Abstract
Microrchidia (MORC) proteins have been described as epigenetic regulators and plant immune mediators in Arabidopsis. Typically, plant and animal MORC proteins contain a hallmark GHKL-type (Gyrase, Hsp90, Histidine kinase, MutL) ATPase domain in their N-terminus. Here, 356 and 83 MORC orthologues were identified in 60 plant and 27 animal genomes. Large-scale MORC sequence analyses revealed the presence of a highly conserved motif composition that defined as the MORC domain. The MORC domain was present in both plants and animals, indicating that it originated in the common ancestor before the divergence of plants and animals. Phylogenetic analyses showed that MORC genes in both plant and animal lineages were clearly classified into two major groups, named Plants-Group I, Plants-Group II and Animals-Group I, Animals-Group II, respectively. Further analyses of MORC genes in green plants uncovered that Group I can be subdivided into Group I-1 and Group I-2. Group I-1 only contains seed plant genes, suggesting that Group I-1 and I-2 divergence occurred at least before the emergence of spermatophytes. Group I-2 and Group II have undergone several gene duplications, resulting in the expansion of MORC gene family in angiosperms. Additionally, MORC gene expression analyses in Arabidopsis, soybean, and rice revealed a higher expression level in reproductive tissues compared with other organs, and showed divergent expression patterns for several paralogous gene pairs. Our studies offered new insights into the origins, phylogenetic relationships, and expressional patterns of MORC family members in green plants, which would help to further reveal their functions as plant epigenetic regulators.
Keywords: MORC domain, gene duplication, molecular evolution, MORC gene family
Introduction
The Microrchidia (MORC) gene is a highly conserved nuclear protein-coding gene, which was originally identified in mice and associated with the formation of mouse spermatid cells and with the completion of the meiotic process (Watson et al. 1998; Inoue et al. 1999). Generally, MORC proteins contain several characteristic conserved domain features: at the N-terminus, they usually contain the conserved GHKL (Gyrase, Hsp90, Histidine kinase, MutL) and S5 fold domains, which together form a catalytically active ATPase module; at the other end, one or more undetermined coiled-coil domain can be found (Dutta and Inouye 2000; Iyer, Abhiman, et al. 2008). Previous studies showed that the GHKL-type ATPase is prevalent in both prokaryotic and eukaryotic proteins, and is associated with DNA structure rearrangement (Gyrase or topoisomerase; G), heat shock (HSP90; H), signal transduction (Histidine kinase; K), and DNA mismatch repair (MutL; L) (Dutta and Inouye 2000; Iyer, Abhiman, et al. 2008). Currently, MORC genes have been found in both prokaryotic and eukaryotic organisms (Iyer, Abhiman, et al. 2008; Iyer, Anantharaman, et al. 2008). There are 4 and 7 canonical MORC gene family members being identified in humans and Arabidopsis thaliana, respectively (Inoue et al. 1999; Iyer, Abhiman, et al. 2008; Kang and Klessig 2008; Kang et al. 2008; Li et al. 2012; 2013). Iyer et al. (2008) found that several prokaryotic MORC proteins play a vital role in bacterial restriction-modification system. Accordingly, they proposed that MORC proteins in eukaryotes may affect the chromatin structure by manipulating the superstructure of DNA molecules in response to some epigenetic regulatory signals, such as DNA methylation or histone modifications (Iyer, Abhiman, et al. 2008). Unlike the MORC proteins in animals which contains an extra CW-type zinc finger domain at their C-terminal that can specifically recognize the epigenetic regulatory markers of histone H3K4me2 and H3K4me3 (He et al. 2010; Eberl et al. 2013), plant MORCs usually contain only one conserved coiled-coil domain that forms a putative basic leucine zipper motif at their C-terminal, which is thought to be involved in the regulation of protein–protein interactions (Langen et al. 2014).
Recent studies indicated that MORC proteins not only participate in pathogen resistance in plant multiple layered immunity system (Kang and Klessig 2008; Kang et al. 2010, 2012) but they may also be involved in the transcriptional gene silencing of some genes and transposable elements (TEs) (Lorkovic et al. 2012; Moissiard et al. 2012; Brabbs et al. 2013). The first report on a plant MORC gene, namely Compromised Recognition of TCV (CRT1), was initially identified in a forward genetics screening of Arabidopsis mutants compromised in recognition of the Turnip crinkle virus (TCV) (Kang and Klessig 2008; Kang et al. 2008). The analysis of CRT1 sequence revealed that it contains the GHKL ATPase and S5 fold domains, which is the hallmark of MORC proteins and thus the gene was renamed as AtMORC1 (Kang et al. 2010; Moissiard et al. 2012). Further studies showed that Arabidopsis MORC genes form a family composed of 7 members. Among these, silencing of AtMORC1 and its closely related paralogous, AtMORC2, lead to a compromised pathogen resistance, including nonhost resistance, basal resistance, PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI) (Kang and Klessig 2008; Kang et al. 2010, 2012). In addition, MORC1 is one of very few proteins that physically interacted with many immune components, including at least 12 plant resistance proteins (R) and the PAMP recognition receptor FLS2 (Kang and Klessig 2008; Kang et al. 2010, 2012; Langen et al. 2014). Other studies in barley and Solanaceae species revealed that MORC1 can positively or negatively modulate plant immunity (Langen et al. 2014; Manosalva et al. 2015). In potato (S. tuberosum), a mutation of MORC1 orthologous (StMORC1) negatively affected the basal resistance in plant immunity, whereas silencing of MORC1 in the closely related species tomato (SlMORC1) and Nicotiana benthamiana (NbMORC1) or in the monocot barley (HvMORC1) resulted in an increased disease resistance (Langen et al. 2014; Manosalva et al. 2015). Transient expression of tomato and potato MORC1 genes in N. benthamiana revealed that their ability to suppress or promote plant immunity is related to the differences of MORC1 proteins themselves, rather than the cellular environment where they act. Furthermore, domain-swapping and mutational analyses indicated that the C-terminal region plays important roles in both determining and modulating the species-specific effects of the MORC1 proteins in Solanaceae (Manosalva et al. 2015).
Apart from these functions in plant immunity, accumulating experimental evidence suggested that MORC proteins are vital epigenetic regulators of transcriptional gene silencing. Recently, three independent forward-genetic studies in Arabidopsis revealed that mutations in AtMORC1 or AtMORC6 sequences lead to the derepression of transposable elements (TEs) and silenced-genes expression in plants (Lorkovic et al. 2012; Moissiard et al. 2012; Brabbs et al. 2013). These observations led to an upsurge in epigenetic studies related to the MORC-mediated transcriptional gene silencing. However, whether AtMORC1 and AtMORC6 play roles in gene silencing by modulating RNA-dependent DNA methylation (RdDM) (Lorkovic et al. 2012; Brabbs et al. 2013; Liu et al. 2014, 2016; Harris et al. 2016) or by altering the chromatin structure (Moissiard et al. 2012), is still unclear. Moissiard et al. (2014) showed that AtMORC6 can bind to AtMORC1 and its closest homologous AtMORC2. Moreover, AtMORC1 and AtMORC2 act in a partially redundant manner (Moissiard et al. 2014). In contrast to AtMORC1, -2, and -6, which can form heterodimers to mediate TE transcriptional gene silencing, AtMORC4 and AtMORC7 can form discrete homopolymers in the nucleolus region adjacent to chromocenters and act semiredundantly to repress some of the genes involved in disease resistance (Harris et al. 2016). In addition, a genome-wide chromatin accessibility study using both DNase-Seq and ChIP-Seq approaches to investigate the potential role of MORC proteins, and the results indicated that AtMORC1 is preferentially associated with TEs, and suggested that a subset of these TEs may contribute to regulate expression of their proximal defense genes (Bordiya et al. 2016).
