Abstract
Both exfoliation glaucoma (XFG) and primary open-angle glaucoma (POAG) have been linked to decreased conventional outflow of aqueous humor (AH). To better understand the molecular changes in the AH content under such conditions, we analyzed the miRNA profiles of AH samples from patients with POAG and XFG compared to non-glaucoma controls. Individual AH samples (n = 76) were collected from POAG and XFG patients and age-matched controls during surgical procedure. After RNA extraction, the miRNA profiles were individually determined in 12 POAG, 12 XFG and 11 control samples. We identified 205, 295 and 195 miRNAs in the POAG, XFG and control samples, respectively. Our differential expression analysis identified three miRNAs (miR-125b-5p, miR-302d-3p and miR-451a) significantly different between POAG and controls, five miRNAs (miR-122-5p, miR-3144-3p, miR-320a, miR-320e and miR-630) between XFG and controls and one miRNA (miR-302d-3p) between POAG and XFG. While none of these miRNAs have been previously linked to glaucoma, miR-122-5p may target three glaucoma-associated genes: OPTN, TMCO1 and TGF-ß1. Pathway analysis revealed that these miRNAs are involved in potential glaucoma pathways, including focal adhesion, tight junctions, and TGF-ß signaling. Comparison of the miRNA profile in AH to unrelated human serum (n = 12) exposed potential relationships between these two fluids, although they were not significantly correlated. In summary, we have successfully profiled the miRNA expression without amplification in individual human AH samples and identified several POAG or XFG-associated miRNAs. These miRNAs may play a role in pathways previously implicated in glaucoma and act as biomarkers for disease pathogenesis.
Introduction
Glaucoma is a heterogeneous group of neurodegenerative disorders characterized by the loss of retinal ganglion cells (RGC) and is a leading cause of vision loss worldwide (1,2). The most common types of open-angle glaucoma (OAG) are primary open-angle glaucoma (POAG) and exfoliation glaucoma (XFG) (3,4). POAG is defined as having the presence of the glaucomatous optic neuropathy without any identifiable secondary cause (5,6). XFG is a secondary glaucoma that occurs in patients with exfoliation syndrome (XFS, OMIM #177650), an age-related systemic disorder characterized by abnormal fibrillary deposits within the eye and various other organs (7,8). For both XFG and POAG, the only modifiable risk factor is elevated intraocular pressure (IOP) (4,5,9–11).
In the anterior segment of the eye, the production and outflow of aqueous humor (AH) maintains IOP levels. This clear fluid is responsible for maintaining the shape and optical properties of the eye while also providing nutrients and removing waste from the anterior segment tissues. AH is produced by the ciliary epithelium, exiting the eye via two outflow pathways: the conventional pathway and the unconventional pathway (12,13). The conventional pathway consists of the trabecular meshwork (TM) and Schlemm’s canal (SC) tissues and accounts for about 80% of the AH drainage in older adults (13). Decreased outflow through these tissues has been shown to be the main contributor to the elevated IOP levels seen in glaucoma (13,14). With XFG, the abnormal fibrillary material found in the anterior segment can accumulate along the conventional outflow pathway, leading to disorganization and degeneration of the TM and SC and elevation of IOP (15). Such elevated pressure levels are a major risk factor to RGC degeneration, and if left untreated, visual impairment could ensue.
Although family history is another important risk factor for glaucoma, the overwhelming majority of adult onset glaucoma is considered to be a common and complex disorder with no clear inheritance pattern. Though many linkage analyses and genome-wide association studies have attempted to unravel the genetics of glaucoma, these genetic associations account for less than 10% of all glaucoma cases (16–18). Therefore, other genetic factors, such as microRNAs (miRNAs), are likely to play a role in the disease pathogenesis (19,20). A recent study has confirmed the potential impact of epigenetics by identifying a common variant in MIR182 that is significantly associated with POAG (21). miRNAs are approximately 22 nucleotide, non-coding RNAs that regulate gene expression by either inhibiting or degrading mRNA (19). Recent studies have shown that miRNAs are present in human AH, both in solution associated with RNA-binding proteins and contained within extracellular vesicles (EVs) including exosomes (22–25). Since EVs released from the ciliary body may contribute to outflow pathway signaling, miRNAs could also play a role in the outflow pathway (26). The potential impact of circulating miRNAs in various diseases, such as cancer and cardiovascular disorders, has already been established (27,28).
Providing the foundation for such AH experiments, one study recently analyzed the miRNA expression differences in the AH of 10 glaucoma and 10 control patients (25). This study combined POAG and XFG into a single experimental group when performing differential analysis. Though these diseases are both types of OAG, XFG has a dramatically different etiology. Clinically, XFG has different presentations and is considered more severe than POAG, typically exhibiting higher IOP, being more difficult to manage clinically and resulting in greater visual field loss (8,9). Such factors indicate that XFG and POAG are distinctly different diseases (8). By not differentiating between the different glaucoma sub-types, this study is unable to accurately capture the differential expression of miRNAs in POAG and XFG. More recently, another study investigated the differential expression of miRNAs in control and POAG AH samples after pre-amplification, using PCR arrays to identify a few miRNAs consistently differentially expressed (DE) in POAG (29).
To completely analyze the AH miRNA expression changes that occur with POAG and XFG, we compared the miRNA content of AH from 12 POAG and 12 XFG patients, separately, to that of 11 cataract patients. Using the NanoString Human v3 miRNA Expression Assay, which uses direct digital detection to count non-amplified miRNA, we identified a number of DE miRNAs that had not been previously associated with glaucoma. The expression profiles produced by NanoString were further validated in an additional 17 POAG, 14 XFG and 10 control AH samples using droplet digital PCR (ddPCR). The target genes and their associated pathways of these DE miRNAs indicate that they have the potential to play a role in the pathogenesis of glaucoma.
Results
Overall miRNA expression
The miRNA expression of 12 POAG, 12 XFG and 11 non-diseased human AH samples was measured using the NanoString digital, non-amplification technology. The phenotypes of these samples and RNA yields are summarized in Table 1, and individual sample tables can be found in Supplementary Material, Table S1. While the samples were age and sex matched, a variable amount of total RNA was obtained from the different samples. These differences were likely due to sample variations, especially since AH was collected from two different institutions.
Table 1.
Aqueous humor sample phenotypes and RNA yield
| Discovery: NanoString |
Validation: ddPCR |
|||||
|---|---|---|---|---|---|---|
| Control | POAG | XFG | Control | POAG | XFG | |
| (n = 11) | (n = 12) | (n = 12) | (n = 10) | (n = 17) | (n = 14) | |
| Age ± S.E.M. | 69.1 ± 2.3 | 67.8 ± 3.0 | 73.5 ± 2.8 | 67.7 ± 2.6 | 70.5 ± 1.6 | 69.7 ± 1.8 |
| Female, % | 45.5 | 50.0 | 58.3 | 80.0 | 52.9 | 14.3 |
| Male, % | 54.5 | 50.0 | 41.7 | 20.0 | 47.1 | 85.7 |
| African American, % | 18.2 | 8.3 | N/A | N/A | N/A | 14.3 |
| Egyptian, % | 0.0 | 0.0 | N/A | N/A | N/A | 14.3 |
| Hispanic, % | 0.0 | 0.0 | N/A | N/A | N/A | 7.1 |
| Caucasian, % | 81.8 | 91.7 | N/A | N/A | N/A | 64.3 |
| IOP ± S.E.M. | N/A | 18.4 ± 1.7 | N/A | N/A | 18.0 ± 0.8 | 23.3 ± 2.2 |
| miRNA, ng | 9.7 ± 3.1 | 2.3 ± 0.2 | 1.2 ± 0.1 | 4.3 ± 1.1 | 2.8 ± 0.4 | 7.8 ± 0.8 |
Overall, our samples contained 298 unique miRNAs, with 205 miRNAs from POAG samples, 295 from XFG samples and 195 from controls (Supplementary Material, Table S2). Figure 1 contains a Venn diagram showing the overlap of these miRNAs between and within the different sample groups. As seen in the figure, a large number of miRNAs (n = 180) were expressed in all three sample groups. While only 15 miRNAs were expressed only in control and XFG AH and only 22 miRNAs exclusively in POAG and XFG AH, few to no specific miRNAs were in control and POAG AH.
Figure 1.
Venn diagram for the distribution of miRNAs between the control, POAG and XFG samples. The miRNA content of the samples groups was organized into a Venn diagram, with the values in the figure representing the number of unique miRNAs identified within and among each sample group.
