Abstract
Single nucleotide polymorphism (SNP) rs738409 C>G in the patatin‐like phospholipase domain containing 3 (PNPLA3) gene results in an amino acid exchange from isoleucin to methionine at position I148M of PNPLA3. The expression of this loss‐of‐function mutation leads to impaired hepatocellular triglyceride hydrolysis and is associated with the development of liver steatosis, fibrosis, and hepatocellular carcinoma. In contrast to these well‐established associations, the relationship of the PNPLA3 rs738409 variant with other metabolic traits is incompletely understood. We therefore assessed the association of the PNPLA3 rs738409 genotype with relevant metabolic traits in a prospective study of patients at high risk for cardiovascular events, i.e., patients undergoing coronary angiography. In a total of 270 patients, known associations of the PNPLA3 rs738409 GG genotype with nonalcoholic steatohepatitis and liver fibrosis were confirmed. In addition, we found an association of the PNPLA3 rs738409 G allele with the presence of diabetes (22% versus 28% versus 58% for CC versus CG versus GG genotype, respectively; P = 0.02). In contrast to its association with nonalcoholic fatty liver disease, liver fibrosis, and diabetes, the minor G allele of PNPLA3 rs738409 was inversely associated with total serum cholesterol and low‐density lipoprotein serum levels (P = 0.003 and P = 0.02, respectively). Finally, there was a trend toward an inverse association between the presence of the PNPLA3 rs738409 G allele and significant coronary heart disease. Comparable trends were observed for the transmembrane 6 superfamily member 2 (TM6SF2) 167 K variant, but the sample size was too small to evaluate this rarer variant. Conclusion: The PNPLA3 rs738409 G allele is associated with liver disease but also with a relatively benign cardiovascular risk profile. (Hepatology Communications 2018;2:798‐806)
Abbreviations
- CAD
coronary artery disease
- CAP
controlled attenuation parameter
- CHD
coronary heart disease
- HDL
high‐density lipoprotein
- LDL
low‐density lipoprotein
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- PNPLA3
patatin‐like phospholipase domain containing 3
- SNP
single nucleotide polymorphism
- TE
transient elastography
- TM6SF2
transmembrane 6 superfamily member 2
- VLDL
very low‐density lipoprotein
The patatin‐like phospholipase domain‐containing 3 (PNPLA3) gene encodes the PNPLA3 protein (also referred to as adiponutrin), which is highly abundant in the liver and in adipocytes.1 PNPLA3 acts as a hydrolase in the metabolism of triglycerides and retinyl esters, suggesting a role for PNPLA3 in energy usage, lipid metabolism, and retinol synthesis.2 An involvement in energy metabolism is further supported by the finding that glucose and insulin are capable of inducing PNPLA3 gene expression through transcription factors carbohydrate‐response element‐binding protein and sterol regulatory element binding protein 1c.3, 4
A single nucleotide polymorphism (SNP) in the PNPLA3 gene, i.e., rs738409 C>G, results in an amino acid exchange from isoleucin to methionine at position I148M of PNPLA3.2 This common variant represents a missense mutation associated with a loss‐of‐function of the hydrolase activity of the enzyme, promoting triglyceride accumulation in hepatocytes, impaired retinol synthesis in hepatic stellate cells, formation of larger lipid droplets, and reduced secretion of triglyceride‐rich very low‐density lipoprotein (VLDL) both in vitro and in vivo (reviewed in Trepo et al.5). A susceptibility to nonalcoholic fatty liver disease (NAFLD) through reduced VLDL secretion has also been reported for the E167K missense mutation in the transmembrane 6 superfamily member 2 (TM6SF2) protein encoded by the TM6SF2 gene, even though the exact protein function is not completely understood.6
There is clear evidence from a large number of genetic association studies that the minor allele of the PNPLA3 rs738409 C>G variant is strongly associated with NAFLD, with hepatic steatosis and fibrosis in alcoholic and chronic viral hepatitis, as well as with the risk of hepatocellular carcinoma development.7, 8, 9, 10, 11 Although genetic association studies cannot proof causality, the above‐mentioned effects of the PNPLA3 I148M variant on lipid metabolism suggest a causal role for this variant in associated liver traits.
