Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is caused by insufficient epigenetic repression of D4Z4 macrosatellite repeat where DUX4, an FSHD causing gene is embedded. There are two forms of FSHD, FSHD1 with contraction of D4Z4 repeat and FSHD2 with chromatin compaction defects mostly due to SMCHD1 mutation. Previous reports showed DUX4-induced gene expression changes as well as changes in microRNA expression in FSHD muscle cells. However, a genome wide analysis of small noncoding RNAs that might be regulated by DUX4 or by mutations in SMCHD1 has not been reported yet. Here, we identified several types of small noncoding RNAs including known microRNAs that are differentially expressed in FSHD2 muscle cells compared to control. Although fewer small RNAs were differentially expressed during muscle differentiation in FSHD2 cells compared to controls, most of the known myogenic microRNAs, such as miR1, miR133a and miR206 were induced in both FSHD2 and control muscle cells during differentiation. Our small RNA sequencing data analysis also revealed both DUX4- and SMCHD1-specific changes in FSHD2 muscle cells. Six FSHD2 microRNAs were affected by DUX4 overexpression in control myoblasts, whereas increased expression of tRNAs and 5S rRNAs in FSHD2 muscle cells was largely recapitulated in SMCHD1-depleted control myoblasts. Altogether, our studies suggest that the small noncoding RNA transcriptome changes in FSHD2 might be different from those in FSHD1 and that these differences may provide new diagnostic and therapeutic tools specific to FSHD2.
Introduction
Facioscapulohumeral muscular dystrophy (FSHD) is caused by incomplete repression of the D4Z4 macrosatellite repeat array on a disease-permissive haplotype of chromosome 4q that results in the aberrant expression of the Double Homeobox 4 (DUX4) retrogene imbedded within the D4Z4 repeat (1). Incomplete repression of this locus in somatic cells can be caused either by contraction of the D4Z4 repeat array (95% of FSHD cases; FSHD1) or by epigenetic defects mostly through mutation of Structural Maintenance of Chromosomes Hinge Domain Containing 1 (SMCHD1) (5% of FSHD cases; FSHD2) (2). Both FSHD1 and FSHD2 patients exhibit a similar clinical phenotype such as progressive weakness and wasting in the facial, shoulder and upper arm muscles. DUX4 is upregulated in both FSHD1 and FSHD2 muscles, and its expression correlates with severity of the FSHD muscle phenotype (3). Previous studies reported mRNA expression changes in either muscle biopsies or primary myoblast lines derived from FSHD patients and most of the gene expression changes were shown to be caused by aberrant DUX4 expression (4–6).
In addition to altered mRNA abundance, noncoding RNAs have also been implicated in FSHD biology. Various types of small noncoding RNAs (ncRNAs) have important functions in gene regulation and cell metabolism. MicroRNAs (miRNAs) are evolutionarily conserved, typically ∼ 22 nucleotide long noncoding RNA molecules that bind to Argonaute (AGO) protein, a component of RNA-induced silencing complex (RISC). miRNA-RISC binds to the 3′-untranslated region of target mRNA and negatively regulates expression of target genes at the post-transcriptional level either by translation inhibition or by mRNA decay (7,8). In previous studies, expression of known miRNAs was shown to be changed in FSHD muscle, mostly in FSHD1, compared to unaffected controls (9–12). These studies suggested that dysregulation of miRNAs may be correlated with FSHD pathology, causing muscle cell death and muscle differentiation defects. However, only a small number of identified FSHD-related known miRNAs were found to overlap between these multiple studies (Supplementary Material, Table S1); this discrepancy might be explained by different types of sample sources and/or differences in experimental design and data processing. In addition, it has been shown that other types of small RNAs, such as tRNA-derived fragments (tRFs) and snoRNAs, could also play a role in gene regulation and therefore be implicated in human disease (reviewed in 13). As a majority of the previous studies used microarray-based approaches and/or focused mostly on known miRNAs, genome-wide analysis of FSHD-specific changes in expression of other types of small non-coding RNAs has not yet been reported. Therefore, in addition to miRNAs, it is possible that other types of small RNAs might play a role in FSHD, and particularly in FSHD2 because of mutations in epigenetic modifiers. SMCHD1 was identified as one of epigenetic modifiers of transgene variegation through a genome-wide screen (14). SMCHD1 regulates maintenance of X inactivation, hypermethylation of CpG islands on the inactive X and expression of gene clusters, such as clustered protocadherins, on both autosomes and the inactive X chromosome (15–18). SMCHD1 binds to the D4Z4 repeat and regulates the epigenetic repression of the D4Z4 repeat (19). SMCHD1 mutations have been implicated in the aberrant DUX4 expression in FSHD2 and increased severity of FSHD1 (19,20), however the effect of SMCHD1 mutations on the transcriptome of small noncoding RNAs including known miRNAs has not been assessed genome-wide.
In this study, we used next generation small RNA sequencing and showed that several types of small ncRNAs including miRNAs are differentially expressed in FSHD2 muscle cells that carry SMCHD1 mutations. Most of the known myogenic miRNAs that were induced during differentiation in the control cells were also induced in FSHD cells. However, fewer small noncoding RNAs including miRNAs were differentially expressed between myoblasts (MB) and differentiated myotubes (MT) in FSHD2 samples compared to controls, suggesting that defects in in the robustness of differentiation and the expression of differentiation-specific small noncoding RNAs might be associated with FSHD muscle phenotype. Further, FSHD2-specific small RNA gene expression analysis revealed that a subset of known miRNAs seems to be affected by DUX4 misexpression in FSHD, whereas other types of small RNAs including several tRNAs and 5S rRNA might be upregulated specifically in FSHD2 due to SMCHD1 mutation.
Results
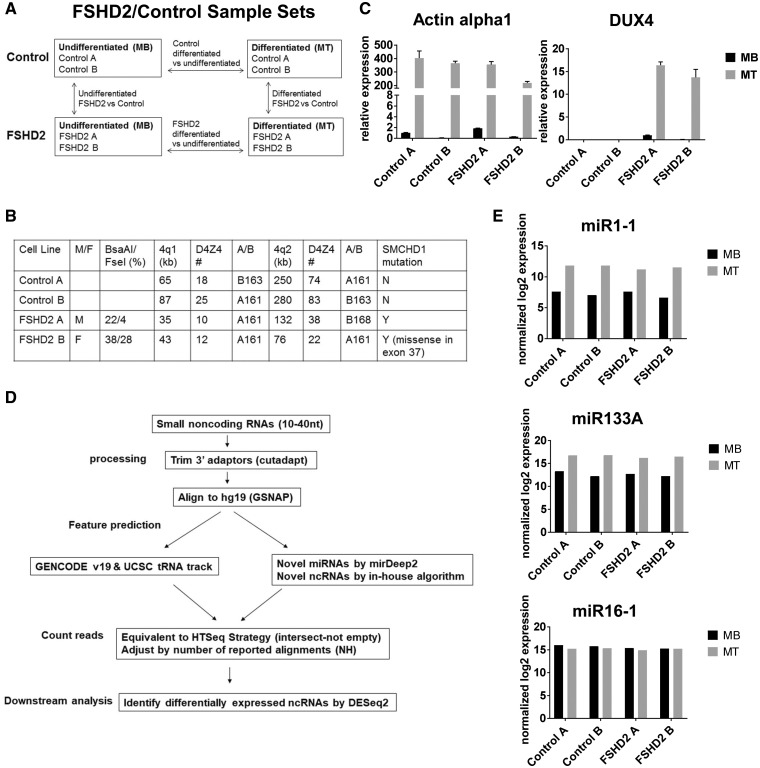
Study design for identifying FSHD2-related small RNAs
Previously several research groups identified miRNAs differentially expressed in FSHD1 muscle cells by using either microarray or small RNA-sequencing (9,10,12). However, there were only a small number of miRNAs that were identified as FSHD-specific in multiple studies and it is still unclear which miRNAs are closely involved in FSHD (Supplementary Material, Table S1). As previous reports mainly focused on miRNA expression changes in FSHD1, and epigenetic changes by SMCHD1 mutation may affect other types of small RNAs in FSHD2, we used next generation sequencing to identify small ncRNAs including miRNAs in two control (control A and control B) and two FSHD2 (FSHD2 A and FSHD2 B) cultured primary muscle cells as undifferentiated myoblasts (MB) and differentiated myotubes (MT) (Fig. 1A). We compared FSHD2 samples with control samples to identify small RNAs that were differentially expressed in FSHD2, and MT samples were compared to MB samples to identify small RNAs differentially expressed during muscle differentiation. FSHD2 muscle cells used in this study were characterized by DNA hypomethylation in the D4Z4 region, presence of SMCHD1 mutations, and a normal range of D4Z4 repeat size (Fig. 1B). Efficient muscle differentiation of each cell line was confirmed by myotube formation and expression of muscle differentiation markers, myogenin and Actin alpha 1 (Actin alpha 1 expression shown at the left panel of Fig. 1C). We also confirmed DUX4 expression in FSHD2 MB/MT samples but not in controls, consistent with the previous studies (the right panel of Fig. 1C).
Figure 1.
