Abstract
Despite the interest aroused by sigma receptors (SRs) in the area of oncology, their role in tumor biology remains enigmatic. The predominant subcellular localization and main site of activity of SRs are the endoplasmic reticulum (ER). Current literature data, including recent findings on the sigma 2 receptor subtype (S2R) identity, suggest that SRs may play a role as ER stress gatekeepers. Although SR endogenous ligands are still unknown, a wide series of structurally unrelated compounds able to bind SRs have been identified. Currently, the identification of novel antiproliferative molecules acting via SR interaction is a challenging task for both academia and industry, as shown by the fact that novel anticancer drugs targeting SRs are in the preclinical-stage pipeline of pharmaceutical companies (i.e., Anavex Corp. and Accuronix). So far, no clinically available anticancer drugs targeting SRs are still available. The present review focuses literature advancements and provides a state-of-the-art overview of SRs, with emphasis on their involvement in cancer biology and on the role of SR modulators as anticancer agents. Findings from preclinical studies on novel anticancer drugs targeting SRs are presented in brief.
Keywords: sigma receptors, anticancer targeted therapies, chaperone activity, endoplasmic reticulum stress, cancer cell proliferation
Introduction
Over the past few decades, sigma receptors (SRs), including sigma 1 and sigma 2 receptor subtypes (S1R and S2R, respectively) have been widely associated with aging- and mitochondria-associated disorders, such as Parkinson’s and Alzheimer’s disease, multiple sclerosis and amyotrophic lateral sclerosis (Martin et al., 1976; Su, 1982; Vaupel, 1983; Quirion et al., 1987; Maurice and Lockhart, 1997; Skuza, 2003; Peviani et al., 2014; Collina et al., 2017b). Although no endogenous SR ligand has ever been found, progesterone has been identified as a potential candidate (Su, 1991; Monnet et al., 1995). This finding, together with a pressing need for new targeted therapeutic options for cancer, has led to important advances in what is known about the molecular structures and biological activities of SRs. However, the specific role played by this orphan receptor family in cell biology has yet to be clarified.
It was recently demonstrated that SRs are localized in plasmatic and subcellular membranes, in particular, the endoplasmic reticulum (ER) where they act as molecular chaperones stabilizing ER membrane proteins (Hayashi, 2015). The ER has a key role in the synthesis, folding, and structural maturation of more than a third of all the proteins produced in the cell, including almost all the secreted proteins (Anelli and Sitia, 2008). When misfolded proteins accumulate above a critical threshold as consequence of stressful conditions, a rapid and coordinated biochemical response involving adaptive signaling pathways [unfolded protein response (UPR)] is triggered (Schubert et al., 2000; Hetz et al., 2015). SR receptors can be considered as gatekeepers of ER stress, a condition that numerous studies have closely correlated with aging-associated diseases including cancer (Xu et al., 2005; Moenner et al., 2007; Brown and Naidoo, 2012; Schonthal, 2012; Yadav et al., 2014). We provide a state-of-the-art overview of S1R and S2R, focusing, in particular, on their involvement in cancer and on their potential role as ER stress gatekeepers. We also report on the compounds showing the greatest potential as biomarkers and effective drugs.
S1R
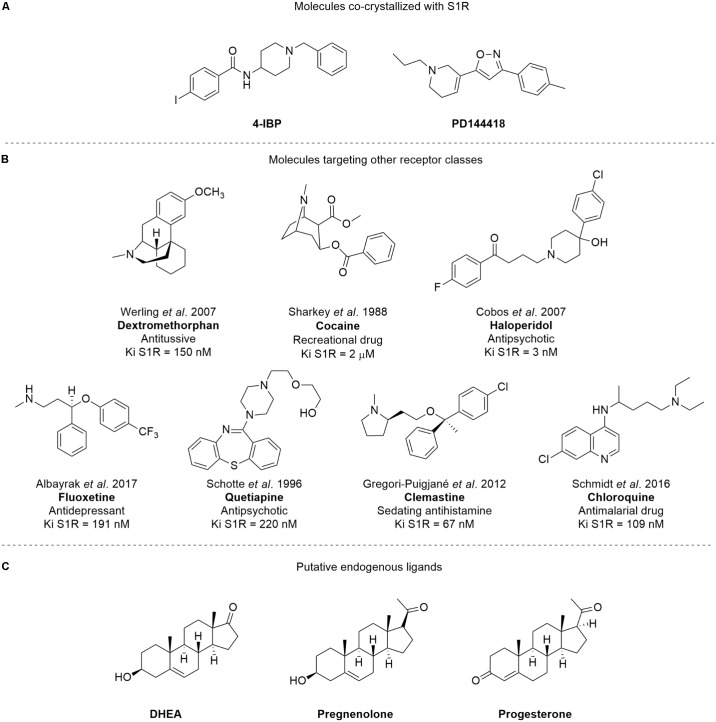
S1R is an integral membrane protein of 26 kDa that is unrelated to any traditional transmembrane receptor (Quirion et al., 1992). Despite its small size, S1R is capable of modulating living systems, regulating the activity of numerous cellular proteins and is, in turn, modulated by a plethora of small molecules. The SIGMAR1 (formerly OPRS1) gene was cloned in 1996, and its protein primary structure has long been known (Hanner et al., 1996). However, the overall three-dimensional structure and topology of its transmembrane architecture was unclear for a long time. Although, several potential structures of S1R have been postulated over the past decade (Hanner et al., 1996; Kekuda et al., 1996; Seth et al., 1998), the crystal structure of S1R [co-crystallized with 4-IBP and PD144418 (Figure 1A), PDB ID: 5HK1 and 5HK2, respectively] was only published in 2016 (Schmidt et al., 2016), revealing a trimeric architecture with a single transmembrane domain in each protomer. The carboxy-terminal domain of the receptor shows an extensive flat, hydrophobic membrane-proximal surface intimately associated with the cytosolic surface of the ER membrane. The domain includes a large, hydrophobic ligand-binding cavity at its center endowed with a remarkable plasticity in ligand recognition (Schmidt et al., 2016). This latter feature is in keeping with the most widely known function of S1R, i.e., a chaperone protein capable of interacting with several client proteins.
FIGURE 1.
Structures of (A) compounds co-crystallized with S1R; (B) SR ligands targeting other receptor classes; (C) putative SR endogenous ligands. The Ki values showed are consistent with those previously reported from literature for Dextromethorphan (Werling et al., 2007), Cocaine (Sharkey et al., 1988), Haloperidol (Cobos et al., 2007), Fluoxetine (Albayrak and Hashimoto, 2017), Quetiapine (Schotte et al., 1996), Clemastine (Gregori-Puigjané et al., 2012), and Chloroquine (Schmidt et al., 2016), respectively.
S1R has been detected at subcellular level primarily in the ER of various cell types in a range of tissues including the CNS, heart, ovaries, kidneys, testes, liver, pancreas, and placenta. High S1R expression has also been observed in embryonic stem cells and during the various stages of embryogenesis (Kekuda et al., 1996; Jbilo et al., 1997; Zamanillo et al., 2000; Ola et al., 2001; Langa et al., 2003; Aydar et al., 2004). In particular, S1R is mainly localized at the mitochondria-associated ER membrane (MAM), an interface between ER and mitochondria considered an important subcellular entity in that it acts as a sort of “tunnel” for lipid transport and Ca2+ signaling between these two organelles and contributes to processes required for cell survival (Hayashi and Su, 2007; Bernard-Marissal et al., 2015; Gregianin et al., 2016; Lewis et al., 2016; Watanabe et al., 2016). In MAM, S1R appears to play an important role as gatekeeper to keep ER stress under control. In brief, under conditions of stress, the level of Ca2+ decreases in the ER and S1R exits from a dormant state induced by its binding with the ER chaperone protein BiP (binding immunoglobulin protein), sustaining the proper conformation of the inositol triphosphate receptor type 3 (IP3), guaranteeing correct Ca2+ signaling from the ER to the mitochondria, and facilitating the synthesis of adenosine triphosphate (ATP; Hayashi and Su, 2007). Moreover, the ER is a unique milieu for the correct three-dimensional conformation of synthesized proteins, the level of which is maintained in a dynamic equilibrium between synthesis and degradation. Under conditions of stress, misfolded or aggregated proteins may accumulate within the ER, activating specific ER stress sensors, one of which is inositol-requiring enzyme 1 (IRE1). IRE1 is predominantly localized at the MAM interface where it is capable of detecting high levels of reactive oxygen species (ROS) produced by the mitochondria. Recent studies have shown that IRE1 is a client of S1R which, activated under ER and oxidative stress, chaperones IRE1, enhances its stability and guarantees the correct transmission of the ER stress signal to the nucleus, increasing the production of antistress and antioxidant proteins (Mori et al., 2013). In addition, S1R attenuates the formation of ROS by enhancing signaling of nuclear factor erythroid two-related factor 2 (Nrf2), a key regulator of antioxidant molecule expression (Wang et al., 2015).
Furthermore, S1R does not only reside in the ER but also in the plasma membrane, cytoplasmic membrane systems and nuclear envelope where it exerts a modulatory activity on different proteins. In particular, upon stimulation from agonists or stressors, S1R translocates to the plasma membrane to interact with ion channels, receptors, and kinases (Chu and Ruoho, 2016). It has also been shown to translocate to the nuclear membrane where it interacts with the nuclear envelope-resident protein emerin, recruiting a series of chromatin-remodeling factors that modulate gene transcription (Tsai et al., 2015).
Notably, S1R stands out from other chaperone proteins because of its unusual and promiscuous binding affinity to a wide series of molecules that target other receptor classes, such as dextromethorphan, cocaine, haloperidol, fluoxetine, quetiapine, clemastine, and chloroquine (Su et al., 2010; Figure 1B). It has been suggested that neurosteroids [e.g., dehydroepiandrosterone (DHEA), pregnenolone, and progesterone; Figure 1C] may be S1R putative endogenous ligands, despite their low binding affinities (0.3–10 μM; Su et al., 1988; Bergeron et al., 1996; Urani et al., 2001).
Overall, these observations point toward S1R being a new class of macromolecules halfway between a chaperone protein and a co-activator of receptors, which are activated by cell machinery to survive under conditions of stress. In support of this, there is growing evidence that S1R is only active in conditions of stress, remaining ‘silent’ in healthy organs (Maurice and Su, 2009; Tsai et al., 2009).
