SUMMARY
Cells migrate by applying rearward forces against extracellular media. It is unclear how this is achieved in amoeboid migration, which lacks adhesions typical of lamellipodia-driven mesenchymal migration. To address this question, we developed optogenetically controlled models of lamellipodia-driven and amoeboid migration. On a 2D surface, migration speeds in both modes were similar. However, when suspended in liquid, only amoeboid cells exhibited rapid migration accompanied by rearward membrane flow. These cells exhibited increased endocytosis at the back and membrane trafficking from back to front. Genetic or pharmacological perturbation of this polarized trafficking inhibited migration. The ratio of cell migration and membrane flow speeds matched the predicted value from a model where viscous forces tangential to the cell-liquid interface propel the cell forward. Since this mechanism does not require specific molecular interactions with the surrounding medium, it can facilitate amoeboid migration observed in diverse microenvironments during immune function and cancer metastasis.
eTOC paragraph
O’Neill et all use optogenetic control of two distinct migration modes to address the question of how propelling forces are generated during adhesion-independent cell migration. They show that rearward plasma membrane flow generates tangential viscous forces at the cell-liquid interface to drive the cell forward.
INTRODUCTION
Cell migration is broadly classified as either mesenchymal or amoeboid. The term “amoeboid” has been used to describe a number of distinct migration modes, including modes with or without pseudopods (Lammermann and Sixt, 2009). Here we use “amoeboid” to refer to cell migration that is characterized by fast speeds, high levels of actomyosin contractility, rounded cell shapes, and little or no adhesion (Paluch, et al., 2016). Amoeboid migration plays important roles in developmental biology and immune system function (Lammermann and Germain, 2014; Richardson and Lehmann, 2010). Additionally, tumor cells can transition between mesenchymal and amoeboid migration modes and thereby increase cancer invasiveness (Wolf, et al., 2003). Integrin-mediated adhesion had long been thought to be essential for cell migration, but recent findings have challenged the need for specific adhesions (Paluch, et al., 2016; Renkawitz, et al., 2009; Lammermann, et al., 2008), and more generally the requirement for direct interaction with a 2D surface or 3D matrix (Barry and Bretscher, 2010).
Forty years ago, the physicist EM Purcell outlined several physically feasible ways for single cells to propel themselves through aqueous solutions (Purcell, 1977). Plausible “swimming” modes for cells differ fundamentally from those found in macroscopic examples that shape our intuition, because viscous forces completely dominate under the low Reynolds number conditions that characterize hydrodynamics at the cellular level (Purcell, 1977). The Reynolds number is a dimensionless number that quantifies the relative contributions of inertial and viscous forces that determine fluid dynamics, and it varies by a factor of roughly 100 million between a human swimmer and a single cell (Purcell, 1977). An intriguing quantitative prediction based on low Reynolds number calculations is that if a cell’s surface flows in one direction at a constant speed, viscous forces generated tangential to the cell surface can propel it through liquid in the opposite direction at nearly the same speed (Leshansky and Kenneth, 2008; Leshansky, et al., 2007). This “surface treadmilling” mechanism has been proposed to contribute to cell migration (Bretscher, 2014; Barry and Bretscher, 2010), but it has not been possible to test it directly due to the lack of a model system where plasma membrane flow can be controlled at will.
Optical methods, including optogenetics and optical control of chemical gradients, provide rapid, directionally reversible control over dynamic cell behaviors (Collins, et al., 2015; Karunarathne, et al., 2015). Subcellular optogenetics in particular can help deconstruct complex cell behaviors such as migration, by targeted activation or inhibition of proteins that control different aspects of the migratory response. For example, migrating cells exhibit polarized intracellular signaling with distinct biochemical events localized to the front or back (Surve, et al., 2016; Krause and Gautreau, 2014). The Rho family GTPases Rac and Cdc42 control leading edge extension, whereas RhoA activity controls actomyosin contractility at the back (O’Neill, et al., 2016; Yang, et al., 2016; Raftopoulou and Hall, 2004). Since actomyosin contractility is thought to generate forces important for migration (Paluch, et al., 2016), we reasoned that optogenetic control of RhoA could be used to generate a model system to study how those forces are leveraged to drive the cell forward.
Surprisingly, we discovered that optogenetic activation of endogenous RhoA at one side of a macrophage generates fast and persistent cell migration that closely mimics amoeboid migration, with the photoactivated side forming the cell rear. Cells in which a GPCR was optically activated demonstrated distinctly different lamellipodial movement. In combination with live cell imaging, genetic and pharmacological perturbations, we found that RhoA drives a cycle of rearward plasma membrane flow coupled to polarized vesicle trafficking. To examine whether this rearward flow of the plasma membrane can support migration in the absence of any adhesion, we suspended the cells in a liquid. Under these conditions adhesion-dependent mechanisms that have been proposed to explain cell migration cannot operate. We found that optically activated cells suspended in liquid migrated faster than cells adhered to a surface. In contrast, the lamellipodial mode of migration lacked retrograde membrane flow and failed to drive cell migration of cells suspended in liquid. The results support the prediction that tangential viscous forces are sufficient to propel a cell forward in the absence of any adhesive forces. Amoeboid cells tend to have high levels of RhoA activity and transitions from mesenchymal to amoeboid migration usually involve increased RhoA activity (Sanz-Moreno, et al., 2008; Gadea, et al., 2007; Sahai and Marshall, 2003). This mechanism is thus likely to play a general role in the amoeboid cell migration that is seen during tumor invasiveness or immune cell function.
RESULTS
Optically driving cell migration from the rear
We performed subcellular optogenetics and live cell imaging using RAW 264.7 macrophages. These cells are capable of chemotaxis (Green, et al., 2009), and their low basal motility facilitates the use of optogenetics to test the effects of activating or inhibiting select proteins on polarity formation, migration initiation, and directional control of migration (O’Neill, et al., 2016; O’Neill and Gautam, 2014; Karunarathne, et al., 2013). Importantly, these cells are capable of the transition between mesenchymal and amoeboid modes of migration (Van Goethem, et al., 2010). To achieve subcellular control over the activation of endogenous RhoA, we optically triggered membrane recruitment of a Rho-selective guanine nucleotide exchange factor (GEF) using an engineered light-inducible dimerization system called iLID (Fig. 1A) (Guntas, et al., 2015). In the iLID system, the interaction between a protein SspB and its peptide binding target SsrA is rendered light-dependent by fusing SsrA to the blue light-sensitive LOV2 domain (Guntas, et al., 2015). Coexpression of a membrane targeted LOV-SsrA fusion (iLID-CaaX) and different GEF domains fused to SspB has been used previously to optically recruit GEFs for monomeric G proteins Cdc42 and Rac to the plasma membrane to trigger G protein activation (O’Neill, et al., 2016; Guntas, et al., 2015). Here we used light-inducible membrane recruitment of the DHPH domain of LARG, a Rho selective GEF, to achieve activation of RhoA. LARG activates RhoA significantly more than RhoB or RhoC and not other Rho family proteins such as Cdc42 or Rac (Jaiswal, et al., 2013). Of the three Rho isoforms, RNAseq data from our lab indicates that RhoA is the most highly expressed in RAW 264.7 cells (Table S1). Similar optogenetic approaches have recently been used to examine the role for RhoA signaling in cytokinesis and tissue deformation (Wagner and Glotzer, 2016) (Valon, et al., 2017).
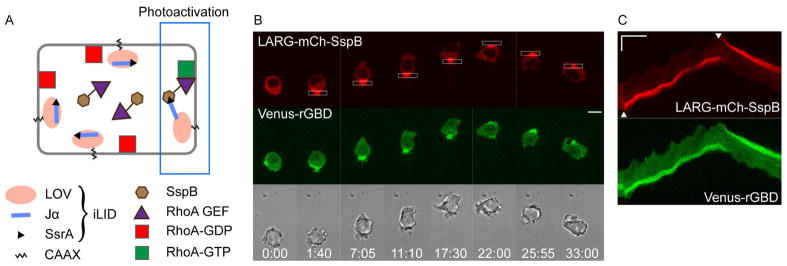
Fig. 1. Localized optical RhoA activation drives cell migration from the rear.
(A) Scheme for optical activation of RhoA using the iLID system.
(B) Optical control of RhoA activation and cell migration in a cell expressing iLID-CaaX, LARG-mCh-SspB (red), and Venus-rGBD (green), a RhoA-GTP sensor. Scale bar is 10 μm.
(C) Kymographs of the same cell showing the accumulation of LARG (red) and RhoA-GTP (green) in the rear of migrating cell. Scale bar is 10 μm (vertical) and 5 min (horizontal). For this and all subsequent kymographs, white arrowheads indicate the time of initial photoactivation and reversal. Image sequences in all Figures use white boxes to show the localization of photoactivation with 445 nm light, and show time in min:sec.
To determine the response to subcellular RhoA activation, we plated cells on a 2D glass surface following transfection with LARG-mCh-SspB, iLID-CaaX, and a RhoA sensor called Venus-rGBD (Fig. 1B). Photoactivation of one side of the cell with blue light resulted in recruitment of LARG-mCh-SspB from the cytosol to the plasma membrane within the photoactivated region. RhoA activation in the same region was detected by membrane translocation of Venus-rGBD, which contains the RhoA-GTP binding domain of rhotekin (Benink and Bement, 2005). Thus optically triggered membrane recruitment of LARG was sufficient to generate localized activation of RhoA.
Remarkably, optical RhoA activation at one side of the cell generated persistent migration in the opposite direction (Fig. 1B; Movie 1). The dynamic changes at cellular and molecular levels are better visualized in the movie here and in the movies accompanying all of the figures below. The corresponding kymograph shows that the migration direction rapidly reversed upon switching the side of optical activation (Fig. 1C). Identical photoactivation conditions did not produce cell migration in cells expressing comparable levels of mCherry-SspB lacking the Rho-GEF domain (Movie 1). RhoA is commonly thought to be important for retraction of the cell rear following extension of the leading edge (Worthylake, et al., 2001), or for preventing the formation of multiple leading edges (Worthylake and Burridge, 2003), so it was surprising to discover that targeted activation of RhoA can initiate, direct, sustain, and reverse cell migration.
Cell migration independent of leading edge signaling
Optical control of cell migration has previously only been achieved by controlling leading edge signaling. This includes optical control of a constitutively active Rac mutant (Wu, et al., 2009), and previous studies in our lab using optical control of endogenous signaling proteins including heterotrimeric G protein subunits, Cdc42, and Rac (O’Neill, et al., 2016; O’Neill and Gautam, 2014; Karunarathne, et al., 2013). All of these approaches lead to the activation of canonical leading edge signaling including Rac activation, actin polymerization and lamellipodia formation, and drive cell migration on 2D substrates in the direction of optical activation. Since optical RhoA activation drives cell migration from the rear, we asked if it activates these same signaling events from across the cell, or if it triggers a distinct mode of migration.
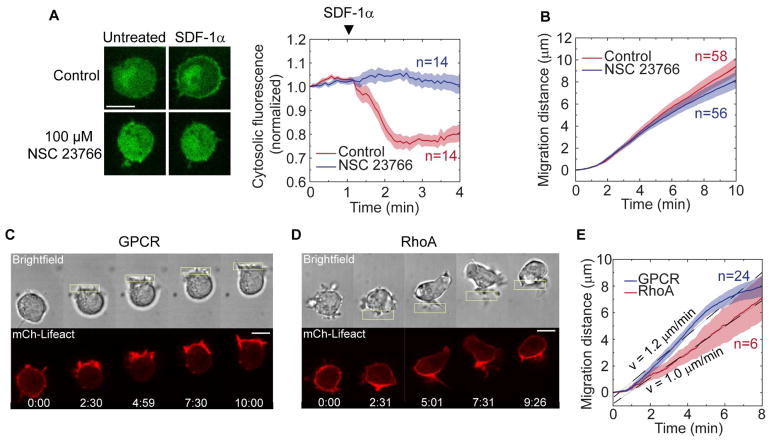
First, we tested if Rac inhibition altered RhoA-driven migration. The small molecule NSC 23766 inhibits Rac1 and Rac2 (Yao, et al., 2011), and our RNAseq data indicate that RAW cells express Rac1 and Rac2, but not Rac3 (Table S1). Treatment with 100 μM NSC clearly inhibited Rac activity measured downstream of the chemoattractant receptor CXCR4 upon stimulation with SDF-1α (Fig. 2A), but the same treatment did not inhibit RhoA-driven migration (Fig. 2B). These results showed that RhoA drives migration from the cell rear through a Rac-independent mechanism.
