Abstract
Despite many hypotheses regarding the roles of fluorescent proteins (FPs), their biological roles and the genetic basis of FP-mediated color polymorphisms in Acropora remain unclear. In this study, we determined the genetic mechanism underlying fluorescent polymorphisms in A. digitifera. Using a high-throughput sequencing approach, we found that FP gene sequences in FP multigene family exhibit presence–absence polymorphism among individuals. A few particular sequences in short-to-middle wavelength emission and middle-to-long wavelength emission clades were highly expressed in adults, and different sequences were highly expressed in larvae. These highly expressed sequences were absent in the genomes of individuals with low total FP gene expression. In adults, presence–absence differences of the highly expressed FP sequences were consistent with measurements of emission spectra of corals, suggesting that presence–absence polymorphisms of these FP sequences contributed to the fluorescent polymorphisms. The functions of recombinant FPs encoded by highly expressed sequences in adult and larval stages were different, suggesting that expression of FP sequences with different functions may depend on the life-stage of A. digitifera. Highly expressed FP sequences exhibited presence–absence polymorphisms in subpopulations of A. digitifera, suggesting that presence–absence status is maintained during the evolution of A. digitifera subpopulations. The difference in FPs between adults and larvae and the polymorphisms of highly expressed FP genes may provide key insight into the biological roles of FPs in corals.
Keywords: reef-building corals, copy number variation, multigene family
Introduction
Reef-building corals (Scleractinia) show diverse coloration (Marshall et al. 2003). It has been proposed that fluorescence is a major determinant of color in corals (Dove et al. 2001; Kelmanson and Matz 2003; Oswald et al. 2007). The sources of fluorescence in corals are fluorescent proteins (FPs), which emit visible light, and chlorophyll-a from symbiotic dinoflagellate algae, which emit near infrared light (Moisan and Mitchell 2001; Mazel and Fuchs 2003). FPs are excited by environmental light and emit a longer wavelength than the excitation spectrum (Johnsen 2012). The color of an FP is determined by its amino acid sequence (Field et al. 2006). The center of the region determining light emission in FPs comprises the tripeptide “–X–Y–G–” (termed a chromophore), where the first peptide X varies among FPs (Henderson and Remington 2005). Based on the emission spectra, FPs are classified into four groups: cyan (CFP), green (GFP), yellow (YFP), and red (RFP) (Labas et al. 2002; Alieva et al. 2008). Nonfluorescent chromoprotein (CP) is also classified as an FP gene family member based on its amino acid sequence similarity (Labas et al. 2002).
FP genes show signatures of diversifying selection, suggesting that FPs with different spectral ranges may have distinct roles in corals (Field et al. 2006). Although many biological roles of FPs have been predicted, the biological roles of FPs with different spectral properties remain unclear. Originally, a photo-protective role of FPs was proposed (Salih et al. 2000), and roles in photo-protection in algal symbionts have subsequently been proposed for RFP (Gittins et al. 2015) and CP (Smith et al. 2013). An antioxidant role has been suggested for CFP, GFP, RFP, and CP; however, FP mutants lacking a chromophore show greater antioxidant activity than that of wild-type FP (Palmer et al. 2009).
Acropora is one of the most abundant coral genera in coral reefs of the Indo-Pacific region (Veron 2000) and the whole genome of Acropora digitifera has been decoded (Shinzato et al. 2011). The natural variation of FPs provides an opportunity to analyze the biological role of FPs. FP-mediated color polymorphisms within Acropora species have been reported. For example, the photo-inhibitory effect is thought to be substantially weaker in Acropora palifera individuals with the fluorescent phenotype than in those with the nonfluorescent phenotype (Salih et al. 2000). A. aspera with the blue phenotype is less sensitive to changes in light than A. aspera with the light blue or cream phenotype at normal temperatures, but A. aspera with the blue phenotype is more sensitive to increases in temperature (Dove 2004). In A. nobilis, individuals with the blue phenotype show greater zooxanthellae and photosynthetic pigment concentrations than those of individuals with the brown phenotype (Smith et al. 2013). In A. valida, individuals with the purple phenotype show a weaker photo-inhibitory effect than individuals with the brown phenotype (Smith et al. 2013). In vitro analyses have demonstrated that CP affects the algal chlorophyll excitation spectrum, supporting a role for CP in the photo-protection of zooxanthellae (Smith et al. 2013). However, CP expression is not vital for shallow-water corals, as evidenced by the adjacent growth of individuals with different color phenotypes in a shallow-water reef (Smith et al. 2013). In A. millepora, a fluorescence polymorphism has been observed in both larval and adult stages (Kenkel et al. 2011; Gittins et al. 2015; Strader et al. 2016). Redder A. millepora larvae exhibit diapause-like physiological characteristics and are less responsive to settlement cues than greener larvae, suggesting that red fluorescence might be a marker of long-range larval dispersal (Kenkel et al. 2011; Strader et al. 2016). Adult A. millepora exhibit high, medium, and low redness levels under the same light levels in shallow water, and the expression levels of RFP are correlated with reduced photo-damage of zooxanthellae under light stress (Gittins et al. 2015). These expression levels are correlated with the number of particular RFP sequences with a particular promoter type (Gittins et al. 2015). Although color polymorphisms mediated by different FPs have been reported in many Acropora species, the genetic differences have only been analyzed for RFP sequences in A. millepora.
Several FP sequences have been reported for a single species in the genus Acropora (Alieva et al. 2008; D’Angelo et al. 2008; Gittins et al. 2015). In A. digitifera, the FP multigene family has been comprehensively described. More than 30 FP gene copies per individual have been reported in A. digitifera, and these copies can be separated into short-/middle-wavelength emission (S/MWE including CFP and GFP), middle-/long-wavelength emission (M/LWE including GFP and RFP), and chromoprotein (CP) clades (Takahashi-Kariyazono et al. 2016). All known FPs from Acropora species belong to one of these three clades in the FP multigene family (Takahashi-Kariyazono et al. 2016). Although FP gene copy numbers and several FP sequences have been reported in A. digitifera, the sequences in this family have not been comprehensively identified. It is necessary to determine all FP gene sequences in each individual to understand the genetic basis of fluorescence polymorphisms and the biological roles of FPs.
In this study, we focused on exon 3 of the FP gene, because it is the longest exon and contains nucleotides coding for a chromophore. We examined all exon 3 sequences from FP genes and their expression patterns in adult and larval A. digitifera. The presence and absence of highly expressed FP sequences contributed to the total expression differences among individuals.
Materials and Methods
Specimen Collection and Species Identification
A branch fragment was collected from each of 11 A. digitifera colonies in Sesoko, Okinawa, Japan. In total, 11 coral fragments (sample ID: S1601–08 and S1610–12) were preserved in RNAlater (Thermo Scientific, Waltham, MA). In addition, five different colonies of A. digitifera (sample ID: S1401–05) were collected from the field and subsequently maintained at the Sesoko Station aquarium (Tropical Biosphere Research Center, University of the Ryukyus). Bundles of gametes from each of five colonies of A. digitifera were mixed to allow fertilization. Larvae were reared by daily transfer to fresh seawater and maintained at ∼26°C. A single larva was preserved in RNAlater (Thermo Scientific). Species were identified based on morphology. This study was approved by the Aquaculture Agency of Okinawa Prefecture (permit numbers 26-9 and 28-31).
DNA Extraction and Preparation of DNA Libraries for Hybridization
Genomic DNAs were extracted from 12 coral fragments of A. digitifera (sample ID: S1403, S1601–08, and S1610–12) using DNeasy Blood & Tissue Kits (QIAGEN, Hilden, Germany). Genomic DNAs were fragmented to 500 bp using the Covaris M220 (Covaris, Inc., Woburn, MA). Following the manufacturer’s instructions, DNA libraries of 11 A. digitifera samples (sample ID: S1601–08 and S1610–12, supplementary table S1, Supplementary Material online) were constructed using the TruSeq Nano DNA Library Preparation Kit (Illumina, Inc., San Diego, CA).
Preparation of Hybridization Probes for FP Gene Capture
The FP gene family in A. digitifera can be divided into three clades (S/MWE, M/LWE, and CP) (Takahashi-Kariyazono et al. 2016). FP gene sequences were amplified using TaKaRa Ex Taq (Takara, Shiga, Japan) and the following primer sets: MiA_MWEe2_F1 and AdiFP2XbaI_R_L for the FP gene in the S/MWE clade, MiA_MLWEe2_F3 and MiA_MLWe5_R2 for the FP gene in the M/LWE clade, and AdiFP8L_KpnIF_L and AdiFP8XbaI_R2_L for the FP gene in the CP clade. All primer sequences are shown in supplementary table S2, Supplementary Material online. Genomic DNAs of A. digitifera (sample ID: S1403) and Acropora sp. were used as templates. PCR was performed using the GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA). PCR conditions for the amplification of FP gene sequences were as follows: denaturation for 3 min at 94°C, followed by 35 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 50°C, and extension for 3 min at 72°C.
