Abstract
Developmental eye defects often severely reduce vision. Despite extensive efforts, for a substantial fraction of these cases the molecular causes are unknown. Recessive eye disorders are frequent in consanguineous populations and such large families with multiple affected individuals provide an opportunity to identify recessive causative genes. We studied a Pakistani consanguineous family with three affected individuals with congenital vision loss and progressive eye degeneration. The family was analyzed by exome sequencing of one affected individual and genotyping of all family members. We have identified a non-synonymous homozygous variant (NM_001128918.2: c.1708C > G: p.Arg570Gly) in the MARK3 gene as the likely cause of the phenotype. Given that MARK3 is highly conserved in flies (I: 55%; S: 67%) we knocked down the MARK3 homologue, par-1, in the eye during development. This leads to a significant reduction in eye size, a severe loss of photoreceptors and loss of vision based on electroretinogram (ERG) recordings. Expression of the par-1 p.Arg792Gly mutation (equivalent to the MARK3 variant found in patients) in developing fly eyes also induces loss of eye tissue and reduces the ERG signals. The data in flies and human indicate that the MARK3 variant corresponds to a loss of function. We conclude that the identified mutation in MARK3 establishes a new gene-disease link, since it likely causes structural abnormalities during eye development and visual impairment in humans, and that the function of MARK3/par-1 is evolutionarily conserved in eye development.
Introduction
Developmental eye birth defects are of clinical significance due to severely reduced visual impairment (VI), yet the genetic cause of only a small number of gene variants associated with developmental eye defects has been identified (1). VI causes a significant burden in the life of individuals and has a strong impact on their psychological and socioeconomic experiences. The elucidation of the genomic causality provides a better understanding of the molecular pathophysiology of the various forms of VI, enhances the diagnostic possibilities and provides targets and possible strategies for therapeutic interventions (2,3). Mendelian disorders provide a direct way to identify genes and variants of high phenotypic impact (4). Large consanguineous families with multiple affected individuals are an obvious source to identify pathogenic genetic variants in recessive disorders (5–8). It has been estimated that more than 10% of marriages occur among close relatives worldwide (9); in some countries, this reaches almost 70%, and this is the case in Pakistan (www.consang.net). We argue that a considerable number of additional recessive genes implicated in VI could be identified in these populations (https://sph.uth.edu/retnet/).
Here, we report a Pakistani consanguineous family of three affected individuals with severe VI and progressive eye loss. The family was analyzed by combining whole exome sequencing and homozygosity mapping, which is a powerful and cost-effective methodology to identify candidate genes in undiagnosed families with likely autosomal recessive disorders (10–12). We identified a homozygous, likely pathogenic missense variant in MARK3 (microtubule-associated protein/microtubule affinity-regulating kinase 3) gene (OMIM* 602678) that is likely to cause VI and the progressive phthisis bulbi in patients after birth. Modeling of the variant in the Drosophila homologue of the MARK3 gene leads to reduced eye development and dysfunction, supporting the pathogenicity of the MARK3 variant.
Results
Phenotypes
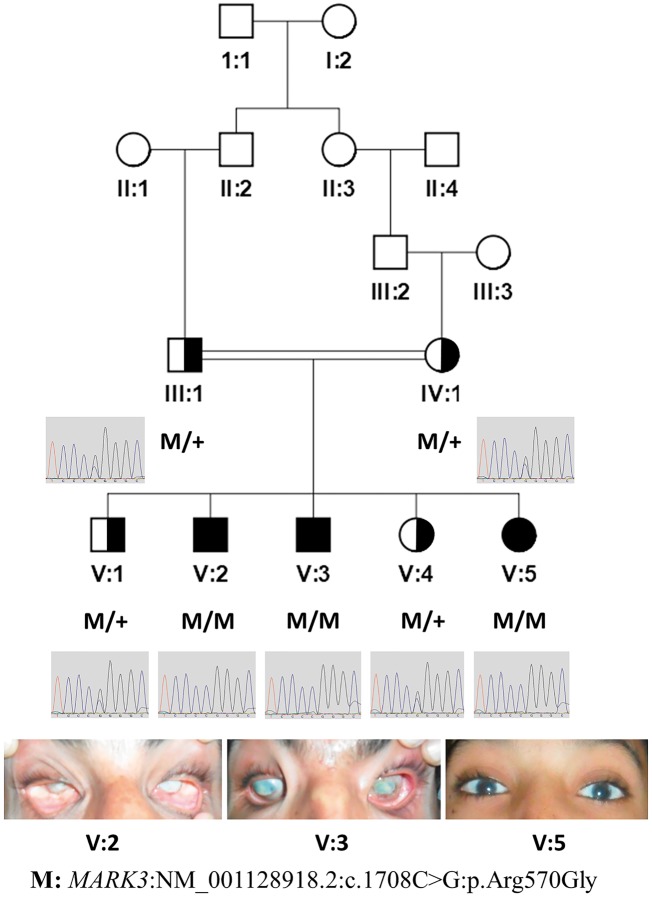
Family F105 with three affected individuals (Fig. 1), is of Pakistani origin from the area of Jamshoro, Sindh, Pakistan. Parents are first cousins once removed. All three probands had poor vision at birth. Individuals V: 2 and V: 3, who are 30- and 18-years old, respectively, developed eye phthisis by adulthood. Individual V: 5, a 15-years old female, has poor vision with low acuity along with hazy cornea (Figs 1 and 2).