Fig. 2. RhoA-driven migration is Rac-independent and does not polarize actin to the leading edge or generate lamellipodia.
(A) Verification of Rac inhibition by NSC 23766. Cells were transfected with a Rac biosensor (green) and chemoattractant receptor CXCR4. Cells were treated with 100 μM NSC 23766 for 10 min or untreated (control). Cytosolic Rac biosensor fluorescence was continuously measured for 4 min and 10 μg of SDF-1α was added at t=1 min.
(B) RhoA-driven migration with and without the Rac inhibitor NSC 23766. In this and all other graphs, n is the number of cells, solid lines represent mean values and shaded areas show SEM.
(C) Actin dynamics during GPCR-driven migration in a cell expressing mCh-Lifeact (red) and parapinopsin.
(D) Actin dynamics during RhoA-driven migration in a cell expressing LARG-mTurq-Sspb, iLID-CaaX, and mCh-Lifeact (red).
(E) Comparison of migration speeds for RhoA versus GPCR-driven migration on a 2D glass surface. Cells were transfected with mCh-Lifeact, Venus-KRasCT, and their respective optogenetic construct. Localized photoactivation was initiated at t=30 s.
All scale bars are 10 μm.
We then compared RhoA-driven migration to GPCR-driven migration for cells on a 2D glass surface. We used lamprey parapinopsin as the light-activated GPCR (Koyanagi, et al., 2004). Previously we have used human blue opsin to optically drive cell migration (Karunarathne, et al., 2013). Similar to blue opsin, parapinopsin also activates heterotrimeric Gi proteins, but it has the additional advantage of being a bistable opsin. Both receptors bind 11-cis retinal, and photoactivation with blue light converts the chromophore to all-trans retinal, resulting in receptor activation. Whereas blue opsin rapidly releases all-trans retinal and requires a new 11-cis retinal molecule to bind before it can be reactivated by subsequent photostimulation, parapinopsin remains bound to all-trans retinal and can convert it back to 11-cis in response to green/yellow light (Koyanagi, et al., 2004). In cell migration experiments where the receptor must remain activated for long periods of time, this feature of parapinopsin helps overcome the deleterious effects of exhausting the supply of 11-cis retinal in solution.
As expected for polarized GPCR signaling, subcellular parapinopsin activation generated actin polymerization and pronounced lamellipodia at the front of the cell (Fig. 2C; Movie 2). In contrast, optical RhoA activation caused an increase in filamentous actin at the cell rear, and failed to generate lamellipodia at the leading edge (Fig. 2D; Movie 2). Despite the striking differences in actin polarization and cell morphology, these distinct migration modes generated very similar cell translocation speeds when adhered to a surface (Fig. 2E).
Blebbing has been reported as an alternative mechanism for leading edge extension in the absence of lamellipodia (Charras and Paluch, 2008). During RhoA-driven migration, we found that some cells generated blebs at the leading edge (Movie 1). However, blebbing was not essential for RhoA-driven migration, as other cells exhibited efficient migration without blebbing (Movie 2).
These results show that RhoA signaling at the cell rear can drive the cell forward independent of leading edge protrusion. Consistent with our findings, leukocytes that cannot generate lamellipodia because they lack the Rac effector WAVE actually migrate faster than wild-type cells (Leithner, et al., 2016). Similarly, Walker carcinoma cells increase their migration speed upon pharmacological inhibition of lamellipodia formation (Keller, et al., 2002), and rounded breast cancer cells can migrate through 3D matrices independent of lamellipodia extension or blebbing (Poincloux, et al., 2011). Altogether, these findings suggest that optical RhoA activation provides a useful approach to study how diverse cell types can migrate using a mechanism that does not involve leading edge extension.
Rearward membrane flow and uropod formation
How can a cell propel itself forward without extending the leading edge? The fundamental physical requirement is simply that the cell must exert rearward-directed forces on some extracellular medium, whether it is a 2D surface, 3D matrix, or the surrounding liquid. One way this can be achieved is if the surface of the cell flows backward and pushes against an extracellular surface or liquid.
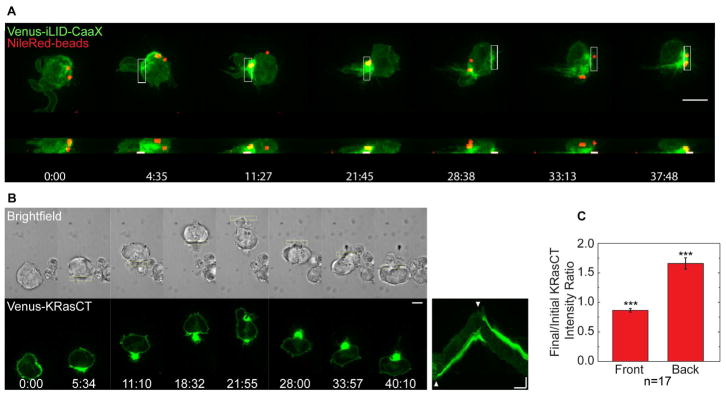
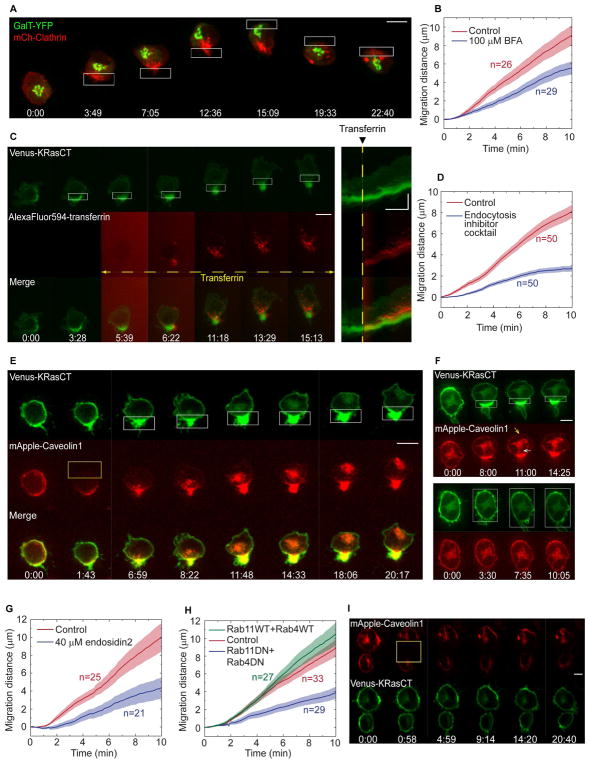
To test for rearward plasma membrane flow during RhoA-driven migration, we imaged fluorescent beads bound to the outer surface of the plasma membrane. The beads moved along the cell surface to the back of the cell, and moved again to the new cell rear when the migration direction was optically reversed (Fig. 3A, Movie 3). We also imaged the following genetically encoded constructs containing different plasma membrane targeting motifs (Fig. 3B, Fig. S1A): (1) Venus-KRasCT, which combines a covalently attached farnesyl lipid with proximal positively charged residues to localize to the intracellular surface of the plasma membrane (Fig. 3B), (2) Venus-PH-PLCδ, which binds to PI(4,5)P2 in the plasma membrane (Fig. S1B), (3) YFP-GL containing a consensus N-glycosylation (GL) site fused to a GPI-attachment signal to attach to the exterior leaflet of the plasma membrane (Fig. S1A, top panel), (4) β2AR, a GPCR, which is a transmembrane protein (Fig. S1A, middle panel), and (5) β1 integrin, also a transmembrane protein (Fig. S1A, bottom panel). All of these plasma membrane markers rapidly accumulated at the back of the cell upon optical RhoA activation. We analyzed the intensity change in the front and back half of the cell for Venus-KRasCT and found that the increased intensity at the back was accompanied by an intensity decrease at the front (Fig. 3C). Together these results suggest that the entire plasma membrane flows towards the back of the cell during RhoA-driven migration.
Fig. 3. Cells undergoing RhoA-driven migration exhibit rearward membrane flow.
(A) Movement of surface bound beads to the cell rear during RhoA-driven migration in a cell expressing LARG-mTurq-SspB and Venus-iLID-CaaX (green). Microbeads are in red. Images shown are top and side view maximum intensity projections from an 8-plane z-stack.
(B) Accumulation of Venus-KRasCT (green) at the cell rear during RhoA-driven migration in a cell coexpressing LARG-mCh-SspB and iLID-CaaX. The corresponding kymograph is shown on the right. Scale bar for image sequences is 10 μm. Kymograph scale bars are 10 μm (vertical) and 5 min (horizontal).
(C) Fractional change in KRasCT fluorescence intensity in the front or back half of the cell, for n=17 cells. Error bars are SEM. The fractional change is significantly <1 at the cell front, and >1 at the cell rear (*** one sample t-test with p<0.001).
Upon optical RhoA activation, the plasma membrane markers typically condensed into a uropod-like structure. Consistent with uropods in immune cells migrating in a chemoattractant gradient, the RhoA-generated uropods were enriched in myosin, ezrin, moesin, β1-integrin, and PI(4,5)P2 (Figs. 5A, Fig. S1A, B, Movie 4) (Martinelli, et al., 2013; Lokuta, et al., 2007). Once a pronounced uropod had formed, switching the side of optical RhoA activation most often caused the existing uropod to swing around to the other side of the cell (Movie 4). Interestingly, the migration direction reversed immediately upon switching the side of photoactivation, before the established uropod had repositioned to the new cell rear (Movie 4). Our results suggest that RhoA-mediated retrograde membrane flow helps to drive uropod formation, and that once formed the uropod passively flows along with the membrane toward the region of highest RhoA activity. It is not the location of the uropod at a given instant, but rather the location of RhoA activity and direction of membrane flow that determines the migration direction.
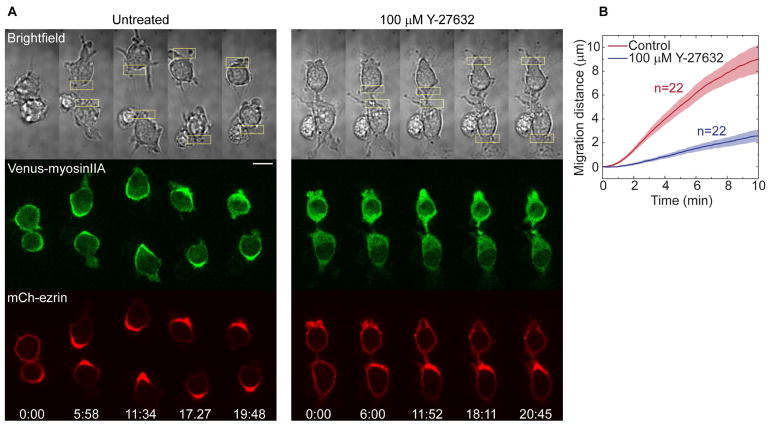
Fig. 5. RhoA-driven migration is ROCK-dependent.
(A) Effect of ROCK inhibition on RhoA-driven migration in cells expressing LARG-mTurq-SspB, iLID-CaaX, Venus-myosinIIA (green), and mCh-ezrin (red). Optical activation was applied before and after 10 min incubation with 100 μM Y-27632.
(B) Migration speeds of cells with and without ROCK inhibition. Localized photoactivation was initiated at t=30 s.
Scale bar: 10 μm.
Coupling of membrane flow to polarized vesicle trafficking
If the plasma membrane continually flows toward the back of the cell, then how does the cell maintain its size and shape? This could be achieved through a polarized endocytic cycle that traffics lipids from the back to the front of the cell. We combined live cell imaging with pharmacological and genetic perturbations to determine if intracellular membrane trafficking is polarized and if it plays a role in RhoA-driven migration.