Considering the roles that MORC proteins play in pathogen resistance and the important role of MORCs as epigenetic regulators in plant transcriptional gene silencing, the functional study of these genes may help to further understand the molecular mechanism by which epigenetic regulation and disease resistance is acquired. Although the functions of MORC protein are gradually being unveiled, their research is still limited to a few model species, such as humans, mice, Arabidopsis, barley, and some Solanaceae species, and their precise molecular mechanism remains to be clarified. At the same time, studies on the evolution of MORC gene family in plants or other organisms are also very limited. A phylogenetic study showed that MORC proteins are widely distributed among eukaryotes, from the higher lineages (except fungi) to the lower apicomplexans and heteroloboseans, indicating that the MORC gene in eukaryotes has a relative early origin (Iyer, Abhiman, et al. 2008; Iyer, Anantharaman, et al. 2008). Another study on plant immune system showed that there are 5 MORC genes in barley and 6 MORC genes in Solanaceae species (Langen et al. 2014; Manosalva et al. 2015). Further phylogenetic analysis revealed that the MORC gene family in several angiosperm plants could be finely divided into three clades (Langen et al. 2014; Manosalva et al. 2015). However, these studies are only based on the analysis of MORC gene family in a few species, and the evolutionary origin and phylogenetic relationships with other species are still not very clear.
In this study, the MORC genes were preliminarily identified in major plant and animal lineages based on many sequenced plant and animal genomes and relevant plant transcriptome data. Phylogenetic analyses and other bioinformatics tools were used to explore the evolutionary history and the origin of MORC genes in plants and animals. Our results showed that the MORC genes in plant and animal lineages could be classified into two major groups, respectively, named Plants-Group I, Plants-Group II and Animals-Group I, Animals-Group II. The origin of MORC genes could be traced back to the common ancestor before the divergence of plants and animals. Additionally, we found that MORC proteins exhibited some highly conserved motif compositions among plants and animals, and we defined them as the MORC domain that spanned ∼370 amino acids in length. We also deeply explored the evolutionary relationships of MORC gene family in green plants, and the results showed that some subfamily of MORC genes underwent distinct gene duplication patterns during the evolution of angiosperms.
Materials and Methods
Data Sources and Sequence Retrieval
In total, 60 sequenced plant genomes, 27 animal genomes and 56 plant transcriptome data sets were collected and screened for MORC sequences in order to obtain a representation as large as possible of the major plant and animal lineages (supplementary tables S1 and S2, Supplementary Material online). Among them, 43 plants genomes were retrieved from Phytozome v11.0 (Goodstein et al. 2012) (https://phytozome.jgi.doe.gov/; last accessed May 6, 2017), whereas the remaining ones were obtained from other databases (Chen F, et al. 2018) listed in supplementary table S1, Supplementary Material online. The water lily (Nymphaea colorata) genome came from our own genome project (Zhang L et al., in preparation). Transcriptome data were derived from the 1,000 plants database (OneKP or 1KP) (https://db.cngb.org/onekp/; last accessed May 6, 2017) (supplementary tables S2 and S3, Supplementary Material online). The animal genomes were derived from the Ensembl database (http://ensemblgenomes.org/; last accessed May 6, 2017). First, the protein sequence of Arabidopsis AtMORC1 was downloaded from the Arabidopsis Information Resource (TAIR10) and used as a query for a local BLASTP search against several proteome data sets with an e-value cut-off of <1e-10. Sequences were manually adjusted in multiple sequence alignment, and those sequences with obvious errors were excluded from subsequent analyses. The longest transcript isoform was selected if two or more transcripts are annotated for the same gene from alternative splicing. The screening for MORC sequences in transcriptome databases used the online BLASTP tool of the OneKP database. Likewise, the AtMORC1 protein sequence was used as query according to default parameters.
Multiple Sequence Alignment
To explore the phylogenetic relationships of the MORC genes in plant and animal lineages, MORC protein sequences identified in both plant and animal genomes were used to conduct multiple sequence alignment (MSA) analysis. Preparatory MSA was performed using MUSCLE v3.8.31 (Edgar 2004) with default parameters. Then a preliminary approximate Maximum likelihood (ML) tree was generated based on the alignments using FastTree v2.1 (Price et al. 2009) with default parameters. Additionally, a second round of MSA was carried out independently for each subgroup identified in the tree by using MUSCLE.
Phylogenetic and Synteny Analysis
Phylogenetic analyses were conducted using two different methods: the Maximum Likelihood and the Bayesian method. The ML tree was constructed using RaxML-HPC tool from the online CIPRES Science Gateway (https://www.phylo.org; last accessed July 16, 2017). The bootstrap value was set to 1,000 repetitions whereas other parameters were set to default. Similarly, the Bayesian trees were constructed using the MrBayes tool from the online CIPRES Science Gateway, running for 5×107 generations with 100 Markov chains and all other parameters set to default. In addition, duplicate gene pairs were searched for cross-genome syntenic relationships using the Plant Genome Duplication Database (Lee et al. 2013) (PGDD).
Motif Analysis and MORC Domain Definition
In order to further identify novel conserved motif composition that might not be recorded in public database, the software Multiple Em for Motif Elicitation (MEME) (Bailey and Elkan 1994; Bailey 2009) was employed using the following parameters: the maximum width of a motif was 100aa and the number of motifs was 20, the other parameters set to default. The conserved motif patterns were then redrew and visualized by the TBtools (Chen C, et al. 2018). In addition, given the highly conserved motif compositions of MORC proteins, we defined the MORC domain for MORC proteins. We used the MUSCLE program to carry out multiple sequence alignment and the highly conserved motif regions that were trimmed by the trimAL (Capella-Gutierrez et al. 2009) software with a gap threshold value of 0.8 were selected for constructing the MORC domain seed model. The seed model was built by Hmmbuild embedded in HMMER (Eddy 1998) program, and then using preconstructed data sets to test this MORC domain model (supplementary fig. S5, Supplementary Material online). We also inferred the 3D structure of MORC1 protein and MORC domain in Arabidopsis, rice, human and mice based on the protein homology modeling on the Swiss-model database (Arnold et al. 2006; Guex et al. 2009; Kiefer et al. 2009; Biasini 2014). The mice MORC3 protein crystal structure was selected as the template for homologous modeling process and the best hit with the highest scores was chose as the predicted structures. The conserved logo of MORC domain was generated using the Weblogo webserver (Schneider and Stephens 1990; Crooks et al. 2004).
Gene Structure and Subcellular Localization Analysis
We used the Gene Structure Displayer Server (GSDS) (Hu et al. 2015) website to identify the exon–intron organization of partial MORC genes in representative species. Firstly, we downloaded the GFF annotation files from Phytozome and then retrieved the annotated information of corresponding species MORC gene, and uploaded the annotated information to the GSDS website based on the default parameters for deducing MORC gene exon–intron structures. The detailed information of some MORC genes was listed in supplementary table S4, Supplementary Material online. The subcellular localization of MORC proteins was predicted by using the CELLO v2.5 (Yu et al. 2006, 2010) webserver.
Expression Profile Analyses
RNA-Seq data were derived from open source data sets downloaded from public websites and were standardized with Fragments per kilobase of exon per million reads mapped (FPKM) for quantifying the gene expression level (supplementary table S5, Supplementary Material online). Among them, the Arabidopsis RNA-Seq data (Zhang and Ma 2012; Zhang et al. 2014) included the following tissues: seeding, stage 4 flower, stage 1–9 flower, stage 12 flower, inflorescence meristem (IM) and meiosis; Soybean (Libault et al. 2010) transcriptome data contained the following tissues: leaves, root, nodules, stem, root hairs, shoot apical meristem, seed, flower, and pod; rice RNA-Seq data (Sakai et al. 2011) contained the following tissues: leaf, root, shoot, callus, panicle before flowering, panicle after flowering, and seed. Finally, heat maps of MORC gene expression profile in different species were obtained using the R pheatmap (https://CRAN.R-project.org/package=pheatmap; last accessed July 16, 2017) package.