Figure 1.
MARK3 variant segregation in the Pakistani family pedigree F105 and eye phenotype. Genotypes of the variant (M), also shown by chromatograms (reverse complement), in the pedigree diagram of a consanguineous family of three affected and two unaffected individuals. Photographs of eyes of the affected individuals are shown in the lower panel. Half-filled symbols denote a carrier for the pathogenic variant. Filled symbols denote affected individuals.
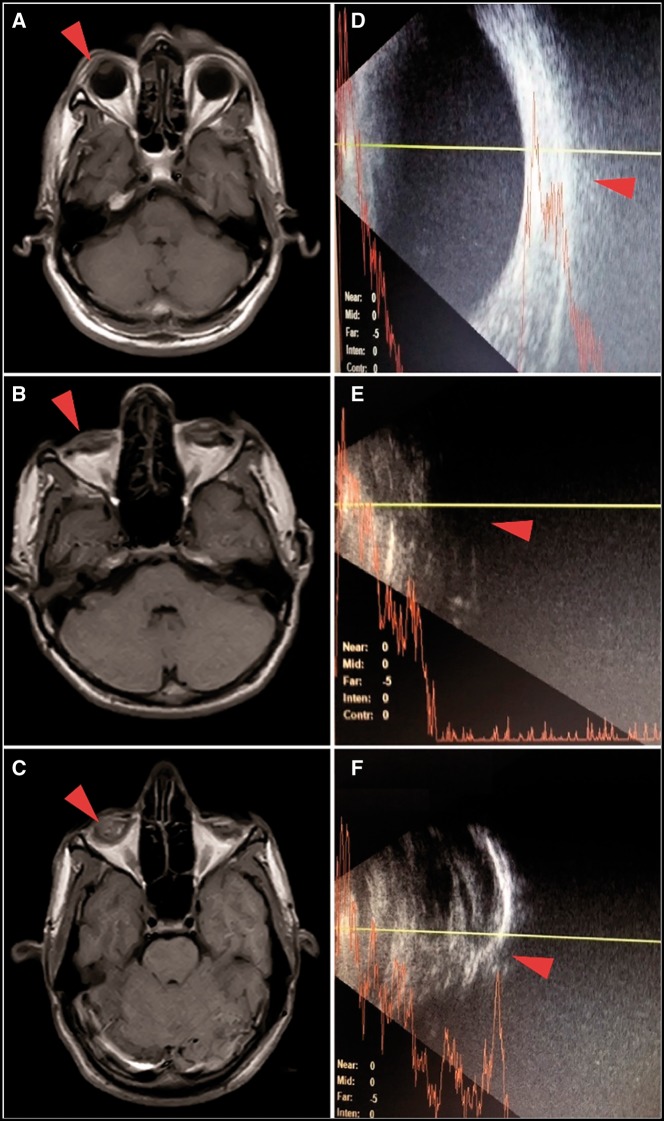
The magnetic resonance imaging (MRI) findings revealed that bilaterally the eye globe was deformed and smaller in size in both probands V: 2 and V: 3. In addition, there was a loss of the normal structural differentiation between the anterior and posterior chambers, including aqueous and vitreous humor. The right and left eye globes were measured as 1.7× 1.4 cm and 1.7×1.5 cm, respectively, in V: 2; while in V: 3 they were 2.0×1.8 cm and 2.0×1.7 cm (normal =2.42 ± 0.96×2.41 ± 0.97 cm) (13) (Fig. 2). B-scan and A-scan ultrasonography of affected individual V: 3 showed that the posterior wall was thick along with vitreous opacity, and the interior–posterior chamber was small with an axial diameter of about 14 mm (normal =24 mm). No posterior eye wall was detected in patient V: 2 on the B-scan (Fig. 2). The ophthalmologic examination of patient V: 5 showed hypermetropia with corneal plana and ptosis. The best corrected visual acuity was 2/60 and 6/60 in the right and left eye, respectively. Fundoscopic examination revealed a clear fundus with a normal cup and disk ratio and the tonometry indicated normal intraocular pressure (IOP) in both eyes (17 mm/Hg OD, 12 mm/Hg OS).
Figure 2.
MRI and B-scans of two affected individuals (V: 2 and V: 3). B and C: T1 weighted MRI images of patients V: 2 and V: 3, respectively, arrows indicate the shrunken eye globes; A is the representative MRI image with normal eye globes. E and F are the B-scan images of patients V: 2 and V: 3, respectively, showing abnormal eye globe; D is a representative B-scan of a normal eye.
Genetic analysis
Exome sequencing was performed in one affected individual (V: 3) of the family, and 95.3% of the target exome regions were covered by more than 10 reads. We did not find any potential likely pathogenic variant by considering all of the 261 genes that have been documented to affect vision, including those in the RetNet (https://sph.uth.edu/retnet/) database. Subsequently, we used a 720K SNP-array to genotype all five siblings (V: 1, V: 2, V: 3, V: 4 and V: 5) and both parents (III: 1 and IV: 1) of family F105. By combining both the exome sequence and genotyping data through CATCH (14) analysis, we identified two candidate variants that segregated in the family with a recessive pattern of inheritance; the first was in the MARK3 gene (NM_001128918.2: c.1708C > G: p.Arg570Gly) on chromosome 14 (OMIM* 602678), and the second in the PAH gene (NM_000277.2: c.355C > T: p.Pro119Ser) on chromosome 12 (OMIM *612349). The segregation of both variants was confirmed through Sanger sequencing in all members of the family (Fig. 1 and Supplementary Material, Fig. S1).
