Abstract
Inter-organelle membrane contact sites (MCSs) serve as unique microenvironments for the sensing and exchange of cellular metabolites and lipids. Though poorly defined, ER-endolysosomal contact sites are quickly becoming recognized as centers for inter-organelle lipid exchange and metabolic decision-making. Here, we review the discovery and current state of knowledge of ER-endolysosomal MCSs with particular focus on the molecular players that establish and/or utilize these contact sites in metabolism. We also discuss associations of ER-endolysosomal MCS-associated proteins in human disease, as well as the therapeutic promise these contact sites hold in modulating cellular physiology.
Keywords: Membrane contact site (MCS), Inter-organelle, Endoplasmic reticulum, Lysosome, Endosome
10.1. Introduction: ER-Endolysosomal Membrane Contact Sites in Lipid Metabolism and Disease
No organelle is an island, and to maintain cellular homeostasis, organelles must constantly communicate their needs and exchange lipids and other metabolites. The mechanisms that govern this inter-organelle metabolic exchange have, until recently, been poorly defined. Early studies attributed most organelle-organelle lipid exchange through vesicular pathways. Indeed, many lipids produced by the endoplasmic reticulum (ER), the major anabolic organelle of lipid metabolism, can be trafficked along with proteins to other places within the cell such as the plasma membrane (PM) through the formation of coated vesicles (Novick et al. 1980). However, it has been established that even in the absence of ER-derived vesicle trafficking, the transfer of sterol between the ER and PM persists (Baumann et al. 2005).
How, then, does non-vesicular inter-organelle lipid exchange occur? Recent studies suggest that at least one pathway relies on lipid exchange at sites of close contact between different membrane-bound organelles, sites now established as specific membrane contact sites (MCSs) (reviewed in (Levine and Loewen 2006; Elbaz and Schuldiner 2011; Prinz 2014; Henne et al. 2015a; Phillips and Voeltz 2016)). Often observed but generally ignored, these organelle-organelle contact sites are quickly becoming recognized as hotspots for lipid metabolism and metabolic decision-making. Elegant studies in both budding yeast and mammalian cells have identified key proteins that act as molecular “tethers” connecting different organelles (Manford et al. 2012; Zhang et al. 2012; Giordano et al. 2013; Tavassoli et al. 2013). Perhaps the most characterized of these are contact sites formed between the ER and the PM. Numerous studies demonstrate that ER-PM contact sites serve as major regulators for the exchange of the PM-manufactured phospholipid PtdIns4P with ER-derived phosphatidylserine (PtdSer) (Stefan et al. 2011; Moser von Filseck et al. 2015). Other functions for ER-PM contact sites continue to be revealed, including roles in the regulation of cell signaling and Ca2+ signaling (Liou et al. 2007; Omnus et al. 2016).
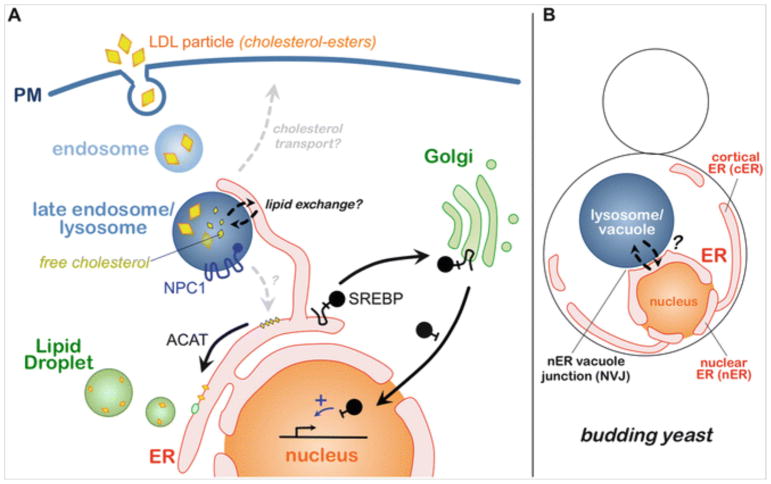
The PM is constantly internalized via endocytosis, thus initiating the endolysosomal pathway—the network of endosomes and lysosomes that constitutes the principle intracellular trafficking pathway for cell surface proteins and environmentally derived metabolites like cholesterol. In the human body, most cells rely on retrieving extracellular cholesterol through the internalization of LDL particles via their co-internalization with the LDL receptor in so-called receptor-mediated endocytosis (Goldstein 2007) (Fig. 10.1). Following their internalization, LDL particles are trafficked through endosomes to the lysosome, where hydrolases breakdown LDL cholesterol-esters (CE) and triacylglycerides (TAGs) into free cholesterol and fatty acids. Where these lipid metabolites go from here is much less clear. Approximately 80% of LD-derived cholesterol must, eventually, traffic back to the cell surface, and this process requires lysosomal proteins Npc1 and Npc2 (so-called Niemann-Pick proteins) that scavenge cholesterol from the lysosome lumen (Infante et al. 2008).
Fig. 10.1.
Trafficking pathways in human and yeast ER-endolysosomal systems. (a) Receptor-mediated endocytosis of LDL particles (containing sterol-esters) and their trafficking through the endosomal pathway to late endosomes/lysosomes, where cholesterol-esters within LDL particles are degraded into free cholesterol. Free cholesterol must then leave the lysosome lumen in an NPC1-dependent manner and traffic to the plasma membrane (PM) and/or the endoplasmic reticulum (ER). At the ER, the SREBP system senses local cholesterol levels. Low ER cholesterol induces the translocation and proteolytic cleavage of SREBP-2 at the Golgi, producing a soluble transcription factor which enters the nucleus. (b) The ER-endolysosomal system of budding yeast Saccharomyces cerevisiae. The ER is partitioned into distal cortical ER (cER) and nuclear ER (nER), which makes direct contact with the vacuole/lysosome via the nER vacuole junction (NVJ)
Following its removal from the lysosome, much of this free cholesterol must also enter the ER, where it is sensed by the sterol regulatory element-binding protein (SREBP) system that serves as the principle nutrient-sensing pathway for cellular sterols (Brown and Goldstein 1997). The SREBP pathway is principally composed of two integral membrane proteins, SREBP-1a and SREBP-2, which function both as sterol sensors and transcriptional regulators of cellular sterol metabolism. When ER-resident cholesterol drops below ~5 mole %, SREBP-2 is efficiently trafficked to the Golgi via COPII vesicles, where it is proteolytically cleaved into a soluble “mature” form by Golgi-resident proteases (Fig. 10.1). This liberates SREBP-2 from membranes, allowing it to translocate into the nucleus where it can initiate a transcriptional response aimed at upregulating sterol biogenesis (Brown and Goldstein 1997).