Although NAFLD can be considered a hepatic manifestation of the metabolic syndrome, the strong association between PNPLA3 rs738409 variants with NAFLD is contrasted by inconsistent results regarding the impact of this variant on other determinants of the metabolic syndrome, such as serum lipid profiles or insulin resistance.12, 13, 14, 15 In the present study, we therefore aimed to further characterize the relationship of PNPLA3 rs738409 variants with metabolic traits in a cohort of individuals who are at high risk of cardiovascular events, i.e., patients undergoing coronary angiography.
Patients and Methods
PATIENTS
Patients were selected from a prospective monocenter study (https://clinicaltrials.gov/ct2/show/NCT01638832) that included 576 consecutive patients undergoing elective coronary angiography at the cardiology department of the University Hospital Frankfurt between January 2012 and October 2014. Patients were excluded from the primary study if they had an acute myocardial infarction, ascites, were pregnant or breastfeeding, younger than 18 years old, or unable to provide full legal capacity. The primary goal of this study was to characterize the epidemiological association between coronary heart disease (CHD) and NAFLD; details of the primary study are described in Friedrich‐Rust et al.16 Clinical and demographic variables as well as laboratory test results were obtained directly before coronary angiography.
In the present subanalysis, all patients from the primary study were included if they had given written informed consent for genetic testing and if DNA samples for genetic testing were available. The study protocol was approved by the ethical committee of the University of Frankfurt.
Elective coronary angiography was performed by an experienced cardiologist. Indications for coronary angiography were suspected first‐time manifestation of coronary artery disease (CAD) or suspected progression of known CAD in >88% of patients. Clinically significant CAD was defined as presence of relevant coronary stenosis (i.e., ≥75% stenosis or the vessel diameter).
Liver fibrosis was assessed by transient elastography (TE) using a FibroScan 502 touch device (Echosens, Paris, France) with the FibroScan M‐Probe (3.5 MHz, shear wave frequency 50 Hz, depth of measurement 25‐65 mm). To detect liver fibrosis, a TE cutoff value of 7.9 kPa was chosen. In addition, serologic testing for liver fibrosis was performed using the FibroTest.17 Controlled attenuation parameter (CAP) was used to quantify hepatic steatosis. A CAP cutoff value of 234 dB/m was used to define the presence of steatosis.18 Nonalcoholic steatohepatitis (NASH) was suspected in case of a CAP value of ≥324 dB/m in combination with a TE value of ≥7.9 kPa in the absence of congestive or right‐sided heart failure and in the absence of alcohol consumption >10 or 20 g/day in female and male study participants, respectively. Diabetes was diagnosed according to current German and European clinical practice guidelines by the responsible physician.
PNPLA3 AND TM6SF2 GENOTYPING
Genomic DNA was extracted from peripheral blood using proteinase K digestion of white blood cells and isopropanol precipitation. DNA concentration and quality were assessed by NanoDrop 2000 (PeqLab). Genotyping for the PNPLA3 rs738409 and TM6SF2 rs58542926 SNPs was performed using TaqMan SNP Genotyping Assays (C_7241_10 and C_89463510_10, respectively, Cat. #4351379; Thermo Fisher Scientific) on a StepOnePlus Real‐Time Polymerase Chain Reaction System (Applied Biosciences) according to the manufacturer's recommendations.
STATISTICAL ANALYSES
Associations of the PNPLA3 rs738409 or TM6SF2 rs58542926 genotype with continuous or dichotomic variables were assessed in linear and logistic regression models, respectively. After univariate analyses, multivariate analyses were performed for significant associations. Multivariate models were obtained by backward selection using a P value >0.15 for removal from the model. SNPs were analyzed using an additive model of inheritance (none, one, or two copies of the minor allele were coded 0, 1, and 2, respectively, assuming greater effect with increased copy number of the minor allele), unless otherwise specified. Group differences were assessed by means of χ2 contingency tables or Wilcoxon‐Mann‐Whitney U tests, as appropriate.