Study design, sample validation and data analysis. (A) Undifferentiated myoblasts (MB) and differentiated myotubes (MT) from two unaffected and two FSHD2 individuals were used to identify small RNAs differentially expressed in FSHD2 during muscle differentiation. (B) Control and FSHD2 muscle cells used in this study were characterized in terms of D4Z4 DNA methylation (Bsa A1/Fse1), D4Z4 repeat size and presence of SMCHD1 mutations. (C) qRT-PCR analysis of the control and FSHD2 MB and MT samples used for small RNA seq showed that the muscle differentiation marker, Actin alpha1, was induced in both control and FSHD2 MT samples and that DUX4 was expressed in FSHD2 samples but not in controls. Error bars in the graph show the SD of the mean for technical triplicates. (D) The flowchart depicting main steps of our small RNA sequencing data analysis. (E) Small RNA libraries were validated by comparing normalized log2 read counts for myogenic and ubiquitous miRNAs between MT and MB samples. Levels of expression of myogenic miRNAs (myomiRs), miR1–1 and miR133A were induced both in control and FSHD2 MT compared to MB, but expression of miR16–1, a ubiquitous miRNA, was not changed during muscle differentiation as expected.
The small RNA libraries for control and FSHD2 MB/MT samples were generated from total RNA, and cDNA fragments corresponding to 10–40 nt long small RNAs were captured for sequencing, generating 25∼30 million reads per sample. The flow chart in Figure 1D shows overall small RNA sequencing data analysis. After adapter sequences were trimmed using Cutadapt, sequence reads were aligned to hg19 using Genomic Short-read Nucleotide Alignment Program (GSNAP) allowing up to 2 mismatches (21). Finally, the aligned sequence reads were annotated against genome. Annotated gene features were obtained from the integrated GENCODE V19 and UCSC tRNA Table Browser. Additional features were predicted using miRDeep2 for novel miRNAs (22) based on miRBASE 19 and in-house algorithm for novel ncRNAs (see Materials and Methods for details). Note that our small RNA sequence reads when aligned to the D4Z4 region (AF117653) and then filtered to remove all the sequences aligned somewhere else in the genome, including multiple hits corresponding to simple repeats, did not reveal any previously reported unique D4Z4-derived small RNA transcripts (23,24), possibly due to low levels of expression in the whole cell lysate since most of the D4Z4 small RNAs were identified in the chromatin-associated fraction (23,24) and/or difficulties with the repetitive sequence alignment.
Genome-wide alignment and annotation of different types of small RNAs present in the small RNA libraries showed that miRNAs comprised 30–40% of the total small RNA reads with the major peaks of miRNAs at 20–25nt, rRNAs/tRNAs at 30–35nt and unknown (‘other’ or ‘protein coding’) small RNAs contributed to both 25 and 35nt peaks. As 5′ and 3′ adapters were added to small RNAs with 5′ phosphate and 3′ hydroxyl group (mostly miRNAs processed by Drosha/Dicer1) during library preparation process, small RNAs derived from tRNAs, rRNAs, snoRNAs with the same 5′ and 3′ group still could be processed for small RNA library construction (25–27). Differential expression of small RNAs in FSHD2 condition was analyzed using DESeq2 (28) (see Materials and Methods for details).
To validate our small RNA libraries and sequencing process, we compared normalized read numbers for myogenic microRNAs (myomiRs), miR1–1 and miR133a, and confirmed that the expression levels of these myomiRs were increased in both control and FSHD2 myotube (MT) samples, compared to myoblast (MB) samples. In contrast, expression of miR16, a ubiquitously and highly expressed miRNA (29,30), was not changed during muscle differentiation in both control and FSHD2 samples (Fig. 1E).
Identification of small RNAs that were differentially expressed between control and FSHD2 muscle cells (FSHD2 versus control)
Previously many miRNAs, including myogenic miRNAs (myomiRs), were shown to be dysregulated in FSHD1 muscle cells (9,10). To test whether small RNA expression is also affected in FSHD2, we first performed an unsupervised hierarchical clustering analysis of small RNA expression in four control samples (Control A MB/MT, Control B MB/MT) and four FSHD2 samples (FSHD2 A MB/MT, FSHD2 B MB/MT). This unbiased analysis revealed two main clusters corresponding to myoblast (MB) and myotube (MT) samples for all types of small RNAs (Fig. 2A), suggesting that the most prominent differences in small RNA expression among these samples are attributed to muscle differentiation. However, FSHD2 samples also grouped separately from the control samples within the major MB and MT clusters, suggesting FSHD2-specific small RNA expression changes.
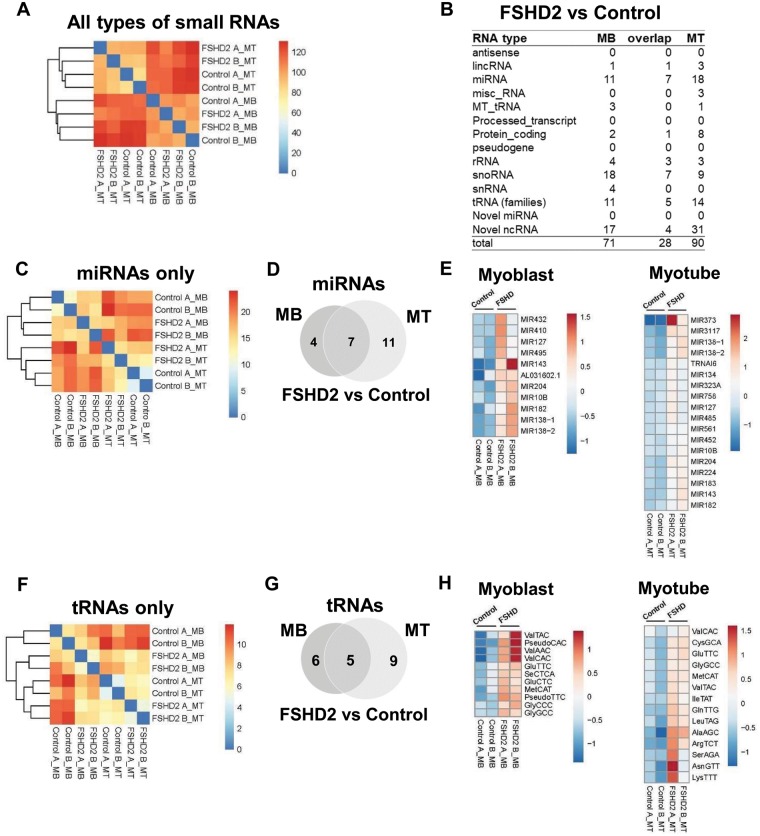
Figure 2.
Small RNAs differentially expressed in FSHD2 cells (FSHD2 versus Control). (A) Unbiased hierarchical analysis for all types of small RNAs revealed two major clusters corresponding to MB and MT samples with FSHD2 samples grouped separately from the control samples within the major MB and MT clusters. (B) Small RNAs differentially expressed in FSHD2 samples in comparison to controls identified by the DESeq2 analysis performed on all types of small RNAs (Padj < 0.05) (Supplementary Material, Table S2). miRNAs, tRNAs and novel noncoding RNAs (ncRNAs) were the major types of FSHD2-specific small RNAs. (C) Unbiased hierarchical clustering analysis for miRNAs only. (D and E) The Venn diagram and heatmaps show 11 known miRNAs differentially expressed in FSHD2 MB compared to controls and 18 known miRNAs differentially expressed in FSHD2 MT compared to controls. About 7 miRNAs were differentially expressed in both FSHD2 MB and MT. (F) Unbiased hierarchical clustering analysis for tRNAs only. (G and H) The Venn diagram and heatmaps show 11 tRNAs differentially expressed in FSHD2 MB compared to controls and 14 tRNAs—in FSHD2 MT compared to controls (5 FSHD2-specific tRNAs overlapped between MB and MT).
To identify small RNAs differentially expressed in FSHD2 myoblasts or myotubes compared to controls, we performed differential gene expression analysis using DESeq2 [adjusted P-value (Padj)<0.05]. miRNAs and tRNAs were two prominent types of small RNAs among differentially expressed small RNAs in FSHD2 muscle cells (Fig. 2B andSupplementary Material, Table S2). Unsupervised hierarchical clustering analysis exclusive for miRNAs revealed the same clustering pattern as for all types of small RNAs, with control and FSDH2 samples sub-clustering within the two major MB and MT clusters (Fig. 2C). Compared to control samples, 11 and 18 known miRNAs were differentially expressed in FSHD2 myoblasts and myotubes, respectively. About 7 miRNAs (miR10b, miR127, miR138–1, miR138–2, miR143, miR182 and miR204) showed elevated expression in both FSHD2 MB and MT (Fig. 2B, D and E). In contrast to the previous report by Dmitriev et al. (10), our data did not show increased expression of known myogenic miRNAs (myomiRs), such as miR-1, miR133A and miR206, in FSHD2 undifferentiated myoblasts; moreover most of these myomiRs were robustly upregulated during muscle differentiation in both control and FSHD2 cells as described below and in Figure 1E. In addition, we did not observe differential expression of miR411, a previously identified miRNA that was upregulated in FSHD myoblasts and when over-expressed in vitro repressed myogenic factors, such as MyoD and myogenin (31). These discrepancies between our results and previous reports might be due to different FSHD mutation types (FSHD2 versus FSHD1), different sample source and stages of muscle differentiation or different gene expression analysis (microarray versus RNA-seq) and data processing methods.
