Summary
Glycolysis is linked to the rapid recall capacity of memory CD8+ T cells, but the pathways that glucose fuels and the molecular and subcellular structural elements enabling enhanced glucose metabolism in nascent activated memory CD8+ T cells are not known. We found that mitochondria–ER contact sites are immunometabolic hubs that integrate mTORC2-initiated signaling with glucose metabolism and mitochondrial respiration in memory CD8+ T cells. Specifically, in this subcellular compartment, mTORC2, Akt and Gsk-3β were present by default. Rapid activation of Akt by mTORC2 led to inhibition of Gsk-3β at mitochondria–ER junctions, enabling recruitment of hexokinase I (HK-I) to the mitochondrial channel, VDAC. Binding of HK-I to VDAC promoted cellular respiration by facilitating metabolite flux into mitochondria. Glucose tracing pinpointed pyruvate oxidation in mitochondria, which was the metabolic requirement for rapid generation of IFN-γ in memory T cells. Subcellular organization of key signaling events enabling HK-I recruitment to VDAC thus underpins the metabolic reprogramming needed for memory CD8+ T cells to rapidly acquire effector function.
Introduction
After successful control of intracellular infection most effector CD8+ T cells are removed by apoptosis, whereas a small subset develops to become memory cells. Memory CD8+ T cells form the cellular basis for accelerated protection upon re-exposure to the same pathogen, which is a hallmark of adaptive immunity (Harty and Badovinac, 2008). Expansion of T cells following activation –and subsequent differentiation into effector and memory populations– is accompanied by reprogramming of metabolic pathway usage (MacIver et al., 2013). Importantly, survival and function of differentiated T cell subsets is underpinned by specific metabolic repertoires. Our group, and others, previously reported that memory CD8+ T cells possess unique metabolic signatures compared to naïve (NV) and effector T cells (Balmer et al., 2016; Buck et al., 2016; Gubser et al., 2013; O'Sullivan et al., 2014; van der Windt et al., 2012; van der Windt et al., 2013). EM CD8+ T cells have increased respiratory capacity compared to naïve counterparts (Gubser et al., 2013; van der Windt et al., 2012). This distinct mitochondrial feature of memory CD8+ T cells has been linked to increased fatty acid oxidation, which contributes to memory CD8+ T cell differentiation and function (O'Sullivan et al., 2014). For example, exogenous short chain fatty acids enhance IFN-γ production in memory CD8+ T cells by increasing glycolysis via acetylation of GAPDH (Balmer et al., 2016).
Effector memory (EM) cells constitute an important memory cell subset, circulating in the blood from where they are recruited to inflamed tissues or organs (Sallusto et al., 2004; Thome et al., 2014). Upon exposure to cognate antigen, EM CD8+ T cells are able to rapidly produce key effector molecules, such as IFN-γ (Sallusto et al., 1999). The capacity of memory T cells to produce effector molecules with more rapid kinetics is a defining element of their protective capacity, and hence biology. This innate-like ability to elaborate effector function during the immediate early phase of the recall response has been linked to distinct biochemical, signaling, epigenetic signatures (Barski et al., 2017; Farber, 2009; Kannan et al., 2012; Weng et al., 2012). We previously demonstrated that a key metabolic feature of EM CD8+ T cells is the induction of Warburg metabolism (aerobic glycolysis) following stimulation (Gubser et al., 2013). This rapid and stable activation-induced glycolysis of EM CD8+ T cells, as reflected by enhanced lactate production, is dependent on mTORC2–Akt signaling but not mTORC1, which primarily exerts its control on metabolism of activated effector cells (Gubser et al., 2013). The capacity of EM CD8+ T cells to rapidly produce effector molecules, such as IFN-γ, is intricately tied to the fast induction of glycolysis by the mTORC2–Akt pathway (Bantug et al., 2017; Gubser et al., 2013). Stable glycolysis drives rapid IFN-γ production in memory CD8+ T cells at the epigenetic level, by promoting H3K9 acetylation at the IFNG promoter (Gubser et al., 2013). Recently, IFN-γ production by effector Th1 cells was also shown to be dependent on glycolysis through epigenetic regulation of the Ifng promoter (Peng et al., 2016).
Mitochondrial mass and activity are greater in memory T cells as compared to naïve counterparts. However, the impact of mitochondrial metabolism and glucose-derived pyruvate oxidation in the mitochondria during the immediate-early phase of the recall response has not been investigated in any detail. Moreover, a unifying cellular and molecular model integrating mitochondrial function, glucose metabolism and signaling events in nascent activated EM CD8+ T cells has not been described.
