly how the deletions affect chromatin structure and histone modifications is currently unclear. Do the deletions have positional and (or) length effects related to the regulation of C. rubella FLC chromatin structure state and histone modifications? In the future, it will be intriguing to further determine the extent to which epigenetic modifications could account for natural phenotypic variation and adaptation in a species with extremely low genetic diversity.
In conclusion, our results provide strong evidence that parallel evolution of independent deletions at the same 5′ UTR region shapes phenotype variation in plants. It will be of great interest to clarify the extent to which parallel evolution shapes variation in other traits among natural populations of this species and, more broadly, the role of parallel evolution in diverse taxa in general.
METHODS
Plant Materials and Growth Conditions
The 34 Capsella rubella accessions used in this study were described previously (Guo et al., 2009) and are listed in Supplemental Table 1. FRIsf2 flc-3 Arabidopsis thaliana plants, which harbor a nonfunctional FLC allele and a functional FRI allele in the reference Col-0 background, were previously reported by Michaels and Amasino (1999). For this study, plants were grown on the soil mix (soil:vermiculite = 1:1) under long-day conditions (16 h light/8 h dark) at 20°C. Trays of plants were moved to random positions in the growth rooms every 2 d to reduce positional effects. Flowering time was measured as leaf number and DTF. DTF was defined as the number of days from planting to the day of the first flower anthesis. The flowering times of 86IT1 × MTE and 762 × MTE F2 plants, together with their parents, were measured in 2012 and 2013 (Figures 1A and 1B; Supplemental Figure 7). The flowering times of 34 C. rubella accessions was measured in year 2016 (Supplemental Figure 10A and Supplemental Table 1).
Mapping by Sequencing
DNA was extracted from pooled leaves of early-flowering F2 plants using a CTAB (cetyltrimethyl ammonium bromide) protocol (Doyle and Doyle, 1987). Paired-end reads (100 bp) were sequenced using the Illumina HiSeq 2000 with an insert size of ∼300 bp. In total, 159,506,270 and 105,233,658 reads were obtained for 86IT1 × MTE and 762 × MTE F2 populations, respectively. For 86IT1 × MTE, short reads with ∼68.9-fold coverage were mapped to the MTE genome (Phytozome; http://www.phytozome.net/capsella/), and SNPs were called using SHORE (Ossowski et al., 2008). By applying SHORE’s scoring matrix approach to optimize heterozygous SNP detection and strict filtering (uniqueness of reads, coverage ≥10, minor allele frequency ≥0.2, and SNP quality score ≥25), 110,091 SNPs were retained. For the 762 × MTE combination, short reads with ∼54.5-fold coverage were mapped to a preliminary assembly of the MTE genome and SNPs were called using SHORE. In total, 297,682 SNPs were retained after filtering according to the following parameters: coverage ≥4 and minor allele frequency ≥0.15. The SNPs from each pooled population were used as markers to identify regions with an excess of homozygous alleles across the genome using SHOREmap (Schneeberger et al., 2009).
DNA Sequence Analysis
The FLC upstream region (∼2.7 kb) from C. rubella accessions and related species and an ∼9.5-kb genomic DNA fragment of FLC in MTE, 86IT1, and 762 were obtained by PCR. Purified PCR products were sequenced using the ABI 3730 automated sequencer (Applied Biosystems). Lasergene Seqman (DNASTAR) was used to assemble sequences. Primer sequences are listed in Supplemental Table 6.
Plasmid Construction and Plant Transformation
To generate C. rubella FLC transgenes, an ∼9.5-kb genomic DNA fragment of the C. rubella FLC gene from three accessions (including ∼2.7 kb upstream of the ATG start codon and ∼1.0 kb downstream of the stop codon) with or without SNPs was cloned into the pCAMBIA1300 vector (Clontech) between the KpnI and SalI sites. To produce the Pro(32+86IT1):gMTE-FLC construct, the ∼2.7-kb upstream sequence of FLC from two different accessions (86IT1 and MTE) and the full FLC genomic fragment from ATG to TAG (∼5.8 kb) plus 1 kb of sequence from 3′ downstream amplified from MTE were ligated together and cloned into pCAMBIA1300 first, then we inserted the 32-bp region deleted in 86IT1 plus its around sequences, beginning from −76 bp to +64 bp (the number was counted according to the position of ATG is defined as +1), at the beginning of the promoter region by the KpnI site.