When looking at the counts significantly different than background, the average number of normalized counts were 22 ± 0.95, 22 ± 1.0 and 21 ± 1.1 for control, POAG and XFG, respectively. The most abundant miRNAs (95th percentile) for each sample group are shown in Table 2. As seen in the table, a majority of these miRNAs were highly expressed in both glaucoma and control samples. Figure 2 shows the average normalized counts of these abundantly expressed miRNAs, with several of these miRNAs showing significantly different expression (P ≤ 0.05) between at least two of the sample groups. It is worth noting that even though miR-451a appears to be highly expressed in XFG samples, this miRNA was not found to be significantly different than background level. This is likely because of the high level of variation between the different XFG samples, with one sample having more than 10-fold higher expression of miR-451a than the others.
Table 2.
Most abundant (95th percentile) miRNAs per sample group
| Control |
POAG |
XFG |
|||
|---|---|---|---|---|---|
| miRNA | No. of countsa | miRNA | No. of countsa | miRNA | No. of countsa |
| hsa-miR-122-5p | 151 ± 27 | hsa-miR-1246 | 162 ± 32 | hsa-miR-122-5p | 266 ± 17 |
| hsa-miR-1246 | 122 ± 22 | hsa-miR-122-5p | 148 ± 17 | hsa-miR-1246 | 159 ± 34 |
| hsa-miR-3065-5p | 78 ± 6.1 | hsa-miR-3065-5p | 86 ± 5.0 | hsa-miR-3065-5p | 102 ± 11 |
| hsa-miR-3144-3p | 43 ± 11 | hsa-miR-3144-3p | 46 ± 4.9 | hsa-miR-3144-3p | 67 ± 3.3 |
| hsa-miR-612 | 42 ± 3.8 | hsa-miR-574-5p | 43 ± 3.0 | hsa-miR-574-5p | 48 ± 3.5 |
| hsa-miR-2682-5p | 41 ± 2.7 | hsa-miR-612 | 41 ± 2.5 | hsa-miR-149-5p | 43 ± 2.5 |
| hsa-miR-125b-5p | 40 ± 4.8 | hsa-miR-451a | 39 ± 14 | hsa-miR-320e | 42 ± 7.1 |
| hsa-miR-149-5p | 39 ± 1.9 | hsa-miR-149-5p | 38 ± 2.4 | hsa-let-7b-5p | 39 ± 1.8 |
| hsa-let-7b-5p | 39 ± 2.1 | hsa-miR-192-5p | 35 ± 1.6 | hsa-miR-2682-5p | 38 ± 3.6 |
| hsa-miR-574-5p | 36 ± 1.6 | hsa-miR-155-5p | 34 ± 3.1 | hsa-miR-125b-5p | 36 ± 1.6 |
| hsa-miR-155-5p | 34 ± 1.7 | hsa-miR-21-5p | 33 ± 1.6 | hsa-miR-155-5p | 35 ± 1.4 |
| hsa-miR-192-5p | 34 ± 3.1 | hsa-miR-548ah-5p | 33 ± 1.5 | hsa-miR-192-5p | 34 ± 1.0 |
| hsa-miR-548ah-5p | 31 ± 1.6 | hsa-miR-320e | 33 ± 4.0 | hsa-miR-451a | 33 ± 22.4 |
| hsa-miR-21-5p | 27 ± 1.6 | hsa-let-7b-5p | 32 ± 1.6 | hsa-miR-21-5p | 31 ± 1.6 |
The mean number of normalized counts for each sample group ± S.E.M.
Figure 2.
Expression of the most abundantly expressed miRNAs in the control, POAG and XFG samples. The most abundant miRNAs were defined as those within the top 95th percentile. Represented here are the mean number of normalized counts per sample group for each of abundantly expressed miRNA, with * indicating a P-value ≤ 0.05.
Differential expression
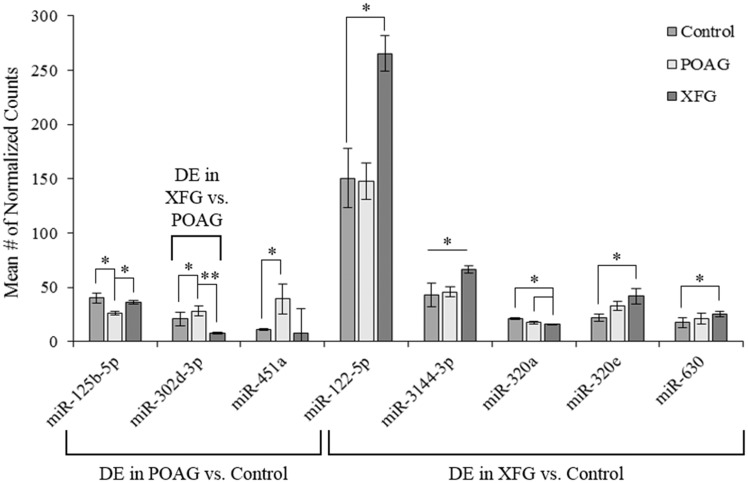
Through differential analysis using the limma Bioconductor package, we were able to identify several DE miRNAs between our samples. Probes were defined as significant based on stringent significance criteria: an absolute difference between the mean number of counts (ΔCounts) ≥ 5, an absolute difference in log 2 fold change (log 2FC) ≥ 0.6 and a P-value ≤ 0.05. Such analyses showed that three miRNAs were differentially expressed between POAG and control, five between XFG and control and one between XFG and POAG (Fig. 3). These miRNAs and their associated ΔCounts, log 2FC and P-value are shown in Table 3.
Figure 3.
Expression of the DE miRNAs in POAG, XFG, and between XFG and POAG. Differential analyses indicated that three miRNAs were DE in POAG, five in XFG and one between XFG and POAG. The mean number of normalized counts per sample group for the eight DE miRNAs are represented in this figure, with * denoting P ≤ 0.05 and ** for P < 0.001.
Table 3.
Significant differentially expressed miRNAs
| miRNA | ID | ΔCountsa | Log 2FCb | P-value | |
|---|---|---|---|---|---|
| POAG versus control | hsa-miR-125b-5p | MIMAT0000423 | 14 | −0.63 | 0.0013 |
| hsa-miR-302d-3p | MIMAT0000718 | 7 | 1.69 | 0.0006 | |
| hsa-miR-451a | MIMAT0001631 | 28 | 1.37 | 0.0038 | |
| XFG versus control | hsa-miR-122-5p | MIMAT0000421 | 115 | 0.70 | 0.0095 |
| hsa-miR-3144-3p | MIMAT0015015 | 24 | 1.48 | 0.0020 | |
| hsa-miR-320a | MIMAT0000510 | 5 | −0.84 | 0.0001 | |
| hsa-miR-320e | MIMAT0015072 | 20 | 0.97 | 0.0010 | |
| hsa-miR-630 | MIMAT0003299 | 8 | 0.78 | 0.0176 | |
| XFG versus POAG | hsa-miR-302d-3p | MIMAT0000718 | 20 | −2.98 | < 0.0001 |
The absolute difference between the mean numbers of normalized counts for the comparison groups.
The log 2FC of the mean number of normalized counts of the comparison groups.
DE miRNA pathway analyses
Using the miRTarBase online database, experimentally validated high and low confidence gene targets, as defined previously, of the eight DE miRNAs in POAG or XFG were identified (Supplementary Material, Table S3) (30,31). Overall, the three DE POAG miRNAs targeted 103 high confidence gene targets. Of these 103 genes, four were targeted by at least two of these DE miRNAs: AKT1, BCL2, IL6R and KLF13. Using WEB-based GEne SeT AnaLysis Toolkit (Webgestalt), 62 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were identified to be significantly represented, with significance defined as having a Benjamini-Hochberg (BH) adjusted P-value of 0.05 and containing at least two target genes. A full list of these pathways and their associated gene targets and adjusted P-values can be found in the Supplementary Material, Table S4. Figure 4 represents the biologically relevant pathways as a heatmap, with shades of gray representing the number of gene targets of down-regulated DE miRNAs and shades of blue for the gene targets of up-regulated DE miRNAs. Biologically relevant was defined as pathways that have potential to play a role in glaucoma pathogenesis based upon currently available information. After correcting for pathway biases, 5 of the 65 pathways were found to be significantly represented (P-value < 0.001) in the POAG high confidence target genes: focal adhesion, long-term potentiation, progesterone-mediated oocyte maturation, tight junction and toll-like receptor signaling.