Despite our understanding of LDL particle internalization and endolysosomal trafficking, how cholesterol exits the lysosome and enters the ER remains largely unclear. However, recent studies implicate ER-lysosome MCSs in the non-vesicular transfer of cholesterol and other lipids between these two organelles, although the proteins and mechanisms that govern this remain poorly defined (Murley et al. 2015; Eden et al. 2016; Hariri et al. 2016). Addressing how cholesterol moves between the ER and endolysosomal system is of major biomedical importance, as numerous genetic and acquired diseases are associated with the pathological accumulation of cholesterol within the lysosome. Most notable is Niemann-Pick type C, a homozygous recessive pediatric neurological disease (one of more than 50 so-called lysosomal storage diseases) associated with loss-of-function alleles in genes NPC1 and NPC2. Both encode lysosomal proteins Npc1 and Npc2 that are essential in the initial steps of cholesterol efflux from the lysosomal lumen (Ko et al. 2003; Infante et al. 2008). Loss of either causes the pathological accumulation of free cholesterol and sphingolipids within lysosomes, eventually blocking endolysosomal trafficking and autophagy and leading to progressive neuronal cell death. Numerous acquired diseases may also be associated with poor endolysosomal cholesterol homeostasis. Diabetes and general metabolic syndrome are closely tied to cholesterol levels and insulin signaling, which in turn is affected by the cholesterol trafficking burden along the endolysosomal pathway. As such, defects in the retrograde trafficking of cholesterol and other lipids out of lysosomes can contribute to several metabolic syndromes and hypercholesterolemia (high blood cholesterol) that in turn may contribute to pathologies including atherosclerosis and coronary heart disease (for further details, see reference (Reiner et al. 2014)).
The purpose of this chapter is to briefly discuss the current knowledge of ER-endolysosomal MCSs and to outline the importance of understanding this MCS in human health and disease. Many of the discoveries in this field come from elegant studies using both yeast and mammalian model systems, and these will be discussed with respect to their impacts in the basic and biomedical understanding. Finally, we will conclude by providing an assessment of the current trajectory of biomedical research on ER-endolysosomal MCSs, and how current and future studies will pave the way for potentially ground-breaking therapeutic strategies for metabolic diseases.
10.2. Discovery and Imaging of ER-Endolysosomal MCSs
Modern cell biology began in the 1950s with the introduction of transmission electron microscopy (TEM) as a high-resolution visualization tool for biologists. Using this approach, the pioneering work of George Palade and Keith Porter revealed the subcellular architecture of individual cells. Their studies highlighted how crowded cells were, and how organelles often maintained close contact with one another. They also unexpectedly observed the first inter-organelle MCSs including those formed between the ER and PM, as well as the ER and mitochondria (Porter and Palade 1957; Porter and Machado 1960). The ER-PM contact sites of muscle cells were closely noted (so called PM-sarcoplasmic reticulum contact sites that are important for Ca2+ regulated muscle contractions).
ER-endosome, ER-late endosome (LE), and ER-lysosome MCSs were not noted during these initial TEM studies, but more recent microscopy work shows them to be highly dynamic and common within mammalian cells. Elegant TEM work from Clare Futter’s group showed that the ER makes close contact with LEs and multi-vesicular bodies (MVBs) in mammalian cells (Eden et al. 2010). These studies showed that PTP1B, an ER-localized protein phosphatase, utilized the close inter-organelle contacts provided by ER-LE MCSs to regulate the trafficking and signaling of the EGF receptor, an important oncogene often targeted in anti-cancer therapeutics. Phospho-regulation of EGF receptor trafficking via PTP1B triggered its recognition by the endosomal sorting complex required for transport (ESCRT) pathway and thus its downregulation via delivery to the lysosome for degradation (Eden et al. 2010). Thus the first function for an ER-endolysosomal MCS—receptor regulation—was proposed.
Subsequent work using live-cell imaging further demonstrated that ER-endosome and ER-LE MCSs are highly dynamic and change as an endosome matures. Studies from Gia Voeltz’s group showed that the ER makes contact with endosomes shortly after they are formed (Friedman et al. 2013). Intriguingly, the degree of ER-endosome contact increases as early endosomes mature into LEs (monitored by exchanging the early endosome marker Rab5 for Rab7). This suggests that the ER may sense and actively participate in endosomal maturation. This idea was further supported by the observation that ER-LE MCSs appeared to define sites of vesicle scission at the LE from which small tubules sprouted and vesicles budded. This again suggested an active role for the ER in endosomal dynamics and potentially that the ER may contribute mechanical force to the vesicle scission reaction. How this is achieved is currently unclear, but is consistent with the previously proposed role of ER tubules in defining sites of mitochondrial division at ER-mitochondrial MCSs (Friedman et al. 2011). An intriguing possibility is that ER-mitochondrial and ER-LE MCSs create unique lipid microenvironments that promote spontaneous vesicle fission.
10.3. Molecular Identification of ER-Endolysosomal Tethering Proteins
10.3.1. Nvj1 and the NVJ
Early work on mammalian ER-endolysosomal MCSs is built upon pioneering work using the budding yeast Saccharomyces cervisiae to study ER-endolysosomal contact sites. These studies conducted in David Goldfarb’s group identified the first proteins that functioned as molecular tethers to establish MCSs between the ER and yeast lysosome, also known as the vacuole (Pan et al. 2000). Notably, the yeast vacuole serves as both a “late endosome” and “lysosome” for the cell, in that it is a Rab7-positive organelle that also contains lumenal hydrolases dedicated to catabolic metabolism. ER-vacuole MCSs may then, by extension, serve as model systems for the study of ER-endosome and ER-lysosome MCSs.