Following the manufacturer’s instructions, FP gene sequences in all three clades were labeled with biotin using the Label IT Biotin Labeling Kit (Takara). Biotin-labeled DNA fragments were used as hybridization probes.
FP Gene Capture
FP gene capture was performed using the SeqCap EZ Hybridization and Wash Kit (Roche, Basel, Switzerland), SeqCap EZ Accessory Kit v2 (Roche), SeqCap HE-Oligo Kit (Roche), and SeqCap EZ Pure Capture Bead Kit (Roche) following the manufacturer’s instructions for SeqCap EZ Library SR (Roche), with minor modifications. Briefly, Biotin-labeled FP-gene fragments were used as hybridization probes, instead of the SeqCap EZ library (Roche). Equal amounts of all libraries (in total, 1 µg) were mixed with 135 ng of Biotin-labeled FP-gene fragments and were hybridized at 46°C for 72 h. Other procedures were performed in accordance with the manufacturer’s instructions.
Sequencing of Captured DNA Libraries and Extraction of Exon 3 Sequences
Short DNA sequences (paired-end, 250 bp) were determined from the captured libraries using the Illumina Hiseq2500 platform. The nucleotide sequences were deposited in the DDBJ Sequenced Read Archive under accession number DRR120550–DRR120560. Since the average insert size of the libraries was 500 bp, the overlapping paired reads were merged into one sequence read using CLC Genomics Workbench (https://www.qiagenbioinformatics.com/; last accessed June 30, 2018), after the removal of the adaptor sequences and low-quality reads (quality score <20). When the paired reads did not overlap, both reads were used, without merging. Merged and unmerged reads were used for sequence identification.
The longest exon containing nucleotides encoding a chromophore, that is, exon 3 (e3), was used for subsequent analyses. To identify e3 sequences in the FP gene, the ancestral sequences of FP genes (explained below) were used as a reference for mapping. To construct the ancestral sequences, full-length coding regions of FP genes of Acropora species were collected from the NCBI database. The accession numbers of the collected sequences are shown in supplementary table S3, Supplementary Material online. Using these FP sequences, the ancestral sequences of two subclades in the S/MWE, M/LWE, and CP clades (Takahashi-Kariyazono et al. 2016) were estimated by the maximum likelihood method with a preset tree topology using MEGA ver. 7 (Kumar et al. 2016). The ancestral sequences are listed in the supplementary material online.
Short DNA reads from each sample were mapped to the reference sequences composed of the e3 ancestral sequences of two subclades in the S/MWE, M/LWE, and CP clades using CLC Genomics Workbench. Reads showing similarity (>80%) to the e3 sequence (S/MWE and M/LWE clades: length, 225 bp [accessions: BR000963 and XM_015914911]; CP clade: length, 219 bp; supplementary fig. S1, Supplementary Material online) were mapped to reference sequences. Reads mapped to the ancestral sequences of each of the three clades were separately extracted and assembled using ATGC (GENETYX CORPORATION, Tokyo, Japan) (matching percentage = 100%), and unassembled reads were removed. Assembled contigs for each of the three clades were aligned, and contigs covering the full length of e3 were selected. Sequences in intronic regions were removed from contigs. For each sample, DNA reads (merged reads and other reads) were mapped (complete match) to these assembled e3 contigs using CLC Genomics Workbench. The e3 contigs in which at least two reads covered the full length were identified as e3 sequences, those containing insertions or deletions that cause frameshifts and/or premature stop codons were regarded as pseudogene (pseudo-e3) sequences. Among all e3 sequences isolated from 11 individuals, identical sequences were removed. The phylogenetic tree of e3 sequences was constructed using the maximum likelihood method based on the Kimura 2-parameter mode with gamma distributed rates and invariant sites using MEGA ver. 7 (Kumar et al. 2016). Bootstrap support for each clade was obtained based on 1,000 replicates. E3 sequences were deposited in GenBank under accession numbers LC349488–LC349725.
Validation of the Absence of Highly Expressed Exon 3 Sequences
Following the manufacturer’s instructions, DNA libraries of three A. digitifera samples (sample ID: S1601, S1603, and S1606) were constructed using the TruSeq DNA PCR-Free Library Preparation Kit (Illumina, Inc.). Short DNA sequences (paired-end, 125 bp) were determined from these three and from the DNA libraries of eight A. digitifera samples (sample ID: S1602, S1604–5, S1607–8, and S1610–S1612) described under Materials and Methods (DNA extraction and preparation of DNA libraries for hybridization) by the Illumina HiSeq2500 platform. The nucleotide sequences were deposited in the DDBJ Sequenced Read Archive under accession numbers DRR108003–DRR108012 and DRR108024. To verify for the absence of the highly expressed sequences in the genomes of individuals in which these sequences were not isolated, short reads from the genomes of each individual were mapped to all isolated e3 sequences. Short DNA reads showing similarity (100%) with 80% read lengths were mapped to e3 sequences using CLC Genomics Workbench. When the full length of e3 was covered, with at least one coverage without any mismatches, the e3 sequence was regarded as positive in the genome of the individual. When any reads mapped to the e3 without any mismatches, the e3 sequence was regarded as negative in the genome of the individual. If a few or several reads were mapped to e3, but the full length of e3 was not covered, the e3 sequences were classified by the second mapping condition. The second mapping was performed under the same criteria as the moderate mapping condition (similarity 100% with 40% of read length) using the same short DNA reads and e3 sequences. When the full length of e3 was covered without any mismatches under the moderate mapping condition, the e3 sequence was regarded as positive. The remaining e3 sequences were categorized as unclassified.
Estimation of A. digitifera FP Gene Copy Numbers
FP gene copy numbers in the A. digitifera genome were estimated by quantitative PCR (qPCR) using 11 A. digitifera specimens. FP gene copy number estimation was performed using previously described methods (Takahashi-Kariyazono et al. 2016) with minor modifications. Briefly, two new reverse primers (MLWE_qPCR_R1-3 for M/LWE and MiA_CP_e3_R1-3 for CP) were used for qPCR. Primer sequences are shown in supplementary table S2, Supplementary Material online. qPCR was performed using the Thermal Cycler Dice TP800 (Takara). Genomic DNAs of 11 samples (S1601–8, S1610–12) were used as templates.
RNA Extraction and Sequencing
Total RNAs were extracted from 11 adult coral fragments of A. digitifera, the same individuals used for DNA extraction, and a single A. digitifera larva using TRIzol reagent (Thermo Fisher Scientific). cDNAs were synthesized from total RNA using the PrimeScript II 1st Strand cDNA Synthesis Kit (Takara). RNAs from 11 adults were used to construct libraries for high-throughput RNA sequencing using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Bio Labs, Ipswich, MA). Short DNA sequences (paired-end, 125 bp) were determined from the libraries using the Illumina HiSeq2500 platform. The nucleotide sequences were deposited in the DDBJ Sequenced Read Archive under accession numbers DRR108013-DRR108023.
Expression Levels of Exon 3 Sequences of FP Genes
After the removal of the adaptor sequences and low-quality reads (quality score <20), RNA-seq reads from 11 individuals were mapped to A. digitifera genome assembly ver.1.1. In addition to the 11 adult individuals, RNA-seq reads from A. digitifera larvae (12.1 Gbp, accession: SRX1534820) were downloaded from a public database (DDBJ) and mapped to A. digitifera genome assembly ver.1.1. Reads showing high sequence similarity (>90%, >112 bp) were mapped to query sequences and expression values were calculated using CLC genomic workbench. RPKM (Reads Per Kilobase of exon model per Million mapped reads) were used to normalize expression values to identify candidate housekeeping genes. First, house-keeping genes that showed stable expression among individuals and developmental stages were selected as internal controls for normalization of FP gene expression levels. In particular, the variance indexes for RPKM values (SD/average RPKM) were calculated for all genes among 11 adults and larvae. Thirty-four genes with average RPKM values among 11 adults and larvae of ≥50 and with a variance index of RPKM among 11 adults and larvae of <1.0 were selected as house-keeping genes (supplementary table S4, Supplementary Material online).
