Abstract
Several lines of evidence indicate that during transformation epithelial cancer cells can acquire mesenchymal features via a process called epithelial-to-mesenchymal transition (EMT). This process endows cancer cells with increased invasive and migratory capacity, enabling tumour dissemination and metastasis. EMT is associated with a complex metabolic reprogramming, orchestrated by EMT transcription factors, which support the energy requirements of increased motility and growth in harsh environmental conditions. The discovery that mutations in metabolic genes such as FH, SDH and IDH activate EMT provided further evidence that EMT and metabolism are intertwined. In this review, we discuss the role of EMT in cancer and the underpinning metabolic reprogramming. We also put forward the hypothesis that, by altering chromatin structure and function, metabolic pathways engaged by EMT are necessary for its full activation.
Background
In the last decades, cancer research uncovered the many enabling features of tumours cells [1]. Among these, activation of epithelial-to-mesenchymal transition (EMT), a process where epithelial cancer cells acquire mesenchymal features, is emerging as key determinant of cancer cell invasion and metastasis [2–4]. To metastasise, cancer cells acquire the ability to erode the extracellular matrix, the motility to extravasate into the blood stream, and the plasticity to grow in a different tissue. In all these phases, nutrient supply can be limited and cancer cells experience varying degree of stress [5]. Accordingly, metastatic cells fine-tune their metabolism to adapt to the ever-changing environment [6, 7]. In line with this observation, part of the genetic reprogramming orchestrated by EMT affects the expression of metabolic genes, regulating glucose, lipids, glutamine, and nucleotide metabolism. Yet, to what extent EMT rewires the metabolic network is still unclear. The recent discovery that oncogenic mutations of metabolic enzymes such as fumarate hydratase (FH), succinate dehydrogenase (SDH) and isocitrate dehydrogenase (IDH) drive EMT [8–10] indicates that the connection between EMT and metabolism is deeper than anticipated. Indeed, these works revealed that components of the metabolic network can directly affect chromatin structure and function, impinging on signalling cascades required for the full activation of EMT [11, 12]. In this review, we describe the role of EMT in tumorigenesis, how EMT affects metabolism, and how, in turn, dysregulation of metabolic genes affect the execution of EMT.
The epithelial-to-mesenchymal transition in cancer
In tissues, epithelial cells are organised in compact layers anchored to the basal lamina. During transformation, some of these cells lose their epithelial features and acquire a mesenchymal phenotype through a process defined as epithelial-to-mesenchymal-transition (EMT). This process is characterised by profound transcriptional [13] and epigenetic changes [14, 15] that lead to the loss of cell-to-cell junctions and the acquisition of a motile and migratory phenotype, enabling the invasion of the basal lamina, which eventually may lead to metastasis. At the molecular level, EMT is dictated by a network of transcription factors (EMT-TFs) that directly or indirectly represses one of the key epithelial markers, E-Cadherin [13, 16]. These EMT-TFs belongs to various family of chromatin interacting family of proteins, including Snail (Snai1 and Snai2), bHLH (Twist1 and Twist2), and zinc finger and E-box binding (Zeb1 and Zeb2). Cross-activation of EMT by other oncogenic stimuli and the identification of non-canonical EMT-TFs such as Kruppel-like-factor (KLF8), the homebox proteins goosecoid (GSC) or fork-head protein (FOXC2), contributes to the great complexity of EMT regulation [13, 16]. Moreover, recent evidence has shown that microRNAs are also potent regulators of EMT, affecting the expression of multiple targets of this cascade [17].
The role of the EMT-TFs in invasion and metastasis has been extensively investigated [16]. In vivo experiments using a spontaneous squamous cell carcinoma mouse model showed that the expression of the EMT-TF Twist1 is sufficient to trigger EMT and the subsequent dissemination of cancer cells into the blood stream. Interestingly, the colonisation of target tissues by these metastatic cells is driven by a mesenchymal-to-epithelial transition (MET) and requires suppression of Twist1 [18]. Other works identified a primary role of the EMT in breast cancer progression. For instance, Twist1 controls the ability of aggressive breast 4T1 cells to migrate in vitro and to metastasise to the lung in vivo [19]. The role of Twist1 in early dissemination and metastasis was also corroborated in human epidermal growth factor receptor 2 (Her2)-positive mammary cancer cells. It was shown that in early lesions in mouse breast, a subpopulation of cells that express high levels of Twist1, low levels of E-cadherin, and markers of Wnt signalling activation, invade the adjacent tissue and lead to early dissemination and subsequent mestastasis [20]. Moreover, in mouse skin squamous cell carcinoma, Twist1 is required in both early and late stages of tumour progression in a gene dosage- dependent manner [21]. Other EMT-TF are directly involved in breast cancer metastasis. For instance, the expression of Snai1 in a mouse model of breast cancer activates the dissemination of cancer cells and its deletion dramatically impairs the formation of metastasis [22]. The impact of SNAI1 activation in the malignancy of breast tumours has been further confirmed by the discovery that the discoidin domain receptor 2 (DDR2), a protein expressed in ductal breast carcinomas, drives invasion in vitro and metastasis in vivo through the nuclear stabilisation of Snai1, via phosphorylation mediated by extracellular related kinase 2 (ERK2) [23]. Even though a series of convincing works established the involvement of EMT in metastasis formation, its real importance in tumour evolution is still questioned. For instance, two groups recently showed that the EMT is dispensable for metastasis in a model of pancreatic [24] and breastcancer [25]. These results suggest that the role of EMT in cancer progression is likely tissue-specific and that it might be implicated in other features of cancer. Indeed, it has recently emerged that EMT, via the expression of EMT-TFs, enables stemness in cancer cells [2, 16]. For instance, an orchestrated signal mediated by SNAI2 and SOX9 induces a stem state and promotes tumorigenesis in mammary luminal cells [26], while the ectopic expression of TWIST1 or SNAI1 results in the expression of stem markers in human immortalised mammary cells [27]. Moreover, ZEB1-mediated suppression of miR200 favours the expression of polycomb repressor protein Bmi1 [28, 29] and Suz12 [30], two regulators of self-renewal and stemness in breast cells. Further work showed that the acquisition of stem-like properties through EMT activation is involved, at least in part, in both chemoresistance [31, 32] and tumour dormancy [31, 33–35]. These two prominent features of cancer therapy may be interlinked. Seminal work using an elegant in vivo model to trace EMT lineage during metastasis showed that EMT-positive cells are responsible for recurrence of lung metastasis after chemotherapy with cyclophosphamide, suggesting that chemoresistance, EMT and dormancy may be part of the same pathway [25].
EMT activation induces a metabolic rewiring
Recent findings indicate that mesenchymal cancer cells have different metabolic needs compared their epithelial counterparts, to satisfy the metabolic demands of increased motility and invasion. Yet, how EMT regulates metabolism is still poorly understood. In the effort to corroborate this link, Shaul and colleagues analysed the expression of metabolic genes in high-grade carcinomas expressing mesenchymal markers using publically available data from almost 1000 cancer cell lines. They found that these mesenchymal cells exhibit high expression levels of 44 metabolic genes. These genes were found upregulated also upon induction of EMT by expression of Twist1 in human mammary epithelial cells. Among these enzymes, Dihydropyrimidine dehydrogenase (DPYD), an enzyme involved in pyrimidine catabolism, was required for EMT, both in vitro and in vivo [36] (Figure 1). Importantly, exogenous dihydropyrimidines are sufficient to rescue EMT after silencing of DPYD, suggesting that these metabolites are a limiting factor during the EMT. However, the how they regulate EMT is currently unknown.