The A. thaliana mutant accession with FRIsf2 and flc-3 alleles in the Col-0 background was transformed using Agrobacterium tumefaciens strain GV3101 with the floral dipping method. Seeds from T0 transgenic plants were selected on MS medium supplemented with 50 mg/L hygromycin 10 d; positive lines were then transferred to soil and the presence of the transgene was confirmed by PCR.
Analysis of Gene Transcript Levels
RNA was extracted from whole 14-d-old seedlings grown on MS medium for C. rubella accessions and leaves for T1 transgenic plants before blooming. Total RNA was extracted using the Plant RNA Kit (Omega) with an OnColumn DNase I digestion treatment. Spectrophotometry and gel electrophoresis were performed to detect RNA quality. To synthesize cDNA, 1 µg of RNA was used in the SuperScript III First-Strand Synthesis System (Invitrogen). RT-qPCR analysis (56°C, 45S and 45 cycles) was performed using SYBR Premix Ex TaqII mix (TaKaRa) on the ViiA 7 Real-time PCR System (Life Technologies). Each experiment was repeated with three independent biological replicates, and RT-qPCR reactions were performed with three technical replicates for each sample. The expression level was calculated as 2ΔΔCT and then normalized to C. rubella/A. thaliana TUBLIN. All primers used are listed in Supplemental Table 6.
5′ RACE and 3′ RACE
Full-length FLC sense and antisense cDNA were obtained by 5′ and 3′ RACE with the SMARTer RACE cDNA amplification kit (Clontech) following the manufacturer’s instruction. One microgram of RNA was used to synthesis RACE-Ready cDNAs. Gene-specific primers for 5′ RACE are listed in Supplemental Table 1. After indentifying the band(s) of interest, DNA was extracted from the gel using the StarPrep Gel Extraction Kit (Genestar; D205-01), and the purified fragments were ligated into the Peasy-Blunt vector (Transgene; CB111-01). At least 15 clones for each fragment were sequenced.
Cordycepin Treatment
Plants were grown on MS plates containing 0.3% sucrose (w/v) at 20°C. Approximately 100 14-d-old seedlings were transferred to incubation buffer (1 mM PIPES, pH 6.25, 1 mM sodium citrate, 1 mM potassium chloride, and 15 mM sucrose) and incubated for 30 min with shaking. Cordycepin (C-3394; Sigma-Aldrich) was then added to a final concentration of 150 µg/mL and vacuum infiltration was performed for 30 s. Seedlings representing ∼0.5 g of material were collected at various time points and used for RNA extraction. RT-qPCR was processed to analyze the expression data. Eukaryotic initiation factor-4A (eiF4A) and the homolog of AT3G45970 in C. rubella named Carubv10017847m.g were used as controls to measure high and low mRNA stability levels, respectively (Golisz et al., 2013). The expression level at each time point (C. rubella FLC/CrTUBULIN) was normalized to the expression at time 0 and is presented on a binary logarithm scale. Three biological replicates for each accession were used.
Measurement of Splicing Efficiency
Splicing efficiency was measured as described previously (Mahrez et al., 2016). Briefly, a primer corresponding to an exon was combined with a primer corresponding to a neighboring intron (for the unspliced transcript) or covering the splicing junction (for the spliced transcript). RT controls were always included to confirm the absence of genomic DNA contamination.