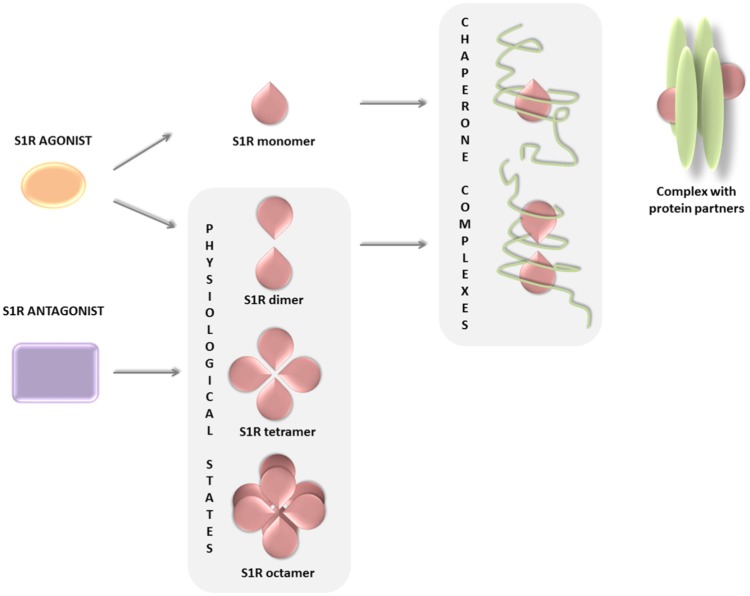
The biological response following ligand binding remains only partially understood and appears to be related to the oligomerization properties of S1R. The mechanistic models proposed (Mishra et al., 2015; Chu and Ruoho, 2016) suggest that the receptor changes its oligomerization status after binding with its ligands, some stabilizing the formation or the stabilization of S1R monomeric, dimeric, and higher oligomeric complexes. Thus, dimer and monomer forms may be functional chaperone states, whereas higher oligomeric complexes of S1R may act as a repository for the active forms. In addition, the S1R monomer is known to bind to protein partners on the plasma membrane, forming a functional unit potentially indicative of a secondary function of S1R and independent of its chaperone activity (Figure 2; Gromek et al., 2014; Mavlyutov et al., 2015; Bolshakova et al., 2016). In conclusion, the equilibrium of S1R in different states of oligomerization, i.e., monomers, dimers, or higher oligomeric forms, may explain its multiple interactions with such a wide number of heterogeneous classes of proteins.
FIGURE 2.
S1R ligands and their ability to change the oligomerization status of the receptor.
S2R
Over the years, pharmacological, chemical, and biological papers have demonstrated that S2R is a potential therapeutic target for several diseases including neurodegenerative disorders and cancer (Vilner and Bowen, 1993; Vilner et al., 1995; Bowen, 2000; Wheeler et al., 2000; Crawford and Bowen, 2002; Kashiwagi et al., 2009; Hornick et al., 2010; Guo and Zhen, 2015). Based on indirect evidence of S2R overexpression in peripheral and cerebral tumors, it has been hypothesized as a potential target for anticancer therapy (Crawford and Bowen, 2002; Crawford et al., 2002; Rui et al., 2016), and S2R radiotracers have been developed to image tumors (Tu et al., 2005, 2007, 2010; Mach and Wheeler, 2009; Mach et al., 2009). However, despite the numerous studies performed to date in this setting, the unknown molecular identity of the receptor has limited biological investigations and hindered the search for new drugs that act via the S2R pathway.
Xu et al. (2013) hypothesized that S2R is a part of the progesterone receptor membrane component 1 (PGRMC1) complex and since numerous articles have been published based on this supposition (Yang et al., 2002; Suchanek et al., 2005; Peluso et al., 2006, 2010; Cahill, 2007; Rohe et al., 2009; Intlekofer and Petersen, 2011; Szczesna-Skorupa and Kemper, 2011; Ahmed et al., 2012; Mir et al., 2012, 2013; Bali et al., 2013; Izzo et al., 2014a,b). However, the hypothesis, albeit appealing, had some serious weaknesses including the discrepancy between the molecular weight of PGRMC1 and S2R, and the low-binding affinity of PGRMC1 for haloperidol, the latter considered a characteristic signature of S2R (Hellewell and Bowen, 1990; Walker et al., 1990; Pal et al., 2007; Abate et al., 2015; Chu et al., 2015; Van Waarde et al., 2015).
In a recent paper, Alon et al. (2017) purified the putative S2R from calf liver tissue and attributed its identity to TMEM97, a relatively unknown protein belonging to the TMEM (transmembrane) gene family, resident in ER, involved in cholesterol homeostasis (Bartz et al., 2009) and in Niemann–Pick type C disease as NPC1-interacting protein (Ebrahimi-Fakhari et al., 2015). Cellular cholesterol homeostasis is a process of central importance and highly regulated. Dysregulation of the biosynthesis and uptake of cholesterol and cellular lipid accumulation has been correlated with ER stress and activation of the UPR (Colgan et al., 2011). The authors showed that the pharmacologic profile of TMEM97 is the same as that of S2R and that TMEM97 ligands bind S2R (Alon et al., 2017). The 3D structure of TMEM97/S2R, once understood, could shed fundamental light on its biological functions and their potential involvement in a broad spectrum of driver pathways of cancer and neurodegenerative diseases, thus facilitating the development of novel effective drugs.
Er Stress and Srs
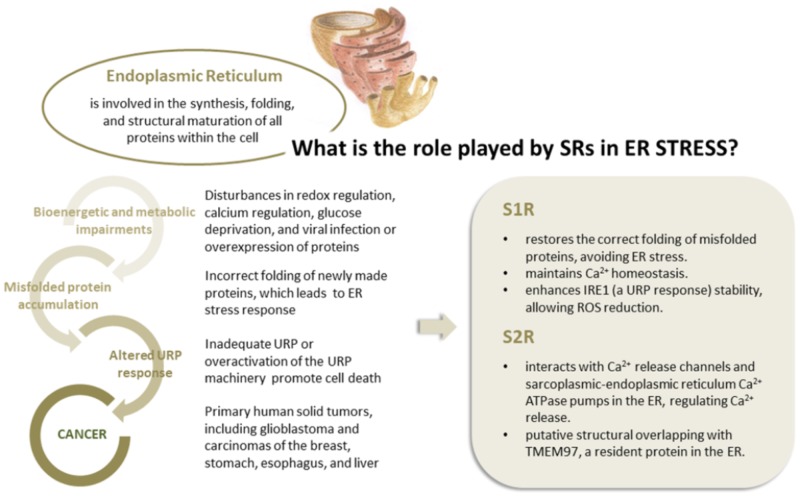
Current literature data, including recent findings on S2R, suggest that SRs are not a receptor family, a hypothesis further supported by the lack of endogenous ligands and by the SR capability to bind different proteins. Some experimental evidences suggest the pivotal role of SRs in ER stress response. First, SRs are predominantly expressed in ER and over expressed in several pathological conditions (i.e., cancer and neurodegenerative diseases). Furthermore, the S2R identity with TMEM97, a transmembrane protein of ER involved in cholesterol homeostasis has been demonstrated. Under physiological conditions, chaperones resident in the cytosol and ER lumen ensure precise folding of newly synthesized native proteins. ER stress due to accumulating misfolded proteins triggers a signaling reaction referred to as UPR (Hetz et al., 2015) that is committed to restoring ER protein homeostasis (or proteostasis) by increasing protein-folding capacity to ensure cell survival and normal functioning (Walter and Ron, 2011). However, in the event that UPR fails to restore a physiological protein equilibrium, the same ER sensors trigger an alternative response known as “terminal UPR,” leading to cell death (Shore et al., 2011; Oakes and Papa, 2015).
There is now evidence that ER stress is a driver of physiological and pathological brain aging (proteinopathies or protein misfolding disorders) and that neuronal UPR influences global proteostasis at the whole organism level (Martínez et al., 2017). Furthermore, numerous authors have demonstrated a high activation of the UPR machinery in several human solid tumors, including glioblastoma and breast, stomach, esophageal, and liver cancer (Fernandez et al., 2000; Shuda et al., 2003; Moenner et al., 2007). This is hardly surprising as cancer cells often spread to unfavorable environments characterized by hypoxia, low pH, high levels of ROS and inadequate glucose and amino acid supply, all of which may compromise ER protein folding (Ma and Hendershot, 2004; Koumenis, 2006; Lee and Hendershot, 2006; Moenner et al., 2007). Moreover, intrinsic stresses common to many tumor cells due to their genomic instability may lead to increased protein synthesis and secretory activity (Tollefsbol and Cohen, 1990; Ruggero, 2013; Dejeans et al., 2014; Horne et al., 2014).
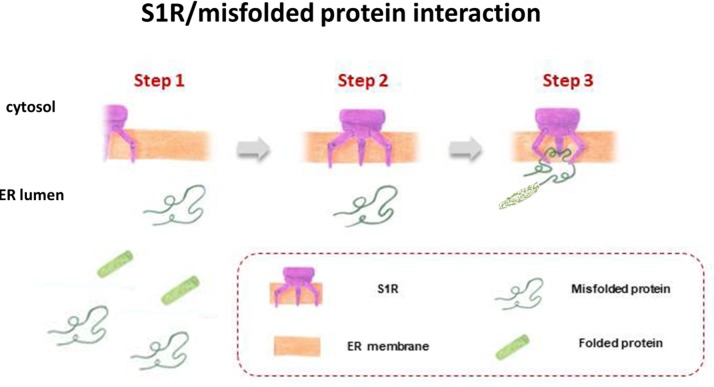
S1R, integrated into UPR machinery, may act as a chaperone protein to restore the correct folding of misfolded proteins (mainly ion channels, but also transcription factors and kinases), providing an escape route for chronically damaged cells that would otherwise die in response to ER stress (Figure 3). The same may be true for S2R/TMEM97, the biological role of which has yet to be clarified, but indirect evidence points toward its being one of the key factors in ER stress management. This may be because S2R belongs to the TMEM protein superfamily, a group of about 310 different proteins considered constituents of cell membranes such as ER, mitochondrial membranes, and lysosomal and Golgi apparatus. The function of the majority of TMEM proteins has yet to be clarified, principally because of problems in extracting and purifying transmembrane proteins (the same difficulty encountered in the unveiling of the molecular identity of S2R; Gebreselassie and Bowen, 2004; Palmer et al., 2007). However, some of these proteins are thought to be involved in conditions of ER stress, e.g., transmembranous anion channels (ANO1; Fuller, 2012), molecules responsible for oncosis (TMEM123; Ma et al., 2001), protein glycosylation (TMEM165; Foulquier et al., 2012), pathogen intoxication (TMEM181; Carette et al., 2009), and innate immunity response (TMEM173; Ishikawa and Barber, 2008). This last protein is currently arousing great interest in the area of cancer research (Harding, 2017). In addition, the S2R/TMEM97 may exert antiproliferative effects in actively proliferating tumor cells, attenuating ER stress. This assumption is probably correlated to the ability of TMEM97 in modulating cholesterol homeostasis, since a deregulation of this sophisticated mechanism leads to the activation of UPR machinery to restore the physiological conditions (Figure 4; Colgan et al., 2011).
FIGURE 3.
S1R acts as a chaperone protein to restore the correct folding of misfolded proteins.
FIGURE 4.
The correlation between ER stress and cancer conditions. The role played by S1R and S2R in modulating ER stress.