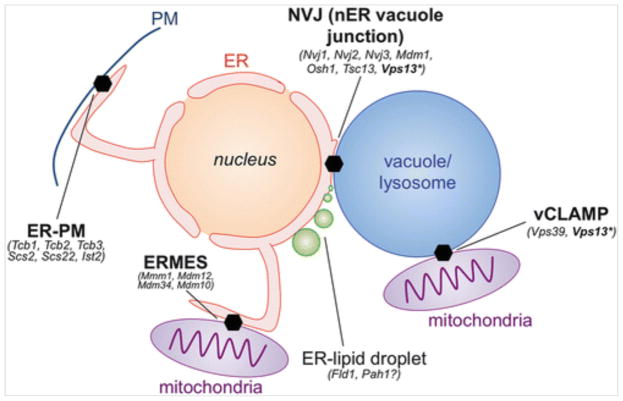
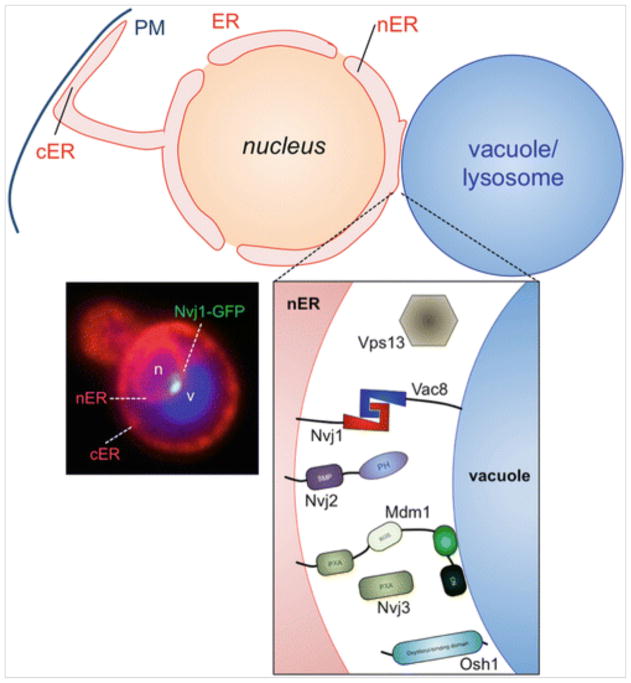
Yeast usually exhibit a single ER-vacuole MCS that forms as a circular “patch” juxtaposed between the vacuole and outer nuclear envelope, a region of ER continuous with the rest of the ER network often referred to as the nuclear ER (nER). Yeast nER-vacuole contacts are thus denoted nER-vacuole junctions (NVJs) (Fig. 10.2). Due to their size, and the close physical proximity of the nucleus and vacuole within yeast, NVJs are observed to be stable structures that exist for most of the cell’s lifetime. This is in contrast to the dynamic, short-lived ER-endosome MCSs seen in mammalian cells.
Fig. 10.2.
Guide to yeast inter-organelle contact sites and their resident proteins
Pioneering work by David Goldfarb’s group revealed the molecular machinery that establishes the NVJ. NVJs form through the direct interaction of ER-resident protein Nvj1 with the vacuole surface protein Vac8, which had previously been identified in vacuolar inheritance screens (Pan et al. 2000). This Nvj1:Vac8 hetero-dimerization thus defines the first molecular “tethers” that establish ER-endolysosomal MCSs.
10.3.2. Other NVJ Proteins
Subsequent studies on the NVJ demonstrated it as a site of lipid metabolism and membrane remodeling. During nitrogen starvation, the NVJ was shown to dramatically invaginate into the vacuole interior where it was eventually engulfed to form intra-lumenal vesicles within the vacuole lumen. Although the reason for this process is still unclear, it appears to represent a specific form of micro-autophagy and was thus termed piecemeal microautophagy of the nucleus (PMN) (Roberts et al. 2003). Other functions for the NVJ, including a site for the production of ceramide via fatty acid processing enzymes like Tsc13, have been proposed for the NVJ (Kvam et al. 2005). Consistent with this, other lipid metabolism proteins such as Nvj2, Osh1, and have also been identified as resident NVJ proteins, suggesting that NVJs function in many different lipid metabolic processes (Fig. 10.3) (Toulmay and Prinz 2012).
Fig. 10.3.
The nER vacuole junction (NVJ) resident proteins and their architectures. The fluorescence micrograph depicts a living yeast with the ER network labeled in red (DsRed-HDEL protein), the vacuole/lysosome in blue (CMAC dye), and endogenous Nvj1 tagged with GFP (Nvj1-GFP)
Recently, a study by the Will Prinz lab identified a protein domain found to reside specifically at inter-organelle MCSs: the synaptotagmin-like mitochondrial lipid-binding protein (SMP) domain (Toulmay and Prinz 2012). Nvj2 contains a bone vide SMP domain. Since a crystal structure of the SMP domain suggests it may physically bind to lipids with a hydrophobic cavity within the domain, it is postulated that SMP domain-containing proteins function in the non-vesicular transfer of lipids between different organelles (AhYoung et al. 2015; Reinisch and De Camilli 2016; Schauder et al. 2014).