To calculate expression values for e3 sequences, RNA-seq reads from 11 adults and larvae (accession: SRX1534820 and DRR054773) were mapped to all isolated e3 sequences using CLC Genomics Workbench. Reads showing high similarity to corresponding e3 sequences (100%, >119 bp) were mapped to those sequences. To normalize mapped reads, the total numbers of reads mapped to the 34 house-keeping genes were used as internal controls. The normalized expression values for FP sequences were calculated as follows: (Mapping coverage of the FP e3 sequence/Mapped reads on the internal control) × 100,000, where the mapping coverage of the FP e3 sequence was estimated as the reads mapped to the e3 sequence divided by the e3 length (bp) and mapped reads on the internal control indicates reads mapped to the 34 house-keeping genes. The e3 sequences of FPs in the S/MWE, M/LWE, and CP clades were 225,225, and 219 bp in length, respectively.
Identification of Symbiodinium Clades in Each Sample
To identify the clades of Symbiodinium in each sample, RNA-seq reads of 11 individuals were mapped to the nucleotide sequences of nuclear 28S (nr28S) and chloroplast 23S (cp23S) ribosomal DNA from eight clades of Symbiodinium (Pochon et al. 2012) using CLC Genomics Workbench. The accession numbers for each sequence are shown in supplementary table S5, Supplementary Material online.
Expression Analyses of Symbiodinium Genes
To analyze gene expression in symbiotic algae living in all coral individuals, RNA-seq reads of 11 coral individuals showing high similarity (>80%) and lengths of >100 bp were mapped to the transcriptome of Symbiodinium goreaui (clade C) (Davies et al. 2018) using CLC Genomics Workbench. The following comparisons were evaluated according to gene expression and gene presence–absence patterns: 1) high expression of S/Me3_a_hi1 and the absence of this sequence in the genome; 2) presence and absence of S/Me3_a_hi1 in the genome; 3) expression of M/L-De3_a and the absence of this sequence in the genome; 4) expression and no expression of M/LWE clade TYG-type sequences; 5) high (expression values ≥10) and low expression (expression values ≤5) of total CP sequences. The contigs with significant expression differences (p < 0.05 with FDR correction) between two groups were selected.
Identification and Cloning of FP cDNA Sequences, Purification, and Spectroscopic Analyses of Recombinant FP Proteins
Full-length cDNAs of FP genes were amplified by PCR. Using the cDNA of an A. digitifera larva as a template, S/MWE-FP full-length cDNAs were amplified using PrimeSTAR GXL DNA Polymerase (Takara). M/LWE-FP full-length cDNAs were amplified using PrimeSTAR GXL DNA Polymerase (Takara) from the cDNAs of an A. digitifera larva and an adult (sample ID: S1603). CP full-length cDNAs were amplified using TaKaRa Ex Taq from the cDNAs of an A. digitifera larva and an adult (sample ID: S1606). PCR conditions for the amplification of full-length cDNAs were as follows: denaturation for 3 min at 94°C, followed by 30 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 55°C, and extension for 1 min at 72°C. All primer sets and primer sequences are shown in supplementary tables S2 and S6, Supplementary Material online. PCR products were cloned into the T-Vector pMD20 vector (Takara), and the sequences were verified using the Applied Biosystems Automated 3130xl Sequencer (Foster City, CA).
To construct recombinant FP proteins, vectors with cloned FP sequences were used as templates for subcloning into expression vectors. Full-length FP cDNAs were subcloned into the pCold I expression vector (Takara) and then used to transform BL21 Escherichia coli cells (Takara). Each clone was grown in 20 mL of LB medium supplemented with ampicillin and IPTG overnight, and the recombinant proteins were extracted by sonication and purified using TALON beads with poly-histidine tags (Takara).
Emission and excitation spectra of purified recombinant FP proteins in 50 mmol/L phosphate buffer solution with 500 mmol/L imidazole, pH 7.0, were measured using the RF-6000 spectro-fluorophotometer (SHIMAZU, Kyoto, Japan). Absorption spectra of purified recombinant CP proteins were measured using a UV-1800 spectrophotometer (SHIMAZU). Measurements were obtained at least three times for each FP.
Identification of FP Genes in the Three A. digitifera Subpopulations
The publicly available genomic DNA reads of A. digitifera collected from the southern Ryukyu Archipelago located in southwestern Japan were used (supplementary fig. S2, Supplementary Material online). These individuals have been separated into four subpopulations; Okinawa, Kerama, Yaeyama-South, and Yaeyama-North (Shinzato et al. 2016). Individuals with mapping read coverage ≥6 were selected for our analysis. The mapping read coverages were based on those in a previous report (Shinzato et al. 2016). A total of 91 sets of genomic DNA reads from 40 individuals belonging to Okinawa, 39 individuals belonging to Kerama, and 12 individuals belonging to Yaeyama-North were downloaded. A subpopulation (Yaeyama-South) was excluded from our analysis, because there were only two individuals with mapping read coverage ≥6 in this subpopulation. The accession numbers of downloaded genomic DNA reads are shown in supplementary table S7, Supplementary Material online. Validation of the existence of highly expressed e3 sequences in each individual from the three subpopulations was performed using the method explained under Materials and Methods (Validation of the absence of highly expressed exon 3 sequences).
Live Coral Fluorescence Measurements
Light emission, including reflectance and fluorescence spectra, was measured from each of 11 A. digitifera colonies that were used for DNA and RNA extraction. Visual observation did not reveal clear color differences among 11 A. digitifera colonies. An LED source with a peak wavelength of 448 nm was used as the excitation light source. The distances from the excitation light and measurement probe to objects were 6 cm, and spectra were recorded using the Jaz Spectrometer (Ocean Optics, Dunedin, FL) in the dark. For all measurements, light emitted at longer than 660 nm was not attributed to coral fluorescence because chlorophyll a from the symbiotic dinoflagellate algae living within the coral tissues emit light with a primary peak wavelength ∼685 nm and a secondary peak at 730 nm (Moisan and Mitchell 2001; Mazel and Fuchs 2003). Fluorescence was estimated by excitation (LED)–emission spectral subtraction.
Results
Copy Number Variation and Sequence Diversity in the FP Gene Family
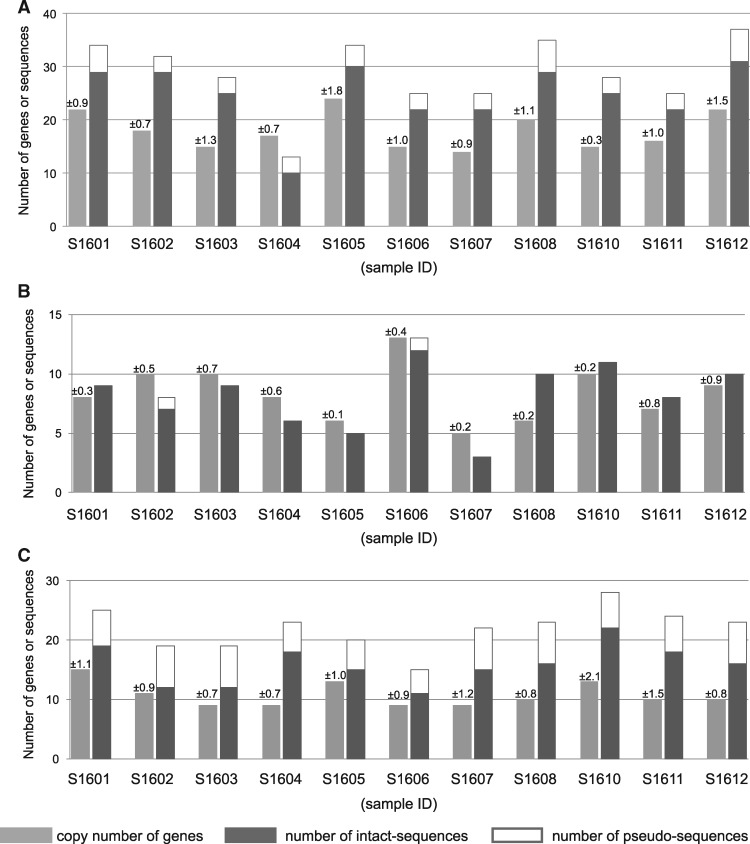
We quantified FP gene copy numbers in 11 A. digitifera genomes by qPCR. We detected 14–24 FP gene copies in the S/MWE clade, 5–13 in the M/LWE clade, and 9–15 in the CP clade (fig. 1), indicating copy number variation among individuals.
Fig. 1.
—FP gene copy numbers and total numbers of sequences obtained in each clade. FP gene copy numbers and exon 3 sequence numbers in the MWE (A), M/LWE (B), and CP (C) clades from 11 adult colonies of Acropora digitifera are given. Copy numbers are shown in light gray with SDs obtained from three replicates. Numbers of intact exon 3 sequences are shown in dark gray and pseudo-exon 3 sequences are shown in white. S1601–S1612 indicate sample IDs.