Results
Memory CD8+ T cells rapidly increase respiration upon activation and are enriched for mitochondria–ER appositions
Distinct metabolic features of memory T cells underpin their ability to rapidly respond to cognate antigen, which is a defining aspect of adaptive immunity. Directly ex vivo, memory CD8+ T cells possess more mitochondrial mass and greater respiratory capacity than their naïve counterparts (Gubser et al., 2013; van der Windt et al., 2012). How respiration of CD8+ T cell subsets is altered early following activation remains largely unexplored. Thus, we first examined mitochondrial respiration in non-activated vs. nascent activated NV and EM CD8+ T cells, in vitro. Specifically, human NV and EM CD8+ T cells were cultured for 12 hours in presence or absence of α-CD3/α-CD28 monoclonal antibodies (mAb), and mitochondrial respiration and glycolysis were assessed by metabolic flux analysis. Under non-activated conditions, as previously demonstrated, both maximal and spare respiratory capacities were greater in EM than in NV CD8+ T cells (Fig. 1A) (Gubser et al., 2013; van der Windt et al., 2012). Following activation, a noticeable increase in basal and maximal respiration, as well as spare respiratory capacity, was observed in activated EM CD8+ T cells but not in NV counterparts (Fig. 1A). Glycolysis was elevated in both activated CD8+ T cell subsets, with EM cells displaying a more prominent increase than their NV counterparts (Fig. S1A).
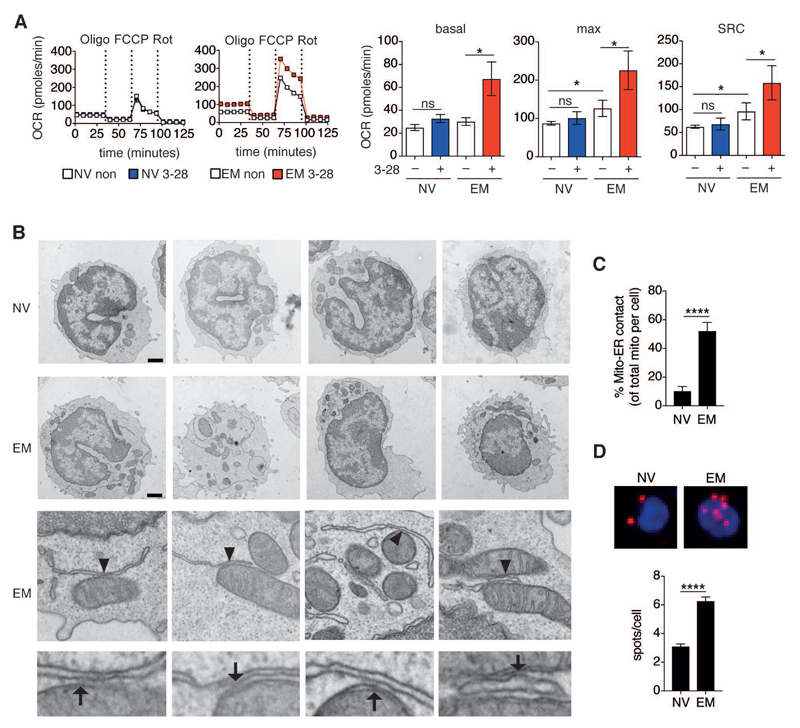
Figure 1. Effector memory CD8+ T cells selectively increase respiration upon activation and possess abundant mitochondria-associated ER membranes.
(A) Left, representative mitochondrial perturbation assays of naïve (NV) and effector memory (EM) human CD8+ T cells left non-activated (non) or activated for 12 h with plate bound α-CD3 (5 μg/ml) and soluble α-CD28 (5 μg/ml) mAb (3-28). Mitochondrial perturbation was performed by sequential treatment with oligomycin (Oligo, 1 μM), FCCP (2 μM) and rotenone (Rot, 130 nM). Oxygen consumption rate (OCR) was measured by metabolic flux analysis. Right, Summary bar graphs showing calculated basal respiration, maximal respiration (max) and spare respiratory capacity (SRC) in non-activated (–) or activated (+) NV and EM human CD8+ T cells (n = 6 donors).
(B) Transmission electron microscopy images of human NV (top panel) and EM (middle and bottom panels) CD8+ T cells. Arrowheads mark mitochondria and ER contact sites that were digitally magnified in the bottom panel (representative of n = 3 donors). Arrows show electron dense regions of mitochondria–ER junctions. Scale bars = 1000 nm.
