Abstract
Box H/ACA RNAs are a group of small RNAs found in abundance in eukaryotes (as well as in archaea). Although their sequences differ, eukaryotic box H/ACA RNAs all share the same unique hairpin-hinge-hairpin-tail structure. Almost all of them function as guides that primarily direct pseudouridylation of rRNAs and spliceosomal snRNAs at specific sites. Although box H/ACA RNA-guided pseudouridylation has been extensively studied, the detailed rules governing this reaction, especially those concerning the guide RNA-substrate RNA base-pairing interactions that determine the specificity and efficiency of pseudouridylation, are still not exactly clear. This is particularly relevant given that the lengths of the guide sequences involved in base-pairing vary from one box H/ACA RNA to another. Here, we carry out a detailed investigation into guide-substrate base-pairing interactions, and identify the minimum number of base pairs (8), required for RNA-guided pseudouridylation. In addition, we find that the pseudouridylation pocket, present in each hairpin of box H/ACA RNA, exhibits flexibility in fitting slightly different substrate sequences. Our results are consistent across three independent pseudouridylation pockets tested, suggesting that our findings are generally applicable to box H/ACA RNA-guided RNA pseudouridylation.
Keywords: RNA-guided pseudouridylation, box H/ACA RNA, guide-substrate base-pairing, pseudouridylation pocket
INTRODUCTION
Pseudouridine (Ψ) is the most abundant post-transcriptionally modified nucleotide and is found in a wide range of RNAs, including mRNA and noncoding RNAs (such as tRNA, rRNA, and snRNA) (Maden 1990; Bjork 1995; Grosjean et al. 1995; Auffinger and Westhof 1998; Massenet et al. 1998; Ofengand and Fournier 1998; Reddy and Busch 1988; Sprinzl et al. 1998; Hopper and Phizicky 2003; Yu and Meier 2014). Ψ has chemical properties that are distinct from those of uridine and any other known nucleotides (Cohn 1959; Charette and Gray 2000). Thus, it is expected that Ψ provides unique contributions to RNA function. Indeed, over the years, a large amount of work has indicated that Ψs in rRNAs and snRNAs play an important role in protein translation and pre-mRNA splicing, respectively (King et al. 2003; Zhao and Yu 2004; Yang et al. 2005; Liang et al. 2009; Jack et al. 2011; Yu and Meier 2014; Wu et al. 2016).
Ψ is derived from uridine via an isomerization reaction called pseudouridylation (Ge and Yu 2013). Pseudouridylation can be catalyzed by stand-alone protein enzymes (PseudoUridine Synthases) or by a family of RNA-protein complexes termed box H/ACA RNPs (Fig. 1). The former is an RNA-independent reaction; the latter involves RNA (the box H/ACA RNA), and therefore is an RNA-dependent reaction.
FIGURE 1.
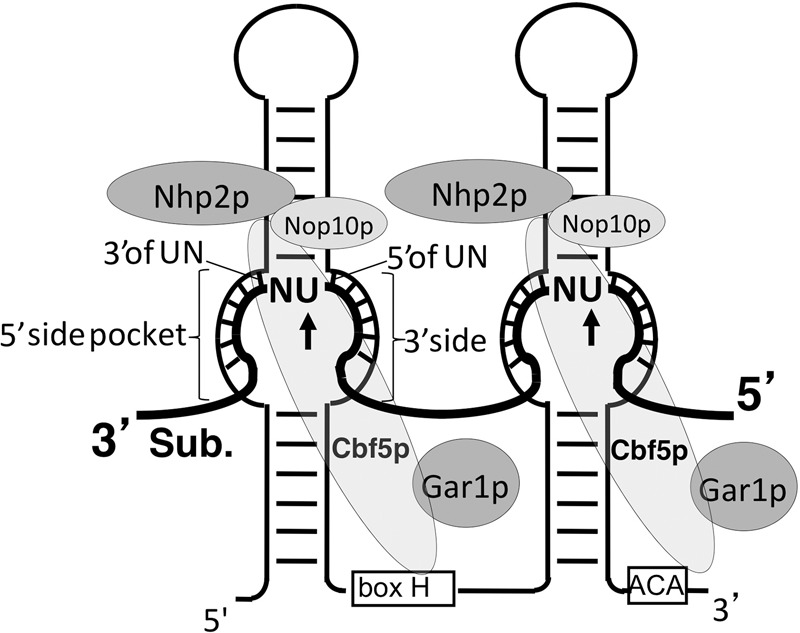
Box H/ACA RNP bound (base-paired) with its substrate RNA. Box H/ACA RNA (thin line) folds into a hairpin-hinge (containing an H box)-hairpin-tail (containing an ACA box) structure. The substrate RNA (thick line) is also shown. The internal loop (pseudouridylation pocket) of each hairpin base pairs with the substrate RNA, positioning the target U (arrow) and its immediate 3′ nucleotide (N) at the base of the upper stem (the dinucleotide is left unpaired). The four core box H/ACA RNP proteins, Nhp2, Nop10, Gar1, and Cbf5 (pseudouridylase), are also shown. Further illustrated is the internal loop (pseudouridylation pocket) within the 5′ hairpin, where the 5′ side and 3′ side of the pseudouridylation pocket as well as the base pairs immediately 5′ and 3′ of the unpaired dinucleotide UN (substrate) are indicated.
Box H/ACA RNPs, which are found in abundance in the nucleoli and Cajal bodies of eukaryotic cells, mainly catalyze pseudouridylation of rRNA and snRNA at specific sites (Ganot et al. 1997a; Ni et al. 1997; Yu et al. 2005). Each box H/ACA RNP consists of one unique box H/ACA RNA (with a unique sequence) and four common core proteins, including Nhp2, Nop10, Gar1, and Cbf5/NAP57/Dyskerin (a pseudouridylase) (Fig. 1). Although they have different sequences, all eukaryotic box H/ACA RNAs fold into an identical hairpin-hinge-hairpin-tail structure (Fig. 1). The hinge and tail regions of the RNA contain the evolutionarily conserved Box H (ANANNA) and Box ACA, respectively (Balakin et al. 1996; Ganot et al. 1997b). In each hairpin, there is an internal loop (the pseudouridylation pocket), which functions as a guide by base-pairing with the RNA substrate. As a consequence of this base-pairing, the target uridine, along with its 3′ immediately adjacent nucleotide, is precisely positioned at the base of the upper stem of the hairpin, and pseudouridylated by Cbf5/NAP57/Dyskerin (Fig. 1).
Over the years, box H/ACA RNA-guided RNA pseudouridylation has been extensively studied, and three sequence/structure elements necessary for efficient pseudouridylation have been identified (Bortolin et al. 1999; Xiao et al. 2009; De Zoysa and Yu 2018). First, it is necessary to have two stable hairpins, each harboring a pseudouridylation pocket. Second, it is important to have a fixed distance of ∼15 nucleotides (nt) between the target uridine (at the base of the upper stem) and the box H or box ACA (Bortolin et al. 1999; Xiao et al. 2009). Third, sufficient base-pairing interactions between the guide sequence in the pseudouridylation pocket and the substrate sequence are also required for efficient pseudouridylation (Ganot et al. 1997a; Ni et al. 1997; Xiao et al. 2009). However, the exact natures of these three elements, especially the rules governing guide RNA-substrate interactions, are still not exactly clear. This is particularly relevant given that the number of nucleotides involved in base-pairing with the substrate RNA varies from one box H/ACA RNA to another. Specifically, while some of the box H/ACA RNAs form extensive base-pairing interactions with their substrates, some others form only a limited number of base pairs with their substrates. The minimum number of base pairs between guide and substrate required for pseudouridylation has remained unclear.