Results
Identification of MORC Genes in Plant and Animal Lineages
To identify MORC genes in different plant and animal species, several comprehensive data sets that contain selected representative plants and animals were constructed and a genome-wide search was performed using the Arabidopsis AtMORC1 (AT4G36290) amino acid sequence as the query for a local BLASTP search against these data sets. In all, 356 candidate MORC sequences were retrieved from 60 sequenced plant genomes including dicots (36 species: 222 sequences), monocots (15:108), basal angiosperms (2:7), Pteridophytes (1:6), Bryophytes (2:9), Charophyta (1:1), and Chlorophyta (3:3) (table 1 and supplementary table S1, Supplementary Material online). Similarly, 83 candidate MORC sequences were also detected in 27 representative animal species based on the BLASTP searching results (table 1 and supplementary table S1, Supplementary Material online). The copy number of MORC genes varied considerably among the major lineages of plants, ranging from the single copy detected in several algae such as Klebsormidium flaccidum, Ostreococcus lucimarinusand, Ostreococcus RCC809, and Micromonas pusilla CCMP1545, to the 23 sequences identified in Triticum aestivum. Preliminary statistic results showed that at least 4 copies of MORC genes are present in eudicots with the exception of Cajanus cajan and Phaseolus vulgaris which possess only three MORC members. Among the monocots, Spirodela polyrhiza was the only plant with three MORC genes while all other species contained at least 5 members. In the basal angiosperms, three and 4 MORC genes were detected in Amborella trichopoda and Nymphaea colorata, respectively. While, there were 6 copies of MORC genes in the lycophytes Selaginella moellendorffli and 7 copies in the moss Physcomitrella patens, 2 copies in Sphagnum fallax. However, it is worth noting that only one copy of MORC gene was found in unicellular algae. MORC genes were also widespread in animals that spanned from simple invertebrates to higher mammals. The gene copy number in animals was expectantly stable and most species contained 4 MORC members, with the highest number of 5 in mouse and rat. Further investigation revealed that the multiple copies in animals were mainly resulted from the gene duplication in vertebrates.
Table 1.
The Distribution of MORC Genes in Representative Genome-Sequenced Species
| Taxonomy | Class | Species name | Abbr. | Group I |
Group II | Total | |
|---|---|---|---|---|---|---|---|
| Group I-1 | Group I-2 | ||||||
| Angiosperms | Eudicots | Brassica napus | Bna | 2 | 4 | 10 | 16 |
| Brassica rapa | Bra | 1 | 1 | 5 | 7 | ||
| Brassica oleracea | Bol | 0 | 1 | 5 | 6 | ||
| Thellungiella halophila | Thhalv | 1 | 1 | 3 | 5 | ||
| Arabidopsis thaliana | AT | 3 | 1 | 3 | 7 | ||
| Arabidopsis lyrata | Aly | 3 | 1 | 3 | 7 | ||
| Capsella rubella | Carubv | 1 | 1 | 4 | 6 | ||
| Lepidium meyenii | maca | 4 | 3 | 11 | 18 | ||
| Carica papaya | evm | 1 | 2 | 1 | 4 | ||
| Gossypium hirsutum | Gh | 2 | 2 | 5 | 9 | ||
| Gossypium raimondii | Gorai | 1 | 1 | 3 | 5 | ||
| Gossypium arboreum | Goarb | 1 | 1 | 3 | 5 | ||
| Theobroma cacao | Thecc | 1 | 2 | 1 | 4 | ||
| Citrus clementina | Ciclev | 1 | 1 | 2 | 4 | ||
| Citrus sinensis | Orange | 1 | 1 | 2 | 4 | ||
| Eucalyptus grandis | Eucgr | 1 | 2 | 2 | 5 | ||
| Prunus mume | Pm | 1 | 4 | 2 | 7 | ||
| Prunus persica | Prupe | 1 | 2 | 2 | 5 | ||
| Pyrus bretschneideri | Pbr | 0 | 3 | 6 | 9 | ||
| Malus domestica | MDP | 1 | 3 | 7 | 11 | ||
| Fragaria vesca | mrna | 0 | 2 | 2 | 4 | ||
| Glycine max | Glyma | 2 | 2 | 2 | 6 | ||
| Phaseolus vulgaris | Phvul | 1 | 1 | 1 | 3 | ||
| Cajanus cajan | C.cajan | 0 | 2 | 1 | 3 | ||
| Medicago truncatula | Medtr | 1 | 2 | 1 | 4 | ||
| Citrullus lanatus | Cla | 1 | 2 | 2 | 5 | ||
| Cucumis sativus | Cucsa | 2 | 1 | 2 | 5 | ||
| Manihot esculenta | Manes | 1 | 2 | 2 | 5 | ||
| Ricinus communis | Rco | 1 | 1 | 2 | 4 | ||
| Populus trichocarpa | Potri | 1 | 2 | 3 | 6 | ||
| Linum usitatissimum | Lus | 2 | 3 | 3 | 8 | ||
| Vitis vinifera | VIT | 1 | 2 | 1 | 4 | ||
| Solanum lycopersicum | Solyc | 1 | 2 | 3 | 6 | ||
| Mimulus guttatus | Migut | 1 | 1 | 5 | 7 | ||
| Aquilegia coerulea | Aquca | 1 | 1 | 2 | 4 | ||
| Nelumbo nucifera | NNU | 1 | 1 | 2 | 4 | ||
| Monocots | Aegilops tauschii | EMT | 1 | 4 | 1 | 6 | |
| Triticum aestivum | Traes | 9 | 11 | 3 | 23 | ||
| Hordeum vulgare | MLOC | 1 | 4 | 1 | 6 | ||
| Brachypodium distachyon | Bradi | 1 | 3 | 2 | 6 | ||
| Oryza sativa | LOC_Os | 1 | 3 | 2 | 6 | ||
| Panicum virgatum | Pavir | 2 | 6 | 4 | 12 | ||
| Panicum hallii | Pahal | 1 | 3 | 2 | 6 | ||
| Setaria italica | Si | 1 | 3 | 2 | 6 | ||
| Zea mays | GRMZM/AC | 1 | 4 | 2 | 7 | ||
| Sorghum bicolor | Sobic | 1 | 4 | 2 | 7 | ||
| Ananas. comosus | Aco | 1 | 2 | 2 | 5 | ||
| Phoenix dactylifera | PDK | 1 | 2 | 2 | 5 | ||
| Musa acuminata | GSMUA | 2 | 2 | 1 | 5 | ||
| Phalaenopsis equestris | PEQU | 1 | 2 | 2 | 5 | ||
| Spirodela polyrhiza | Spipo | 1 | 1 | 1 | 3 | ||
| Basal angiosperms | Nymphaea colorata | Nym | 1 | 1 | 2 | 4 | |
| Amborella trichopoda | Scaffold | 1 | 1 | 1 | 3 | ||
| Pteridophyta | Lycophytes | Selaginella moellendorffii | Smo | 1 | 5 | 6 | |
| Bryophyta | Mosses | Physcomitrella patens | Phpat | 0 | 1 | 6 | 7 |
| Sphagnum fallax | Sphfal | 0 | 1 | 1 | 2 | ||
| Charophyta | Charophytes | Klebsormidium flaccidum | Kfl | 0 | 0 | 1 | 1 |
| Chlorophyta | Chlorophytes | Ostreococcus lucimarinus | Olu | 1 | 1 | ||