C) Percentage of mitochondria–ER contact sites per cell in NV (n = 15) and EM (n = 13) CD8+ T cells (n = 3 donors).
(D) Top, representative proximity ligation assay (PLA) images of freshly sorted NV (left) and EM (right) CD8+ T cells probed with α-IP3R1 and α-VDAC1 antibodies. Red spots indicate contact sites between mitochondria (α-VDAC) and ER (α-IP3R). Cells were counterstained with DAPI (blue). Bottom, quantitative analysis of PLA from 3 donors; NV (n = 72 cells) and EM (n = 73 cells).
Data are presented as mean±SEM. Two-tailed paired Wilcoxon signed rank tests (A) and two-tailed unpaired Student’s t test (C,D) were used to compare groups. * P < 0.05, **** P < 0.0001, ns = not significant. See also Figure S1.
Since mitochondrial shape is intricately linked to T cell effector maturation and memory cell survival (Buck et al., 2016; Klein Geltink et al., 2017), we also analyzed mitochondrial morphology in human NV and EM CD8+ T cells by electron and confocal microscopy. Electron microscopy images revealed disparities in mitochondrial morphology, with EM cells displaying long, tubular mitochondria, whereas mitochondria in naïve cells were more rounded (mean aspect ratio –major axis/minor axis length– of NV vs. EM cells: 1.6 and 2.5, respectively) (Fig. S1B). Morphometric quantification of confocal microscopy images further demonstrated that mitochondria in EM cells possessed longer branch length than naïve counterparts (median mitochondrial branch length of NV vs. EM cells: 0.88 vs. 1.07 μm, respectively) (Fig. S1C). The mitochondrial membrane potential, which was higher in EM cells under basal conditions (Fig. S1D) was not altered in both subpopulations following stimulation (Fig. S1E).
Endoplasmic reticulum (ER)–mitochondria interactions modulate mitochondrial respiration and bioenergetics in non-immune cells (Theurey and Rieusset, 2017). In this specific subcellular region, ER and mitochondrial membranes are physically tethered to each other (Bravo et al., 2011; Cardenas et al., 2010; Giacomello and Pellegrini, 2016; Raturi and Simmen, 2013). In yeast, tethering of both organelles is mediated by the ER–mitochondria encounter structure (ERMES) complex (Murley and Nunnari, 2016). However, in metazoans, the molecular composition of mitochondria–ER tethering complexes is not fully defined (Murley and Nunnari, 2016). Nonetheless, metazoan mitochondria–ER contacts have been shown to function as cellular hubs in various physiologic and cellular stress-related conditions. In search of ultra-structural features that may offer a biologic basis for the contrasting activation-induced respiration phenotypes of NV vs. EM CD8+ T cells (beyond mitochondrial content and morphology), we evaluated the abundance of mitochondria–ER contact sites in both subsets. We observed that in naïve cells, mitochondria in close association to the ER were sparse (Fig. 1B, top panel), with only around 10% of mitochondria per cell in close proximity to ER membranes (Fig. 1C). In EM cells, by contrast, the frequency of mitochondria–ER contacts per cell (as observed by electron microscopy) was higher (around 50% of mitochondria per cell with contact to ER) (Fig. 1B, middle panels, and Fig. 1C). The average distance between apposed ER and mitochondrial membranes in EM CD8+ T cells was 21 nm (data not shown). Electron dense zones between the mitochondrial outer membrane and ER at contact sites indicated that these organellar appositions were distinct subcellular structures (Fig. 1B, bottom panel). To further elaborate on these findings we performed proximity ligation assays (PLA) to quantify the abundance of mitochondria–ER junctions in NV and EM CD8+ T cell subsets. PLA is a powerful tool to detect and quantify co-localization of proteins in situ (Bagchi et al., 2015). Mitochondria–ER contacts were detected and enumerated by using α-VDAC1 (voltage dependent anion channel 1, mitochondria) and α-IP3R1 (inositol triphosphate receptor 1, ER) antibodies as probes (Tubbs et al., 2014). Once again, the abundance of mitochondria–ER contact sites was found to be markedly lower in NV compared to EM CD8+ T cells (Fig. 1D). Lastly, NV and EM CD8+ T cells were also labeled with mitochondria (mitotracker deep red) and ER (α-KDEL motif antibody) markers, and the proximity of both organelles was assessed by confocal microscopy. Co-localization of mitochondria with ER was greater in EM cells than naïve counterparts (Fig. S1F). In all these data identified (i) a rapid increase in mitochondrial respiration as a distinct functional feature of EM CD8+ T cells, and (ii) increased abundance of mitochondrial–ER contacts as a defining structural characteristic of EM CD8+ T cells.