Further substantial evidence of the close relation between S2R and ER activity is the ability of S2R ligands to influence the release of Ca2+ from this organelle. Although the mechanism behind this behavior is not fully understood, S2R seems to interact, directly and indirectly, with the Ca2+ release channels-IP3-gated Ca21 channel (IP3 receptor), ryanodine-gated Ca2+ channel (ryanodine receptors), and the sarcoplasmic-ER Ca2+ ATPase (SERCA) pumps in the ER, regulating the release of Ca2+ (Vilner and Bowen, 2000; Mach et al., 2013; Figure 4).
SRs and Cancer
Several studies suggest that the deregulation of S1R may be involved in several human diseases, including cancer. In fact, S1R overexpression is associated with an invasive and metastatic phenotype in many human tumors, whereas low expression levels are found in normal cells (Bem et al., 1991; Spruce et al., 2004; Wang et al., 2004; Aydar et al., 2006; Skrzycki and Czeczot, 2013; Xu et al., 2014; Gueguinou et al., 2017; Kim and Maher, 2017). In the last decade, the correlation between S1R and cancer cells has been extensively studied, leading to hypothesize its functions in tumor biology and to investigate its therapeutic implications in cancer (see, as example, Achison et al., 2007; Palmer et al., 2007; Crottés et al., 2013; Kim and Maher, 2017). In response to environmental conditions occurring in tumor tissue, S1R may activate different adaptation mechanisms on the basis of the client protein present in a given cancer cell type (Crottés et al., 2013).
Moreover, radioligand binding assays highlighted a high density of S1R in neuronal and non-neuronal tumors (i.e., surgical specimens of renal and colorectal carcinoma and sarcoma), leading to the hypothesis of an important role of S1R in cancers (Kim and Maher, 2017).
Of note, S1R modulates the activity of several ion channels, promoting cell proliferation and survival. Several studies indeed suggested that ion channels constitute one of the main client protein families for S1R (Carnally et al., 2010; Crottès et al., 2011; Balasuriya et al., 2012; Kourrich et al., 2012). Notably, ion channels have long been considered involved in key aspects of cancer progression, including mitosis, migration, apoptosis, adhesion to the extracellular matrix (ECM) angiogenesis, homing, and drug resistance (Lang et al., 2004; Weber et al., 2006; Pillozzi et al., 2007; Gillet et al., 2009; Becchetti and Arcangeli, 2010; Pillozzi and Arcangeli, 2010; Becchetti, 2011). This may explain why cancer cells are capable of adapting to adverse metabolic conditions present in tumor tissue (Wulff and Castle, 2009; Prevarskaya et al., 2010; Arcangeli, 2011). Although the modulation of ion channel expression in cancer cells is still not fully understood, it is thought to be a consequence of the acquisition of an undifferentiated phenotype. Indeed, it is acknowledged that tumors, unlike healthy tissue, often show high levels of ion channels and transporters (Peruzzo et al., 2016).
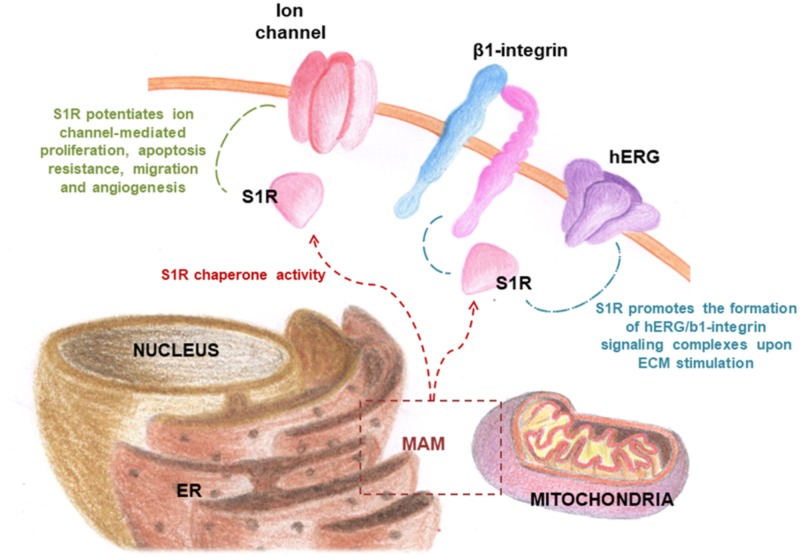
In this respect, it has been hypothesized that S1R may be involved in the remodeling of cancer cell electrical properties, potentiating ion channel function associated with proliferation, cell death resistance, invasion, and angiogenesis (Figure 4; Crottés et al., 2013). Recently, it was shown that S1R influences cancer cell behavior by modulating membrane electrical characteristics in response to the ECM properties and stimuli (Crottès et al., 2011), thus activating the PI3K/AKT pathway, cell motility, and VEGF secretion (Figure 5). In vivo, S1R expression increased tumor aggressiveness by enhancing invasion and angiogenesis, and reducing survival (Crottés et al., 2016).
FIGURE 5.
S1R chaperone modulates the activity of numerous ion channels in cancer cells.
In addition, studies analyzing the promoter region of the S1R gene highlighted the presence of a number of binding sites for several transcription factors. Some are frequently involved in cancer cell proliferation, including two nuclear factor NF-κB, activator proteins (AP-1 and AP-2), globin transcription factor 1 (GATA-1), interleukin six responsive element (IL6RE), and steroid-response elements (Prasad et al., 1998). Of note, it was shown that the use of S1R antagonists prevented nuclear translocation of androgen receptor (AR), induced proteasomal degradation of AR and its splice variant, ARV7 (frequently detected in castrate-resistant prostate cancer), and consequently suppressed their transcriptional activity (Thomas et al., 2017). Within this context, S1R probably acts as a chaperone or scaffolding protein that coordinates the maturation and transport of client proteins crucial for physiological AR function.
Like S1R, S2R is also highly expressed in proliferating tumor cells, whereas low expression is observed in normal quiescent cells (Mach et al., 1997; Wheeler et al., 2000; Colabufo et al., 2006). Data currently available on S2R mechanisms of action derive from pharmacological experiments aimed at evaluating the impact of S2R-selective ligands on tumor cell biology. Although findings indicate that S2R plays a pivotal role in regulating tumor cell proliferation, survival, and invasion, its mechanisms of action and biochemical role in intracellular signaling pathways are still unclear. Within this context, several experiments have been performed to elucidate the molecular cascades behind S2R activation, including caspase-mediated apoptosis, autophagy, and cell cycle impairment (Zeng et al., 2012). Over the years, numerous studies have been performed to unambiguously define the mechanism of action linked to S2R. Česen et al. (2013) showed that siramesine (Figure 6), the gold standard of S2R agonists (Ostenfeld et al., 2005; Groth-Pedersen et al., 2007; Rui et al., 2016), triggers cancer cell death through mitochondria destabilization. In brief, siramesine induces ROS generation, which leads to the peroxidation of cardiolipin and the release of cytochrome C from the mitochondria. Thus, the effect of siramesine on mitochondrial membranes may functionally disable the mitochondria and alter cell homeostasis, thereby initiating cell death. Another study reported on the ability of S2R agonists to regulate the sphingolipid metabolic cascade. Sphingolipids are essential molecules in the process of cell proliferation and differentiation, and impairment of their biosynthetic pathway may lead to apoptosis and cellular motility (Crawford et al., 2002). A p53- and caspase-independent apoptotic pathway differing from that activated by alkylating, antiblastic drugs has also been ascribed to S2R selective agonists. This distinctive cytotoxic effect of S2R agonists could thus be useful to treat metastatic cancer (Crawford and Bowen, 2002). Moreover, Hornick et al. (2010) found that cell death can be induced by S2R agonists through early permeabilization of the lysosomal membrane and protease translocation, which trigger downstream effectors of apoptosis. Conversely, other authors observed that some S2R ligands are capable of mobilizing intracellular calcium ions and modulating potassium channels (Vilner and Bowen, 2000; Cassano et al., 2006, 2009), leading to an incorrect ionic balance and ultimately to cell death. All of these studies showed that the multiple pathways triggered by S2R are dependent on both the selected S2R ligand and the cell line under evaluation. Summing up, both S1R and S2R seem to be involved in cancer progression.
FIGURE 6.
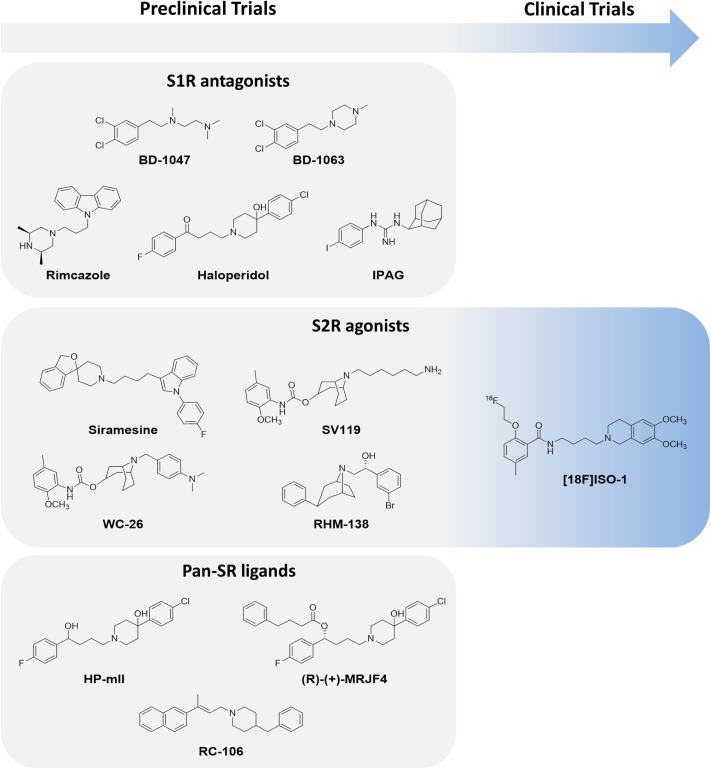
S1R and/or S2R modulators in preclinical or clinical trials.
SR Modulators and Cancer
Interest in SRs has increased substantially over the years, as documented by the high number of research papers and reviews published, patents registered (Bowen, 2000; Bourrie et al., 2004; Collina et al., 2013, 2017a; Georgiadis et al., 2017), as well as by the presence of SR modulators in the pipelines of pharmaceutical industries1,2,3 (Accuronix Therapeutics, 2018; Anavex Pipeline, 2018; Context Therapeutics, 2018).
Modulators of SRs have historically been categorized as antagonists or agonists basing on their ability to activate or inactivate the receptors and therefore on their pharmacological behavior (Martin et al., 1976; Mei and Pasternak, 2002; Mégalizzi et al., 2007; Su et al., 2010; Nguyen et al., 2015).
Regarding S1R subtype, it has been evidenced that S1R antagonists are able to prevent the activation of specific S1R-ion channel pathways; this represents a straightforward strategy against tumors (Crottés et al., 2013). Within this context, numerous anticancer molecules endowed with S1R antagonist behavior have been introduced into the pharmacological arena. BD-1047 and BD-1063 (Figure 6) are two such molecules that emerged by virtue of their ability to reduce proliferation in breast cancer cell lines (Hajipour et al., 2010). However, despite promising preclinical profiles, neither has reached the clinical experimentation phase.
