Abstract
The co-occurrence of a copy number variant (CNV) and a functional variant on the other allele may be a relevant genetic mechanism in schizophrenia. We hypothesized that the cumulative burden of such double hits—in particular those composed of a deletion and a coding single-nucleotide variation (SNV)—is increased in patients with schizophrenia. We combined CNV data with coding variants data in 795 patients with schizophrenia and 474 controls. To limit false CNV-detection, only CNVs called by two algorithms were included. CNV-affected genes were subsequently examined for coding SNVs, which we termed “CNV–SNVs.” Correcting for total queried sequence, we assessed the CNV–SNV-burden and the combined predicted deleterious effect. We estimated P-values by permutation of the phenotype. We detected 105 CNV–SNVs; 67 in duplicated and 38 in deleted genic sequence. Although the difference in CNV–SNVs rates was not significant, the combined deleteriousness inferred by CNV–SNVs in deleted sequence was almost 4-fold higher in cases compared with controls (nominal P = 0.009). This effect may be driven by a higher number of CNV–SNVs and/or by a higher degree of predicted deleteriousness of CNV–SNVs. No such effect was observed for duplications. We provide early evidence that deletions co-occurring with a functional variant may be relevant, albeit of modest impact, for the genetic etiology of schizophrenia. Large-scale consortium studies are required to validate our findings. Sequence-based analyses would provide the best resolution for detection of CNVs as well as coding variants genome-wide.
Introduction
In the past fifteen years an increasing number of specific genetic variants conferring risk for schizophrenia are being identified. Although these findings start to substantiate the observed heritability of schizophrenia, the emerging picture also suggests that the underlying genetic architecture of this illness is complex. Thus far, there is substantial evidence for the role of variants that occur frequently in the population [minor allele frequency (MAF) of 5% or more] and are associated with a small risk effect. The cumulative effect of such common variants could account for a substantial part—approximately a third—of the observed genetic susceptibility for schizophrenia (1,2). At present 108 loci of common risk variants for schizophrenia have been identified (3). In addition, rare variants also contribute to genetic risk of schizophrenia; these are alleles that occur infrequently in the population (e.g. MAF < 1%) but may be associated with relatively large risk effects in the individual carrier. Recent studies have demonstrated the role of rare single-nucleotide variants (SNVs) in schizophrenia (4–7) although thus far no specific SNV has been unequivocally associated with the disorder. In contrast, the first studies to imply copy number variants (CNVs) as risk factors for schizophrenia appeared already in 2008 (8–10). This finding was replicated in subsequent studies (11–13), identifying a number of recurrent CNVs consistently associated with schizophrenia (14).
CNVs, together with other structural variants (e.g. inversions), explain a substantially larger proportion of variation in the human genome than single nucleotide polymorphisms (SNPs) (15,16). The pathogenic impact of a CNV is highly variable and amongst others related to variant type (e.g. deletions versus duplications), size (large versus small), gene content (genic versus non-genic), transmission status (de novo versus inherited) and frequency in the population (rare versus common), with pathogenic impact more likely in each first example cited (17).
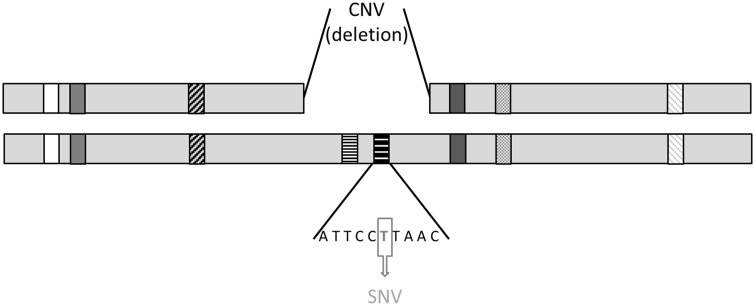
A large proportion of the CNVs associated with schizophrenia reported thus far are de novo (8). This is noteworthy, because although each genome contains on average well over 1000 CNVs (15), de novo CNVs are estimated to occur in the genome at a rate of only 0.01–0.02 per generation (17). Recently, novel methods have increased the ability to detect CNVs of smaller size (e.g. <500 bp), generating higher estimates of mutation rates (18). Although heritability characteristics of these smaller CNVs are still under study, it is estimated that of the larger CNVs an estimated 99% detected in any individual are inherited (19). Importantly, although there is strong evidence for the pathogenicity of de novo CNVs, to date there is no formal evidence against a pathogenic role of inherited CNVs. It can be argued that most of such inherited CNVs—with the exception of very large CNVs (13)—are not likely to exert strong pathogenic effects in general because of their common occurrence in the population. However, one particular mechanism, which could render an otherwise neutral CNV into a pathogenic genetic event is the possible co-occurrence with a functional mutation on the other allele, a phenomenon that can be referred to as “unmasking” or a specific type of “compound heterozygosity” (see Fig. 1). The psychiatry genetics literature provides precedents for this mechanism; several case studies report the co-occurrence of an inherited deletion and a functional variant on the remaining allele in probands with autism (20–22) and in schizophrenia (23). Also, the rate of a slightly different type of compound heterozygosity, i.e. two rare loss of function SNVs co-occurring at the same locus, was found to be significantly increased in autism compared with controls (24). However, the same event was not found to be increased in schizophrenia (25).