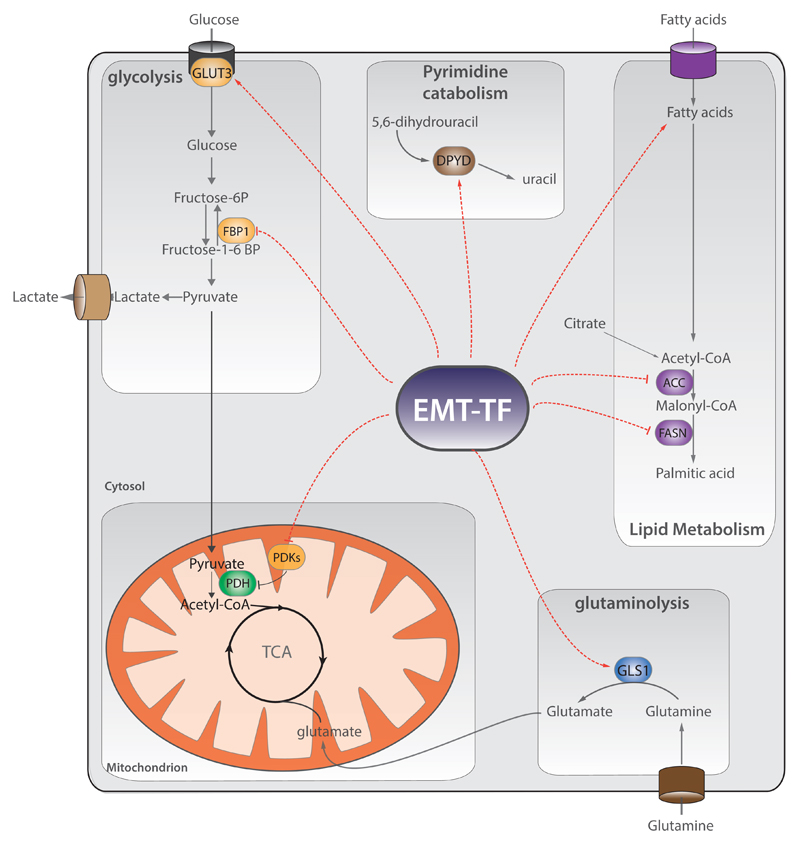
Fig.1. EMT controls metabolic reprogramming.
EMT transcription factors (EMT-TFs) control the expression of metabolic genes of different pathways such as glycolysis, lipid metabolism, and mitochondrial metabolism, and glutaminolysis. Specifically, EMT-TFs suppress the expression of fructose-1,6-bisphosphatase 1 (FBP1), fatty acid synthase (FASN), acetyl-coA carboxylase (ACC), nucleoside transporter, and pyruvate dehydrogenase kinase 4 (PDK4), whilst enhance the expression of dihydropyrimidine dehydrogenase (DPYD), glutaminase 1 (GLS1), enzymes of glutathione metabolism, cytochrome P450, aldehyde dehydrogenases, and glucose transporter 3 (GLUT3). Red dashed arrows indicate the metabolic nodes regulated by EMT-TFs. TCA=tricarboxylic acid cycle.
Overall, these results suggested that metabolic rewiring is required to complete the reprogramming orchestrated by EMT. In further support of these findings, it was found that SNAI1 expression represses the glycolytic enzyme fructose-1,6-bisphosphatase 1 (FBP1), favouring glucose uptake and the diversion of glycolytic carbons towards biosynthetic pathways, including the pentose phosphate shunt (Figure 1). Interestingly, FBP1 loss impairs respiration and the activity of respiratory chain complex I [37]. Activation of glycolysis by EMT was also observed in breast and prostate cancer cells, where it is required for both cytoskeleton remodelling and increasing cell traction [38]. Glycolysis is targeted by EMT also in non-small cell lung cancer cells (NSCLC), where ZEB1 activate the expression of glucose transporter 3 (GLUT3) [39]. However, the metabolic reprogramming upon EMT in NSCLC is controversial. For instance, the treatment of NSCLC with TGF-β induces a shift from glycolysis to oxidative phosphorylation (OXPHOS) and leads to an overall increase in amino acids, in particular in glutamate, via a higher flux of carbons through the Tricarboxylic acid (TCA) cycle. Mechanistically, this shift from glycolysis to OXPHOS is achieved by a selective repression of pyruvate dehydrogenase kinase 4 (PDK4) during EMT [40]. Finally, EMT induction by TGF-β in colon cancer cells elicits the nuclear translocation of pyruvate kinase M2 (PKM2) and the silencing of PKM2 prevents EMT triggering by TGF-β in these cells [41] (Figure 1).
Other metabolic pathways are targeted during EMT, including lipid metabolism (Figure 1). For example, EMT activation by either TNFα or TGF-β favours the accumulation of unsaturated triacylglycerides in DU145 prostate cancer cells [42]. Furthermore, the activation of EMT by overexpression of SNAI1 suppresses transcriptional regulators of the lipogenesis carbohydrate-responsive element binding protein (ChREBP) leading to the silencing of both fatty acid synthase (FASN) and acetyl-CoA carboxylase (ACC) [43]. Finally, another pathway required during EMT is glutaminolysis (Figure 1): lung cancer cells that undergo an EMT become increasingly sensitive to Glutaminase-1 (GLS1) inhibitors [44].
As discussed above, EMT activation is involved in both chemoresistance and tumour dormancy. Even though the role of metabolism in these processes is largely unknown, recent works suggest that metabolic rewiring can be important in both chemoresistance and tumour dormancy. For instance, EMT-positive breast cells that are responsible for recurrent lung metastasis after chemotherapy increased the expression of metabolic enzymes such as drug transporters, aldehyde dehydrogenase (ALDHs), cytochrome P450s, and enzymes of glutathione metabolism [25] (Figure 1). Likely, these metabolic changes protect the cells from oxidative stress experienced during therapy. Furthermore, deletion of Twist1 or Snai1 in chemoresistant pancreatic cancer cells increase the expression of a nucleosides transporter, which leads to increase uptake of the anticancer drug gemcitabine [24]. The link between EMT, metabolic alterations, and tumour dormancy remains mainly indirect. It is widely known that during tumour dormancy, cancer cells undergo proliferative arrest and enter quiescence [34]. Therefore, it not surprising that this change in proliferation rate is accompanied by a metabolic rewiring. For instance, pancreatic ductal cancer cells surviving after oncogene ablation acquire stem-like traits and are dependent on oxidative phosphorylation for survival [45]. In addition, quiescent leukaemia stem cells (LSC) rely on mitochondrial metabolism: targeting the oxidative phosphorylation through BCL-2 inhibition is sufficient to eradicate LSC population [46]. However, the impact of EMT-TFs in regulating these metabolic alterations during dormancy is largely unknown and it might be related to the dynamic shift between EMT and MET that occurs on tumour circulating cells [47].