10.3.3. Protrudin
Although they are essential for the formation of a proper NVJ, surprisingly, Nvj1 and Vac8 are not conserved in metazoans. How then are the ER and endosomes/lysosomes “tethered” in mammalian cells? Several recent studies have identified potential tethering proteins. Harold Stenmark’s group identified Protrudin, an integral ER protein with a PtdIns3P-binfing FYVE domain, as a potential tether connecting the ER and Rab7-positive LEs (Raiborg et al. 2015). Protrudin is enriched at ER-endosome MCSs, and its overexpression induced the formation of ER-endosome MCS in mammalian cells. These Protrudin-positive MCSs promoted the migration of LEs to the cell periphery in a microtubule-dependent manner. ER-LE MCSs are thus linked to a novel function: endosomal positioning and migration. This fucntion is supported by previous studies from the Neefjes group, which showed that ORP1L senses endosomal sterol levels and regulates the positioning endosomes within the cytoplasm (Johansson et al. 2007). When endosomal sterol levels are low, ORP1L binding to endosomes is reduced, promoting the formation of ER-LE MCSs in a VAP-dependent manner (Rocha et al. 2009). When endosomal cholesterol levels increase, such as in Niemann-Pick type C, ORP1L associates tightly to endosomes and promotes their trafficking to the perinuclear cell interior. Collectively, Protrudin and ORP1L represent two critical regulators of ER-LE MCSs that govern both endosome positioning and sterol metabolism in mammalian cells.
10.3.4. Mdm1 Family Proteins
Recent work on the yeast NVJ has revealed new, highly conserved proteins that regulate ER-endolysosomal MCSs. Yeast protein Mdm1 and its paralog Nvj3 were recently shown to localize to the NVJ (Henne et al. 2015b). Unlike Nvj1 and Vac8, Mdm1 is highly conserved in metazoans as a member of the Sorting Nexin (SNX) protein family. The SNX family is defined by the presence of a PtdIns3P-binding Phox homology (PX) domain (Yu and Lemmon 2001). Humans encode four Mdm1 homologs: Snx13, 14, 19, and 25 with near-identical domain architecture to Mdm1. All feature an N-terminal transmembrane region and signal sequence that appears to anchor them in the ER. On their C-terminal half, they contain the PX that directly binds the phospholipid PtdIns3P on the vacuole/lysosome surface, thus establishing ER-vacuole/lysosome tethering (Henne et al. 2015b). Though it requires Mdm1 for its NVJ localization, Nvj3 is soluble and contains a PX-Associated (PXA) that is unique to this protein family. The structure and function of the PXA domain is currently unclear, but implicated in lipid metabolism as its overexpression causes cells to become hypersensitized to drugs that perturb sphingolipid metabolism. In mammalian cells, Mdm1 homolog Snx14 is associated with a form of pediatric cerebellar ataxia and appears phenomenologically similar to lysosomal storage diseases (Thomas et al. 2014; Akizu et al. 2015). Indeed, SNX14-deficient cells manifest defects in lysosome homeostasis, suggesting that Snx14 plays some role in lysosome function. A role for Mdm1 family proteins in lipid metabolism is further supported by studies on Snazarus (Snz), the Drosophila melanogaster ortholog of Mdm1. These studies link Snz to aging and fat metabolism by showing that fruit flies deficient for Snz displayed extended lifespan (Suh et al. 2008). Intriguingly, Snz was found to be highly expressed in the fly fat body, a metabolic center of insect lipid metabolism akin to both the mammalian liver and adipocyte. Further studies on Snz and its homologs will no doubt continue to reveal roles for this protein family in lipid metabolism and inter-organelle cross talk.
10.3.5. Vps13
In addition to Mdm1 and Nvj3, other yeast proteins with clear metazoan homologs have been implicated in ER-endolysosomal MCSs. Vps13, first identified in the vacuolar protein sorting (vps) screen conducted in Scott Emr’s group in the 1980s, is 3,144 amino acids in length and one of the largest proteins encoded in the yeast genome (Bankaitis et al. 1986). Several metazoan homologs exist but are poorly characterized. Using elegant yeast genetics, Benoit Kornmann’s group recently discovered that gain-of-function Vps13 mutants were capable of rescuing the viability of yeast with a defective ERMES complex, an ER-mitochondrial tethering factor required for growth on non-fermentable carbon media (Lang et al. 2015). They then used complementation assays to discover that Vps13 can localize to a previously identified mitochondria-vacuole MCS (also called vacuole and mitochondria patch, vCLAMP) when grown in fermentable (dextrose) conditions (Elbaz-Alon et al. 2014; Honscher et al. 2014). Strikingly, Vps13 re-localized to the NVJ when grown in non-fermentable (glycerol) conditions (Lang et al. 2015). Indeed, Vps13 appears highly dynamic and has been observed to localize at vCLAMPs, ER-endosome, and NVJ MCSs in yeast depending upon growth conditions (Park et al. 2016). This suggests that Vps13 can localize to multiple MCSs, and also that the cell can actively remodel the composition of MCSs in response to changes in nutrient status and demand.
10.3.6. Lam/Ltc Family Proteins
The idea that different MCSs can dynamically change in response to changes in the cell’s metabolic status is supported by recent yeast work linking members of the ‘lipid transfer protein anchored at membrane contact sites’ (Lam) protein family to ER-endolysosomal MCSs. Lam6/Ltc1, the most studied member within this protein family, has been observed to localize to ER-mitochondria (ERMES), NVJ, and vCLAMP MCSs within the same cell, suggesting it may function in multiple metabolic processes (Murley et al. 2015; Elbaz-Alon et al. 2015; Gatta et al. 2015).
What, then, are the functions of Lam proteins at MCSs? At least two functions have been proposed. Overexpression of GFP-Lam6 was found to expand both the NVJ and ERMES MCSs, suggesting Lam proteins may regulate and possibly help extend inter-organelle contact sites in response to changes in cellular nutrient status (Elbaz-Alon et al. 2015). Another potential function is the direct movement of sterols between organelles. This could be achieved via direct binding and shuttling of a sterol molecule within the Lam protein tertiary fold. Lam family members contain a VASt/StART domain that binds sterols and may facilitate this inter-organelle sterol exchange (Murley et al. 2015; Elbaz-Alon et al. 2015). How this occurs, and how these proteins regulate lipid exchange and MCS size remains to be understood.