Figure 1.
Schematic representation of a compound heterozygous double hit. In the normal situation each gene on the autosomes is present in two copies (diploidy). In this example both horizontally dashed genes are present in only one copy (haploidy). In addition, the horizontally dashed gene on the right is affected by a coding single nucleotide variant (SNV). The compound heterozygous double hit consists of the co-occurrence of a CNV (in this example a deletion) on the one allele and a coding SNV on the remaining allele.
Here, we hypothesized that in patients with schizophrenia co-occurrence of CNVs and functional point mutations at the same locus occur more frequently compared with controls. We tested this hypothesis by assessing the number of these events, as well as the predicted collective deleterious functional effect they infer, comparing between a group of patients with schizophrenia and a group of healthy control individuals. To this end, we used the results of a whole genome CNV study in patients with schizophrenia and controls—which have been reported in a previous study (11)—and combined this dataset with whole exome SNP data obtained in the same sample.
Results
The entire sample set in which both CNV and SNV data were available consisted of 1269 individuals (795 cases and 474 controls). We observed 905 deletions (involving 109, 1 Mb of genic sequence) and 1069 duplications (involving 201, 5 Mb genic sequence). These results have previously been reported in detail (11). In this CNV dataset, we identified a total of 105 compound heterozygous events consisting of a concurrent CNV and an SNV at the same locus (CNV–SNVs; 38 in deleted sequence, 67 in duplicated sequence). Given that genomic regions differ with regard to gene density, we used the total genic sequence affected by either deletions or duplications (see dataset characteristics in Table 1 and CNV_SNV characteristics in the online Supplementary Material) as the basis for correction of our further findings.
Table 1.
Sample set characteristics
| Cases | Controls | Total | ||
|---|---|---|---|---|
| n | 795 | 474 | 1269 | |
| % Male | 74% | 51% | ||
| Number of identified CNV–SNVs | All CNVs | 71 | 34 | 105 |
| Del | 27 | 11 | 38 | |
| Dup | 44 | 23 | 67 | |
| Total of genic sequence queried (Mb) | All CNVs | 375, 7 | 226, 6 | 602, 3 |
| Del | 236, 2 | 140, 9 | 377, 1 | |
| Dup | 139, 4 | 85, 7 | 225, 2 | |
CNV–SNVs; compound heterozygous events characterized by the co-occurrence of a CNV and a non-synonymous point mutation at the same locus.
In cases compared with controls, we observed 18.9 CNV–SNVs versus 15.0 CNV–SNVs per 100 Mb queried genic sequence (deletions and duplications together), the difference was not significant (see Table 2). However, the cumulative deleterious impact, i.e. the sum of individual predicted deleteriousness scores inferred by all CNV–SNVs in deleted sequence, was approximately 4-fold higher in cases compared with controls (5.81 in cases versus 1.51 in controls, nominal, permutation-based P = 0.009) whereas the cumulative deleterious impact of CNV–SNVs in duplicated sequence was virtually identical (9.49 in cases versus 9.16 in controls, P = 0.369). Finally, we observed a trend-level difference for the average predicted deleteriousness per CNV–SNV in deleted sequence (0.51 in cases, 0.19 in controls, P = 0.074) but not in duplicated sequence (respectively 0.30 and 0.34, P = 0.533).