Overall, these results suggest that metabolic reprogramming is instrumental to the phenotypic shift observed during the EMT. Whether these metabolic changes are simply required to fulfil the energy requirements of more aggressive cells or to support some of the signalling cascades involved in this process is still unknown.
Metabolic reprogramming activates the epithelial- to-mesenchymal transition
Recent evidence suggests that the link between EMT and metabolism is mutual and, in some circumstances, alterations of metabolism can drive EMT. The next part of the review describes how the dysregulation of metabolic pathways is associated with EMT induction. These findings are summarised in Figure 2.
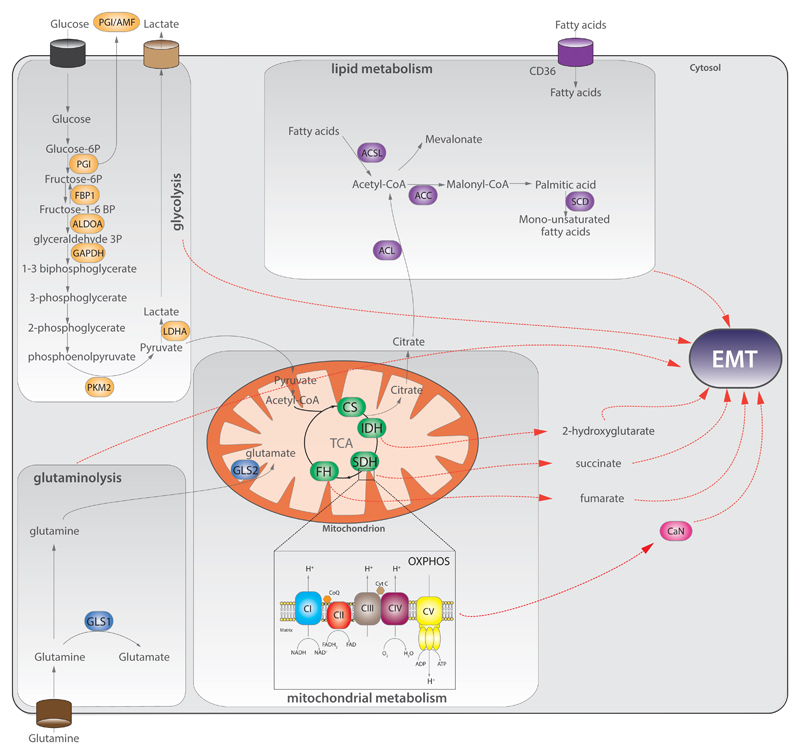
Fig.2. Metabolic genes control EMT.
Aberrant expression of metabolic enzymes of glycolysis (orange), lipid metabolism (purple), glutaminolysis (blue), mitochondrial metabolism (green), leads to EMT. Red dashed arrows indicate the link between specific metabolic pathway/metabolites and EMT. ACC=acetyl-CoA carboxylase; ACL=ATP citrate lyase; ACSL=acetyl-CoA synthetase; ALDOA=aldolase A; CaN=calcineurin A; CI-CV=respiratory chain complexes I-V; CoQ=coenzyme Q; CS=citrate synthase; CytC=cytochrome C; FBP1=fructose-1,6-bisphosphatase 1; FH=fumarate hydratase; GAPDH=glyceraldehyde-3-phosphate dehydrogenase; GLS=glutaminase; IDH=isocitrate dehydrogenase; LDHA=lactic dehydrogenase A; PGI =phosphoglucose isomerase; PKM2=pyruvate kinase M2; SCD=steroyl-CoA desaturase; SDH=succinate dehydrogenase.
Glycolysis
Aerobic glycolysis is the most distinctive metabolic alteration of cancer cells [1, 48] but the role of glycolytic enzymes in the induction of EMT has emerged only in the last years. Phosphoglucose isomerase (PGI) is a glycolytic enzyme that converts glucose-6P to fructose 6-P. This enzyme was found to be secreted by cancer cells and to act as cytokine, taking the name of autocrine motility factor (AMF). Overexpression of PGI/AMF causes a NF-kB-dependent stabilisation of ZEB1 and ZEB2 in breast cancer cells [49] and ectopic expression in normal epithelial breast MCF10A triggers EMT [50]. Importantly, suppression of PGI/AMF leads to reverse MET in lung fibrosarcoma [51] and endometrial cancer cells [52]. As described above, the expression of the glycolytic enzyme fructose-1,6-biphosphatase (FBP1) blocks the induction of EMT mediated by SNAI1 in luminal breast cells. The silencing of FBP1 favours EMT also in gastric cells in vitro [53]. Other glycolytic enzymes are involved in EMT induction. For instance, the silencing of Aldolase A (ALDOA), an enzyme that converts fructose-1,6-bisphosphate to glyceraldehydes-3-phosphate and hydroxy-acetone, impairs lung squamous carcinoma cell motility and tumorigenesis and this phenomenon is associated with repression of mesenchymal markers [54]. Furthermore, silencing of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) inhibits EMT by repressing SNAI1 in colon cancer [55]. Finally, overexpression of lactate dehydrogenase (LDH), the enzyme that converts pyruvate to lactate, leads to increased migration and invasion in bladder cancer cells [56].
Mitochondrial metabolism
Mitochondrial dysfunction is a key feature of cancer and has been frequently associated with increased aggressiveness and metastatic potential [57, 58]. Yet, the mechanistic link between mitochondrial dysfunction and EMT have only recently been investigated. In 2014 it was shown that mitochondrial dysfunction induced by depletion of mitochondrial DNA in breast cells leads to profound morphological and molecular changes that resembles EMT, including increased expression of EMT-TFs, metalloproteases and suppression of E-cadherin, triggered by a Calcineurin A (CaN)-dependent mechanism [59]. In support of this finding, we recently found that the downregulation of mitochondrial genes is a common feature of highly aggressive cancers, and that it significantly correlates with the activation of EMT across 21 different types of cancer [60]. More recently, we and others have demonstrated that EMT is a key signature of tumours harbouring mutations in the TCA cycle enzymes FH, SDH and IDH [8–10].
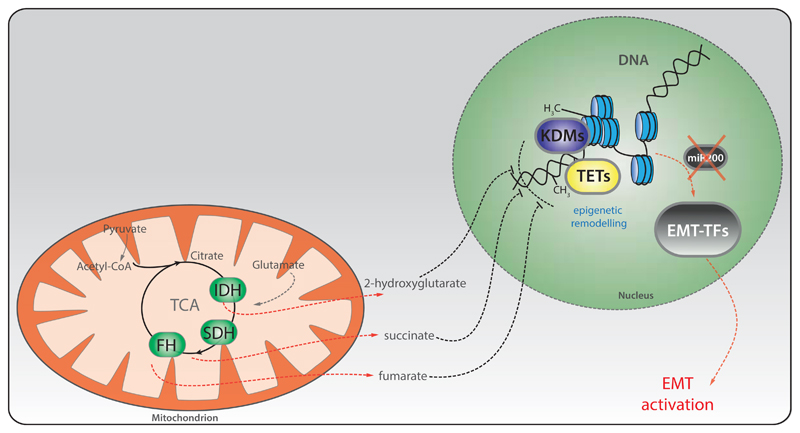
Fumarate hydratase is the enzyme that converts fumarate to malate. Mutations of this enzyme lead to Hereditary Leiomyomatosis and Renal Cell Cancer (HLRCC) [61] and other tumour types, including paragangliomas and pheochromocytomas [62, 63], whilst FH deletions have been found in neuroblastoma [64]. FH-mutant renal tumours are highly aggressive and metastasise even when small [65]. However, the mechanisms underpinning this aggressiveness are still under investigation. We recently demonstrated that FH-deficient cells exhibit a striking mesenchymal phenotype, linked with the expression of an EMT signature [8]. The link between FH and EMT was also observed in nasopharyngeal carcinoma, where FH is transcriptionally repressed by the lymphoid-specific helicase (LSH) [66]. Mechanistically, we found that fumarate, which accumulates in FH-deficient cells and tumours, is responsible for the induction of EMT by inhibiting the Ten-Eleven Translocation (TET)-dependent demethylation of the anti-metastatic microRNA miR200 [8], known inhibitors of both SNAI2 [67] and ZEB1 [68] (Figure 3).