| Ostreococcus RCC809 | OstRCC809_2 | 1 | 1 | ||||
| Micromonas pusilla CCMP1545 | MicpuC | 1 | 1 | ||||
| Metazoa | Vertebrates | Pan troglodytes | ENSPTRG | 2 | 2 | 4 | |
| Homo sapiens | ENSG | 2 | 2 | 4 | |||
| Gorilla gorilla | ENSGGOG | 2 | 2 | 4 | |||
| Pongo pygmaeus | ENSPPYG | 2 | 2 | 4 | |||
| Rattus norvegicus | ENSRNOG | 3 | 2 | 5 | |||
| Mus musculus | ENSMUSG | 3 | 2 | 5 | |||
| Sus scrofa | ENSSSCG | 2 | 1 | 3 | |||
| Canis familiaris | ENSCAFG | 2 | 2 | 4 | |||
| Equus caballus | ENSECAG | 2 | 2 | 4 | |||
| Bos taurus | ENSBTAG | 2 | 2 | 4 | |||
| Loxodonta africana | ENSLAFG | 2 | 2 | 4 | |||
| Meleagris gallopavo | ENSMGAG | 1 | 1 | 2 | |||
| Gallus gallus | ENSGALG | 1 | 1 | 2 | |||
| Taeniopygia guttata | ENSTGUG | 1 | 1 | 2 | |||
| Ornithorhynchus anatinus | ENSOANG | 0 | 2 | 2 | |||
| Xenopus tropicalis | ENSXETG | 2 | 1 | 3 | |||
| Latimeria chalumnae | ENSLACG | 1 | 2 | 3 | |||
| Takifugu rubripes | ENSTRUG | 1 | 2 | 3 | |||
| Tetraodon nigroviridis | ENSTNIG | 1 | 2 | 3 | |||
| Gasterosteus aculeatus | ENSGACG | 1 | 2 | 3 | |||
| Danio rerio | ENSDARG | 1 | 3 | 4 | |||
| Petromyzon marinus | ENSPMAG | 0 | 1 | 1 | |||
| Invertebrates | Branchiostoma floridae | Brafl | 2 | 2 | 4 | ||
| Sea urchin | Sea | 2 | 0 | 2 | |||
| Ciona intestinalis | ENSCING | 1 | 0 | 1 | |||
| Ciona savignyi | ENSCSAVG | 1 | 0 | 1 | |||
| Nematostella vectensis | Nemve | 1 | 1 | 2 | |||
To further explore the earlier evolutionary relationships of MORC genes in green plants, we also searched for MORC protein sequences in the OneKP transcriptome database by means of the online BLASTP tool. MORC protein sequences were identified in gymnosperms, monilophytes, lycophytes, liverwort, moss, and green algae. Finally, 154 nonredundant putative MORC protein sequences were retrieved from 56 nonflowering plants listed in supplementary table S2, Supplementary Material online. In detail, 50 MORC sequences were identified in gymnosperms (16 species), 58 in pteridophytes, 40 in bryophyte (16 species), and 6 in green algae. The copy number of MORC protein in these earlier evolved species was also considerably different. Among them there were 8 MORC members in Adiantum tenerum with the highest number, followed by 7 members in Sceptridum dissectum, and at least one copy in several lower species and unicellular algae.
Definition and Origin of MORC Domain
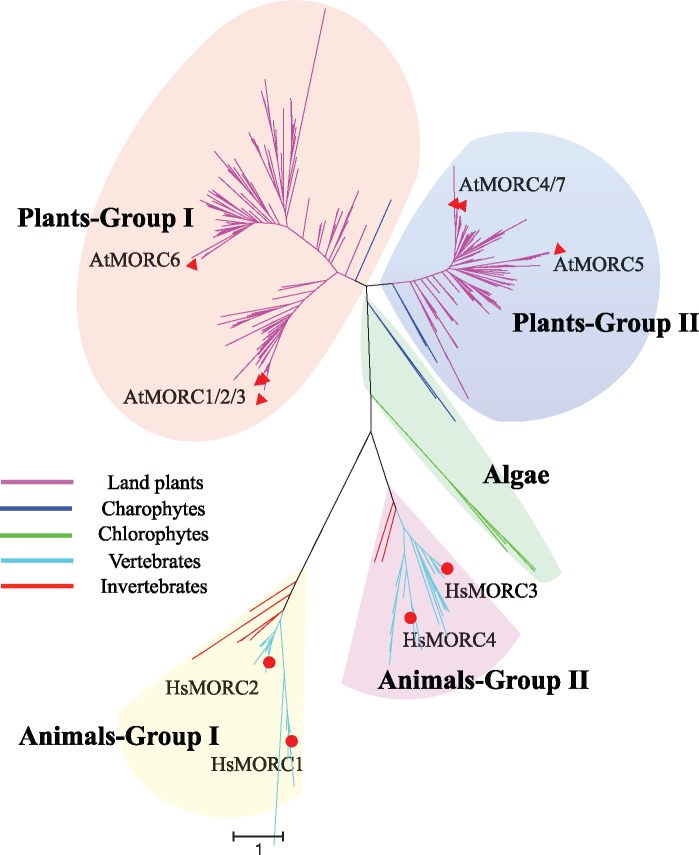
To unveil the evolutionary relationships and history of MORC genes among plant and animal lineages, we constructed a phylogenetic tree with Maximum Likelihood method. Topology of ML tree showed that MORC genes clearly cluster into two major groups in plants and animals, respectively (fig. 1 and supplementary fig. S1, Supplementary Material online), which was designed as Plants-Group I, Plants-Group II and Animals-Group I, Animals-Group II. Within the animal clade, each group contained both vertebrate and invertebrate species, and the vertebrate clade underwent a gene duplication event forming two small branches. In the plant clade, each group possesses the whole land plant lineages and Charophyta algae while the Chlorophyta algae are located in the base of these two plant groups. These results suggested that the MORC genes were present in a common ancestor before the divergence of plants and animals and had underwent several events of gene duplication during the course of long-term evolution, thus generating multiple copies in many plants and animals.
Fig. 1.
—Phylogenetic classification of MORC genes in plant and animal lineages. Phylogenetic tree was constructed using the ML method implemented in RaxML-HPC2 through the online CIPRES Science Gateway website. The topology of ML tree showed that MORC genes in plants and animals can be clearly classified into two major groups, respectively, which is designed as Plants-Group I, Plants-Group II and Animals-Group I, Animals-Group II. The green algae were located in the base of these two plant groups. Seven and four canonical MORC genes in Arabidopsis and human were labeled with red triangle and circle.
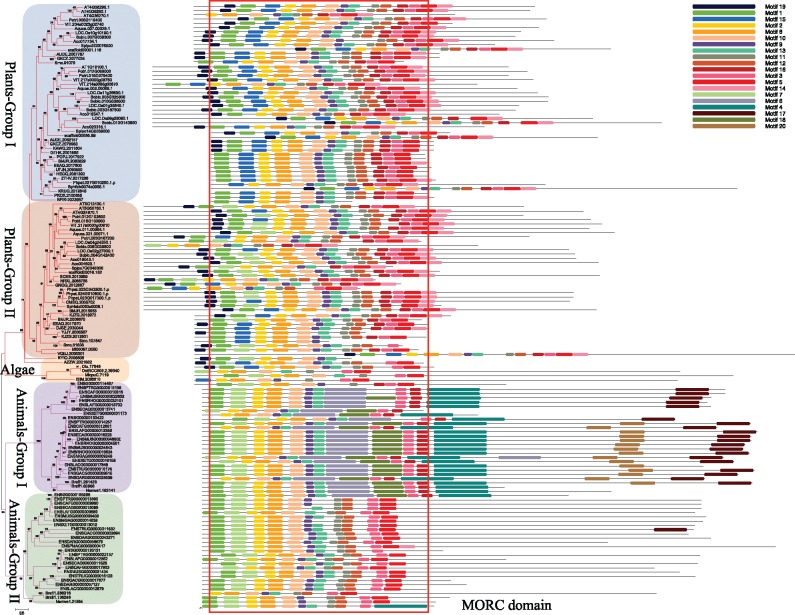
The MORC proteins generally present several typical conserved domain features: the N-term usually contains the conserved GHKL and S5 fold domains, which together form a functional module with ATPase activity, whereas the C-term possess one or more conserved coiled-coil domains (Dutta and Inouye 2000; Iyer, Abhiman, et al. 2008). To identify possible novel conserved motifs in MORC proteins, we used the local Multiple Em for Motif Elicitation (MEME) tool to analyze all candidate MORC protein sequences identified in our study. Apart from the typical conserved motifs preserved in above known domains, we also identified several additional conserved motifs within the N-terminal region along with the ATPase domain. The composition of these newly specific successive motifs spanned ∼370 amino acids in length and was defined as the MORC domain (fig. 2 and supplementary figs. S2–S4, Supplementary Material online). Multiple sequence alignment (MSA) of MORC proteins in some representative species was conducted and the conserved MORC domain region was selected to constructing the training data set of MORC domain seed model. The MORC domain seed model was tested by using several protein data sets and results showed this model can be used as query simplifying the identification of MORC genes in other species.