Table 2.
Results of comparison CNV–SNVs in cases and controls
| Cases | Controls | P-value | ||
|---|---|---|---|---|
| Number of CNV–SNVs per 108 bp | All CNVs | 18.9 | 15.0 | 0.167 |
| Del | 11.4 | 7.8 | 0.172 | |
| Dup | 31.6 | 26.8 | 0.292 | |
| Cumulative predicted deleteriousness per 108 bp | All CNVs | 7.17 | 4.40 | 0.071 |
| Del | 5.81 | 1.51 | 0.009 | |
| Dup | 9.49 | 9.16 | 0.369 | |
| Average predicted deleteriousness per CNV–SNV | All CNVs | 0.38 | 0.29 | 0.141 |
| Del | 0.51 | 0.19 | 0.074 | |
| Dup | 0.30 | 0.34 | 0.533 |
P-values were obtained by 10 000 random permutations of case-control status.
Post hoc we reiterated the same analytical steps in the same sample, however, this time with a dataset generated from low-stringency CNV calling (i.e. all CNVs were called by PennCNV only), the result of which did not demonstrate the case-control differences observed in our original analysis (results not shown).
Discussion
Findings of our study suggest that the cumulative burden of deleterious impact inferred by CNV–SNVs in deleted sequence is increased in patients with schizophrenia compared with controls.
This effect may not only be driven by a higher number of CNV–SNVs in cases, but also independently by an on average higher degree of deleteriousness of CNV–SNVs identified in cases. It is worth noting that both of these effects were not detected for CNV–SNVs in duplicated sequence, consistent with the observed stronger phenotypic impact of deletions in other studies (16). Conceivably, in the scenario of a double hit, the duplication of a normal allele could act compensatory to the deleterious impact of a functional mutation on the non-duplicated allele. We have previously reported an increased SNV burden in schizophrenia patients in the same dataset; a significant difference was apparent when comparing SNVs unique to cases with SNVs unique to controls (4). Of note, an overall exome-wide association of SNVs was not detectable in our previous study, as was to be expected given the low minor allele frequencies and modest effect sizes. These results indicate that the observed increased deleteriousness in the current study is specific to CNV–SNVs—in particular of SNVs in deleted sequence—and cannot be attributed to a global exome-wide difference of SNVs between cases and controls.
Our inability to repeat the findings when using a reduced stringency of CNV calling could indicate a type I error; i.e. in reality there is no increased burden of double hits in schizophrenia patients. Alternatively, it is possible that more false than true positive signals were introduced when relying on CNV calling by one algorithm instead of two (QuantiSNP and PennCNV). Indeed, large-scale variability between the output of different CNV methods has been reported previously (26). Our own previously reported analyses in this dataset are consistent with this notion; only 16% of the total of gene-containing CNVs called by either QuantiSNP or PennCNV, is called by both (11).
The tests reported here are not independent from each other. Nevertheless, if we assume three separate hypotheses (number of CNV_SNVs, cumulative predicted deleteriousness and average predicted deleteriousness per CNV–SNV) the corrected alpha would be 0.017, indicating that our main finding, i.e. the difference in cumulative deleterious impact of CNV–SNVs in deleted sequence, remains statistically significant after this correction (P = 0.009).
Despite their relatively strong risk effects, invariable full penetrance of CNVs is uncommon. Therefore, it is likely that additional risk factors of environmental or of genetic origin are required (27). The latter includes parent-of-origin and imprinting effects, modifying variation in the remainder of the genome and factors that influence expression levels of the region affected by the CNV (28). Here, we provide tentative evidence for a modest role of genetic variation on the remaining allele in the variable penetrance of deletions in schizophrenia.