Results
PATIENT CHARACTERISTICS, PNPLA3 rs738409 ALLELE, AND GENOTYPE FREQUENCIES
The inclusion criteria for the present study were met by 270 patients. Of those, 262 and 8 individuals were of Caucasian and Asian ethnicity, respectively. Epidemiological and clinical characteristics of patients based on the PNPLA3 rs738409 genotype are summarized in Table 1. Allele frequencies of the PNPLA3 rs738409 major C and minor G alleles were 76% and 24%, respectively. The distribution of homozygous and heterozygous carriers corresponded to the expectations from Hardy‐Weinberg equilibrium (P > 0.25). Frequencies of rs738409 CC, CG, and GG genotypes were 57%, 39%, and 4%, respectively. We also genotyped the TM6SF2 E167K variant, which, however, had a much lower frequency of the minor T allele that did not allow a detailed analysis of associations with metabolic traits in our relatively low sample size (frequency of rs58542926 CC, CT, and TT genotypes, 92%, 8%, and 0%, respectively; frequency of C and T alleles, 96% and 4%, respectively; Hardy‐Weinberg equilibrium P > 0.25).
Table 1.
Baseline Characteristics of Included Patients
|
Genotype CC (n = 154) |
Genotype CG (n = 104) |
Genotype GG (n = 12) |
|
|---|---|---|---|
| Male sex, n (%) | 124 (81) | 83 (80) | 10 (83) |
| Age [years], mean (SD) | 63 (12) | 64 (11) | 66 (10) |
| BMI [kg/m2], mean (SD) | 27.51 (4.52) | 27.47 (4.7) | 29.23 (5.68) |
| Waist circumference [cm], mean (SD) | 99.77 (11.74) | 100.9 (12.64) | 107.5 (13.08) |
| Nicotine consumption [py], mean (SD) | 25 (30) | 31 (33) | 22 (23) |
| EtOH consumption [g/day], mean (SD) | 6.3 (9.02) | 6.98 (10.22) | 6.09 (8.26) |
| Significant CHD [presence], n (%) | 122 (79) | 82 (79) | 8 (67) |
| Type I diabetes mellitus, n (%) | 2 (1) | 3 (3) | 0 (0) |
| Type II diabetes mellitus, n (%) | 31 (20) | 26 (25) | 7 (58) |
| Statins, n (%) | 98 (64) | 74 (71) | 10 (83) |
| Heart rate, mean (SD) | 70 (15) | 72 (16) | 69 (17) |
| Systolic blood pressure, mean (SD) | 127 (21) | 126 (23) | 123 (20) |
| Diastolic blood pressure, mean (SD) | 75 (11) | 75 (11) | 71 (13) |
| Triglycerides [mg/dL], mean (SD) | 144.1 (91.49) | 154.29 (106.84) | 108.36 (23.29) |
| Total cholesterol [mg/dL], mean (SD) | 183.99 (50.61) | 171.68 (43.96) | 140.45 (57.19) |
| LDL [mg/dL], mean (SD) | 102.32 (41.7) | 91.76 (34.87) | 88.91 (38.11) |
| HDL [mg/dL], mean (SD) | 51.81 (19.77) | 49.02 (19.65) | 45.89 (15.79) |
| Total bilirubin [mg/dL], mean (SD) | 0.58 (0.4) | 0.60 (0.38) | 0.75 (0.39) |
| GGT [U/L], mean (SD) | 70 (130) | 61 (75) | 49 (46) |
| AST [U/L], mean (SD) | 32 (31) | 29 (12) | 37 (24) |
| ALT [U/L], mean (SD) | 30 (21) | 31 (26) | 36 (23) |
| Hemoglobin [g/dL], mean (SD) | 13.35 (1.82) | 13.4 (1.75) | 13.03 (2.02) |
| Platelets [/nL], mean (SD) | 230 (83) | 222 (66) | 211 (41) |
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; EtOH, ethanol; GGT, gamma‐glutamyl transferase; py, pack years.
PNPLA3 rs738409 GENOTYPE AND PRESENCE OF NAFLD AND LIVER FIBROSIS
The total number of patients with suspected NASH was relatively low in the present cohort (39 out of 258 patients with available CAP values, 15%). Therefore, a dominant model of inheritance was calculated in addition to the classical additive model for the assessment of associations between the PNPLA3 rs738409 genotype and presence of NASH or liver fibrosis. Frequencies of NASH were 15%, 14%, and 33% in patients with rs738409 CC, CG, and GG genotypes, respectively (P = 0.3 [additive] and P = 0.07 [dominant]; Fig. 1). Mean CAP values were 265, 269, and 281 dB/m in patients with rs738409 CC, CG, and GG genotypes, respectively (not significant). Mean TE values were significantly higher in patients with the PNPLA3 rs738409 GG genotype compared to patients with the PNPLA3 rs738409 CC and CG genotypes (8.5 versus 7.5 and 7.0 kPA, P = 0.2 [additive] and P = 0.03 [dominant]; Fig. 1). In addition, the mean FibroTest category was higher in patients with the PNPLA3 rs738409 GG genotype compared to patients with the PNPLA3 rs738409 CC and CG genotypes (4.4 versus 2.9 versus 2.8; P = 0.2 [additive] and P = 0.04 [dominant]; Fig. 1).