The missense variant in MARK3, segregated with the disease phenotype in a recessive fashion in the seven members of the family studied, with the LOD score of 1.45 (Fig. 1). The frequency of this variant in the gnomAD database (http://gnomad.broadinstitute.org/) is 0.000052 and the probability of LoF intolerance (pLI) for MARK3 in the ExAC database (http://exac.broadinstitute.org/) (15) is zero, suggesting that null mutations in one allele of MARK3 are not uncommon and that a disease associated with the gene would likely be recessive (MARRVEL.org) (16). The MARK3 variant is predicted to be pathogenic by various prediction algorithms [DANN (17) score: 0.99]. MARK3 Arg570 is very well conserved down to Drosophila (MARRVEL.org) (16) (Supplementary Material, Fig. S2).
The non-synonymous variant PAH p.Pro119Ser has been classified as likely pathogenic in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) because in a compound heterozygote P119S/IVS12 + 1G > A case, the phenotype was a mild phenylketonuria (PKU) with phenylalanine levels of 440 µmol/l (18). Furthermore, the p.Pro119Ser variant is categorized as tetrahydrobiopterin (BH4) responsive type of PKU (19). The phenotype in the subjects V: 2, V: 3 and V: 5 was not compatible with the PKU; however, we have measured phenylalanine and tyrosine levels from Guthrie cards. Individual V: 2 has phenylalanine 117 µmol/l and tyrosine 36 µmol/l and individual V: 3 has 148 µmol/l phenylalanine and 46 µmol/l tyrosine (normal levels are phenylalanine: 26–98 µmol/l and tyrosine: 35–107 µmol/l). These results confirm that the phenylalanine levels were slightly increased but not in the PKU disease range. We conclude that the homozygosity of the PAH p.Pro119Ser variant does not result in PKU in this family. This ‘incidental’ finding, which is not related to the eye phenotype, provides additional information in the PKU field related to the interpretation of this variant. It helps populate public databases with genomic variants from consanguineous individuals so that homozygosity for likely pathogenic variants can be evaluated against certain phenotypes.
Drosophila model analysis
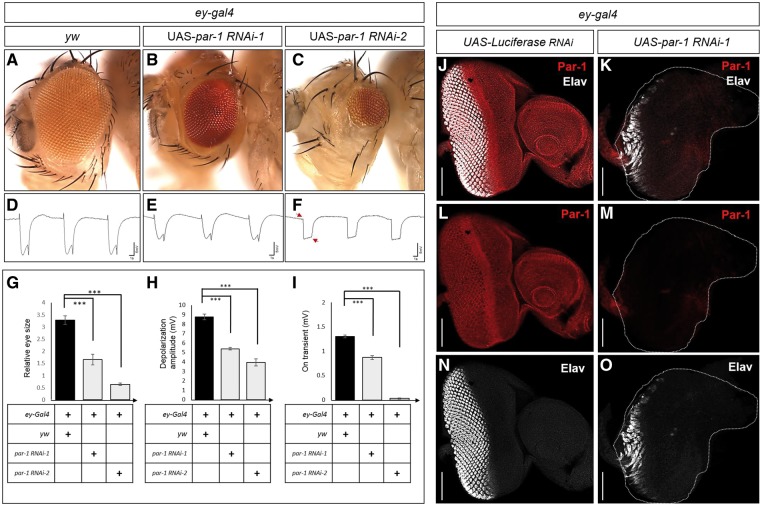
The homologue of MARK3 with the highest DIOPT score (11/12) in Drosophila is par-1, indicating that par-1 is the orthologue of MARK3 (16). We therefore used two independent RNAis to reduce Par-1 expression in the developing eye by driving the RNAis with the eyeless GAL4 (ey-GAL4) driver (20) which is expressed throughout eye development. As shown in Figure 3, this leads to a significant reduction in eye size (Fig. 3A–C) and a reduced amplitude of the electroretinograms (ERGs) (Fig. 3D–F). Control RNAi experiments do not affect the eye morphology (Supplementary Material, Fig. S3). The most severe phenotype associated with RNAi-2 causes not only a severe reduction in amplitude, arguing that the retinal function is severely impaired, but also a loss of on-off transients (arrows in Fig. 3F, H and I), indicating a loss of vision. We confirmed that the ey > par-1 RNAi successfully knocked down the Par-1 protein level in developing eye using a polyclonal Par-1 antibody (Fig. 3J and K). Importantly, the number of developing photoreceptors marked by Elav antibody which labels neurons/photoreceptors, is severely reduced in eye imaginal discs when compared to control discs (Fig. 3N and O). In summary, the data show that Par-1 is required for proper eye development and photoreceptor differentiation and that its loss leads to vision loss.
Figure 3.