Recent studies suggest that a valid alternative to the development of novel drug candidates against S1R could be the re-evaluation of well-established S1R modulators such as rimcazole, haloperidol, and IPAG (1-(4-Iodophenyl)-3-(2-adamantyl)guanidine; Schrock et al., 2013; Figure 6). These compounds can be defined as inducers of apoptosis involving NF-κB pathway in lung, Hodgkin’s lymphoma, and breast cancer cell lines (Spruce et al., 2000) or regulating ER stress, ROS production and translational repression (Kim et al., 2012; Happy et al., 2015). Moreover, they promote antiproliferative and antiangiogenic mechanisms in breast carcinoma xenograft (Spruce et al., 2001; Gilmore et al., 2004). Of note, it was recently suggested that S1R is involved in antitumor immunity mediated by the PD-1/PD-L1 checkpoint pathway, a known mechanism that allows tumor cells to escape immune surveillance and circumvent the generation of an immune response against the tumor. Maher et al. (2018) provided evidence of the ability of S1R to co-localize and physically bind to PD-L1, also demonstrating that the pharmacologic inhibition of S1R decreases PD-L1 cell surface expression and immune checkpoint activity in in vitro models. In particular, the authors reported that the exposure of cancer lines to the S1R modulator IPAG (1-(4-Jodophenyl)-3-(2-adamantyl) guanidine) induced selective autophagic degradation of PD-L1. This suggests that S1R modulators could be potential therapeutic agents in strategies aimed at inducing an immune response against cancer cells. In conclusion, a growing body of evidence points toward the potential of S1R ligands as anticancer therapeutic agents. However, the multiple roles of this protein in cancer biology need to be better clarified through further research.
There is also evidence that S2R modulators may be promising drugs against cancer, even if some functional and structural aspects of SR2 have yet to be elucidated (Wheeler et al., 2000; Zeng et al., 2012). Moreover, the putative overlapping of the pharmacological activity and ligand binding profile of S2R with TMEM97, a protein overexpressed in some tumor types, reinforces the idea that this SR subtype could be a marker for tumorigenesis (Alon et al., 2017). The definition of the crystal structure of TMEM97/S2R, as yet unknown, will be crucial in designing new chemical entities with a high affinity for this receptor. So far, effective S2R agonists have been discovered using a ligand-based approach (Ostenfeld et al., 2005; Groth-Pedersen et al., 2007; Chu et al., 2009; Tu et al., 2010; Petersen et al., 2013; Rhoades et al., 2013). Of note, some molecules belonging to different chemical classes have been extensively investigated in experimental studies. Recent research carried out on mouse breast cancer, human, or murine pancreatic cancer and human melanoma cell lines has shown that some S2R agonists, i.e., siramesine, SV119, WC-26, and RHM-138 (Figure 6), exert a cytotoxic effect at very low concentrations (Kashiwagi et al., 2007; Zeng et al., 2012). However, although S2R agonists are promising pharmaceutical/therapeutic tools, there is still a long way to go before they can be implemented into clinical practice. Only one compound, [18F]ISO-1, a potential PET marker of cell proliferation, is currently being evaluated in a phase I clinical trial (Dehdashti et al., 2013).
An innovative strategy that has also been successfully applied to the area of SRs and could potentially constitute an effective treatment for cancer is the use of dual target molecules. Given the high potential of this therapeutic approach, research has been focused on identifying compounds endowed with an S1R antagonist/S2R agonist profile, defined by Rui and colleagues as “pan-SR ligands” (Rui et al., 2016; Rossi et al., 2017). Marrazzo, the forefather of pan-SR therapy, showed that a haloperidol metabolite HP-mII (Figure 6) was effective against both SR subtypes and induced a modest antiproliferative activity in LNCaP and PC3 prostate cancer cells and in rat C6 glioma cells (Kashiwagi et al., 2007; Marrazzo et al., 2011). Recently, HP-mII was totally synthesized and functionalized with 4-phenylbutanoyl chloride, accessing the prodrug (R)-(+)-MRJF4 (Figure 6). This molecule possesses a more pronounced ability to reach the CNS and induce the death of rat C6 glioma cells than the original HP-mII molecule (Sozio et al., 2015). RC-106 (Figure 6), belonging to the pan-SR category, was recently evaluated in a panel of cancer cell lines (i.e., pancreas, breast, prostate, and glioblastoma) and showed a cytotoxic effect at the micromolar range (Rui et al., 2016). In the light of current evidence, pan-SR ligands could represent a new anticancer frontier capable of modulating different molecular cascades.
Conclusion
In this review, we reported on recent advances in research into SRs, focusing in particular on ER stress and cancer. Both S1R and S2R potentially play a key role in tumor biology as ER stress gatekeepers and are highly expressed in proliferating cancer cells. S1R expression enhances tumor cell aggressiveness by potentiating invasion and angiogenesis, whereas S2R is closely involved in regulating cell proliferation, survival, and invasion. Over the years, numerous compounds have been identified that are capable of binding both receptor types and are endowed with promising anticancer activity. However, further studies are needed to better characterize these enigmatic proteins and unravel their function in cancer biology. Although several potential structures of S1R have been postulated in the last decade, the S1R structure was only elucidated in 2016, thus permitting the design of new chemical entities with a high affinity for this receptor. Conversely, the design of new S2R modulators remains a challenge for researchers because the three-dimensional structure is still unknown. Despite the recent hypothesis that S2R is identical to TMEM97, the 3D structure of the latter has yet to be discovered. Of all the compounds investigated to date, those with an S1R antagonist/S2R agonist profile (i.e., the pan-SR ligands) endowed with excellent anticancer effects, represent a promising strategy to counteract cancer.
In conclusion, we strongly believe that the development of pan-SR drugs is destined to occupy a prominent position in the drug discovery arena and could open up new avenues for the treatment of cancer.
Author Contributions
AT and SC conceived the research topic, the design of the review, and wrote the manuscript. MR critically contributed to discuss the potential role of SR modulators in cancer therapy. All authors performed the literature review and participated in the drafting and revision of the manuscript, thus making a direct and intellectual contribution to the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
- Abate C., Niso M., Infantino V., Menga A., Berardi F. (2015). Elements in support of the “non-identity” of the PGRMC1 protein with the σ2 receptor. Eur. J. Pharmacol. 758 16–23. 10.1016/j.ejphar.2015.03.067 [DOI] [PubMed] [Google Scholar]
- Accuronix Therapeutics (2018). Our Approach. Available at: https://accuronix.com/approach/ [Google Scholar]
- Achison M., Boylan M. T., Hupp T. R., Spruce B. A. (2007). HIF-1 alpha contributes to tumour-selective killing by the sigma receptor antagonist rimcazole. Oncogene 26 1137–1146. 10.1038/sj.onc.1209890 [DOI] [PubMed] [Google Scholar]
- Ahmed I. S. A., Chamberlain C., Craven R. J. (2012). S2R Pgrmc1: the cytochrome-related sigma-2 receptor that regulates lipid and drug metabolism and hormone signaling. Expert Opin. Drug Metab. Toxicol. 8 361–370. 10.1517/17425255.2012.658367 [DOI] [PubMed] [Google Scholar]
- Albayrak Y., Hashimoto K. (2017). Sigma-1 receptor agonists and their clinical implications in neuropsychiatric disorders. Adv. Exp. Med. Biol. 964 153–161. 10.1007/978-3-319-50174-1_11 [DOI] [PubMed] [Google Scholar]
- Alon A., Schmidt H. R., Wood M. D., Sahn J. J., Martin S. F., Kruse A. C. (2017). Identification of the gene that codes for the σ2 receptor. Proc. Natl. Acad. Sci. U.S.A. 114 7160–7165. 10.1073/pnas.1705154114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anavex Pipeline (2018). Anavex Life Science Corp. Available at: http://anavex.com/pipeline/ [Google Scholar]
- Anelli T., Sitia R. (2008). Protein quality control in the early secretory pathway. EMBO J. 27 315–327. 10.1038/sj.emboj.7601974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangeli A. (2011). Ion channels and transporters in cancer. 3. Ion channels in the tumor cell-microenvironment cross talk. Am. J. Physiol. Cell. Physiol. 301 C762–C771. 10.1152/ajpcell.00113.2011 [DOI] [PubMed] [Google Scholar]
- Aydar E., Onganer P., Perrett R., Djamgoz M. B., Palmer C. P. (2006). The expression and functional characterization of sigma (sigma) 1 receptors in breast cancer cell lines. Cancer Lett. 242 245–257. 10.1016/j.canlet.2005.11.011 [DOI] [PubMed] [Google Scholar]
- Aydar E., Palmer C. P., Djamgoz M. B. A. (2004). Sigma receptors and cancer: possible involvement of ion channels. Cancer Res. 64 5029–5035. 10.1158/0008-5472.CAN-03-2329 [DOI] [PubMed] [Google Scholar]
- Balasuriya D., Stewart A. P., Crottès D., Borgese F., Soriani O., Edwardson J. M. (2012). The sigma-1 receptor binds to the Nav1.5 voltage-gated Na+ channel with 4-fold symmetry. J. Biol. Chem. 287 37021–37029. 10.1074/jbc.M112.382077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bali N., Arimoto J. M., Morgan T. E., Finch C. E. (2013). Progesterone antagonism of neurite outgrowth depends on microglial activation via Pgrmc1/S2R. Endocrinology 154 2468–2480. 10.1210/en.2012-2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz F., Kern L., Erz D., Zhu M., Gilbert D., Meinhof T., et al. (2009). Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 10 63–75. 10.1016/j.cmet.2009.05.009 [DOI] [PubMed] [Google Scholar]
- Becchetti A. (2011). Ion channels and transporters in cancer. 1. Ion channels and cell proliferation in cancer. Am. J. Phiosiol. Cell Physiol. 301 255–265. 10.1152/ajpcell.00047.2011 [DOI] [PubMed] [Google Scholar]