Fig. 2.
—Phylogenetic relationships and motif compositions of some representative MORC genes in plants and animals. The phylogenetic tree was reconstructed using some representative species based on the ML method implemented in RaxML-HPC2 through the online CIPRES Science Gateway website. For major nodes, ML bootstrap values >70% are labeled based on 1,000 replications. Motifs conserved across MORC proteins were identified through a MEME analysis and each motif composition is presented in supplementary figure S3, Supplementary Material online. Based on their highly conserved motif composition, we proposed a characteristic MORC domain that was marked in red box on the right.
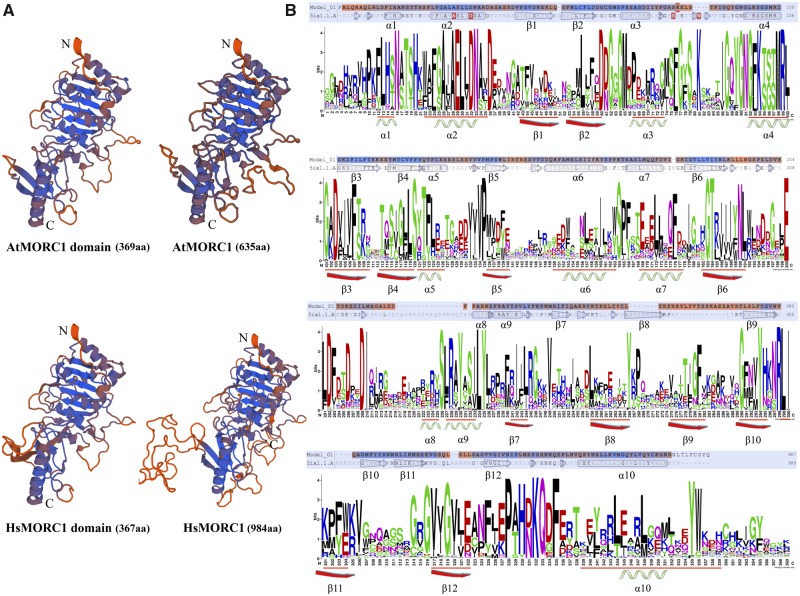
Moreover, to further resolve the putative 3D structure of MORC domain, we predicted the MORC1 protein and its MORC domain structure in Arabidopsis, rice, human, and mouse by means of protein homology modeling based on mouse MORC3 crystal structure in Swiss-Model database. Among the output results, the structure with highest score was chosen as the best for both MORC1 protein and MORC domain putative 3D structures. Homologous modeling results showed that the MORC domain is highly conserved in MORC proteins and constitutes the major component of MORC proteins (fig. 3 and supplementary fig. S6, Supplementary Material online). The MORC domain shares a very similar 3D structure in both plants and animals. In Arabidopsis and humans, the AtMORC1 and HsMORC1 proteins were composed of 635 and 984 amino acids in length and the MORC domain spanned ∼370 amino acids at their N-terminal. Furthermore, we also found that there were some highly conservative secondary structure elements like 10 conserved α-helixes and 12 β-sheets existed in the MORC domain.
Fig. 3.
—Homologous modeling of MORC1 protein and MORC domain in several representative species. The 3D structure of MORC1 protein and MORC domain in Arabidopsis and human were predicted based on the Swiss-model database according to mice MORC3 protein crystal structure (A). Sequence logo of MORC domain was generated on the Weblogo website using all aligned MORC domain sequences (B). Some conserved secondary structures in MORC domain were labeled (on the logo bottom) according to human MORC1 domain modeling result (on the logo top).
Phylogenetic Classification of MORC Gene Family into Two Major Groups in Green Plants
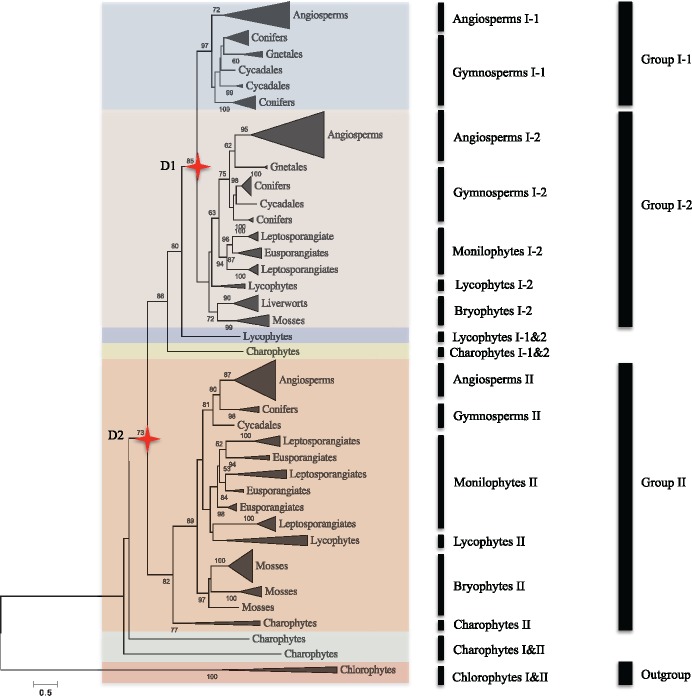
To explore the early history and evolution of the MORC gene family in green plants, we conducted phylogenetic analyses by aligning the full-length sequence of some MORC proteins considered as representative of green plant lineages, using both the ML and Bayesian methods. Phylogenetic analyses revealed that the MORC gene family in green plants clustered into two main groups, designated as Group I and Group II, where the Group I can be further subdivided into Group I-1 and Group I-2 (fig. 4 and supplementary figs. S7–S9, Supplementary Material online). The topology of the ML tree clearly separated the unicellular or complex green algae (Chlorophyta and Charophyta) from other embryophyta and suggested that the MORC gene family was subjected to two ancient gene duplication events (D1 and D2) during the evolution of extant terrestrial and seed plants. These duplication events eventually resulted in the generation of three clades of extant seed plants, which corresponds to the former classification of MORC genes into three groups based on the analysis of relatively limited angiosperm species (Langen et al. 2014; Manosalva et al. 2015).
Fig. 4.
—Phylogenetic relationships of MORC gene family in major groups of green lineage. The phylogenetic tree was built using the ML method implemented in RaxML-HPC2 through the online CIPRES Science Gateway and the Chlorophyta was rooted as outgroup. Two major groups (Group I-1 and 2 and Group II) and two major gene duplication events (D1 and D2) are recognized. Numbers above each branch indicates bootstrap support percentage values based on 1,000 replications and values >50% are shown. Gene lineages composed of angiosperms, gymnosperms, monilophytes, lycophytes, bryophytes, charophyta, and chlorophyta are labeled on the right.
In Group I, MORC genes underwent an ancient gene duplication event (D1) that formed two subclades of Group I-1 and Group I-2. The Group I-1 only contains seed plants whereas the Group I-2 contains the whole land plant lineages. Therefore, both ferns and bryophyta branches might be lost in Group I-1. However, gymnosperms formed several branches in Group I-1, indicating that at least one gene duplication event occurred in the ancestor of gymnosperms. In Group II, MORC genes expanded in both bryophytes and ferns, resulting in the formation of two monophyletic groups in mosses, whereas three monophyletic groups were formed in the Eusporangiate and Leptosporangiate monilophytes.