Dissociation of mitochondria–ER junctions attenuates mitochondrial respiration in T cells
In HeLa cells, stable interaction between mitochondria and the ER has been shown to require microtubule polymerization (Bravo et al., 2011). To determine whether mitochondria–ER junctions were similarly dependent on microtubule polymerization in memory CD8+ T cells, we again used PLA. Activated EM CD8+ T cells exposed to nocodazole, a microtubule polymerization inhibitor, displayed a marked reduction in abundance of mitochondria–ER junctions (Fig. 2A, and Fig. S2A). We then examined whether these interactions were required for enhancement of mitochondrial respiration in activated memory CD8+ T cells. Nocodazole treatment attenuated the activation-induced increase in mitochondrial respiration (Fig. 2B, and Fig. S2B). In contrast, nocodazole did not significantly affect glycolysis (Fig. 2B). To further verify these results, we perturbed mitochondria–ER appositions by treating activated EM CD8+ T cells with the ER stress blocker, 4-PBA, since reduction of ER stress has been shown to reduce mitochondria–ER interaction in non-immune cells (Prasad et al., 2016). Indeed, mitochondria–ER junctions were reduced in 4-PBA treated cells (Fig. S2C), which again led to a reduction in mitochondrial respiration (Fig. S2D). Congruent with the above findings, nocodazole treatment also reduced abundance of mitochondria–ER contact in Jurkat T cells (Fig. 2C). Importantly, mitochondrial respiration was likewise diminished in nocodazole treated Jurkat T cells, whereas glycolysis remained unaffected (Fig. 2D). Although there were alterations in spare respiratory capacity (SRC) in both CD8+ memory T cells and Jurkat T cells following nocodazole treatment, the biologic significance of SRC is difficult to interpret (Olenchock et al., 2017), and challenging to compare between both cell types. We therefore focused our analysis on basal respiration. Lastly, the presence of mitochondria–ER appositions –as well as their sensitivity to dissociation with nocodazole– was also verified in bona fide murine memory CD8+ T cells sorted from mice that had been exposed to Listeria monocytogenes (Fig. S2E).
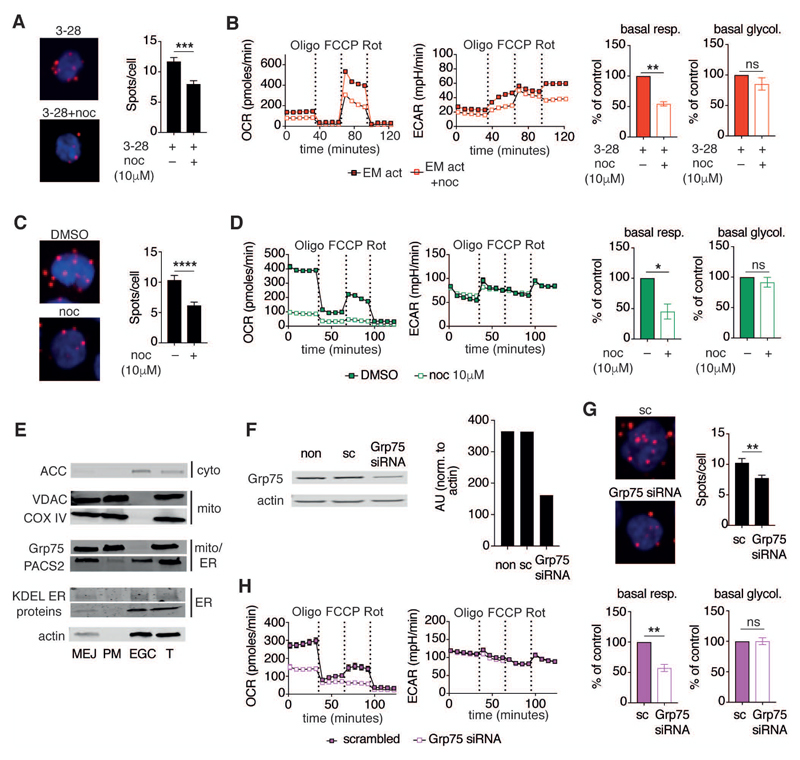
Figure 2. Dissociation of mitochondria–ER contacts diminishes mitochondrial respiration but not glycolysis.