Here, using the yeast system, we carried out a detailed analysis to clarify the rules for box H/ACA RNA-guided RNA pseudouridylation. We found that a minimum of 8 base pairs (bp) between the guide and the substrate are necessary for RNA-guided pseudouridylation. Interestingly, we also found that the pseudouridylation pocket, specifically the base of the upper stem of the hairpin that normally harbors two unpaired nucleotides, is rather flexible such that it can tolerate as many as four (perhaps even more) unpaired nucleotides.
RESULTS
It has widely been believed that proper, and sometimes extensive, base-pairing interactions between guide RNA and substrate RNA are key to substrate recognition, and that disruption of base pairs, especially the one immediately 5′ to the target uridine (Fig. 1), would result in a reduction or complete loss of pseudouridylation at the target site. To experimentally test this hypothesis and to understand the rules for RNA-guided RNA pseudouridylation, we decided to dissect in detail how guide-substrate base-pairing interactions contribute to this reaction.
Mutations in the branch site recognition region of yeast U2 snRNA have no significant effects on snR81-guided U2 pseudouridylation at position 42
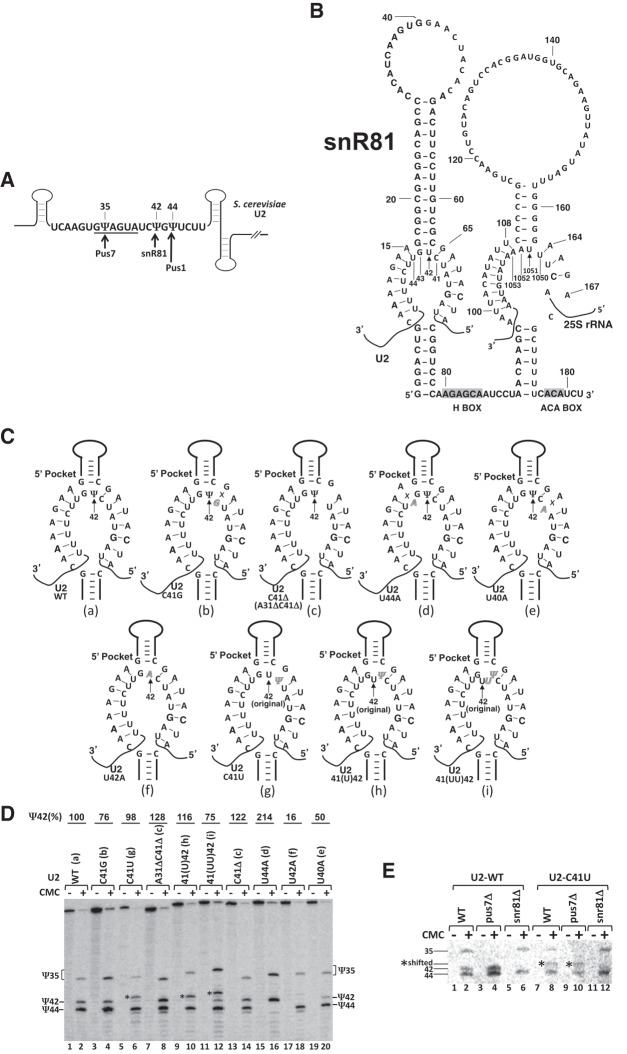
There are three naturally occurring Ψs in the branch site recognition region of yeast U2 snRNA. They are Ψ35, Ψ42, and Ψ44 (Fig. 2A). While the formation of Ψ35 and Ψ44 is catalyzed by stand-alone PUS enzymes (Pus7 and Pus1, respectively), the formation of Ψ42 is catalyzed by the 5′ pseudouridylation pocket of snR81 box H/ACA RNP (Fig. 2A,B; Massenet et al. 1999; Ma et al. 2003, 2005; Yu and Meier 2014). It is well documented that in order to recognize and specify the target uridine (U42), the 5′ pocket guide sequence of snR81 RNA has to form stable base-pairing interactions with the U2 sequence flanking the dinucleotide U42G43, thus positioning U42 (target nucleotide) and G43 at the base of the upper stem of the hairpin (Fig. 2B). There are 7 perfect and uninterrupted bp on each side of the pocket between the wild-type guide sequence and wild-type U2 (a total of 14 bp). Surprisingly, when the branch site recognition region of yeast U2 snRNA was targeted for mutations, which altered the base-pairing interactions between the 5′ guide pocket of snR81 and U2 snRNA, pseudouridylation was not significantly affected at position 42, as judged by the standard pseudouridylation assay, CMC modification followed by primer extension (Fig. 2C,D). Specifically, when C41 of U2 snRNA, located immediately 5′ adjacent to the unpaired target uridine (U42), was either mutated to G (C41G) (Fig. 2C, b) or deleted (C41Δ) (Fig. 2C, c), virtually no change was observed in Ψ42 formation (Fig. 2D, lanes 3 and 4; lanes 13 and 14) compared to the wild-type U2 (Fig. 2C, a; Fig. 2D, lanes 1 and 2). Consistently, deletion of both C41 and A31 (Fig. 2C, c) did not result in reduction of Ψ42 level either (Fig. 2D, lanes 7 and 8). Likewise, when U44, located immediately 3′ of the unpaired dinucleotide U42G43, was mutated to A44 (U44A) (Fig. 2C, d), pseudouridylation at position 42 remained unchanged, although Ψ44 (modified by Pus1) was lost as expected (Fig. 2D, lanes 15 and 16). Although the Ψ42 level was reduced when U40 was mutated to A40 (U40A) (Fig. 2C, e), the formation of Ψ42 was still clearly detected (Fig. 2D, lanes 19 and 20). Only when U42 was mutated to A42 (U42A) (Fig. 2C, f), was the Ψ42 signal completely lost (Fig. 2D, lanes 17 and 18).
FIGURE 2.
Effects of mutations of U2 snRNA on Ψ42 formation within U2 snRNA. (A) Schematic representation of S. cerevisiae U2. The sequence of branch site recognition region, including the branch site recognition motif (underlined), is shown. The three pseudouridines, Ψ35, Ψ42, and Ψ44, and their respective enzymes, Pus7, snR81, and Pus1, are indicated. Pus7 and Pus1 are stand-alone proteins, and snR81 is a box H/ACA RNP. (B) Sequence and structure of snR81 box H/ACA RNA. The primary sequences of snR81 and its substrates, U42 (indicated by the arrow) of U2 (5′ pocket) and U1051 (indicated by the arrow) of 25S rRNA (3′ pocket), are shown. The H and ACA boxes are shaded. (C) Base-pairing interactions between the snR81 5′ pocket and U2. Depicted are base-pairing interactions between the guide sequence of the 5′ pocket and its U2 substrate sequences, either wild-type (a) or mutants (b–i). Italicized letters represent mutated nucleotides. Crosses (Xs) indicate disrupted base-pairing interactions. In (g–i), the original U42 is also indicated. (D) Detection of Ψs in yeast U2 using CMC-modification followed by primer-extension. Wild-type U2 (lanes 1 and 2) and various mutant (lanes 3–20) U2 snRNAs (illustrated in C, a–i) were assayed for pseudouridylation. Signals corresponding to Ψ35, Ψ42, and Ψ44 are indicated. Shifted Ψ42 bands are indicated by asterisks (*). The formation of Ψ42 was quantified using the formula Ψ42/(Ψ35 + Ψ42 + Ψ44). The mutants (mutant lanes) were then normalized to the wild-type control. The final pseudouridylation efficiency numbers are shown at the top of each lane. Note, in lane 12, although there is still some formation of Ψ42 at the original position, pseudouridylation is mostly shifted to the 5′-most uridine site. (E) U42-to-Ψ42 conversion by snR81. Pseudouridylation of wild-type U2 (lanes 1–6) and C41U mutant U2 (lanes 7–12) was analyzed in wild-type (lanes 1 and 2, and lanes 7 and 8), pus7-deletion (lanes 3 and 4, and lanes 9 and 10), and snr81-deletion (lanes 5 and 6, and lanes 11 and 12) S. cerevisiae strains. Signals corresponding to Ψ35, Ψ42, and Ψ44 are indicated. Shifted Ψ42 bands are indicated by asterisks (*).