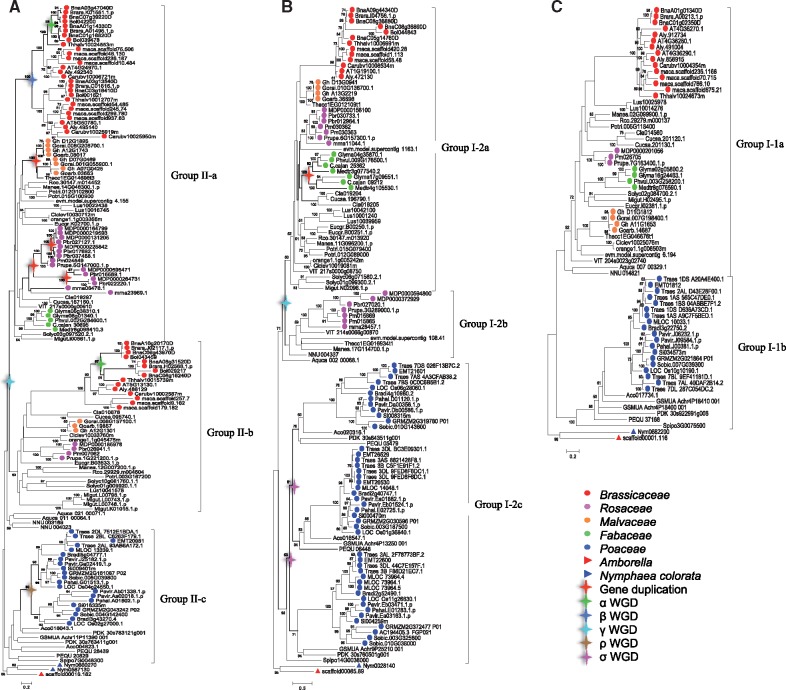
To elucidate in detail the phylogenetic relationships between the MORC gene families in angiosperms, we reconstructed a ML phylogenetic tree with 337 MORC protein sequences from 53 sequenced species. MORC genes are widely distributed among angiosperms, and their copy number is increased in most species. There are three copies of MORC gene in the basal angiosperms A. trichopoda, which are distributed in each clade base. While another basal angiosperm N.colorata had 4 copies, which have two copies in Group II (supplementary fig. S10, Supplementary Material online). MORC genes within each group were also investigated separately by reconstructing its ML phylogenetic tree, and the results showed that the MORC gene family expanded considerably in Group I-2 and Group II, while in Group I-1 it maintained the number of MORC members relatively stable (fig. 5). In Group II, MORC genes clustered into three subclades and there were two small clades with Group II-a and Group II-b in eudicots while one clade with Group II-c was grouped in monocots (fig. 5A). In Group I-2, MORC genes in eudicots were also subdivided into two subclades with Group I-2a and Group I-2b, while it formed three small branches in monocots with Group I-2c (fig. 5B). In Group I-1, MORC genes were grouped in Group I-1a and Group I-1b within eudicots and monocots. Except for several species possessing multiple MORC gene members, most species had only single copy in Group I-1 (fig. 5C). To further validate the preliminary classification of MORC genes in green plants based on the phylogenetic analyses, we also analyzed the MORC genes exon–intron structure from several representative plant species. MORC gene exon–intron organization and intron phase can be divided into three models: Group I-1 phase: 0220-2210-0000-0210-1, Group I-2 phase: 0220-2210-0000-0212-01, Group II phase: 0220-2210-0000-0212-010. The distribution pattern of intron phase was conserved at the 5′-end in most MORC genes, all of which were 0220-2210-0000, suggesting that MORC genes maintained a relatively conservative evolutionary model in the long-term evolutionary course (supplementary fig. S11, Supplementary Material online). At their 3′-end, the exon and intron distribution of MORC genes varies from 0210-1 in Group I-1 to 0212-01 in Group I-2 and 0212-010 in Group II. The number of exons in each subgroup gradually increased correspondingly, thus forming the three distinctive clades of Group I-1, Group I-2, and Group II.
Fig. 5.
—Maximum likelihood tree of detailed phylogenetic relationships among each group in representative angiosperm MORC genes. Phylogenetic trees of the Group II (A), Group I-2 (B), and Group I-1 (C) from representative angiosperms were reconstructed by ML method using in RaxML-HPC2 program through the online CIPRES Science Gateway with default parameters. For each clade, numbers above branches indicate bootstrap support percentage values based on 1,000 replications. The red asterisks indicate the family-specific gene duplication event and other different color asterisks represent several ancient whole genome duplication (WGD) events.
Expression Analysis of Plant MORC Genes
To explore the expression of MORC genes at different plant developmental stages, we compared available RNA-Seq data for MORC genes in three representative plant species. In all species here analyzed, we found that different MORCs were differentially expressed at different developmental stages, suggesting their involvement in a vast range of physiological processes during plant development (supplementary fig. S12, Supplementary Material online). We also found that the expression of MORC genes in reproductive tissues, such as flowers and seeds, was relatively higher than that in vegetative ones, indicating that the MORC genes might involve in the development of plant reproductive organs. Based on Arabidopsis RNA-seq data (Zhang and Ma 2012; Zhang et al. 2014), the expression of MORC genes was significantly higher in F1–9 (flowering stages 1 to 9), but decreased relatively in F4 (flowering stages 4) and F12 (flowering stages 12) (supplementary fig. S12A, Supplementary Material online). Interestingly, AtMORC5 was highly expressed during the meiosis process, whereas its expression level was lower in other developmental stages. Similarly, AtMORC7 appeared to be specifically expressed in F1–9 stage. In addition, the AtMORC1, AtMORC2, and AtMORC3 genes, belonging to Group I-1, were generated by tandem repeat duplication in Arabidopsis, in which AtMORC1 and AtMORC2 gene had the highest sequence similarity, but their expression pattern was clearly different. The expression of AtMORC1 in near all developmental stages was considerably higher than that of AtMORC2 gene, indicating that this paralogous gene pairs might have experienced transcriptional subfunctionalization (supplementary fig. S13A, Supplementary Material online).
Similarly, the expression level of MORC genes in reproductive tissues of soybean and rice was also higher than that in vegetative tissues. The expression pattern of Group II members GmMORC7a and GmMORC7b was similar with a higher level of transcript accumulation in flower tissues (supplementary fig. S12B, Supplementary Material online). The expression level of GmMORC6a and GmMORC6b, both belonging to Group I-2, was high in seeds and shoot apical meristem (SAM). Moreover, GmMORC6a gene was expressed relatively higher than GmMORC6b (supplementary fig. S13B, Supplementary Material online). Consistently, the expression pattern of GmMORC1a and GmMORC1b in Group I-1 was also different. The expression level of GmMORC1b was considerably higher than that of GmMORC1a, indicating that these paralogous genes may be subjected to different selection pressures after duplication. In rice, the OsMORC7 gene was highly expressed in seed while OsMORC4 was specifically expressed in callus (supplementary fig. S12C, Supplementary Material online). The expression of OsMORC6a and OsMORC6b are also considerable different. OsMORC6a is highly expressed in all tissues, while OsMORC6b only shows some degree of expression in seeds, suggesting that these genes might also have functional divergence after the duplication events (supplementary fig. S13C, Supplementary Material online).