Despite the high estimated copy numbers, only ten FP genes (including nine partial genes) were identified in the assembled genome sequence of A. digitifera (Shinzato et al. 2012). This inconsistency may reflect the difficulty of multigene family assembly (Mariano et al. 2015). To identify sequence diversity in FP genes, we determined the exon 3 (e3) sequences of FP genes without de novo assembly. We chose e3 because it has the longest exon among FP genes (those of the S/MWE, M/LWE, and CP clades were 225, 225, and 219 bp in length, respectively) and contains the region that encodes a chromophore. DNA fragments containing FP genes in libraries were condensed by FP gene capture, and short DNA sequences (paired-end, 250 bp) were determined (2,141,708–2,850,976 reads) from 11 samples (supplementary table S1, Supplementary Material online). These reads from each library were mapped to e3 of S/MWE FP (1,069–1,532 reads), M/LWE FP (215–655 reads), and CP (628–1,698 reads) genes. Using mapped reads, e3 sequences of FP genes were determined from 11 A. digitifera genomes (fig. 1). We detected different e3 nucleotide sequences from 11 individuals, including 111 intact and 19 pseudosequences in the S/MWE clade, 29 intact and one pseudosequence in the M/LWE clade, and 49 intact and 29 pseudosequences in the CP clade (table 1). The intact e3 sequences were translated into amino acid sequences. The number of amino acid sequences were 67, 17, and 38 in the S/MWE, M/LWE, and CP clades, respectively. Based on the first peptide of the chromophore, we divided the e3 sequences in the M/LWE clade into two types, TYG-type and DYG-type. The phylogenetic trees based on the nucleotide sequences of e3 are shown in supplementary figure S3A–C, Supplementary Material online. Among the three clades, we detected the most e3 sequences at both the nucleotide and amino acid sequence levels in the S/MWE clade.
Table 1.
Number of Exon 3 Sequences Isolated in Three Clades
| Clade | Intact exon 3 |
Pseudo-exon 3 |
|
|---|---|---|---|
| Nucleotide | Amino Acid | Nucleotide | |
| S/MWE | 111 | 67 | 19 |
| M/LWE | 29 | 17 | 1 |
| CP | 49 | 38 | 29 |
Expression Differences for Exon 3 Sequences of FP Genes among Adult Individuals
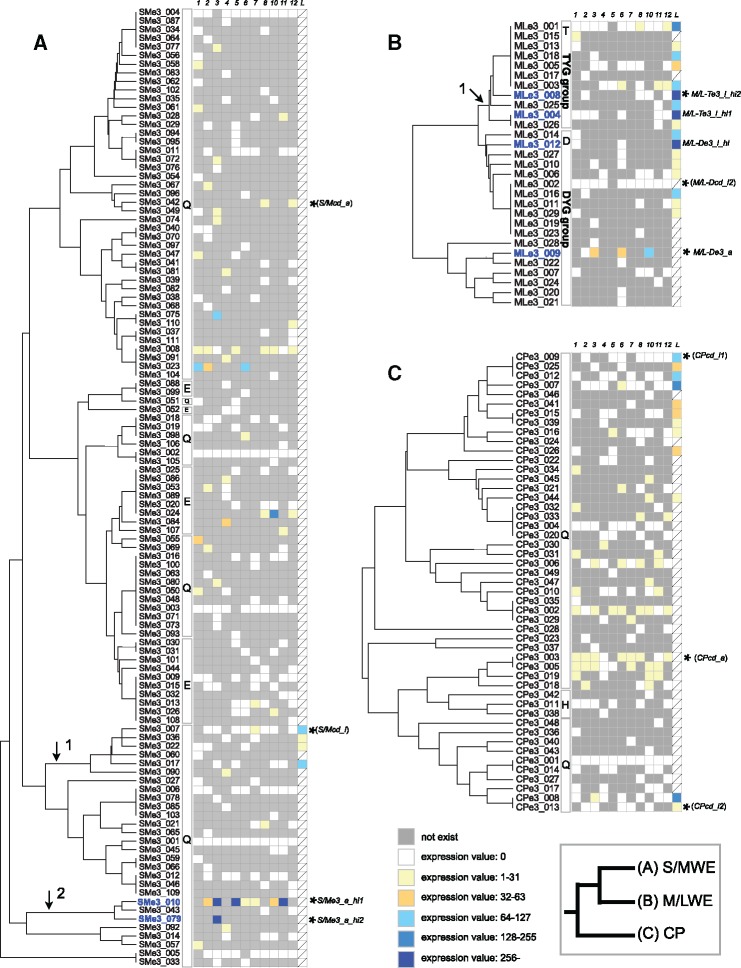
We examined the expression levels of each e3 sequence based on RNA-seq reads from 11 adult individuals (41,007,102–109,525,174 reads). In RNA-seq read mapping, 36–50% of RNA-seq reads were mapped to the A. digitifera genome ver. 1.1 (supplementary table S9, Supplementary Material online). Expression levels of e3 sequences were normalized by the expression levels of 34 house-keeping genes and categorized into six levels (fig. 2). The expressed e3 sequences in larval (see below) and adult stages are summarized in table 2.
Fig. 2.
—Expression of each FP sequence in adults and larvae. Rows indicate the exon 3 sequences belonging to S/MWE clade (A), M/LWE clade (B), and CP clade (C). Columns indicate the normalized expression values for FP sequences calculated by the expression of the internal control genes (see Materials and Methods) in 11 adults and larvae. Sample IDs S1601–S1612 are indicated as 01–12. L indicates larvae. A gray box indicates the absence of the sequence in a genome of a sample and a slash mark indicates that the sequence data were not available. Exon 3 sequences were aligned based on the phylogenetic trees in supplementary figure S2, Supplementary Material online (UPGMA based on amino acid differences) to show amino acid differences. Black arrow indicates a branch supported by the ML tree in supplementary figure S3, Supplementary Material online. The first peptide of the chromophore for each exon 3 sequences is shown in a rectangle. An asterisk indicates the exon 3 sequence used for the recombinant protein analysis.
Table 2.
Number of Expressed Sequences in the Larval and Adult Stages
| Clade | Adults and Larvae | Only Adults | Only Larvae |
|---|---|---|---|
| S/MWE | 1 | 34 | 3 |
| M/LWE_TYG | 2 | 1 | 7 |
| M/LWE_DYG | 0 | 1 | 8 |
| CP | 4 | 16 | 8 |
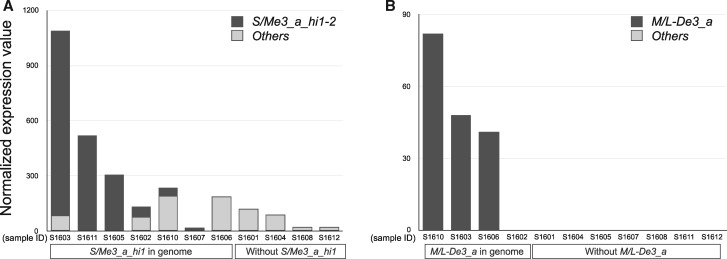
We detected variation in total expression levels in each clade or type of M/LWE among 11 individuals (table 3). In the S/MWE clade, among the individuals with high overall FP gene expression (table 3, total expression of the S/MWE clade sequences in S1603, S1605, and S1611), one e3 sequence (SMe3_010) was highly expressed (fig. 2A). We refer to this e3 sequence as “S/Me3_a_hi1”; it was also detected in four other individuals (fig. 2A: S1602, S1606–7, and S1610), but at lower levels than those in the three high-expression individuals (table 3). In the other four individuals (fig. 2A: S1601, S1604, S1608, and S1612), we did not detect S/Me3_a_hi1 in genomic DNA (250 bp reads) or RNAseq reads. In addition to S/Me3_a_hi1, the other e3 sequence (e3 ID: SMe3_079) in a single individual (fig. 2A: S1603) showed high sequence similarity with S/Me3_a_hi1 and high expression. We refer to this e3 sequence as “S/Me3_a_hi2.” Among three individuals with high overall FP gene expression in the S/MWE clade, the expression of S/Me3_a_hi1 and S/Me3_a_hi2 sequences accounted for >90% of the total expression (fig. 3A). The absence of S/Me3_a_hi1 and S/Me3_a_hi2 in the genomes of individuals shown by the 250 bp reads from genomic DNA was verified using short reads (paired-end, 125 bp). These results indicate that each of the S/Me3_a_hi1 and S/Me3_a_hi2 sequences were present in one individual and absent from another individual, thus, there were presence–absence polymorphisms in these 11 individuals.