Our DESeq2 analysis was originally done on all small RNA gene features and the results for each subtype of small RNAs were presented separately (Fig. 2 and Supplementary Material, Table S2). To compare our data with previously published reports, we also performed the DESeq2 analysis on miRNAs only (miRBASE 19). This analysis revealed more known miRNAs that were significantly upregulated or downregulated in FSHD2 cells (Padj < 0.05): 26 miRNAs in FSHD2 MB compared to control MB and 28 miRNAs in FSHD2 MT compared to control MT, with 12 miRNAs (miR10b, miR129–1, miR129–2, miR134, miR138–1, miR138–2, miR143, miR145, miR182, miR204, miR3117, miR4421) that showed differential expression in both FSHD2 MB and MT (Supplementary Material, Fig. S1 and Table S3). Importantly, these included most of the FSHD2-specific miRNAs identified in the original small RNA DEseq2 analysis: 10 out of 11 miRNAs differentially expressed in FSHD2 MB and 13 out of 18 miRNAs differentially expressed in FSHD2 MT overlapped with the results of the miRNA only DEseq2 analysis (Fig. 2B/Supplementary Material, Table S2 versus Supplementary Material, Fig. S1/Supplementary Material, Table S3). The discrepancy between these two DESeq2 analyses might be partially due to using different reference gene annotations (GENCODE19 versus miRBASE19). Additional miRNAs that showed significant expression changes in FSHD2 cells included miR411, miR372 and miR432. Therefore, depending on the analysis, our FSHD2-specific miRNA lists share either two miRNAs (miRNAs from Supplementary Material, Table S2) or 10 miRNAs (Supplementary Material, Table S3) with the lists of miRNAs differentially expressed in FSHD shown by previous reports (summarized in Supplementary Material, Table S1).
Although unsupervised hierarchical clustering of tRNAs only again showed two major clusters corresponding to MB and MT (Fig. 2F), the DESeq2 analysis performed on all small RNAs identified several tRNAs differentially expressed in FSHD2 compared to controls (Fig. 2B, G and H). [Note that tRNA genes with the same anticodon (tRNA families) have almost identical mature tRNA sequences. Thus, we use the term ‘tRNA’ to refer to a family of tRNA genes that have the same anticodon]. About 11 and 14 tRNAs were upregulated in FSHD2 myoblasts and myotubes compared to control MB and MT, respectively (Fig. 2G and H). Five tRNAs (ValTAC, ValCAC, GluTTC, MetCAT and GlyGCC) were upregulated in both FSHD2 myoblasts and myotubes. snoRNAs were another prominent type of small ncRNAs that showed differential expressions in FSHD2 muscle cells. About 18 and 9 snoRNAs were upregulated in FSHD2 MB and MT compared to control MB and MT, respectively (Fig. 2B).
For small RNAs that did not align to any known features, we used miRDeep2 algorithm to predict novel miRNAs (22), and none of these novel miRNAs exhibited differential expression in FSHD2 relative to control samples (Fig. 2B). For small RNA reads that were not aligned to known features and did not meet criteria for novel miRNAs, we used an in-house algorithm (see Materials and Methods for more information) to predict potential novel small ncRNAs. Most of the novel ncRNAs differentially expressed in FSHD2 relative to controls were located in introns of encoding genes (7 out of 17 for MB and 24 out of 31 for MT) (Fig. 2B andSupplementary Material, Table S2), suggesting that these sequence reads might be generated from long nascent or noncoding transcripts spanning gene introns.
Our differential expression analysis for all small RNAs revealed various types of small RNAs, mostly miRNAs, tRNAs, and novel small ncRNAs that were differentially expressed between control and FSHD2 muscle cells (Fig. 2B). Since the FSHD2 primary muscle cells used in our study for small RNA sequencing originated from patients carrying SMCHD1 mutations and exhibiting aberrant DUX4 expression (Fig. 1B and C), the observed FSHD2-specific changes in small RNA expression may be due to both aberrant DUX4 expression and SMCHD1–related defects in chromatin compaction.
Identification of subsets of differentially expressed small RNAs affected by DUX4 overexpression (DUX4 signature)
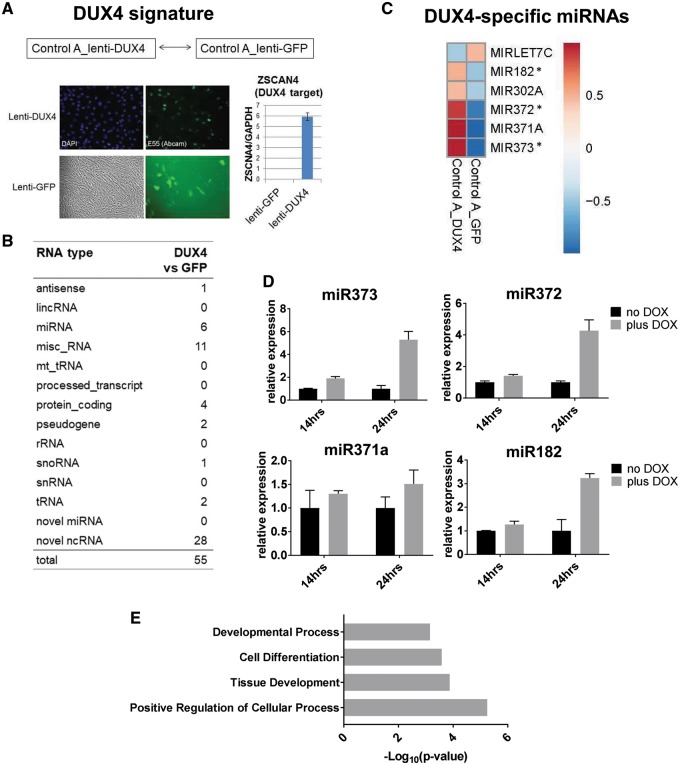
To test whether any of the observed FSHD2-specific changes in small RNA expression were due to aberrant DUX4 expression, we prepared two additional control samples for small RNA-seq: control myoblasts (Control A) transduced with the previously validated lentiviral vector constitutively expressing either DUX4 (lenti-DUX4) or GFP (lenti-GFP) as a negative control (6,32). Overexpression of the exogenous DUX4 in Control A MB transduced with lenti-DUX4 but not lenti-GFP was confirmed by measuring the expression of ZSCAN4, a DUX4 target gene (Fig. 3A). As P-values for differential gene expression analysis could not be calculated due to a small sample number and lack of replicates (DUX4 versus GFP), we used DESeq2 regularized-logarithm (rlog) function to compute the log transformation which allowed us to reduce large magnitude of fold-change for low-count features and to rank genes based on their differential expression (rlog2FC). In contrast to the normal log2FC expression analysis where the noisiest weak genes could appear at the top, the rlog2FC ranking would place at the top genes that are strongly upregulated in one sample compared to another. We selected differentially expressed small RNA genes with rlog2FC cutoff > 0.8. Our DESeq2 data analysis for these samples identified only a small number of small RNAs differentially expressed in control myoblasts transduced with DUX4 expressing lentivirus (rlog2FC >0.8) (Fig. 3B andSupplementary Material, Table S4), suggesting that only a subset of FSHD2-specific small RNAs are directly affected by aberrant DUX4 expression. Five known miRNAs (miR182, miR302A, miR371a, miR372 and miR373) were upregulated and one miRNA (let-7C) was downregulated in control MB transduced with DUX4 lentivirus compared to GFP control (Fig. 3C). Only three miRNAs, miR182, miR372 and miR373 overlapped with the miRNAs differentially expressed in FSHD MB or MT (Padj <0.05) (Fig. 2E, Supplementary Material, Fig. S1B and Tables S2 and S3). However, all five DUX4-specific miRNAs that were upregulated in lenti-DUX4 transduced control MB showed the same trend of expression, increased expression in FSHD2 MB/MT in comparison to control MB/MT (Note that two out of five miRNAs were not included in the FSHD2 differentially expressed miRNA lists due to the P-value cut-off, Supplementary Material, Table S4).
Figure 3.