We started by simultaneously imaging fluorescent clathrin and a Golgi marker during RhoA-driven migration (Fig. 4A, Movie 5). Clathrin is a key molecular component of the endocytic machinery, and its spatial distribution has been used to identify polarized endocytosis (Kirchhausen, 2009). Vesicle trafficking from the trans-Golgi apparatus to the plasma membrane is a major mechanism for delivering lipids to the cell surface, and this process can be enhanced in cellular regions where the Golgi is closer to the plasma membrane. Prior to photoactivation, mCherry-clathrin appeared as fluorescent puncta present uniformly throughout the cell. Following photoactivation at one side of the cell, clathrin polarized toward the side of RhoA activation (12 of 13 cells). In contrast, the trans-Golgi marker YFP-GalT (galactosyltransferase) moved forward in the cell until it was just behind the leading edge (45 of 58 cells) (Fig. 4A, Fig. S2B, Movie 5). Upon optical reversal of the migration direction, clathrin and the trans-Golgi rapidly switched sides, thereby maintaining clathrin at the new back and the trans-Golgi at the new front. Thus both markers dynamically positioned themselves in a manner consistent with the direction of polarized membrane trafficking that is required to maintain the rearward plasma membrane flow. Furthermore, the small molecule brefeldin-A, which inhibits Golgi to plasma membrane transport (Miller, et al., 1992), inhibited the migration (Fig. 4B, Fig. S3, Movie 6). The partial inhibition suggested that trafficking pathways independent of the Golgi were also involved in supporting this form of migration.
Fig. 4. RhoA-driven migration requires polarized vesicle trafficking.
(A) Subcellular dynamics of mCh-clathrin (red) and a trans-Golgi marker (GalT-YFP, green) during RhoA-driven migration. Cells are coexpressing LARG-mTurq-SspB and iLID-CaaX. Images are maximum intensity projections from an 8-plane z-stack.
(B) Effect of brefeldin-A on RhoA-driven migration. Cells expressing LARG-mCh-SspB, iLID-CaaX, and Venus-KRasCT were incubated with 100 μM brefeldin-A or DMSO (control) for 30–45 min prior to initiation of migration. See also Fig S5, Movie 8.
(C) Polarized endocytosis of transferrin during RhoA-driven migration. Cells are expressing LARG-mTurq-SspB, iLID-CaaX, and Venus-KRasCT (green). AlexaFluor594-transferrin (red) was added 5 min after initiating photoactivation. Corresponding kymographs are shown on the right. See also Movie 9.
(D) Effect of endocytosis inhibition on RhoA-driven migration in cells expressing LARG-mCh-SspB, iLID-CaaX, and Venus-KRasCT. Prior to initiation of migration, cells were incubated for 30–45 min with either 0.41% DMSO (control) or an endocytosis inhibitor cocktail consisting of 10 μM secinH3, 25 μM pitstop2, and 100 μM dynasore. See also Fig S5, Movie 12.
(E) Polarized trafficking of caveolin vesicles during RhoA-driven migration in a cell expressing LARG-mTurq-SspB, iLID-CaaX, Venus-KrasCT (green) and mApple-caveolin1 (red). mApple-caveolin1 was photobleached with 594 nm laser light in half of the cell (yellow rectangle) at t=1:43. Localized photoactivation of RhoA (white rectangles) was initiated immediately after photobleaching. See also Movie 11.
(F) Localized (top) and global (bottom) RhoA photoactivation of a cell expressing LARG-mTurq-SspB, iLID-CaaX, Venus-KRasCT, (green) and mApple-caveolin1 (red). Note the formation of caveolin vesicles in the cell rear (white arrow) and the decrease in caveolin at the plasma membrane (yellow arrow) in response to localized RhoA photoactivation. See also Movie 10.
(G) Effect of exocytosis inhibition by endosidin2 on RhoA-driven migration. Prior to initiation of migration, cells were incubated for 30 min with 40 μM endosidin2. See also Fig S5, Movie 13.
(H) Effect of dominant negative Rab4 and Rab11 on RhoA-driven migration. Cells were transfected with LARG-mTurq-SspB, iLID-CaaX, and mCh-KRasCT alone (control), or together with both wild-type YFP-Rab4 and YFP-Rab11, or both dominant negative mutants (YFP-Rab4DN, GFP-Rab11DN). See also Fig S5, Movie 14.
(I) Caveolin dynamics in the absence of RhoA photoactivation. mApple-caveolin1 (red) was photobleached with 594 nm laser light (yellow rectangle). Cells are coexpressing LARG-mTurq-SspB, iLID-CaaX, and Venus-KRasCT (green). Image sequence scale bars are 10 μm. Kymograph scale bars are 10 μm (vertical) and 5 min (horizontal).
To more directly demonstrate that endocytosis is polarized we used dye-labeled transferrin, a commonly used model endocytic cargo (Fig. 4C, Movie 5). Optical RhoA activation was applied to initiate migration, then fluorescent transferrin was added and imaged as it internalized into the migrating cell. Shortly after addition, internalized transferrin was strongly polarized to the cell rear. The kymographs of individual migrating cells show that over time some of the internalized transferrin moved away from the cell rear suggesting that endocytosed vesicles can traffic from the back to the front of the cell (Fig. 4C).
We then examined whether polarized endocytosis during migration is a direct result of RhoA activation, or a result of the consequent rearward membrane flow. The distinct properties of caveolin, a reporter for non-clathrin mediated endocytosis (Le Roy and Wrana, 2005), facilitated these experiments. mApple-caveolin is predominantly bound to the plasma membrane in the basal state of the cell. When caveolin is endocytosed there is a detectable decrease in mApple-caveolin in the plasma membrane apart from the intracellular increase, so both plasma membrane intensity and intracellular fluorescence serve as reporters of endocytosis. We first globally activated RhoA in mApple-caveolin expressing cells and examined endocytosis. There was no detectable increase in caveolin internalization or plasma membrane reduction (Fig. 4F (bottom panel), Movie 7). This showed that RhoA activation by itself does not lead to increased caveolin-reported endocytosis. In contrast, subsequently initiating migration in the same cell by localized activation of RhoA triggered a caveolin fluorescence intensity decrease at the plasma membrane and increase in endocytosis at the back of the cell (Fig. 4F (top panel), Movie 7). This served as an internal control to verify that the optogenetic RhoA activation system was working in these cells.
These results indicate that polarized endocytosis is a result of retrograde plasma membrane flow. Rearward membrane flow is expected to lead to a decrease in membrane tension at the cell rear. A decrease in membrane tension is known to stimulate endocytosis (Echarri and Del Pozo, 2015). The increase in endocytosis restricted to the back of the migrating cells is thus consistent with expectations. In our model, endocytosis at the rear is not essential for plasma membrane flow. It is, however, essential for migration, because rearward plasma membrane flow has to be supported by internal trafficking in the opposite direction to conserve cell size and maintain migration.
We then performed FRAP based experiments to examine whether directional trafficking of the endocytosed membranes at the back of the cell occurred during RhoA driven migration. To follow any potential movement of the endocytosed membranes across the cell during the progress of migration we photobleached one half of each cell expressing mApple-caveolin and then induced RhoA-driven migration. Over a period of time as the cell migrated, caveolin-labeled membranes moved consistently towards the bleached front part of the cell and were localized close to the plasma membrane (Fig. 4E, Movie 7). In a control experiment where mApple-caveolin in half the cell was bleached without any subsequent localized photoactivation of RhoA, there was no detectable increase in intracellular membranes containing mApple-caveolin (Fig. 4I, Movie 7). This confirmed that endocytosis and trafficking require the migratory response to be initiated. These results suggest that membranes that accumulate at the back of the cell through endocytosis, traffic towards the front during migration, consistent with the existence of an exocytic pathway to the plasma membrane at the cell front.
We then tested if the polarized membrane trafficking described above is functionally important for RhoA-driven migration. Cells use both clathrin-dependent and independent pathways for endocytosis, so we inhibited endocytosis using a cocktail of small molecule inhibitors, including a clathrin inhibitor (pitstop2), a dynamin inhibitor (dynasore), and an inhibitor of the Arf6 GEF cytohesin (secinH3). This cocktail significantly inhibited endocytosis measured by fluorescent transferrin internalization in unpolarized cells (Fig. S2A). Consistent with a role for the polarized endocytosis in driving migration, the cocktail significantly inhibited cell migration (Fig. 4D, Fig. S3, Movie 6).
We also tested the effect of inhibiting exocytosis and endosomal recycling on RhoA-driven migration. We used endosidin-2, an inhibitor of the Exo70 subunit of the exocyst complex that has been shown to inhibit exocytosis and endosomal recycling in plant and human cells (Zhang, et al., 2016). Treatment with endosidin-2 significantly inhibited RhoA-driven migration relative to control cells (Fig. 4G, Fig. S3, Movie 6). Coexpression of dominant negative mutants of Rab4 and Rab11, two proteins involved in endosomal recycling, also inhibited the migration (Fig. 4H, Fig. S3, Movie 6). Altogether these results show that RhoA activity at the cell rear polarizes intracellular membrane trafficking and this is essential for its ability to drive cell migration. Interestingly, endocytosis has also been reported to polarize to the cell rear in T lymphocytes, and the polarization was prevented by inhibition of RhoA or ROCK (Samaniego, et al., 2007). Our results suggest that RhoA can play a general role in generating polarized membrane trafficking in migrating cells.
Cortex-membrane interactions in membrane flow generation
How does polarized RhoA signaling generate directional membrane flow? We thought that RhoA might leverage forces generated by myosin motor proteins to orchestrate the membrane flow, because it is well known to control actomyosin contractility through myosin phosphorylation. RhoA activity could localize myosin activity to the cell rear, causing it to pull the actin cortex backward. Direct interactions between the cortex and plasma membrane could then generate the rearward membrane flow. Amoeboid migration has been shown to be based on retrograde cortical flow (Liu, et al., 2015; Ruprecht, et al., 2015).
Consistent with this hypothesis, myosin IIA polarized to the back of the cell during RhoA-driven migration (Fig. 5A), and filamentous actin increased at the cell rear (Fig. 2D). To test whether polarized myosin activity is important for RhoA-driven migration, we examined the effect of inhibiting ROCK, a Rho-associated kinase that controls myosin phosphorylation. The small molecule ROCK inhibitor Y-27632 prevented myosin polarization and inhibited the migration (Fig. 5A, B). Thus RhoA’s control over localized myosin activity is essential for its ability to generate migration. Ezrin-radixin-moesin (ERM) family proteins are known to act as links between the cortex and plasma membrane by simultaneously binding to both cortical actin and PI(4,5)P2 lipids in the plasma membrane (Roubinet, et al., 2012). All three, ezrin, moesin, and PIP2 accumulated at the cell rear during RhoA-driven migration, as would be expected for coupled cortical-membrane flow (Fig. 5A, Fig. S1B, Movie 4).
Membrane flow as an adhesion independent mechanism for cell migration
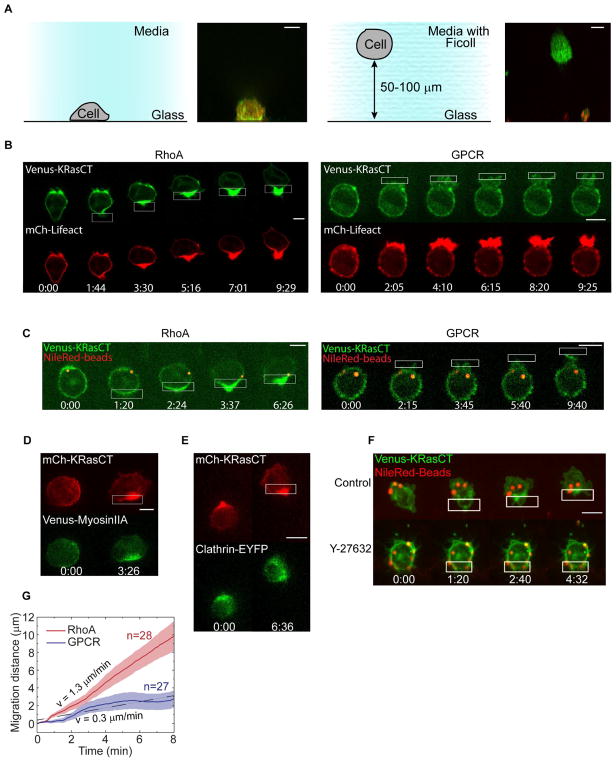
Unlike other migration mechanisms that require direct surface contacts, retrograde membrane flow is predicted to be capable of driving cell migration using forces generated at the interface between the cell and the surrounding liquid (Purcell, 1977). We therefore tested if optical RhoA activation could generate directional migration of cells suspended in liquid (Fig. 6). To prevent sedimentation and adhesion of the cells to the surface, we suspended them in an iso-dense medium containing 23% Ficoll. The appropriate concentration of Ficoll was determined empirically.