Drosophila Par-1 is required for proper eye development and photoreceptor differentiation. (A–C) Adult heads with eyes in which various transgenes were expressed: ey-Gal4 in a y w background (A), ey-Gal4 driving UAS-par-1 RNAi-1 (B), or UAS-par-1 RNAi-2 (C). Expression of either par-1 RNAi-1 (BL#35342) or par-1 RNAi-2 (BL#32410) by ey-Gal4 causes small eyes (B and C). (D–F) ERGs of flies shown above. Arrows indicate the loss of on- and off-transients of ERGs. Quantification of eye sizes (G) (n=10; error bars represent ±SEM, ∗∗∗P<0.001, t-test) (H and I). Quantification of the ERG amplitude (H) and on-transients (I) of ERG traces in (D–F) (n=13; error bars represent ±SEM, ∗∗∗P<0.001, t-test). (J–O) Expression of par-1 RNAi-1 driven by ey-Gal4 significantly reduces the levels of Par-1 in eye discs. (J and K) Eye imaginal disc is stained for Par-1 (red) and the pan-neural marker Elav (white). Par-1 is ubiquitously expressed in the developing eye disc (L). The expression of Par-1 is severely reduced in eye disc of ey > UAS-par-1 RNAi-1 (M). (N and O) Knockdown of Par-1 results in a dramatic reduction in photoreceptor numbers. Elav staining in wild-type control discs (N). Knockdown of Par-1 severely reduces the number the photoreceptors in developing eye disc. Scale bars, 50 μm (J–O).
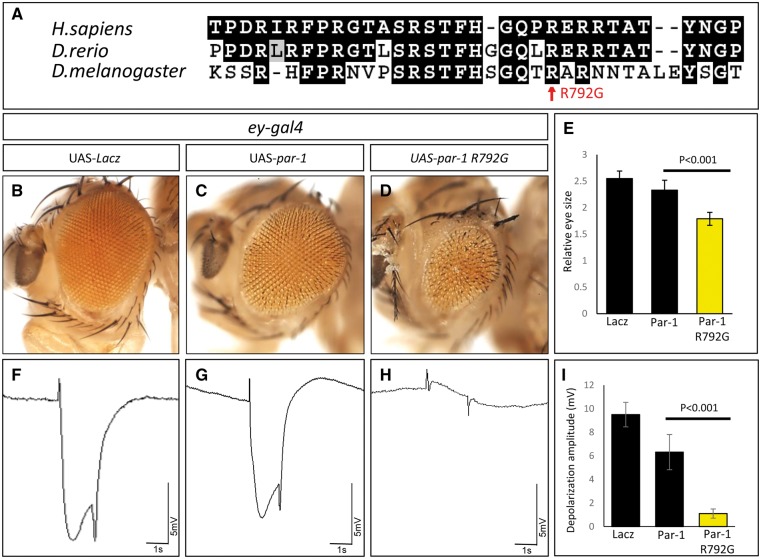
To investigate the functional effects of the identified MARK3 variants, we created the equivalent Drosophila par-1 mutation (p.Arg792Gly) and generated transgenic animals harboring this variant cDNA (UAS-par-1 p.Arg792Gly) as well as the wild-type Drosophila cDNA (UAS-par-1 WT). We again used ey-GAL4 to express these cDNAs in the developing eye (21). As a negative control, we expressed the lacZ gene with ey-GAL4 and observed a normal eye morphology and ERG (Fig. 4B and F). Expression of the wild-type Par-1 protein in the developing eye causes a mild defect in eye morphology and a subtle reduction in ERG amplitude (Fig. 4C and G). In contrast, expression of the Par-1 p.Arg792Gly variant induces severely reduced eyes that are very rough and causes a near complete loss of amplitude of the ERG (Fig. 4D, H and I). In summary, these data suggest that Par-1 p.Arg792Gly acts as a dominant negative mutation in flies when overexpressed.
Figure 4.
Par-1 p.Arg792Gly, the Drosophila mutation homologous to the human MARK3 p.Arg570Gly variant, affects eye development and function. (A) Multiple-sequence alignment of the protein domain of MARK3 that contains the p.Arg570Gly shows that this domain is evolutionarily conserved from flies to human. (B–D) Adult fly heads expressing UAS-lacZ (B), UAS-par-1 WT (C), or UAS-par-1 p.Arg792Gly (D), when driven by ey-Gal4. Expression of Par-1 WT causes a mild defect in eye morphology (C), whereas expression of the p.Arg792Gly protein causes severely reduced eyes that are rough (D). Quantification of the eye sizes (E) (n=10; error bars represent ±SEM, P<0.001, t-test). (F–H) ERGs of flies carrying ey-Gal4 with UAS-lacZ (F), UAS-par-1 WT (G), or UAS-par-1 p.Arg792Gly (H). Quantification of the ERGs shows that expression of the Par-1 p.Arg792Gly mutation causes a near complete loss of ERG amplitude (I) (n=10; error bars represent ±SEM, P<0.001, t-test).