Table 3.
Expression Values for Specific Exon 3 Sequences
| Sample | S/MWE |
M/LWE (DYG) |
M/LWE (TYG) |
CP |
||||
|---|---|---|---|---|---|---|---|---|
| S/Me3_a_hi1-2 | Other Than S/Me3_a_hi1-2 | Total | M/L-De3_a | Other Than M/L-De3_a | Total | Total | Total | |
| S1601 | 0 | 120 | 120 | 0 | 0 | 0 | 1 | 13 |
| S1602 | 56 | 77 | 133 | 0 | 0 | 0 | 0 | 8 |
| S1603 | 1,002 | 86 | 1,088 | 48 | 0 | 48 | 0 | 6 |
| S1604 | 0 | 89 | 89 | 0 | 0 | 0 | 0 | 3 |
| S1605 | 304 | 5 | 309 | 0 | 0 | 0 | 1 | 4 |
| S1606 | 1 | 188 | 189 | 41 | 0 | 41 | 1 | 43 |
| S1607 | 16 | 2 | 18 | 0 | 0 | 0 | 0 | 5 |
| S1608 | 0 | 24 | 24 | 0 | 0 | 0 | 1 | 5 |
| S1610 | 46 | 191 | 237 | 83 | 0 | 83 | 0 | 10 |
| S1611 | 514 | 5 | 519 | 0 | 0 | 0 | 1 | 8 |
| S1612 | 0 | 21 | 21 | 0 | 0 | 0 | 2 | 7 |
| Larvae | 0 | 239 | 239 | 0 | 1,236 | 1,236 | 1,186 | 830 |
Fig. 3.
—Proportions of highly expressed exon 3 sequences in the S/MWE and M/LWE clades. The normalized expression values for FP sequences calculated by the expression of the internal control genes (see Materials and Methods) in the (A) S/MWE clade (S/Me3_a_hi1 and S/Me3_a_hi2) and (B) M/LWE clade (M/L-De3_a) and the expression of all FP sequences in each clade, except highly expressed sequences shown in black and gray, respectively. The y axis indicates the normalized expression value. S1601–S1612 indicate sample IDs.
In the M/LWE clade, we observed that 4 of 29 e3 nucleotide sequences were expressed in at least one adult. In four adult individuals (fig. 2B: S1602, S1604, S1605, and S1607), we did not detect the expression of any e3 sequence. In adults, only one DYG-type sequence (e3 ID: MLe3_009) was expressed in three samples (fig. 2B: S1603, S1606, and S1610), and little expression of TYG-type sequences was observed. We refer to this expressed DYG-type e3 sequence as “M/L-De3_a”; we identified this sequence in four individuals and verified these findings using short reads (paired-end, 125 bp) of genomic DNA from these individuals (supplementary table S7, Supplementary Material online). Four individuals had M/L-De3_a, among which one individual (fig. 3B; ID: S1602) did not express this sequence. The other six individuals did not possess this sequence in their genome (supplementary table S7, Supplementary Material online), indicating a presence–absence polymorphism of M/L-De3_a.
In the CP clade, we detected the expression of 20 out of 49 e3 nucleotide sequences in at least one adult. In one sample (ID: S1606), total expression in the CP clade was relatively high (table 3), and this expression was attributed to five e3 sequences (fig. 2C). We did not identify an e3 sequence with high expression in the CP clade (fig. 2C).
Expression Differences in Exon 3 Sequences of FP Genes between Adults and Larvae
We compared the expression of each e3 sequence between planula larvae and adults using publicly available RNA-seq reads of larvae (accession: SRX1534820) and RNA-seq reads of 11 adults, respectively. In the S/MWE clade, we observed larval expression of four closely related e3 sequences, and we did not detect the expression of these four e3 sequences in adults, except for low expression in one individual (fig. 2A), indicating differences in the regulation of FP gene expression between adults and larvae. The total expression of sequences in this clade was lower in larvae than in the adults with the three highest expression levels (table 3).
In the M/LWE clade, we observed higher total expression of both TYG-type and DYG-type sequences in larvae than in adults (table 3). Although we observed lower expression of TYG-type than DYG-type sequences in 11 adults, we detected similar total expression values for TYG-type and DYG-type sequences in larvae. We observed high expression of two (fig. 2B: MLe3_004, and MLe3_008) and one (fig. 2B: MLe3_012) TYG-type and DYG-type e3 sequences, respectively. Hereafter, we refer to these e3 sequences as “M/L-Te3_l_hi1,” “M/L-Te3_l_hi2,” and “M/L-De3_l_hi.” Among 11 adult individuals, we isolated M/L-Te3_l_hi1, M/L-Te3_l_hi2, and M/L-De3_l_hi from six, four, and two individuals, respectively (fig. 2B), and the presence and absence of these sequences in the genome were verified by short reads (paired-end, 125 bp) of genomic DNA (supplementary table S7, Supplementary Material online). These results indicate that highly expressed sequences in larvae showed presence–absence polymorphisms in adult individuals.
In the CP clade, we observed higher total expression of CP clade sequences in larvae than in adults (table 3). We detected the expression of 12 e3 sequences in larvae, and no or low expression of these e3 sequences in 11 adults (fig. 2C). These results indicate differences in CP sequences expression between adults and larvae.
The genetic differences in FP genes (copy number variation and presence–absence polymorphisms) in individuals used as “adult samples” and “larval samples” might affect the expression patterns of FP genes at different life stages. To examine this possibility, we compared two independent RNA-seq data sets of A. digitifera larvae (accessions: SRX1534820 and DRR054773). The results showed that two RNA-seq data sets from larvae had similar expression patterns, and that the patterns were different from those of adults (supplementary fig. S5, Supplementary Material online). This expression pattern difference was most likely the result of a difference in expression between adults and larvae rather than the result of a difference in genetic background.
In all three clades, some e3 sequences exhibited no expression in both adults and larvae (fig. 2A–C). It is possible that these unexpressed sequences are parts of pseudogenes and/or are expressed at different life stages (e.g., developmental stages) or in specific conditions.
Functional Differences among FPs
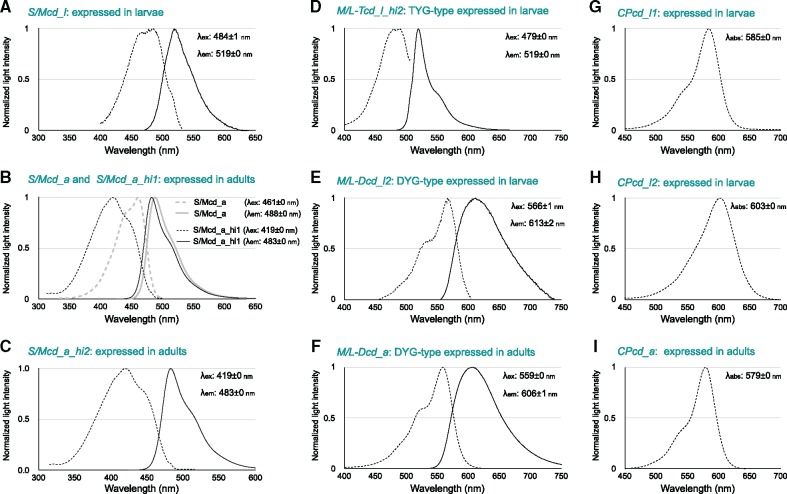
To evaluate functional differences among FP e3 sequences, we cloned full-length FP coding regions, constructed recombinant FPs, and measured emission and excitation or absorption spectra. In the S/MWE clade, we cloned four different full-length cDNAs of FP genes, that is, S/Mcd_l expressed in larvae, S/Mcd_a expressed in adults, and S/Mcd_a_hi1 and S/Mcd_a_hi2, which are highly expressed in adults. The e3 sequences of full-length cDNAs of FP genes were identical to the e3 sequences we determined, and the correspondence between full-length cDNA names and e3 sequence IDs is shown in supplementary table S6, Supplementary Material online. The emission peak of recombinant S/Mcd_l expressed in larvae was 519 ± 0 nm (fig. 4A), whereas the emission peaks of two recombinant proteins expressed in adults (S/Mcd_a and S/Mcd_a_hi1) were 488 ± 0 nm and 483 ± 0 nm, respectively (fig. 4B). Thus, recombinant FPs encoded by FP sequences highly expressed in larvae and adults exhibited different emission peaks.
Fig. 4.