Fig. 6. Directional membrane flow, but not polarized lamellipodia formation, drives migration of cells suspended in liquid.
(A) Schematic showing the different conditions for cells on a 2D glass surface (left) or suspended in Ficoll (right). Images are side-view maximum intensity projections from z-stacks.
(B) Comparison of migration and actin responses to optical RhoA or GPCR activation in suspended cells. Cells express Venus-KRasCT (green), mCh-Lifeact (red), and either LARG-mTurq-SspB and iLID-CaaX (left) or untagged parapinopsin (right). Images are single confocal slices. See also Movies 15–16.
(C) Localized activation of RhoA, but not GPCR, in suspended cells induces rearward membrane flow. Cells were transfected with Venus-KRasCT (green) and either LARG-mTurq-SspB and iLID-CaaX (left) or untagged parapinopsin (right). NileRed-containing beads (red) were bound to the extracellular part of transmembrane integrins. Images are single confocal slices.
(D) Polarized myosin response during RhoA-driven migration for suspended cell expressing LARG-mTurq-SspB, iLID-CaaX, mCh-KRasCT (red) and Venus-myosinIIA (green). Images are top-view maximum intensity projections from z-stacks.
(E) Polarized clathrin distribution during RhoA-driven migration in a suspended cell expressing LARG-mTurq-SspB, iLID-CaaX, mCh-KRasCT (red) and Clathrin-EYFP (green). Images are top-view maximum intensity projections from z-stacks.
(F) Bead and cell movement during RhoA driven migration in suspended cells with or without the ROCK inhibitor Y-27632. Cells are expressing LARG-mTurq-SspB, iLID-CaaX, and Venus-KRasCT (green). Cells were incubated with 100 μM Y-27632 for 10 min prior to resuspension in Ficoll also containing 100 μM Y-27632. Images are top-view maximum intensity projections from z-stacks.
(G) Speed of RhoA-driven migration and GPCR-driven migration in suspended cells expressing Venus-KRasCT and either parapinopsin (for GPCR-driven migration) or LARG-mTurq-SspB and iLID-CaaX (for RhoA-driven migration). Localized photoactivation was initiated at t=30 s. All scale bars are 10 μm.
We used a Ficoll solution because it has Newtonian properties and has been widely used as a solution of molecules for a long time (Martinez, et al., 2014; Barry and Bretscher, 2010; Berg and Turner, 1979). There is extensive evidence that even at high concentrations, molecules diffuse at ~10−7cm2/s in a Ficoll solution (Lavrenko, 1986). The viscosity of these solutions increases monotonically with concentration (Busch, 2000). At concentrations used here, Ficoll molecules are globular, not linear – they are thus incapable of forming a lattice (Bohrer, 1984). Finally consistent with the properties of a liquid, the macrophage cells in our experiments sink in Ficoll solutions even very slightly below the concentration that we found to be iso-dense.
For optically driven migration studies we selected cells that were at least 50 μm above the glass surface. Side-views of z-stack images provide further verification that the cells imaged in Ficoll were free in suspension, because the suspended cells had fully round cross-sections whereas cells adhered to the glass surface were flat on the bottom like hemispheres (Fig. 6A).
Remarkably, optical RhoA activation in suspended cells generated persistent, directional cell migration (Fig. 6B, Movie 8). The migration was similar to that of RhoA-driven migration on a 2D surface at both the cellular and molecular levels. The cells formed uropods and migrated without lamellipodia. Accumulation of the plasma membrane marker KRasCT at the cell rear and retrograde movement of beads bound to the cell surface demonstrated rearward membrane flow (Fig. 6C, Fig. S4). Myosin and filamentous actin polarized toward the cell rear, as did clathrin (Fig. 6B, D, E). On average, RhoA-driven migration was approximately 30% faster in suspension than in 2D (Fig. 6G, 2E). These results showed that when freed from the constraints of adhesion to a surface the cells actually moved faster. Overall these results show that the mechanism behind RhoA-driven migration does not require any direct contact with a solid surface, consistent with a surface treadmilling mechanism.
To further test the contribution of retrograde membrane flow, we asked if a different mode of optically driven cell migration that does not generate membrane flow could also drive migration of suspended cells. We tested optical activation of GPCR signaling using parapinopsin in suspended cells. Above we showed that optical GPCR activation generated migration speeds comparable to optical RhoA activation for cells on a 2D glass surface (Fig. 2, Movie 2). In contrast to optical RhoA activation, optical GPCR activation failed to drive migration of suspended cells despite producing strongly polarized lamellipodia (Fig. 6B, Fig. 6G, Movie 8). It also failed to generate rearward plasma membrane flow (Fig. 6C). These results show that whereas migration modes based on lamellipodia or membrane flow can both drive migration in 2D, only retrograde membrane flow can generate forces required to propel suspended cells forward through liquid.
To further test the link between cortical flow, plasma membrane flow, and migration, we examined the effect of ROCK enzyme inhibition on plasma membrane flow and migration in suspended cells. We found that ROCK inhibition abrogated both RhoA directed cell migration and integrin-bound bead movement in suspended cells (Fig. 6F). In contrast, in control cells both cellular and bead movement were elicited. This result is consistent with a role for membrane flow in driving the migration of suspended cells. It is also consistent with our model where the flow of the actin cortex drives the plasma membrane flow.
Sufficiency of viscous forces for cell propulsion
Since RhoA activation generates plasma membrane flow, we asked if the membrane flow is fast enough to account for the observed cell translocation speeds through surface treadmilling alone. To put this question into context, first consider an object moving by surface treadmilling in contact with a solid substrate. This could be for example a tank moving along the ground. The rearward speed of the tank track relative to the tank is referred to as the “treading speed”. As long as the tank track does not slip along the ground, the track must have zero velocity with respect to the ground while in contact with it. This condition can only be achieved if the forward speed of the tank relative to the ground equals the treading speed. The treading speed gives an upper bound on the tank’s forward speed, because any slipping of the track on the ground will reduce the tank’s speed. The same upper bound holds for a cell moving on a 2D surface, regardless of whether it is driven by retrograde actin flow coupled through integrins to the surface or by plasma membrane flow coupled to the surface through non-specific friction.
For an object moving through liquid, the relationship between the treading speed and the forward translocation speed is less intuitive and strongly depends on the Reynolds number. Because our intuition of swimming mechanisms is largely influenced by macroscopic examples governed by high Reynolds number hydrodynamics, it may be tempting to assume that surface treadmilling would be very inefficient in liquid, and that cell protrusions resembling paddles would be needed for propulsion. However, fluid dynamics calculations performed at low Reynolds number previously revealed that for a spherical cell with no shape changes, surface treadmilling generates forward translocation at a speed equal to 67% of the treading speed (Leshansky, et al., 2007). To arrive at this result, the authors derived a general formula for the propulsion speed of ellipsoidal objects moving by surface treadmilling within an incompressible fluid characterized by low Reynolds number. Their derivation assumes the so-called “no-slip boundary condition”, meaning that the surface of the object and the liquid touching the object move at the same speed. Measurements on lipid bilayers using a dynamic force apparatus showed that aqueous fluid flow over lipid bilayers satisfies the no-slip boundary condition (Cross, 2006), indicating that it is a reasonable assumption when considering the fluid dynamics of cells suspended in liquid. For an ellipsoid with zero elongation, i.e. a sphere, their formula simplifies to , where U is the propulsion speed and us is the surface treadmilling speed. This analytical result suggests that the dominance of viscous forces at the cellular scale makes surface treadmilling a viable mechanism for self-propulsion through liquid. It should be emphasized that this relationship between surface treading and forward translocation speeds is independent of viscosity within the low Reynolds number regime (Leshansky, et al., 2007). Thus the change in viscosity due to the addition of Ficoll to the cell culture medium has little impact on this mechanism.
We sought to determine if the relationship between the forward cell translocation speed and the rearward membrane flow speed during RhoA-driven migration agrees with the above prediction. Minor deviations from the predicted value can be expected, given that the theoretical ratio of 0.67 was derived assuming constant, uniform directional membrane flow over the surface of a perfect sphere, whereas the details of the membrane flow will be more complex for real cells. On the other hand, if the cell speed were orders of magnitude faster than expected from the above theoretical results, it would strongly suggest that a different mechanism must contribute to the migration.
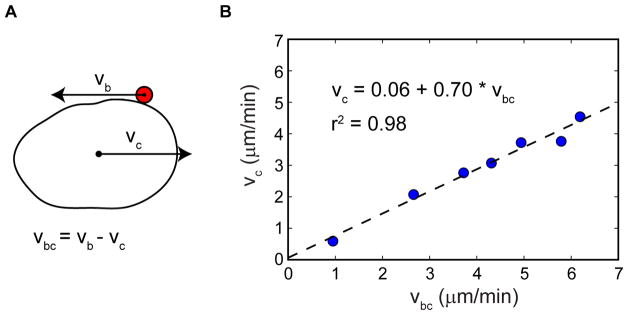
To quantify the ratio of cell translocation to membrane flow speeds, we measured the cell translocation speeds and the movement of membrane bound fluorescent beads upon optogenetic RhoA activation in suspended cells (Fig. 7). As detailed in the methods section, here the speed was measured only during periods when the cell speed and bead movement could be stringently analyzed. This is in contrast to other data shown above, where cell migration speeds were averaged over large numbers of cells and comparably longer time periods. This explains why Fig. 7 includes cells travelling at speeds up to nearly 5 μm/min, whereas the average cell speed in Ficoll was 1.3 μm/min. The measured ratio of bead to cell speed at 0.70 agrees well with the predicted value of 0.67. This suggests that viscous forces generated by the liquid in response to the retrograde membrane flow are in principle sufficient to generate cell migration at speeds observed here.
Fig. 7. Relative speeds of membrane flow and cell translocation suggest that viscous forces tangential to the cell surface suffice to drive the migration.
(A) Side view schematic of a migrating cell with a bead attached to the outer plasma membrane. vc is the speed of the cell. vb is the speed of the bead which is moving rearward along with the membrane. vbc is the speed of the bead with respect to the cell, which corresponds to the speed of rearward membrane flow.
(B) Scatter plot of vc and vbc during RhoA-driven migration. Of 24 cells imaged, 7 cells met the criteria required to allow quantification of both cell and membrane flow speeds as detailed in the Methods section. The linear fit yields vc/vbc = 0.70 (dashed line). The predicted value for a perfect sphere with uniform, constant membrane flow and no cell shape changes is 0.67.
Cell shape changes might also contribute to the adhesion-independent migration. For example, small protrusions can be seen moving rearward during RhoA driven migration of suspended cells (Movie 8). Such protrusions have been proposed to explain migration of Dictyostelium cells in suspension, although membrane flow was not examined in that context (Bae and Bodenschatz, 2010). Potential contributions from cell shape changes cannot be ruled out here due to the lack of an experimental perturbation that can prevent cell shape changes while leaving membrane flow intact. Nonetheless, the quantitative results in Fig. 7 suggest that membrane flow is sufficient to support amoeboid migration.