More surprisingly, when C41 was mutated to U41 (C41U) (Fig. 2C, g), we observed a shift in target uridine: U41, instead of U42, became pseudouridylated (Fig. 2D, lanes 5 and 6). To further confirm this observation, we inserted one (Fig. 2C, h) or two (Fig. 2C, i) uridines in between C41 and U42 in U2 snRNA and left them unpaired; in each test, the 5′-most unpaired uridine, instead of the original targeted uridine, became modified (Fig. 2D, lanes 9 and 10, and lanes 11 and 12). This shifting of modification can be clearly identified in denaturing urea-PAGE, as the original pseudouridine signal (the original Ψ42 band) disappeared or greatly diminished while a new pseudouridine signal, corresponding to the 5′-most inserted uridine, appeared (Fig. 2D, lanes 6, 10, and 12).
To ensure that snR81 box H/ACA RNP was responsible for the above altered pseudouridylation at position 42 (and the 5′-most uridine), we performed the same U2 mutational analysis in the snR81-deletion strain. As shown in Figure 2E, no un-shifted or shifted Ψ42 bands were observed when snR81 was deleted (lanes 11 and 12), suggesting that the Ψ formation observed above (at position U42 or the 5′-most unpaired uridine) was indeed catalyzed by snR81 box H/ACA RNP. These unexpected but interesting results prompted us to investigate, in more detail, the base-pairing interactions between the guide RNA and substrate RNA.
There exists a minimum guide-substrate base-pairing requirement for RNA-guided pseudouridylation
Given the fact that pseudouridylation activity tolerated one mismatch immediately 5′ or 3′ of the unpaired dinucleotide (U42G43), we reasoned that perhaps stability between the guide and substrate RNA could be maintained even with one less base pair compared to wild-type U2. If true, we speculated that there might be a minimum number of base pairs (between a guide and substrate RNA) needed to maintain pseudouridylation activity. At the minimum number of base pairs, the mismatch immediately 5′ or 3′ of the unpaired dinucleotide U42G43 would become intolerable (or in other words, the minimum number of base pairs would create the need for base-pairing involving the nucleotide immediately 5′ or 3′ of the unpaired dinucleotide U42G43).
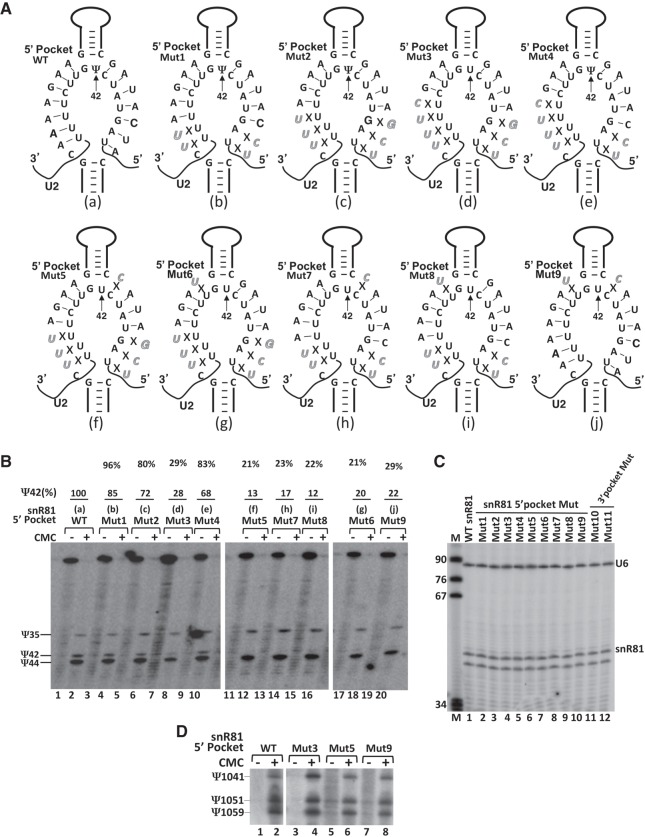
We first tested the same 5′ guide pocket of yeast snR81 that targets U42 in U2 snRNA. Rather than mutating U2 (which is not easy to do given that most mutations introduced into this region of U2 are lethal), we mutated the snR81 guide sequence (involved in guide-substrate base-pairing) from distal to proximal relative to the base of the upper stem where the dinucleotide U42G43 is positioned, thus disrupting the base-pairing interactions between guide and substrate one at a time (Fig. 3A, and also refer to Fig. 2B). After transforming the snR81-deletion strain with various snR81 mutant plasmids, we assayed U2 snRNA pseudouridylation at position 42. As shown in Figure 3A,B, when the number of base pairs was cut down to 5 (Fig. 3A, Mut1; b) or 4 (Fig. 3A, Mut2; c) on both sides of the pocket (a total of 10 or 8, respectively), Ψ42 formation was still well detected (Fig. 3B, lanes 3 and 4, and lanes 5 and 6, respectively). However, when an additional mismatch was introduced (Fig. 3A, Mut3; d), making a total of 7 bp (3 on the 5′ side [left side] of the pocket and 4 on the 3′ side [right side]), the Ψ42 signal almost completely disappeared (only a background level was detected) (Fig. 3B, lanes 7 and 8). When 1 bp was restored on the 3′ side (Fig. 3A, Mut4; e), thus restoring the total number of 8 bp (3 on the 5′ side of the pocket and 5 on the 3′ side), the formation of Ψ42 was rescued (Fig. 3B, lanes 9 and 10).
FIGURE 3.
Effects of mutations of snR81 (within the 5′ pocket) on Ψ42 formation within U2 snRNA. (A) Schematic representation of base-pairing interactions between the 5′ pocket of snR81 and U2. Depicted are base-pairing interactions between the wild-type (a) or various mutant (b–j) 5′ pockets of snR81 and the wild-type U2 substrate sequence. Italicized letters represent mutated nucleotides. Crosses (Xs) indicate disrupted base-pairing interactions. (B) Detection of Ψs in yeast U2 using CMC-modification followed by primer-extension. U2 pseudouridylation was assayed in the context of wild-type (lanes 1 and 2) and various mutant (lanes 3–20) snR81 (illustrated in A, a–j). Signals corresponding to Ψ35, Ψ42, and Ψ44 are indicated. The formation of Ψ42 was quantified using the formula Ψ42/(Ψ35 + Ψ42 + Ψ44). The mutants (mutant lanes) were then normalized to the wild-type control. The final pseudouridylation efficiency numbers (Ψ42%) are shown at the top of each lane. Note, in the Mut3 lane (lane 8), the small amount of “Ψ42” band is probably due to RNA degradation in this specific sample (as hinted by the CMC-minus lane––lane 7). (C) The levels of wild-type and mutant snR81 box H/ACA RNAs in the cell. Total RNA was recovered and primer-extension was used to measure the levels of wild-type (lane 1) and various mutant (lanes 2–12) snR81 RNAs. The U6 level was also measured as an internal control. The snR81 and U6 primer-extension products are indicated. The band below the snR81 band is a nonspecific primer-extension product. Lane M is a size marker. (D) Independent pseudouridylation pockets. The wild-type snR81 (lanes 1 and 2) and the 5′ pocket-mutated snR81 RNAs (lanes 3–8), which failed to guide U2 pseudouridylation at position 42 (see A and B), were tested for their ability (using their 3′ pocket) to guide 25S rRNA pseudouridylation at position 1051. Ψ1051, and its neighboring pseudouridines, Ψ1041 and Ψ1059, are indicated.