Our findings require replication. The relatively low number of identified CNV–SNVs was not to be unexpected given our conservative CNV calling and the use of a 250k SNP arrays data. Although providing reliable data, this approach limited our a priori statistical power to detect the hypothesized effects. We performed a power calculation, assuming a low rate of observed CNV–SNVs in controls (0.034) and the same analytical strategy (i.e. combining CNV data with exomeSNP data). Depending on the expected difference of CNV–SNVs the required sample size varies greatly. Approximately 48 thousand subjects (cases and controls) would be required to achieve >80% ability to detect a 20% difference in CNV–SNV rates. In contrast, to detect a 2-fold difference the sample size of the current study would be sufficient. Such effect size may not be unrealistic given the 2-fold increased rate of two rare loss of function SNVs at the same locus in autism cases, with baseline rates in controls comparable to the rate of observed CNV–SNVs in controls in our study (24). However, we argue that with the increasing availability of sequencing data the most suitable follow up study would be to combine high-resolution CNV data with whole genome or exome sequencing data, while applying frequency filters on both CNVs and SNVs, and prioritization strategies on the basis of objective metrics of variant pathogenicity such as available through the Exome Aggregation Consortium (ExAC) database (29). Because of the large numbers of variants that can thus be queried, these studies will be much better powered to detect a possible difference.
In summary, our findings suggest that the co-occurrence of a deletion with a deleterious SNV on the remaining allele may be a relevant, albeit modest, mechanism involved in the etiology of schizophrenia. This type of double hit event can be considered an example of compound heterozygosity, a mechanism involved in a number of recessive traits (e.g. (30–32)). Our results provide early evidence that this mechanism may also play a role in schizophrenia. In terms of explaining heritability, the impact of this mechanism is expected to be low. However, similar to other rare causative genetic events it may contribute to the identification of specific genes involved in this illness. A potential clinical implication may exist in some instances where a putatively pathogenic deletion in an individual with schizophrenia is inherited from a healthy parent. In those cases it could be informative to sequence the remaining allele in the proband to screen for additional SNVs as described in this study.
Materials and Methods
Acquired in the same sample set, we merged whole genome CNV data (from 834 cases and 672 controls) with whole exome SNP data (from 1042 cases and 961 controls) to obtain a combined CNV-SNP dataset of 795 patients diagnosed with schizophrenia and 474 unrelated healthy controls.
Samples were recruited by the Genetic Risk and Outcome of Psychosis (GROUP) Consortium from the Netherlands. Cases were patients fulfilling DSM-IV criteria for a diagnosis of schizophrenia or any other non-affective psychotic disorder, assessment was done using the Comprehensive Assessment of Symptoms and History (CASH) or the Schedules for Clinical Assessment for Neuropsychiatry (SCAN 2.1). Controls were volunteers without a (lifetime) diagnosis of any (affective or non-affective) psychotic disorder. Both cases and control subjects were of Dutch descent (with at least three of four grandparents of Dutch ancestry). Ethical approval was obtained from the Ethical Review Board of the University Medical Centre Utrecht as well as from the UCLA Institutional Review Board and all participants gave written informed consent. Detailed assessment methods of the GROUP cohort have been reported previously (11,33).
CNV calling
Genomic DNA of all participants was hybridized to the HumanHap550v3 BeadArray (Illumina, San Diego, CA) at UCLA Neurosciences Genomics Core according to standard protocols. CNV calling was performed with two algorithms, QuantiSNP (34) and PennCNV (35). Only gene-containing CNVs with length >50 kb, called by both algorithms were retained for the primary analysis. By including only overlapping CNVs, we made an effort to limit the false positive rate of CNV detection (36). All CNVs—including both rare and common—retained by this method were used for subsequent analysis, i.e. no further selection was applied. A detailed description of this method, including quality control steps as well as a complete list of results in this dataset have been published previously (11). To match the build of the exome array, the genomic coordinates of the CNVs were updated to hg19 using LiftOver (http://genome.ucsc.edu/cgi-bin/hgLiftOver; date last accessed May 28, 2018). A post hoc analysis was performed using CNVs of length [50–300 kb] called by PennCNV only to measure the robustness of our first observation under low stringency conditions of CNV calls.
SNV calling
For this purpose all samples were genotyped at UCLA Neurosciences Genomics Core using the Illumina HumanExome BeadChip. This array was conceptualized as midway between exome sequencing and common SNP arrays, allowing the query of more than 250, 000 relatively rare (MAF of 0.01% or more) putatively functional coding SNPs as explained at http://genome.sph.umich.edu/wiki/Exome_Chip_Design; date last accessed May 28, 2018. Quality control was performed using PLINK (v1.08p) (37) and has previously been described in detail for this dataset (4). In particular, using a set of common independent variants, we excluded ethnic outliers on the basis of the first two multidimensional scaling components (4).