- Becchetti A., Arcangeli A. (2010). Integrins and ion channels in cell migration: implications for neuronal development, wound healing and metastatic spread. Adv. Exp. Med. Biol. 674 107–123. 10.1007/978-1-4419-6066-5_10 [DOI] [PubMed] [Google Scholar]
- Bem W. T., Thomas G. E., Mamone J. Y., Homan S. M., Levy B. K., Johnson F. E., et al. (1991). Overexpression of sigma receptors in nonneural human tumors. Cancer Res. 51 6558–6562. [PubMed] [Google Scholar]
- Bergeron R., de Montigny C., Debonnel G. (1996). Potentiation of neuronal NMDA response induced by dehydroepiandrosterone and its suppression by progesterone: effects mediated via sigma receptors. J. Neurosci. 16 1193–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard-Marissal N., Medard J. J., Azzedine H., Chrast R. (2015). Dysfunction in endoplasmic reticulum-mitochondria crosstalk underlies SIGMAR1 loss of function mediated motor neuron degeneration. Brain 138 875–890. 10.1093/brain/awv008 [DOI] [PubMed] [Google Scholar]
- Bolshakova A. V., Kukanova E. O., Gainullina A. N., Zhemkov V. A., Korban S. A., Bezprozvanny I. B. (2016). Sigma-1 receptor as a potential pharmacological target for the treatment of neuropathology. St. Petersburg Polytechnical Univ. J.: Phys. Math. 2 31–40. 10.1016/j.spjpm.2016.03.003 [DOI] [Google Scholar]
- Bourrie B., Bribes E., Derocq J.-M., Vidal H., Casellas P. (2004). Sigma receptor ligands: applications in inflammation and oncology. Curr. Opin. Investig. Drugs 5 1158–1163. [PubMed] [Google Scholar]
- Bowen W. D. (2000). Sigma receptors: recent advances and new clinical potentials. Pharm. Acta Helv. 74 211–218. [DOI] [PubMed] [Google Scholar]
- Brown M. K., Naidoo N. (2012). The endoplasmic reticulum stress response in aging and age-related diseases. Front. Physiol. 3:263 10.3389/fphys.2012.00263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill M. A. (2007). Progesterone receptor membrane component 1: an integrative review. J. Steroid Biochem. Mol. Biol. 105 16–36. 10.1016/j.jsbmb.2007.02.002 [DOI] [PubMed] [Google Scholar]
- Carette J. E., Guimaraes C. P., Varadarajan M., Park A. S., Wuethrich I., Godarova A., et al. (2009). Haploid genetic screens in human cells identify host factors used by pathogens. Science 326 1231–1235. 10.1126/science.1178955 [DOI] [PubMed] [Google Scholar]
- Carnally S. M., Johannessen M., Henderson R. M., Jackson M. B., Michael Edwardson J. (2010). Demonstration of a direct interaction between sigma-1 receptors and acid-sensing Ion channels. Biophys. J. 98 1182–1191. 10.1016/j.bpj.2009.12.4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassano G., Gasparre G., Contino M., Niso M., Berardi F., Perrone R., et al. (2006). The sigma-2 receptor agonist PB28 inhibits calcium release from the endoplasmic reticulum of SK-N-SH neuroblastoma cells. Cell Calcium 40 23–28. 10.1016/j.ceca.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Cassano G., Gasparre G., Niso M., Contino M., Scalera V., Colabufo N. A. (2009). F281, synthetic agonist of the sigma-2 receptor, induces Ca2+ efflux from the endoplasmic reticulum and mitochondria in SK-N-SH cells. Cell Calcium 45 340–345. 10.1016/j.ceca.2008.12.005 [DOI] [PubMed] [Google Scholar]
- Česen M. H., Repnik U., Turk V., Turk B. (2013). Siramesine triggers cell death through destabilisation of mitochondria, but not lysosomes. Cell Death Dis. 4:e818. 10.1038/cddis.2013.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu U., Ruoho A. E. (2016). Biochemical pharmacology of the sigma-1 receptor. Mol. Pharmacol. 89 142–153. 10.1124/mol.115.101170 [DOI] [PubMed] [Google Scholar]
- Chu U. B., Mavlyutov T. A., Chu M. L., Yang H., Schulman A., Mesangeau C., et al. (2015). The sigma-2 receptor and progesterone receptor membrane component 1 are different binding sites derived from independent genes. EBioMedicine 2 1806–1813. 10.1016/j.ebiom.2015.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W., Xu J., Zhou D., Zhang F., Jones L. A., Wheeler K. T., et al. (2009). New N-substituted 9-azabicyclo[3.3.1]nonan-3α-yl phenylcarbamate analogs as σ2 receptor ligands: synthesis, in vitro characterization, and evaluation as PET imaging and chemosensitization agents. Bioorg. Med. Chem. 17 1222–1231. 10.1016/j.bmc.2008.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos E. J., Del Pozo E., Baeyens J. M. (2007). Irreversible blockade of sigma-1 receptors by haloperidol and its metabolites in guinea pig brain and SH-SY5Y human neuroblastoma cells. J. Neurochem. 102 812–825. 10.1111/j.1471-4159.2007.04533.x [DOI] [PubMed] [Google Scholar]
- Colabufo N. A., Berardi F., Contino M., Ferorelli S., Niso M., Perrone R., et al. (2006). Correlation between sigma 2 receptor protein expression and histopathologic grade in human bladder cancer. Cancer Lett. 237 83–88. 10.1016/j.canlet.2005.05.027 [DOI] [PubMed] [Google Scholar]
- Colgan S. M., Hashimi A. A., Austin R. C. (2011). Endoplasmic reticulum stress and lipid dysregulation. Expert Rev. Mol. Med. 13:e4. 10.1017/S1462399410001742 [DOI] [PubMed] [Google Scholar]
- Collina S., Bignardi E., Rui M., Rossi D., Gaggeri R., Zamagni A., et al. (2017a). Are sigma modulators an effective opportunity for cancer treatment? A patent overview (1996-2016). Expert Opin. Ther. Pat. 27 565–578. 10.1080/13543776.2017.1276569 [DOI] [PubMed] [Google Scholar]
- Collina S., Gaggeri R., Marra A., Bassi A., Negrinotti S., Negri F., et al. (2013). Sigma receptor modulators: a patent review. Expert Opin. Ther. Pat. 23 597–613. 10.1517/13543776.2013.769522 [DOI] [PubMed] [Google Scholar]
- Collina S., Rui M., Stotani S., Bignardi E., Rossi D., Curti D., et al. (2017b). Sigma receptor modulators a weapon against Multiple Sclerosis disease? Future Med. Chem. 9 2029–2051. 10.4155/fmc-2017-0122 [DOI] [PubMed] [Google Scholar]
- Context Therapeutics (2018). Available at: http://www.contexttherapeutics.com/our-science/ [Google Scholar]
- Crawford K. W., Bowen W. D. (2002). Sigma-2 receptor agonists activate a novel apoptotic pathway and potentiate antineoplastic drugs in breast tumor cell lines. Cancer Res. 62 313–322. [PubMed] [Google Scholar]
- Crawford K. W., Coop A., Bowen W. D. (2002). Sigma-2 receptors regulate changes in sphingolipid levels in breast tumor cells. Eur. J. Pharmacol. 443 207–209. 10.1016/S0014-2999(02)01581-9 [DOI] [PubMed] [Google Scholar]
- Crottés D., Guizouarn H., Martin P., Borgese F., Soriani O. (2013). The sigma-1 receptor: a regulator of cancer cell electrical plasticity? Front. Physiol. 4:175. 10.3389/fphys.2013.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crottès D., Martial S., Rapetti-Mauss R. L., Pisani D. F., Loriol C., Pellissier B., et al. (2011). Sig1R protein regulates hERG channel expression through a post-translational mechanism in leukemic cells. J. Biol. Chem. 286 27947–27958. 10.1074/jbc.M111.226738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crottés D., Rapetti-Mauss R., Alcaraz-Perez F., Tichet M., Gariano G., Martial S., et al. (2016). SIGMAR1 Regulates membrane electrical activity in response to extracellular matrix stimulation to drive cancer cell invasiveness. Cancer Res. 76 607–618. 10.1158/0008-5472.CAN-15-1465 [DOI] [PubMed] [Google Scholar]
- Dehdashti F., Laforest R., Gao F., Shoghi K. I., Aft R. L., Nussenbaum B., et al. (2013). Assessment of cellular proliferation in tumors by PET using 18F-ISO-1. J. Nucl. Med. 54 350–357. 10.2967/jnumed.112.111948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejeans N., Manié S., Hetz C., Bard F., Hupp T., Agostinis P., et al. (2014). Addicted to secrete - novel concepts and targets in cancer therapy. Trends Mol. Med. 20 242–250. 10.1016/j.molmed.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D., Wahlster L., Bartz F., Werenbeck-Ueding J., Praggastis M., Zhang J., et al. (2015). Reduction of TMEM97 increases NPC1 protein levels and restores cholesterol trafficking in niemann-pick type C1 disease cells. Hum. Mol. Genet. 25 3588–3599. 10.1093/hmg/ddw204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez P. M., Tabbara S. O., Jacobs L. K., Manning F. C., Tsangaris T. N., Schwartz A. M., et al. (2000). Overexpression of the glucose-regulated stress gene GRP78 in malignant but not benign human breast lesions. Breast Cancer Res. Treat. 59 15–26. [DOI] [PubMed] [Google Scholar]
- Foulquier F., Amyere M., Jaeken J., Zeevaert R., Schollen E., Race V., et al. (2012). TMEM165 deficiency causes a congenital disorder of glycosylation. Am. J. Hum. Genet. 91 15–26. 10.1016/j.ajhg.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller C. M. (2012). Time for TMEM? J. Physiol. 590 5931–5932. 10.1113/jphysiol.2012.245563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebreselassie D., Bowen W. D. (2004). Sigma-2 receptors are specifically localized to lipid rafts in rat liver membranes. Eur. J. Pharmacol. 493 19–28. 10.1016/j.ejphar.2004.04.005 [DOI] [PubMed] [Google Scholar]
- Georgiadis M.-O., Karoutzou O., Foscolos A.-S., Papanastasiou I. (2017). Sigma receptor (σR) ligands with antiproliferative and anticancer activity. Molecules 22:E1408. 10.3390/molecules22091408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet L., Roger S., Besson P., Lecaille F., Gore J., Bougnoux P., et al. (2009). Voltage-gated sodium channel activity promotes cysteine cathepsin-dependent invasiveness and colony growth of human cancer cells. J. Biol. Chem. 284 8680–8691. 10.1074/jbc.M806891200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore D. L., Liu Y., Matsumoto R. R. (2004). Review of the pharmacological and clinical profile of rimcazole. CNS Drug Rev. 10 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregianin E., Pallafacchina G., Zanin S., Crippa V., Rusmini P., Poletti A., et al. (2016). Loss-of-function mutations in the SIGMAR1 gene cause distal hereditary motor neuropathy by impairing ER-mitochondria tethering and Ca2+ signalling. Hum. Mol. Genet. 25 3741–3753. 10.1093/hmg/ddw220 [DOI] [PubMed] [Google Scholar]