Discussion
MORC Domain Is Highly Conserved in Both Plant and Animal Lineages
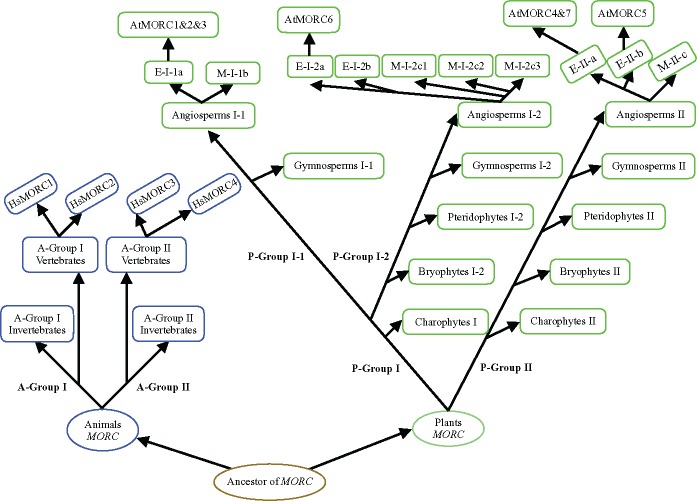
Our analysis showed that the MORC genes are widely distributed among plant and animal lineages, and the copy number of MORC gene considerably varies between plants and animals. We identified 20 conserved motifs among the MORC gene family in both plant and animal lineages. Among them, four motifs 1-15-2-8 or 1-7-2-8 constituted the well-known GHKL ATPase domain and S5 fold domain was composed of the motif 5 and motif 14. The combined GHKL ATPase and S5 fold domain can together constitute a catalytically active ATPase module that involved in ATP binding and hydrolysis (Dutta and Inouye 2000). Based on the composition of the highly conserved motif, we defined the MORC domain spanning ∼370 amino acids, which contained 10 conserved α-helixes and 12 β-sheets in plant and animal, and shares a very similar 3D structure in plant and animal. The MORC domain is highly conserved in both plants and animals, suggesting that it might originate from a common ancestor before the divergence of plants and animals. Recent studies have revealed that both the plant and animal MORC proteins exhibited many similar functions involved in diseases resistance and epigenetic gene silencing (Koch et al. 2017). The MORC domain as the characteristic feature conserved in MORC proteins may take charge of their functional correlations among these proteins. We proposed a model for the evolution of MORC genes in plant and animal lineages. According to this proposed model, the MORC gene origin can be traced back to the common ancestor before the divergence of plants and animals. Subsequently several genome-wide duplication events occurred, thus leading to the formation of actual multigene families detected in both plants and animals (fig. 6). In animals, both the A-Group I and II have encountered gene duplications in the evolution of vertebrates, thus generating 4 MORC members in humans. Interestingly, MORC genes in plants experienced a large-scale expansion in their long-term evolutionary history. In P-Group II, there are three and two monophyletic groups in monilophyte and moss, respectively, indicating that at least two gene duplication events have occurred in these plant lineages. Moreover, a eudicots-shared WGD event was also happened in P-Group II dicots lineages. Noteworthy, P-Group I-1 MORC genes are only detected in seed plants whereas all land plant lineages are represented in P-Group I-2, suggesting that the P-Group I-1 divided at least before the divergence of seed plants, and the monilophytes and bryophytes might have lost MORCs belonging to this group in the long-term evolution. Additionally, among the P-Group I-2 angiosperm lineages, there have encountered one and two rounds of WGD events in dicots and monocots, resulting in large expansion of MORC genes in angiosperms.
Fig. 6.
—A proposed model for the evolution of MORC genes in plants and animals. Based on sequence similarity, exon–intron organization and MORC domain, MORC genes in plants and animals can be divided into two major groups, respectively. The MORC gene is present in both plants and animals and its origin can be traced back to the common ancestor before the divergence of plants and animals. Animal MORCs underwent a gene duplication during the evolution of vertebrates in each subgroups and thus produced multiple gene copies in vertebrate lineages. Plant MORCs encountered several rounds of gene duplication events in the long-term evolutionary courses, eventually generating large-scale expansion in most species.
Contrasting Evolution Patterns among Different MORC Subfamilies in Angiosperms
The MORC gene family is widely distributed among the green plant lineage and are organized in multiple copies in higher angiosperms, whereas appear to be present in single copy in unicellular algae, indicating that the MORC gene family in higher plants underwent several round of gene duplication events in the long-term evolutionary history. The copy number of MORC members differs between species. In higher plants, MORC gene family was divergent in angiosperms. For example, the Group II can be subdivided into two small clades with Group II-a and Group II-b in eudicots. Further synteny analysis revealed that Group II-a and Group II-b were likely generated by large-scale segmental duplications or whole genome duplications (WGDs), which corresponded to theγWGD event shared by core eudicots (supplementary fig. S14, Supplementary Material online). From the phylogenetic tree, the number of MORC genes in Group II-a was relatively higher compared with that in Group II-b, suggesting that some MORC genes might have been lost in Group II-b. For instance, the Fabaceae species were absent in the Group II-b. In Group II-a, MORC genes were duplicated in Brassicaceae, Malvaceae, and Rosaceae families. Synteny analysis indicated that these MORC genes were expanded mainly because of family or species-specific WGD events (supplementary fig. S14, Supplementary Material online). Additionally, most of MORC genes had two or more copies in Group II-c, indicating that in monocots MORC genes had also experienced gene duplication. Further investigation revealed this duplication is consistent with theρWGD event within Poaceae. Consistently with the evolutionary pattern of Group II, Group I-2 also underwent theγWGD event, thus forming two subclades with Group I-2a and Group I-2b in eudicots, which is further supported by the syntenic analysis (supplementary fig. S14, Supplementary Material online). Many genes were also lost in Group I-2b, such is the case in Brassicaceae and Fabaceae species. Interestingly, there were three subclades of MORC genes in Group I-2c of monocot branch, which may be caused by two ancient σ WGD events in the older monocot. Our results indicated that MORC genes in Group I-1 have strikingly different evolution patterns of gene duplication compared with other groups. The Group I-1 MORC genes appear to be relatively more conserved compared with Group I-2 and Group II. Generally, most species in Group I-1 considered have only a single copy of MORC gene. The low-copy numbers of MORC genes in this group suggested a functional conservation during the long-term evolution. However, it is noteworthy that Arabidopsis possess three Group I-1 MORC genes: AtMORC1 (AT4G36290), AtMORC2 (AT4G36280), and AtMORC3 (AT4G36270). These paralogous genes were generated by tandem repeat duplication (Harris et al. 2016). AtMORC1 (also known as CRT1) and its closet paralogous AtMORC2 (CHR1) play an important role in the resistance to various pathogens in the plant multilayered immune systems (Kang and Klessig 2008; Kang et al. 2010, 2012). This gene was first identified in Arabidopsis by a screen for mutants with compromised recognition of the Turnip Crinkle Virus (Kang and Klessig 2008). Further studies in Arabidopsis showed that AtMORC1 was not only involved in the regulation of the plant multilayer immune system (Kang and Klessig 2008; Kang et al. 2010, 2012) but also in transcriptional gene silencing (Lorkovic et al. 2012; Moissiard et al. 2012; Brabbs et al. 2013). Similar functions were also observed in other plants, suggesting that the MORC1 roles may be evolutionarily conserved within Group I-1. Unlike the Group I-1, both the Group I-2 and Group II have encountered multiple rounds of WGD events, leading to the increase of MORC gene members in angiosperms. In eudicots, two important events took place: an eudicot-shared ancient gene duplication in Group I-2 and Group II, and the subsequent loss of most genes in one clade. Similarly, a ρ WGD occurred in Group II Poaceae while two older σ WGD events in Group I-2 monocots. These gene duplications increased the quantity of genetic material and improved the chance of functional innovations during the long-term evolutionary course.