Small RNAs affected by exogenous DUX4 expression in control myoblasts (DUX4 signature). (A) Control myoblasts (Control A) transduced with DUX4 or GFP expressing lentivirus previously described in (32) were generated to identify small RNAs affected by DUX4 overexpression. Immunofluorescence analysis shows expression of the exogenous DUX4 (E55 DUX4 antibody, Abcam) in lenti-DUX4 transduced control MB and live fluorescent image shows GFP expression in lenti-GFP transduced MB. qRT-PCR analysis shows that ZSCAN4, the DUX4 target gene, was induced in lenti-DUX4 transduced cells but not in lenti-GFP cells. (B) DESeq2 analysis of small RNAs expressed in lenti-DUX4 and lenti-GFP myoblasts revealed six known miRNAs differentially expressed in the DUX4-expressing myoblasts compared to GFP control (Supplementary Material, Table S4), whereas most of tRNAs that were induced in FSHD2 muscle cells did not show increased expression in DUX4-overexpressing myoblasts. (C) Heatmap for the six DUX4-specific miRNAs. Asterisks mark DUX4-specific miRNAs that were significantly upregulated in FSHD2 muscle cells compared to controls in DESeq2 analyses performed on all small RNAs (Fig. 2E and Supplementary Material, Table S2) or on miRNAs only (Supplementary Material, Fig. S1B, Fig. S1 and Table S3). (D) qRT-PCR analysis of the DUX4-specific miRNA expression in control myoblasts stably transduced with Doxycycline (DOX)-inducible DUX4 lentivirus previously described in (4). Error bars in the graph show the SD of the mean for real-time PCR technical triplicates. DUX4-specific miRNAs, miR373, miR372, miR371a and miR182, were induced in DOX-treated DUX4 expressing MB (plus DOX: 14 and 24 h) compared to the untreated MB (no DOX). (E) GO term analysis showed that positive regulation of cellular processes, developmental processes and cell differentiation were overrepresented GO categories for predicted DUX4-specific miRNAs target genes that were downregulated (on mRNA level) in DUX4-expressing myoblasts.
To confirm DUX4-specific miRNA expression, we used qRT-PCR to measure the levels of expression of mature miR371a, miR372, miR373 and miR182 in an independent DUX4 overexpression system, previously generated control myoblast line stably transduced with doxycycline (DOX)-inducible DUX4 lentivirus (4). This recently developed inducible DUX4 expression system has been extensively characterized and shown to accurately and reproducibly re-capitulate DUX4-associated transcriptional changes in FSHD muscle cells (4). Indeed, the DUX4-specific miRNAs were induced in DOX-treated samples (plus DOX: 14 and 24 h) compared to the untreated sample (no DOX), suggesting that DUX4 regulates expression of these miRNAs either directly or indirectly (Fig. 3D).
Predicted and/or validated target genes of DUX4-induced miRNAs
To study the potential function of DUX4-induced miRNAs and correlation with the aberrant DUX4 expression in FSHD2, we searched predicted target genes for the DUX4-specific miRNA using Targetscan (Targetscan.org; date last accessed May 16, 2018) (33). The predicted target genes for each DUX4-induced miRNA is listed Supplementary Material, Table S5. As miRNAs bind to the 3′-UTR of their target genes and decrease their mRNA level through deadenylation and/or RNA decay or the protein level through the translation inhibition, we analyzed our previously published mRNA-seq datasets for control myoblasts transduced with the DUX4-expressing lentivirus (exogenous DUX4) and FSHD myoblasts expressing endogenous DUX4 (4) to identify genes robustly downregulated in both exogenous and endogenous DUX4 expressing myoblasts. Then we compared 280 downregulated genes with the predicted target genes of the DUX4-induced miRNAs (miR371A, miR372, miR373, miR182 and miR302A). Interestingly, 58 out of 280 genes overlapped with the predicted target genes of the five DUX4-specific miRNAs (Supplementary Material, Table S5). Gene Ontology (GO) analysis of the 58 predicted miRNA target genes showing downregulated mRNA levels in DUX4-expressing myoblasts by using PATHER GO biological process (34) showed that they are involved in positive regulation of cellular process (35 genes), tissue development (19 genes), cell differentiation (27 genes) and developmental process (34 genes) (Fig. 3E andSupplementary Material, Table S6), suggesting that aberrant DUX4 expression may deregulate developmental process and differentiation through DUX4-induced miRNAs in FSHD.
Differentially expressed small RNAs affected by SMCHD1 mutation (SMCHD1 signature)
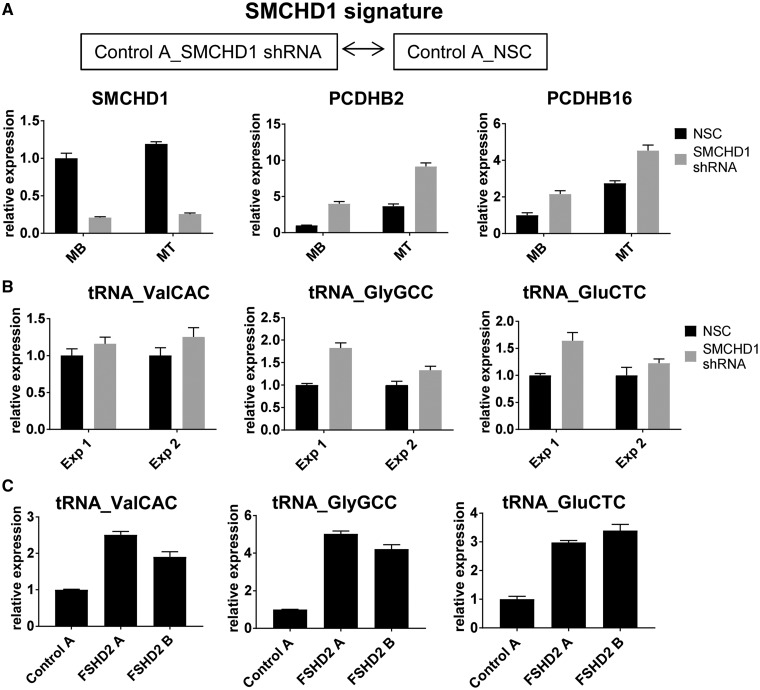
Most of tRNAs differentially expressed in FSHD2 MB and MT samples compared to controls (Fig. 2F) did not show significant changes in control myoblasts transduced with lenti-DUX4 (Fig. 3B), suggesting that differential expression of tRNAs might be due to epigenetic defects caused by SMCHD1 mutations. Interestingly, it has been shown that SMCHD1 binds to tRNA clusters located on chromosome 1 that are hypomethylated in FSHD2, and tRNAs within these clusters were also upregulated in FSHD2 muscle cells (35). To test whether SMCHD1 is involved in regulation of tRNA expression, we knocked down SMCHD1 in control muscle cells (Control A) using a previously validated shRNA targeting SMCHD1 (19). SMCHD1 depletion and upregulation of the known SMCHD1 target genes, such as protocadherin B2 (PCDHB2) and B16 (PCDHB16) (16,36), was confirmed by qRT-PCR (Fig. 4A). We tested the expression of three tRNAs (GlyGCC, GluCTC and ValCAC) that were found to be differentially expressed in our FSHD2 muscle cells (Fig. 2H, Supplementary Material, Table S2) and are located on chromosome 1. In two independent SMCHD1 knock down experiments, we observed increased levels of expression of these tRNAs in SMCHD1 depleted control muscle cells compared to non-silencing control (Fig. 4B), suggesting that differential expression of these tRNAs in FSHD2 may be due to SMCHD1 mutations. FSHD2-specific upregulation of these tRNAs was also validated by qRT-PCR analysis of control and FSHD2 myoblasts, consistent with our small RNA-seq data (Fig. 4C).
Figure 4.
Small RNAs affected by SMCHD1 knockdown in control muscle cells (SMCHD1 signature). (A) SMCHD1 was depleted in control (Control A) myoblasts (MB) and myotubes (MT) using a previously validated SMCHD1 shRNA (19) and a non-silencing control (NSC) shRNA (used as a negative control) in two independent experiments. qRT-PCR analysis of the SMCHD1-depleted cells confirmed SMCHD1 downregulation and induction of the SMCHD1 target genes, PCDHB6 and PCDHB16 shown for a single representative experiment. (B) qRT-PCR analysis of tRNA expression in two independent SMCHD1 knock down experiments (Exp 1 and Exp 2 presented as biological replicates). Three FSHD2-specific tRNAs (tRNA_ValCAC, tRNA_GlyGCC and tRNA_GluCTC) identified in our DESeq2 analysis (Fig. 2H and Supplementary Material, Table S2) were also induced in SMCHD1-depleted control myoblasts, suggesting that differential expression of tRNAs in FSHD2 may be due to SMCHD1 mutations. (C) FSHD2-specific upregulation of the tRNAs tested in (B) was confirmed by qRT-PCR analysis of control and FSHD2 muscle cells. Expression levels normalized to those of GAPDH in the same sample are presented for each experiment as mean ± SD for real-time PCR triplicates.
In addition to tRNAs, we observed increased expression of four 5S rRNA genes in FSHD2 muscle cells compared to controls (Fig. 2B andSupplementary Material, Table S2), and notably one of them (RNA5SP19: RNA 5S ribosomal Pseudogene 19) is located at the region of chromosome 1 (chr1: 228,743,045–228,782,516) found to be hypomethylated in FSHD2 samples (35).