- Gregori-Puigjané E., Setola V., Hert J., Crews B. A., Irwin J. J., Lounkine E., et al. (2012). Identifying mechanism-of-action targets for drugs and probes. Proc. Natl. Acad. Sci. U.S.A. 109 11178–11183. 10.1073/pnas.1204524109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromek K. A., Suchy F. P., Meddaugh H. R., Wrobel R. L., LaPointe L. M., Chu U. B., et al. (2014). The oligomeric states of the purified sigma-1 receptor are stabilized by ligands. J. Biol. Chem. 289 20333–20344. 10.1074/jbc.M113.537993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth-Pedersen L., Ostenfeld M. S., Høyer-Hansen M., Nylandsted J., Jäättelä M. (2007). Vincristine induces dramatic lysosomal changes and sensitizes cancer cells to lysosome-destabilizing siramesine. Cancer Res. 67 2217–2225. [DOI] [PubMed] [Google Scholar]
- Gueguinou M., Crottès D., Chantôme A., Rapetti-Mauss R., Potier-Cartereau M., Clarysse L., et al. (2017). The SigmaR1 chaperone drives breast and colorectal cancer cell migration by tuning SK3-dependent Ca2+ homeostasis. Oncogene 36 3640–3647. 10.1038/onc.2016.501 [DOI] [PubMed] [Google Scholar]
- Guo L., Zhen X. (2015). Sigma-2 receptor ligands: neurobiological effects. Curr. Med. Chem. 22 989–1003. [DOI] [PubMed] [Google Scholar]
- Hajipour A. R., Fontanilla D., Chu U. B., Arbabian M., Ruoho A. E. (2010). Synthesis and characterization of N, N-dialkyl and N-alkyl-N-aralkyl fenpropimorph-derived compounds as high affinity ligands for sigma receptors. Bioorg. Med. Chem. 18 4397–4404. 10.1016/J.BMC.2010.04.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanner M., Moebius F. F., Flandorfer A., Knaus H.-G., Striessnig J., Kempnert E., et al. (1996). Purification, molecular cloning, and expression of the mammalian sigma,-binding site. Pharmacology 93 8072–8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happy M., Dejoie J., Zajac C. K., Cortez B., Chakraborty K., Aderemi J., et al. (2015). Sigma 1 Receptor antagonist potentiates the anti-cancer effect of p53 by regulating ER stress, ROS production, Bax levels, and caspase-3 activation. Biochem. Biophys. Res. Commun. 456 683–688. 10.1016/j.bbrc.2014.12.029 [DOI] [PubMed] [Google Scholar]
- Harding S. M. (2017). Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548 466–470. 10.1038/nature23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T. (2015). Sigma-1 receptor: the novel intracellular target of neuropsychotherapeutic drugs. J. Pharmacol. Sci. 127 2–5. 10.1016/j.jphs.2014.07.001 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Su T. P. (2007). Sigma-1 receptor chaperones at the ER- mitochondrion interface regulate Ca2+ signaling and cell survival. Cell 131 596–610. 10.1016/j.cell.2007.08.036 [DOI] [PubMed] [Google Scholar]
- Hellewell S. B., Bowen W. D. (1990). A sigma-like binding site in rat pheochromocytoma (PC12) cells: decreased affinity for (+)-benzomorphans and lower molecular weight suggest a different sigma receptor form from that of guinea pig brain. Brain Res. 527 244–253. [DOI] [PubMed] [Google Scholar]
- Hetz C., Chevet E., Oakes S. A. (2015). Proteostasis control by the unfolded protein response. Nat. Cell Biol. 17 829–838. 10.1038/ncb3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne S. D., Chowdhury S. K., Heng H. H. Q. (2014). Stress, genomic adaptation, and the evolutionary trade-off. Front. Genet. 5:92 10.3389/fgene.2014.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornick J. R., Xu J., Vangveravong S., Tu Z., Mitchem J. B., Spitzer D., et al. (2010). The novel sigma-2 receptor ligand SW43 stabilizes pancreas cancer progression in combination with gemcitabine. Mol. Cancer 9:298. 10.1186/1476-4598-9-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer K. A., Petersen S. L. (2011). Distribution of mRNAs encoding classical progestin receptor, progesterone membrane components 1 and 2, serpine mRNA binding protein 1, and progestin and ADIPOQ receptor family members 7 and 8 in rat forebrain. Neuroscience 172 55–65. 10.1016/j.neuroscience.2010.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Barber G. N. (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455 674–678. 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo N. J., Staniszewski A., To L., Fa M., Teich A. F., Saeed F., et al. (2014a). Alzheimer’s therapeutics targeting amyloid beta 1-42 oligomers I: abeta 42 oligomer binding to specific neuronal receptors is displaced by drug candidates that improve cognitive deficits. PLoS One 9:e111898. 10.1371/journal.pone.0111898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo N. J., Xu J., Zeng C., Kirk M. J., Mozzoni K., Silky C., et al. (2014b). Alzheimer’s therapeutics targeting amyloid beta 1-42 oligomers II:sigma-2/PGRMC1 receptors mediate abeta 42 oligomer binding and synaptotoxicity. PLoS One 9:e111899. 10.1371/journal.pone.0111899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jbilo O., Vidal H., Paul R., Nys N., De Bensaid M., et al. (1997). Purification and characterization of the human SR 31747A-binding protein. J. Biol. Chem. 272 27107–27115. [DOI] [PubMed] [Google Scholar]
- Kashiwagi H., McDunn J. E., Simon P. O., Goedegebuure P. S., Vangveravong S., Chang K., et al. (2009). Sigma-2 receptor ligands potentiate conventional chemotherapies and improve survival in models of pancreatic adenocarcinoma. J. Transl. Med. 7:24. 10.1186/1479-5876-7-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi H., McDunn J. E., Simon P. O., Goedegebuure P. S., Xu J., Jones L., et al. (2007). Selective sigma-2 ligands preferentially bind to pancreatic adenocarcinomas: applications in diagnostic imaging and therapy. Mol. Cancer 6:48. 10.1186/1476-4598-6-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kekuda R., Prasad P. D., Fei Y.-J., Leibach F. H., Ganapathy V. (1996). Cloning and functional expression of the human type 1 sigma receptor (hSigmaR1). Biochem. Biophys. Res. Commun. 229 553–558. [DOI] [PubMed] [Google Scholar]
- Kim F. J., Maher C. M. (2017). Sigma1 pharmacology in the context of cancer. Handb. Exp. Pharmacol. 244 237–308. 10.1007/164_2017_38 [DOI] [PubMed] [Google Scholar]
- Kim F. J., Schrock J. M., Spino C. M., Marino J. C., Pasternak G. W. (2012). Inhibition of tumor cell growth by Sigma1 ligand mediated translational repression. Biochem. Biophys. Res. Commun. 426 177–182. 10.1016/j.bbrc.2012.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumenis C. (2006). ER stress, hypoxia tolerance and tumor progression. Curr. Mol. Med. 6 55–69. [DOI] [PubMed] [Google Scholar]
- Kourrich S., Su T. P., Fujimoto M., Bonci A. (2012). The sigma-1 receptor: roles in neuronal plasticity and disease. Trends Neurosci. 35 762–771. 10.1016/j.tins.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang F., Gulbins E., Szabo I., Lepple-Wienhues A., Huber S. M., Duranton C., et al. (2004). Cell volume and the regulation of apoptotic cell death. J. Mol. Recognit. 17 473–480. 10.1002/jmr.705 [DOI] [PubMed] [Google Scholar]
- Langa F., Codony X., Tovar V., Lavado A., Gimenez E., Cozar P., et al. (2003). Generation and phenotypic analysis of sigma receptor type I (sigma1) knockout mice. Eur. J. Neurosci. 18 2188–2196. 10.1046/j.1460-9568.2003.02950.x [DOI] [PubMed] [Google Scholar]
- Lee A. S., Hendershot L. M. (2006). ER stress and cancer. Cancer Biol. Ther. 5 721–722. [DOI] [PubMed] [Google Scholar]
- Lewis A., Tsai S. Y., Su T. P. (2016). Detection of isolated mitochondria-associated ER membranes using the sigma-1 receptor. Methods Mol. Biol. 1376 133–140. 10.1007/978-1-4939-3170-5_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Zhang C., Prasad K. V., Freeman G. J., Schlossman S. F. (2001). Molecular cloning of Porimin, a novel cell surface receptor mediating oncotic cell death. Proc. Natl. Acad. Sci. U.S.A. 98 9778–9783. 10.1073/pnas.171322898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Hendershot L. M. (2004). The role of the unfolded protein response in tumour development: friend or foe? Nat. Rev. Cancer 4 966–977. 10.1038/nrc1505 [DOI] [PubMed] [Google Scholar]
- Mach R. H., Dehdashti F., Wheeler K. T. (2009). PET radiotracers for imaging the proliferative status of solid tumors. PET Clin. 4 1–15. 10.1016/j.cpet.2009.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach R. H., Smith C. R., al-Nabulsi I., Whirrett B. R., Childers S. R., Wheeler K. T. (1997). Sigma 2 receptors as potential biomarkers of proliferation in breast cancer. Cancer Res. 57 156–161. [PubMed] [Google Scholar]
- Mach R. H., Wheeler K. T. (2009). Development of molecular probes for imaging sigma-2 receptors in vitro and in vivo. Cent. Nerv. Syst. Agents Med. Chem. 9 230–245. 10.2174/1871524910909030230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach R. H., Zeng C., Hawkins W. G. (2013). The σ2 receptor: a novel protein for the imaging and treatment of cancer. J. Med. Chem. 56 7137–7160. 10.1021/jm301545c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher C. M., Thomas J. D., Haas D. A., Longen C. G., Oyer H. M., Tong J. Y., et al. (2018). Small-molecule sigma1 modulator induces autophagic degradation of PD-L1. Mol. Cancer Res. 16 243–255. [DOI] [PubMed] [Google Scholar]
- Marrazzo A., Fiorito J., Zappalà L., Prezzavento O., Ronsisvalle S., Pasquinucci L., et al. (2011). Antiproliferative activity of phenylbutyrate ester of haloperidol metabolite II [( ± )-MRJF4] in prostate cancer cells. Eur. J. Med. Chem. 46 433–438. 10.1016/j.ejmech.2010.10.012 [DOI] [PubMed] [Google Scholar]
- Martin W. R., Eades C. G., Thompson J. A., Huppler R. E., Gilbert P. E. (1976). The effects of morphine- and nalorphine- like drugs in the nondependent and morphine-dependent chronic spinal dog. J. Pharmacol. Exp. Ther. 197 517–532. [PubMed] [Google Scholar]
- Martínez G., Duran-Aniotz C., Cabral-Miranda F., Vivar J. P., Hetz C. (2017). Endoplasmic reticulum proteostasis impairment in aging. Aging Cell 16 615–623. 10.1111/acel.12599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice T., Lockhart B. P. (1997). Neuroprotective and anti-amnesic potentials of sigma (σ) receptor ligands. Prog. Neuropsychopharmacol. Biol. Psychiatry 21 69–102. 10.1016/S0278-5846(96)00160-1 [DOI] [PubMed] [Google Scholar]