At the minimum number of base pairs (total of 8), the base pair immediately 5′ or 3′ of the unpaired dinucleotide U42G43 became necessary. Disruption of the base pair either 5′ (Fig. 3A, Mut5, f) or 3′ (Fig. 3A, Mut6, g) of the unpaired dinucleotide U42G43, thus leaving a total of only 7 bp, resulted in complete loss of Ψ42 (Fig. 3B, lanes 11 and 12, and lanes 17 and 18, respectively). This result suggests that although neither of these 2 bp is absolutely required when there is a large number of base pairs between the guide and the substrate (see Fig. 2D, lanes 3, 4, 13–16), they become necessary when the number of base pairs between the guide and the substrate approaches 8, the minimum number required for pseudouridylation. Perhaps more interestingly, disruption of the base pair immediately 5′ (Fig. 3A, Mut7, h) or 3′ (Fig. 3A, Mut8, i) of the unpaired dinucleotide U42G43, while maintaining the total number of base pairs at 9 (4 on the 5′ side of the pocket and 5 on the 3′ side, or 5 on the 5′ side of the pocket and 4 on the 3′ side), also led to almost total loss of Ψ42 (Fig. 3B, lanes 13–16). Finally, simultaneous disruption of the base pairs 5′ and 3′ of the unpaired dinucleotide U42G43, while keeping other nucleotides unchanged (thus maintaining a total of 12 bp) (Fig. 3A, Mut9, j), again led to almost total loss of Ψ42 (Fig. 3B, lanes 19 and 20). These results suggest that these base pairs do not just contribute to guide-substrate interactions. Rather, they appear to play some additional role(s) in guiding pseudouridylation; they appear to be more important than the base pairs at the distal ends of the duplex.
To ensure that all snR81 RNAs, including the wild-type and all mutants, were equally expressed, we carried out primer-extension analysis to measure the snR81 level in each strain. As shown in Figure 3C, the wild-type and various mutant snR81 RNAs were expressed at the same level. In addition, we also found that the two pseudouridylation pockets in the same box H/ACA RNA were functionally independent, as the same snR81 mutants whose 5′ pockets failed to guide Ψ42 formation were fully functional in catalyzing, using their 3′ pseudouridylation pockets, the formation of Ψ1051 of 25S rRNA (the natural target of snR81's 3′ pocket) (Fig. 3D).
The pseudouridylation pocket is flexible enough to fit more than two unpaired nucleotides (U42G43) at the base of upper stem
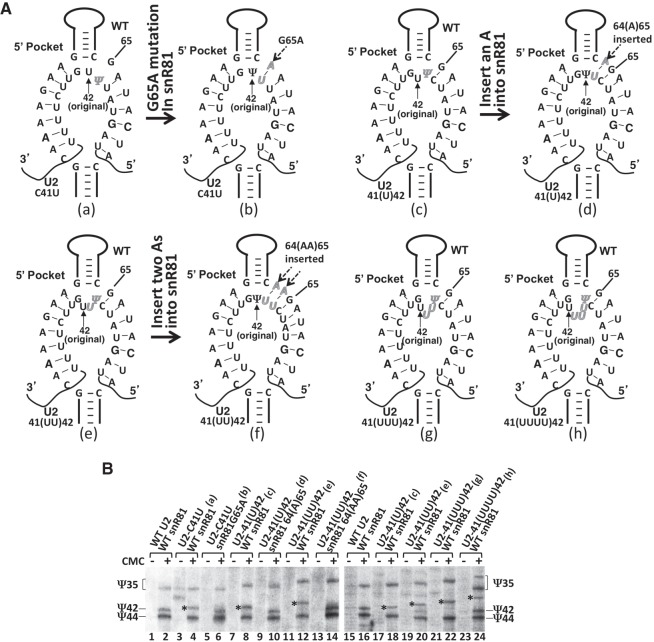
Our initial results also indicated that even though pseudouridylation activity was not affected when C41 was changed to U41, or when one or two uridines were inserted between C41 and U42, in both of these scenarios, the specificity of the target uridine changed: Pseudouridylation shifted to the 5′-most uridine (Fig. 2C, g, h, i; Fig. 2D, lanes 6, 10, and 12). These results prompted us to hypothesize that the guide pocket is flexible, allowing it to fit different lengths of unpaired nucleotides at the base of the upper stem. In this scenario, if the 5′-most uridine is forced to pair with the guide sequence, it may no longer be the target for pseudouridylation. To test this hypothesis, we changed G65 to A65 in snR81 to restore base-pairing with U41 in the C41U U2 mutant (Fig. 4A, a and b). When C41U U2 was tested, we observed that U41 was no longer the pseudouridylation target (it was not pseudouridylated) and that the modification shifted back to position 42 (U42 was converted to Ψ42) (Fig. 4B, compare lane 6 with lane 4). Similarly, we created snR81 mutants where one (Fig. 4A, c and d) or two (Fig. 4A, e and f) adenosines were inserted between C64 and G65 to base pair with the uridine(s) inserted in the C41(U)U42 and C41(UU)U42 U2 mutants. Again, we observed that these now paired uridines were no longer the targets, and pseudouridylation specificity shifted back to the original target uridine (Fig. 4B, compare lane 10 with lane 8, and lane 14 with lane 12).
FIGURE 4.
Flexibility of the pseudouridylation pocket. (A) Schematic representations of the 5′ pocket of snR81 base-paired with its substrate. Shown are base-pairing between C41U-U2 and wild-type snR81 (a), between C41U-U2 and G65A-snR81 (compensatory mutant) (b), between 41(U)42-U2 and wild-type snR81 (c), between 41(U)42-U2 and 64(A)65-snR81 (compensatory mutant) (d), between 41(UU)42-U2 and wild-type snR81 (e), between 41(UU)42-U2 and 64(AA)65-snR81 (compensatory mutant) (f), between 41(UUU)42-U2 and wild-type snR81 (g), and between mutant 41(UUUU)42-U2 and wild-type snR81 (h). (B) Pseudouridylation assay in the context of snR81 and U2 RNAs shown in (A). Lanes 3 and 4 are mutant U2 (C41U) and wild-type snR81; lanes 5 and 6 are mutant U2 (C41U) and compensatory mutant snR81 (G65A); lanes 7 and 8 and lanes 17 and 18 are mutant U2 [41(U)42] and wild-type snR81; lanes 9 and 10 are mutant U2 [41(U)42] and compensatory mutant snR81 [64(A)65]; lanes 11 and 12 and lanes 19 and 20 are mutant U2 [41(UU)42] and wild-type snR81; lanes 13 and 14 are mutant U2 [41(UU)42] and compensatory mutant snR81 [64(AA)65]; lanes 21 and 22 are mutant U2 [41(UUU)42] and wild-type snR81; lanes 23 and 24 are mutant U2 [41(UUUU)42] and wild-type snR81; lanes 1 and 2 and lanes 15 and 16 are wild-type U2 and wild-type snR81 (controls). Ψ35, Ψ42, and Ψ44 are indicated. The asterisks mark the shifted Ψ42 bands.