Figure 4.
Heatmap of the KEGG pathways enriched for the target genes of the POAG DE miRNAs. This heatmap illustrates the number of target genes of the POAG DE miRNAs involved in each enriched pathway, with shades of blue for the up-regulated miRNAs and shades of gray for the down-regulated miRNAs. The pathways are clustered based on their representations in the target genes.
The same pathways analysis pipeline was performed using the miRNAs DE between XFG and control, and 81 high confidence gene targets were identified for these miRNAs. Of these high confidence targets, three were targets of more than one of the XFG DE miRNAs: BCL2L2, IGF1R and RAC1. Together the high confidence target genes were significantly represented in 63 different KEGG pathways. A full list of these pathways and their associated genes and adjusted P-values can be found in the Supplementary Material, Table S5. Figure 5 is a heatmap illustrating the allocation of gene targets between miRNAs and biologically relevant pathways, similar to that created for XFG DE miRNAs. Biases correction indicated that none of the high confidence gene target pathways were significantly represented (P-value < 0.001) for the XFG DE miRNAs.
Figure 5.
Heatmap of the KEGG pathways enriched for the target genes of the XFG DE miRNAs. This heatmap illustrates the number of target genes of the XFG DE miRNAs involved in each enriched pathway, with shades of blue for the up-regulated miRNAs and shades of gray for the down-regulated miRNAs. The pathways are clustered based on their representations in the target genes.
Individual targets genes, many of which were included in the significant pathways, could be considered biologically relevant. Most substantially, three glaucoma-associated genes (OPTN, TGF-ß1 and TMCO1) are targeted by miR-122-5p, which was differentially expressed in XFG. Most of the target genes were defined as being relevant based on their potential role in disease pathogenesis or glaucoma-related pathways (Table 4). Twenty-three of the DE miRNA target genes had been identified previously as being DE in TM cells from patients with POAG (Supplementary Material, Table S6a), and many of the other target genes are expressed at the protein level in glaucoma-relevant tissues or in circulating biofluids, of which 15 are differentially expressed in the AH of POAG patients (Supplementary Material, Table S6b) (6,32–37).
Table 4.
Biologically relevant gene targets in DE miRNAs
| Target gene | POAG DE miRNAs | XFG DE miRNAs |
|---|---|---|
| AKT1 (69,70) | miR-125b-5p, miR-302d-3p, miR-451a | — |
| ATXN1 (69,70) | miR-125b-5p | miR-320a, miR-630 |
| BAK1 (71) | miR-125b-5p | — |
| BCL2 (71) | miR-125b-5p, miR-451a | miR-630 |
| BCL2L2 (71) | miR-125b-5p, miR-302d-3p | miR-122-5p, miR-630 |
| C3 (72) | — | miR-122-5p |
| CDK4 (73) | — | miR-122-5p, miR-320a |
| ESR1 (74) | miR-302d-3p | — |
| FBN1 (75) | miR-125b-5p | — |
| HSPD1 (76) | miR-125b-5p | — |
| IL6R (77) | miR-125b-5p, miR-451a | — |
| LAMA5 (78) | — | miR-320a |
| MAPK1 (79) | — | miR-320a, miR-122-5p |
| MAPK14 (79) | miR-125b-5p | — |
| MCL1 (71) | miR-125b-5p | miR-320a |
| MMP13 (80–82) | miR-125b-5p | — |
| MMP2 (80–82) | miR-451a | — |
| MMP9 (80–82) | miR-451a | — |
| OPTN (83) | — | miR-122-5p |
| PIK3CD (69,70) | miR-125b-5p | — |
| SELE (84) | — | miR-630 |
| SOD2 (85) | — | miR-3144-3p |
| STAT3 (86) | miR-125b-5p | — |
| TFRC (87) | — | miR-320a |
| TGFB1 (88) | — | miR-122-5p |
| TGFBR2 (89) | — | miR-630 |
| TMCO1 (89,90) | — | miR-122-5p |
| TP53 (91,92) | miR-125b-5p | — |
| VEGFA (93,94) | miR-302d-3p | — |
| ZEB2 (94) | — | miR-320a |
Numbers in parentheses refer to the citations for biological relevance of the target genes.
Comparison of AH and serum expression profiles
Using the same analysis pipeline as used for the AH samples, the miRNA expression profile of 12 commercially available human serum samples were analyzed. Of the 289 miRNAs found in the serum, 158 were contained in both serum and AH, and therefore, 140 of the miRNAs from our profile were specific to AH. Overall, while the AH had more miRNAs represented, the serum profile showed a larger range and higher expression than AH, without correcting for starting volume of RNA. The miRNA profiles were sorted based on the average number of normalized counts, and the most abundant (95th percentile) were identified (Table 5). Of the most abundant miRNAs, four were found to be contained in both the AH and serum, as indicated in Table 5. Comparing this profile to the miRNAs DE in AH, miR-302d-3p and miR-320a were not found to be present in the serum. miR-122-5p, miR-320e and miR-451a were among the most abundant (90th percentile) for both groups, while miR-125b-5p, miR-3144–3p and miR-630 exhibited higher abundance in the AH than the serum. Correlation analyses between all the miRNAs (r = 0.035) and between only miRNAs above background level in either serum or AH (r = 0.0049) indicated that, overall, the serum and AH miRNA profiles were not significantly correlated.
Table 5.
Most abundant miRNAs in normal AH and serum
| Aqueous humor |
Serum |
||
|---|---|---|---|
| miRNA | No. of countsb | miRNA | No. of countsb |
| hsa-miR-122-5pa | 189 ± 15 | hsa-miR-451a | 5275 ± 998 |
| hsa-miR-1246a | 149 ± 18 | hsa-miR-16-5p | 288 ± 43 |
| hsa-miR-3065-5pa | 89 ± 4.8 | hsa-miR-144-3p | 244 ± 39 |
| hsa-miR-3144-3p | 52 ± 4.4 | hsa-miR-223-3p | 233 ± 33 |
| hsa-miR-574-5p | 43 ± 1.9 | hsa-miR-320ea | 201 ± 29 |
| hsa-miR-149-5p | 40 ± 1.4 | hsa-miR-126-3p | 197 ± 20 |
| hsa-miR-612 | 37 ± 2.0 | hsa-miR-1246a | 190 ± 40 |
| hsa-miR-2682-5p | 37 ± 1.9 | hsa-miR-23a-3p | 187 ± 23 |
| hsa-let-7b-5p | 36 ± 1.2 | hsa-miR-142-3p | 185 ± 22 |
| hsa-miR-155-5p | 34 ± 1.3 | hsa-miR-25-3p | 169 ± 15 |
| hsa-miR-192-5p | 34 ± 1.2 | hsa-miR-150-5p | 136 ± 16 |
| hsa-miR-125b-5p | 34 ± 2.0 | hsa-miR-122-5pa | 127 ± 11 |
| hsa-miR-320ea | 32 ± 3.3 | hsa-let-7a-5p | 89 ± 11 |
| hsa-miR-548ah-5p | 32 ± 0.9 | hsa-miR-3065-5pa | 80 ± 4.0 |
miRNAs abundantly present in both AH and serum.
The mean number of normalized counts for each sample group ± S.E.M.
Validation of miRNA profile
To validate the miRNA profiles obtained using the NanoString technology, we used ddPCR to measure the expression of the DE miRNAs. The individual patient samples used for the ddPCR experiments were different than those used for NanoString, as indicated in Table 1. We successfully validated the differential expression of miR-320a and miR-451a (Fig. 6). These results in Figure 6 indicate that the general trend between the sample groups within a miRNA observed in the ddPCR data were comparable to our previously attained miRNA profiles. While the relationships between the different miRNAs stayed relatively consistent, we did observe that the overall expression level of miR-122-5p was much lower for all sample groups than expected based on the NanoString results. This lower expression of miR-122-5p was also seen in the human serum samples (n = 4), while maintaining the same relationship between serum and AH expression, as shown in Figure 7. We also found that the amount of miR-122-5p and miR-125b-5p from ddPCR was not significantly different between our sample groups, unlike with NanoString. Expression of miR-320e from ddPCR, though, did not produce a similar relationship between the sample groups. For miR-302d-3p, miR-3065-5p, miR-3144–3p and miR-630, ddPCR showed no measurable expression, maybe due to limited amount of available AH RNA.
Figure 6.