Identification of small RNAs differentially expressed during muscle differentiation in control and FSHD2 cells (MT versus MB)
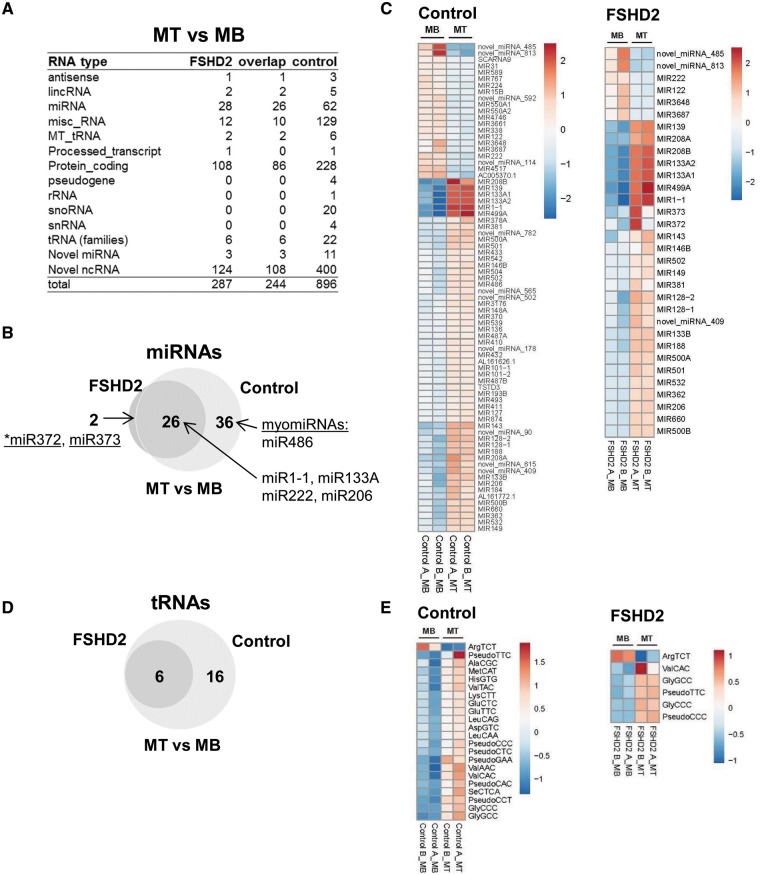
It has recently been reported that miRNA regulation is disrupted in FSHD with most changes affecting cell cycle and muscle-specific miRNAs (9,10). To determine small RNAs that are differentially expressed in FSHD2 cells during muscle differentiation, we compared myotube samples with the corresponding myoblast samples (control MT versus control MB and FSHD2 MT versus FSHD2 MB). Consistent with the previous report by Colangelo et al. (9), differential expression analysis (DESeq2) of all small RNAs in our MT samples compared to MB samples revealed that fewer known miRNAs were differentially expressed during muscle differentiation in FSHD2 cells (28 miRNAs) in comparison to controls (62 miRNAs) (Padj < 0.05) (Fig. 5A andSupplementary Material, Table S2). Out of 28 miRNAs differentially expressed in FSHD2 MT compared to MB, 26 overlapped with miRNAs differentially expressed in control MT compared to MB (Fig. 5B). These 26 miRNAs included only 9 out of the 14 previously characterized myogenic miRNAs (myomiRs) (37,38) that were differentially expressed during muscle differentiation in our control cells (miR1–1, miR31, miR101, miR128b, miR133b, miR206, miR222, miR362, miR432, miR486, miR500, miR502, miR550 and miR660) (Fig. 5B and C andSupplementary Material, Table S2). Consistent with our original DESeq2 analysis for all small RNAs, the DESeq2 analysis performed on miRNAs only also revealed fewer known miRNAs that were differentially expressed during muscle differentiation in FSHD2 muscle cells (32 miRNAs) in comparison to controls (134 miRNAs) (Padj <0.05) (Supplementary Material, Fig. S2 and Table S3), suggesting that the reduced expression of known miRNAs including myomiRs in FSHD2 myotubes could be related to muscle differentiation defects in FSHD.
Figure 5.
Small RNAs differentially expressed in control and FSHD2 cells during muscle differentiation (MT versus MB). (A) Small RNAs differentially expressed in control MT compared to MB and in FSHD2 MT compared to MB identified by the DESeq2 analysis performed on all types of small RNAs (Padj < 0.05) (Supplementary Material, Table S2). (B and C) The Venn diagram and heatmaps show miRNAs differentially expressed in FSHD2 MT compared to MB and control MT compared to MB. Although, 26 miRNAs including major myogenic miRNAs were differentially expressed during muscle differentiation (MT versus MB) in both control and FSHD2 samples, fewer miRNAs showed significant expression changes in FSHD2 samples compared to controls (28 versus 62 miRNAs), suggesting potential defects in the FSHD2 muscle differentiation program. In addition to the known miRNAs, the heatmaps in (C) also included novel miRNAs predicted by miRDeep2. (D and E) The Venn diagram and heatmaps show tRNAs differentially expressed in MT samples compared to MB samples. Similar to miRNAs, fewer tRNAs were differentially expressed during muscle differentiation (MT versus MB) in FSHD2 samples than in controls (6 versus 18 tRNAs).
Among known myomiRs, miR486 exhibited statistically significant upregulation of expression only in controls MT but not in FSHD2 MT (Fig. 5B and C andSupplementary Material, Table S2). miR486 has been shown to be directly regulated by MyoD, SRF and MKL1 as well as to accelerate muscle differentiation by downregulating Pax7 translation through binding to its 3′-UTR (37,39). The previous report on small RNA-seq analysis of FSHD1 muscle cells also showed that miR486 was not induced in FSHD myotubes (9). Interestingly, miR486 is downregulated in Duchenne muscular dystrophy (DMD) patient muscles and muscles of dystrophin-deficient mice (Dmdmdx-5Cv mice) and overexpression of miR486 improves muscle phenotypes in Dmdmdx-5Cv muscle (40). In addition, two miRNAs, miR372 and miR373, that were significantly upregulated during muscle differentiation in FSHD2 cells but not in controls (Fig. 5B and C) were also induced by DUX4 overexpression in control myoblasts (DUX4 signature) (Fig. 3C), suggesting that miR372 and miR373 might be upregulated in FSHD2 cells due to aberrant DUX4 expression.
In addition to known miRNAs, we identified 11 novel miRNAs (predicted by miRDeep2, see Materials and Methods section for details) that were differentially expressed during muscle differentiation. Three novel miRNAs were differentially expressed in both control and FSHD2 cells and eight novel miRNAs were differentially expressed only in control samples (Fig. 5A and C). All of the 11 novel miRNAs were located either in exons or introns of known genes (Supplementary Material, Table S2). One of these miRNAs, novel_miRNA_485, that was downregulated during muscle differentiation in both control and FSHD2 samples, is identical to miR7974 listed at miRBASE v21 as a miRNA with unknown function. As we used GENCODE V19 and miRBASE v19 to predict features for our sequence reads, information about newly registered miRNAs, like miR7974, was missing in our data analysis. Interestingly, another identified novel_miRNA_565 is located in the Ankyrin1 intron and processed from its antisense transcript, whereas myomiR miR486 is expressed from the sense transcript at the same location, similar to the relation between iab-4 and iab-8 miRNAs in Drosophila (41). Both miR486 and novel_miRNA_565 were upregulated during muscle differentiation only in control samples, suggesting their potential involvement in muscle differentiation defects caused by FSHD condition.
In general, there were fewer small noncoding RNAs differentially expressed during muscle differentiation in FSHD2 cells in comparison to controls (Fig. 5A andSupplementary Material, Table S2). About 22 tRNAs, for example, were differentially expressed in control MT samples compared to MB and only 6 out of these 22 tRNAs were differentially expressed in FSHD2 MT compared to MB (Fig. 5D and E).
Discussion
It is important to identify dysregulated gene expression in a disease condition in order to understand the mechanisms by which the disease occurs as well as to develop efficient therapeutic interventions. Previous studies suggested a correlation between changes in known miRNA expression and muscle diseases, including FSHD. In this study, in addition to known miRNAs, we identified other types of small ncRNAs that were differentially expressed in FSHD2 muscle cells. Interestingly, only few of our FSHD2-related miRNAs were identified by previous studies (see Supplementary Material, Table S1). The discrepancies among FSHD miRNA studies might be due to usage of different cell sources (e.g. FSHD1 versus FSHD2) and/or different experimental methods/data analyses. Although, most myogenic miRNAs were still induced in differentiated FSHD2 muscle cells, many small ncRNAs showed overall damped expression level in FSHD2 muscle compared to control, suggesting a potential connection with muscle differentiation defects in FSHD2. Our study also revealed that some expression changes in small RNAs in FSHD2 muscle might be connected with either aberrant DUX4 expression or mutations in SMCHD1.
We found different types of small noncoding RNAs differentially expressed during muscle differentiation including known myogenic miRNAs (myomiRs). We did not observe dysregulation of major myomiRs such as miR-1–1, miR-133a and miR-206 in FSHD2 muscle cells during differentiation. This result is consistent with the recent report that shows the expression pattern of myomiRs was not changed in FSHD1 human fetal muscle biopsies compared to control (12). However, compared to control cells, many myotube-related miRNAs did not show differential expression during muscle differentiation in FSHD2. In future studies, it will be worth testing whether modulation of these miRNAs might affect the muscle differentiation program in control muscle and contribute to muscle differentiation defects in FSHD.