Next, we assessed the extent of the flexibility of the guide pocket. To this end, we inserted both three and four uridines between the C41 and U42 in the U2 snRNA, creating C41(UUU)U42 (Fig. 4A, g) and C41(UUUU)U42 (Fig. 4A, h) two new mutants, respectively. While replacement of wild-type U2 with C41(UUU)U42 U2 did not have a significant effect on cell growth, substitution of wild-type U2 with the four-uridine insertion U2, C41(UUUU)U42, resulted in a very slow growth phenotype. To facilitate cell growth and hence the analysis of pseudouridylation, the wild-type U2 was retained in cells expressing the four-uridine insertion U2 (two copies of U2, wild-type and mutant, were coexpressed). As shown in Figure 4B, when the three-uridine insertion U2 was tested, pseudouridylation was again shifted to the 5′-most uridine (lanes 21 and 22). When the four-uridine insertion U2 was expressed, we also observed a shift of target to the 5′-most uridine (lanes 23 and 24). Because the wild-type U2 was coexpressed, we also detected the formation of Ψ35 and Ψ42 in the wild-type U2 (lanes 23 and 24). These results suggest that the pseudouridylation pocket is indeed flexible and it can fit more than two unpaired nucleotides at the base of the upper stem.
The guide-substrate base-pairing requirement can be generalized to other pseudouridylation pockets
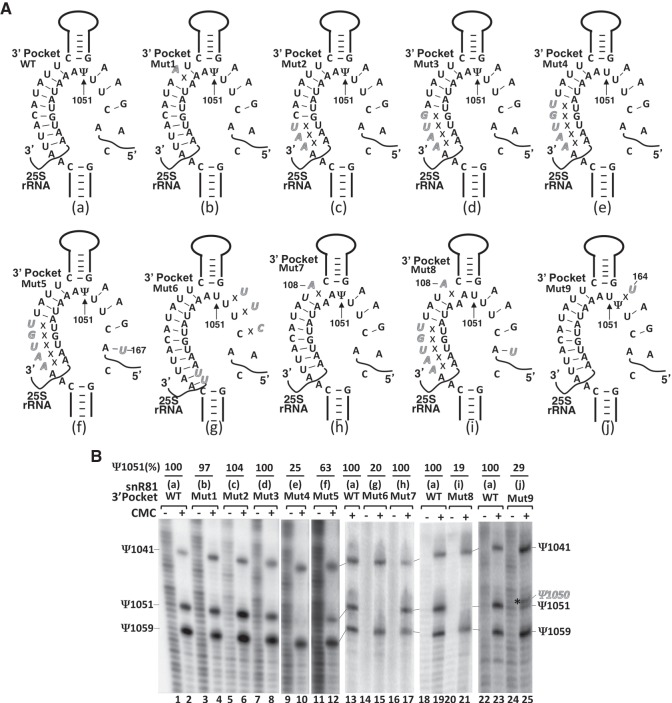
To determine whether the guide-substrate base-pairing requirement and the flexibility of the pseudouridylation pocket discussed above can be generally applied to box H/ACA RNA-guided pseudouridylation, we tested two additional box H/ACA RNA pseudouridylation pockets. Since there are two pseudouridylation pockets (5′ and 3′) in snR81, and only the 5′ pocket (guiding Ψ42 formation in U2) had been tested (Fig. 4), we turned our focus to the 3′ pocket, which guides 25S rRNA pseudouridylation at position 1051 (Ψ1051) (Fig. 2B). There are a total of 12 bp between the wild-type 3′ pocket and the wild-type 25S rRNA (9 on the 5′ side of the pocket, and 3 on the 3′ side), a number far greater than the previously determined minimum number of 8 bp. Mutations were introduced into the 3′ pocket of snR81 to disrupt/alter base pairs between the guide sequence (in the 3′ pocket) and the substrate sequence (in 25S rRNA) (Fig. 5A). As expected, disruption of 1 bp in the middle of the duplex (Fig. 5A, Mut1, b) did not affect pseudouridylation at position 1051 (Ψ1051) (Fig. 5B, lanes 3 and 4). When the number of base pairs was cut down to 9 (6 on the 5′ side and 3 on the 3′ side) (Fig. 5A, Mut2, c) or 8 (5 on the 5′ side and 3 on the 3′ side) (Fig. 5A, Mut3, d), no obvious changes in Ψ1051 formation was observed (Fig. 5B, lanes 5 and 6, and lanes 7 and 8). However, when the number of base pairs was cut down further to 7 (4 on the 5′ side and 3 on the 3′ side) (Fig. 5A, Mut4, e), pseudouridylation at position 1051 was completely abolished (Fig. 5B, lanes 9 and 10). Introduction of an appropriate point mutation into snR81 at position 167 (A167-to-U167 change), which formed a new base pair interaction with A147 of 25S rRNA and restored the total number of base pairs to 8 (4 on each side) (Fig. 5A, Mut5, f), resulted in the rescue of Ψ1051 formation (Fig. 5B, lanes 11 and 12). We created another 3′ pocket mutant in which the guide-substrate base pairs were completely disrupted on one side (the 3′ side) while the base pairs on the other side (5′ side) were expanded, keeping a total of 11 base-pairs (Fig. 5A, Mut6, g). This mutant failed to guide Ψ1051 formation (Fig. 5B, lanes 14 and 15), suggesting that one-sided anchoring/recognition (base pair interactions) is not sufficient for RNA-guided pseudouridylation.
FIGURE 5.
Effects of mutations of snR81 (within the 3′ pocket) on Ψ1051 formation within the 25S rRNA. (A) Schematic representations of the 3′ pocket of snR81 base-paired with 25S rRNA. Depicted are base-pairing interactions between the wild-type (a) or various mutant (b–j) 3′ pockets of snR81 and the wild-type 25S rRNA substrate sequence. Italicized letters represent mutated nucleotides. Crosses (Xs) indicate disrupted base-pairing interactions. (B) Detection of Ψs in yeast 25S rRNA using CMC-modification followed by primer-extension. Pseudouridylation of 25S rRNA was carried out in the context of wild-type (lanes 1 and 2, lane 13, lanes 18 and 19, and lanes 22 and 23) and various mutant (lanes 3–12,14–17,20,21,24,25) snR81 (illustrated in A, a–j). Signals corresponding to Ψ1041, Ψ1051 and Ψ1059 are indicated. The asterisk marks the shifted pseudouridylation band (Ψ1050). Ψ1051 formation was quantified using the formula Ψ1051/(Ψ1041 + Ψ1051 + Ψ1059). The mutants (mutant lanes) were then normalized to the wild-type control. The final pseudouridylation efficiency numbers (Ψ1051%) are shown at the top of each lane. The number shown at the top of lanes 24 and 25 reflects the efficiency of Ψ1050 formation (shifted band).
To assess the importance of the base pair immediately next to the unpaired dinucleotide at the base of the upper stem, we introduced a point mutation into the 3′ pocket at position 108. Although this mutation resulted in the disruption of the base pair 3′ of the unpaired dinucleotide (Fig. 5A, Mut7, h), the formation of Ψ1051 was not affected (Fig. 5B, lanes 16 and 17). However, when more mutations were introduced into the 3′ pocket to allow a total of 7 (one less than the minimum number) base pairs between the guide and the substrate (Fig. 5A, Mut8, i), we observed a total loss of Ψ1051 (Fig. 5B, lanes 20 and 21).