Merging CNV and SNV data
Following CNV calling, in each subject the RefSeq gene content of each region affected by a deletion or a duplication was identified using the gene prediction track of the UCSC genome browser (hg19, http://genome.ucsc.edu/; date last accessed May 28, 2018) and applying a 50 kb fuzzy border at the CNV boundaries. Any gene within these pre-defined boundaries, including those partly overlapping a CNV-boundary, were considered as “CNV-affected” genes and included in the subsequent step. We then examined the exonic regions of all “CNV-affected” genes for the occurrence of SNVs with MAF < 0.05, generating a library of compound heterozygous events consisting of a concurrent CNV and an SNV at the same locus (CNV–SNVs).
Prediction of deleterious impact
We used a previously reported algorithm, CONsensus DELeteriousness (CONDEL) (38), to calculate the level of deleterious effect caused by the amino-acid substitutions of the SNVs. CONDEL is an assessment tool of deleteriousness, primarily on the basis of knowledge from studies of Mendelian traits; it combines the prediction output of five bioinformatics tools (SIFT, Polyphen2, MAPP, LogR and Pfam E-value) into a continuous consensus score between 0 and 1 for each nonsynonymous SNV. Because the array also includes splice site and stop-altering SNVs that are not scored by the algorithm, we applied, as previously described (4), an augmented version of the CONDEL, which adds both classes by assigning a maximal deleteriousness score (i.e. 1).
Statistical methods
We compared the total number of CNV–SNVs as well as the cumulative burden of deleterious effect inferred by CNV–SNVs between cases and controls. Here, we define “cumulative burden of deleterious effect” as the sum of all predicted deleteriousness scores inferred by CNV–SNVs identified in a sample of individuals. We compared total number of CNV–SNVs as well as the cumulative burden of deleterious effect between cases and controls, while controlling for the total amount of DNA sequence queried in each subgroup. To estimate significance of the observed case control differences, we performed 10 000 permutations of the phenotype, i.e. randomizing case control status. Nominal P-values were then determined by comparing the observed value for a given test against the distribution of values obtained by permutation. Significance threshold corrected for multiple testing was set at 0.017 (0.05/3, correcting for three tests). All bioinformatics and statistical procedures were performed with R version 3.1.2 (https://www.r-project.org).
Power calculation
On the basis of the number of gene-containing deletions called by both algorithms and the number of rare variants observed using the array, we assess power to identify difference in proportion of deletions that have at least one low-frequency variant (MAF < 5%) in a gene overlapping the CNV. In our sample, the proportion of CNVs with such rare variants is 0.034 in controls. At our current sample size we were well powered to pick up effect sizes corresponding roughly to a 2-fold increased rate of double hits. Much larger samples are required to detect smaller effect sizes. For instance, one would need two samples with >17 000 deletions to reliably detect a difference in cumulative burden of 20% at α = 0.05 with 80% power. Given the increased burden of deletions in schizophrenia cases (in our sample controls have on average 0.73 deletions called by both algorithms, compared with 0.67 in controls), this corresponds to approximately 25 000 controls and 23 000 cases. Note that this power calculation neither incorporates the subtle increased burden of rare variants (4), nor does it take into account the functionality of included variants. Moreover, these estimates are on the basis of very stringent CNV calling and a sparse genotyping array.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are grateful for the generosity of time and effort by the patients, their families and healthy subjects. GROUP investigators are: Behrooz Z. Alizadeha, Agna A. Bartels-Velthuisa, Nico J. van Beverenb,c,d, Richard Bruggemana, Wiepke Cahne, Lieuwe de Haanf, Philippe Delespaulg, Carin J. Meijerf, Inez Myin-Germeysh, Frederike Schirmbeckf, Claudia J.P. Simonsg,i, Neeltje E. van Harene, Jim van Osg,j, Ruud van Winkelg,h
aUniversity of Groningen, University Medical Center Groningen, University Center for Psychiatry, Groningen, The Netherlands; bAntes Center for Mental Health Care, Rotterdam, The Netherlands; cErasmus MC, Department of Psychiatry, Rotterdam, The Netherlands; dErasmus MC, Department of Neuroscience, Rotterdam, The Netherlands; eUniversity Medical Center Utrecht, Department of Psychiatry, Brain Centre Rudolf Magnus, Utrecht, The Netherlands; fAcademic Medical Center, University of Amsterdam, Department of Psychiatry, Amsterdam, The Netherlands; gMaastricht University Medical Center, Department of Psychiatry and Psychology, School for Mental Health and Neuroscience, Maastricht, The Netherlands; hKU Leuven, Department of Neuroscience, Research Group Psychiatry, Center for Contextual Psychiatry, Leuven, Belgium; i GGzE, Institute for Mental Health Care Eindhoven and De Kempen, Eindhoven, The Netherlands.