DISCUSSION
Several decades have passed since the physicist EM Purcell reasoned that it was physically plausible that cells or microorganisms could propel themselves through liquid using tangential movement of their surface without shape changes (Purcell, 1977). His “toroidal swimmer” example has inspired models of cell migration based on “surface treadmilling”, where retrograde plasma membrane flow over the entire cell surface is coupled to polarized endocytic trafficking (Bretscher, 2014; Bretscher, 1996; Bretscher, 1984). However, a direct demonstration of this mechanism has been lacking. Various examples of membrane flow or polarized membrane trafficking have been reported (Fletcher and Rappoport, 2010; Samaniego, et al., 2007; Bretscher and Aguado-Velasco, 1998; Bretscher, 1984), but it has been difficult to experimentally examine their contributions to migration directly.
To test if membrane flow can drive cell migration, we developed an optogenetic system that provides dynamic control over cell migration and plasma membrane flow. We also developed the ability to direct stereotypical migratory responses in cells suspended in a liquid. This is essential to determine the potential role of viscous forces at the cell-liquid interface in the absence of any contributions from direct molecular links between the actin cortex and a solid substrate through intermediate molecules. In this context it is important to emphasize that models that require adhesion such as a “transmembrane clutch” mechanism, while providing an explanation for migration under substrate attached conditions, are not applicable under conditions where a cell is suspended in a liquid.
We found that optical activation of RhoA induced migration with all the characteristics of amoeboid migration. The migration was accompanied by collective rearward flow of lipids and proteins comprising the plasma membrane. In independent experiments we used well established reporters that bind to (a) the lipid PIP2, (b) phospholipids at the inner leaflet of the plasma membrane or (c) lipids on the outer leaflet of the plasma membrane. Each of these reporters demonstrated retrograde flow during amoeboid cell migration demonstrating that lipids on the plasma membrane flow backwards when a cell moves forward. We also used integrin-functionalized beads and fluorescent protein tagged transmembrane receptors to observe the rearward flow of proteins on the plasma membrane. The results showed consistent retrograde movement of the proteins during migration. Optogenetic control provides the ability to almost instantaneously switch the location of optical activation and introduce rapid directional changes to the migratory trajectory. The flow of both lipids and proteins was highly correlated with the direction of migration. Reversal of migration induced reversal of plasma membrane flow. Membrane flow during amoeboid migration has been directly confirmed very recently using a complementary technique involving photobleaching of fluorescent lipids (Tanaka, et al., 2017).
Three main results support a model where viscous forces generated at the cell-liquid interface by rearward plasma membrane flow drive an adhesion-independent mode of amoeboid cell migration. First, RhoA activation was able to generate directed migration for cells suspended in liquid, and the migration speeds achieved were significantly faster than those of cells on a 2D surface. The forces that propel the cell forward in a liquid when in suspension must be independent of adhesions or any type of solid substrate interaction.
Second, an alternative migration mode lacking rearward membrane flow is incapable of propelling cells suspended in liquid. This alternative mode, triggered by optical GPCR activation, generated polarized lamellipodia formation but no detectable retrograde membrane flow. It was able to drive cell migration on 2D glass substrates at speeds comparable to RhoA activation, but failed to propel cells suspended in a liquid. This clear difference suggests that the retrograde membrane flow is essential for RhoA’s ability to generate substrate-independent directional migration.
Third, low Reynolds number hydrodynamic calculations reveal that retrograde surface movement in a sphere can support forward translocation of that body at a speed that is 67% of the surface flow speed (Leshansky, et al., 2007). The quantitative relationship between the plasma membrane flow speed and the cell translocation speed that we determined here experimentally is in strikingly close agreement with this prediction. This agreement indicates that viscous forces generated by the surrounding liquid in response to the rearward membrane flow are sufficient to produce the observed migration speeds.
Despite the plasma membrane flowing backwards during migration, the size and shape of the cells are conserved consistent with recent results where Dictyostelium cell size is similarly conserved (Tanaka, et al., 2017). This suggested that internal trafficking plays a role in maintaining membrane homeostasis. The polarization of markers for endosomes and active endocytosis from back to front as well as the movement of a trans Golgi marker to the cell front during migration support the prediction that directional intracellular vesicle trafficking supports the plasma membrane flow. Furthermore, inhibition of motility by pharmacological and genetic perturbations of endocytosis and exocytosis provide additional support for polarized trafficking underlying amoeboid migration in the absence of adhesion.
It is worth noting that a net flux of membrane to the front of the cell could even be achieved if polarized endocytosis at the rear were accompanied by spatially uniform exocytosis. Either way, this directional trafficking would balance the rearward flow of the plasma membrane. However, our results do suggest that trafficking is directional from back to the front of the cell. Our results also suggest that the increased endocytosis that occurs at the back during RhoA directed migration is likely due to the lowered tension that arises when the plasma membrane flows backwards.
Overall, our results suggest that RhoA stimulated actomyosin contractility at the back generates retrograde plasma membrane flow through the coupling of the plasma membrane to the flow of the actin cortex. Retrograde actin flows are widely observed in amoeboid migration (Liu, et al., 2015; Maiuri, et al., 2015), and faster actin flow is correlated with increased directional persistence and faster migration speeds (Maiuri, et al., 2015). Inhibiting the ROCK enzyme results in inhibition of both cell migration and movement of beads attached to the cell surface. Since ROCK is activated by RhoA and mediates a pathway involved in myosin contraction and resultant actin cortical flow, this result suggests that RhoA directed migration is governed by myosin induced actin flow. Although many examples of adhesion-independent migration are known, their mechanistic basis has remained unclear (Bergert, et al., 2015; Renkawitz and Sixt, 2010; Lammermann, et al., 2008). Our results highlight how coupling actin flow to surface membrane flow provides cells with a means of adhesion-independent migration that does not sacrifice migration speed. Given the extensive coupling of the actomyosin cortex with the plasma membrane in various cell types (Kapus and Janmey, 2013), this is likely to be a widely used mechanism.
Interestingly, low Reynolds number hydrodynamics calculations show that propulsion by surface treadmilling is much more energetically efficient than other modes of swimming, such as those involving rotating flagella, because it minimizes momentum transfer to the surrounding liquid resulting in reduced viscous dissipation (Leshansky, et al., 2007). Furthermore, the cell recruits native trafficking without expending energy in synthesizing components for flagella or cilia. This suggests that the amoeboid migratory mode, which is used by immune cells and tumor cells with invasive capabilities, is highly efficient (Liu, et al., 2015). While membrane flow may be sufficient to drive adhesion-independent migration on its own, it can also act synergistically with mechanisms that depend on cell shape changes, such as intercalation of cellular protrusions into gaps in the extracellular matrix that allows the cell to push off matrix fibers (Paluch, et al., 2016), to direct non-adhesive amoeboid migration in 3D matrices.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, N Gautam (ngautam@wustl.edu).
Experimental Model and Subject Details
RAW 264.7 is a macrophage cell line that was established from an ascites of a tumour induced in a male mouse by intraperitoneal injection of Abselon Leukaemia Virus (Raschke, et al., 1978).
Method Details
DNA Constructs
The following constructs were obtained from Addgene: mVenus-myosinIIA (#56389, Michael Davidson), GFP-rGBD (#26732, William Bement), mCherry-ezrin (#55043, Michael Davidson), mCherry-moesin (#55103, Michael Davidson), Venus-iLID-CaaX (#60411, Brian Kuhlman), mRFP-FKBP12-5ptpase (#67516, Tamas Balla), mRFP-FKBP12 (#67514, Tamas Balla), EYFP-Clathrin-19 (#56584, Michael Davidson), mCherry-Lifeact-7 (#54491, Michael Davidson), mApple-caveolin (#54872, Michael Davidson), pUASP-YFP-Rab4 (#37690, Matthew Scott), pUASP-YFP-Rab4DN (#37691, Matthew Scott).
The Rac biosensor is a modified version of a previously published biosensor (Moshfegh, et al., 2014) and was kindly provided by Louis Hodgson (Albert Einstein College of Medicine of Yeshiva University, New York City, NY). Lck-FRB-ECFP was kindly provided by Carsten Schultz (EMBL Heidelberg, Germany). Parapinopsin was kindly provided by Akihisa Terakita (Osaka City University, Japan). β2AR-YFP was kindly provided by Michael Bruchas (Washington University in St. Louis, USA). YFP-Rab11 and GFP-Rab11DN were kindly provided by Jennifer Stow (University of Queensland, Australia).
The cDNA clone for CXCR4 was obtained from the cDNA Resource Center (www.cdna.org). iLID-CaaX was previously described (O’Neill, et al., 2016). The DHPH domain of LARG was kindly provided by Reza Ahmadian (Universität Düsseldorf), Germany). LARG-mCh-SspB-R73Q was made by ligating a PCR fragment containing the DHPH domain of LARG (KpnI and BamH1) with an mCherry-SspB-R73Q fragment (BamH1 and XbaI) into pcDNA3.1 (KpnI and XbaI). LARG-mTurq-SspB-R73Q was made by replacing the mCherry in LARG-mCh-SspB-R73Q with mTurquoise (synthesized as a Gblock by IDT) flanked by BspE1 at both 5′ and 3′ ends. Venus-rGBD was made by ligating Venus (HindIII-KpnI), rGBD(KpnI–EcoR1) and pcDNA3.1 (HindIII-EcoR1). Venus-KRasCT and mCh-KRasCT were synthesized as Gblocks (HindIII-XbaI) and cut and ligated with pcDNA3.1 (HindIII-XbaI). PCR fragments of YFP Rab 4/Rab4DN (NheI-NotI) were transferred from the original Addgene constructs (#37690 and #37691) into pcDNA3.1(NheI-NotI).
Cell culture
RAW 264.7 cells were obtained from the Washington University Tissue Culture Support Center and ATCC (Manassas, VA). Cells were cultured in high-glucose DMEM (D6429; Sigma Aldrich) supplemented with 10% dialyzed fetal bovine serum (Atlanta Biologicals), and 1% penicillin-streptomycin at 37°C in a humidified atmosphere of 5% CO2. RAW cells ranging from passage 4 to 16 were used for experiments.
Transfection
RAW cells were transfected by electroporation in Amaxa Nucleofector Solution V using the T-020 setting on an Amaxa Nucleofector 2b device. Each electroporation was performed on 2–3 million cells in 100 μl of Nucleofector solution, followed immediately be addition of 500 μl of warm culture medium. For experiments performed on glass, the electroporated cells were incubated in suspension at 37°C and 5% CO2 for ten minutes, then plated in glass bottom dishes at 50–100 μl per dish. The dishes were kept at 37°C and 5% CO2 for 1 h to allow cells to adhere to the surface, followed by addition of 2 ml of warmed culture medium. Imaging was performed 4–10 h after electroporation.
For experiments performed in Ficoll, cells were plated in a T25 flask immediately after electroporation and media was exchanged for fresh RAW media after 1 h. Prior to imaging, cells were incubated with EDTA (17-711E; Lonza) for 10 min to detach them from the flask, spun for 90 s at 1173 rpm, and resuspended in 100 μL of media containing 23% Ficoll400 (F8016; Sigma Aldrich). A 4×5 mm glass cylinder (7030304; Bioptechs) was fixed to a glass bottom dish using silicon grease and 25 μL of cell suspension in 23% Ficoll was pipetted into the cylinder. The following layers were added on top to create a Ficoll gradient: 20 μl 10% Ficoll, 20 μl 5% Ficoll, and 20 μl mineral oil to prevent evaporation. This protocol was adapted from previously published work (Barry and Bretscher, 2010). Imaging was performed 4–10 h after electroporation.
Chemicals
Stock solutions of Y-27632, NSC 23766, and SDF-1α were prepared in H2O. Stock solutions of secinH3, dynasore, pitstop2, and brefeldin A were prepared in DMSO. The reagents were kept at −20°C until use. Dilutions were made in warm Hank’s Balanced Salt Solution (HBSS) containing 1 mg/l of glucose. Stock solution of 9-cis-retinal was prepared in ethanol and kept at −80°C until use. Dilutions were made in HBSS with 1 mg/l of glucose. All retinal preparation was performed with minimum light exposure. Cells were incubated with 10 μM of retinal for at least 20 min prior to imaging.