—Emission and absorption spectra of FP recombinant proteins. Absorption (dotted line) and emission (solid line) spectra of recombinant proteins belonging to the S/MWE clade (A, S/Mcd_l; B, S/Mcd_a and S/Mcd_a_hi1; and C, S/Mcd_a_hi2), M/LWE clade (D, M/L-Tcd_i_hi2; E, M/L-Dcd_l2; and F, M/L-Dcd_a), and CP clade (G, CPcd_a; H, CPcd_l1; I, CPcd_l2) are shown. Horizontal and vertical axes indicate wavelengths (nanometers) and normalized light intensities, respectively.
Furthermore, we detected similar emission peaks for recombinant S/Mcd_a_hi1 (highly expressed) and S/Mcd_a (fig. 4B). Recombinant S/Mcd_a_hi1 was excited by light under 380 nm, whereas S/Mcd_a was not highly excited by light in this range. The excitation and emission spectra of recombinant S/Mcd_a_hi1 (shown in black in fig. 4B) and S/Mcd_a_hi2 (fig. 4C) were identical, indicating that these two highly expressed e3 sequences in adults share the same function in the wavelength of excitation and emission spectra of FP.
In the M/LWE clade, we cloned three different full-length cDNAs of FP genes, that is, M/L-Tcd_l_hi2 (TYG-type expressed in larvae), M/L-Dcd_l2 (DYG-type with identical amino acid sequences expressed in larvae), and M/L-Dcd_a (DYG-type expressed in adults). Emission peaks for the recombinant M/L-Tcd_l_hi2, M/L-Dcd_l2, and M/L-Dcd_a were 519 ± 0 nm, 613 ± 2 nm, and 606 ± 1 nm, respectively (fig. 4D–F).
In the CP clade, we cloned three different full-length cDNAs of FP genes, that is, CPcd_l1 expressed in larvae, CPcd_l2 expressed in larvae, and CPcd_a expressed in adults. Absorption peak values for recombinant CPcd_l1, CPcd_l2, and CPcd_a were 585 ± 0 nm, 603 ± 0 nm, and 579 ± 0 nm, respectively (fig. 4G–I). These CPs absorbed long wavelength light.
Identification of Highly Expressed FP Sequences in Three A. digitifera Subpopulations
In S/MWE, we detected high S/Me3_a_hi1 and S/Me3_a_hi2 expression in three adult individuals with high total FP gene expression. Additionally, in the M/LWE clade DYG-type, expression differences among 11 adult individuals were caused by differences in M/L-De3_a expression. In larvae, M/L-Te3_l_hi1, M/L-Te3_l_hi2, and M/L-De3_l_hi in the M/LWE clade were highly expressed.
To confirm whether these sequences existed in A. digitifera subpopulations in the southern Ryukyu Archipelago, we examined 102 individuals from three subpopulations (Okinawa, Kerama, and Yaeyama-North) (Shinzato et al. 2016). We detected the presence of each of S/Me3_a_hi1, M/L-De3_a, M/L-Te3_l_hi1, M/L-Te3_l_hi2, and M/L-De3_l_hi in the genomes of individuals from the three subpopulations (table 4 and supplementary table S7, Supplementary Material online). We detected the presence and absence of each of S/Me3_a_hi1, M/L-Te3_l_hi2, and M/L-De3_l_hi in the genomes of individuals from the three subpopulations (table 4 and supplementary table S7, Supplementary Material online), indicating that the presence–absence polymorphic state of these sequences was shared by the three subpopulations. We found S/Me3_a_hi2 in two subpopulations, but not in the Yaeyama-North subpopulation containing six individuals without this sequence or with partially covered (categorized as unclassified, see Materials and Methods). In addition, we found only individuals with M/L-Te3_l_hi1 or unclassified individuals in Kerama and Yaeyama-North. In these cases, we were unable to determine whether or not S/Me3_a_hi2 and M/L-Te3_l_hi1 were presence–absence polymorphic in these subpopulations. We found individuals with M/L-De3_a, but no individual without M/L-De3_a or unclassified individuals in Yaeyama-North, suggesting that this sequence is fixed or exists at high frequencies in this subpopulation. However, since the number of individuals in Yaeyama-North used in this analysis was smaller than in the other two subpopulations, it could have occurred by chance.
Table 4.
Number of Individuals That Possess or Did Not Possess Specific Exon 3 Sequences in Their Genome
| FP e3 Sequence | Subpopulation | Number of Individuals |
Polymorphic State | ||
|---|---|---|---|---|---|
| Positive | Negative | Unclassified | |||
| S/Me3_a_hi1 | Okinawa (n=51) | 28 | 7 | 16 | Polymorphic |
| Kerama (n=39) | 27 | 4 | 8 | Polymorphic | |
| Yaeyama_North (n=12) | 3 | 5 | 4 | Polymorphic | |
| S/Me3_a_hi2 | Okinawa (n=51) | 1 | 11 | 39 | Polymorphic |
| Kerama (n=39) | 3 | 10 | 26 | Polymorphic | |
| Yaeyama_North (n=12) | 0 | 6 | 6 | Unknown | |
| M/L-De3_a | Okinawa (n=51) | 13 | 27 | 11 | Polymorphic |
| Kerama (n=39) | 9 | 23 | 7 | Polymorphic | |
| Yaeyama_North (n=12) | 12 | 0 | 0 | (Presence-fixed) | |
| M/L-Te3_l_hi1 | Okinawa (n=51) | 31 | 5 | 15 | Polymorphic |
| Kerama (n=39) | 32 | 0 | 7 | Unknown | |
| Yaeyama_North (n=12) | 4 | 0 | 8 | Unknown | |
| M/L-Te3_l_hi12 | Okinawa (n=51) | 13 | 3 | 35 | Polymorphic |
| Kerama (n=39) | 11 | 2 | 26 | Polymorphic | |
| Yaeyama_North (n=12) | 2 | 3 | 7 | Polymorphic | |
| M/L-De3_l_hi | Okinawa (n=51) | 13 | 14 | 24 | Polymorphic |
| Kerama (n=39) | 19 | 6 | 14 | Polymorphic | |
| Yaeyama_North (n=12) | 4 | 4 | 4 | Polymorphic | |
Differences of FP Gene Expression Were Consistent with Differences in Fluorescence Emitted from Coral Individuals
The emission spectra (including reflectance and fluorescence) from 11 A. digitifera colonies excited by LED (448 nm spectrum peak) were measured in the sea. In individuals with high expression of S/Me3_a_hi1, fluorescence spanned ∼475–570 nm (supplementary fig. S4A–C, Supplementary Material online).
We did not observe fluorescence from four individuals without S/Me3_a_hi1 in their genomes (supplementary fig. S4D–G, Supplementary Material online). Two of the four individuals with low expression of S/Me3_a_hi1 (ID: S1607, 1610) did not show fluorescence (supplementary fig. S4J and K, Supplementary Material online). The remaining two individuals (ID: S1602, 1606) with low expression of S/Me3_a_hi1 showed fluorescence with a peak at 516 nm (supplementary fig. S4H and I, Supplementary Material online). The fluorescence spanning ∼500–590 nm in one individual (ID: 1606) (supplementary fig. S4I, Supplementary Material online) may have been emitted by different FPs, because the emission peak of the FP encoded by S/Me3_a_hi1 was 483 nm (fig. 4B). The fluorescence from one individual (ID: 1602) (supplementary fig. S4H, Supplementary Material online) may have been emitted by a mixture of FPs encoded by S/Me3_a_hi1 and other sequences. These results show that fluorescence polymorphisms existed among 11 A. digitifera; individuals with fluorescence spanning ∼475–570 nm, one individual with fluorescence spanning ∼500–590 nm, and individuals with no fluorescence. Coral fluorescence spanning ∼475–570 nm was consistent with the high expression of S/Me3_a_hi1 and absence of S/Me3_a_hi1 in the genome.
FP Expression Does Not Affect Gene Expression in Symbiotic Algae
Photo-protective effects of FPs on photosynthetic algal symbionts have been proposed (Salih et al. 2000; Smith et al. 2013; Gittins et al. 2015). However, we observed differences in FP expression among individuals collected from the same shallow water light environment. To compare the conditions of symbiotic algae living in each coral individual, we evaluated the levels of all expressed genes in symbiotic algae by RNA-seq.
First, we identified the clade of symbiotic algae in the 11 individuals that we collected using the mapping results for RNA-seq reads to the nucleotide sequence list of nr28S and cp23S ribosomal DNA from eight clades of algae. In all individuals, RNA-seq reads were mapped to nr28S and cp23S of the clade C, and few reads were mapped to sequences of other clades (supplementary table S5, Supplementary Material online). This result indicates that most symbiotic algae living in the 11 individuals of A. digitifera belonged to clade C.