Furthermore, we would like to thank all research personnel involved in the GROUP project, in particular: Joyce van Baaren, Erwin Veermans, Ger Driessen, Truda Driesen, Karin Pos, Erna van ’t Hag, Jessica de Nijs, Atiqul Islam, Wendy Beuken and Debora Op ’t Eijnde.
Conflict of Interest statement. None declared.
Funding
Brain and Behavior Research Foundation (2010 Young Investigator Award to J.A.S.V.), the Geestkracht programme of the Dutch Health Research Council [Zon-Mw, grant number 10–000-1001 for the infrastructure for the GROUP study and NIH/NIMH R21 MH092783 (to R.A.O.)]. Additional funding for the GROUP study was obtained from participating pharmaceutical companies (Lundbeck, AstraZeneca, Eli Lilly, Janssen Cilag) and universities and mental health care organizations (Amsterdam: Academic Psychiatric Centre of the Academic Medical Center and the mental health institutions: GGZ Ingeest, Arkin, Dijk en Duin, GGZ Rivierduinen, Erasmus Medical Centre, GGZ Noord Holland Noord. Groningen: University Medical Center Groningen and the mental health institutions: Lentis, GGZ Friesland, GGZ Drenthe, Dimence, Mediant, GGNet Warnsveld, Yulius Dordrecht and Parnassia psycho-medical center The Hague. Maastricht: Maastricht University Medical Centre and the mental health institutions: GGZ Eindhoven en De Kempen, GGZ Breburg, GGZ Oost-Brabant, Vincent van Gogh voor Geestelijke Gezondheid, Mondriaan, Virenze riagg, Zuyderland GGZ, MET ggz, Universitair Centrum Sint-Jozef Kortenberg, CAPRI University of Antwerp, PC Ziekeren Sint-Truiden, PZ Sancta Maria Sint-Truiden, GGZ Overpelt, OPZ Rekem. Utrecht: University Medical Center Utrecht and the mental health institutions Altrecht, GGZ Centraal and Delta).
Contributor Information
GROUP investigators:
Behrooz Z Alizadeh, Agna A Bartels-Velthuis, Nico J van Beveren, Richard Bruggeman, Wiepke Cahn, Lieuwe de Haan, Philippe Delespaul, Carin J Meijer, Inez Myin-Germeys, Frederike Schirmbeck, Claudia J P Simons, Neeltje E van Haren, Jim van Os, Ruud van Winkel, Joyce van Baaren, Erwin Veermans, Ger Driessen, Truda Driesen, Karin Pos, Erna van ’t Hag, Jessica de Nijs, Atiqul Islam, Wendy Beuken, and Debora Op ’t Eijnde
References
- 1. Lee S.H., DeCandia T.R., Ripke S., Yang J., Schizophrenia Psychiatric Genome-Wide Association Study C., International Schizophrenia C., Molecular Genetics of Schizophrenia C., Sullivan P.F., Goddard M.E., Keller M.C.. et al. (2012) Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet., 44, 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ripke S., O'Dushlaine C., Chambert K., Moran J.L., Kahler A.K., Akterin S., Bergen S.E., Collins A.L., Crowley J.J., Fromer M.. et al. (2013) Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet., 45, 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schizophrenia Working Group of the Psychiatric Genomics, C. (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature, 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Loohuis L.M.O., Vorstman J.A.S., Ori A.P., Staats K.A., Wang T., Richards A.L., Leonenko G., Walters J.T., DeYoung J., Cantor R.M.. et al. (2015) Genome-wide burden of deleterious coding variants increased in schizophrenia. Nat. Commun., 6, 7501.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ambalavanan A., Girard S.L., Ahn K., Zhou S., Dionne-Laporte A., Spiegelman D., Bourassa C.V., Gauthier J., Hamdan F.F., Xiong L.. et al. (2016) De novo variants in sporadic cases of childhood onset schizophrenia. Eur. J. Hum. Genet., 24, 944–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Girard S.L., Gauthier J., Noreau A., Xiong L., Zhou S., Jouan L., Dionne-Laporte A., Spiegelman D., Henrion E., Diallo O.. et al. (2011) Increased exonic de novo mutation rate in individuals with schizophrenia. Nat. Genet., 43, 860–863. [DOI] [PubMed] [Google Scholar]