Detection of Open Chromatin by Mononuclease Digestion
Open chromatin was detected according to previous studies (Shu et al., 2013; Zhang et al., 2017). About 0.3 g of 10-d-old seedlings of pooled T2 transgenic plants frozen in liquid nitrogen were ground to a powder and suspended in 10 mL of ice-cold nuclei isolation buffer (1 M hexylene glycol, 20 mM PIPES-KOH, pH 7.6, 10 mM MgCl2, 1 mM EGTA, 15 mM NaCl, 0.5 mM spermidine, 0.15 mM spermine, 0.5% Triton X-100, 10 mM β-mercaptoethanol, and 1× protease inhibitor cocktail [Roche]) with gentle rotation for 15 min. The suspension was filtered through 30-nm CellTrics, and the elute was centrifuged for 10 min at 1500g at 4°C. The pellet was resuspended as the crude nuclei extract with digestion buffer (40 mM Tris-HCl, pH 7.9, 0.3 M Suc, 10 mM MgSO4, 1 mM CaCl2, and 1× protease inhibitor cocktail [Roche]). Equal aliquots of nuclei extract were subjected to digestion with increasing amounts of mononuclease at 30°C for 15 min, and DNA was extracted using phenol:chloroform:isoamyl alcohol (25:24:1, v/v) treatment followed by ethanol precipitation for the PCR analysis.
Bisulfite Sequencing
Approximately 500 ng of genomic DNA was used for bisulfite conversion using the EZ DNA Methylation-Gold Kit (ZYMO Research) according to the manufacturer’s instructions. Bisulfite-treated DNA (1 µL) was then amplified by PCR in a 25-μL reaction using ZymoTaq DNA polymerase (ZYMO Research) and degenerate primers (Supplemental Table 6). PCR products were resolved on a 1% agarose gel, and then excised, purified, and cloned into a pGEM-T vector (Promega) for sequencing. For each plant genotype, 20 independent top-strand clones were sequenced. For tested regions, bisulfite DNA sequences were analyzed, and the levels of DNA methylation were calculated using the online Kismeth program (Gruntman et al., 2008). For each genotype, the percentage of cytosine methylation in each context (CG, CHG, or CHH) was calculated.
Chromatin Immunoprecipitation
For the chromatin immunoprecipitation (ChIP) assay, we started with ∼600 μL T2 transgenic seed for each genotype (these being transgenic lines transformed with full-length FLC alleles of MTE-FLC, 86IT1-FLC, or 762-FLC transferred into the FRISf-2 flc-3 background; see Figure 2B). These starting pools of seed were divided randomly into three samples of ∼200 μL seed per genotype, and germinated into three sets of 10-d-old seedlings to create three biological replicates per genotype (at least 3 g seedling tissue, equal to more than 500 seedlings, was used for each replicate). Each ∼3 g replicate tissue sample was cross-linked with 1% formaldehyde in a vacuum for 20 min and then ground to a powder in liquid nitrogen. The chromatin complexes were isolated and sonicated, and then incubated with various antibodies. A total of 10 µg of anti-AcH3 (06-599; Millipore), anti-H3K36me3 (ab9050; Abcam), and anti-H3K27me3 (07-449; Millipore) antibodies was used in a 10-μL volume for immunoprecipitation. An equal amount of sample without antibody was used as a mock control. The precipitated DNA was recovered and analyzed by RT-qPCR with SYBR Premix Ex TaqII Mix (Takara) using specific primers listed in Supplemental Table 6. Each ChIP value was normalized to its respective input DNA value. All ChIP-qPCR experiments were independently performed in triplicate.
Phylogenetic Analysis of C. rubella FLC
A neighbor-joining tree of 19 C. rubella samples based on 1,795,299 SNPs matrix across the whole genome was constructed using PHYLIP (version 3.695; Feisenstein, 1989) (Supplemental File 1) and based on C. rubella FLC genomic sequences (Supplemental File 2) was aligned and constructed using Mega 6 (Tamura et al., 2013). Topological robustness of the phylogenetic tree was assessed by bootstrapping with 100 replicates for whole genome data and 1000 replicates for FLC genomic sequences data, respectively. The C. rubella resequencing data were reported in a previous study (Agren et al., 2014).
Statistical Analyses
All the statistical analyses were performed in R (http://www.r-project.org/).
Accession Numbers
The genome sequence of the pooled F2 population of MTE × 86IT1 and MTE × 762 has been deposited in the GenBank database with accession number PRJNA305780. Promoter sequences of FLC produced from PCR products were submitted to GenBank with accession numbers KU297662, KU297663, KU297666 to KU297672, and KU356733 to KU356752. The accession numbers of all genes identified or used in this study list as follows: CrFLC (Carubv10003343m), CrFT (Carubv10021034m), CrSOC1 (Carubv10024046m), CrTUBULIN (Carubv10017235m), CrEIF4A (Carubv10013828m), AtFLC (AT5G10140), AtFT (AT1G65480), AtSOC1 (AT2G45660), and AtTUBULIN (AT5G62690).