Figure 1.
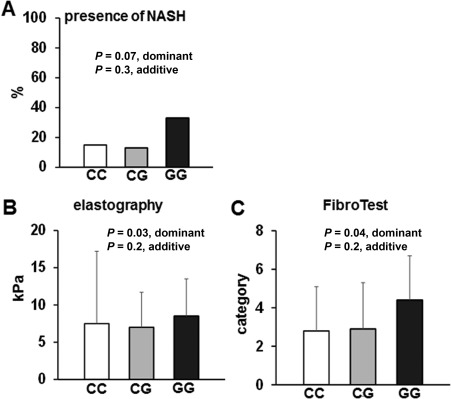
Association of the PNPLA3 rs738409 genotype with NAFLD and liver fibrosis. (A) Prevalence of NASH based on the PNPLA3 rs738409 genotype. (B,C) Association between the PNPLA3 rs738409 genotype and degree of liver fibrosis, assessed by (B) transient elastography (FibroScan) and (C) FibroTest. Mean values and standard deviations are shown for B and C.
PNPLA3 rs738409 GENOTYPE AND METABOLIC TRAITS
The presence of diabetes increased significantly in individuals carrying the PNPLA3 rs738409 minor allele (22% versus 28% versus 58% for CC versus CG versus GG genotypes, P = 0.02; Fig. 2). In addition, there was a progressive increase of mean waist circumference in individuals with the PNPLA3 rs738409 CC versus CG versus GG genotype, but this association was not significant (99 cm versus 101 cm versus 108 cm, P = 0.07).
Figure 2.
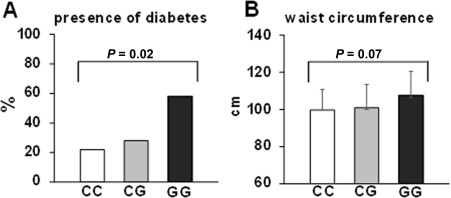
Association of the PNPLA3 rs738409 genotype and metabolic traits. (A) The presence of diabetes; (B) waist circumference. Mean values and standard deviations are shown for B.
In contrast to its association with NAFLD, liver fibrosis, and diabetes, the minor G allele of PNPLA3 rs738409 appeared to have an opposing effect on serum cholesterol levels. A progressively significant decrease in serum levels of total cholesterol as well as of LDL and high‐density lipoprotein (HDL) was observed in patients with rs738409 CC versus CG versus GG genotypes (Fig. 3). The association between the PNPLA3 rs738409 genotype and LDL serum concentration remained significant in multivariate analysis (P = 0.04), together with age (P = 0.026), male sex (P = 0.023), and CAP value (P = 0.02) (Table 2). Furthermore, the association between the PNPLA3 rs738409 genotype and total cholesterol and LDL serum concentration remained significant in the subgroup of patients who were not treated with statins at the time of study inclusion (total cholesterol beta = –33.89, SD beta = 10.79, P = 0.0025; LDL beta = –18.54, SD beta = 8.94, P = 0.04).
Figure 3.
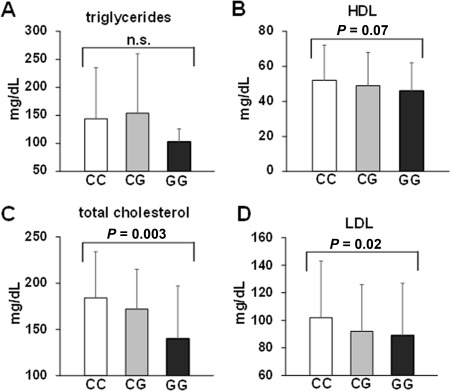
Association of the PNPLA3 rs738409 genotype with serum lipid concentrations. Serum concentrations of (A) triglycerides, (B) HDL, (C) total cholesterol, and (D) LDL are shown based on the PNPLA3 rs738409 genotype. Mean values and standard deviations are shown for A through D.