Fig.3. EMT activation by mutations in FH, SDH and IDH requires epigenetic reprogramming.
Schematic representation of how mitochondrial metabolites accumulated upon mutation of the indicated TCA cycle enzymes activate the EMT. A common pathway affected by these metabolites is the epigenetic suppression of a family of antimetastatic microRNAs, miR200, via the inhibition of histone demethylases (KDMs) and DNA demethylases (TETs). Of note, in the case of 2HG, the suppression of miR200 is indirect, and occurs via activation of Zeb1/2. See the text for more details. FH=fumarate hydratase; SDH=succinate dehydrogenase; IDH=isocitrate dehydrogenase.
Another TCA cycle enzyme implicated in EMT is Succinate dehydrogenase (SDH), a component of the respiratory chain that converts succinate to fumarate. SDH mutations have been described in pheochromocytomas and paragangliomas [69–72], sporadic renal cancer [73] and gastrointestinal stromal tumours [74, 75]. A recent study revealed that human metastatic pheochromocytomas and paragangliomas harbouring SDHB mutations are invasive and exhibit activation of EMT–TFs such as SNAI1 and SNAI2, suggesting the induction of EMT in these tumours [10]. Consistently, it was shown that loss of SDHB in chromaffin cells induces these EMT-TFs and leads to the epigenetic silencing of keratin-19 [76, 77]. Importantly, the migratory phenotype of these cells is reversed by the use of a DNA methylation inhibitor, decitabine. The link between SDHB deficiency and EMT was also shown in colorectal cancer, where the silencing of SDHB promotes cell migration and invasion in a TGF-β/SNAI1-mediated-process [78], and also in ovarian cancer [79]. Finally, loss of the assembly factor SDH5 [80], induces EMT in lung cancer cells and metastasis in-vivo through activation of a glycogen-synthase kinase (GSK-3β)-β-catenin axis [81]. Although these studies did not focus on the accumulation of succinate as a mediator of EMT, we recently found that succinate, similarly to fumarate, can induce the epigenetic suppression of miR200 and subsequent EMT induction in Sdhb-deficient epithelial kidney cells [8] (Figure 3).
Other TCA cycle enzymes recently appeared in the spotlight of cancer biology and EMT are Isocitrate Dehydrogenases (IDHs), enzymes involved in the oxidative decarboxylation of isocitrate to alpha-ketoglutarate (aKG). Three isoforms of IDH have been identified: cytosolic IDH1 and mitochondrial IDH2 are NADP+-dependent enzymes, while mitochondrial IDH3 is a NAD+-dependent protein. Heterozygous mutations in either IDH1 or IDH2 have been found in gliomas and leukaemia [82–84]. IDH1 and IDH2 mutations are neomorphic and lead to the production of 2-hydroxyglutarate (2HG), which was shown to induce EMT. Similar to what was described for FH and SDH deficient cells, EMT in IDH-mutant cells is driven by alterations of the miR200-Zeb1 axis (Figure 3). This phenomenon was observed in breast tumours [9], and in colorectal cancer cells [9, 85].
Finally, another TCA cycle enzyme associated with EMT is citrate synthase (CS), the enzyme that catalyses the first committed step of the TCA cycle. Silencing of CS induces morphological and molecular changes in human cervical carcinoma cells that resemble EMT, and promotes metastasis in vivo. The molecular mechanisms responsible for this phenotype are not clear, but it is possible that the mitochondrial dysfunction observed in these cells is involved [86]. However, more recent experiments indicate that CS is upregulated in other tumour types such as ovarian cancers and that its silencing impairs both motility and invasion of tumour cells in vitro [87]. Therefore, the role of CS in tumour progression is still unclear and it might be tissue-dependent.
Lipid metabolism
Several recent reports support the connection between lipid metabolism and EMT. For instance, the overexpression of acetyl-CoA synthetase (ACSL1 and ACSL4) and steroyl-CoA desaturase (SCD) can activate EMT in colorectal cancer, leading to increased migration, invasion and colony formation in vitro. Importantly, the expression of these three enzymes is associated with poor prognosis in stage II colorectal cancer patients [88]. In addition, elevated fatty acid uptake via CD36 activates a Wnt-dependent EMT in hepatocellular carcinoma (HCC) [89]. Of note, in human oral cancer cells CD36-positive cells are responsible for cancer initiation and metastasis in vivo. However, in the latter model the EMT is not involved in the formation of metastasis [90]. Other enzymes of lipid metabolism have been identified as EMT regulators. For instance, silencing of ATP citrate lyase (ACL) reverses EMT in lung cancer and impairs stemness in both lung and breast cells by SNAI1 repression [91]. Moreover, silencing of acetyl-CoA carboxylase 2 (ACC2) reverted the EMT transition triggered by glucose stress, triglyceride deposit and malonyl-CoA accumulation in kidneys [92]. Interestingly, treatment of cancer cells with fatty acids such as arachidonic or linoleic acid elicits an EMT that is downstream of the oncogenic cascades mediated by SRC, NF-kB and FAK [93, 94].
Glutaminolysis
Most cancer cells depend on glutamine utilisation [48], and the role of glutaminolysis in EMT has been recently investigated. The inhibition of glutaminolysis by targeting GLS1 impairs in vivo metastasis through repression of SNAI1 [95]. On the contrary, the expression of GLS2, the mitochondrial isoform of glutaminase, inversely correlates with stage, tumour size, and prognosis in HCC. However, this phenomenon is independent of GLS2 glutaminase activity and involves the GLS2-mediated stabilisation of the EMT-related microRNA miR-34a via the Dicer complex [96]. These results suggest that the effects of glutamine catabolism on EMT might be context-dependent and more work is necessary to elucidate the importance of glutaminolysis in this process.