Beads
Carboxyl-functionalized beads with an average diameter of 1.31 μm containing the fluorophore Nile Red (FCM-1056-2, Spherotech) were conjugated to the integrin-binding peptide GRGDS (G4391; Sigma Aldrich) by incubating 30 μl of 1% w/v bead solution for 4 h with 50 μg/ml GRGDS in phosphate buffered saline (PBS) containing 5 mg/ml of the carboxyl-activating agent N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) (E7750; Sigma Aldrich). The solution was then buffer exchanged into 1 ml of fresh PBS by centrifuging twice at 5,000 rpm for 5 min in order to remove excess peptide and EDC.
For experiments on glass, cells were incubated with 50 μl of peptide-functionalized bead solution per dish for 1 h, then washed with warm media to remove unbound beads. For experiments in Ficoll, the beads were added to the cells 1 h prior to detaching the cells from the T25 flask.
Live cell imaging and optogenetic control of cell migration. Imaging and optical activation were performed using an Andor Revolution spinning disk confocal imaging system. The system consisted of a Leica DMI6000B microscope unit with adaptive focus control, a Yokogawa CSU-X1 spinning disk unit, an Andor iXon electron-multiplying charge-coupled device (EMCCD) camera, a laser combiner with 445, 488, 515, and 594 nm solid state lasers, and an Andor FRAPPA unit for photoactivation of manually selected regions of the sample in real time. All components were controlled using Andor iQ2 software. For optical activation, the 445 nm laser was used at 145 nW power and scanned across the selected region at a rate of 0.9 ms/μm2. Photoactivation and image capture were performed once every 5 s for single plane imaging. Z-stacks contained 6–9 confocal planes. Laser wavelengths of 515 and 594 nm were used to excite Venus and mCherry, respectively. Emission filters were Venus 528/20 nm and mCherry 628/20 nm (Semrock). All images were acquired using a 63x, 1.4 NA oil immersion objective (Leica). Imaging was performed inside a temperature-controlled chamber held at 37°C and 5% CO2.
Migration trajectory analysis
Using a custom script written in Python, and using scikit-image to load and save image data (van der Walt, et al., 2014), multi-image TIF files of migrating cells were converted to kymographs where the x and y axes represent time and cell position, respectively. The cell outline depicted in the kymographs was manually traced in ImageJ (Schneider, et al., 2012), and a second Python script was used to create trajectories from the traces based on the position of the cell rear. The displacement values for each time point were averaged for n number of cells to create a mean trajectory and shaded regions representing SEM.
Quantitative comparison of membrane and cell speeds
Image sequences of cells undergoing RhoA driven migration in suspension were acquired as z-stacks with 8 planes per cell. Of 24 cells imaged, 7 satisfied the following criteria and were used for quantitative analysis: (i) The retrograde movement of at least one integrin-bound bead on the cell surface followed a trajectory that stays within a given xy-plane to facilitate quantitative analysis of its movement. (ii) The cell did not exhibit sideways drift – this could be detected by drift of unbound beads within the field of view. (iii) The number of beads on the cell surface was low enough to track an individual bead. (iv) The cell retained a rounded shape during the migration, with no dramatic changes in the cell shape that could complicate measurement of the cell velocity or the membrane velocity.
Coordinates of the center of the bead and the back of the cell were recorded at each time point using the ImageJ plugin MtrackJ (Meijering, et al., 2012). Remaining analysis was performed using a custom-written Python script as follows. First the bead coordinates were converted from the lab frame to the frame of the moving cell by subtracting the cell coordinates at each time point. The membrane and cell speeds were calculated over the time window corresponding to clear retrograde movement of the bead along the cell surface. The speeds were computed as the sum of individual displacements between time points, divided by the total time. The linear fit to the ratio of velocities was performed using the Scipy function for linear regression.
Quantification and Statistical Analysis
Data presented in the text are expressed as the mean ± standard error of the mean. Statistical significance was determined using one-sample t-test as indicated in the legend for Figure 3.
Data and Software Availability
For software for image analysis, see Key Resources Table.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| GRGDS peptide | Sigma Aldrich | G4391 |
| EDTA | Lonza | 17-711E |
| High-glucose DMEM | Sigma Aldrich | D6429 |
| Dialyzed fetal bovine serum | Atlanta Biologicals | S12850 |
| Penicillin-streptomycin | Sigma Aldrich | B21210 |
| Y-27632 dihydrochloride | Enzo | ALX-270-333-M001 |
| NSC 23766 | Tocris Bioscience | 2161 |
| SDF-1α | Sigma Aldrich | SRP3276 |
| SecinH3 | Tocris Bioscience | 2849 |
| DMSO | Sigma Aldrich | D2438 |
| Hank’s Balanced Salt Solution (HBSS) | Corning | 21-023-CM |
| Glucose | Sigma Aldrich | G 7021 |
| 9-cis-retinal | Sigma Aldrich | R5754 |
| Dulbecco’s phosphate buffered saline (PBS) | Sigma Aldirch | D1408 |
| N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) | Sigma Aldrich | E7750 |
| Dynasore | Sigma Aldrich | D7693 |
| Pitstop2 | Abcam | ab120687 |
| Brefeldin A | Abcam | ab12029 |
| Endosidin2 | Sigma Aldrich | SML1681 |
| Ficoll400 | Sigma Aldrich | F8016 |
| Transferrin from human serum, Alexa Fluor 594 conjugate | Life Technologies | T13343 |
| Critical Commercial Assays | ||
| Amaxa Cell Line Nucleofector Kit V | Lonza | VCA-1003 |
| Experimental Models: Cell Lines | ||
| RAW 264.7 cells | Washington University Tissue Culture Support Center | N/A |
| RAW 264.7 cells | ATCC | TIB-71 |
| Oligonucleotides | ||
| LARG sense primer with KpnI (GCGGGTACCATGCCC AAC TGGCAGCAGCTTGTTAGTCGAG) | IDT | N/A |
| LARG antisense primer with BamHI (CGCGGATCCGG ATT GCTCCTTCACTGATGCAG) | IDT | N/A |
| mTurquoise GBlock sequence with BspeI on 5′ and 3′ ends | IDT | N/A |
| rGBD sense primer with KpnI (CGGGGTACCATCCTGG AG GACCTCAATATGCTC) | IDT | N/A |
| rGBD antisense primer with EcoRI (CGCGAATTCCTAG CCT GTCTTCTCCAGCACCTGGG) | IDT | N/A |
| YFP-Rab4 sense primer NheI (GCGCTAGCTAGCATGA GTA AAGGAGAAGAACTTTTC) | IDT | N/A |
| YFP-Rab4 antisense primer with NotI (ATAGTTTAGCG GCC GCTTAAACGCGACAGGTGCAATCCGGCTTATTGATGC) | IDT | N/A |
| Venus-KRasCT Gblock sequence with HindIII and XbaI | IDT | N/A |
| mCh-KRasCT Gblock sequence with HindIII and XbaI | IDT | N/A |
| Recombinant DNA | ||
| mVenus-myosinIIA-C-18 | Michael Davidson https://www.addgene.org/fluorescent-proteins/davidson/ | Addgene #56389 |
| GFP-rGBD | Benink and Bement, 2005 | Addgene #26732 |
| mCherry-ezrin-N-14 | https://www.addgene.org/fluorescent-proteins/davidson/ | Addgene #55043 |
| mCherry-moesin-C-14 | https://www.addgene.org/fluorescent-proteins/davidson/ | Addgene #55103 |
| Venus-iLID-CaaX | Guntas et al., 2015 | Addgene #60411 |
| EYFP-Clathrin-19 | Rizzo et al., 2009 | Addgene #56584 |
| mCherry-Lifeact-7 | https://www.addgene.org/fluorescent-proteins/davidson/ | Addgene #54491 |
| mApple-caveolin | https://www.addgene.org/fluorescent-proteins/davidson/ | Addgene #54872 |
| pUASP-YFP-Rab4 | Zhang et al., 2007 | Addgene #37690 |
| pUASP-YFP-Rab4DN | Zhang et al., 2007 | Addgene #37691 |
| Rac biosensor | Louis Hodgson (Albert Einstein College of Medicine of Yeshiva University, New York City, NY) | N/A |
| Parapinopsin | Akihisa Terakita (Osaka City University, Japan) | N/A |
| β2AR-YFP | Michael Bruchas (Washington University in St. Louis, USA) | N/A |
| YFP-Rab11 | Jennifer Stow (University of Queensland, Australia) | N/A |
| GFP-Rab11DN | Jennifer Stow (University of Queensland, Australia) | N/A |
| cDNA clone for CXCR4 | cDNA Resource Center (www.cdna.org) | N/A |
| DHPH domain of LARG | Reza Ahmadian (Universität Düsseldorf) | N/A |
| LARG-mCh-SspB-R73Q in pcDNA3.1 | This paper | N/A |
| LARG-mTurq-SspB-R73Q in pcDNA3.1 | This paper | N/A |
| Venus-rGBD in pcDNA3.1 | This paper | N/A |
| Venus-KRasCT in pcDNA3.1 | This paper | N/A |
| mCh-KRasCT in pcDNA3.1 | This paper | N/A |
| YFP-Rab4 in pcDNA3.1 | This paper | N/A |
| YFP-Rab4DN in pcDNA3.1 | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij |
| ImageJ plugin MtrackJ | Erik Meijering | https://imagescience.org/meijering/software/mtrackj/ |
| Scikit-image | van der Walt et al., 2014 | scikit-image.org |
| Other | ||
| Carboxyl-functionalized beads with Nile Red | Spherotech | FCM-1056-2 |
| RNA seq data for RAW 264.7 cells | Genome Technology Access Center, Washington University | N/A |
Supplementary Material
Optically triggered RhoA activation and cell migration, related to Figure 1.
Part 1: Cell transfected with LARG-mCh-SspB (red), iLID-CaaX, and Venus-rGBD (green).
Part 2: Optical activation with and without the RhoA GEF. Cells transfected with iLID-CaaX, Venus-myosinIIA, and either LARG-mCh-SspB (left) or mCh-SspB (right). White rectangle represents the area of photoactivation. Time is in min:sec.
Distinct migration modes driven by RhoA vs GPCR activation, related to Figure 2.
Part 1: Cells transfected with Venus-KRasCT (green), mCh-Lifeact, and either parapinopsin (GPCR) or LARG-mTurq-SspB and iLID-CaaX (RhoA). White rectangle represents the area of photoactivation.
Part 2: RhoA-driven migration without blebbing. Cell transfected with LARG-mTurq-SspB, iLID-CaaX, mCh-PIPK1γ661, and YFP. Yellow rectangle represents the area of photoactivation. Time is in min:sec.
Rearward plasma membrane flow during RhoA-driven migration, related to Figure 3.
Cell transfected with LARG-mTurq-SspB and Venus-iLID-CaaX (green). NileRed-containing beads (red) were bound to cellular integrins. Both top-view (above) and side-view (below) maximum intensity projections of z-stacks are shown. White rectangle represents the area of photoactivation. Time is in min:sec.
Moesin and PIP2 responses during RhoA-driven migration, related to Figure 5. Cell transfected with LARG-mTurq-SspB, iLID-CaaX, mCh-moesin (red) and Venus-PH-PLCδ (green). White rectangle represents the area of photoactivation. Time is in min:sec.
Polarized endocytosis and Golgi localization during RhoA-driven migration, related to Figure 4.
Part 1: Clathrin and trans-Golgi dynamics during RhoA-driven migration. Cell transfected with LARG-mTurq-SspB, iLID-CaaX, GalT-YFP (green) and mCh-clathrin (red). Images are maximum intensity projections from a z-stack.
Part 2: Polarized transferrin endocytosis during RhoA-driven migration. Cell transfected with LARG-mTurq-SspB, iLID-CaaX, and Venus-KRasCT (green). 0.1 μg/ml of AlexaFluor594-transferrin (red) was added at the time point indicated. White rectangle represents the area of photoactivation. Time is in min:sec.