(A) Left, representative PLA images of EM CD8+ T cells activated for 12 h with α-CD3/α-CD28 (3-28) mAb as in Figure 1A and concomitantly treated with DMSO (top) or nocodazole (10 μM, bottom). Cells were stained as in Figure 1D. Right, representative summary bar graph of PLA from 1 of 3 donors treated with DMSO (n = 28 cells) or nocodazole (n = 23 cells).
(B) Left, representative mitochondrial perturbation assay of EM CD8+ T cells activated as in Figure 1A or activated in the additional presence of nocodazole (10 μM). Right, bar graphs of basal respiration and glycolysis (n = 4 donors).
(C) Left, representative PLA images of Jurkat T cells cultured in the presence of DMSO (top) or nocodazole (10 μM, bottom) for 12 h. Right, bar graph showing the number of detected mitochondria–ER junctions per cell in DMSO (n = 17) and nocodazole (n = 17) treated cells. Representative of n = 2 independent experiments.
(D) Left, representative mitochondrial perturbation assay of Jurkat T cells cultured as in (C). Right, bar graphs of basal respiration and glycolysis of control and nocodazole treated Jurkat T cells (n = 3 independent experiments).
(E) Immunoblot of cellular fractions and total cell lysates (T) from Jurkat T cells. Cells were separated into fractions containing mitochondria–ER junctions (MEJ), pure mitochondria (PM), and remaining supernatant from MEJ (containing the majority of ER, Golgi, and cytoplasm – EGC). Blots were probed with antibodies targeting ACC, VDAC, Cox iv, Grp75, PACS2, KDEL motifs, and actin.
(F) Left, immunoblot analysis of lysates from non-transfected (non), scrambled siRNA (sc) transfected, and Grp75 siRNA (siRNA) transfected Jurkat T cells. Cells were cultured for 72 h after transfection. Blots were probed with Grp75 and actin antibodies. Right, quantification of Grp75 abundance in control and Grp75 siRNA transfected lysates normalized to actin (representative of n = 3 independent experiments).
(G) PLA images (left) and quantification of mitochondria–ER contact abundance (right) of control (sc, n = 32) and Grp75 siRNA (n = 63) transfected cells (n = 2 independent experiments).
(H) Left, representative mitochondrial perturbation assay of Jurkat T cells cultured for 72 h after transfection with scrambled (sc) and Grp75 targeted siRNA. Right, bar graphs of basal respiration and glycolysis from control (sc) and Grp75 siRNA transfected Jurkat T cells (n = 3 independent experiments).
Data are presented as mean±SEM. Two-tailed unpaired (A,C,G), and paired (B,D,H) Student’s t tests were used to compare groups. * P < 0.05, ** P < 0.01, *** P < 0.001 **** P < 0.0001, ns = not significant. See also Figure S2.
Lymphocytes have fewer mitochondria–ER contacts than most cell types, making it technically challenging to study this subcellular compartment at a biochemical level in T cells. Therefore, to examine the molecular composition of mitochondria–ER contact sites in T cells, we modified an established fractionation protocol (Wieckowski et al., 2009), and enriched for subcellular fractions containing mitochondria as well as ER membranes that are localized to mitochondria–ER junctions (MEJ fraction). To validate the technique, we assessed the abundance of proteins that were previously found to be enriched at mitochondria–ER contact sites, and also probed for mitochondrial-, ER- and cytoplasmic proteins in pelleted MEJ fractions from Jurkat T cells, relative to supernatants from these fractions which contain most of the ER, Golgi and cytoplasm (EGC). In addition, we also compared the MEJ fraction to pure mitochondria (PM) that were isolated using a different technique (Krippner et al., 1996). As an additional control, the presence of all examined proteins was also confirmed in unfractionated total cell lysates (T). ACC, a cytoplasmic protein, was enriched in the EGC fraction but absent from mitochondria containing fractions (i.e. MEJ and PM) (Fig. 2E). As expected, the mitochondrial proteins VDAC and Cox iv were similarly present in MEJ and PM fractions (Fig. 2E). GRP75, a protein that acts as a molecular tether between mitochondria and ER by associating with VDAC and IP3R, was detected in both mitochondria containing fractions (i.e. MEJ and PM), indicating that GRP75 is primarily associated with mitochondria (Szabadkai et al., 2006) (Fig. 2E). By contrast, PACS2, another mitochondria–ER tethering protein that is predominantly located on the ER-side of the junction was more abundant in the MEJ fraction than in pure mitochondria (Myhill et al., 2008) (Fig. 2E). Both mitochondria markers, as well as GRP75, were not detected in the EGC fraction, whilst PACS2 and proteins containing the KDEL motif –which is a retention-signal for ER localized proteins– were enriched in this ER containing fraction (Fig. 2E). In addition, the abundance of KDEL motif containing proteins, as well as actin, was lower in the MEJ fraction compared to the EGC fraction, indicating that the MEJ fraction is indeed enriched for ER membranes located at contact sites with mitochondria (Fig. 2E). Together, these experiments established these MEJ fractions as a valid source to probe for proteins localized to mitochondria–ER junctions in cells with low abundance of mitochondria–ER contact sites.