We also tested which small RNAs were affected by DUX4 overexpression (recapitulating aberrant DUX4 expression in FSHD1 and FSHD2 muscles) or SMCHD1 depletion (recapitulating SMCHD1 mutations in FSHD2 muscles) in control muscle cells. We identified a group of miRNAs induced by DUX4 overexpression. Interestingly, consistent with previous reports on DUX4 expression in germ lines, human embryonic stem cells (hESCs), and induced pluripotent cells (iPSCs) (24,42), most DUX4-induced miRNAs (miR-302–367 and miR-371–373 clusters) are hESC-specific miRNAs that are highly expressed in hESCs and downregulated during differentiation (43). These ESC-specific miRNA clusters have been shown to maintain rapid proliferation, to regulate pluripotency and to promote iPSC reprogramming process (44–49). DUX4 might have roles in regulation of pluripotency through DUX4-specific microRNAs in early development. Misexpression of these miRNAs by DUX4 in FSHD muscle might cause defects in muscle differentiation program and/or cell cytotoxicity.
Our data also suggested that induced tRNA and 5S rRNA expression in FSHD2 might be mainly due to loss of SMCHD1. Indeed, hypomethylation of the tRNA and 5S rRNA clusters located on chromosome 1 has been shown in FSHD2 muscle cells (35), and these clusters overlapped with the tRNAs and 5S rRNAs differentially expressed in our FSHD2 samples. However, a number of the differentially expressed FSHD2-specific small RNAs, mostly novel small RNAs, showed differential expression either in both MB and MT (28 out of 133) or in MT only (62 out of 133) (Fig. 2B), which is unlikely to be related to SMCHD1 mutations since the SMCHD1 protein levels are known to be decreased during muscle differentiation (50).
Our data analysis showed that expression of several tRNAs were induced in FSHD2 muscle cells compared to controls. Considering our small RNA reads are up to 40 bp long and the size of mature tRNAs are 76 ∼ 90 nucleotides, our small RNA seq data suggest potential expression of tRNA-derived small RNAs (tRNA fragments) in FSHD2 muscle. A previous extensive analysis of tRNA derived fragments revealed that they represent a distinct evolutionarily conserved class of smallnoncoding RNAs that, similar to miRNAs, associate with AGO proteins and recognize specific RNA targets (51). Recent studies demonstrated tRNA fragments were implicated in cancers and neurodegenerative disease, but involvement of these small noncoding RNAs in FSHD pathology has not been studied (52,53). Like tRNA fragments, several small nucleolar RNAs (snoRNAs) were shown to be induced in FSHD2 muscle cells, suggesting that these small RNA reads might be potential snoRNA-derived RNAs (sdRNAs). All snoRNAs differentially expressed in FSHD2 muscle cells were from two clusters of snoRNA genes (SNORD113 with 9 copies and SNORD114 with 31 copies) located in the imprinted domains in 14q32 locus (54). This region was not defined as one of hypomethylated loci in FSHD2 muscle cells regulated by SMCHD1 and differential expression of these snoRNAs in FSHD2 condition has not been studied (35), however, snoRNA-derived small RNAs, such as sno-miRNAs, might also have a role in gene regulation (55).
Thus, our current study demonstrated the expression changes of several types of small ncRNAs as well as known miRNAs in FSHD2 muscle cells with SMCHD1 mutations. Most myogenic miRNAs were still differentially expressed in differentiated FSHD2 muscle cells. However, many small ncRNAs showed overall dampened expression in FSHD2 muscle compared to control, suggesting a potential connection with muscle differentiation defects in FSHD2. Further, some expression changes were associated with either aberrant DUX4 expression or SMCHD1 mutation, suggesting distinct signatures in FSHD2 and FSHD1 small ncRNA transcriptomes.
Materials and Methods
Myoblast cultures
The Fred Hutchinson Cancer Research Center approved the use of de-identified human cells for this study. Primary myoblasts from two unaffected (NR135 designated as Control A and NR209 as Control B) and two FSHD2 (2062 designated as FSHD2 A and 2305 as FSHD2 B) individuals were obtained through the Fields Center at the University of Rochester (http://www.urmc.rochester.edu/fields%2Dcenter/; date last accessed May 16, 2018). The myoblasts were derived from a needle biopsy of the vastus lateralis (http://www.urmc.rochester.edu/fields-center/protocols/needle-muscle-biopsy.cfm; date last accessed May 16, 2018) and established as primary cultures through dispase and collagenase dispersion (http://www.urmc.rochester.edu/fields-center/protocols/myoblast-cell-cultures.cfm; date last accessed May 16, 2018). Primary myoblast cell lines were routinely maintained in F-10 Medium (Invitrogen) supplemented with 20% fetal bovine serum (Atlanta Biologicals, GA), 1% Pen/Strep, 10 ng/ml hrFGF (Promega) and 1μM Dexamethasone (Sigma) (Growth media). Myoblasts were induced to differentiation to myotubes with DMEM, 1% heat-inactivated horse serum, 10 μg/ml insulin (Sigma) and 10 μg/ml transferrin (Sigma) (Differentiation media) for 48 h. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2.
Lentiviral transduction of control myoblasts
DUX4- or GFP-expressing lentivirus previously described in (32) or lentivirus (pGIPZ) with the previously validated SMCHD1 shRNA (#3881) (19) were generated by transfection of the appropriate lentiviral vector into 293T cells, along with the packaging and envelope plasmids pMD2.G and psPAX2 using lipofectamine 3000 reagent (ThermoFisher). Control myoblasts (Control A) were transduced with the DUX4 or GFP lentivirus in the presence of polybrene for 24 h. Generation of the control myoblasts (Control 1) stably transduced with the doxycycline (DOX)-inducible DUX4 lentivirus was previously described in (4). The stably transduced cells were treated with 1μg/ml DOX for 14 and 24 h before RNA extraction.
To knock down SMCHD1, control myoblasts (Control A) were transduced with SMCHD1 shRNA lentivirus for 24 h, followed by puromycin selection (2 μg/ml) for a week before harvesting RNA.
RNA isolation
Total RNA from myoblasts (MB) and myotubes (MT) from two control and two FSHD2 individuals was isolated using TRIZOL (Invitrogen) according to the manufacturer’s instructions. The isolated RNA was treated with DNase twice to remove contaminating DNA prior cDNA generation. First, isolated RNA was treated with RNase-free DNaseI (Sigma-Aldrich, #04716728001 ROCHE) followed by acidic phenol/chloroform extraction, and then treated with TURBO RNase-free DNase (Ambion) followed by inactivation reagent treatment. The DNaseI-treated RNA was tested for genomic DNA contamination by PCR and RNA quality by RNA gel. Please note that a recent study on isolation of small noncoding RNAs reported that a number of miRNAs with low GC content can be selectively lost when extracted from a small number of cells using TRIZOL (56). To avoid this bias, our primary myoblasts (MB) were cultured to 70–80% confluence prior to RNA extraction and to 90–100% confluence prior to induction to differentiation to myotubes (MT). Indeed, most of the miRNAs reported sensitive to cell density during TRIZOL extraction, including miR-301a, miR-106b, miR-34a, miR-29b, miR-21 and miR-15a (56), showed robust expression levels in our MB and MT samples from control and FSHD2 individuals (Supplementary Material, Table S3).
RNA processing for high-throughput sequencing: small RNA library preparation
To ensure high standard and consistency in small RNA libraries preparation, the eight total RNA preps from FSHD2 and control MB and MT samples were further processed at the FHCRC Genomics Core for quality control (QC) analysis and small RNA libraries preparation using Illumina TruSeq small RNA protocol. Briefly, the RNA samples were QC analyzed and small RNA-specific 3′-adapter and 5′-adapter linker sequences were ligated to RNA samples. RNA was then reverse transcribed (RT) to create single stranded cDNA, followed by cDNA amplification with a common primer corresponding to the 5′-adapter sequence and a 3′-adapter primer containing a 6-nt unique index (barcode) sequence to allow multiplex sequencing. Eight small RNA libraries (each labeled with a unique ‘barcode’ sequence) were pooled for size selection, and 130–160 bp cDNA fragments corresponding to 10–40 nt long small RNAs were captured and sequenced in 2 lanes of a 50-cycle single read run using the Illumina HiSeq 2000 system with expected 25–35 million sequence reads per library.
The data is available through GEO accession number GSE113133.
cDNA generation
cDNA was generated with Superscript II (Invitrogen) under the following conditions. About 1 μl of Oligo-dT primer or 250ng of random primer (Invitrogen) was added to 1 μg of RNA and incubated at 65°C for 5 min in a thermocycler. The RNA/primer mixture was chilled on ice. Next, the reaction mixture, nuclease free H2O, 5× buffer, RNaseOut (Invitrogen), dNTPs, DTT and RT was added to the RNA/primer mixture and incubated for 1 h at 45°C and for 10 min at 50°C. The RT was inactivated at 70°C for 15 min. Synthesis of cDNA was verified and normalized by amplification of the control transcripts GAPDH. no-RT reaction was used as a control for DNA contamination and primers were verified by amplification of cDNA.