Validation of AH miRNA expression profiles using ddPCR. Measuring miRNA concentration of POAG (n = 17), XFG (n = 14), and control (n = 10) AH with ddPCR (A) gave a similar relative expression profile to that produced with the NanoString technology (B) for five of the DE miRNAs: miR-122-5p, miR-125b-5p, miR-320a, miR-320e and miR-451a, with * denoting P ≤ 0.05.
Figure 7.
Validation of serum miR-122-5p expression using ddPCR. Expression of miR-122-5p in non-diseased serum (n = 4) and control AH (n = 10) were measured using ddPCR (A) and the NanoString technology (B). Similar trends in expression levels were seen between the two techniques.
Discussion
In this study, we profiled the miRNA in individual human AH samples from 11 cataract, 12 POAG and 12 XFG patients using the NanoString non-amplification, digital detection technology. Through differential analysis we found three miRNAs and five miRNAs with significantly different expression in POAG and XFG, respectively, compared to controls. Of these, miR-302d-3p was the only one differentially expressed between POAG and XFG. A number of the target genes of these miRNAs have been previously implicated in glaucoma, either through differential expression or potential involvement in disease pathogenesis or glaucoma-related pathways. Pathway analyses of the high confidence gene targets, using WebGestalt, indicated that many of these genes, and potentially their miRNA regulators, are involved in pathways possibly involved in glaucoma.
Previously three studies have been conducted to analyze the miRNA content of human AH from cataract patients, looking either at all miRNAs in solution or specifically the miRNAs contained in exosomes (22–24). Another two studies analyzed the DE miRNAs in glaucomatous AH, using cataract patients as controls (25,29). Upon comparing our cataract AH sample profiles to those from these studies using the profiles supplied by Jayaram et al., we found approximately 7–36% overlap between the miRNA lists (Supplementary Material, Table S7), with 4–73% of the miRNAs from the studies not being included in the NanoString assay (29). Comparisons to the other glaucoma AH studies found no similarities between our lists of DE miRNAs, though 76% of the DE miRNAs from Tanaka et al. were not measured with the NanoString assay. This limited overlap between our profiles is likely because of differences in techniques, with amplification-based methods, such as PCR or sequencing, having been shown to produce different miRNA profiles than non-amplification-based profiling techniques (38,39). The limit of our array to a specific set of 800 miRNAs, the stringency of our background cut-off level and DE qualification standards, the use of individual samples rather than pooled samples, and the variations in individual patients and sampling techniques probably accounts for the differences between our glaucoma miRNA profiles.
To determine the potential specificity of the AH miRNAs, we compared the AH expression profile to that of human serum from a commercial supplier. While we found similarities between the two data sets, the comparison showed that many of the AH miRNAs were not contained in serum, including two of the DE miRNAs (miR-302d-3p and miR-320a), suggesting the unique exposure of TM and SC cells in the anterior segment to these specific miRNAs. In contrast, three of the DE miRNAs were found to be abundantly present in both the serum and the AH, all of which were differentially expressed in XFG. Since XFS is a systemic disorder, further analysis of whether these miRNAs are differentially deposited in serum of XFG patients could be beneficial, especially miR-451a. This miRNA was the most abundant in serum and also significantly DE in XFG AH. Because of its potential role in disease pathogenesis and the proven stability of miRNAs in circulating biofluids, miR-451a could be used as a disease marker in serum, which is more accessible than AH (28). To better study the potential of using this miRNA as a biomarker, it would be beneficial to measure the expression in AH and serum samples collected from the same patient.
These DE miRNAs were found to target a number of genes implicated in the disease, either through gene expression or involvement in disease-associated pathways. Of these genes, many were found to be involved in the TGF-ß pathway, with KEGG pathway analysis showing this pathway to be significantly represented by target genes of both POAG and XFG DE miRNAs (Supplementary Material, Table S3). This signaling pathway is thought to be involved in mediating extracellular matrix secretion and deposition, and TGFß-1 and TGFß-2 have elevated expression in XFG and POAG, respectively (40). Though the exact role our DE miRNAs could play in this pathway is unclear, their changes in expression indicate an involvement in the accumulation and structural changes in extracellular matrix that have been implicated in glaucoma disease pathogenesis.
Many of the individual target genes and significant pathways were related to neural functions, specifically neuroprotection or apoptosis of RGC s through pathways such as the AKT signaling and the Bcl-2-regulated apoptosis pathway, which is highly targeted by the DE miR-125b-5p (41). Besides the neural-related proteins, the DE miRNAs were found to target many genes with proteins expressed in anterior segment tissues, some even DE in glaucomatous AH. Many of these proteins being previously implicated in glaucoma-associated pathways, such as metabolism and cell structure, adhesion, and motility, further highlight the possible involvement of our DE miRNAs in glaucoma.
Because of the complex nature of miRNAs, pathway analyses of their gene targets often give an inaccurate representation of the actual pathway involvement (42,43). The unequal representation of a gene in relation to its functional assignment and pathway representation, due to insufficient study in comparison to other genes, only acts to enhance these biases (42,43). Therefore, the assumption of uniform sampling, which is critical to most pathway analyses pipelines, is not applicable. To correct for these inherent biases, we compared our pathway results to those of 100 sets of randomly selected miRNAs.
This pathway correction resulted in five of the POAG and none of the XFG high confidence gene target pathways showing highly significant correlation to our miRNA gene targets. Of these significant POAG pathways, two are of particular interest to glaucoma: focal adhesion and tight junction. Both of these processes affect the structure of the ECM, which impacts the stiffness of the TM and SC. Such fundamental mechanisms have the potential to regulate AH outflow (44). Unlike POAG, none of the XFG pathways were significant after bias correction. This lack of significance is most likely because two of the five DE miRNAs had no high confidence gene targets, therefore, causing an increase in the likelihood that randomly selected miRNAs would be more represented in any particular pathway. When analyzing the specific relationship between miRNAs and their gene targets, it is important to note that a single miRNA can target many genes, and one gene may be regulated by multiple miRNAs. Overall, up-regulation of a miRNA, though, should correlate with down-regulation of the gene target.
To examine the validity of our miRNA expression profiles obtained using NanoString, we measured the miRNA concentration of the DE miRNAs using ddPCR. In the miRNAs with measurable expression levels, we saw a similar expression trend both between groups and between miRNAs, with the exception of miR-320e. Though producing similar relationships between sample groups as NanoString, miR-122-5p showed lower expression than expected compared to the other miRNAs. This trend was replicated with serum, and the relationship between non-diseased serum and AH miR-122-5p expression levels was the similar between the two techniques. In miR-303d-3p, miR-3065-5p, miR-3144–3p and miR-630, ddPCR produced no measurable expression. This difference between ddPCR and NanoString could be attributed to variability in the affinity of the assay probes for the miRNA, as well as differences in sensitivity between the techniques (38). The inability to detect some of the miRNAs with ddPCR could be because ddPCR was not sensitive to detect the low concentrations of these miRNAs in extremely limited amount (< 1 ng) of AH RNA samples, especially without amplification, which is not included in ddPCR. Another factor contributing to the dissimilar results between ddPCR and NanoString could be the large variability in miRNA expression between AH samples, as seen the large error bars of Figure 6. This is especially true considering that we did not use samples from the same patients for the two techniques.
Though pathway and gene target analyses indicated possible biological relevance of our miRNAs in glaucoma pathogenesis, not enough is known to draw any conclusions. One main contributor to the complicated role of AH miRNAs in the anterior chamber is the uncertainty of the source or recipient of the miRNAs in solution, and AH expression could indicate a response to elevated IOP, rather than a cause of changes in outflow dynamics. Without knowing this, the potential effect of the miRNAs on the anterior segment tissues is uncertain. The potential role of glaucoma medical therapies on the miRNA profile also needs to be taken into consideration. Since most of the glaucoma patients in this study were under medical treatment at the time of AH sample collection, this limitation could not be currently explored. Future studies could better explore this concern by comparing the miRNA profile of individual patients before and after treatment. This could be achieved by collecting AH at both the time of glaucoma surgery, when patients are under treatment, and then again during cataracts surgery, when the patient is no longer under treatment. Because this study was designed to be exploratory, treatment, along with other factors such as phenotype and disease severity, were not included. Future studies would benefit from including these analyses, though. It is also worth noting that these AH samples were collected at one time point, therefore, do not represent the miRNA expression fluctuations that may occur with changes in IOP over a period of time. With our small sample size, this study is also unable to address whether the DE miRNAs are correlated to glaucoma phenotypes or severity.