Table 2.
Linear Regression Analyses of LDL Serum Concentration
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| beta (SD beta) | P | beta (SD beta) | P | |
| Age [years] | –0.44 (0.21) | 0.036 | –0.47 (0.21) | 0.026 |
| Male sex | 12.91 (6.41) | 0.045 | 14.54(6.36) | 0.023 |
| BMI [kg/m2] | 0.59 (0.67) | 0.385 | ||
| GGT [U/L] | 0.05 (0.02) | 0.037 | 0.04 (0.02) | 0.088 |
| FibroScan [kPa] | –0.05 (0.32) | 0.12 | ||
| CAP [dB/m] | 0.07 (0.046) | 0.13 | 0.09 (0.38) | 0.02 |
| PNPLA3 rs738409 G allele | –10.33 (5.00) | 0.04 | –10.00 (5.00) | 0.04 |
Abbreviations: BMI, body mass index; GGT, gamma‐glutamyl transferase.
Collectively, the PNPLA3 G allele appears to be associated with traits promotional (NAFLD, diabetes, HDL concentration) and protective (lower LDL concentration) for the occurrence of CHD. We therefore performed logistic regression analysis for the presence of clinically relevant CHD, one of the most crucial sequelae of the metabolic syndrome. In univariate analysis, there was a trend for an inverse association between the presence of the PNPLA3 rs738409 G allele and the risk of significant CHD (P = 0.10). Yet, the significance level increased in the multivariate model after correction for age, sex, use of statins, and HDL serum levels (odds ratio, 0.21; 95% confidence interval, 0.05‐0.88; P = 0.03, for the presence of significant CHD based on the presence of the PNPLA3 G versus C allele) (Table 3).
Table 3.
Logistic Regression Analyses of CHD
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | |
| Age [years] | 1.02 (0.99‐1.05) | 0.14 | 1.03 (1.00‐1.06) | 0.09 |
| Male sex | 0.31 (0.15‐0.66) | 0.002 | 0.41 (0.18‐0.98) | 0.04 |
| Waist circumference [cm] | 1.03 (1.00‐1.06) | 0.08 | ||
| Nicotine consumption [py] | 1.01 (1.00‐1.02) | 0.2 | ||
| Presence of diabetes mellitus | 1.22 (0.79‐1.87) | 0.4 | ||
| Use of statins | 2.96 (1.46‐6.00) | 0.003 | 2.85 (1.34‐6.06) | 0.006 |
| LDL [mg/dL] | 1.00 (0.99‐1.01) | 0.4 | ||
| HDL [mg/dL] | 0.98 (0.96‐0.99) | 0.006 | 0.98 (0.96‐1.00) | 0.04 |
| FibroScan [kPa] | 1.03 (0.97‐1.09) | 0.4 | ||
| CAP [dB/m] | 1.00 (0.99‐1.01) | 0.2 | ||
| PNPLA3 rs738409 G allele | 0.34 (0.09‐1.23) | 0.10 | 0.21 (0.05‐0.88) | 0.03 |
Abbreviations: CI, confidence interval; OR, odds ratio.
Numerically comparable but statistically not significant trends were observed for the TM6SF2 167 K variant (Table 4), but the sample size of our study was too small to firmly assess this rarer variant.
Table 4.
Associations Between TM6SF2 E167K Variantsa and Metabolic Traits
|
Homozygous CC (n = 247) |
Heterozygous CT (n = 21) |
P value | |
|---|---|---|---|
| Waist circumference [cm], (SD) | 100.16 (12.18) | 104.95 (12.48) | 0.12 |
| Triglycerides [mg/dL], (SD) | 148.49 (98.24) | 130.00 (68.50) | 0.52 |
| Total cholesterol [mg/dL], (SD) | 178.06 (50.10) | 165.52 (38.96) | 0.33 |
| LDL [mg/dL], (SD) | 98.22 (39.55) | 88.42 (36.37) | 0.38 |
| HDL [mg/dL], (SD) | 50.33 (19.97) | 49.89 (11.67) | 0.62 |
| Diabetes, n (%) | 57 (24) | 7 (33) | 0.34 |
TM6SF2 E167K genotyping failed in 2 patients.