Quantitative RT-PCR
Quantification of mRNA or tRNA levels in control and FSHD2 myoblasts and myotubes was carried out by qRT-PCR on the automated ABI 7900 PCR machine (Applied Biosystems) using iTaq Univer SYBR Green (Bio-Rad) with ROX passive reference dye added. cDNA was generated with OligodT or RNA random primers using 1 µg RNA, then diluted 1:1 with distilled water. One microliter of cDNA was used as template for the real-time reaction. PCR cycling was performed at [94°/2 min, (94°/30 s, 60°/30 s, 72°/30 s) ×40] with an additional ramping step added after cycling to calculate dissociation curves and confirm that fluorescence detected was due to full size PCR product and not PCR artifacts. The standard curve assay as described by Applied Biosystems was used for absolute quantification or delta-delta-Ct method was used for relative expression to reference gene. The values calculated for transcripts levels were normalized to those of GAPDH in the same samples. The primers used for real time RT-PCR are listed in Supplementary Material, Table S7. Expression data for each representative experiment were presented as mean ± SD for real-time PCR triplicates.
qRT-PCR analysis of microRNA expression
Quantification of miRNA levels in control myoblasts stably transduced with the DOX-inducible DUX4 lentivirus (4) was carried out by qPT-PCR by following the protocol developed by Dr. Busk (57,58). qPCR primers for DUX4-specific miRNAs were designed to amplify mature miRNA sequences using miRprimer program (see PCR primers in Supplementary Material, Table S7). Briefly, cDNA samples were generated with specific RT primers with tag sequence (5′-CAGGTCCAGTTTTTTTTTTTTTTTVN, where V is A, C and G and N is A, C, G and T) in the presence of poly(A) polymerase (New England Biolabs). Then, qPCR was carried out as described earlier with specific miRNA primers designed by miRprimer program (https://sourceforge.net/projects/mirprimer/; date last accessed May 16, 2018). Depending on the alignment of small RNA-seq reads of miRNA genes, we selected either 5p- (5′ side of pre-miRNA/hairpin) or 3p- (3′ side of pre-miRNA/hairpin) sequences of mature miRNAs for primer design. Please note that out of the five DUX4-specific miRNAs (182, 302a, 371a, 372 and 373), we were unable to design primers to 302a that passed QC. miRNA expression levels were normalized to expression levels of 7SL scRNA (59) measured in the same sample using primers listed in Supplementary Material, Table S7. mRNA expression data were presented as mean ± SD for real-time PCR triplicates.
Sequencing data analysis
Bioinformatics pipeline
Each of our small RNA libraries consisted of ∼ 25–35 million reads and the reads length after trimming the adapters ranged from 10 to 40 nt. The pipeline for preparing data for downstream analysis included three major steps: preprocessing, feature annotation/prediction and counting reads in features.
For preprocessing, sequence reads were clustered, de-convoluted, trimmed from 3′ prime adapters and assessed for quality control and size distribution; finally, the reads were aligned to the human reference genome (hg19) using Genomic Short-read Nucleotide Alignment Program (GSNAP, version 2014-12-29) (21) allowing up to 2 mismatches per read, maximum mismatch score was set to 0.07. To mitigate the effects of PCR amplification bias introduced during sample preparation, only one copy of any duplicated read was retained for further analysis.
The genomic features of interest were collected from the following sources. The aligned reads were annotated against the integrated GENCODE V19 and UCSC tRNA track. The miRDeep2 program (22) was used to predict novel miRNAs based on miRBASE 19. Only novel miRNAs that were not already annotated by GENCODE V19 were kept. Subsequently, the aligned reads that were not mapped to the annotated features and novel miRNAs were collected and we used an in-house algorithm to predict novel non-coding RNAs. Using these reads, we clustered neighbors of reads within 200 bp to form novel ncRNA. Overall, our list of genomic features was a collection of annotated and predicted coding and noncoding RNAs, including known miRNAs, tRNAs, novel miRNAs and novel non-coding RNAs.
The challenge of the counting of reads in our study was that there were many overlapping regions between noncoding RNAs and exons of protein-coding features. The reads that mapped to these regions were kept, because they were likely originated from noncoding RNAs. Therefore, the precedence was given to the noncoding features, and the reads mapped to the disjoint regions of the noncoding features were assigned counts first. Then the remaining reads that were not mapped to the noncoding features were assigned to the protein-coding features. The assigned counts were adjusted by the number of reported multiple alignments of each read.
Downstream analysis
The DESeq2 package (28) was used to perform differential analysis comparing expression between two biological conditions (Supplementary Material, Tables S2–S4). In the case when replicates were not available, the rlog function from DESeq2 was used to compute the log transformation which allowed us to reduce large magnitude of fold-change for low-count features (Supplementary Material, Table S4).
Software
The counting hits algorithm was adapted and modified from the summarizeOverlaps function from the Bioconductor Genomic Alignments package. The Bioconductor 3.2 packages (compatible to R 3.2) was used for RNA-seq-related data analysis and mirDeep2 (22) was used for novel miRNA prediction.
miRNA target prediction and gene ontology analysis
Predicted target genes for five DUX4-specific miRNAs were searched from targetscan database (www.targetscan.org; date last accessed May 16, 2018) and only putative target genes of broadly conserved miRNAs were selected for further analysis (Supplementary Material, Table S5). GO category analysis for predicted target genes was conducted using the PANTHER classification system (http://pantherdb.org/geneListAnalysis.do; date last accessed May 16, 2018) using the statistical overrepresentation test against all human genes, using the complete GO biological process annotation (Supplementary Material, Table S6).
Supplementary Material
Supplementary Material is available at HMG online.
Funding
This work was supported by NINDS P01NS069539 (S.J.T., G.N.F., S.M.v.d.M., D.G.M., R.T.), NIAMS R56 AR070778 (D.G.M.) and Friends of FSH Research (J.W.L., G.N.F., D.G.M., S.J.T.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Conflict of Interest statement. None declared.
Supplementary Material
References
- 1. Tawil R., van der Maarel S.M., Tapscott S.J. (2014) Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skelet. Muscle, 4, 12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Daxinger L., Tapscott S.J., van der Maarel S.M. (2015) Genetic and epigenetic contributors to FSHD. Curr. Opin. Genet. Dev., 33, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van der Maarel S.M., Miller D.G., Tawil R., Filippova G.N., Tapscott S.J. (2012) Facioscapulohumeral muscular dystrophy: consequences of chromatin relaxation. Curr. Opin. Neurol., 25, 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jagannathan S., Shadle S.C., Resnick R., Snider L., Tawil R.N., van der Maarel S.M., Bradley R.K., Tapscott S.J. (2016) Model systems of DUX4 expression recapitulate the transcriptional profile of FSHD cells. Hum. Mol. Genet., 25, 4419–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rickard A.M., Petek L.M., Miller D.G. (2015) Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet., 24, 5901–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yao Z., Snider L., Balog J., Lemmers R.J., Van Der Maarel S.M., Tawil R., Tapscott S.J. (2014) DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet., 23, 5342–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ha M., Kim V.N. (2014) Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol., 15, 509–524. [DOI] [PubMed] [Google Scholar]
- 8. Jonas S., Izaurralde E. (2015) Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet., 16, 421–433. [DOI] [PubMed] [Google Scholar]
- 9. Colangelo V., Francois S., Solda G., Picco R., Roma F., Ginelli E., Meneveri R. (2014) Next-generation sequencing analysis of miRNA expression in control and FSHD myogenesis. PLoS One, 9, e108411.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dmitriev P., Stankevicins L., Ansseau E., Petrov A., Barat A., Dessen P., Robert T., Turki A., Lazar V., Labourer E.. et al. (2013) Defective regulation of microRNA target genes in myoblasts from facioscapulohumeral dystrophy patients. J. Biol. Chem., 288, 34989–35002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eisenberg I., Eran A., Nishino I., Moggio M., Lamperti C., Amato A.A., Lidov H.G., Kang P.B., North K.N., Mitrani-Rosenbaum S.. et al. (2007) Distinctive patterns of microRNA expression in primary muscular disorders. Proc. Natl. Acad. Sci. U. S. A., 104, 17016–17021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Portilho D.M., Alves M.R., Kratassiouk G., Roche S., Magdinier F., de Santana E.C., Polesskaya A., Harel-Bellan A., Mouly V., Savino W.. et al. (2015) miRNA expression in control and FSHD fetal human muscle biopsies. PLoS One, 10, e0116853.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martens-Uzunova E.S., Olvedy M., Jenster G. (2013) Beyond microRNA–novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett., 340, 201–211. [DOI] [PubMed] [Google Scholar]
- 14. Ashe A., Morgan D.K., Whitelaw N.C., Bruxner T.J., Vickaryous N.K., Cox L.L., Butterfield N.C., Wicking C., Blewitt M.E., Wilkins S.J.. et al. (2008) A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development. Genome Biol., 9, R182.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blewitt M.E., Gendrel A.V., Pang Z., Sparrow D.B., Whitelaw N., Craig J.M., Apedaile A., Hilton D.J., Dunwoodie S.L., Brockdorff N.. et al. (2008) SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet., 40, 663–669. [DOI] [PubMed] [Google Scholar]