- Maurice T., Su T.-P. (2009). The pharmacology of sigma-1 receptors. Pharmacol. Ther. 124 195–206. 10.1016/j.pharmthera.2009.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov T. A., Guo L. W., Epstein M. L., Ruoho A. E. (2015). Role of the sigma-1 receptor in amyotrophic lateral sclerosis (ALS). J. Pharmacol. Sci. 127 10–16. 10.1016/j.jphs.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mégalizzi V., Mathieu V., Mijatovic T., Gailly P., Debeir O., De Neve N., et al. (2007). 4-IBP, a σ1 receptor agonist, decreases the migration of human cancer cells, including glioblastoma cells, in vitro and sensitizes them in vitro and in vivo to cytotoxic insults of proapoptotic and proautophagic drugs. Neoplasia 9 358–369. 10.1593/neo.07130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J., Pasternak G. W. (2002). σ1 receptor modulation of opioid analgesia in the mouse. J. Pharmacol. Exp. 300 1070–1074. 10.1124/jpet.300.3.1070 [DOI] [PubMed] [Google Scholar]
- Mir S. U., Ahmed I. S., Arnold S., Craven R. J. (2012). Elevated progesterone receptor membrane component 1/sigma-2 receptor levels in lung tumors and plasma from lung cancer patients. Int. J. Cancer 131 E1–E9. 10.1002/ijc.26432 [DOI] [PubMed] [Google Scholar]
- Mir S. U., Schwarze S. R., Jin L., Zhang J., Friend W., Miriyala S., et al. (2013). Progesterone receptor membrane component 1/ Sigma-2 receptor associates with MAP1LC3B and promotes autophagy. Autophagy 9 1566–1578. 10.4161/auto.25889 [DOI] [PubMed] [Google Scholar]
- Mishra A. K., Mavlyutov T., Singh D. R., Biener G., Yang J., Oliver J. A., et al. (2015). The sigma-1 receptors are present in monomeric and oligomeric forms in living cells in the presence and absence of ligands HHS public access. Biochem. J. 1 263–271. 10.1042/BJ20141321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moenner M., Pluquet O., Bouchecareilh M., Chevet E. (2007). Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 67 10631–10634. 10.1158/0008-5472.CAN-07-1705 [DOI] [PubMed] [Google Scholar]
- Monnet F. P., Mahé V., Robel P., Baulieu E. (1995). Neurosteroids, via cr receptors, modulate the [3lHlnorepinephrine release evoked by N-methyl-D-aspartate in the rat hippocampus. Proc. Natl. Acad. Sci. U.S.A. 92 3774–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T., Hayashi T., Hayashi E., Su T. P. (2013). Sigma-1 receptor chaperone at the ER-mitochondrion interface mediates the mitochondrion-ER-nucleus signaling for cellular survival. PLoS One 8:e76941. 10.1371/journal.pone.0076941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L., Lucke-Wold B. P., Mookerjee S. A., Cavendish J. Z., Robson M. J., Scandinaro A. L., et al. (2015). Role of sigma-1 receptors in neurodegenerative diseases. J. Pharmacol. Sci. 127 17–29. 10.1016/j.jphs.2014.12.005 [DOI] [PubMed] [Google Scholar]
- Oakes S. A., Papa F. R. (2015). The role of endoplasmic reticulum stress in human pathology. Annu. Rev. Pathol. Mech. Dis. 10 173–194. 10.1146/annurev-pathol-012513-104649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ola M. S., Moore P., El-Sherbeny A., Roon P., Agarwal N., Sarthy V. P., et al. (2001). Expression pattern of sigma receptor 1 mRNA and protein in mammalian retina. Brain Res. Mol. Brain Res. 95 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostenfeld M. S., Fehrenbacher N., Høyer-Hansen M., Thomsen C., Farkas T., Jäättelä M. (2005). Effective tumor cell death by sigma-2 receptor ligand siramesine involves lysosomal leakage and oxidative stress. Cancer Res. 65 8975–8983. 10.1158/0008-5472.CAN-05-0269 [DOI] [PubMed] [Google Scholar]
- Pal A., Hajipour A. R., Fontanilla D., Ramachandran S., Chu U. B., Mavlyutov T., et al. (2007). Identification of Regions of the sigma-1 receptor ligand binding site using a novel photoprobe. Mol. Pharmacol. 72 921–933. 10.1124/mol.107.038307.other [DOI] [PubMed] [Google Scholar]
- Palmer C. P., Mahen R., Schnell E., Djamgoz M. B. A., Aydar E. (2007). Sigma-1 receptors bind cholesterol and remodel lipid rafts in breast cancer cell lines. Cancer Res. 67 11166–11175. 10.1158/0008-5472.CAN-07-1771 [DOI] [PubMed] [Google Scholar]
- Peluso J. J., Liu X., Gawkowska A., Lodde V., Wu C. A. (2010). Progesterone inhibits apoptosis in part by PGRMC1-regulated gene expression. Mol. Cell. Endocrinol. 320 153–161. 10.1016/j.mce.2010.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso J. J., Pappalardo A., Losel R., Wehling M. (2006). Progesterone membrane receptor component 1 expression in the immature rat ovary and its role in mediating progesterone’s antiapoptotic action. Endocrinology 147 3133–3140. 10.1210/en.2006-0114 [DOI] [PubMed] [Google Scholar]
- Peruzzo R., Biasutto L., Szabò I., Leanza L. (2016). Impact of intracellular ion channels on cancer development and progression. Eur. Biophys. J. 45 685–707. 10.1007/s00249-016-1143-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen N. H. T., Olsen O. D., Groth-Pedersen L., Ellegaard A.-M., Bilgin M., Redmer S., et al. (2013). Transformation-associated changes in sphingolipid metabolism sensitize cells to lysosomal cell death induced by inhibitors of acid sphingomyelinase. Cancer Cell 24 379–393. 10.1016/j.ccr.2013.08.003 [DOI] [PubMed] [Google Scholar]
- Peviani M., Salvaneschi E., Bontempi L., Petese A., Manzo A., Rossi D., et al. (2014). Neuroprotective effects of the Sigma-1 receptor (S1R) agonist PRE-084, in a mouse model of motor neuron disease not linked to SOD1 mutation. Neurobiol. Dis. 62 218–232. 10.1016/j.nbd.2013.10.010 [DOI] [PubMed] [Google Scholar]
- Pillozzi S., Arcangeli A. (2010). Physical and functional interaction between integrins and hERG1 channels in cancer cells. Adv. Exp. Med. Biol. 674 55–67. [DOI] [PubMed] [Google Scholar]
- Pillozzi S., Brizzi M. F., Bernabei P. A., Bartolozzi B., Caporale R., Basile V., et al. (2007). VEGFR-1 (FLT-1), beta1 integrin, and hERG K+ channel for a macromolecular signaling complex in acute myeloid leukemia: Role in cell migration and clinical outcome. Blood 110 1238–1250. 10.1182/blood-2006-02-003772 [DOI] [PubMed] [Google Scholar]
- Prasad P. D., Li H. W., Fei Y. J., Ganapathy M. E., Fujita T., Plumley L. H., et al. (1998). Exon-intron structure, analysis of promoter region, and chromosomal localization of the human type 1 sigma receptor gene. J. Neurochem. 70 443–451. [DOI] [PubMed] [Google Scholar]
- Prevarskaya N., Skryma R., Shuba Y. (2010). Ion channels and the hallmarks of cancer. Trends Mol. Med. 16 107–121. 10.1016/j.molmed.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Quirion R., Bowen W. D., Itzhak Y., Junien J. L., Musacchio J. M., Rothman R. B., et al. (1992). A proposal for the classification of sigma binding sites. Trends Pharmacol. Sci. 13 85–86. [DOI] [PubMed] [Google Scholar]
- Quirion R., Chicheportiche R., Contreras P. C., Johnson K. M., Lodge D., William Tam S., et al. (1987). Classification and nomenclature of phencyclidine and sigma receptor sites. Trends Neurosci. 10 444–446. 10.1016/0166-2236(87)90094-4 2469203 [DOI] [Google Scholar]
- Rhoades D., Kinder D., Mahfouz T. (2013). A comprehensive ligand based mapping of the σ2 receptor binding pocket. Med. Chem. 10 98–121. 10.2174/1573406409999131119103621 [DOI] [PubMed] [Google Scholar]
- Rohe H. J., Ahmed I. S., Twist K. E., Craven R. J. (2009). PGRMC1 (progesterone receptor membrane component 1): a targetable protein with multiple functions in steroid signaling, P450 activation and drug binding. Pharmacol. Ther. 121 14–19. 10.1016/j.pharmthera.2008.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D., Rui M., Di Giacomo M., Schepmann D., Wünsch B., Monteleone S., et al. (2017). Gaining in pan-affinity towards sigma 1 and sigma 2 receptors. SAR studies on arylalkylamines. Bioorg. Med. Chem. 25 11–19. 10.1016/j.bmc.2016.10.005 [DOI] [PubMed] [Google Scholar]
- Ruggero D. (2013). Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 5 1–27. 10.1101/cshperspect.a012336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui M., Rossi D., Marra A., Paolillo M., Schinelli S., Curti D., et al. (2016). Synthesis and biological evaluation of new aryl-alkyl(alkenyl)-4-benzylpiperidines, novel sigma receptor (SR) modulators, as potential anticancer-agents. Eur. J. Med. Chem. 124 649–665. 10.1016/j.ejmech.2016.08.067 [DOI] [PubMed] [Google Scholar]
- Schmidt H. R., Zheng S., Gurpinar E., Koehl A., Manglik A., Kruse A. C. (2016). Crystal structure of the human σ1 receptor. Nature 532 527–530. 10.1038/nature17391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonthal A. H. (2012). Targeting endoplasmic reticulum stress for cancer therapy. Front. Biosci. 4 412–431. [DOI] [PubMed] [Google Scholar]
- Schotte A., Janssen P. F., Gommeren W., Luyten W. H., Van Gompel P., Lesage A. S., et al. (1996). Risperidone compared with new and reference antipsychotic drugs: in vitro and in vivo receptor binding. Psychopharmacology 124 57–73. [DOI] [PubMed] [Google Scholar]
- Schrock J. M., Spino C. M., Longen C. G., Stabler S. M., Marino J. C., Pasternak G. W., et al. (2013). Sequential cytoprotective responses to Sigma1 ligand-induced endoplasmic reticulum stress. Mol. Pharmacol. 84 751–762. 10.1124/mol.113.087809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U., Antón L. C., Gibbs J., Norbury C. C., Yewdell J. W., Bennink J. R. (2000). Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404 770–774. 10.1038/35008096 [DOI] [PubMed] [Google Scholar]
- Seth P., Fei Y.-J., Li H. W., Huang W., Leibach F. H., Ganapathy V. (1998). Cloning and functional characterization of a σ receptor from rat brain. J. Neurochem. 70 922–931. 10.1046/j.1471-4159.1998.70030922.x [DOI] [PubMed] [Google Scholar]
- Sharkey J., Glen K. A., Wolfe S., Kuhar M. J. (1988). Cocaine binding at sigma receptors. Eur. J. Pharmacol. 149 171–174. [DOI] [PubMed] [Google Scholar]