- 7. Purcell S.M., Moran J.L., Fromer M., Ruderfer D., Solovieff N., Roussos P., O'Dushlaine C., Chambert K., Bergen S.E., Kahler A.. et al. (2014) A polygenic burden of rare disruptive mutations in schizophrenia. Nature, 506, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu B., Roos J.L., Levy S., van Rensburg E.J., Gogos J.A., Karayiorgou M. (2008) Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet., 40, 880–885. [DOI] [PubMed] [Google Scholar]
- 9. Walsh T., McClellan J.M., McCarthy S.E., Addington A.M., Pierce S.B., Cooper G.M., Nord A.S., Kusenda M., Malhotra D., Bhandari A.. et al. (2008) Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science, 320, 539–543. [DOI] [PubMed] [Google Scholar]
- 10. International Schizophrenia, C. (2008) Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature, 455, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buizer-Voskamp J.E., Muntjewerff J.W., Genetic R., Outcome in Psychosis Consortium M., Strengman E., Sabatti C., Stefansson H., Vorstman J.A., Ophoff R.A. (2011) Genome-wide analysis shows increased frequency of copy number variation deletions in Dutch schizophrenia patients. Biol. Psychiatry, 70, 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Szatkiewicz J.P., Neale B.M., O'Dushlaine C., Fromer M., Goldstein J.I., Moran J.L., Chambert K., Kahler A., Magnusson P.K., Hultman C.M.. et al. (2013) Detecting large copy number variants using exome genotyping arrays in a large Swedish schizophrenia sample. Mol. Psychiatry, 18, 1178–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kirov G., Grozeva D., Norton N., Ivanov D., Mantripragada K.K., Holmans P., International Schizophrenia C., Wellcome Trust Case Control C., Craddock N., Owen M.J.. et al. (2009) Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum. Mol. Genet., 18, 1497–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rees E., Walters J.T., Georgieva L., Isles A.R., Chambert K.D., Richards A.L., Mahoney-Davies G., Legge S.E., Moran J.L., McCarroll S.A.. et al. (2014) Analysis of copy number variations at 15 schizophrenia-associated loci. Br. J. Psychiatry, 204, 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Hsi-Yang Fritz M.. et al. (2015) An integrated map of structural variation in 2,504 human genomes. Nature, 526, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sudmant P.H., Mallick S., Nelson B.J., Hormozdiari F., Krumm N., Huddleston J., Coe B.P., Baker C., Nordenfelt S., Bamshad M.. et al. (2015) Global diversity, population stratification, and selection of human copy-number variation. Science, 349, aab3761.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Malhotra D., Sebat J. (2012) CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell, 148, 1223–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kloosterman W.P., Francioli L.C., Hormozdiari F., Marschall T., Hehir-Kwa J.Y., Abdellaoui A., Lameijer E.W., Moed M.H., Koval V., Renkens I.. et al. (2015) Characteristics of de novo structural changes in the human genome. Genome Res., 25, 792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Acuna-Hidalgo R., Veltman J.A., Hoischen A. (2016) New insights into the generation and role of de novo mutations in health and disease. Genome Biol., 17, 241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vorstman J.A., van Daalen E., Jalali G.R., Schmidt E.R., Pasterkamp R.J., de Jonge M., Hennekam E.A., Janson E., Staal W.G., van der Zwaag B.. et al. (2011) A double hit implicates DIAPH3 as an autism risk gene. Mol. Psychiatry, 16, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siu W.K., Lam C.W., Gao W.W., Vincent Tang H.M., Jin D.Y., Mak C.M. (2016) Unmasking a novel disease gene NEO1 associated with autism spectrum disorders by a hemizygous deletion on chromosome 15 and a functional polymorphism. Behav. Brain Res., 300, 135–142. [DOI] [PubMed] [Google Scholar]