To compare gene expression, RNA-seq reads from 11 adult individuals were mapped to the transcriptome of S. goreaui (clade C) (Davies et al. 2018). Approximately 20–34% of RNA-seq reads were mapped to the contigs (supplementary table S9, Supplementary Material online). For all comparisons, the expression levels of algal genes were strongly correlated between groups (r = 0.98–1.00, supplementary table S8, Supplementary Material online). In the comparisons based on presence and absence of S/Me3_a_hi1 (comparisons #1 and 2) and the expression level of M/L-De3_a (comparison #3), no gene showed significant differences in expression. In the comparisons based on the expression level of M/LWE TYG-type sequences (comparison #4), we detected significant differences (p < 0.05 with FDR correction) in the expression levels of one contig annotated as cytochrome b6-f complex subunit four between groups (supplementary table S10, Supplementary Material online). In the comparisons based on the expression level of CP sequences (comparison #5), we detected significant differences (p < 0.05 with FDR correction) in the expression levels of one unannotated contig between groups (supplementary table S10, Supplementary Material online).
Discussion
Genetic Mechanism Underlying the Fluorescence Polymorphism among Individuals of A. digitifera
Acropora FPs are thought to be involved in the protection of photosynthetic algal symbionts against sunlight (Salih et al. 2000; Smith et al. 2013; Gittins et al. 2015). However, intraspecific FP-mediated color polymorphisms have been found within the same light environment (Smith et al. 2013; Gittins et al. 2015). This natural variation raises questions about the importance of FPs for photo-protection. To address this question, it is necessary to determine the mechanism that generates the intraspecific fluorescence polymorphism.
The expression of FP genes in adult Acropora individuals is influenced by external stimuli, such as light, heat, and injury. CFP, GFP, RFP, and CP levels increase according to the light intensity (D’Angelo et al. 2008; Roth et al. 2010), and CFP is down-regulated in dark stress (DeSalvo et al. 2012). Heat stress down-regulates GFP (Roth and Deheyn 2013). The expression of CFP increases in response to injury (D’Angelo et al. 2012). However, these factors cannot explain variation in fluorescence in corals living in the same environment, without injury.
Previously, we reported variation in the sequences and total copy number of particular FP genes in A. digitifera (Takahashi-Kariyazono et al. 2016); therefore, we analyzed copy number variation in 11 adult individuals of A. digitifera, living in the same light environment in shallow water. We detected copy number variation for FP genes from each of three clades (S/MWE, M/LWE, and CP). The numbers of e3 sequences and the e3 sequence repertories in each of three clades also differed among individuals. The numbers of e3 sequences were generally higher than the estimated gene copy numbers in the S/MWE and CP clades, suggesting that FP genes in these clades exhibit high allelic diversity. Hence, there were large genetic differences in FP genes in each of two clades among individuals of A. digitifera.
In each of three clades, we detected variation in the total expression levels of FP genes among 11 adult individuals. In the S/MWE clade, one sequence (S/Me3_a_hi1) showed a presence–absence polymorphism, and high expression of this sequence contributed to the total expression levels in the individuals with the top three overall expression levels of FP sequences in the S/MWE clade. In this clade, four individuals possessed S/Me3_a_hi1, but the expression levels were lower than those of the other three individuals with high expression. This lower expression may be due to differences in gene regulation or promotor sequence differences. In the M/LWE clade DYG-type, expression differences among 11 adult individuals were explained by the expression of one sequence (M/L-De3_a) in three individuals that possessed this sequence. These results suggest that particular FP sequences contribute to differences in total FP gene expression among individuals. In the CP clade, we did not observe a highly expressed sequence in adult samples. A comparative analysis of the emission spectra and expression of FP sequences indicated that fluorescence emission and nonemission of corals were consistent with high expression of S/Me3_a_hi1 and the absence of S/Me3_a_hi1 in the genome, respectively. According to these results, we conclude that the presence–absence differences of particular FP sequences with high expression contribute to the fluorescence differences among individuals in A. digitifera.
FP Gene Expression Differences between Adults and Larvae
The overall expression of FP genes in each clade differed substantially between adults and larvae, especially for both DYD- and TYG-type in the M/LWE clade and in the CP clade. High expression of the M/LWE clade in larvae was consistent with the high proportion of RFP in the larval transcriptome of A. millepora (Meyer et al. 2011). These results suggest that FPs in the M/LWE and CP clades have important roles in larvae. However, several adult individuals did not possess the FP sequences with high expression in larvae in the M/LWE and CP clades. This result suggests that the individuals without highly expressed sequences in larvae did not express these sequences when they were larvae. In other words, the intensity of fluorescence and the expression of FP genes by highly expressed sequences might be different among larvae. Indeed, a difference in fluorescence has been reported among larvae of A. millepora (Kenkel et al. 2011; Strader et al. 2016).
In the S/MWE clade, the expressed FP sequences were different between adults and larvae. In the S/MWE clade, the FP sequence with high expression in larval stage was absent in several adult individuals, suggesting that the expression of this FP gene might differ among larval individuals. Hence, the expression of FPs was different between adults and larvae.
Hypothetical Biological Roles of FPs in A. digitifera
The biological roles of FPs have been proposed based on natural variation in fluorescence in corals (Salih et al. 2000; Smith et al. 2013; Gittins et al. 2015). However, observed fluorescence variation can be explained by genetic differences or by differences in the regulation of gene expression among individuals. In this study, we demonstrated FP genetic variation among individuals of A. digitifera. Based on genetic differences, it was possible to separate individuals into different groups. Therefore, we examined two proposed biological roles of FPs using genetic differences and RNA-seq data containing reads from algal symbionts.
FPs in adult Acropora species have a proposed role in the photo-protection of algal symbionts (Salih et al. 2000; Smith et al. 2013; Gittins et al. 2015). A supportive role in photosynthesis by algal symbionts has also been proposed (Salih et al. 2000). These two biological roles predict that algal photosynthesis for individuals with high FP gene expression is more active than that for individuals with very low expression. In cultured symbiotic algae (genus Symbiodinium), up-regulation of a transcript encoding intrinsic membrane-bound light harvesting proteins is caused by the high intensity of light illumination, although most of transcripts encoding genes related to photosynthesis did not show light intensity-regulated expression (Xiang et al. 2015).
We compared the expression of symbiotic algae within a coral host (in hospite) with respect to the presence and absence of highly expressed genes in the S/MWE and M/LWE (TYG- and DYG-type) clades in their genomes. In the case of CP, the groups were categorized by their expression values. In comparisons of S/MWE, M/LWE DYG-type, and CP clades, no gene related to algal photosynthesis showed differential expression, suggesting that FP gene expression in the S/MWE, M/LWE DYG-type, and CP clades observed in this study did not have strong effects on transcriptional regulatory processes of genes related to algal photosynthesis. A contig annotated as cytochrome b6-f complex subunit 4 was significantly more highly expressed (p < 0.05 with FDR correction) in individuals without M/LWE TYG-type expression than in individuals with M/LWE TYG-type expression. However, genes encoding the other subunits of cytochrome b6-f complex did not show differential expression (supplementary table S11, Supplementary Material online). Therefore, we could not conclude the effects of M/LWE TYG-type on algal photosynthesis based on the differential expression of only one subunit of the cytochrome b6-f complex. In this study, we only studied 11 individuals; thus, it is possible that RNA-seq analysis with larger data sets might detect some more subtle differences.
One finding in our study was that there was a difference in FP function between adults and larvae. Adult individuals with fluorescence emission mainly expressed FP sequences of the S/MWE clade excited by short wavelength light, including UV light, and emitted fluorescence with a peak under 490 nm. Larvae mainly expressed FP genes in the S/MWE clade excited by short wavelength light (visible light, no UV) and emitted fluorescence with a peak at a longer wavelength (519 nm). Among the FP recombinants that we analyzed, both the excitation and emission spectra of larval FPs had longer wavelengths than those of adult FPs. The same shift in fluorescence has been observed in Seriatopora hystrix (Roth et al. 2013), suggesting that FPs may have similar roles in adults and larvae of distantly related coral species. In addition to the S/MWE clade, larvae mainly expressed FP genes in the M/LWE and CP clades. The excitation and emission wavelengths were middle (TYG-type) and long (DYG-type) in the M/LWE clade. CPs absorbed long wavelength light. In summary, it is possible to hypothesized that adults utilized UV to short wavelength light and larvae utilized middle to long wavelength light via FPs, based on the functions of the highly expressed sequences in adults and larvae. These different light usages may provide insight into the biological roles of FPs.