Effects of pharmacological and genetic inhibition of trafficking on RhoA-driven migration, related to Figure 4.
Part 1: Effect of Brefeldin A on RhoA-driven migration. Cells transfected with LARG-mCh-SspB, iLID-CaaX, and Venus-KRasCT (green). Cells were incubated with 100 μM brefeldin-A or DMSO (control) for 30–45 min prior to initiation of migration. White rectangle represents the area of photoactivation. Part 2: Pharmacological inhibition of endocytosis during RhoA-driven migration. Cells transfected with LARG-mCh-SspB, iLID-CaaX, and Venus-KRasCT (green). Cells were incubated with an endocytosis inhibitor cocktail (10 μM secinH3, 25 μM pitstop2, and 100 μM dynasore) or DMSO (control) for 30–45 min prior to initiation of migration. White rectangle represents the area of photoactivation. Part 3: Effect of endosidin2 on RhoA-driven migration. Cells transfected with LARG-mCh-SspB, iLID-CaaX, and Venus-KRasCT (green). Cells were incubated for 30 min with 40 μM endosidin2 or DMSO (control) prior to initiation of migration. Part 4: Effect of dominant negative Rab4 and Rab11 on RhoA-driven migration. Cells transfected either with LARG-mTurq-SspB, iLID-CaaX, and mCh-KRasCT (red) alone, or together with dominant negative mutants (YFP-Rab4DN and GFP-Rab11DN). White rectangle represents the area of photoactivation. Time is in min:sec.
Caveolin endocytosis during uniform and polarized optical RhoA activation, related to Figure 4.
Part 1: Effect of global versus local RhoA activation on endocytosis. Cell transfected with LARG-mTurq-SspB, iLID-CaaX, Venus-KRasCT (green), and mApple-caveolin1 (red).
Part 2: Polarized trafficking of caveolin vesicles during RhoA-driven migration. Cells transfected with LARG-mTurq-SspB, iLID-CaaX, Venus-KRasCT (green), and mApple-caveolin1 (red). In half of the cell, mApple was photobleached with 594 nm laser light (yellow rectangle) prior to initiation of migration.
Part 3: Lack of polarized trafficking in the absence of RhoA-driven migration. Cells transfected with LARG-mTurq-SspB, iLID-CaaX, Venus-KRasCT (green), and mApple-caveolin1 (red). In half of the cell, mApple was photobleached with 594 nm laser light (yellow rectangle). No blue light photoactivation was performed. White rectangle represents the area of photoactivation in parts 1 and 2. Time is in min:sec.
Optical RhoA vs GPCR activation in suspended cells, related to Figure 6.
Part 1: Optical RhoA activation in a suspended cell. Cell transfected with LARG-mTurq-SspB, iLID-CaaX, Venus-KRasCT (green) and mCh-Lifeact (red). The cell is suspended in 23% Ficoll. Images are maximum intensity projections from a z-stack.
Part 2: Optical GPCR activation in a suspended cell. Cell transfected with parapinopsin, Venus-KRasCT (green), and mCh-Lifeact (red). The cell is suspended in 23% Ficoll. White rectangle represents the area of photoactivation. Time is in min:sec.
Highlights.
Optogenetic RhoA or GPCR activation drives amoeboid or lamellipodial cell migration
Only the amoeboid mode exhibits rearward plasma membrane flow
Both modes propel adherent cells, but only the amoeboid mode propels suspended cells
Tangential viscous forces at the cell surface drive adhesion-independent migration
Acknowledgments
This work was funded by the NIH through NIGMS grants GM069027, GM107370, and GM122577. We thank Reza Ahmadian, Louis Hodgson, Carsten Schultz, Jennifer Stow, Akihisa Terakita, and Michael Bruchas for kindly providing DNA constructs. We thank Thomas O’Reilly-Pol (Washington University) for help with interpreting RNAseq data. We thank Sean Collins (UC Davis) and John Royer (University of Edinburgh) for useful discussions.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
Conceptualization, P.R.O and N.G.; Investigation, P.R.O, J.A.C., X.M.; Resources, K.M., V.K.; Formal Analysis, P.R.O., J.A.C., X.M.; Writing – Original Draft, P.R.O. and N.G.; Writing – Review and Editing, P.R.O., J.A.C, X.M., V.K. and N.G.; Visualization: P.R.O, J.A.C. and X.M.; Funding Acquisition, N.G.; Supervision, N.G.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bae AJ, Bodenschatz E. On the swimming of Dictyostelium amoebae. Proc Natl Acad Sci USA. 2010;107:E165–6. doi: 10.1073/pnas.1011900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry NP, Bretscher MS. Dictyostelium amoebae and neutrophils can swim. Proc Natl Acad Sci USA. 2010;107:11376–80. doi: 10.1073/pnas.1006327107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benink HA, Bement WM. Concentric zones of active RhoA and Cdc42 around single cell wounds. J Cell Biol. 2005;168:429–39. doi: 10.1083/jcb.200411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC, Turner L. Movement of microorganisms in viscous environments. Nature. 1979;278:349–51. doi: 10.1038/278349a0. [DOI] [PubMed] [Google Scholar]
- Bergert M, Erzberger A, Desai RA, Aspalter IM, Oates AC, Charras G, Salbreux G, Paluch EK. Force transmission during adhesion-independent migration. Nat Cell Biol. 2015;17:524–9. doi: 10.1038/ncb3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohrer MPP, GD, Carroll PJ. Hindered diffusion of Dextran and Ficoll in microporous membranes. Macromolecules. 1984;17:1170–1173. [Google Scholar]
- Bretscher MS. Endocytosis: relation to capping and cell locomotion. Science. 1984;224:681–6. doi: 10.1126/science.6719108. [DOI] [PubMed] [Google Scholar]
- Bretscher MS. Getting membrane flow and the cytoskeleton to cooperate in moving cells. Cell. 1996;87:601–6. doi: 10.1016/s0092-8674(00)81380-x. [DOI] [PubMed] [Google Scholar]
- Bretscher MS. Asymmetry of single cells and where that leads. Annu Rev Biochem. 2014;83:275–89. doi: 10.1146/annurev-biochem-060713-035813. [DOI] [PubMed] [Google Scholar]
- Bretscher MS, Aguado-Velasco C. Membrane traffic during cell locomotion. Curr Opin Cell Biol. 1998;10:537–41. doi: 10.1016/s0955-0674(98)80070-7. [DOI] [PubMed] [Google Scholar]
- Busch NAK, TV, Bloomfield A. Tracer Diffusion of Proteins in DNA Solutions. 2. Green Fluorescent Protein in Crowded DNA Solutions. Macromolecules. 2000;33:5932–5937. [Google Scholar]
- Charras G, Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nat Rev Mol Cell Biol. 2008;9:730–6. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- Collins SR, Yang HW, Bonger KM, Guignet EG, Wandless TJ, Meyer T. Using light to shape chemical gradients for parallel and automated analysis of chemotaxis. Mol Syst Biol. 2015;11:804. doi: 10.15252/msb.20156027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross BSA, Cottin-Bizonne C, Rieu J-P, Charlaix E. Boundary flow of water on supported phospholipid films. Europhys Lett. 2006;73:390–395. [Google Scholar]
- Echarri A, Del Pozo MA. Caveolae - mechanosensitive membrane invaginations linked to actin filaments. J Cell Sci. 2015;128:2747–58. doi: 10.1242/jcs.153940. [DOI] [PubMed] [Google Scholar]
- Fletcher SJ, Rappoport JZ. Moving forward: polarised trafficking in cell migration. Trends Cell Biol. 2010;20:71–8. doi: 10.1016/j.tcb.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Gadea G, de Toledo M, Anguille C, Roux P. Loss of p53 promotes RhoA-ROCK-dependent cell migration and invasion in 3D matrices. J Cell Biol. 2007;178:23–30. doi: 10.1083/jcb.200701120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CE, Liu T, Montel V, Hsiao G, Lester RD, Subramaniam S, Gonias SL, Klemke RL. Chemoattractant signaling between tumor cells and macrophages regulates cancer cell migration, metastasis and neovascularization. Plos One. 2009;4:e6713. doi: 10.1371/journal.pone.0006713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, Kuhlman B. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci USA. 2015;112:112–7. doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal M, Dvorsky R, Ahmadian MR. Deciphering the molecular and functional basis of Dbl family proteins: a novel systematic approach toward classification of selective activation of the Rho family proteins. J Biol Chem. 2013;288:4486–500. doi: 10.1074/jbc.M112.429746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapus A, Janmey P. Plasma membrane--cortical cytoskeleton interactions: a cell biology approach with biophysical considerations. Compr Physiol. 2013;3:1231–81. doi: 10.1002/cphy.c120015. [DOI] [PubMed] [Google Scholar]
- Karunarathne WK, Giri L, Patel AK, Venkatesh KV, Gautam N. Optical control demonstrates switch-like PIP3 dynamics underlying the initiation of immune cell migration. Proc Natl Acad Sci USA. 2013;110:E1575–83. doi: 10.1073/pnas.1220755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunarathne WK, O’Neill PR, Gautam N. Subcellular optogenetics - controlling signaling and single-cell behavior. J Cell Sci. 2015;128:15–25. doi: 10.1242/jcs.154435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller H, Zadeh AD, Eggli P. Localised depletion of polymerised actin at the front of Walker carcinosarcoma cells increases the speed of locomotion. Cell Motil Cytoskeleton. 2002;53:189–202. doi: 10.1002/cm.10063. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Imaging endocytic clathrin structures in living cells. Trends Cell Biol. 2009;19:596–605. doi: 10.1016/j.tcb.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi M, Kawano E, Kinugawa Y, Oishi T, Shichida Y, Tamotsu S, Terakita A. Bistable UV pigment in the lamprey pineal. Proc Natl Acad Sci USA. 2004;101:6687–91. doi: 10.1073/pnas.0400819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–90. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- Lammermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Soldner R, Hirsch K, Keller M, Forster R, Critchley DR, Fassler R, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–5. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- Lammermann T, Germain RN. The multiple faces of leukocyte interstitial migration. Semin Immunopathol. 2014;36:227–51. doi: 10.1007/s00281-014-0418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammermann T, Sixt M. Mechanical modes of ‘amoeboid’ cell migration. Curr Opin Cell Biol. 2009;21:636–44. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Lavrenko PNM, OI, Didenko SA. Hydrodynamic properties and the shape of the molecules of the polysaccharide Ficoll-400 in solution. Polymer Science. 1986;28:576–584. [Google Scholar]
- Le Roy C, Wrana JL. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat Rev Mol Cell Biol. 2005;6:112–26. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- Leithner A, Eichner A, Muller J, Reversat A, Brown M, Schwarz J, Merrin J, de Gorter DJ, Schur F, Bayerl J, et al. Diversified actin protrusions promote environmental exploration but are dispensable for locomotion of leukocytes. Nat Cell Biol. 2016;18:1253–1259. doi: 10.1038/ncb3426. [DOI] [PubMed] [Google Scholar]
- Leshansky AM, Kenneth O. Surface tank treading: Propulsion of Purcell’s toroidal swimmer. Phys Fluids. 2008;20:063104. [Google Scholar]
- Leshansky AM, Kenneth O, Gat O, Avron JE. A frictionless microswimmer. New J Phys. 2007;9:145. [Google Scholar]
- Liu YJ, Le Berre M, Lautenschlaeger F, Maiuri P, Callan-Jones A, Heuze M, Takaki T, Voituriez R, Piel M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 2015;160:659–72. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Lokuta MA, Senetar MA, Bennin DA, Nuzzi PA, Chan KT, Ott VL, Huttenlocher A. Type Igamma PIP kinase is a novel uropod component that regulates rear retraction during neutrophil chemotaxis. Mol Biol Cell. 2007;18:5069–80. doi: 10.1091/mbc.E07-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri P, Rupprecht JF, Wieser S, Ruprecht V, Benichou O, Carpi N, Coppey M, De Beco S, Gov N, Heisenberg CP, et al. Actin Flows Mediate a Universal Coupling between Cell Speed and Cell Persistence. Cell. 2015;161:374–86. doi: 10.1016/j.cell.2015.01.056. [DOI] [PubMed] [Google Scholar]