Supplemental Data
Supplemental Figure 1. 5′ UTR Sequence of FLC Identified in the MTE, 86IT1, and 762 Accessions.
Supplemental Figure 2. Leaf Number of Three Transgenic Plants Expressing Different C. rubella FLC Alleles in 20°C LD Conditions.
Supplemental Figure 3. RT-qPCR Analyses of FT and SOC1 Expression Level in Transgenic Plants Expressing Different C. rubella FLC Alleles.
Supplemental Figure 4. Leaf Number ofTransgenic Plants in Figure 3 under 20°C LD Conditions.
Supplemental Figure 5. RT-qPCR Analyses of FT and SOC1 Expression Level in Transgenic Plants Expressing Different C. rubella FLC Alleles with or without Del-32.
Supplemental Figure 6. RT-qPCR Analyses of FT and SOC1 Expression Level in Transgenic Plants Expressing Different C. rubella FLC Alleles with or without Del-54.
Supplemental Figure 7. Deletions in the 5′ UTR of FLC May Be Essential for Variation in Vernalization Response in C. rubella.
Supplemental Figure 8. The Location of 32-bp Region Deleted in 86IT1-FLC 5′-UTR Is Important for Early Flowering.
Supplemental Figure 9. DNA Methylation Levels in the Regions Surrounding the Transcription Start and the End of the First Intron of C. rubella FLC in MTE, 86IT1, and 762.
Supplemental Figure 10. Distribution of Flowering Time and Geographic Locations of C. rubella Accessions.
Supplemental Figure 11. Sequence Variation of the FLC Promoter Region from −528 to −1 among Six Congeners and within Three C. rubella Accessions.
Supplemental Table 1. Capsella rubella Accessions Used in This Study.
Supplemental Table 2. Marker Analysis of Early Flowering Plants in the F2 Generation of 86IT1 × MTE.
Supplemental Table 3. Marker Analysis of Early Flowering Plants in the F2 Generation of 762 × MTE.
Supplemental Table 4. Candidate Genes Included in the 120-kb Region Containing the Causal Locus Responsible for Variation of Flowering Time between 86IT1 and MTE.
Supplemental Table 5. Candidate Genes Included in the 13-kb Region Containing the Causal Locus Responsible for Variation of Flowering Time between 762 and MTE.
Supplemental Table 6. Primers Used in This Study.
Supplemental File 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Figure 5C.
Supplemental File 2. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Figure 5D.
Acknowledgments
We thank Song Ge, Xiaofeng Cao, Jia-Wei Wang, Yong-Xiu Liu, and Rongcheng Lin for helpful comments and discussions, as well as Lianwei Zhu for assistance with the ChIP experiments. This work was supported by grants from the National Natural Science Foundation of China (91731306 and 31470331 to Y.-L.G. and 31600187 to L.Y.) and by the CAS/SAFEA International Partnership Program for Creative Research Teams (Y.-L.G.).
AUTHOR CONTRIBUTIONS
L.Y., H.-N.W., and Y.-L.G. designed the experiments. L.Y., H.-N.W., X.-H.H., X.-M.N., T.-S.H., and J.Z. performed experiments. L.Y., Y.-P.Z., Z.Z., M.T., S.B., and Y.-L.G. analyzed and interpreted the experimental data. L.Y., H.-N.W., S.B., and Y.-L.G. wrote the manuscript with input from all the other authors.
Footnotes
Articles can be viewed without a subscription.