- Shore G. C., Papa F. R., Oakes S. A. (2011). Signaling cell death from the endoplasmic reticulum stress response. Curr. Opin. Cell Biol. 23 143–149. 10.1016/j.ceb.2010.11.003.Signaling [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M., Kondoh N., Imazeki N., Tanaka K., Okada T., Mori K., et al. (2003). Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. J. Hepatol. 38 605–614. 10.1016/S0 [DOI] [PubMed] [Google Scholar]
- Skrzycki M., Czeczot H. (2013). Altered expression level of Sigma1 receptor gene in human colorectal cancer. J. Recept. Signal Transduct. Res. 33 313–318. 10.3109/10799893.2013.822891 [DOI] [PubMed] [Google Scholar]
- Skuza G. (2003). Potential antidepressant activity of sigma ligands. Pol. J. Pharmacol. 55 923–934. [PubMed] [Google Scholar]
- Sozio P., Fiorito J., Di Giacomo V., Di Stefano A., Marinelli L., Cacciatore I., et al. (2015). Haloperidol metabolite II prodrug: asymmetric synthesis and biological evaluation on rat C6 glioma cells. Eur. J. Med. Chem. 90 1–9. 10.1016/j.ejmech [DOI] [PubMed] [Google Scholar]
- Spruce B. A., Campbell L. A., McTavish N., Cooper M. A., Appleyard M. V. L., O’Neill M., et al. (2004). Small molecule antagonists of the σ-1 receptor cause selective release of the death program in tumor and self-reliant cells and inhibit tumor growth in vitro and in vivo. Cancer Res. 64 4875–4886. 10.1158/0008-5472.CAN-03-3180 [DOI] [PubMed] [Google Scholar]
- Spruce B. A., Eccles S., Dexter M. (2001). Sigma Receptor ligands and their medical uses. International Patent No WO 01/74359. [Google Scholar]
- Spruce B. A., Perkins N. D., Samson J., McTavish N. (2000). Materials and methods relating to the induction of apoptosis in target cells. International Patent No WO 00/00599. [Google Scholar]
- Su T. P. (1982). Evidence for sigma opioid receptor: binding of [3H]SKF-10047 to etorphine-inaccessible sites in guinea-pig brain. J. Pharmacol. Exp. Ther. 223 284–290. [PubMed] [Google Scholar]
- Su T.-P. (1991). sigma receptors. Putative links between nervous, endocrine and immune systems. Eur. J. Biochem. 200 633–642. 10.1111/j.1432-1033.1991.tb16226.x [DOI] [PubMed] [Google Scholar]
- Su T. P., Hayashi T., Maurice T., Buch S., Ruoho A. E. (2010). The sigma-1 receptor chaperone as an inter-organelle signaling modulator. Trends Pharmacol. Sci. 31 557–566. 10.1016/j.tips.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T. P., London E. D., Jaffe J. H. (1988). Steroid binding at sigma receptors suggests a link between endocrine, nervous, and immune systems. Science 240 219–221. [DOI] [PubMed] [Google Scholar]
- Suchanek M., Radzikowska A., Thiele C. (2005). Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat. Methods 2 261–267. 10.1038/nmeth752 [DOI] [PubMed] [Google Scholar]
- Szczesna-Skorupa E., Kemper B. (2011). Progesterone receptor membrane component 1 inhibits the activity of drug-metabolizing cytochromes P450 and binds to cytochrome P450 reductase. Mol. Pharmacol. 79 340–350. 10.1124/mol.110.068478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. D., Longen C. G., Oyer H. M., Chen N., Maher C. M., Salvino J. M., et al. (2017). Sigma1 targeting to suppress aberrant androgen receptor signaling in prostate cancer. Cancer Res 77 2439–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefsbol T. O., Cohen H. J. (1990). The protein synthetic surge in response to mitogen triggers high glycolytic enzyme levels in human lymphocytes and occurs prior to DNA synthesis. Biochem. Med. Metab. Biol. 44 282–291. 10.1016/0885-4505(90)90073-A [DOI] [PubMed] [Google Scholar]
- Tsai S.-Y., Hayashi T., Mori T., Su T.-P. (2009). Sigma-1 receptor chaperones and diseases. Cent. Nerv. Syst. Agents Med. Chem. 9 184–189. 10.1016/j.biotechadv.2011.08.021.Secreted [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S.-Y. A., Chuang J.-Y., Tsai M.-S., Wang X.-F., Xi Z.-X., Hung J.-J., et al. (2015). Sigma-1 receptor mediates cocaine-induced transcriptional regulation by recruiting chromatin-remodeling factors at the nuclear envelope. Proc. Natl. Acad. Sci. U.S.A. 112 E6562–E6570. 10.1073/pnas.1518894112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z., Dence C. S., Ponde D. E., Jones L., Wheeler K. T., Welch M. J., et al. (2005). Carbon-11 labeled sigma2 receptor ligands for imaging breast cancer. Nucl. Med. Biol. 32 423–430. 10.1016/j.nucmedbio.2005.03.008 [DOI] [PubMed] [Google Scholar]
- Tu Z., Xu J., Jones L. A., Li S., Dumstorff C., Vangveravong S., et al. (2007). Fluorine-18-labeled benzamide analogues for imaging the σ 2 receptor status of solid tumors with positron emission tomography. J. Med. Chem. 50 3194–3204. 10.1021/jm0614883 [DOI] [PubMed] [Google Scholar]
- Tu Z., Xu J., Jones L. A., Li S., Zeng D., Kung M.-P., et al. (2010). Radiosynthesis and biological evaluation of a promising σ 2 - receptor ligand radiolabeled with fluorine-18 or iodine-125 as a PET/SPECT probe for imaging breast cancer. Appl. Radiat. Isot. 68 2268–2273. 10.1016/j.apradiso.2010.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urani A., Cois Roman F. J., Phan A., Maurice T. (2001). The antidepressant-like effect induced by sigma(1)-receptor agonists and neuroactive steroids in mice submitted to the forced swimming test. J. Pharmacol. Exp. Ther. 298 1269–1279. [PubMed] [Google Scholar]
- Van Waarde A., Rybczynska A. A., Ramakrishnan N. K., Ishiwata K., Elsinga P. H., Dierckx R. A. (2015). Potential applications for sigma receptor ligands in cancer diagnosis and therapy. Biochim. Biophys. Acta 1848 2703–2714. 10.1016/j.bbamem.2014.08.022 [DOI] [PubMed] [Google Scholar]
- Vaupel D. B. (1983). Naltrexone fails to antagonize the sigma effects of PCP and SKF 10,047 in the dog. Eur. J. Pharmacol. 92 269–274. [DOI] [PubMed] [Google Scholar]
- Vilner B. J., Bowen W. D. (1993). Sigma receptor-active neuroleptics are cytotoxic to C6 glioma cells in culture. Eur. J. Pharmacol. 244 199–201. [DOI] [PubMed] [Google Scholar]
- Vilner B. J., Bowen W. D. (2000). Modulation of cellular calcium by sigma-2 receptors: release from intracellular stores in human SK-N-SH neuroblastoma cells. J. Pharmacol. Exp. Ther. 292 900–911. [PubMed] [Google Scholar]
- Vilner B. J., John C. S., Bowen W. D., Thomsen C., Farkas T., Jäättelä M. (1995). Sigma-1 and sigma-2 receptors are expressed in a wide variety of human and rodent tumor cell lines. Cancer Res. 55 408–413. [PubMed] [Google Scholar]
- Walker J. M., Bowen W. D., Walker F. O., Matsumoto R. R., De Costa B., Rice K. C. (1990). Sigma receptors: biology and function. Pharmacol. Rev. 42 355–402. [PubMed] [Google Scholar]
- Walter P., Ron D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334 1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Wang B., Rouzier R., Albarracin C. T., Sahin A., Wagner P., Yang Y., et al. (2004). Expression of sigma 1 receptor in human breast cancer. Breast Cancer Res. Treat. 87 205–214. [DOI] [PubMed] [Google Scholar]
- Wang J., Shanmugam A., Markand S., Zorrilla E., Ganapathy V., Smith S. B. (2015). Sigma 1 receptor regulates the oxidative stress response in primary retinal Müller glial cells via NRF2 signaling and system x, the Na-independent glutamate-cystine exchanger. Free Radic. Biol. Med. 86 25–36. 10.1016/j.freeradbiomed.2015.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Ilieva H., Tamada H., Nomura H., Komine O., Endo F., et al. (2016). Mitochondria-associated membrane collapse is a common pathomechanism in SIGMAR1- and SOD1-linked ALS. EMBO Mol. Med. 8 1421–1437. 10.15252/emmm.201606403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C., De Queiroz F. M., Downie B. R., Suckow A., Stühmer W., Pardo L. A. (2006). Silencing the activity and proliferative properties of the human EagI potassium channel by RNA interference. J. Biol. Chem. 281 13030–13037. 10.1074/jbc.M600883200 [DOI] [PubMed] [Google Scholar]
- Werling L. L., Keller A., Frank J. G., Nuwayhid S. J. (2007). A comparison of the binding profiles of dextromethorphan, memantine, fluoxetine and amitriptyline: treatment of involuntary emotional expression disorder. Exp. Neurol. 207 248–257. 10.1016/j.expneurol.2007.06.013 [DOI] [PubMed] [Google Scholar]
- Wheeler K. T., Wang L. M., Wallen C. A., Childers S. R., Cline J. M., Keng P. C., et al. (2000). Sigma-2 receptors as a biomarker of proliferation in solid tumours. Br. J. Cancer 82 1223–1232. 10.1054/bjoc.1999.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff H., Castle N. (2009). Voltage-gated potassium channels as therapeutic targets. Nat. Rev. Drug Discov. 8 982–1001. 10.1038/nrd2983.Voltage-gated [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Bailly-Maitre B., Reed J. C. (2005). Endoplasmic reticulum stress: cell life and death decisions. J. Clin. Invest. 115 2656–2664. 10.1172/JCI26373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Yi W., Chen Y., Ma L., Wang J., Yu G. (2014). Overexpression of Sig1R is closely associated with tumor progression and poor outcome in patients with hilar cholangiocarcinoma. Med. Oncol. 31:261. 10.1007/s12032-014-0261-8 [DOI] [PubMed] [Google Scholar]
- Xu J., Zeng C., Chu W., Pan F., Rothfuss J. M., Zhang F., et al. (2013). Identification of the PGRMC1 protein complex as the putative sigma-2 receptor binding site. Nat. Commun. 2:380. 10.1038/ncomms1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R. K., Chae S.-W., Kim H.-R., Chae H. J. (2014). Endoplasmic reticulum stress and cancer. J. Cancer Prev. 19 75–88. 10.15430/JCP.2014.19.2.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., et al. (2002). Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110 489–500. 10.1016/S0092-8674(02)00872-3 [DOI] [PubMed] [Google Scholar]
- Zamanillo D., Andreu F., Ovalle S., Perez M. P., Romero G., Farre A. J., et al. (2000). Up-regulation of sigma 1 receptor mRNA in rat brain by a putative atypical antipsychotic and sigma receptor ligand. Neurosci. Lett. 282 169–172. [DOI] [PubMed] [Google Scholar]
- Zeng C., Rothfuss J., Zhang J., Chu W., Vangveravong S., Tu Z., et al. (2012). Sigma-2 ligands induce tumour cell death by multiple signalling pathways. Br. J. Cancer 106 693–701. 10.1038/bjc.2011.602 [DOI] [PMC free article] [PubMed] [Google Scholar]