We also tested whether the changing target uridine phenomenon would also occur in the context of the 3′ pocket of snR81 and the 25S RNA substrate. Because the nucleotide immediately 5′ of U1051 is also a uridine (U1050), we simply introduced a point mutation into snR81 at position 164 (A-to-U change) such that U1050 of the substrate was left unpaired (Fig. 5A, Mut9, j). Upon expression of this snR81 mutant, a shift of target uridine was detected: The Ψ1051 signal was reduced close to the background level, and concurrently a new signal corresponding to Ψ1050 appeared (Fig. 5B, lanes 24 and 25).
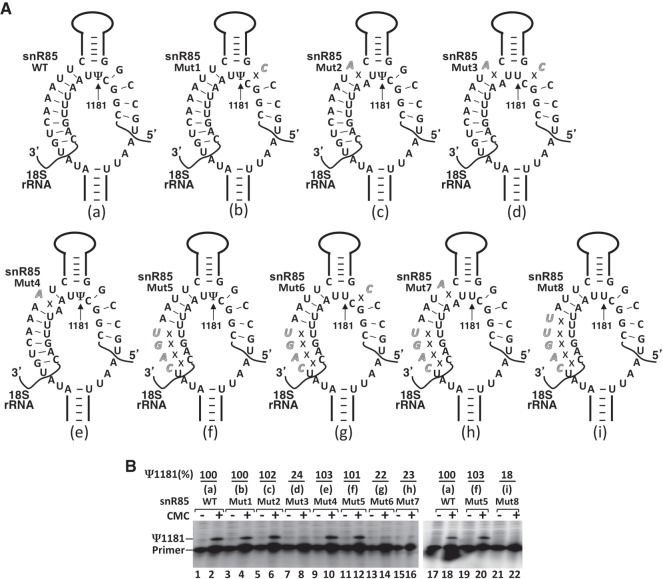
We next tested another experimentally verified pseudouridylation pocket: the 5′ pocket of snR85, which guides the formation of Ψ1181 of 18S rRNA. There are a total of 12 bp between wild-type snR85 and wild-type 18S rRNA (8 on the 5′ side, and 4 on the 3′ side) (Fig. 6A, a). We again manipulated the number of base pairs by introducing mutations into the 5′ pocket of snR85. As shown in Figure 6, disruption of a base pair in the middle of the 5′ duplex (Fig. 6A, Mut4, e) had no impact on the formation of Ψ1181 (Fig. 6B, lanes 9 and 10). Likewise, disruption of the base pair 5′ (Fig. 6A, Mut1, b) or 3′ (Fig. 6A, Mut2, c) of the unpaired dinucleotide (U1181U1182) at the base of the upper stem did not have any effect on Ψ1181 formation (Fig. 6B, lanes 3 and 4, and lanes 5 and 6, respectively). However, simultaneous disruption of both of these base pairs (Fig. 6A, Mut3, d) resulted in the loss of Ψ1181 (Fig. 6B, lanes 7 and 8). When the number of base pairs was cut down to 8 (4 on each side) (Fig. 6A, Mut5, f), the Ψ1181 signal was still detected (Fig. 6B, lanes 11 and 12, and lanes 19 and 20). However, when the number of base pairs was cut down further to 7 (3 on the 5′ side and 4 on the 3′ side) (Fig. 6A, Mut8, i), Ψ1181 was completely lost (Fig. 6B, lanes 21 and 22). Likewise, at the minimum number of 8 bp, disruption of the base pair 5′ (Fig. 6A, Mut6, g) or 3′ (Fig. 6A, Mut7, h) of the unpaired dinucleotide at the base of the upper stem resulted in complete loss of Ψ1181 (Fig. 6B, lanes 13 and 14, and lanes 15 and 16, respectively). Taken together, our results indicate that the rules for governing RNA-guided pseudouridylation, such as the guide-substrate base-pairing requirement as well as the flexibility of the pseudouridylation pocket, are generally applicable to different pseudouridylation pockets.
FIGURE 6.
Effects of mutations of snR85 (within the 5′ pocket) on Ψ1181 formation within the 18S rRNA. (A) Schematic representations of the 5′ pocket of snR81 base-paired with 18S rRNA. Depicted are base-pairing interactions between the wild-type (a) or various mutant (b–i) 5′ pockets of snR85 and the wild-type 18S rRNA substrate sequence. Italicized letters represent mutated nucleotides. Crosses (Xs) indicate disrupted base-pairing interactions. (B) Detection of Ψs in yeast 18S rRNA using CMC-modification followed by primer-extension. Pseudouridylation of 18S rRNA was carried out in the context of wild-type (lanes 1 and 2, and lanes 17 and 18) and various mutant (lanes 3–16, and lanes 19–22) snR85 (illustrated in A, b–i). Signal corresponding to Ψ1181 is indicated. Ψ1181 formation was quantified using the formula Ψ1181/(Ψ1181 + primer). The mutants (mutant lanes) were then normalized to the wild-type control. The final pseudouridylation efficiency numbers (Ψ1181%) are shown at the top of each lane.
DISCUSSION
Although box H/ACA RNA-guided RNA pseudouridylation has been widely discussed and extensively studied, the rules that govern this reaction are not exactly clear. In the current work, we have investigated the base-pairing interactions between the guide sequence in the pseudouridylation pocket and the substrate sequence. By performing mutational analysis on three different pseudouridylation pockets and their substrates, we have demonstrated that there is a minimal guide-substrate base-pairing requirement (8 bp) for RNA-guided pseudouridylation. We also show that the pseudouridylation pockets are flexible in accommodating different substrate sequences at the base of the upper stem. It appears that the rules identified here can be generalized to box H/ACA RNA-guided pseudouridylation. Of note is that we only introduce mutations into the guide-substrate duplex region. It is therefore unlikely that these mutations affect the binding of box H/ACA RNP proteins. Thus, we believe that each box H/ACA RNA construct assembles into functional box H/ACA RNP.
In order to catalyze a reaction, an enzyme must first recognize its substrate. In box H/ACA RNP-catalyzed pseudouridylation, box H/ACA RNA serves as a guide that recognizes its substrate RNA, and binds to it via base-pairing interactions. It is therefore conceivable that there is a minimum binding affinity or minimum base-pairing requirement for the reaction. Here, we show that this minimum base-pairing requirement is 8 bp, which can be either equally or unevenly distributed to both sides (5′ and 3′) of the pseudouridylation pocket. This number of base pairs are perhaps required for keeping the substrate bound. Although it is desirable to further dissect base-pairing requirements in the context of C-G and A-U pairs, which are known to contribute differently to base-pairing interactions, substrate mutation intolerance (in the case of U2) or difficulty in changing substrate sequences (in the case of rRNA) precludes us from carrying out further mutational analysis. Nonetheless, our data (generated primarily through mutations of guide sequences of three independent pseudouridylation pockets provide consistent evidence that 8 bp are necessary for box H/ACA RNA-guided pseudouridylation.
We also note that interactions between the guide and substrate are not regular Watson–Crick A-form helix. Instead, when pairing, the substrate comes in from one side to contact the guide without threading through the guide pocket to form an A-form helix (Jin et al. 2007; Wu and Feigon 2007). This kind of base-pairing is therefore unusual and may be suboptimal. We attempted to calculate the free energy (ΔG values) of base-pairing interactions between guides and substrates, but failed to find an appropriate calculation algorithm (currently available algorithms are only suited for standard Watson–Crick base-pairing). To generate meaningful free energy (ΔG) values for such unusual base-pairing interactions, further algorithm development is necessary.