Discussion
We propose here that the biallelic non-synonymous p.Arg570Gly variant in the MARK3 gene likely causes phthisis bulbi and VI in the affected individuals of the Pakistani consanguineous family F105. MARK3 is involved in the phosphorylation of various microtubule-associated proteins MAP2 (OMIM *157130), MAP4 (OMIM *157132) and tau/MAPT (OMIM *157140) (22). Loss of MAPT protein results in defects in photoreceptor development in Drosophila (23), and a small eye phenotype was observed in transgenic rats bearing a human MAPT gene (24). Also, MARK3 interacts with Mitf (25), which is known to cause the COMMAD syndrome with severe microphthalmia (26). MARK3 together with DLG5 function as negative regulators of the Hippo pathway: the MARK3/DLG5 complex inhibits the kinase activity of MST1/2 and LATS1 by inhibiting the MST1/2 mediated phosphorylation of LATS1 (22). The MARK3 knockout mouse presented with resistance to weight gain under high fat diet and exhibited defects in gluconeogenesis (27). However, these mice were not studied for phenotypes related to vision. This colony of mice does no longer exists and was not available for further studies.
We studied the par-1 (ortholog of MARK3) gene in Drosophila. We found that the down-regulation of par-1 by RNAi specifically in the eye leads to a very severe loss of photoreceptors in the developing fly eye and causes a near or complete loss of visual function based on ERG (Fig. 3). Moreover, expression of Par-1 p.Arg792Gly, the equivalent of human variant p. Arg570Gly, results in a small, non-functional eye which closely resembles the ey > par-1 RNAi phenotype. These data strongly suggest that the MARK3 p.Arg570Gly variant is pathogenic.
MARK3/par-1 is a Ser/Thr kinase initially identified as an essential polarity gene for a proper asymmetric division in Caenorhabditis elegans embryos (28). Prior studies in invertebrates and vertebrates have shown that the MARK3/par-1 homologs are necessary for cell polarity and that loss-of-function mutations are associated with a range of developmental defects (29–32). In Drosophila, Par-1 is necessary for anterior–posterior axis formation, and par-1 loss-of-function embryos exhibit severe patterning defects (33). Moreover, loss of Par-1 in various imaginal discs leads to a mild reduction in tissue size, induces some apoptotic cell death during development and affects Hippo signaling (34).
In Xenopus, early dorsoanterior development of the embryo is disrupted by the loss of par-1. This causes a loss of Wnt signaling (32). Interestingly, in the same study, knocking down of par-1 expression resulted in a lack of eye development as well as a decrease in body axis length. Wnt signaling is involved in eye development and retinal angiogenesis (35–37). Other genes involved in Wnt signaling have been implicated in eye diseases in humans. For instance, individuals with pathogenic mutations in FZD4, another core component of the Wnt pathway, disrupts retinal angiogenesis (38,39). Furthermore, individuals with mutations in the FZD4 binding partner, NDP, are affected by Norrie disease (40). These mutations cause blindness as well as phenotypes that are very similar to those observed in the patients with the MARK3 p.Arg570Gly variant. Hence, MARK3/par-1 is evolutionarily critical for early eye development and may affect Wnt signaling.
The study of the incidental finding in the PAH gene emphasizes the notion that the pathogenicity of variants for recessive disorders should be evaluated in the context of the other allele. In the case of the family F105 presented here, the PAH p.Pro119Ser variant in homozygosity cause a mild increase of the phenylalanine level but not in the PKU range. On the other hand, the same PAH p.Pro119Ser variant in combination with a ‘null’ splicing error causes mild PKU that requires treatment (18). The further development of databases of recessive variants would be essential for the interpretation of their phenotypic relationship and the genetic counseling regarding the at-risk carrier status.
We did not find other cases of mutations in MARK3 by sharing our findings in Genematcher (41). Additional families with MARK3 pathogenic variants and similar eye phenotype will be necessary to establish causality. Patients with Norrie-like phenotype but with an autosomal mode of inheritance should be evaluated for MARK3 pathogenic variants if possible. We conclude that the MARK3 homozygous missense variant is an excellent candidate to cause the progressive phthisis of the eyes and VI at birth observed in our patients.
Materials and Methods
Patients and study consent
The family was enrolled at the Molecular Biology & Genetics Department, Medical Research Center, Liaquat University of Medical & Health Sciences, Jamshoro, Pakistan, and was studied at the Department of Genetic Medicine and Development, University of Geneva, Geneva, Switzerland. This study was approved by the Bioethics Committee of the University Hospitals of Geneva (Protocol number: CER 11–036), and by the ethical committee of Liaquat University of Medical & Health Sciences, Jamshoro, Pakistan. Informed consent was obtained from the members of this family.
Clinical investigations
The detailed clinical investigations were performed on all affected individuals to characterize the variability of the phenotype. MRI of orbit was performed on patients V: 2 and V: 3 of Figure 1, and T1, T2 weighted images were obtained without contrast in axial, coronal and sagittal planes. Bilateral B and A scans were also performed on patients V: 2 and V: 3. The Patient V: 5 was subjected to fundoscopy, tonometry and visual acuity assessments.