Discussion
The present study reveals divergent associations of the PNPLA3 rs738409 variant with metabolic traits in a patient cohort at particular high risk for cardiovascular events. As expected, the PNPLA3 rs738409 minor G allele is associated with liver disease, i.e., with NAFLD and higher degrees of liver fibrosis in this special patient population. Moreover, we could show an association between the PNPLA3 rs738409 G allele and the presence of diabetes. However, the PNPLA3 rs738409 G allele was associated with lower levels of LDL, an important determinant of overall cardiovascular risk, and there was a trend toward a protective association between the PNPLA3 rs738409 G allele and lower rates of significant CHD.
The association between the PNPLA3 rs738409 genotype and hepatic steatosis/liver fibrosis is well established for various types of liver disease, including metabolic, alcoholic, and viral hepatitis.5 It is therefore not surprising that this association was observed in the present cohort of individuals at high risk for cardiovascular events as well. Interestingly, results from animal models have suggested an interaction between the PNPLA3 I148M variant and environmental factors in the progression of liver steatosis. Neither PNPLA3 –/– knockout mice nor mice in which the wild‐type PNPLA3 variant was overexpressed in the liver had a relevant change of liver fat content compared to controls.19, 20 However, knockin mice expressing the PNPLA3 I148M protein showed increased hepatic fat accumulation compared to wild‐type mice if they were at high metabolic risk, i.e., if they were fed with a high‐sucrose diet, while no difference was observed in mice on normal chow.21 In this regard, the main inclusion criteria of our study, i.e., a given indication for coronary angiography, might be a significant strength to detect associations between PNPLA3 variants and metabolic traits as it has resulted in the selection of patients who are at high risk for cardiovascular events.
From a clinical point of view, our data indicate that the PNPLA3 rs738409 G allele increases the risk of liver disease and insulin resistance but, due to an association with lower LDL serum levels, may be associated with a relatively benign cardiovascular outcome of these traits. Indeed, there is growing evidence that the PNPLA3 variant is associated with metabolic traits, such as insulin resistance and serum lipid concentrations.12, 13, 14, 15, 22, 23 In particular, the results of our study are in line with a recently published, large, exome‐wide genome‐wide association study showing that the PNPLA3 rs738409 G allele is associated with type 2 diabetes but inversely with LDL, HDL, and triglyceride serum levels.24 Additional studies have confirmed an association between the PNPLA3 rs738409 G allele and diabetes but not with CAD or arteriosclerosis in the absence of diabetes.25, 26 Importantly, a moderate inverse association between the PNPLA3 rs738409 G allele and CAD was also shown in a large meta‐analysis of genome‐wide association study data for CAD, a finding that is in line with our results.27 Finally, a large Chinese study has shown an inverse association between serum aminotransferase levels and CHD, although in that study no direct association between the PNPLA3 rs738409 G allele and serum lipid parameters was observed.28 Overall, the results of these previous studies and of our study suggest that the PNPLA3 rs738409 G allele is associated with hepatic fat accumulation, liver disease, and diabetes but, due to the association with lower cholesterol levels, not with a higher net risk for cardiovascular disease. This hypothesis is further supported by in vitro studies using recombinant PNPLA3‐I148M proteins, which have shown that this variant decreases hepatocellular efflux of fatty acids and VLDL secretion while increasing hepatocellular triglyceride content.2, 29, 30 Comparable findings have been reported for the rarer TM6SF2 E167K variant,31 which, however, could not be thoroughly assessed in our study due to the limited sample size. Based on these findings, we also consider the sample size of our study as being too small to thoroughly assess the membrane‐bound O‐acyltransferase domain containing 7‐transmembrane channel‐like 4 (MBOAT7‐TMC4) rs641738 variant, a third genetic variant that has recently been identified as a genetic risk factor for NALFD.32
Our study has several limitations. First, the overall patient number and in particular the number of patients with liver disease were relatively small because that was not a selection criteria for this study. Furthermore, a significant number of patients of our primary study gave no permission for genetic testing, further limiting the sample size of the present analyses. Moreover, no liver biopsy data were available, and the diagnosis of NAFLD, NASH, and liver fibrosis was based on noninvasive test results. Yet, the association between PNPLA3 rs738409 and liver disease is well established and the primary goal of this study was the confirmation of associations between PNPLA3 rs738409 variants and other metabolic traits, for which the study appears adequately powered.