MORC Genes Have a Role in Development of the Reproductive Tissue
Our results showed that several MORC genes in three representative species (e.g., AtMORC1, AtMORC7, GmMORC7a, OsMORC7, OsMORC6a) are expressed in almost every tissues examined, suggesting that they might be involved in a wide range of biological processes. On the contrary, some other MORCs appeared to be highly expressed exclusively in reproductive tissues, indicating they might be involved in the development of plant reproductive organs. Expression analyses also revealed several instances of tissue-specific expression that suggested roles in specific developmental programs. For instance, Arabidopsis AtMORC5 transcript is preferentially accumulated during meiosis but nearly absent in other stages and vegetative tissues. Another example is rice OsMORC4, which is preferentially expressed in callus. Duplicated genes might undergo different evolutionary fates including nonfunctionalization, subfunctionalization, and neofunctionalization (Liu, Li, et al. 2014). Changes in the expression pattern of duplicated genes usually indicate functional divergence after duplication events. In our study, we found that some paralogs showed divergent expression patterns among the three species, like the AtMORC1/AtMORC2 in Arabidopsis, the GmMORC1a/GmMORC1b, GmMORC6a/GmMORC6b in soybean, and the OsMORC6a/OsMORC6b in rice. These gene pairs exhibited clearly divergent expression patterns in different tissues, indicating that they are diverged under selection pressure during the evolution. It was demonstrated that the expression divergence is often the first step in functional divergence between duplicate genes, thereby increasing the chance of duplicate genes to be retained in the genome (Zhang 2003). Our study revealed that the MORC genes are relatively highly expressed in reproductive tissues during the plant development, and the results had provided a basis for further investigating the functional role of different MORC genes in plant development.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81502437), and a start-up fund from Fujian Agriculture and Forestry University to LZ. F.C. is supported by a grant from State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops (SKB2017004) and a grant from natural science foundation of Fujian Province (2018J01603). We acknowledge OneKP (Gane Ka-Shu Wong from University of Alberta) and other databases for the sequence data supporting.
Literature Cited
- Arnold K, Bordoli L, Kopp J, Schwede T.. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 222:195–201. [DOI] [PubMed] [Google Scholar]
- Bailey TL. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37(Web Server issue):W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C.. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 2:28–36. [PubMed] [Google Scholar]
- Biasini M. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42(Web Server issue):W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordiya Y, et al. , 2016. Pathogen infection and MORC protein affect chromatin accesibility of transposable elements and expression of their proximal genes in Arabidopsis. Mol Plant Microbe Interact. 299:674–687. [DOI] [PubMed] [Google Scholar]
- Brabbs TR, et al. , 2013. The stochastic silencing phenotype of Arabidopsis morc6 mutants reveals a role in efficient RNA-directed DNA methylation. Plant J. 755:836–846. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2515:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Xia R, Chen H, He Y.. 2018. TBtools, a Toolkit for Biologists integrating various HTS-data handing tools with a user-friendly interface. bioRxiv 289660; doi: 10.1101/289660. [DOI]
- Chen F, et al. , 2018. The sequenced angiosperm genomes and genome databases. Front Plant Sci. 9:418.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE.. 2004. WebLogo: a sequence logo generator. Genome Res. 146:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R, Inouye M.. 2000. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 251:24–28. [DOI] [PubMed] [Google Scholar]
- Eberl HC, Spruijt CG, Kelstrup CD, Vermeulen M, Mann M.. 2013. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol Cell 492:368–378. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 149:755–763. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, et al. , 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40(Database issue):D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC, Schwede T.. 2009. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30(S1):S162–S173. [DOI] [PubMed] [Google Scholar]
- Harris CJ, et al. , 2016. Arabidopsis AtMORC4 and AtMORC7 form nuclear bodies and repress a large number of protein-coding genes. PLoS Genet. 125:e1005998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He FH, et al. , 2010. Structural insight into the Zinc Finger CW domain as a histone modification reader. Structure 189:1127–1139. [DOI] [PubMed] [Google Scholar]
- Hu B, et al. , 2015. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 318:1296–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue N, et al. , 1999. New gene family defined by MORC, a nuclear protein required for mouse spermatogenesis. Hum Mol Genet. 87:1201–1207. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Abhiman S, Aravind L.. 2008. MutL homologs in restriction-modification systems and the origin of eukaryotic MORC ATPases. Biol Direct 3:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Anantharaman V, Wolf MY, Aravind L.. 2008. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int J Parasitol. 381:1–31. [DOI] [PubMed] [Google Scholar]
- Kang HG, et al. , 2010. Endosome-associated CRT1 functions early in resistance gene-mediated defense signaling in Arabidopsis and tobacco. Plant C 223:918–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, et al. , 2012. CRT1 is a nuclear-translocated MORC endonuclease that participates in multiple levels of plant immunity. Nat Commun. 3:1297.. [DOI] [PubMed] [Google Scholar]
- Kang HG, Klessig DF.. 2008. The involvement of the Arabidopsis CRT1 ATPase family in disease resistance protein-mediated signaling. Plant Signal Behav. 39:689–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Kuhl JC, Kachroo P, DF K.. 2008. CRT1, an Arabidopsis ATPase that interacts with diverse resistance proteins and modulates disease resistance to Turnip Crinkle Virus. Cell Host Microbe 31:48–57. [DOI] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T.. 2009. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37(Database issue):D387–D392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A, et al. , 2017. MORC proteins: novel players in plant and animal health. Front Plant Sci. 8:1720.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen G, et al. , 2014. The compromised recognition of turnip crinkle virus1 subfamily of microrchidia ATPases regulates disease resistance in barley to biotrophic and necrotrophic pathogens. Plant Physiol. 1642:866–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Tang HB, Wang XY, Paterson AH.. 2013. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41(Database issue):D1152–D1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DQ, et al. , 2012. MORC2 signaling integrates phosphorylation-dependent, ATPase-copuled chromatin remodeling during the DNA damage response. Cell Rep. 26:1657–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DQ, Nair SS, Kumar R.. 2013. The MORC family: new epigenetic regulators of transcription and DNA damage response. Epigenetics 87:685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, et al. , 2010. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 631:86–99. [DOI] [PubMed] [Google Scholar]
- Liu W, et al. , 2014. Genome-wide survey and expression analysis of calcium-dependent protein kinase in Gossypium raimondii. PLoS One 96:e98189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z-W, et al. , 2014. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 101:e1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z-W, et al. , 2016. Two components of the RNA-Directed DNA methylation pathway associate with MORC6 and silence loci targeted by MORC6 in Arabidopsis. PLoS Genet. 125:10: e1006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic ZJ, Naumann U, Matzke AJ, Matzke M.. 2012. Involvement of a GHKL ATPase in RNA-directed DNA methylation in Arabidopsis thaliana. Curr Biol. 2210:933–938. [DOI] [PubMed] [Google Scholar]
- Manosalva P, Manohar M, Kogel K-H, Kang H-G, Klessig DF.. 2015. The GHKL ATPase MORC1 modulates species-specific plant immunity in Solanaceae. Mol Plant Microbe Interact. 288:927–942. [DOI] [PubMed] [Google Scholar]
- Moissiard G, et al. , 2012. MORC family ATPases required for heterochromatin condensation and gene silencing. Science 3366087:1448–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard G, et al. , 2014. Transcriptional gene silencing by Arabidopsis microrchidia homologues involves the formation of heteromers. Proc Natl Acad Sci USA. 11120:7474–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 267:1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, et al. , 2011. Retrogenes in rice (Oryza sativa L. ssp. japonica) exhibit correlated expression with their source genes. Genome Biol Evol. 3:1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM.. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1820:6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ML, et al. , 1998. Identification of morc (microrchidia), a mutation that results in arrest of spermatogenesis at an early meiotic stage in the mouse. Proc Natl Acad Sci USA. 9524:14361–14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C-S, Chen Y-C, Lu C-H, Hwang J-K.. 2006. Prediction of protein subcellular localizations. Proteins Struct Funct Bioinformatics 643:643.. [DOI] [PubMed] [Google Scholar]
- Yu CS, Lin CJ, Hwang JK.. 2004. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 135:1402–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol Evol. 186:292–298. [Google Scholar]
- Zhang L, et al. , 2014. Analysis of Arabidopsis floral transcriptome: detection of new florally expressed genes and expansion of Brassicaceae-specific gene families. Front Plant Sci. 5:802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ma H.. 2012. Complex evolutionary history and diverse domain organization of SET proteins suggest divergent regulatory interactions. New Phytol. 1951:248–263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.