Laboratory and computational analysis
Exome sequencing was performed using the SureSelect Human All Exon kit v5 (Agilent Technologies, Santa Clara, CA, USA) on an Illumina HiSeq4000. The data analysis was done by using a customized pipeline, based on Burrows–Wheeler aligner tool (BWA) (42), SAMtools (42), PICARD (http://broadinstitute.github.io/picard/) and GATK (43). The alignment was performed by using the reference human genome (GRCh37/hg19) (44). Genotyping of samples from all available family members was done using the 720 K SNP array (HumanOmniExpress Bead Chip by Illumina Inc®, San Diego, CA, USA). PLINK (45) was used to perform homozygosity mapping by taking into account a window of 50 consecutive homozygous SNPs, allowing a maximum of one mismatch in a homozygous region. Runs of homozygosity (ROH) were defined as homozygous regions separated by the first contiguous heterozygous SNPs. The family was analyzed by using CATCH (14), an in-house program which combines the information of family pedigree, ROH, and data of exome sequence. This program identifies variants in the ROHs of patients but not in normal individuals of a family. Subsequently, the variants were filtered as described in previous studies (11,12). All the candidate variants were further verified by Sanger sequencing.
Cloning and transgenesis
Site-directed mutagenesis was performed for full-length cDNA of par-1 in the pENTR223 Gateway donor vector (RE47050 from Drosophila Genomics Resource Center). To generate a par-1 p.Arg792Gly (R792G) [equivalent to human MARK3 p.Arg570Gly (R570G)], we performed site-directed mutagenesis using the following primers; FW: 5′-CTTTCACTCTGGTCAAACCGGAGCACGAAACAACACAGCG-3′ RV: 5′-CGCTGTGTTGTTTCGTGCTCCGGTTTGACCAGAGTGAAAG-3′. The fully sequenced pENTR223 par-1 WT and pENTR223 par-1 R792G clones were subcloned into pGW-attB-HA (46) vector to generate pGW-attB-HA-par-1 WT and pGWattB-HA-par-1 R792G. These constructs were injected into y, w,ΦC31; VK33 embryos (47) and transgenic flies were selected by the w+ marker on pGW-attB-HA vector.
ERG recording of fly eye
ERG recordings were performed as described by Verstreken et al. (2003) (48). In brief, flies were glued to a glass slide. A recording electrode filled with 3 M NaCl was placed on the eye, and another reference electrode on the fly head. During the recording, a 1 s pulse of light stimulation was given, and the ERG traces of five flies for each genotype were recorded and analyzed using WinWCP V5.3.3 software.
Drosophila genetics
The following stocks were obtained from the Bloomington Drosophila Stock Center in Indiana University (BDSC). w[*]; P{w[+mC]=UAS-lacZ.B}Bg4–2-4b (RRID: BDSC_1777), ey-gal4 (on II), y1 sc* v1; P{TRiP.HMS00405}attP2 (RRID: BDSC_35342), y1 sc* v1; P{TRiP.GL00253}attP2 (RRID: BDSC_32410). All flies were maintained at 25°C. For measuring eye size, we used ImageJ.
Immunohistochemistry
Eye discs were dissected from third instar larvae, fixed in periodate-lysine-paraformaldehyde (PLP) fixative for 15 min at room temperature and incubated with primary antibodies overnight at 4°C. Primary antibodies were as follows: α-Elav (1: 200, 7E8A10, DSHB, Iowa City, IA) (49), α-Par-1 (1: 200) (50). Secondary antibodies conjugated with Cy3, Cy5 or Alexa 488 were from Jackson ImmunoResearch and were incubated for imaging in Vectashield (Vector Laboratories) with 4′, 6-diamidino-2-phenylindole. Fluorescent images were acquired using a Zeiss LSM880 confocal microscope.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank the ProVisu Foundation for provided a grant to S.E.A., and the Swiss Government for the Scholarships of Excellence program that provided competitive funding to M.A. We are thankful to all members of the family for the participation in this study. We thank Dr. I. Kern for advice, Dr. M. Garieri for helping in editing figures and Michael Wangler and Karen Schulze for comments on the manuscript.
Conflict of Interest statement. None declared.
Funding
Part of the study was funded by Higher Education Commission, Pakistan, NRPU Grant No: 2835. H.J.B. is supported by the NIH (R24OD022005) and is an Investigator of the Howard Hughes Medical Institute. Confocal microscopy was performed in the Neurovisualization core of the BCM IDDRC (supported by the NICHD U54HD083092). Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) at Indiana University were used in this study. Monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD and maintained at The University of Iowa, Department of Biology.