10.3.7. Annexin A1
Recently, other mammalian ER-endosome “tethers” have been identified that play roles in lipid and sterol metabolism. Using TEM and biochemistry, Emily Eden found that Annexin A1 mediates a specific subtype of ER-LE MCS formed between the ER and MVBs that downregulate the EGF receptor (Eden et al. 2016). Depletion of Annexin A1 reduced ER-MVB MCSs and perturbed EGR receptor signaling. Surprisingly, they find that ER-derived cholesterol is necessary for EGF receptor downregulation at the MVB and suggest that these ER-MVB MCSs function to transfer this cholesterol to the MVB limiting membrane, where cholesterol is required for the formation of intra-lumenal vesicles. How this cholesterol transfer is achieved is still unclear, but appears regulated by ORP1L.
10.4. ER-Endolysosomal MCSs in Human Health and Disease
Numerous ER-endolysosomal MCS proteins are linked to inherited or acquired diseases, indicating important roles in cellular metabolism and development. These connections underlie the important role for ER-endolysosomal cross talk in cellular homoeostasis in metazoans. Refer to Table 10.1 for further details.
Table 10.1.
ER-endolysosomal proteins and associated diseases
| ER-Endolysosomal contact site proteins in disease | ||||
|---|---|---|---|---|
| Protein | Localization(s) | Disease(s) associated | References | |
| Yeast | Human | |||
| Vps13 | VPS13A-D | ER-endosome, ER-vacuole (NVJ), ERMES? | VPS13A: chorea-acanthocytosis (CHAC); VPS13B: Cohen Syndrome (COH) | Ueno et al. (2001) and Kondo et al. (2005) |
| Scs2/22 | Vap-A,B (Als8) | ER-PM, ER-vacuole(?) | ALS disease | Teuling et al. (2007) |
| Tsc13 | TECR | ER-vacuole (NVJ) | Autosomal recessive non-syndromic intellectual disability | Nolan et al. (2008) |
| Mdm1 | Snx13, 14, 19, 25 | ER-vacuole (NVJ) | Snx14: autosomal recessive cerebellar ataxia with intellectual disability; Snx19: insulin secretion regulation | Thomas et al. (2014), Akizu et al. (2015) and Harashima et al. (2012) |
| Osh1 | OSBPs/ORPs | ER-vacuole (NVJ) | Linked to cholesterol-related metabolic disregulation | Ngo and Ridgway (2009) and Bouchard et al. (2009) |
| n/a | Protrudin | ER-endosome | Heriditary spastic paraplegia (HSP) | Hashimoto et al. (2014) |
| n/a | Annexin A1 | ER-late endosome/Multi-vescicular endosome | Links to pancreatic cancer progression | Belvedere et al. (2016) |
10.4.1. Neurological Disease
The VAP proteins VAP-A and VAP-B (Scs2 and Scs22 in yeast) have been linked to ER-PM MCSs, but have recently been implicated in controlling ER-endosome contact and endosomal migration via ORP1L (Rocha et al. 2009). A mutant form of VAP-A (containing the P56S mutation) is linked to familial amyotropic lateral sclerosis (ALS) in humans (Teuling et al. 2007). This mutation likely causes the protein to aggregate, resulting in general ER stress and cytotoxicity. Since a number of proteins also interact with VAP proteins via FFAT motifs, this general aggregation likely leads to general protein dysfunction and progressive cell toxicity (Prosser et al. 2008).
Vps13 and Mdm1, both residents of the yeast NVJ, have homologs linked to inherited neurological diseases in humans. Vps13 has at least two human orthologs with clear links to human disease: VPS13A, which is associated with chorea-acanthocytosis (CHAC), and VPS13B, linked to Cohen syndrome (COH) (Ueno et al. 2001; Kondo et al. 2005). Both are progressive neurological conditions associated with muscle dystonia and developmental delay. CHAC is highly variable in phenotype and can include abnormal red blood cell shape, as well as epilepsy. Mdm1 exhibits four human orthologs, two of which have clear mammalian disease links. SNX14-deficiency is associated with pediatric cerebellar ataxia and intellectual disability (Thomas et al. 2014; Akizu et al. 2015). A SNX13-deficient (also called RGS-PX1) mouse was found to exhibit severe developmental defects, delayed neural tube closure, and embryonic lethality (Zheng et al. 2006). Tsc13, the enoyl-CoA reductase found at the yeast NVJ, is also highly conserved in mammals as TECR, which is linked to an autosomal recessive non-syndromic intellectual disability (Nolan et al. 2008).
The Protrudin-encoding gene ZFYVE27 is mutated in individuals with hereditary spastic paraplegia (HSP), an inherited disease associated with muscle weakness in the lower half of the body (Hashimoto et al. 2014). This disease is generally associated with ER stress, suggesting that loss-of-function alleles may affect protein stability and trigger general ER stress response.
10.4.2. Cancer
ER-LE MCSs were observed in mammalian cells while studying PTP1B, an ER-localized protein phosphatase that regulates EGF receptor signaling, an oncogene product commonly targeted in anti-cancer therapies (Eden et al. 2010). As ER-LE MCSs may regulate the duration of EGF receptor signaling and other receptor signaling events, this may tie them closely to cancer progression. This is consistent with the proposed roles for Protrudin in cell migration, as cancer cells often display aggressive cell migration during tissue invasion and metastasis (Raiborg et al. 2015). Consistent with this, Annexin A1, which has been proposed as an ER-LE tether, is directly linked to pancreatic cancer progression (Belvedere et al. 2016).
10.4.3. Aging and Metabolism
The Drosophila ortholog of yeast Mdm1, Snz, is associated with lifespan extension in insects as well as obesity in mammals, as the expression of SNX13 and SNX14 was shown to increase in ob/ob mouse models (Suh et al. 2008). The connection between this aging phenotype and lipid metabolism is currently not clear, but Snz is highly expressed in the fly fat body, the central organ of insect fat metabolism analogous to the liver and adipocytes of mammals. This suggests that Snz may play a role in regulating fat storage and/or mobilization. Numerous connections exist between lipid storage mobilization and lifespan and generally show that caloric restriction may extend lifespan through the inhibition of TORC signaling and subsequent activation of autophagy (McCormick et al. 2011).