In summary, the miRNA expression profile of AH from POAG and XFG patients was explored in comparison to cataract controls. Several miRNAs were identified as being significantly differentially expressed between the POAG, XFG and control. Many of these miRNAs were shown to target genes and to be significantly associated with pathways possibly involved in glaucoma. While more investigation is needed, this study highlights potential key miRNAs differentially expressed in glaucoma, which could be possible targets for therapy or biomarkers.
Materials and Methods
AH extraction and RNA isolation
The research conducted in this study adhered to the tenets of the Declaration of Helsinki. AH samples (50–100 μl) from patients with cataract, POAG and XFG were collected at the Duke University Eye Center and the Vanderbilt University Medical Center using protocols approved by the respective Institutional Review Board. AH samples were acquired in the same fashion at the Duke University and the Vanderbilt University as described previously (22,45). The criteria for cataract control and diagnosis of POAG and XFG were harmonized between Duke and Vanderbilt sites as we reported previously (21,22,46–49). XFG was defined by (a) age ≥50 years old at time of recruitment; (b) documentation of characteristic ocular exfoliation material at the pupil margin or surface of the ocular lens, either through clinical exam or medical records; (c) glaucomatous optic neuropathy and (d) visual field loss. POAG was defined as characteristic visual field defects consistent with glaucomatous optic neuropathy. Elevated IOP (>21 mmHg) was not used as a criterion for POAG or XFG. Controls were also older than age 50 and had no evidence of any type of glaucoma, including XFG and POAG, by clinical exam or medical records, as well as an IOP of less than 21 mmHg. Clinical examination, for both cases and controls, included measurement of visual acuity and IOP, slit lamp biomicroscopy, dilated examination of the lens and fundus, and fundoscopy. Cases also had visual field assessment primarily using the Humphrey automated visual fields. Individuals were excluded if other types of glaucoma, such as pigment dispersion, steroid induced or uveitis, were evident on exam. After collection, samples were immediately frozen and stored at −80°C. Overall, 21 cataract samples, 29 POAG samples and 26 XFG samples from both Caucasian and African American individuals were collected, with the cataract samples acting as a non-diseased control. Of these samples, 11 control, 12 POAG and 12 XFG samples were used for NanoString miRNA expression measurements, and the remaining samples were used for ddPCR miRNA expression measurements. One additional control sample was used in the NanoString miRNA expression measurements but failed quality control (QC) checks, therefore, it was not included in the study. Phenotypes of these samples, per group, can be found in Table 1, and Supplementary Material, Table S1 gives individual sample phenotypes. IOP represented the untreated IOP prior to the glaucoma treatment. A small number of AH samples had certain phenotypes missing due to inability to access information related to HIPAA regulations/consenting issues.
Total RNA was isolated from the AH samples using the miRCURY RNA Isolation Kit for Cell and Plant (Exiqon, Woburn, MA), according to the manufacturer’s instructions and as described previously (21,22). A final volume of 50 µL of RNA was collected from the RNA isolation columns using the supplied elution buffer. This RNA was dried and reconstituted with 3.5 μl of the elution buffer. The total RNA concentration was measured with the RNA 6000 Pico Kit using the Agilent 2100 Bioanalyzer (Santa Clara, CA), and each sample contained between 0.5 and 12 ng of RNA.
miRNA expression analysis
The miRNA expression of the AH RNA samples was measured using the NanoString nCounter Human v3 miRNA Expression Assay (Seattle, WA) at the Georgia Esoteric & Molecular Diagnostic Laboratory at the Medical College of Georgia of Augusta University (http://www.augusta.edu/mcg/pathology/gem-lab.php; date last accessed February 8, 2018), which includes hybridization tags for 800 miRNAs from miRBase v21, according to the manufacturer’s instructions. The coverage of this assay is 800 out of 2588 known mature miRNAs, according to miRBase v21. This technology has been previously shown to be a viable miRNA profiling method, even with limited amounts of miRNA (38,39,50). Since one NanoString cartridge is able to process up to 12 samples, we measured the expression of our data in three different batches. Batches 1 and 2 each contained a mix of control and POAG samples, in which batch 3 included all XFG samples.
Using an analysis pipeline based on recommendations by NanoString, raw data RCC files produced by the nCounter were initially processed using the NanoString nSolver 3.0 software (Seattle, WA). Before analysis, each sample was run through several QC checks, examining the quality of imaging, binding density, positive control linearity and positive control limit of detection. With the nSolver software, the raw miRNA counts were normalized based on RNA content using the trimmed geometric mean, which entailed calculating the average number of counts for each sample using the miRNA with the median 40% of the counts. This normalization technique was chosen for its similarity to the trimmed mean of M (TMM) method, which has previously been shown to be successful in normalizing miRNA counts (51). The miRNA counts were further normalized to account for technology-associated sources of variation using the geometric mean of the positive control probes POS_C, POS_D and POS_E. Because their number of counts was substantially outside the range of our data set, positive control probes POS_A, POS_B and POS_F were excluded.
After normalization, the data was exported from nSolver into CSV files and then imported into the R Language and Environment for Statistical Computing (52). Within R, the background level probes for the control, POAG and XFG samples were identified using NanoString’s recommended methods. In summary, the negative control probes for each sample were extracted and multiplied by the corresponding normalization factors produced by nSolver. Then for each probe, a Welch’s t-test was performed, comparing the normalized negative control counts to the sample counts separately for control, POAG and XFG samples. The counts for a specific probe were considered background if they were not found to be significantly different than the negative controls (P > 0.05).
Once the background level probes were identified for each sample type, the limma Bioconductor package was used to perform the differential analysis (53–58). Before running the analysis, a principal component analysis (PCA) plot was constructed to identify any outliers or batch differences, especially since samples were processed with nCounter in three different batches. All PCA plots were created using the ggPlot2 package in R (59). This PCA plot (Supplementary Material, Fig. S1A) indicated that substantial differences existed between batch 1 and batches 2 and 3. To counter this differences, batch effects were adjusted for using the limma package, grouping batches 2 and 3 in the same category. These batches were grouped together because batch 3 included all of the XFG samples, so adjusting for batch effects with batches 2 and 3 separately would force the XFG counts to conform to those of the POAG and control samples. Also, batches 2 and 3 showed little differences in the PCA plot compared to batch 1.
Another PCA plot was made (Supplementary Material, Fig. S1B) after adjusting for batch effects, verifying that batch effects were removed, and then differential analyses were performed, comparing POAG to control, XFG to control and XFG to POAG using limma, which has been previously shown to work with NanoString data (60). Further data analysis was conducted using Microsoft Excel 2013 (Microsoft Corp., Seattle, WA). Only probes with an absolute log 2 fold change (log 2FC) ≥ 0.6, a P-value ≤ 0.05 and an absolute difference in counts (ΔCount) ≥ 5 were considered to be differentially expressed. All heatmaps were created using the Heatplus package in R (61).
Pathway analysis of gene targets
Gene targets of differentially expressed miRNAs were identified using miRTarBase, an online database of experimentally validated targets of miRNA and their corresponding experimental documentation (31). Only high confidence target genes were selected for pathway analyses, as defined previously (30). WebGestalt, an online pathway analysis tool, was used to analyze the KEGG pathways of these gene targets, with significance defined as having a Benjamini-Hochberg adjusted P-value of 0.05 and containing at least two target genes (62–64). Since the assumption of uniform sampling cannot be made when analyzing the pathways of miRNA target genes, these biases in the miRNA pathway analyses were corrected for using the technique described by Bleazard et al. (42). The bias analysis was done using the R Language and Environment for Statistical Computing, and KEGG data was extracted using the KEGGREST Bioconductor package (53,65). Sets of random miRNAs were selected without replacement and with 100 iterations. Benjamini-Hochberg adjusted P-values ≥0.01 were considered significant (64).