References
- 1. Slavotinek A.M. (2011) Eye development genes and known syndromes. Mol. Genet. Metab., 104, 448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Graw J. (2003) The genetic and molecular basis of congenital eye defects. Nat. Rev. Genet., 4, 876–888. [DOI] [PubMed] [Google Scholar]
- 3. Pierce E.A., Bennett J. (2015) The status of RPE65 gene therapy trials: safety and efficacy. Cold Spring Harb. Perspect. Med., 5, a017285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Antonarakis S.E., Beckmann J.S. (2006) Mendelian disorders deserve more attention. Nat. Rev. Genet., 7, 277–282. [DOI] [PubMed] [Google Scholar]
- 5. Waryah A.M., Narsani A.K., Sheikh S.A., Shaikh H., Shahani M.Y. (2013) The novel heterozygous Thr377Arg MYOC mutation causes severe Juvenile Open Angle Glaucoma in a large Pakistani family. Gene, 528, 356–359. [DOI] [PubMed] [Google Scholar]
- 6. Zelinger L., Banin E., Obolensky A., Mizrahi-Meissonnier L., Beryozkin A., Bandah-Rozenfeld D., Frenkel S., Ben-Yosef T., Merin S., Schwartz S.B.. et al. (2011) A missense mutation in DHDDS, encoding dehydrodolichyl diphosphate synthase, is associated with autosomal-recessive retinitis pigmentosa in Ashkenazi jews. Am. J. Hum. Genet., 88, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Estrada-Cuzcano A., Neveling K., Kohl S., Banin E., Rotenstreich Y., Sharon D., Falik-Zaccai T.C., Hipp S., Roepman R., Wissinger B.. et al. (2012) Mutations in C8orf37, encoding a ciliary protein, are associated with autosomal-recessive retinal dystrophies with early macular involvement. Am. J. Hum. Genet., 90, 102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu H., Kahrizi K., Musante L., Fattahi Z., Herwig R., Hosseini M., Oppitz C., Abedini S.S., Suckow V., Larti F.. et al. (2018) Genetics of intellectual disability in consanguineous families. Mol. Psychiatry, doi: 10.1038/s41380-017-0012-2. [DOI] [PubMed] [Google Scholar]
- 9. Bittles A.H., Black M.L. (2010) Evolution in health and medicine Sackler colloquium: consanguinity, human evolution, and complex diseases. Proc. Natl. Acad. Sci. U S A, 107, 1779–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Makrythanasis P., Kato M., Zaki M.S., Saitsu H., Nakamura K., Santoni F.A., Miyatake S., Nakashima M., Issa M.Y., Guipponi M.. et al. (2016) Pathogenic variants in PIGG cause intellectual disability with seizures and hypotonia. Am. J. Hum. Genet., 98, 615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Makrythanasis P., Nelis M., Santoni F.A., Guipponi M., Vannier A., Bena F., Gimelli S., Stathaki E., Temtamy S., Megarbane A.. et al. (2014) Diagnostic exome sequencing to elucidate the genetic basis of likely recessive disorders in consanguineous families. Hum. Mutat., 35, 1203–1210. [DOI] [PubMed] [Google Scholar]
- 12. Ansar M., Riazuddin S., Sarwar M.T., Makrythanasis P., Paracha S.A., Iqbal Z., Khan J., Assir M.Z., Hussain M., Razzaq A.. et al. (2017) Biallelic variants in LINGO1 are associated with autosomal recessive intellectual disability, microcephaly, speech and motor delay. Genet. Med., doi: 10.1038/gim.2017.113. [DOI] [PubMed] [Google Scholar]
- 13. Augusteyn R.C., Nankivil D., Mohamed A., Maceo B., Pierre F., Parel J.M. (2012) Human ocular biometry. Exp. Eye Res., 102, 70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Santoni F.A., Makrythanasis P., Antonarakis S.E. (2015) CATCHing putative causative variants in consanguineous families. BMC Bioinformatics, 16, 310.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O'Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B.. et al. (2016) Analysis of protein-coding genetic variation in 60, 706 humans. Nature, 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang J., Al-Ouran R., Hu Y., Kim S.Y., Wan Y.W., Wangler M.F., Yamamoto S., Chao H.T., Comjean A., Mohr S.E.. et al. (2017) MARRVEL: integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am. J. Hum. Genet., 100, 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Quang D., Chen Y., Xie X. (2015) DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics, 31, 761–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lindner M., Steinfeld R., Burgard P., Schulze A., Mayatepek E., Zschocke J. (2003) Tetrahydrobiopterin sensitivity in German patients with mild phenylalanine hydroxylase deficiency. Hum. Mutat., 21, 400.. [DOI] [PubMed] [Google Scholar]
- 19. Pey A.L., Stricher F., Serrano L., Martinez A. (2007) Predicted effects of missense mutations on native-state stability account for phenotypic outcome in phenylketonuria, a paradigm of misfolding diseases. Am. J. Hum. Genet., 81, 1006–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hazelett D.J., Bourouis M., Walldorf U., Treisman J.E. (1998) decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development, 125, 3741–3751. [DOI] [PubMed] [Google Scholar]
- 21. Brand A.H., Perrimon N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 22. Kwan J., Sczaniecka A., Heidary Arash E., Nguyen L., Chen C.C., Ratkovic S., Klezovitch O., Attisano L., McNeill H., Emili A.. et al. (2016) DLG5 connects cell polarity and Hippo signaling protein networks by linking PAR-1 with MST1/2. Genes Dev., 30, 2696–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bolkan B.J., Kretzschmar D. (2014) Loss of Tau results in defects in photoreceptor development and progressive neuronal degeneration in Drosophila. Dev. Neurobiol., 74, 1210–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goto K., Yasuda M., Sugawara A., Kuramochi T., Itoh T., Azuma N., Ito M. (2006) Small eye phenotypes observed in a human tau gene transgenic rat. Curr. Eye Res., 31, 107–110. [DOI] [PubMed] [Google Scholar]