References
- Agren J.A., Wang W., Koenig D., Neuffer B., Weigel D., Wright S.I. (2014). Mating system shifts and transposable element evolution in the plant genus Capsella. BMC Genomics 15: 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani M.C., Castaings L., Wötzel S., Mateos J.L., Wunder J., Wang R., Reymond M., Coupland G. (2012). PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering. PLoS Genet. 8: e1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt J., Reznick D. (2008). Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol. Evol. (Amst.) 23: 26–32. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S., Sureshkumar S., Agrawal M., Michael T.P., Wessinger C., Maloof J.N., Clark R., Warthmann N., Chory J., Weigel D. (2006). The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat. Genet. 38: 711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomer R.H., Dean C. (2017). Fine-tuning timing: natural variation informs the mechanistic basis of the switch to flowering in Arabidopsis thaliana. J. Exp. Bot. 68: 5439–5452. [DOI] [PubMed] [Google Scholar]
- Caicedo A.L., Stinchcombe J.R., Olsen K.M., Schmitt J., Purugganan M.D. (2004). Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. USA 101: 15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.F., et al. (2010). Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327: 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo P.F., Hosemann K.E., Balabhadra S., Villarreal G. Jr., Dickson M., Grimwood J., Schmutz J., Myers R.M., Schluter D., Kingsley D.M. (2005). Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307: 1928–1933. [DOI] [PubMed] [Google Scholar]
- Csorba T., Questa J.I., Sun Q., Dean C. (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA 111: 16160–16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L. (1987). A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem. Bull. 19: 11–15. [Google Scholar]
- Elmer K.R., Meyer A. (2011). Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends Ecol. Evol. (Amst.) 26: 298–306. [DOI] [PubMed] [Google Scholar]
- Feisenstein J. (1989). PHYLIP-phylogeny inference package (version 3.2). Cladistics 5: 164–166. [Google Scholar]
- Foxe J.P., Slotte T., Stahl E.A., Neuffer B., Hurka H., Wright S.I. (2009). Recent speciation associated with the evolution of selfing in Capsella. Proc. Natl. Acad. Sci. USA 106: 5241–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani S., Gendall A.R., Lister C., Dean C. (2003). Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132: 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golisz A., Sikorski P.J., Kruszka K., Kufel J. (2013). Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res. 41: 6232–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntman E., Qi Y., Slotkin R.K., Roeder T., Martienssen R.A., Sachidanandam R. (2008). Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.L., Bechsgaard J.S., Slotte T., Neuffer B., Lascoux M., Weigel D., Schierup M.H. (2009). Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc. Natl. Acad. Sci. USA 106: 5246–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.L., Todesco M., Hagmann J., Das S., Weigel D. (2012). Independent FLC mutations as causes of flowering-time variation in Arabidopsis thaliana and Capsella rubella. Genetics 192: 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. (2009). Control of the transition to flowering by chromatin modifications. Mol. Plant 2: 554–564. [DOI] [PubMed] [Google Scholar]
- He G., Elling A.A., Deng X.W. (2011). The epigenome and plant development. Annu. Rev. Plant Biol. 62: 411–435. [DOI] [PubMed] [Google Scholar]
- He Y., Michaels S.D., Amasino R.M. (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302: 1751–1754. [DOI] [PubMed] [Google Scholar]
- Hepworth J., Dean C. (2015). Flowering Locus C's lessons: conserved chromatin switches underpinning developmental timing and adaptation. Plant Physiol. 168: 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra H.E. (2006). Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity (Edinb.) 97: 222–234. [DOI] [PubMed] [Google Scholar]
- Hu G.L., Hu Z.L., Li Y., Gu F., Zhao Z.P., Chen G.P. (2011). A splicing site mutation in BrpFLC1 and repressed expression of BrpFLC genes are associated with the early flowering of purple flowering stalk. Russ. J. Plant Physiol. 58: 431–438. [Google Scholar]
- Jiang D., Yang W., He Y., Amasino R.M. (2007). Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19: 2975–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Wang Y., Wang Y., He Y. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS One 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., West J., Lister C., Michaels S., Amasino R., Dean C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347. [DOI] [PubMed] [Google Scholar]