Profiling serum miRNA expression
Human serum samples (n = 12) were purchased from BioreclamationIVT (Hicksville, NY). These 12 samples were from 6 males (3 Caucasian and 3 African American individuals) and 6 females (2 Caucasian and 4 African American individuals), which was similar to the ethnicity background of AH donors. None of these serum donors were reported to have glaucoma or other common ocular diseases. The mean age of these donors were 47.8 ± 3.8 years (from 26 to 64 years old), which is approximately 20 years younger than that from AH donors. Serum EVs were isolated from 200 µl serum using polyethylene glycol-based precipitation as reported previously (66–68). Briefly, the supernatant was collected after a 3000×g centrifugation for 15 min and was mixed with PEG stock buffer (50% PEG-8000, 0.5 m NaCl) in a 5:1 volume ratio. After incubating at 4˚C for 1 h, the mixture was spun at 1500×g for 30 min. The pellet was washed with 1× PBS buffer and spun at 1500×g for 5 min. EV pellets were resuspended in 50 µl 1× PBS. Serum exosomal RNA was isolated using the miRCURY RNA Isolation kit (Catalog # 300110) from Exiqon (67). The miRNA expression was profiled using the same pipeline analysis as that used for the AH, except that batch correction was not necessary for the serum samples.
Validation of miRNA expression
To validate the miRNA expression data, we measured the concentration of the DE miRNAs in 17 POAG, 14 XFG and 10 non-diseased human AH samples using ddPCR. As described previously, the ddPCR assays were performed using the QX200 droplet digital PCR system from Bio-Rad (Hercules, CA). For miR-122-5p, miR-125b-5p, miR-320a, miR-320e and miR-451a, 0.5 ng of the concentrated RNA samples were reverse transcribed using the TaqMan Advanced MicroRNA cDNA Synthesis Kit (Applied Biosystems, Grand Island, NY) according the manufacturer’s instructions. Since limited amount of RNA was available, 10 μl of the reverse transcription product was used for the final miR-Amp step instead of the recommended 5 μl. For ddPCR, 3 µl of the synthesized cDNA was used per reaction. TaqMan Advanced MicroRNA Assays (Catalog # A25576) from Applied Biosystems and QX200 ddPCR Supermix for Probes (no dUTP) (Bio-Rad) were used for the PCR reaction mix. The resulting data were analyzed using the associated Bio-Rad QuantaSoft software and Microsoft Excel.
For the remaining DE miRNAs (miR-302d-3p, miR-3065–5p, miR-3144–3p and miR-630), the Advanced TaqMan MicroRNA Assays were unsuccessful; therefore, the TaqMan MicroRNA Assays (Catalog # 4427975, Applied Biosystems) were used instead. Reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (Catalog # 4366596, Applied Biosystems) with 0.8 ng of RNA according to the manufacturer’s instructions. Then, 5 μl of this cDNA was used per ddPCR reaction, which was performed as described previously. Serum miR-122-5p expression was validated in a similar manner. For QC, all samples were run with no-template controls containing water instead of cDNA, ensuring that the reagents were not contaminated. The differences of miRNA expression in all the examined AH samples of control, POAG and XFG from ddPCR were analyzed using a Student’s t-test with the assumption of unequal variance.
Availability of data and materials
The data sets supporting the conclusions of this paper are available in the NCBI Gene Expression Omnibus and are accessible through the GEO Series accession number GSE105269, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE105269; date last accessed February 8, 2018).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We sincerely thank all the donors for their ocular samples. This study would not be feasible without these precious samples. We thank Drs. Ravindra Kolhe and Ashis K. Mondal in the Department of Pathology at Augusta University for technical support on NanoString technology.
Conflict of Interest statement. None declared.
Funding
This work was supported by The Glaucoma Foundation (TGF), The Glaucoma Research Foundation (GRF), The National Glaucoma Research at BrightFocus Foundation, NIH R01EY023242, R01EY027746, R01EY020894, R01EY023287, and the Startup Fund from the Medical College of Georgia at Augusta University to YL.
References
- 1. Quigley H.A. (1996) Number of people with glaucoma worldwide. Br. J. Ophthalmol., 80, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thylefors B., Negrel A.D., Pararajasegaram R., Dadzie K.Y. (1995) Global data on blindness. Bull. World Health Organ., 73, 115–121. [PMC free article] [PubMed] [Google Scholar]
- 3. Ritch R. (1994) Exfoliation syndrome-the most common identifiable cause of open-angle glaucoma. J. Glaucoma., 3, 176–177. [PubMed] [Google Scholar]
- 4. Weinreb R.N., Khaw P.T. (2004) Primary open-angle glaucoma. Lancet, 363, 1711–1720. [DOI] [PubMed] [Google Scholar]
- 5. Kwon Y.H., Fingert J.H., Kuehn M.H., Alward W.L. (2009) Primary open-angle glaucoma. N. Engl. J. Med., 360, 1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Y., Allingham R.R., Qin X., Layfield D., Dellinger A.E., Gibson J., Wheeler J., Ashley-Koch A.E., Stamer W.D., Hauser M.A. (2013) Gene expression profile in human trabecular meshwork from patients with primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci., 54, 6382–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mitchell P., Wang J.J., Hourihan F. (1999) The relationship between glaucoma and pseudoexfoliation—The Blue Mountains Eye Study. Arch. Ophthalmol., 117, 1319–1324. [DOI] [PubMed] [Google Scholar]
- 8. Challa P. (2009) Genetics of pseudoexfoliation syndrome. Curr. Opin. Ophthalmol., 20, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leske M.C., Heijl A., Hussein M., Bengtsson B., Hyman L., Komaroff E. and Early Manifest Glaucoma Trial, G. (2003) Factors for glaucoma progression and the effect of treatment: the early manifest glaucoma trial. Arch. Ophthalmol., 121, 48–56. [DOI] [PubMed] [Google Scholar]
- 10. Liu Y., Allingham R.R. (2011) Molecular genetics in glaucoma. Exp. Eye Res., 93, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. The A.I. (2000) The Advanced Glaucoma Intervention Study (AGIS): 7. The relationship between control of intraocular pressure and visual field deterioration. Am. J. Ophthalmol., 130, 429–440. [DOI] [PubMed] [Google Scholar]
- 12. Coca-Prados M., Escribano J. (2007) New perspectives in aqueous humor secretion and in glaucoma: the ciliary body as a multifunctional neuroendocrine gland. Prog. Retin. Eye Res., 26, 239–262. [DOI] [PubMed] [Google Scholar]
- 13. Goel M., Picciani R.G., Lee R.K., Bhattacharya S.K. (2010) Aqueous humor dynamics: a review. Open Ophthalmol. J., 4, 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stamer W.D., Acott T.S. (2012) Current understanding of conventional outflow dysfunction in glaucoma. Curr. Opin. Ophthalmol., 23, 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schlotzer-Schrehardt U., Naumann G.O. (2006) Ocular and systemic pseudoexfoliation syndrome. Am. J. Ophthalmol., 141, 921–937. [DOI] [PubMed] [Google Scholar]
- 16. Wang R., Wiggs J.L. (2014) Common and rare genetic risk factors for glaucoma. Cold Spring Harb. Perspect. Med., 4, a017244.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shastry B. (2013) Emerging Concept of Genetic and Epigenetic Contribution to the Manifestation of Glaucoma. InTech, London, UK. [Google Scholar]
- 18. Liu Y., Allingham R.R. (2017) Major review: molecular genetics of primary open-angle glaucoma. Exp. Eye Res., 160, 62–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonzalez P., Li G., Qiu J., Wu J., Luna C. (2014) Role of microRNAs in the trabecular meshwork. J. Ocul. Pharmacol. Ther., 30, 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jayaram H., Lozano D.C., Johnson E.C., Morrison J.C. (2018) Investigation of microRNA expression in experimental glaucoma. Methods Mol. Biol., 1695, 287–297. [DOI] [PubMed] [Google Scholar]
- 21. Liu Y., Bailey J.C., Helwa I., Dismuke W.M., Cai J., Drewry M., Brilliant M.H., Budenz D.L., Christen W.G., Chasman D.I.. et al. (2016) A common variant in MIR182 is associated with primary open-angle glaucoma in the NEIGHBORHOOD consortium. Invest. Ophthalmol. Vis. Sci., 57, 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dismuke W.M., Challa P., Navarro I., Stamer W.D., Liu Y. (2015) Human aqueous humor exosomes. Exp. Eye Res., 132, 73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dunmire J.J., Lagouros E., Bouhenni R.A., Jones M., Edward D.P. (2013) MicroRNA in aqueous humor from patients with cataract. Exp. Eye Res., 108, 68–71. [DOI] [PubMed] [Google Scholar]
- 24. Wecker T., Hoffmeier K., Plotner A., Gruning B.A., Horres R., Backofen R., Reinhard T., Schlunck G. (2016) MicroRNA profiling in aqueous humor of individual human eyes by next-generation sequencing. Invest. Ophthalmol. Vis. Sci., 57, 1706–1713. [DOI] [PubMed] [Google Scholar]