Interestingly, the base pair immediately 5′ or 3′ of the unpaired dinucleotide at the base of the upper stem of each hairpin does not seem to be important if there is an extensive guide-substrate base-pairing (Fig. 2D, lanes 3 and 4, lanes 15 and 16; Fig. 5B, lanes 16 and 17; Fig. 6B, lanes 3 and 4, lanes 5 and 6). However, it becomes necessary when the number of guide-substrate base pairs approaches the minimum number of 8. That said, we note that the base pair 5′ or 3′ of the unpaired dinucleotide at the base of the upper stem is not an ordinary base pair (e.g., the base pairs at the 5′ or 3′ end of the duplex). Our results show that the base pair is still necessary for pseudouridylation even when the number of guide-substrate base pairs is above the minimum number (Fig. 3B, lanes 13–16). Even at an extensive number of base pairs, simultaneous disruption of both base pairs 5′ and 3′ of the unpaired dinucleotide consistently resulted in ablation of Ψ formation (Fig. 3B, lanes 19 and 20; Fig. 6B, lanes 7 and 8). Taken together, these results suggest that the base pairs 5′ and 3′ of the unpaired dinucleotide play a special role in substrate recognition: Not only do they contribute to the stability of guide-substrate binding, but they also play some other role(s), perhaps helping orient the target uridine at the catalytic center. Our results are consistent with the most recent report by the Gall laboratory, in which they show that the base pair 5′ of the unpaired dinucleotide plays an important role in directing pseudouridylation (Deryusheva and Gall 2018).
To better understand guide-substrate interactions, we have inspected base-pairing interactions between all known yeast box H/ACA RNAs and their natural substrates. In most cases, there are extensive base pairs (>10), although the exact number of base pairs varies. This raises a question: Why are there extensive base-pairing interactions, if only 8 bp seem sufficient for pseudouridylation? One possibility is that less extensive base pairs may result in less efficient pseudouridylation, which was not precisely reflected in our data given that the pseudouridylation assay we used, CMC-modification followed by primer-extension, is not fully quantitative. Another possibility is that different numbers of base pairs (and hence different binding affinities) may have different tolerances to stress. For instance, extensive base-pairing may allow pseudouridylation to occur at both normal and higher temperatures. In contrast, the minimum or close to minimum number of base pairs may allow pseudouridylation only at normal temperature; at higher temperature (stress), binding between the guide and substrate may not be sufficient, leading to loss of pseudouridylation. It may therefore be desirable to have extensive base pairs to ensure proper pseudouridylation even at high temperatures or under other stress conditions.
Having identified the minimum number of base pairs, the natural question is: Is there also a base pair ceiling beyond which pseudouridylation activity is restricted? We speculate that this hypothetical ceiling may in fact exist. It can be envisioned that although excessively high affinity between guide and substrate may not affect pseudouridylation activity, it will likely impact the release of the modified substrate, with potentially deleterious consequences. Along these lines, our results also show that individual mismatches in the middle of the guide-substrate duplex are tolerated, as long as the guide-substrate duplex as a whole is sufficiently stable. In contrast, two mismatches abolish pseudouridylation activity under normal conditions, as previously reported (Wu et al. 2011). Interestingly, however, two mismatches do allow pseudouridylation under certain stress conditions (Wu et al. 2011). Together, these results suggest that this low-specificity relationship between the guide pocket and the substrate may serve to allow different substrates access to the same guide pocket.
It is unexpected that the pseudouridylation pocket can fit substrates with a stretch of unpaired nucleotides at the base of the upper stem of the hairpin (Fig. 4). We have shown that as many as four (and perhaps more) unpaired nucleotides can be positioned at the base of the upper stem without compromising pseudouridylation activity. However, pseudouridylation always occurs at the 5′-most unpaired uridine. While it is not exactly clear as to how the 5′-most uridine is preferred, crystal structure data from archaea shows that there is some extra space in the pseudouridylation pocket and catalytic center of pseudouridylase Cbf5 (Duan et al. 2009). This extra space would allow a long stretch of unpaired nucleotides to loop out without altering the structure or geometry of the pseudouridylation pocket and the catalytic center of Cbf5, thus keeping the enzyme active. With this flexibility, any given pseudouridylation pocket might have more than one substrate. Thus, for each known box H/ACA RNA, there are perhaps additional substrates that are yet to be identified.
MATERIALS AND METHODS
S. cerevisiae strains
46GB - MATa ade2, his3, trp1, leu2, met15, GAL+, cup1:: ura3, snR81::KAN, U2::loxP, [pCAU2 URA]
42GB - MATa ade2, his3, trp1, leu2, met15, GAL+, cup1:: ura3, U2::loxP, [pCAU2 URA]
DD-4741-1 - MATa his3, leu2, met15, ura3, snR85::KAN
Construction of mutant box H/ACA RNA and mutant U2 plasmids
Box H/ACA RNA genes (wild-type and mutant snR81 and snR85) were generated through four piece overlapping PCR with Sal1 and Pst1 restriction sites at the 5′ end and 3′ end, respectively (Wu et al. 2015). After PCR (30 cycles), the DNA products were purified using PCA [phenol/chloroform/isoamyl alcohol (25:24:1)] extraction and ethanol precipitation. The purified PCR products were digested with restriction enzymes and subsequently gel purified. The digested and purified PCR products were inserted into the Sal1 and Pst1 sites of the high-copy vector with a GPD promoter (YEPlac181, LEU 2 µm). Mutant yeast U2 plasmids were generated by site-directed mutagenesis using a plasmid containing the wild-type U2 snRNA (CEN4, HIS) as the template. The plasmids were transformed into E. coli (XL-10 Gold) and multiplied. After being isolated from E. coli, the plasmids were subsequently transformed into yeast cells.
Yeast cell transformation
The E. coli plasmids containing mutant snR81and snR85 box H/ACA RNAs were transformed into 46GB (ΔsnR81) and DD-4741-1 (ΔsnR85) yeast strains, respectively. Transformants were selected on synthetic solid media lacking leucine. The plasmids containing mutant U2 were transformed into yeast strains (42GB) where the SNR20 gene is deleted (lacking U2 snRNA) and wild-type U2 snRNA is supplied by a plasmid (pCAU2-URA). The wild-type U2 snRNA plasmid was shuffled out from each transformant containing a mutant U2 plasmid by 5-FOA selection.
RNA isolation
Ten milliliters of yeast cells growing in YPD (yeast extract, peptone, dextrose) or synthetic dextrose (SD) media lacking the corresponding amino acid were harvested at OD 2 for RNA isolation. RNA was isolated using TRIzol (Invitrogen). Briefly, 100 µL worth 0.5-mm acid washed glass beads from BioSpec products (cat. no. 11079105) were added to yeast cell pellets in 2 mL screw cap tubes. One milliliter of TRIzol was added, and the tubes were vigorously vortexed using beadbeater (30 sec, four times on ice). The lysates were spun at 15,600g for 5 min and the supernatants were added to 0.2 volume of chloroform. After vigorous vortexing (>1 min) and phase separation, the upper phase was extracted using 200 µL of PCA twice. RNA in the aqueous phase was then precipitated with 100% ethanol. After washing the RNA pellet with 75% ethanol, the pellet was air dried and dissolved in appropriate volume of double distilled H2O.