Whether Snz plays a role in autophagy and/or lipid mobilization in insects remains to be determined. However, ChIP-seq data from mammalian studies suggested that Snx13, the rodent homolog of Snz, is upregulated in response to cholesterol starvation in an SREBP-2-dependent manner (Seo et al. 2011). Among other genes upregulated were autophagy genes, suggesting Snx13 may be utilized during lipid mobilization in response to cholesterol starvation. In a separate example, human Mdm1 homolog Snx19 has been linked to insulin secretion pancreatic β-cells (Harashima et al. 2012).
10.4.4. Cholesterol Metabolism
The oxysterol-binding protein Osh1 localizes to the NVJ in yeast and is highly conserved in mammals as the oxysterol-binding proteins (OSBPs). OSBP upregulation is correlated with high blood pressure and hypertension, which are risk factors associated with coronary heart disease (Ngo and Ridgway 2009). OSBPL11 is also upregulated in patients exhibiting morbid obesity (Bouchard et al. 2009).
Although the movement of cholesterol between LEs/lysosomes and the ER is not well defined, it must require the Niemann-Pick proteins Npc1 and Npc2. Nps2 resides within the lysosome lumen and binds to the free cholesterol produced from the breakdown of CE within LDL particles. Upon binding, Npc2 transfers this free cholesterol to Npc1, a transmembrane protein that bridging the inner and outlet monolayers of the lysosome membrane (Infante et al. 2008). Through a poorly defined mechanism, Npc1 then transfers free cholesterol from the lysosome interior to the cytoplasmic leaflet of the lysosome. How this free cholesterol leaves the lysosome to travel to other organelles remains a major question in the lipid biology field. However, loss of either Npc1 or Npc2 is associated with Niemann-Pick type C, an autosomal recessive neurological disease characterized by progressive neuronal cell death.
10.5. Closing: The Therapeutic Promise of ER-Endolysosomal MCS Biology
Cholesterol metabolism has arguably been at the center of some of the greatest biopharmaceutical endeavors of the twentieth century. Existing at the crossroads between genetic and acquired disease, modulating human blood cholesterol levels can simultaneously mitigate symptoms associated with obesity, heart disease, diabetes, and an array of inherited genetic diseases afflicting cellular metabolism. Pioneering work by Mike Brown and Joe Goldstein elucidated not only how LDL-derived cholesterol enters cells, but also how the cell senses and responds to cholesterol burden (Brown and Goldstein 1997). These studies revealed the SREBP system: en elaborate sterol-sensing pathway that can modulate cellular cholesterol levels though an elegant feedback mechanism. In times of cholesterol deprivation, when ER-localized cholesterol reaches <5 mole % of the total ER lipid content, the ER protein SREBP-2 is efficiently trafficked to the Golgi, where it is proteolytically cleaved into a soluble transcription factor. It can then traffic into the nucleus and initiate a transcriptional response to this cholesterol need, increasing expression of sterol biosynthesis proteins including HMG-CoA Reductase.
Capitalizing on this metabolic feedback loop, several groups developed statins, which pharmacologically inhibit HMG-CoA reductase, allowing for the acute modulation of cellular cholesterol production, and by extension blood cholesterol levels, thus reducing the risk of heart disease for millions of patients in the Western world. This major biomedical achievement is the product of meticulous basic research coupled with ambitious biomedical application.
One emerging theme of MCS research is that sites of ER-endolysosomal contact may serve as “metabolic platforms” for regulating inter-organelle lipid trafficking. As such, the proteins that function at MCSs may represent a new class of therapeutic targets. MCS-enriched proteins currently fall under at least two general classes: organelle “tethers” (which help establish and maintain MCSs) and lipid “exchangers” (which utilize MCSs for inter-organelle lipid exchange). By pharmacologically targeting either of these two protein classes, it may be possible to modulate rates of lipid exchange and/or production of specific lipid classes within cells. This, by extension, may tune metabolic pathways or the formation of pathological debris that form the basis for pathophysiology in numerous metabolic diseases. Thus, continued focus on the molecular mechanisms that govern MCS formation and regulation are at the center of a new round of impactful biomedical discoveries.
In closing, the field of inter-organelle cross talk is at an exciting time. New proteins and protein complexes that establish organelle-organelle junctions continue to be discovered, providing new and exciting details regarding how MCSs are formed and function in cellular metabolism. Among all MCSs discovered, those formed between the ER and endolysosomal system appear to be highly utilized by cells to control their lipid metabolism. This is not surprising, as ER-endolysosomal MCSs serve as the very interface—both physically and conceptually—that connect catabolic and anabolic metabolism. The ER serves as a major anabolic lipid producer, and endosomes/lysosomes serve as major lipid “sinks” driving the catabolic breakdown of nutrient lipids and proteins received via endocytosis. Mechanisms of cross talk between these two organelle systems will undoubtedly continue to be discovered as we probe the nature of ER-endolysosomal MCSs.
Acknowledgments
We would like to thank members of the Henne Lab for their helpful thoughts and suggestions during the preparation of this book chapter. WM Henne is supported by grants from the NIH NIGMS (R35GM119768-01), the Searle Foundation (SSP-2016-1482), The American Federation for Aging Research (AFAR) (A15198), the Welch Foundation (I-1873), and the UT Southwestern Endowed Scholars Program.