- 25. Tanaka Y., Tsuda S., Kunikata H., Sato J., Kokubun T., Yasuda M., Nishiguchi K.M., Inada T., Nakazawa T. (2014) Profiles of extracellular miRNAs in the aqueous humor of glaucoma patients assessed with a microarray system. Sci. Rep., 4, 5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lerner N., Avissar S., Beit-Yannai E. (2017) Extracellular esicles mediate signaling between the aqueous humor producing and draining cells in the ocular system. PLoS One, 12, e0171153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De Guire V., Robitaille R., Tetreault N., Guerin R., Menard C., Bambace N., Sapieha P. (2013) Circulating miRNAs as sensitive and specific biomarkers for the diagnosis and monitoring of human diseases: promises and challenges. Clin. Biochem., 46, 846–860. [DOI] [PubMed] [Google Scholar]
- 28. Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O'Briant K.C., Allen A.. et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A., 105, 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jayaram H., Phillips J.I., Lozano D.C., Choe T.E., Cepurna W.O., Johnson E.C., Morrison J.C., Gattey D.M., Saugstad J.A., Keller K.E. (2017) Comparison of microRNA expression in aqueous humor of normal and primary open-angle glaucoma patients using PCR arrays: a pilot study. Invest. Ophthalmol. Vis. Sci., 58, 2884–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Drewry M., Helwa I., Allingham R.R., Hauser M.A., Liu Y. (2016) miRNA profile in three different normal human ocular tissues by miRNA-Seq. Invest. Ophthalmol. Vis. Sci., 57, 3731–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hsu S.D., Tseng Y.T., Shrestha S., Lin Y.L., Khaleel A., Chou C.H., Chu C.F., Huang H.Y., Lin C.M., Ho S.Y.. et al. (2014) miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res., 42, D78–D85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Izzotti A., Longobardi M., Cartiglia C., Sacca S.C. (2010) Proteome alterations in primary open angle glaucoma aqueous humor. J. Proteome Res., 9, 4831–4838. [DOI] [PubMed] [Google Scholar]
- 33. Uechi G., Sun Z., Schreiber E.M., Halfter W., Balasubramani M. (2014) Proteomic view of basement membranes from human retinal blood vessels, inner limiting membranes, and lens capsules. J. Proteome Res., 13, 3693–3705. [DOI] [PubMed] [Google Scholar]
- 34. Goel R., Murthy K.R., Srikanth S.M., Pinto S.M., Bhattacharjee M., Kelkar D.S., Madugundu A.K., Dey G., Mohan S.S., Krishna V.. et al. (2013) Characterizing the normal proteome of human ciliary body. Clin. Proteomics, 10, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Golubnitschaja O., Yeghiazaryan K., Wunderlich K., Schild H.H., Flammer J. (2007) Disease proteomics reveals altered basic gene expression regulation in leukocytes of normal-tension and primary open-angle glaucoma patients. Proteomics Clin. Appl., 1, 1316–1323. [DOI] [PubMed] [Google Scholar]
- 36. Bagnis A., Izzotti A., Centofanti M., Sacca S.C. (2012) Aqueous humor oxidative stress proteomic levels in primary open angle glaucoma. Exp. Eye Res., 103, 55–62. [DOI] [PubMed] [Google Scholar]
- 37. Sacca S.C., Centofanti M., Izzotti A. (2012) New proteins as vascular biomarkers in primary open angle glaucomatous aqueous humor. Invest. Ophthalmol. Vis. Sci., 53, 4242–4253. [DOI] [PubMed] [Google Scholar]
- 38. Mestdagh P., Hartmann N., Baeriswyl L., Andreasen D., Bernard N., Chen C., Cheo D., D'Andrade P., DeMayo M., Dennis L.. et al. (2014) Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat. Methods, 11, 809–815. [DOI] [PubMed] [Google Scholar]
- 39. Kolbert C.P., Feddersen R.M., Rakhshan F., Grill D.E., Simon G., Middha S., Jang J.S., Simon V., Schultz D.A., Zschunke M.. et al. (2013) Multi-platform analysis of microRNA expression measurements in RNA from fresh frozen and FFPE tissues. PLoS One, 8, e52517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fuchshofer R., Tamm E.R. (2012) The role of TGF-beta in the pathogenesis of primary open-angle glaucoma. Cell Tissue Res., 347, 279–290. [DOI] [PubMed] [Google Scholar]
- 41. Sun Y.M., Lin K.Y., Chen Y.Q. (2013) Diverse functions of miR-125 family in different cell contexts. J. Hematol. Oncol., 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bleazard T., Lamb J.A., Griffiths-Jones S. (2015) Bias in microRNA functional enrichment analysis. Bioinformatics, 31, 1592–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Godard P., van Eyll J. (2015) Pathway analysis from lists of microRNAs: common pitfalls and alternative strategy. Nucleic Acids Res., 43, 3490–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Last J.A., Pan T., Ding Y., Reilly C.M., Keller K., Acott T.S., Fautsch M.P., Murphy C.J., Russell P. (2011) Elastic modulus determination of normal and glaucomatous human trabecular meshwork. Invest. Ophthalmol. Vis. Sci., 52, 2147–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuchtey J., Rezaei K.A., Jaru-Ampornpan P., Sternberg P. Jr, Kuchtey R.W. (2010) Multiplex cytokine analysis reveals elevated concentration of interleukin-8 in glaucomatous aqueous humor. Invest. Ophthalmol. Vis. Sci., 51, 6441–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aung T., Ozaki M., Lee M.C., Schlotzer-Schrehardt U., Thorleifsson G., Mizoguchi T., Igo R.P. Jr, Haripriya A., Williams S.E., Astakhov Y.S.. et al. (2017) Genetic association study of exfoliation syndrome identifies a protective rare variant at LOXL1 and five new susceptibility loci. Nat. Genet., 49, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hauser M.A., Aboobakar I.F., Liu Y., Miura S., Whigham B.T., Challa P., Wheeler J., Williams A., Santiago-Turla C., Qin X.. et al. (2015) Genetic variants and cellular stressors associated with exfoliation syndrome modulate promoter activity of a lncRNA within the LOXL1 locus. Hum. Mol. Genet., 24, 6552–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li Z., Allingham R.R., Nakano M., Jia L., Chen Y., Ikeda Y., Mani B., Chen L.J., Kee C., Garway-Heath D.F.. et al. (2015) A common variant near TGFBR3 is associated with primary open angle glaucoma. Hum. Mol. Genet., 24, 3880–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Aung T., Ozaki M., Mizoguchi T., Allingham R.R., Li Z., Haripriya A., Nakano S., Uebe S., Harder J.M., Chan A.S.. et al. (2015) A common variant mapping to CACNA1A is associated with susceptibility to exfoliation syndrome. Nat. Genet., 47, 387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Armstrong D.A., Green B.B., Seigne J.D., Schned A.R., Marsit C.J. (2015) MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol. Cancer, 14, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robinson M.D., Oshlack A. (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol., 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Team R.C. (2015). R Foundation for statistical computing, Vienna, Austria, https://www.R-project.org/.
- 53. Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T.. et al. (2015) Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods, 12, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res., 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Phipson B., Lee S., Majewski I.J., Alexander W.S., Smyth G.K. (2016) Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat., 10, 946–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Law C.W., Chen Y., Shi W., Smyth G.K. (2014) voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol., 15, R29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ritchie M.E., Silver J., Oshlack A., Holmes M., Diyagama D., Holloway A., Smyth G.K. (2007) A comparison of background correction methods for two-colour microarrays. Bioinformatics, 23, 2700–2707. [DOI] [PubMed] [Google Scholar]
- 58. Liu R., Holik A.Z., Su S., Jansz N., Chen K., Leong H.S., Blewitt M.E., Asselin-Labat M.L., Smyth G.K., Ritchie M.E. (2015) Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res., 43, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wickham H. (2009) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York.
- 60. Vaes E., Khan M., Mombaerts P. (2014) Statistical analysis of differential gene expression relative to a fold change threshold on NanoString data of mouse odorant receptor genes. BMC Bioinformatics, 15, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ploner A. (2015) Heatplus: Heatmaps with row and/or column covariates and colored clusters. R package version 2.20.0. https://github.com/alexploner/Heatplus.
- 62. Zhang B., Kirov S., Snoddy J. (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res., 33, W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang J., Duncan D., Shi Z., Zhang B. (2013) WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res., 41, W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Met., 57, 289–300. [Google Scholar]
- 65. Tenenbaum D. (2017) KEGGREST: Client-side REST access to KEGG. R package version 1.14.1.