Pseudouridylation assay
For pseudouridylation assay, CMC modification followed by primer extension method was used. Briefly, 8–12 µg of total cellular RNA extracted above was dissolved in 20 µL of double distilled H2O, and then mixed with 20 µL of a 2× CMC (Sigma, cat. no. C106402) solution containing 0.34 M CMC, 7 M urea, 4 mM EDTA (pH 8.0–8.5), and 50 mM Bicine. The mixture was incubated at 37°C for 30 min. RNA was precipitated and washed with ethanol. The CMC-treated RNA pellet was resuspended in 50 µL of 50 mM sodium carbonate, pH 10.4, and incubated at 37°C for 1.5 h. RNA was again extracted by PCA and precipitated and washed by ethanol. The above RNA pellet was used for primer extension analysis. Briefly, the CMC-modified RNA pellet was resuspended in 4 µL of water, and mixed with 3 µL of annealing buffer (250 mM Tris-HCl, pH 8.3, 300 mM NaCl, 50 mM DTT) and 1 µL (100 nM) of 5′ 32P-labeled primer (5′ GAGTTTCCCCGTGTTGAG 3′, complementary to nucleotides 1190–1207 of 18S rRNA, for mapping Ψs within 18S rRNA; 5′ GTATTGTAACAAATTAAAAGG 3′, complementary to nucleotides 89–109 of U2 snRNA, for mapping Ψs within U2; 5′ GCCCACTAAAAGCTCTTC 3′, complementary to nucleotides 1201–1218 of 25S rRNA, for mapping Ψs within 25S rRNA). The mixture was heated at 90°C for 3 min, and immediately placed at room temperature for 15 min to allow annealing of the primer to RNA. The mixture was then chilled on ice, and 12 μL of reverse transcription (RT) mixture containing 18 mM Tris-HCl, pH 8.3, 21 mM NaCl, 3.6 mM DTT, 11 mM Mg(OAc)2, 0.72 mM dNTPs, and 0.25 unit of AMV reverse transcriptase (Promega), were added. The RT reaction was carried out at 37°C for 10 min followed by another 20 min at 42°C. The reaction was stopped by adding G50 buffer (20 mM Tris·Cl, pH 7.5, 300 mM sodium acetate, 2 mM EDTA, and 0.25% sodium dodecyl sulfate) followed by PCA extraction and ethanol precipitation. The pellet was resuspended in double distilled water and analyzed by denaturing gel electrophoresis and autoradiography.
ACKNOWLEDGMENTS
The authors would like to thank the members of the Yu laboratory for helpful discussions on the manuscript. This work was supported by grant GM104077 from the National Institutes of Health (to Y.-T.Y).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.066837.118.
REFERENCES
- Auffinger P, Westhof E. 1998. Location and distribution of modified nucleotides in tRNA. In Modification and editing of RNA (ed. Grosjean H, Benne R), pp. 569–576. ASM Press, Washington, DC. [Google Scholar]
- Balakin AG, Smith L, Fournier MJ. 1996. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834. [DOI] [PubMed] [Google Scholar]
- Bjork GR. 1995. Biosynthesis and function of modified nucleotides. In tRNA: structure, biosynthesis, and function (ed. Soll D, RajBhandary U), pp. 165–205. ASM Press, Washington, DC. [Google Scholar]
- Bortolin ML, Ganot P, Kiss T. 1999. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J 18: 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette M, Gray MW. 2000. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49: 341–351. [DOI] [PubMed] [Google Scholar]
- Cohn WE. 1959. 5-Ribosyl uracil, a carbon-carbon ribofuranosyl nucleoside in ribonucleic acids. Biochim Biophys Acta 32: 569–571. [DOI] [PubMed] [Google Scholar]
- Deryusheva S, Gall JG. 2018. Orchestrated positioning of post-transcriptional modifications at the branch point recognition region of U2 snRNA. RNA 24: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Zoysa MD, Yu YT. 2018. RNA-dependent pseudouridylation catalyzed by box H/ACA RNPs. Front Biol 3: 1–10. [Google Scholar]
- Duan J, Li L, Lu J, Wang W, Ye K. 2009. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell 34: 427–439. [DOI] [PubMed] [Google Scholar]
- Ganot P, Bortolin ML, Kiss T. 1997a. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809. [DOI] [PubMed] [Google Scholar]
- Ganot P, Caizergues-Ferrer M, Kiss T. 1997b. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev 11: 941–956. [DOI] [PubMed] [Google Scholar]
- Ge J, Yu YT. 2013. RNA pseudouridylation: new insights into an old modification. Trends Biochem Sci 38: 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, Sprinzl M, Steinberg S. 1995. Posttranscriptionally modified nucleosides in transfer RNA: their locations and frequencies. Biochimie 77: 139–141. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Phizicky EM. 2003. tRNA transfers to the limelight. Genes Dev 17: 162–180. [DOI] [PubMed] [Google Scholar]
- Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM, Thompson SR, et al. 2011. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell 44: 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Loria JP, Moore PB. 2007. Solution structure of an rRNA substrate bound to the pseudouridylation pocket of a box H/ACA snoRNA. Mol Cell 26: 205–215. [DOI] [PubMed] [Google Scholar]
- King TH, Liu B, McCully RR, Fournier MJ. 2003. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell 11: 425–435. [DOI] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Fournier MJ. 2009. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 15: 1716–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhao X, Yu YT. 2003. Pseudouridylation (Ψ) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J 22: 1889–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Yang C, Alexandrov A, Grayhack EJ, Behm-Ansmant I, Yu YT. 2005. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J 24: 2403–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden BE. 1990. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol 39: 241–303. [DOI] [PubMed] [Google Scholar]
- Massenet S, Mougin A, Branlant C. 1998. Posttranscriptional modifications in the U small nuclear RNAs. In Modification and editing of RNA (ed. Grosjean H, Benne R), pp. 201–228. ASM Press, Washington, DC. [Google Scholar]
- Massenet S, Motorin Y, Lafontaine DL, Hurt EC, Grosjean H, Branlant C. 1999. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol Cell Biol 19: 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573. [DOI] [PubMed] [Google Scholar]
- Ofengand J, Fournier M. 1998. The pseudouridine residues of rRNA: number, location, biosynthesis, and function. In Modification and editing of RNA (ed. Grosjean H, Benne R), pp. 229–253. ASM Press, Washington, DC. [Google Scholar]
- Reddy R, Busch H. 1988. Small nuclear RNAs: RNA sequences, structure, and modifications. In Structure and function of major and minor small nuclear ribonucleoprotein particles (ed. Birnstiel ML). Springer-Verlag, Berlin, Germany. [Google Scholar]
- Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 26: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Feigon J. 2007. H/ACA small nucleolar RNA pseudouridylation pockets bind substrate RNA to form three-way junctions that position the target U for modification. Proc Natl Acad Sci 104: 6655–6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Xiao M, Yang C, Yu YT. 2011. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J 30: 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Huang C, Yu YT. 2015. Pseudouridine in mRNA: incorporation, detection, and recoding. Methods Enzymol 560: 187–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Adachi H, Ge J, Stephenson D, Query CC, Yu YT. 2016. Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. EMBO J 35: 654–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang C, Schattner P, Yu YT. 2009. Functionality and substrate specificity of human box H/ACA guide RNAs. RNA 15: 176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, McPheeters DS, Yu YT. 2005. ψ35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae. J Biol Chem 280: 6655–6662. [DOI] [PubMed] [Google Scholar]
- Yu YT, Meier UT. 2014. RNA-guided isomerization of uridine to pseudouridine–pseudouridylation. RNA Biol 11: 1483–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Terns RM, Terns MP. 2005. Mechanisms and functions of RNA-guided RNA modification. In Fine-tuning of RNA functions by modification and editing (ed. Grosjean H), pp. 223–262. Springer-Verlag, Berlin, Germany. [Google Scholar]
- Zhao X, Yu YT. 2004. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA 10: 681–690. [DOI] [PMC free article] [PubMed] [Google Scholar]