- 22. Bacchelli E., Ceroni F., Pinto D., Lomartire S., Giannandrea M., D'Adamo P., Bonora E., Parchi P., Tancredi R., Battaglia A.. et al. (2014) A CTNNA3 compound heterozygous deletion implicates a role for alphaT-catenin in susceptibility to autism spectrum disorder. J. Neurodev. Disord., 6, 17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Knight H.M., Pickard B.S., Maclean A., Malloy M.P., Soares D.C., McRae A.F., Condie A., White A., Hawkins W., McGhee K.. et al. (2009) A cytogenetic abnormality and rare coding variants identify ABCA13 as a candidate gene in schizophrenia, bipolar disorder, and depression. Am. J. Hum. Genet., 85, 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lim E.T., Raychaudhuri S., Sanders S.J., Stevens C., Sabo A., MacArthur D.G., Neale B.M., Kirby A., Ruderfer D.M., Fromer M.. et al. (2013) Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron, 77, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruderfer D.M., Lim E.T., Genovese G., Moran J.L., Hultman C.M., Sullivan P.F., McCarroll S.A., Holmans P., Sklar P., Purcell S.M. (2015) No evidence for rare recessive and compound heterozygous disruptive variants in schizophrenia. Eur. J. Hum. Genet., 23, 555–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsuang D.W., Millard S.P., Ely B., Chi P., Wang K., Raskind W.H., Kim S., Brkanac Z., Yu C.E. (2010) The effect of algorithms on copy number variant detection. PLoS One, 5, e14456.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tansey K.E., Rees E., Linden D.E., Ripke S., Chambert K.D., Moran J.L., McCarroll S.A., Holmans P., Kirov G., Walters J.. et al. (2016) Common alleles contribute to schizophrenia in CNV carriers. Mol. Psychiatry, 21, 1085–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bassett A.S., Scherer S.W., Brzustowicz L.M. (2010) Copy number variations in schizophrenia: critical review and new perspectives on concepts of genetics and disease. Am. J. Psychiatry, 167, 899–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B.. et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. De Rosa M., Fasano C., Panariello L., Scarano M.I., Belli G., Iannelli A., Ciciliano F., Izzo P. (2000) Evidence for a recessive inheritance of Turcot's syndrome caused by compound heterozygous mutations within the PMS2 gene. Oncogene, 19, 1719–1723. [DOI] [PubMed] [Google Scholar]
- 31. Dequeker E., Stuhrmann M., Morris M.A., Casals T., Castellani C., Claustres M., Cuppens H., des Georges M., Ferec C., Macek M.. et al. (2009) Best practice guidelines for molecular genetic diagnosis of cystic fibrosis and CFTR-related disorders–updated European recommendations. Eur. J. Hum. Genet., 17, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Knierim E., Hirata H., Wolf N.I., Morales-Gonzalez S., Schottmann G., Tanaka Y., Rudnik-Schoneborn S., Orgeur M., Zerres K., Vogt S.. et al. (2016) Mutations in Subunits of the Activating Signal Cointegrator 1 Complex Are Associated with Prenatal Spinal Muscular Atrophy and Congenital Bone Fractures. Am. J. Hum. Genet., 98, 473–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stefansson H., Rujescu D., Cichon S., Pietilainen O.P., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J.E.. et al. (2008) Large recurrent microdeletions associated with schizophrenia. Nature, 455, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Colella S., Yau C., Taylor J.M., Mirza G., Butler H., Clouston P., Bassett A.S., Seller A., Holmes C.C., Ragoussis J. (2007) QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res., 35, 2013–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F., Hakonarson H., Bucan M. (2007) PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res., 17, 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winchester L., Yau C., Ragoussis J. (2009) Comparing CNV detection methods for SNP arrays. Brief Funct. Genomic Proteomic, 8, 353–366. [DOI] [PubMed] [Google Scholar]
- 37. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J.. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gonzalez-Perez A., Lopez-Bigas N. (2011) Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet., 88, 440–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.