In conclusion, we show that, in addition to known associations with liver steatosis and fibrosis, the PNPLA3 rs738409 minor G allele is associated with the presence of diabetes in a high‐risk population for cardiovascular events but also with lower LDL serum levels and an overall rather neutral risk for significant CAD.
Potential conflict of interest: Dr. Zeuzem consults, advises, and is on the speakers' bureau for AbbVie, Gilead, and Merck. The other authors have nothing to report.
Supported by the Deutsche Forschungsgemeinschaft (LA 2806/2‐1 and LA 2806/5‐1 to C.M.L.).
REFERENCES
- 1. Winberg ME, Motlagh MK, Stenkula KG, Holm C, Jones HA. Adiponutrin: a multimeric plasma protein. Biochem Biophys Res Commun 2014;446:1114‐1119. [DOI] [PubMed] [Google Scholar]
- 2. Huang Y, Cohen JC, Hobbs HH. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem 2011;286:37085‐37093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dubuquoy C, Robichon C, Lasnier F, Langlois C, Dugail I, Foufelle F, et al. Distinct regulation of adiponutrin/PNPLA3 gene expression by the transcription factors ChREBP and SREBP1c in mouse and human hepatocytes. J Hepatol 2011;55:145‐153. [DOI] [PubMed] [Google Scholar]
- 4. Rae‐Whitcombe SM, Kennedy D, Voyles M, Thompson MP. Regulation of the promoter region of the human adiponutrin/PNPLA3 gene by glucose and insulin. Biochem Biophys Res Commun 2010;402:767‐772. [DOI] [PubMed] [Google Scholar]
- 5. Trepo E, Romeo S, Zucman‐Rossi J, Nahon P. PNPLA3 gene in liver diseases. J Hepatol 2016;65:399‐412. [DOI] [PubMed] [Google Scholar]
- 6. Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjaerg‐Hansen A, et al. Exome‐wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2014;46:352‐356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008;40:1461‐1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Valenti L, Al‐Serri A, Daly AK, Galmozzi E, Rametta R, Dongiovanni P, et al. Homozygosity for the patatin‐like phospholipase‐3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology 2010;51:1209‐1217. [DOI] [PubMed] [Google Scholar]
- 9. Tian C, Stokowski RP, Kershenobich D, Ballinger DG, Hinds DA. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet 2010;42:21‐23. [DOI] [PubMed] [Google Scholar]
- 10. Buch S, Stickel F, Trepo E, Way M, Herrmann A, Nischalke HD, et al. A genome‐wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol‐related cirrhosis. Nat Genet 2015;47:1443‐1448. [DOI] [PubMed] [Google Scholar]
- 11. Trepo E, Nahon P, Bontempi G, Valenti L, Falleti E, Nischalke HD, et al. Association between the PNPLA3 (rs738409 C>G) variant and hepatocellular carcinoma: evidence from a meta‐analysis of individual participant data. Hepatology 2014;59:2170‐2177. [DOI] [PubMed] [Google Scholar]
- 12. Palmer CN, Maglio C, Pirazzi C, Burza MA, Adiels M, Burch L, et al. Paradoxical lower serum triglyceride levels and higher type 2 diabetes mellitus susceptibility in obese individuals with the PNPLA3 148M variant. PLoS One 2012;7:e39362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Speliotes EK, Butler JL, Palmer CD, Voight BF, Hirschhorn JN; GIANT Consortium ; MIGen Consortium ; NASH CRN . PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology 2010;52:904‐912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Speliotes EK, Yerges‐Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, et al.; NASH CRN ; GIANT Consortium ; MAGIC Investigators ; GOLD Consortium . Genome‐wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet 2011;7:e1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang L, You W, Zhang H, Peng R, Zhu Q, Yao A, et al. PNPLA3 polymorphisms (rs738409) and non‐alcoholic fatty liver disease risk and related phenotypes: a meta‐analysis. J Gastroenterol Hepatol 2015;30:821‐829. [DOI] [PubMed] [Google Scholar]