References
- AhYoung AP, Jiang J, Zhang J, Khoi Dang X, Loo JA, Zhou ZH, Egea PF. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc Natl Acad Sci USA. 2015;112:E3179–E3188. doi: 10.1073/pnas.1422363112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akizu N, Cantagrel V, Zaki MS, Al-Gazali L, Wang X, Rosti RO, Dikoglu E, Gelot AB, Rosti B, Vaux KK, et al. Biallelic mutations in SNX14 cause a syndromic form of cerebellar atrophy and lysosome-autophagosome dysfunction. Nature genetics. 2015;47:528–534. doi: 10.1038/ng.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Johnson LM, Emr SD. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc Natl Acad Sci USA. 1986;83:9075–9079. doi: 10.1073/pnas.83.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann NA, Sullivan DP, Ohvo-Rekilä H, Simonot C, Pottekat A, Klaassen Z, Beh CT, Menon AK. Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry. 2005;44:5816–5826. doi: 10.1021/bi048296z. [DOI] [PubMed] [Google Scholar]
- Belvedere R, Bizzarro V, Forte G, Dal Piaz F, Parente L, Petrella A. Annexin A1 contributes to pancreatic cancer cell phenotype, behaviour and metastatic potential independently of Formyl Peptide Receptor pathway. Sci Rep. 2016;6:29660. doi: 10.1038/srep29660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard L, Faucher G, Tchernof A, Deshaies Y, Marceau S, Lescelleur O, Biron S, Bouchard C, Pérusse L, Vohl MC. Association of OSBPL11 gene polymorphisms with cardiovascular disease risk factors in obesity. Obesity. 2009;17:1466–1472. doi: 10.1038/oby.2009.71. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Eden ER, White IJ, Tsapara A, Futter CE. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol. 2010;12:267–272. doi: 10.1038/ncb2026. [DOI] [PubMed] [Google Scholar]
- Eden ER, Sanchez-Heras E, Tsapara A, Sobota A, Levine TP, Futter CE. Annexin A1 Tethers Membrane Contact Sites that Mediate ER to Endosome Cholesterol Transport. Dev Cell. 2016;37:473–483. doi: 10.1016/j.devcel.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz Y, Schuldiner M. Staying in touch: the molecular era of organelle contact sites. Trends Biochem Sci. 2011;36:616–623. doi: 10.1016/j.tibs.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell. 2014;30:95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y, Eisenberg-Bord M, Shinder V, Stiller SB, Shimoni E, Wiedemann N, Geiger T, Schuldiner M. Lam6 regulates the extent of contacts between organelles. Cell Rep. 2015;12:7–14. doi: 10.1016/j.celrep.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Dibenedetto JR, West M, Rowland AA, Voeltz GK. Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol Biol Cell. 2013;24:1030–1040. doi: 10.1091/mbc.E12-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta AT, Wong LH, Sere YY, Calderón-Noreña DM, Cockcroft S, Menon AK, Levine TP. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife. 2015;4:e07253. doi: 10.7554/eLife.07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P. PI(4,5)P2-dependent and Ca2+-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 2013;153:1494–1509. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL. History of the discovery of the thymosins. Ann N Y Acad Sci. 2007;1112:1–13. doi: 10.1196/annals.1415.045. [DOI] [PubMed] [Google Scholar]
- Harashima S, Horiuchi T, Wang Y, Notkins AL, Seino Y, Inagaki N. Sorting nexin 19 regulates the number of dense core vesicles in pancreatic β-cells. J Diabetes Investig. 2012;3:52–61. doi: 10.1111/j.2040-1124.2011.00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri H, Ugrankar R, Liu Y, Henne WM. Inter-organelle ER-endolysosomal contact sites in metabolism and disease across evolution. Commun Integr Biol. 2016;9:e1156278. doi: 10.1080/19420889.2016.1156278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Shirane M, Matsuzaki F, Saita S, Ohnishi T, Nakayama KI. Protrudin regulates endoplasmic reticulum morphology and function associated with the pathogenesis of hereditary spastic paraplegia. J Biol Chem. 2014;289:12946–12961. doi: 10.1074/jbc.M113.528687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Liou J, Emr SD. Molecular mechanisms of inter-organelle ER-PM contact sites. Curr Opin Cell Biol. 2015a;35:123–130. doi: 10.1016/j.ceb.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Henne WM, Zhu L, Balogi Z, Stefan C, Pleiss JA, Emr SD. Mdm1/Snx13 is a novel ER-endolysosomal interorganelle tethering protein. J Cell Biol. 2015b;210:541–551. doi: 10.1083/jcb.201503088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, van der Laan M, Cabrera M, Reggiori F, Ungermann C. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev Cell. 2014;30:86–94. doi: 10.1016/j.devcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci USA. 2008;105(40):15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, Olkkonen VM, Neefjes J. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor βlll spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko DC, Binkley J, Sidow A, Scott MP. The integrity of a cholesterol-binding pocket in Niemann-Pick C2 protein is necessary to control lysosome cholesterol levels. Proc Natl Acad Sci USA. 2003;100(5):2518–2525. doi: 10.1073/pnas.0530027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo I, Shimizu A, Asakawa S, Miyamoto K, Yamagata H, Tabara Y, Shimizu N. COH1 analysis and linkage study in two Japanese families with Cohen syndrome. Clinical Genet. 2005;67:270–272. doi: 10.1111/j.1399-0004.2005.00396.x. [DOI] [PubMed] [Google Scholar]
- Kvam E, Gable K, Dunn TM, Goldfarb DS. Targeting of Tsc13p to nucleus-vacuole junctions: a role for very-long-chain fatty acids in the biogenesis of microautophagic vesicles. Mol Biol Cell. 2005;16:3987–3998. doi: 10.1091/mbc.E05-04-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AB, John Peter AT, Walter P, Kornmann B. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J Cell Biol. 2015;210:883–890. doi: 10.1083/jcb.201502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T, Loewen C. Inter-organelle membrane contact sites: through a glass, darkly. Curr Opin Cell Biol. 2006;18:371–378. doi: 10.1016/j.ceb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci USA. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell. 2012;23:1129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- McCormick MA, Tsai SY, Kennedy BK. TOR and ageing: a complex pathway for a complex process. Philos Trans R Soc Lond B Biol Sci. 2011;366:17–27. doi: 10.1098/rstb.2010.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser von Filseck J, Čopič A, Delfosse V, Vanni S, Jackson CL, Bourguet W, Drin G. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 2015;349:432–436. doi: 10.1126/science.aab1346. [DOI] [PubMed] [Google Scholar]