- Kiefer C., Severing E., Karl R., Bergonzi S., Koch M., Tresch A., Coupland G. (2017). Divergence of annual and perennial species in the Brassicaceae and the contribution of cis-acting variation at FLC orthologues. Mol. Ecol. 26: 3437–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Alonso-Blanco C., Vreugdenhil D. (2004). Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55: 141–172. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Ryu H.S., Chung K.S., Posé D., Kim S., Schmid M., Ahn J.H. (2013). Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342: 628–632. [DOI] [PubMed] [Google Scholar]
- Lee J., Yun J.Y., Zhao W., Shen W.H., Amasino R.M. (2015). A methyltransferase required for proper timing of the vernalization response in Arabidopsis. Proc. Natl. Acad. Sci. USA 112: 2269–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen P.H., Remington D.L., Leppälä J., Savolainen O. (2013). Genetic basis of local adaptation and flowering time variation in Arabidopsis lyrata. Mol. Ecol. 22: 709–723. [DOI] [PubMed] [Google Scholar]
- Lempe J., Balasubramanian S., Sureshkumar S., Singh A., Schmid M., Weigel D. (2005). Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 1: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61: 395–420. [DOI] [PubMed] [Google Scholar]
- Liu F., Quesada V., Crevillén P., Bäurle I., Swiezewski S., Dean C. (2007). The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol. Cell 28: 398–407. [DOI] [PubMed] [Google Scholar]
- Lutz U., Nussbaumer T., Spannagl M., Diener J., Mayer K.F., Schwechheimer C. (2017). Natural haplotypes of FLM non-coding sequences fine-tune flowering time in ambient spring temperatures in Arabidopsis. eLife 6: e22114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrez W., Shin J., Muñoz-Viana R., Figueiredo D.D., Trejo-Arellano M.S., Exner V., Siretskiy A., Gruissem W., Köhler C., Hennig L. (2016). BRR2a affects flowering time via FLC splicing. PLoS Genet. 12: e1005924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S., Raitskin O., Wu Z., Liu F., Sun Q., Dean C. (2014). Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 54: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Orgogozo V. (2013). The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution 67: 1235–1250. [DOI] [PubMed] [Google Scholar]
- McGrath P.T., Xu Y., Ailion M., Garrison J.L., Butcher R.A., Bargmann C.I. (2011). Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature 477: 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Vigo B., Martínez-Zapater J.M., Alonso-Blanco C. (2013). The flowering repressor SVP underlies a novel Arabidopsis thaliana QTL interacting with the genetic background. PLoS Genet. 9: e1003289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Clark R.M., Lanz C., Warthmann N., Weigel D. (2008). Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18: 2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S., Fleury D., Mylne J.S., Crevillen P., Inzé D., Avramova Z., Dean C., Grossniklaus U. (2008). ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20: 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posé D., Verhage L., Ott F., Yant L., Mathieu J., Angenent G.C., Immink R.G., Schmid M. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503: 414–417. [DOI] [PubMed] [Google Scholar]
- Protas M.E., Patel N.H. (2008). Evolution of coloration patterns. Annu. Rev. Cell Dev. Biol. 24: 425–446. [DOI] [PubMed] [Google Scholar]
- Qüesta J.I., Song J., Geraldo N., An H., Dean C. (2016). Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 353: 485–488. [DOI] [PubMed] [Google Scholar]
- Raman H., Raman R., Coombes N., Song J., Prangnell R., Bandaranayake C., Tahira R., Sundaramoorthi V., Killian A., Meng J., Dennis E.S., Balasubramanian S. (2016). Genome-wide association analyses reveal complex genetic architecture underlying natural variation for flowering time in canola. Plant Cell Environ. 39: 1228–1239. [DOI] [PubMed] [Google Scholar]
- Salomé P.A., Bomblies K., Laitinen R.A., Yant L., Mott R., Weigel D. (2011). Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics 188: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Bermejo E., Balasubramanian S. (2016). Natural variation involving deletion alleles of FRIGIDA modulate temperature-sensitive flowering responses in Arabidopsis thaliana. Plant Cell Environ. 39: 1353–1365. [DOI] [PubMed] [Google Scholar]
- Sánchez-Bermejo E., Méndez-Vigo B., Picó F.X., Martínez-Zapater J.M., Alonso-Blanco C. (2012). Novel natural alleles at FLC and LVR loci account for enhanced vernalization responses in Arabidopsis thaliana. Plant Cell Environ. 35: 1672–1684. [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A.H., Nielsen K.L., Jørgensen J.E., Weigel D., Andersen S.U. (2009). SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551. [DOI] [PubMed] [Google Scholar]