Conclusions and future perspective
EMT is a fundamental biological process involved in development, fibrosis, and wound healing [4]. Recent evidence indicates that this process is also involved in tumour initiation and metastasis. EMT elicits a complex phenotypic switch that endows cancer cells with ability to survive during invasion, dissemination, and metastasis. This flexibility is achieved at least in part by the rewiring of the metabolic network. As discussed above, EMT, via EMT-TFs, orchestrates profound metabolic changes that allow the cell to sustain the energy needs of a cancer cell in an ever-changing tumour micro environment. Yet, the role of metabolism in EMT seems to go beyond these simple enabling features. Indeed, the observation that dysregulation of cellular metabolism, in some circumstances, drives EMT indicates that parts of the metabolic network could act as a core component of the signalling cascade elicited by the EMT (Figure 4). The data discussed in this review corroborate this hypothesis and indicate that specific metabolic alterations could lead to chromatin changes that are required for the activity of EMT-TFs. Several questions arise. For instance, it is still unclear why different sources of mitochondrial dysfunction converge on EMT. In an interesting parallel, EMT induction is associated with bypass of oncogene –induced senescence [97]. Given that senescence is a common outcome of metabolic stress [98] it is possible that induction of EMT could provide cells with the sufficient plasticity to survive and proliferate in the presence of metabolic defects or under nutrient stress. In this scenario, metastasis could be seen as a strategy to explore novel, and more favourable, metabolic niches, and increased motility the means to this goal. Another outstanding question in the field is to what extent the EMT observed in metabolically-impaired cells contributes to tumorigenesis. The fact that EMT is the most enriched gene signature in FH and SDH-deficient cells seems to support a driving role of EMT in these tumours. It would be important to validate this hypothesis by assessing tumorigenesis in FH- or SDH-deficient models where EMT-TFs are ablated. Finally, the fact that EMT shows unexpected metabolic facets offers interesting therapeutical perspectives (Fig.4). Indeed, EMT could be potentially reverted by targeting specific metabolic enzymes, or targeting the metabolism-dependent epigenetic reprogramming, eventually limiting cancer metastasis. Consistently, inhibitors of mutant IDH were shown to revert glioma cells to a more differentiated state [99], and the DNA methylation inhibitor, decitabine, impairs the invasive phenotype of SDH-deficient cells [77]. Along this strategy, a recent screening was designed to identify small molecules that could revert the mesenchymal phenotype of cancer cells activating E-cadherin transcription. Interestingly, it was found that protein kinase A (PKA) activation by increasing cyclic AMP (cAMP) levels, is sufficient to trigger a mesenchymal-to-epithelial transition (MET) in aggressive breast cancer cells, through activation of the histone demethylases PHF2. cAMP is a key second messenger and its levels are tightly controlled by the energy state of cells [100]. Therefore, it is tempting to speculate that metabolic alterations, through regulation of cAMP levels, are necessary for full EMT activation and that altering metabolism could be a tempting strategy to modify cell phenotype and, more importantly, aggressive features of cancer.
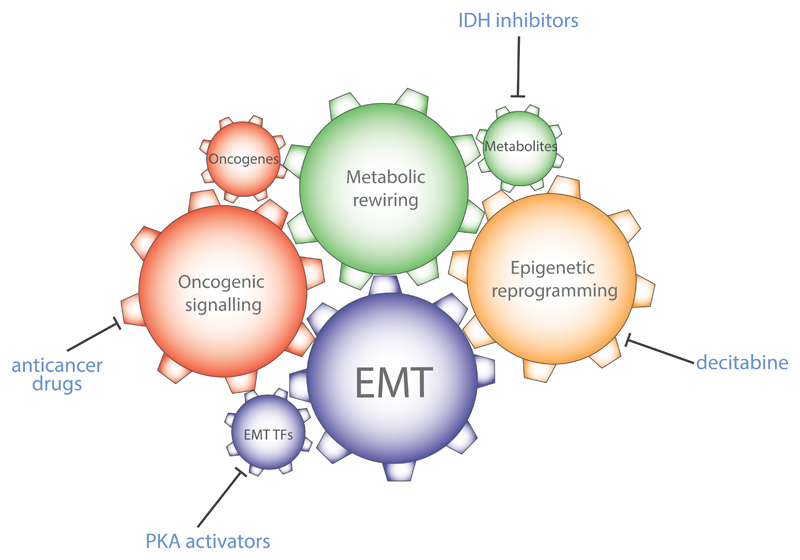
Fig.4. Integration between oncogenic signalling, metabolic transformation, and epigenetic reprogramming during EMT.
EMT requires the coordinated activation of multiple cellular processes, here represented as gears within a clockwork. Each of these components are essential for the full activation of EMT. As consequence, the inhibition of parts of this clockwork hampers the full activation of the EMT. For instance, inhibition of mutant IDH, or activation of PKA can block EMT. PKA=protein kinase A; IDH=isocitrate dehydrogenase.
Overall, in this review we provided compelling evidence that EMT and metabolism are intertwined. Understanding the underpinning molecular determinants of this relation is revealing novel insights into how tumours are formed and disseminate, and will potentially provide novel targets for targeting metastasis, the major killer in cancer.
Footnotes
Competing interests
The authors declare no competing interests.
Authors’ contribution
MS and CF jointly wrote the manuscript.
Authors’ information
MS is a Research Associate in the laboratory of CF. CF is a group leader at the MRC Cancer Unit, University of Cambridge, Cambridge, UK. MS and CF are funded by an MRC Core Funding to the MRC Cancer Unit.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. S0092-8674(11)00127-9 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Ye X, Weinberg RA. Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. S0962-8924(15)00145-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. 39104 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. S0092-8674(16)30796-6 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Vanharanta S, Massague J. Origins of metastatic traits. Cancer Cell. 2013;24:410–421. doi: 10.1016/j.ccr.2013.09.007. S1535-6108(13)00417-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porporato PE, Payen VL, Baselet B, Sonveaux P. Metabolic changes associated with tumor metastasis, part 2: Mitochondria, lipid and amino acid metabolism. Cell Mol Life Sci. 2016;73:1349–1363. doi: 10.1007/s00018-015-2100-2. 10.1007/s00018-015-2100-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payen VL, Porporato PE, Baselet B, Sonveaux P. Metabolic changes associated with tumor metastasis, part 1: tumor pH, glycolysis and the pentose phosphate pathway. Cell Mol Life Sci. 2016;73:1333–1348. doi: 10.1007/s00018-015-2098-5. 10.1007/s00018-015-2098-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sciacovelli M, Goncalves E, Johnson TI, Zecchini VR, da Costa AS, Gaude E, Drubbel AV, Theobald SJ, Abbo SR, Tran MG, et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature. 2016;537:544–547. doi: 10.1038/nature19353. nature19353 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grassian AR, Lin F, Barrett R, Liu Y, Jiang W, Korpal M, Astley H, Gitterman D, Henley T, Howes R, et al. Isocitrate dehydrogenase (IDH) mutations promote a reversible ZEB1/microRNA (miR)-200-dependent epithelial-mesenchymal transition (EMT) J Biol Chem. 2012;287:42180–42194. doi: 10.1074/jbc.M112.417832. M112.417832 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loriot C, Burnichon N, Gadessaud N, Vescovo L, Amar L, Libe R, Bertherat J, Plouin PF, Jeunemaitre X, Gimenez-Roqueplo AP, et al. Epithelial to mesenchymal transition is activated in metastatic pheochromocytomas and paragangliomas caused by SDHB gene mutations. J Clin Endocrinol Metab. 2012;97:E954–962. doi: 10.1210/jc.2011-3437. jc.2011-3437 [pii] [DOI] [PubMed] [Google Scholar]