- 16. Gendrel A.V., Tang Y.A., Suzuki M., Godwin J., Nesterova T.B., Greally J.M., Heard E., Brockdorff N. (2013) Epigenetic functions of smchd1 repress gene clusters on the inactive X chromosome and on autosomes. Mol. Cell. Biol., 33, 3150–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mould A.W., Pang Z., Pakusch M., Tonks I.D., Stark M., Carrie D., Mukhopadhyay P., Seidel A., Ellis J.J., Deakin J.. et al. (2013) Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation. Epigenetics Chromatin., 6, 19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nozawa R.S., Nagao K., Igami K.T., Shibata S., Shirai N., Nozaki N., Sado T., Kimura H., Obuse C. (2013) Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat. Struct. Mol. Biol., 20, 566–573. [DOI] [PubMed] [Google Scholar]
- 19. Lemmers R.J., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R.. et al. (2012) Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet., 44, 1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sacconi S., Lemmers R.J., Balog J., van der Vliet P.J., Lahaut P., van Nieuwenhuizen M.P., Straasheijm K.R., Debipersad R.D., Vos-Versteeg M., Salviati L.. et al. (2013) The FSHD2 gene SMCHD1 is a modifier of disease severity in families affected by FSHD1. Am. J. Hum. Genet., 93, 744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu T.D., Nacu S. (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics, 26, 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Friedlander M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. (2012) miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res., 40, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lim J.W., Snider L., Yao Z., Tawil R., Van Der Maarel S.M., Rigo F., Bennett C.F., Filippova G.N., Tapscott S.J. (2015) DICER/AGO-dependent epigenetic silencing of D4Z4 repeats enhanced by exogenous siRNA suggests mechanisms and therapies for FSHD. Hum. Mol. Genet., 24, 4817–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Snider L., Asawachaicharn A., Tyler A.E., Geng L.N., Petek L.M., Maves L., Miller D.G., Lemmers R.J., Winokur S.T., Tawil R.. et al. (2009) RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet., 18, 2414–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Falaleeva M., Stamm S. (2013) Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. Bioessays, 35, 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Head S.R., Komori H.K., LaMere S.A., Whisenant T., Van Nieuwerburgh F., Salomon D.R., Ordoukhanian P. (2014) Library construction for next-generation sequencing: overviews and challenges. Biotechniques, 56, 61–64, 66, 68, passim. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rother S., Meister G. (2011) Small RNAs derived from longer non-coding RNAs. Biochimie, 93, 1905–1915. [DOI] [PubMed] [Google Scholar]
- 28. Love M.I., Huber W., Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Das M.K., Andreassen R., Haugen T.B., Furu K. (2016) Identification of endogenous controls for use in miRNA quantification in human cancer cell lines. Cancer Genomics Proteomics, 13, 63–68. [PubMed] [Google Scholar]
- 30. Torres A., Torres K., Wdowiak P., Paszkowski T., Maciejewski R. (2013) Selection and validation of endogenous controls for microRNA expression studies in endometrioid endometrial cancer tissues. Gynecol. Oncol., 130, 588–594. [DOI] [PubMed] [Google Scholar]
- 31. Harafuji N., Schneiderat P., Walter M.C., Chen Y.W. (2013) miR-411 is up-regulated in FSHD myoblasts and suppresses myogenic factors. Orphanet. J. Rare Dis., 8, 55.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R.. et al. (2012) DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell, 22, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Agarwal V., Bell G.W., Nam J.W., Bartel D.P. (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife, 4, doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mi H., Poudel S., Muruganujan A., Casagrande J.T., Thomas P.D. (2016) PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res., 44, D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mason A.G., Slieker R.C., Balog J., Lemmers R., Wong C.J., Yao Z., Lim J.W., Filippova G.N., Ne E., Tawil R.. et al. (2017) SMCHD1 regulates a limited set of gene clusters on autosomal chromosomes. Skelet. Muscle, 7, 12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen K., Hu J., Moore D.L., Liu R., Kessans S.A., Breslin K., Lucet I.S., Keniry A., Leong H.S., Parish C.L.. et al. (2015) Genome-wide binding and mechanistic analyses of Smchd1-mediated epigenetic regulation. Proc. Natl. Acad. Sci. U. S. A., 112, E3535–E3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dey B.K., Gagan J., Dutta A. (2011) miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol. Cell. Biol., 31, 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dmitriev P., Barat A., Polesskaya A., O’Connell M.J., Robert T., Dessen P., Walsh T.A., Lazar V., Turki A., Carnac G.. et al. (2013) Simultaneous miRNA and mRNA transcriptome profiling of human myoblasts reveals a novel set of myogenic differentiation-associated miRNAs and their target genes. BMC Genomics, 14, 265.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Small E.M., O’Rourke J.R., Moresi V., Sutherland L.B., McAnally J., Gerard R.D., Richardson J.A., Olson E.N. (2010) Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. U. S. A., 107, 4218–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alexander M.S., Casar J.C., Motohashi N., Vieira N.M., Eisenberg I., Marshall J.L., Gasperini M.J., Lek A., Myers J.A., Estrella E.A.. et al. (2014) MicroRNA-486-dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy-associated symptoms. J. Clin. Invest., 124, 2651–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tyler D.M., Okamura K., Chung W.J., Hagen J.W., Berezikov E., Hannon G.J., Lai E.C. (2008) Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev., 22, 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J.. et al. (2010) Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet., 6, e1001181.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lakshmipathy U., Love B., Goff L.A., Jornsten R., Graichen R., Hart R.P., Chesnut J.D. (2007) MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev., 16, 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Anokye-Danso F., Trivedi C.M., Juhr D., Gupta M., Cui Z., Tian Y., Zhang Y., Yang W., Gruber P.J., Epstein J.A.. et al. (2011) Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell, 8, 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Houbaviy H.B., Murray M.F., Sharp P.A. (2003) Embryonic stem cell-specific MicroRNAs. Dev. Cell, 5, 351–358. [DOI] [PubMed] [Google Scholar]
- 46. Langroudi L., Jamshidi-Adegani F., Shafiee A., Rad S.M., Keramati F., Azadmanesh K., Arefian E., Soleimani M. (2015) MiR-371-373 cluster acts as a tumor-suppressor-miR and promotes cell cycle arrest in unrestricted somatic stem cells. Tumour Biol., 36, 7765–7774. [DOI] [PubMed] [Google Scholar]
- 47. Subramanyam D., Lamouille S., Judson R.L., Liu J.Y., Bucay N., Derynck R., Blelloch R. (2011) Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol., 29, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suh M.R., Lee Y., Kim J.Y., Kim S.K., Moon S.H., Lee J.Y., Cha K.Y., Chung H.M., Yoon H.S., Moon S.Y.. et al. (2004) Human embryonic stem cells express a unique set of microRNAs. Dev. Biol., 270, 488–498. [DOI] [PubMed] [Google Scholar]
- 49. Zhang Z., Hong Y., Xiang D., Zhu P., Wu E., Li W., Mosenson J., Wu W.S. (2015) MicroRNA-302/367 cluster governs hESC self-renewal by dually regulating cell cycle and apoptosis pathways. Stem Cell Rep., 4, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Balog J., Thijssen P.E., Shadle S., Straasheijm K.R., van der Vliet P.J., Krom Y.D., van den Boogaard M.L., de Jong A., F Lemmers R.J.L., Tawil R.. et al. (2015) Increased DUX4 expression during muscle differentiation correlates with decreased SMCHD1 protein levels at D4Z4. Epigenetics, 10, 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kumar P., Anaya J., Mudunuri S.B., Dutta A. (2014) Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol., 12, 78.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kumar P., Kuscu C., Dutta A. (2016) Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem. Sci., 41, 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soares A.R., Santos M. (2017) Discovery and function of transfer RNA-derived fragments and their role in disease. Wiley Interdiscip. Rev. RNA, doi: 10.1002/wrna.1423. [DOI] [PubMed] [Google Scholar]
- 54. Cavaille J., Seitz H., Paulsen M., Ferguson-Smith A.C., Bachellerie J.P. (2002) Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region. Hum. Mol. Genet., 11, 1527–1538. [DOI] [PubMed] [Google Scholar]
- 55. Brameier M., Herwig A., Reinhardt R., Walter L., Gruber J. (2011) Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res., 39, 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim Y.K., Yeo J., Kim B., Ha M., Kim V.N. (2012) Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol. Cell, 46, 893–895. [DOI] [PubMed] [Google Scholar]
- 57. Busk P.K. (2014) A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinformatics, 15, 29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cirera S., Busk P.K. (2014) Quantification of miRNAs by a simple and specific qPCR method. Methods Mol. Biol., 1182, 73–81. [DOI] [PubMed] [Google Scholar]
- 59. Galiveti C.R., Rozhdestvensky T.S., Brosius J., Lehrach H., Konthur Z. (2010) Application of housekeeping npcRNAs for quantitative expression analysis of human transcriptome by real-time PCR. RNA, 16, 450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.