- Martinelli S, Chen EJ, Clarke F, Lyck R, Affentranger S, Burkhardt JK, Niggli V. Ezrin/Radixin/Moesin proteins and flotillins cooperate to promote uropod formation in T cells. Front Immunol. 2013;4:84. doi: 10.3389/fimmu.2013.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez VA, Schwarz-Linek J, Reufer M, Wilson LG, Morozov AN, Poon WC. Flagellated bacterial motility in polymer solutions. Proc Natl Acad Sci USA. 2014;111:17771–6. doi: 10.1073/pnas.1415460111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- Miller SG, Carnell L, Moore HH. Post-Golgi membrane traffic: brefeldin A inhibits export from distal Golgi compartments to the cell surface but not recycling. J Cell Biol. 1992;118:267–83. doi: 10.1083/jcb.118.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshfegh Y, Bravo-Cordero JJ, Miskolci V, Condeelis J, Hodgson L. A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nat Cell Biol. 2014;16:574–86. doi: 10.1038/ncb2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill PR, Gautam N. Subcellular optogenetic inhibition of G proteins generates signaling gradients and cell migration. Mol Biol Cell. 2014;25:2305–14. doi: 10.1091/mbc.E14-04-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill PR, Kalyanaraman V, Gautam N. Subcellular optogenetic activation of Cdc42 controls local and distal signaling to drive immune cell migration. Mol Biol Cell. 2016;27:1442–50. doi: 10.1091/mbc.E15-12-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch EK, Aspalter IM, Sixt M. Focal Adhesion-Independent Cell Migration. Annu Rev Cell Dev Biol. 2016;32:469–490. doi: 10.1146/annurev-cellbio-111315-125341. [DOI] [PubMed] [Google Scholar]
- Poincloux R, Collin O, Lizarraga F, Romao M, Debray M, Piel M, Chavrier P. Contractility of the cell rear drives invasion of breast tumor cells in 3D Matrigel. Proc Natl Acad Sci USA. 2011;108:1943–8. doi: 10.1073/pnas.1010396108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EM. Life at low Reynolds number. Am J Phys. 1977;45:3–11. [Google Scholar]
- Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Raschke WC, Baird S, Ralph P, Nakoinz I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell. 1978;15:261–7. doi: 10.1016/0092-8674(78)90101-0. [DOI] [PubMed] [Google Scholar]
- Renkawitz J, Schumann K, Weber M, Lammermann T, Pflicke H, Piel M, Polleux J, Spatz JP, Sixt M. Adaptive force transmission in amoeboid cell migration. Nat Cell Biol. 2009;11:1438–43. doi: 10.1038/ncb1992. [DOI] [PubMed] [Google Scholar]
- Renkawitz J, Sixt M. Mechanisms of force generation and force transmission during interstitial leukocyte migration. EMBO Rep. 2010;11:744–50. doi: 10.1038/embor.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BE, Lehmann R. Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat Rev Mol Cell Biol. 2010;11:37–49. doi: 10.1038/nrm2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubinet C, Tran PT, Piel M. Common mechanisms regulating cell cortex properties during cell division and cell migration. Cytoskeleton (Hoboken) 2012;69:957–72. doi: 10.1002/cm.21086. [DOI] [PubMed] [Google Scholar]
- Ruprecht V, Wieser S, Callan-Jones A, Smutny M, Morita H, Sako K, Barone V, Ritsch-Marte M, Sixt M, Voituriez R, et al. Cortical contractility triggers a stochastic switch to fast amoeboid cell motility. Cell. 2015;160:673–85. doi: 10.1016/j.cell.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5:711–9. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- Samaniego R, Sanchez-Martin L, Estecha A, Sanchez-Mateos P. Rho/ROCK and myosin II control the polarized distribution of endocytic clathrin structures at the uropod of moving T lymphocytes. J Cell Sci. 2007;120:3534–43. doi: 10.1242/jcs.006296. [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–23. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surve CR, To JY, Malik S, Kim M, Smrcka AV. Dynamic regulation of neutrophil polarity and migration by the heterotrimeric G protein subunits Galphai-GTP and Gbetagamma. Sci Signal. 2016;9:ra22. doi: 10.1126/scisignal.aad8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kikuchi T, Uno H, Okita K, Kitanishi-Yumura T, Yumura S. Turnover and flow of the cell membrane for cell migration. Sci Rep. 2017;7:12970. doi: 10.1038/s41598-017-13438-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valon L, Marin-Llaurado A, Wyatt T, Charras G, Trepat X. Optogenetic control of cellular forces and mechanotransduction. Nat Commun. 2017;8:14396. doi: 10.1038/ncomms14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt S, Schonberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, Gouillart E, Yu T, scikit-image c. scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goethem E, Poincloux R, Gauffre F, Maridonneau-Parini I, Le Cabec V. Matrix architecture dictates three-dimensional migration modes of human macrophages: differential involvement of proteases and podosome-like structures. J Immunol. 2010;184:1049–61. doi: 10.4049/jimmunol.0902223. [DOI] [PubMed] [Google Scholar]
- Wagner E, Glotzer M. Local RhoA activation induces cytokinetic furrows independent of spindle position and cell cycle stage. J Cell Biol. 2016;213:641–9. doi: 10.1083/jcb.201603025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–77. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthylake RA, Burridge K. RhoA and ROCK promote migration by limiting membrane protrusions. J Biol Chem. 2003;278:13578–84. doi: 10.1074/jbc.M211584200. [DOI] [PubMed] [Google Scholar]
- Worthylake RA, Lemoine S, Watson JM, Burridge K. RhoA is required for monocyte tail retraction during transendothelial migration. J Cell Biol. 2001;154:147–60. doi: 10.1083/jcb.200103048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–U111. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HW, Collins SR, Meyer T. Locally excitable Cdc42 signals steer cells during chemotaxis. Nat Cell Biol. 2016;18:191–201. doi: 10.1038/ncb3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao HY, Chen L, Xu C, Wang J, Chen J, Xie QM, Wu X, Yan XF. Inhibition of Rac activity alleviates lipopolysaccharide-induced acute pulmonary injury in mice. Biochim Biophys Acta. 2011;1810:666–74. doi: 10.1016/j.bbagen.2011.03.020. [DOI] [PubMed] [Google Scholar]
- Zhang C, Brown MQ, van de Ven W, Zhang ZM, Wu B, Young MC, Synek L, Borchardt D, Harrison R, Pan S, et al. Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc Natl Acad Sci USA. 2016;113:E41–50. doi: 10.1073/pnas.1521248112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Optically triggered RhoA activation and cell migration, related to Figure 1.
Part 1: Cell transfected with LARG-mCh-SspB (red), iLID-CaaX, and Venus-rGBD (green).
Part 2: Optical activation with and without the RhoA GEF. Cells transfected with iLID-CaaX, Venus-myosinIIA, and either LARG-mCh-SspB (left) or mCh-SspB (right). White rectangle represents the area of photoactivation. Time is in min:sec.
Distinct migration modes driven by RhoA vs GPCR activation, related to Figure 2.
Part 1: Cells transfected with Venus-KRasCT (green), mCh-Lifeact, and either parapinopsin (GPCR) or LARG-mTurq-SspB and iLID-CaaX (RhoA). White rectangle represents the area of photoactivation.
Part 2: RhoA-driven migration without blebbing. Cell transfected with LARG-mTurq-SspB, iLID-CaaX, mCh-PIPK1γ661, and YFP. Yellow rectangle represents the area of photoactivation. Time is in min:sec.
Rearward plasma membrane flow during RhoA-driven migration, related to Figure 3.
Cell transfected with LARG-mTurq-SspB and Venus-iLID-CaaX (green). NileRed-containing beads (red) were bound to cellular integrins. Both top-view (above) and side-view (below) maximum intensity projections of z-stacks are shown. White rectangle represents the area of photoactivation. Time is in min:sec.
Moesin and PIP2 responses during RhoA-driven migration, related to Figure 5. Cell transfected with LARG-mTurq-SspB, iLID-CaaX, mCh-moesin (red) and Venus-PH-PLCδ (green). White rectangle represents the area of photoactivation. Time is in min:sec.
Polarized endocytosis and Golgi localization during RhoA-driven migration, related to Figure 4.
Part 1: Clathrin and trans-Golgi dynamics during RhoA-driven migration. Cell transfected with LARG-mTurq-SspB, iLID-CaaX, GalT-YFP (green) and mCh-clathrin (red). Images are maximum intensity projections from a z-stack.
Part 2: Polarized transferrin endocytosis during RhoA-driven migration. Cell transfected with LARG-mTurq-SspB, iLID-CaaX, and Venus-KRasCT (green). 0.1 μg/ml of AlexaFluor594-transferrin (red) was added at the time point indicated. White rectangle represents the area of photoactivation. Time is in min:sec.
Effects of pharmacological and genetic inhibition of trafficking on RhoA-driven migration, related to Figure 4.
Part 1: Effect of Brefeldin A on RhoA-driven migration. Cells transfected with LARG-mCh-SspB, iLID-CaaX, and Venus-KRasCT (green). Cells were incubated with 100 μM brefeldin-A or DMSO (control) for 30–45 min prior to initiation of migration. White rectangle represents the area of photoactivation. Part 2: Pharmacological inhibition of endocytosis during RhoA-driven migration. Cells transfected with LARG-mCh-SspB, iLID-CaaX, and Venus-KRasCT (green). Cells were incubated with an endocytosis inhibitor cocktail (10 μM secinH3, 25 μM pitstop2, and 100 μM dynasore) or DMSO (control) for 30–45 min prior to initiation of migration. White rectangle represents the area of photoactivation. Part 3: Effect of endosidin2 on RhoA-driven migration. Cells transfected with LARG-mCh-SspB, iLID-CaaX, and Venus-KRasCT (green). Cells were incubated for 30 min with 40 μM endosidin2 or DMSO (control) prior to initiation of migration. Part 4: Effect of dominant negative Rab4 and Rab11 on RhoA-driven migration. Cells transfected either with LARG-mTurq-SspB, iLID-CaaX, and mCh-KRasCT (red) alone, or together with dominant negative mutants (YFP-Rab4DN and GFP-Rab11DN). White rectangle represents the area of photoactivation. Time is in min:sec.
Caveolin endocytosis during uniform and polarized optical RhoA activation, related to Figure 4.
Part 1: Effect of global versus local RhoA activation on endocytosis. Cell transfected with LARG-mTurq-SspB, iLID-CaaX, Venus-KRasCT (green), and mApple-caveolin1 (red).
Part 2: Polarized trafficking of caveolin vesicles during RhoA-driven migration. Cells transfected with LARG-mTurq-SspB, iLID-CaaX, Venus-KRasCT (green), and mApple-caveolin1 (red). In half of the cell, mApple was photobleached with 594 nm laser light (yellow rectangle) prior to initiation of migration.
Part 3: Lack of polarized trafficking in the absence of RhoA-driven migration. Cells transfected with LARG-mTurq-SspB, iLID-CaaX, Venus-KRasCT (green), and mApple-caveolin1 (red). In half of the cell, mApple was photobleached with 594 nm laser light (yellow rectangle). No blue light photoactivation was performed. White rectangle represents the area of photoactivation in parts 1 and 2. Time is in min:sec.
Optical RhoA vs GPCR activation in suspended cells, related to Figure 6.
Part 1: Optical RhoA activation in a suspended cell. Cell transfected with LARG-mTurq-SspB, iLID-CaaX, Venus-KRasCT (green) and mCh-Lifeact (red). The cell is suspended in 23% Ficoll. Images are maximum intensity projections from a z-stack.
Part 2: Optical GPCR activation in a suspended cell. Cell transfected with parapinopsin, Venus-KRasCT (green), and mCh-Lifeact (red). The cell is suspended in 23% Ficoll. White rectangle represents the area of photoactivation. Time is in min:sec.