- 11.Sciacovelli M, Frezza C. Oncometabolites: Unconventional triggers of oncogenic signalling cascades. Free Radic Biol Med. 2016;100:175–181. doi: 10.1016/j.freeradbiomed.2016.04.025. S0891-5849(16)30043-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nowicki S, Gottlieb E. Oncometabolites: tailoring our genes. FEBS J. 2015;282:2796–2805. doi: 10.1111/febs.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. nrm3758 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. nm.3336 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol. 2011;18:867–874. doi: 10.1038/nsmb.2084. nsmb.2084 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. ncb2976 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Lamouille S, Subramanyam D, Blelloch R, Derynck R. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell Biol. 2013;25:200–207. doi: 10.1016/j.ceb.2013.01.008. S0955-0674(13)00009-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. S1535-6108(12)00400-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. S0092867404005768 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R, Avivar-Valderas A, Nagi C, Girnius N, Davis RJ, et al. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature. 2016 doi: 10.1038/nature20609. nature20609 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beck B, Lapouge G, Rorive S, Drogat B, Desaedelaere K, Delafaille S, Dubois C, Salmon I, Willekens K, Marine JC, et al. Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell. 2015;16:67–79. doi: 10.1016/j.stem.2014.12.002. S1934-5909(14)00560-8 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Tran HD, Luitel K, Kim M, Zhang K, Longmore GD, Tran DD. Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Res. 2014;74:6330–6340. doi: 10.1158/0008-5472.CAN-14-0923. 0008-5472.CAN-14-0923 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang K, Corsa CA, Ponik SM, Prior JL, Piwnica-Worms D, Eliceiri KW, Keely PJ, Longmore GD. The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat Cell Biol. 2013;15:677–687. doi: 10.1038/ncb2743. ncb2743 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. nature16064 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. nature15748 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. S0092-8674(12)00165-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. S0092-8674(08)00444-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. S0092-8674(09)00850-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. ncb1998 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–772. doi: 10.1016/j.molcel.2010.08.013. S1097-2765(10)00623-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du B, Shim JS. Targeting Epithelial-Mesenchymal Transition (EMT) to Overcome Drug Resistance in Cancer. Molecules. 2016;21 doi: 10.3390/molecules21070965. E965 [pii] molecules21070965 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vidal SJ, Rodriguez-Bravo V, Galsky M, Cordon-Cardo C, Domingo-Domenech J. Targeting cancer stem cells to suppress acquired chemotherapy resistance. Oncogene. 2014;33:4451–4463. doi: 10.1038/onc.2013.411. onc2013411 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Patel P, Chen EI. Cancer stem cells, tumor dormancy, and metastasis. Front Endocrinol (Lausanne) 2012;3:125. doi: 10.3389/fendo.2012.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giancotti FG. Mechanisms governing metastatic dormancy and reactivation. Cell. 2013;155:750–764. doi: 10.1016/j.cell.2013.10.029. S0092-8674(13)01347-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brabletz T. To differentiate or not--routes towards metastasis. Nat Rev Cancer. 2012;12:425–436. doi: 10.1038/nrc3265. nrc3265 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Shaul YD, Freinkman E, Comb WC, Cantor JR, Tam WL, Thiru P, Kim D, Kanarek N, Pacold ME, Chen WW, et al. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell. 2014;158:1094–1109. doi: 10.1016/j.cell.2014.07.032. S0092-8674(14)00982-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, Lin Y, Yao J, Shi J, Kang T, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–331. doi: 10.1016/j.ccr.2013.01.022. S1535-6108(13)00042-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiraishi T, Verdone JE, Huang J, Kahlert UD, Hernandez JR, Torga G, Zarif JC, Epstein T, Gatenby R, McCartney A, et al. Glycolysis is the primary bioenergetic pathway for cell motility and cytoskeletal remodeling in human prostate and breast cancer cells. Oncotarget. 2015;6:130–143. doi: 10.18632/oncotarget.2766. 2766 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masin M, Vazquez J, Rossi S, Groeneveld S, Samson N, Schwalie PC, Deplancke B, Frawley LE, Gouttenoire J, Moradpour D, et al. GLUT3 is induced during epithelial-mesenchymal transition and promotes tumor cell proliferation in non-small cell lung cancer. Cancer Metab. 2014;2:11. doi: 10.1186/2049-3002-2-11. 2049-3002-2-11 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y, Daemen A, Hatzivassiliou G, Arnott D, Wilson C, Zhuang G, Gao M, Liu P, Boudreau A, Johnson L, et al. Metabolic and transcriptional profiling reveals pyruvate dehydrogenase kinase 4 as a mediator of epithelial-mesenchymal transition and drug resistance in tumor cells. Cancer Metab. 2014;2:20. doi: 10.1186/2049-3002-2-20. 136 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamabe A, Konno M, Tanuma N, Shima H, Tsunekuni K, Kawamoto K, Nishida N, Koseki J, Mimori K, Gotoh N, et al. Role of pyruvate kinase M2 in transcriptional regulation leading to epithelial-mesenchymal transition. Proc Natl Acad Sci U S A. 2014;111:15526–15531. doi: 10.1073/pnas.1407717111. 1407717111 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalmau N, Jaumot J, Tauler R, Bedia C. Epithelial-to-mesenchymal transition involves triacylglycerol accumulation in DU145 prostate cancer cells. Mol Biosyst. 2015;11:3397–3406. doi: 10.1039/c5mb00413f. [DOI] [PubMed] [Google Scholar]