- 16. Friedrich‐Rust M, Schoelzel M, Maier S, Seeger F, Rey J, Fichtlscherer S, et al. Severity of coronary artery disease is associated with non‐alcoholic fatty liver disease: a single‐blinded propsoective monocenter study. PLoS One 2017;12:e0186720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ratziu V, Massard J, Charlotte F, Messous D, Imbert‐Bismut F, Bonyhay L, et al.; LIDO Study Group ; CYTOL study group . Diagnostic value of biochemical markers (FibroTest‐FibroSURE) for the prediction of liver fibrosis in patients with non‐alcoholic fatty liver disease. BMC Gastroenterol 2006;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karlas T, Petroff D, Garnov N, Bohm S, Tenckhoff H, Wittekind C, et al. Non‐invasive assessment of hepatic steatosis in patients with NAFLD using controlled attenuation parameter and 1H‐MR spectroscopy. PLoS One 2014;9:e91987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He S, McPhaul C, Li JZ, Garuti R, Kinch L, Grishin NV, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem 2010;285:6706‐6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen W, Chang B, Li L, Chan L. Patatin‐like phospholipase domain‐containing 3/adiponutrin deficiency in mice is not associated with fatty liver disease. Hepatology 2010;52:1134‐1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smagris E, BasuRay S, Li J, Huang Y, Lai KM, Gromada J, et al. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 2015;61:108‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krarup NT, Grarup N, Banasik K, Friedrichsen M, Faerch K, Sandholt CH, et al. The PNPLA3 rs738409 G‐allele associates with reduced fasting serum triglyceride and serum cholesterol in Danes with impaired glucose regulation. PLoS One 2012;7:e40376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang CF, Dai CY, Yeh ML, Huang CI, Tai CM, Hsieh MH, et al. Association of diabetes and PNPLA3 genetic variants with disease severity of patients with chronic hepatitis C virus infection. J Hepatol 2015;62:512‐518. [DOI] [PubMed] [Google Scholar]
- 24. Liu DJ, Peloso GM, Yu H, Butterworth AS, Wang X, Mahajan A, et al. Exome‐wide association study of plasma lipids in >300,000 individuals. Nat Genet 2017;49:1758‐1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Posadas‐Sanchez R, Lopez‐Uribe AR, Posadas‐Romero C, Perez‐Hernandez N, Rodriguez‐Perez JM, Ocampo‐Arcos WA, et al. Association of the I148M/PNPLA3 (rs738409) polymorphism with premature coronary artery disease, fatty liver, and insulin resistance in type 2 diabetic patients and healthy controls. The GEA study. Immunobiology 2017;222:960‐966. [DOI] [PubMed] [Google Scholar]
- 26. Petta S, Valenti L, Marchesini G, Di Marco V, Licata A, Camma C, et al. PNPLA3 GG genotype and carotid atherosclerosis in patients with non‐alcoholic fatty liver disease. PLoS One 2013;8:e74089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simons N, Isaacs A, Koek GH, Kuc S, Schaper NC, Brouwers M. PNPLA3, TM6SF2, and MBOAT7 Genotypes and Coronary Artery Disease. Gastroenterology 2017;152:912‐913. [DOI] [PubMed] [Google Scholar]
- 28. Xu L, Jiang CQ, Lam TH, Zhang WS, Zhu F, Jin YL, et al. Mendelian randomization estimates of alanine aminotransferase with cardiovascular disease: Guangzhou Biobank Cohort study. Hum Mol Genet 2017;26:430‐437. [DOI] [PubMed] [Google Scholar]
- 29. Pirazzi C, Adiels M, Burza MA, Mancina RM, Levin M, Stahlman M, et al. Patatin‐like phospholipase domain‐containing 3 (PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. J Hepatol 2012;57:1276‐1282. [DOI] [PubMed] [Google Scholar]
- 30. Pingitore P, Pirazzi C, Mancina RM, Motta BM, Indiveri C, Pujia A, et al. Recombinant PNPLA3 protein shows triglyceride hydrolase activity and its I148M mutation results in loss of function. Biochim Biophys Acta 2014;1841:574‐580. [DOI] [PubMed] [Google Scholar]
- 31. Dongiovanni P, Petta S, Maglio C, Fracanzani AL, Pipitone R, Mozzi E, et al. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology 2015;61:506‐514. [DOI] [PubMed] [Google Scholar]
- 32. Mancina RM, Dongiovanni P, Petta S, Pingitore P, Meroni M, Rametta R, et al. The MBOAT7‐TMC4 Variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology 2016;150:1219‐1230.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]