The other key feature of FP genes in A. digitifera was the pattern of polymorphisms. The highly expressed sequences (S/Me3_a_hi1 and M/L-De3_a) in adults from the S/MWE and M/LWE clades were presence–absence polymorphisms in three A. digitifera subpopulations in the southern Ryukyu Archipelago with one exception, M/L-De3_a, in Yeyama-North. In addition, two highly expressed sequences in larvae from the M/LWE clade were presence–absence polymorphisms in all subpopulations, and polymorphisms in fluorescence in larvae were predicted. These polymorphisms suggest that the presence–absence status of highly expressed FP sequences has been maintained in A. digitifera subpopulations, at least in the southern Ryukyu Archipelago. In other words, the presence and absence of fluorescence emission in adults (short wavelengths) and in larvae (middle to long wavelengths) has persisted throughout the evolution of A. digitifera subpopulations. This polymorphic status may be beneficial for the survival of immobile adult corals in various environments, for example, different light conditions, depths, currents, and attached substrates. In larvae, the polymorphic status may be beneficial for different dispersal distances, as suggested in A. millepora (Kenkel et al. 2011; Strader et al. 2016). In summary, the presence–absence polymorphisms of highly expressed FP sequences were a notable feature and will be key to unraveling the biological roles of FPs in corals.
In this study, we demonstrate the genetic basis of differences in fluorescence among individuals in A. digitifera. Comparisons between FP genotypes and the habitats or external environments of corals will provide a basis for understanding the biological roles of FPs in corals in the near future.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by an internal SOKENDAI grant to Y.T., a Center for the Promotion of Integrated Sciences (CPIS) of SOKENDAI grant to Y.T., and a Grant-in-Aid for JSPS Fellows to S.T.-K. We thank Drs Masayuki Hatta (Ochanomizu University) for help with sampling and Mutsumi Nishida (University of the Ryukyus) for arranging the sampling.
Literature Cited
- Alieva NO, et al. , . 2008. Diversity and evolution of coral fluorescent proteins. PLoS One 37:e2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo C, et al. , . 2008. Blue light regulation of host pigment in reef-building corals. Mar Ecol Prog Ser. 364:97–106. [Google Scholar]
- D’Angelo C, et al. , . 2012. Locally accelerated growth is part of the innate immune response and repair mechanisms in reef-building corals as detected by green fluorescent protein (GFP)-like pigments. Coral Reefs 314:1045–1056. [Google Scholar]
- Davies SW, Ries JB, Marchetti A, Castillo KD. (2018) Symbiodinium Functional Diversity in the Coral Siderastrea siderea Is Influenced by Thermal Stress and Reef Environment, but Not Ocean Acidification. Front. Mar. Sci. 5:150. [Google Scholar]
- DeSalvo MK, Estrada A, Sunagawa S, Medina M.. 2012. Transcriptomic responses to darkness stress point to common coral bleaching mechanisms. Coral Reefs 311:215–228. [Google Scholar]
- Dove S. 2004. Scleractinian corals with photoprotective host pigments are hypersensitive to thermal bleaching. Mar Ecol Prog Ser. 272:99–116. [Google Scholar]
- Dove SG, Hoegh-Guldberg O, Ranganathan S.. 2001. Major colour patterns of reef-building corals are due to a family of GFP-like proteins. Coral Reefs 193:197–204. [Google Scholar]
- Field SF, Bulina MY, Kelmanson IV, Bielawski JP, Matz MV.. 2006. Adaptive evolution of multicolored fluorescent proteins in reef-building corals. J Mol Evol. 623:332–339. [DOI] [PubMed] [Google Scholar]
- Gittins JR, D’Angelo C, Oswald F, Edwards RJ, Wiedenmann J.. 2015. Fluorescent protein-mediated colour polymorphism in reef corals: multicopy genes extend the adaptation/acclimatization potential to variable light environments. Mol Ecol. 242:453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JN, Remington SJ.. 2005. Crystal structures and mutational analysis of amFP486, a cyan fluorescent protein from Anemonia majano. Proc Natl Acad Sci USA. 10236:12712–12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen S. 2012. The optics of life. New Jersey: Princeton University Press. [Google Scholar]
- Kelmanson IV, Matz MV.. 2003. Molecular basis and evolutionary origins of color diversity in great star coral Montastraea cavernosa (Scleractinia: faviida). Mol Biol Evol. 207:1125–1133. [DOI] [PubMed] [Google Scholar]
- Kenkel CD, Traylor MR, Wiedenmann J, Salih A, Matz MV.. 2011. Fluorescence of coral larvae predicts their settlement response to crustose coralline algae and reflects stress. Proc R Soc B Biol Sci. 2781718:2691–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 337:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labas YA, et al. , . 2002. Diversity and evolution of the green fluorescent protein family. Proc Natl Acad Sci USA. 99:4256–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariano DC, et al. , . 2015. MapRepeat: an approach for effective assembly of repetitive regions in prokaryotic genomes. Bioinformation 116:276–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NJ, Jennings K, McFarland WN, Loew ER, Losey GS.. 2003. Visual biology of Hawaiian coral reef fishes. III. Environmental light and an integrated approach to the ecology of reef fish vision. Copeia 3:467–480. [Google Scholar]
- Mazel CH, Fuchs E.. 2003. Contribution of fluorescence to the spectral signature and perceived color of corals. Limnol Oceanogr. 48(1part2):390–401. [Google Scholar]
- Meyer E, Aglyamova GV, Matz MV.. 2011. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol Ecol. 20:3599–3616. [DOI] [PubMed] [Google Scholar]
- Moisan TA, Mitchell BG.. 2001. UV absorption by mycosporine-like amino acids in Phaeocystis antarctica Karsten induced by photosynthetically available radiation. Mar Biol. 1381:217–227. [Google Scholar]
- Oswald F, et al. , . 2007. Contributions of host and symbiont pigments to the coloration of reef corals. FEBS J. 2744:1102–1109. [DOI] [PubMed] [Google Scholar]
- Palmer CV, Modi CK, Mydlarz LD.. 2009. Coral fluorescent proteins as antioxidants. PLoS One 410:e7298.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochon X, Putnam HM, Burki F, Gates RD.. 2012. Identifying and characterizing alternative molecular markers for the symbiotic and free-living dinoflagellate genus Symbiodinium. PLoS One 71:e29816.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MS, Deheyn DD.. 2013. Effects of cold stress and heat stress on coral fluorescence in reef-building corals. Sci Rep. 31: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MS, Fan TY, Deheyn DD.. 2013. Life history changes in coral fluorescence and the effects of light intensity on larval physiology and settlement in Seriatopora hystrix. PLoS One 83:e59476.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MS, Latz MI, Goericke R, Deheyn DD.. 2010. Green fluorescent protein regulation in the coral Acropora yongei during photoacclimation. J Exp Biol. 213(Pt 21):3644–3655. [DOI] [PubMed] [Google Scholar]
- Salih A, Larkum A, Cox G, Kühl M, Hoegh-Guldberg O.. 2000. Fluorescent pigments in corals are photoprotective. Nature 4086814:850–853. [DOI] [PubMed] [Google Scholar]
- Shinzato C, et al. , . 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 4767360:320–323. [DOI] [PubMed] [Google Scholar]
- Shinzato C, Mungpakdee S, Arakaki N, Satoh N.. 2016. Genome-wide SNP analysis explains coral diversity and recovery in the Ryukyu Archipelago. Sci Rep. 51:18211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, Shoguchi E, Tanaka M, Satoh N.. 2012. Fluorescent protein candidate genes in the coral Acropora digitifera genome. Zool Sci. 294:260–264. [DOI] [PubMed] [Google Scholar]
- Smith EG, D’Angelo C, Salih A, Wiedenmann J.. 2013. Screening by coral green fluorescent protein (GFP)-like chromoproteins supports a role in photoprotection of zooxanthellae. Coral Reefs 322:463–474. [Google Scholar]
- Strader ME, Aglyamova GV, Matz MV.. 2016. Red fluorescence in coral larvae is associated with a diapause-like state. Mol Ecol. 252:559–569. [DOI] [PubMed] [Google Scholar]
- Takahashi-Kariyazono S, Gojobori J, Satta Y, Sakai K, Terai Y.. 2016. Acropora digitifera encodes the largest known family of fluorescent proteins that has persisted during the evolution of Acropora species. Genome Biol Evol. 811:3271–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veron JEN. 2000. Corals of the world. Townsville (Australia: ): Australian Institute of Marine Science. [Google Scholar]
- Xiang T, Nelson W, Rodriguez J, Tolleter D, Grossman AR.. 2015. Symbiodinium transcriptome and global responses of cells to immediate changes in light intensity when grown under autotrophic or mixotrophic conditions. Plant J. 821:67–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.