- Murley A, Sarsam RD, Toulmay A, Yamada J, Prinz WA, Nunnari J. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J Cell Biol. 2015;209:539–548. doi: 10.1083/jcb.201502033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo M, Ridgway ND. Oxysterol binding protein-related Protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol Biol Cell. 2009;20:1388–1399. doi: 10.1091/mbc.E08-09-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan DK, Chen P, Das S, Ober C, Waggoner D. Fine mapping of a locus for nonsyndromic mental retardation on chromosome 19p13. Am J Med Genet A Part A. 2008;146A:1414–1422. doi: 10.1002/ajmg.a.32307. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Omnus DJ, Manford AG, Bader JM, Emr SD, Stefan CJ. Phosphoinositide kinase signaling controls ER-PM cross-talk. Mol Biol Cell. 2016;27:1170–1180. doi: 10.1091/mbc.E16-01-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Roberts P, Chen Y, Kvam E, Shulga N, Huang K, Lemmon S, Goldfarb DS. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. M Mol Biol Cell. 2000;11:2445–2457. doi: 10.1091/mbc.11.7.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Thorsness MK, Policastro R, McGoldrick LL, Hollingsworth NM, Thorsness PE, Neiman AM. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol Biol Cell. 2016;27:2435–2449. doi: 10.1091/mbc.E16-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter KR, Machado RD. Studies on the endoplasmic reticulum. IV. Its form and distribution during mitosis in cells of onion root tip. J Biophys Biochem Cytol. 1960;7:167–180. doi: 10.1083/jcb.7.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter KR, Palade GE. Studies on the endoplasmic reticulum. III. Its form and distribution in striated muscle cells. J Biophys Biochem Cytol. 1957;3:269–300. doi: 10.1083/jcb.3.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz WA. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J Cell Biol. 2014;205:759–769. doi: 10.1083/jcb.201401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser DC, Tran D, Gougeon PY, Verly C, Ngsee JK. FFAT rescues VAPA-mediated inhibition of ER-to-Golgi transport and VAPB-mediated ER aggregation. J Cell Sci. 2008;121:3052–3061. doi: 10.1242/jcs.028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Wenzel EM, Pedersen NM, Olsvik H, Schink KO, Schultz SW, Vietri M, Nisi V, Bucci C, Brech A, et al. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 2015;520:234–238. doi: 10.1038/nature14359. [DOI] [PubMed] [Google Scholar]
- Reiner Ž, Guardamagna O, Nair D, Soran H, Hovingh K, Bertolini S, Jones S, Ćorić M, Calandra S, Hamilton J, et al. Lysosomal acid lipase deficiency--an under-recognized cause of dyslipidaemia and liver dysfunction. Atherosclerosis. 2014;235:21–30. doi: 10.1016/j.atherosclerosis.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Reinisch KM, De Camilli P. SMP-domain proteins at membrane contact sites: Structure and function. Biochim Biophys Acta. 2016;1861:924–927. doi: 10.1016/j.bbalip.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P, Moshitch-Moshkovitz S, Kvam E, O’Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, Neefjes J. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150Glued and late endosome positioning. J Cell Biol. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder CM, Wu X, Saheki Y, Narayanaswamy P, Torta F, Wenk MR, De Camilli P, Reinisch KM. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 2014;510:552–555. doi: 10.1038/nature13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YK, Jeon TI, Chong HK, Biesinger J, Xie X, Osborne TF. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 2011;13:367–375. doi: 10.1016/j.cmet.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- Suh JM, Stenesen D, Peters JM, Inoue A, Cade A, Graff JM. An RGS-containing sorting nexin controls Drosophila lifespan. PloS one. 2008;3:e2152. doi: 10.1371/journal.pone.0002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavassoli S, Chao JT, Young BP, Cox RC, Prinz WA, de Kroon AI, Loewen CJ. Plasma membrane--endoplasmic reticulum contact sites regulate phosphatidylcholine synthesis. EMBO Rep. 2013;14:434–440. doi: 10.1038/embor.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuling E, Ahmed S, Haasdijk E, Demmers J, Steinmetz MO, Akhmanova A, Jaarsma D, Hoogenraad CC. Motor neuron disease-associated mutant vesicle-associated membrane protein-associated protein (VAP) B recruits wild-type VAPs into endoplasmic reticulum-derived tubular aggregates. J Neurosci. 2007;27:9801–9815. doi: 10.1523/JNEUROSCI.2661-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AC, Williams H, Setó-Salvia N, Bacchelli C, Jenkins D, O’Sullivan M, Mengrelis K, Ishida M, Ocaka L, Chanudet E, et al. Mutations in SNX14 cause a distinctive autosomal-recessive cerebellar ataxia and intellectual disability syndrome. Am J Hum Genet. 2014;95:611–621. doi: 10.1016/j.ajhg.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A, Prinz WA. A conserved membrane-binding domain targets proteins to organelle contact sites. J Cell Sci. 2012;125:49–58. doi: 10.1242/jcs.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S, Maruki Y, Nakamura M, Tomemori Y, Kamae K, Tanabe H, Yamashita Y, Matsuda S, Kaneko S, Sano A. The gene encoding a newly discovered protein, chorein, is mutated in chorea-acanthocytosis. Nature Genet. 2001;28:121–122. doi: 10.1038/88825. [DOI] [PubMed] [Google Scholar]
- Yu JW, Lemmon MA. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J Biol Chem. 2001;276:44179–44184. doi: 10.1074/jbc.M108811200. [DOI] [PubMed] [Google Scholar]
- Zhang D, Vjestica A, Oliferenko S. Plasma membrane tethering of the cortical ER necessitates its finely reticulated architecture. Curr Biol. 2012;22:2048–2052. doi: 10.1016/j.cub.2012.08.047. [DOI] [PubMed] [Google Scholar]
- Zheng B, Tang T, Tang N, Kudlicka K, Ohtsubo K, Ma P, Marth JD, Farquhar MG, Lehtonen E. Essential role of RGS-PX1/sorting nexin 13 in mouse development and regulation of endocytosis dynamics. Proc Natl Acad Sci USA. 2006;103:16776–16781. doi: 10.1073/pnas.0607974103. [DOI] [PMC free article] [PubMed] [Google Scholar]