- 43.Jiang L, Xiao L, Sugiura H, Huang X, Ali A, Kuro-o M, Deberardinis RJ, Boothman DA. Metabolic reprogramming during TGFbeta1-induced epithelial-to-mesenchymal transition. Oncogene. 2015;34:3908–3916. doi: 10.1038/onc.2014.321. onc2014321 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ulanet DB, Couto K, Jha A, Choe S, Wang A, Woo HK, Steadman M, DeLaBarre B, Gross S, Driggers E, et al. Mesenchymal phenotype predisposes lung cancer cells to impaired proliferation and redox stress in response to glutaminase inhibition. PLoS One. 2014;9:e115144. doi: 10.1371/journal.pone.0115144. PONE-D-14-30777 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. nature13611 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, Ashton JM, Pei S, Grose V, O'Dwyer KM, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–341. doi: 10.1016/j.stem.2012.12.013. S1934-5909(12)00755-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. 339/6119/580 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. S1550-4131(15)00621-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmad A, Aboukameel A, Kong D, Wang Z, Sethi S, Chen W, Sarkar FH, Raz A. Phosphoglucose isomerase/autocrine motility factor mediates epithelial-mesenchymal transition regulated by miR-200 in breast cancer cells. Cancer Res. 2011;71:3400–3409. doi: 10.1158/0008-5472.CAN-10-0965. 0008-5472.CAN-10-0965 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Funasaka T, Hogan V, Raz A. Phosphoglucose isomerase/autocrine motility factor mediates epithelial and mesenchymal phenotype conversions in breast cancer. Cancer Res. 2009;69:5349–5356. doi: 10.1158/0008-5472.CAN-09-0488. 0008-5472.CAN-09-0488 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Funasaka T, Hu H, Yanagawa T, Hogan V, Raz A. Down-regulation of phosphoglucose isomerase/autocrine motility factor results in mesenchymal-to-epithelial transition of human lung fibrosarcoma cells. Cancer Res. 2007;67:4236–4243. doi: 10.1158/0008-5472.CAN-06-3935. 67/9/4236 [pii] [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Che Q, Bian Y, Zhou Q, Jiang F, Tong H, Ke J, Wang K, Wan XP. Autocrine motility factor promotes epithelial-mesenchymal transition in endometrial cancer via MAPK signaling pathway. Int J Oncol. 2015;47:1017–1024. doi: 10.3892/ijo.2015.3091. [DOI] [PubMed] [Google Scholar]
- 53.Li J, Wang Y, Li QG, Xue JJ, Wang Z, Yuan X, Tong JD, Xu LC. Downregulation of FBP1 Promotes Tumor Metastasis and Indicates Poor Prognosis in Gastric Cancer via Regulating Epithelial-Mesenchymal Transition. PLoS One. 2016;11:e0167857. doi: 10.1371/journal.pone.0167857. PONE-D-16-34556 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du S, Guan Z, Hao L, Song Y, Wang L, Gong L, Liu L, Qi X, Hou Z, Shao S. Fructose-bisphosphate aldolase a is a potential metastasis-associated marker of lung squamous cell carcinoma and promotes lung cell tumorigenesis and migration. PLoS One. 2014;9:e85804. doi: 10.1371/journal.pone.0085804. PONE-D-13-29290 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu K, Tang Z, Huang A, Chen P, Liu P, Yang J, Lu W, Liao J, Sun Y, Wen S, et al. Glyceraldehyde-3-phosphate dehydrogenase promotes cancer growth and metastasis through upregulation of SNAIL expression. Int J Oncol. 2017;50:252–262. doi: 10.3892/ijo.2016.3774. [DOI] [PubMed] [Google Scholar]
- 56.Jiang F, Ma S, Xue Y, Hou J, Zhang Y. LDH-A promotes malignant progression via activation of epithelial-to-mesenchymal transition and conferring stemness in muscle-invasive bladder cancer. Biochem Biophys Res Commun. 2016;469:985–992. doi: 10.1016/j.bbrc.2015.12.078. S0006-291X(15)31083-4 [pii] [DOI] [PubMed] [Google Scholar]
- 57.Porporato PE, Payen VL, Perez-Escuredo J, De Saedeleer CJ, Danhier P, Copetti T, Dhup S, Tardy M, Vazeille T, Bouzin C, et al. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014;8:754–766. doi: 10.1016/j.celrep.2014.06.043. S2211-1247(14)00527-0 [pii] [DOI] [PubMed] [Google Scholar]
- 58.Chen EI. Mitochondrial dysfunction and cancer metastasis. J Bioenerg Biomembr. 2012;44:619–622. doi: 10.1007/s10863-012-9465-9. [DOI] [PubMed] [Google Scholar]
- 59.Guha M, Srinivasan S, Ruthel G, Kashina AK, Carstens RP, Mendoza A, Khanna C, Van Winkle T, Avadhani NG. Mitochondrial retrograde signaling induces epithelial-mesenchymal transition and generates breast cancer stem cells. Oncogene. 2014;33:5238–5250. doi: 10.1038/onc.2013.467. onc2013467 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaude E, Frezza C. Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival. Nat Commun. 2016;7 doi: 10.1038/ncomms13041. 13041, ncomms13041 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. ng849 [pii] [DOI] [PubMed] [Google Scholar]
- 62.Castro-Vega LJ, Buffet A, De Cubas AA, Cascon A, Menara M, Khalifa E, Amar L, Azriel S, Bourdeau I, Chabre O, et al. Germline mutations in FH confer predisposition to malignant pheochromocytomas and paragangliomas. Hum Mol Genet. 2014;23:2440–2446. doi: 10.1093/hmg/ddt639. ddt639 [pii] [DOI] [PubMed] [Google Scholar]
- 63.Clark GR, Sciacovelli M, Gaude E, Walsh DM, Kirby G, Simpson MA, Trembath RC, Berg JN, Woodward ER, Kinning E, et al. Germline FH mutations presenting with pheochromocytoma. J Clin Endocrinol Metab. 2014;99:E2046–2050. doi: 10.1210/jc.2014-1659. [DOI] [PubMed] [Google Scholar]
- 64.Fieuw A, Kumps C, Schramm A, Pattyn F, Menten B, Antonacci F, Sudmant P, Schulte JH, Van Roy N, Vergult S, et al. Identification of a novel recurrent 1q42.2-1qter deletion in high risk MYCN single copy 11q deleted neuroblastomas. Int J Cancer. 2012;130:2599–2606. doi: 10.1002/ijc.26317. [DOI] [PubMed] [Google Scholar]
- 65.Schmidt LS, Linehan WM. Hereditary leiomyomatosis and renal cell carcinoma. Int J Nephrol Renovasc Dis. 2014;7:253–260. doi: 10.2147/IJNRD.S42097. ijnrd-7-253 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He X, Yan B, Liu S, Jia J, Lai W, Xin X, Tang CE, Luo D, Tan T, Jiang Y, et al. Chromatin Remodeling Factor LSH Drives Cancer Progression by Suppressing the Activity of Fumarate Hydratase. Cancer Res. 2016;76:5743–5755. doi: 10.1158/0008-5472.CAN-16-0268. 0008-5472.CAN-16-0268 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu YN, Yin JJ, Abou-Kheir W, Hynes PG, Casey OM, Fang L, Yi M, Stephens RM, Seng V, Sheppard-Tillman H, et al. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene. 2013;32:296–306. doi: 10.1038/onc.2012.58. onc201258 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. ncb1722 [pii] [DOI] [PubMed] [Google Scholar]
- 69.Muller U, Troidl C, Niemann S. SDHC mutations in hereditary paraganglioma/pheochromocytoma. Familial Cancer. 2005;4:9–12. doi: 10.1007/s10689-004-0621-1. [DOI] [PubMed] [Google Scholar]
- 70.Niemann S, Muller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nature Genetics. 2000;26:268–270. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- 71.Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PEM, Rubinstein WS, Myers EN, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 72.Baysal BE, Maher ER. Genetics and mechanism of pheochromocytoma-paraganglioma syndromes characterized by germline SDHB and SDHD mutations. Endocr-Relat Cancer. 2015;22:T71–T82. doi: 10.1530/Erc-15-0226. [DOI] [PubMed] [Google Scholar]