- 66. Rider M.A., Hurwitz S.N., Meckes D.G. Jr (2016) ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci. Rep., 6, 23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Helwa I., Cai J., Drewry M.D., Zimmerman A., Dinkins M.B., Khaled M.L., Seremwe M., Dismuke W.M., Bieberich E., Stamer W.D.. et al. (2017) A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS One, 12, e0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Klingeborn M., Dismuke W.M., Bowes Rickman C., Stamer W.D. (2017) Roles of exosomes in the normal and diseased eye. Prog. Retin. Eye. Res., 59, 158–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Han H., Wecker T., Grehn F., Schlunck G. (2011) Elasticity-dependent modulation of TGF-beta responses in human trabecular meshwork cells. Invest. Ophthalmol. Vis. Sci., 52, 2889–2896. [DOI] [PubMed] [Google Scholar]
- 70. Huang Y., Cen L.P., Luo J.M., Wang N., Zhang M.Z., van Rooijen N., Pang C.P., Cui Q. (2008) Differential roles of phosphatidylinositol 3-kinase/akt pathway in retinal ganglion cell survival in rats with or without acute ocular hypertension. Neuroscience, 153, 214–225. [DOI] [PubMed] [Google Scholar]
- 71. Nickells R.W., Semaan S.J., Schlamp C.L. (2008) Involvement of the Bcl2 gene family in the signaling and control of retinal ganglion cell death. Prog. Brain Res., 173, 423–435. [DOI] [PubMed] [Google Scholar]
- 72. Liu X., Hu Y., Filla M.S., Gabelt B.T., Peters D.M., Brandt C.R., Kaufman P.L. (2005) The effect of C3 transgene expression on actin and cellular adhesions in cultured human trabecular meshwork cells and on outflow facility in organ cultured monkey eyes. Mol. Vis., 11, 1112–1121. [PubMed] [Google Scholar]
- 73. Wiggs J.L., Yaspan B.L., Hauser M.A., Kang J.H., Allingham R.R., Olson L.M., Abdrabou W., Fan B.J., Wang D.Y., Brodeur W.. et al. (2012) Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet., 8, e1002654.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pasquale L.R., Loomis S.J., Weinreb R.N., Kang J.H., Yaspan B.L., Bailey J.C., Gaasterland D., Gaasterland T., Lee R.K., Scott W.K.. et al. (2013) Estrogen pathway polymorphisms in relation to primary open angle glaucoma: an analysis accounting for gender from the United States. Mol. Vis., 19, 1471–1481. [PMC free article] [PubMed] [Google Scholar]
- 75. Babizhayev M.A., Brodskaya M.W. (1989) Fibronectin detection in drainage outflow system of human eyes in ageing and progression of open-angle glaucoma. Mech. Ageing Dev., 47, 145–157. [DOI] [PubMed] [Google Scholar]
- 76. Tezel G., Hernandez R., Wax M.B. (2000) Immunostaining of heat shock proteins in the retina and optic nerve head of normal and glaucomatous eyes. Arch. Ophthalmol., 118, 511–518. [DOI] [PubMed] [Google Scholar]
- 77. Zhou G., Liu B. (2010) Single nucleotide polymorphisms of metabolic syndrome-related genes in primary open angle glaucoma. Int. J. Ophthalmol., 3, 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Konstas A.G., Marshall G.E., Lee W.R. (1990) Immunogold localisation of laminin in normal and exfoliative iris. Br. J. Ophthalmol., 74, 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Beit-Yannai E., Shmulevich A. (2007) Does the aqueous humor have a role in mitogen-activated protein kinase (MAPK) intracellular signaling in glaucoma? Med. Hypotheses, 68, 299–302. [DOI] [PubMed] [Google Scholar]
- 80. Kumar S., Shah S., Tang H.M., Smith M., Borras T., Danias J. (2013) Tissue plasminogen activator in trabecular meshwork attenuates steroid induced outflow resistance in mice. PLoS One, 8, e72447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ronkko S., Rekonen P., Kaarniranta K., Puustjarvi T., Terasvirta M., Uusitalo H. (2007) Matrix metalloproteinases and their inhibitors in the chamber angle of normal eyes and patients with primary open-angle glaucoma and exfoliation glaucoma. Graefes Arch. Clin. Exp. Ophthalmol., 245, 697–704. [DOI] [PubMed] [Google Scholar]
- 82. Schlotzer-Schrehardt U., Lommatzsch J., Kuchle M., Konstas A.G., Naumann G.O. (2003) Matrix metalloproteinases and their inhibitors in aqueous humor of patients with pseudoexfoliation syndrome/glaucoma and primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci., 44, 1117–1125. [DOI] [PubMed] [Google Scholar]
- 83. Rezaie T., Child A., Hitchings R., Brice G., Miller L., Coca-Prados M., Heon E., Krupin T., Ritch R., Kreutzer D.. et al. (2002) Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science, 295, 1077–1079. [DOI] [PubMed] [Google Scholar]
- 84. Itakura T., Peters D.M., Fini M.E. (2015) Glaucomatous MYOC mutations activate the IL-1/NF-κB inflammatory stress response and the glaucoma marker SELE in trabecular meshwork cells. Mol. Vis., 21, 1071–1084. [PMC free article] [PubMed] [Google Scholar]
- 85. Zenkel M., Kruse F.E., Naumann G.O., Schlotzer-Schrehardt U. (2007) Impaired cytoprotective mechanisms in eyes with pseudoexfoliation syndrome/glaucoma. Invest. Ophthalmol. Vis. Sci., 48, 5558–5566. [DOI] [PubMed] [Google Scholar]
- 86. Keller K.E., Yang Y.F., Sun Y.Y., Sykes R., Acott T.S., Wirtz M.K. (2013) Ankyrin repeat and suppressor of cytokine signaling box containing protein-10 is associated with ubiquitin-mediated degradation pathways in trabecular meshwork cells. Mol. Vis., 19, 1639–1655. [PMC free article] [PubMed] [Google Scholar]
- 87. Nagabhushana A., Chalasani M.L., Jain N., Radha V., Rangaraj N., Balasubramanian D., Swarup G. (2010) Regulation of endocytic trafficking of transferrin receptor by optineurin and its impairment by a glaucoma-associated mutant. BMC Cell Biol., 11, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Koliakos G.G., Schlotzer-Schrehardt U., Konstas A.G., Bufidis T., Georgiadis N., Dimitriadou A. (2001) Transforming and insulin-like growth factors in the aqueous humour of patients with exfoliation syndrome. Graefes Arch. Clin. Exp. Ophthalmol., 239, 482–487. [DOI] [PubMed] [Google Scholar]
- 89. Sethi A., Mao W., Wordinger R.J., Clark A.F. (2011) Transforming growth factor-beta induces extracellular matrix protein cross-linking lysyl oxidase (LOX) genes in human trabecular meshwork cells. Invest. Ophthalmol. Vis. Sci., 52, 5240–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Burdon K.P., Macgregor S., Hewitt A.W., Sharma S., Chidlow G., Mills R.A., Danoy P., Casson R., Viswanathan A.C., Liu J.Z.. et al. (2011) Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat. Genet., 43, 574–578. [DOI] [PubMed] [Google Scholar]
- 91. Fan B.J., Liu K., Wang D.Y., Tham C.C., Tam P.O., Lam D.S., Pang C.P. (2010) Association of polymorphisms of tumor necrosis factor and tumor protein p53 with primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci., 51, 4110–4116. [DOI] [PubMed] [Google Scholar]
- 92. Skarie J.M., Link B.A. (2008) The primary open-angle glaucoma gene WDR36 functions in ribosomal RNA processing and interacts with the p53 stress-response pathway. Hum. Mol. Genet., 17, 2474–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kang J.H., Loomis S.J., Yaspan B.L., Bailey J.C., Weinreb R.N., Lee R.K., Lichter P.R., Budenz D.L., Liu Y., Realini T.. et al. (2014) Vascular tone pathway polymorphisms in relation to primary open-angle glaucoma. Eye (London), 28, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Howe E.N., Cochrane D.R., Richer J.K. (2011) Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res., 13, R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets supporting the conclusions of this paper are available in the NCBI Gene Expression Omnibus and are accessible through the GEO Series accession number GSE105269, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE105269; date last accessed February 8, 2018).