- 25. Schwarz T., Murphy S., Sohn C., Mansky K.C. (2010) C-TAK1 interacts with microphthalmia-associated transcription factor, Mitf, but not the related family member Tfe3. Biochem. Biophys. Res. Commun., 394, 890–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. George A., Zand D.J., Hufnagel R.B., Sharma R., Sergeev Y.V., Legare J.M., Rice G.M., Scott Schwoerer J.A., Rius M., Tetri L.. et al. (2016) Biallelic mutations in MITF cause coloboma, osteopetrosis, microphthalmia, macrocephaly, albinism, and deafness. Am. J. Hum. Genet., 99, 1388–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lennerz J.K., Hurov J.B., White L.S., Lewandowski K.T., Prior J.L., Planer G.J., Gereau RWt., Piwnica-Worms D., Schmidt R.E., Piwnica-Worms H. (2010) Loss of Par-1a/MARK3/C-TAK1 kinase leads to reduced adiposity, resistance to hepatic steatosis, and defective gluconeogenesis. Mol. Cell Biol., 30, 5043–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kemphues K.J., Priess J.R., Morton D.G., Cheng N.S. (1988) Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell, 52, 311–320. [DOI] [PubMed] [Google Scholar]
- 29. Guo S., Kemphues K.J. (1995) par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell, 81, 611–620. [DOI] [PubMed] [Google Scholar]
- 30. Tomancak P., Piano F., Riechmann V., Gunsalus K.C., Kemphues K.J., Ephrussi A. (2000) A Drosophila melanogaster homologue of Caenorhabditis elegans par-1 acts at an early step in embryonic-axis formation. Nat. Cell Biol., 2, 458–460. [DOI] [PubMed] [Google Scholar]
- 31. Cox D.N., Lu B., Sun T.Q., Williams L.T., Jan Y.N. (2001) Drosophila par-1 is required for oocyte differentiation and microtubule organization. Curr. Biol., 11, 75–87. [DOI] [PubMed] [Google Scholar]
- 32. Ossipova O., Dhawan S., Sokol S., Green J.B. (2005) Distinct PAR-1 proteins function in different branches of Wnt signaling during vertebrate development. Dev. Cell, 8, 829–841. [DOI] [PubMed] [Google Scholar]
- 33. Shulman J.M., Benton R., St Johnston D. (2000) The Drosophila homolog of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell, 101, 377–388. [DOI] [PubMed] [Google Scholar]
- 34. Huang H.L., Wang S., Yin M.X., Dong L., Wang C., Wu W., Lu Y., Feng M., Dai C., Guo X.. et al. (2013) Par-1 regulates tissue growth by influencing hippo phosphorylation status and hippo-salvador association. PLoS Biol., 11, e1001620.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gong Y., Slee R.B., Fukai N., Rawadi G., Roman-Roman S., Reginato A.M., Wang H., Cundy T., Glorieux F.H., Lev D.. et al. (2001) LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell, 107, 513–523. [DOI] [PubMed] [Google Scholar]
- 36. Logan C.Y., Nusse R. (2004) The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol., 20, 781–810. [DOI] [PubMed] [Google Scholar]
- 37. Ye X., Wang Y., Nathans J. (2010) The Norrin/Frizzled4 signaling pathway in retinal vascular development and disease. Trends Mol. Med., 16, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paes K.T., Wang E., Henze K., Vogel P., Read R., Suwanichkul A., Kirkpatrick L.L., Potter D., Newhouse M.M., Rice D.S. (2011) Frizzled 4 is required for retinal angiogenesis and maintenance of the blood-retina barrier. Invest. Ophthalmol. Vis. Sci., 52, 6452–6461. [DOI] [PubMed] [Google Scholar]
- 39. Robitaille J., MacDonald M.L., Kaykas A., Sheldahl L.C., Zeisler J., Dube M.P., Zhang L.H., Singaraja R.R., Guernsey D.L., Zheng B.. et al. (2002) Mutant frizzled-4 disrupts retinal angiogenesis in familial exudative vitreoretinopathy. Nat. Genet., 32, 326–330. [DOI] [PubMed] [Google Scholar]
- 40. Yang H., Li S., Xiao X., Guo X., Zhang Q. (2012) Screening for NDP mutations in 44 unrelated patients with familial exudative vitreoretinopathy or Norrie disease. Curr. Eye Res., 37, 726–729. [DOI] [PubMed] [Google Scholar]
- 41. Sobreira N., Schiettecatte F., Valle D., Hamosh A. (2015) GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat., 36, 928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M.. et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pruitt K.D., Tatusova T., Maglott D.R. (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res., 35, D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J.. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bischof J., Bjorklund M., Furger E., Schertel C., Taipale J., Basler K. (2013) A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development, 140, 2434–2442. [DOI] [PubMed] [Google Scholar]
- 47. Venken K.J., He Y., Hoskins R.A., Bellen H.J. (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science, 314, 1747–1751. [DOI] [PubMed] [Google Scholar]
- 48. Verstreken P., Koh T.W., Schulze K.L., Zhai R.G., Hiesinger P.R., Zhou Y., Mehta S.Q., Cao Y., Roos J., Bellen H.J. (2003) Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron, 40, 733–748. [DOI] [PubMed] [Google Scholar]
- 49. Robinow S., White K. (1991) Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol., 22, 443–461. [DOI] [PubMed] [Google Scholar]
- 50. McDonald J.A., Khodyakova A., Aranjuez G., Dudley C., Montell D.J. (2008) PAR-1 kinase regulates epithelial detachment and directional protrusion of migrating border cells. Curr. Biol., 18, 1659–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.