- Schwinn K., Venail J., Shang Y., Mackay S., Alm V., Butelli E., Oyama R., Bailey P., Davies K., Martin C. (2006). A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18: 831–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C., Aranzana M.J., Lister C., Baxter C., Nicholls C., Nordborg M., Dean C. (2005). Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138: 1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu H., Gruissem W., Hennig L. (2013). Measuring Arabidopsis chromatin accessibility using DNase I-polymerase chain reaction and DNase I-chip assays. Plant Physiol. 162: 1794–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Huang H.R., Holm K., Ceplitis A., Onge K.S., Chen J., Lagercrantz U., Lascoux M. (2009). Splicing variation at a FLOWERING LOCUS C homeolog is associated with flowering time variation in the tetraploid Capsella bursa-pastoris. Genetics 183: 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.D., Rausher M.D. (2011). Gene loss and parallel evolution contribute to species difference in flower color. Mol. Biol. Evol. 28: 2799–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A., Schmid M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68: 2013–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.L. (2013). The genetic causes of convergent evolution. Nat. Rev. Genet. 14: 751–764. [DOI] [PubMed] [Google Scholar]
- Stern D.L., Orgogozo V. (2009). Is Genetic Evolution Predictable? Science 323: 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J.F. (2016). Causes of molecular convergence and parallelism in protein evolution. Nat. Rev. Genet. 17: 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureshkumar S., Dent C., Seleznev A., Tasset C., Balasubramanian S. (2016). Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nat. Plants 2: 16055. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl V., Ponnu J., Schlereth A., Arrivault S., Langenecker T., Franke A., Feil R., Lunn J.E., Stitt M., Schmid M. (2013). Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707. [DOI] [PubMed] [Google Scholar]
- Wang R., Farrona S., Vincent C., Joecker A., Schoof H., Turck F., Alonso-Blanco C., Coupland G., Albani M.C. (2009). PEP1 regulates perennial flowering in Arabis alpina. Nature 459: 423–427. [DOI] [PubMed] [Google Scholar]
- Weigel D. (2012). Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol. 158: 2–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner J.D., Borevitz J.O., Uhlenhaut N.H., Ecker J.R., Chory J., Weigel D. (2005). FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170: 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker C., Dean C. (2017). The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33: 555–575. [DOI] [PubMed] [Google Scholar]
- Wood T.E., Burke J.M., Rieseberg L.H. (2005). Parallel genotypic adaptation: when evolution repeats itself. Genetica 123: 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. (1980). The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 286: 854–860. [DOI] [PubMed] [Google Scholar]
- Xu L., Zhao Z., Dong A., Soubigou-Taconnat L., Renou J.P., Steinmetz A., Shen W.H. (2008). Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 28: 1348–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Shen L., Chen Y., Bao S., Thong Z., Yu H. (2014). A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev. Cell 30: 437–448. [DOI] [PubMed] [Google Scholar]
- Yuan W., Luo X., Li Z., Yang W., Wang Y., Liu R., Du J., He Y. (2016). A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 48: 1527–1534. [DOI] [PubMed] [Google Scholar]
- Yuan Y.X., Wu J., Sun R.F., Zhang X.W., Xu D.H., Bonnema G., Wang X.W. (2009). A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J. Exp. Bot. 60: 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., et al. (2017). A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 8: 14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wu Y., Schnable J.C., Zeng Z., Freeling M., Crawford G.E., Jiang J. (2012). High-resolution mapping of open chromatin in the rice genome. Genome Res. 22: 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Yu Y., Meyer D., Wu C., Shen W.H. (2005). Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 7: 1256–1260. [DOI] [PubMed] [Google Scholar]
- Zhou C.M., Zhang T.Q., Wang X., Yu S., Lian H., Tang H., Feng Z.Y., Zozomova-Lihová J., Wang J.W. (2013). Molecular basis of age-dependent vernalization in Cardamine flexuosa. Science 340: 1097–1100. [DOI] [PubMed] [Google Scholar]
- Zou Y.P., et al. (2017). Adaptation of Arabidopsis thaliana to the Yangtze River basin. Genome Biol. 18: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]