- 73.Vanharanta S, Buchta M, McWhinney SR, Virta SK, Peczkowska M, Morrison CD, Lehtonen R, Januszewicz A, Jarvinen H, Juhola M, et al. Early-onset renal cell carcinoma as a novel extraparaganglial component of SDHB-associated heritable paraganglioma. American Journal of Human Genetics. 2004;74:153–159. doi: 10.1086/381054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stratakis CA, Carney JA. The triad of paragangliomas, gastric stromal tumours and pulmonary chondromas (Carney triad), and the dyad of paragangliomas and gastric stromal sarcomas (Carney-Stratakis syndrome): molecular genetics and clinical implications. Journal of Internal Medicine. 2009;266:43–52. doi: 10.1111/j.1365-2796.2009.02110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang YM, Gu ML, Ji F. Succinate dehydrogenase-deficient gastrointestinal stromal tumors. World J Gastroentero. 2015;21:2303–2314. doi: 10.3748/wjg.v21.i8.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loriot C, Domingues M, Berger A, Menara M, Ruel M, Morin A, Castro-Vega LJ, Letouze E, Martinelli C, Bemelmans AP, et al. Deciphering the molecular basis of invasiveness in Sdhb-deficient cells. Oncotarget. 2015;6:32955–32965. doi: 10.18632/oncotarget.5106. 5106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Letouze E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, Janin M, Menara M, Nguyen AT, Benit P, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. S1535-6108(13)00183-9 [pii] [DOI] [PubMed] [Google Scholar]
- 78.Wang H, Chen Y, Wu G. SDHB deficiency promotes TGFbeta-mediated invasion and metastasis of colorectal cancer through transcriptional repression complex SNAIL1-SMAD3/4. Transl Oncol. 2016;9:512–520. doi: 10.1016/j.tranon.2016.09.009. S1936-5233(16)30162-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aspuria PJ, Lunt SY, Varemo L, Vergnes L, Gozo M, Beach JA, Salumbides B, Reue K, Wiedemeyer WR, Nielsen J, et al. Succinate dehydrogenase inhibition leads to epithelial-mesenchymal transition and reprogrammed carbon metabolism. Cancer Metab. 2014;2:21. doi: 10.1186/2049-3002-2-21. 142 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. 1175689 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu J, Gao L, Zhang H, Wang D, Wang M, Zhu J, Pang C, Wang C. Succinate dehydrogenase 5 (SDH5) regulates glycogen synthase kinase 3beta-beta-catenin-mediated lung cancer metastasis. J Biol Chem. 2013;288:29965–29973. doi: 10.1074/jbc.M113.450106. M113.450106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parsons DW, Jones S, Zhang XS, Lin JCH, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma Multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan H, Parsons DW, Jin GL, McLendon R, Rasheed BA, Yuan WS, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 Mutations in Gliomas. New Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. NEJMoa0903840 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Colvin H, Nishida N, Konno M, Haraguchi N, Takahashi H, Nishimura J, Hata T, Kawamoto K, Asai A, Tsunekuni K, et al. Oncometabolite D-2-Hydroxyglurate Directly Induces Epithelial-Mesenchymal Transition and is Associated with Distant Metastasis in Colorectal Cancer. Sci Rep. 2016;6 doi: 10.1038/srep36289. 36289, srep36289 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin CC, Cheng TL, Tsai WH, Tsai HJ, Hu KH, Chang HC, Yeh CW, Chen YC, Liao CC, Chang WT. Loss of the respiratory enzyme citrate synthase directly links the Warburg effect to tumor malignancy. Sci Rep. 2012;2:785. doi: 10.1038/srep00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen L, Liu T, Zhou J, Wang Y, Wang X, Di W, Zhang S. Citrate synthase expression affects tumor phenotype and drug resistance in human ovarian carcinoma. PLoS One. 2014;9:e115708. doi: 10.1371/journal.pone.0115708. PONE-D-14-09107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sanchez-Martinez R, Cruz-Gil S, Gomez de Cedron M, Alvarez-Fernandez M, Vargas T, Molina S, Garcia B, Herranz J, Moreno-Rubio J, Reglero G, et al. A link between lipid metabolism and epithelial-mesenchymal transition provides a target for colon cancer therapy. Oncotarget. 2015;6:38719–38736. doi: 10.18632/oncotarget.5340. 5340 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nath A, Li I, Roberts LR, Chan C. Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma. Sci Rep. 2015;5 doi: 10.1038/srep14752. 14752, srep14752 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pascual G, Avgustinova A, Mejetta S, Martin M, Castellanos A, Attolini CS, Berenguer A, Prats N, Toll A, Hueto JA, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541:41–45. doi: 10.1038/nature20791. nature20791 [pii] [DOI] [PubMed] [Google Scholar]
- 91.Hanai J, Doro N, Sasaki AT, Kobayashi S, Cantley LC, Seth P, Sukhatme VP. Inhibition of lung cancer growth: ATP citrate lyase knockdown and statin treatment leads to dual blockade of mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3-kinase (PI3K)/AKT pathways. J Cell Physiol. 2012;227:1709–1720. doi: 10.1002/jcp.22895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu Y, Huang J, Xin W, Chen L, Zhao X, Lv Z, Liu Y, Wan Q. Lipid accumulation is ahead of epithelial-to-mesenchymal transition and therapeutic intervention by acetyl-CoA carboxylase 2 silence in diabetic nephropathy. Metabolism. 2014;63:716–726. doi: 10.1016/j.metabol.2014.02.010. S0026-0495(14)00049-3 [pii] [DOI] [PubMed] [Google Scholar]
- 93.Martinez-Orozco R, Navarro-Tito N, Soto-Guzman A, Castro-Sanchez L, Perez Salazar E. Arachidonic acid promotes epithelial-to-mesenchymal-like transition in mammary epithelial cells MCF10A. Eur J Cell Biol. 2010;89:476–488. doi: 10.1016/j.ejcb.2009.12.005. S0171-9335(10)00036-1 [pii] [DOI] [PubMed] [Google Scholar]
- 94.Espinosa-Neira R, Mejia-Rangel J, Cortes-Reynosa P, Salazar EP. Linoleic acid induces an EMT-like process in mammary epithelial cells MCF10A. Int J Biochem Cell Biol. 2011;43:1782–1791. doi: 10.1016/j.biocel.2011.08.017. S1357-2725(11)00237-8 [pii] [DOI] [PubMed] [Google Scholar]
- 95.Lee SY, Jeon HM, Ju MK, Jeong EK, Kim CH, Park HG, Han SI, Kang HS. Dlx-2 and glutaminase upregulate epithelial-mesenchymal transition and glycolytic switch. Oncotarget. 2016;7:7925–7939. doi: 10.18632/oncotarget.6879. 6879 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kuo TC, Chen CK, Hua KT, Yu P, Lee WJ, Chen MW, Jeng YM, Chien MH, Kuo KT, Hsiao M, et al. Glutaminase 2 stabilizes Dicer to repress Snail and metastasis in hepatocellular carcinoma cells. Cancer Lett. 2016;383:282–294. doi: 10.1016/j.canlet.2016.10.012. S0304-3835(16)30624-3 [pii] [DOI] [PubMed] [Google Scholar]
- 97.Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. S1535-6108(08)00195-5 [pii] [DOI] [PubMed] [Google Scholar]
- 98.Zheng L, Cardaci S, Jerby L, MacKenzie ED, Sciacovelli M, Johnson TI, Gaude E, King A, Leach JD, Edrada-Ebel R, et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat Commun. 2015;6:6001. doi: 10.1038/ncomms7001. ncomms7001 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. science.1236062 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pattabiraman DR, Bierie B, Kober KI, Thiru P, Krall JA, Zill C, Reinhardt F, Tam WL, Weinberg RA. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science. 2016;351 doi: 10.1126/science.aad3680. aad3680, aad3680 [pii] 351/6277/aad3680 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]