To validate by genetic means the role of mitochondria–ER contacts in controlling mitochondrial function in lymphocytes, Jurkat T cells were transiently transfected with siRNA directed against GRP75, or with a non-targeting scrambled control siRNA. GRP75 knockdown-efficiency was approximately 50% (Fig. 2F), and there was no detectable difference in cell viability (Fig. S2F). In agreement with the effect of microtubule de-polymerization on abundance of mitochondria–ER contact sites in EM CD8+ T cells and Jurkat T cells, mitochondria–ER contacts were significantly reduced in GRP75 depleted cells (Fig. 2G). GRP75 knockdown also resulted in a marked decrease in mitochondrial respiration, while it did not impact glycolysis (Fig. 2H). Together, these results demonstrated that respiration in T cells was dependent on stable contacts between mitochondria and the ER, whereas glycolysis remained unchanged following perturbation of these organellar contacts, indicating that mitochondrial function in newly activated memory T cells, as well as in Jurkat T cells, was intricately tied to their physical association with the ER.
The mTORC2–Akt–Gsk-3β signaling pathway is activated at mitochondria–ER contact sites and critical for early mitochondrial reprogramming in memory CD8+ T cells
We previously reported that mTORC2–Akt signaling is important for the rapid memory CD8+ T cell response and for TCR/CD28 dependent immediate-early glycolytic switch (Gubser et al., 2013). mTORC2 and Akt can localize to mitochondria–ER contact sites in mouse hepatocytes and mouse embryonic fibroblasts (MEF) (Betz and Hall, 2013; Betz et al., 2013). Furthermore, mTORC2 activity in these organellar contacts was enhanced in insulin treated MEF (Betz et al., 2013). We thus hypothesized that mTORC2 and Akt are present in mitochondria–ER contact sites of CD8+ T cells, and that greater mitochondria–ER contact in memory CD8+ T cells directs mTORC2 and Akt signaling through this subcellular structure. To test this hypothesis, we first probed the global expression of mTORC2 components –mTOR, rictor, and Sin1– in human NV and EM CD8+ T cells. We detected no difference in total cellular abundance of these proteins between the subsets (Fig. S3A). Next, we aimed to characterize the localization of mTORC2 and Akt in cellular fractions enriched for mitochondria–ER junctions, by immunoblot analyses. First, using the fractionation protocol described above (Fig. 2E), we probed for mTOR, rictor, raptor, and Akt in MEJ, PM, and EGC fractions, as well as in total cell lysates (T) (Fig. 3A). mTOR, Rictor and Akt were detected in all fractions, with Akt being notably less abundant in pure mitochondria fractions as compared to MEJ fractions (Fig. 3A). We then interrogated whether mTORC2 was also present in the MEJ fraction of sorted NV and EM CD8+ T cells, directly ex vivo. Similar to Jurkat T cells, mTOR and rictor were detected in the MEJ fraction of both EM as well as NV CD8+ T cells (Fig. 3B). This indicated that mitochondria–ER contact sites of naïve CD8+ T cells, while much less abundant (Fig. 1B-D), were not qualitatively different with regards to the presence of mTOR and rictor, and that mTOR and rictor were present by default in this subcellular region. To ensure that mTORC2 remained in the mitochondria–ER junction of activated T cells, we assessed the same subcellular fractions from non-activated vs. α-CD3/α-CD28 mAb activated CD8+ T cells. Due to limitations with regard to available amounts of human cells, these experiments had to be performed with bulk CD8+ T cells. Both mTOR and rictor were similarly detected in the mitochondria–ER fraction prior to- and 2 hours post stimulation (Fig. 3C).
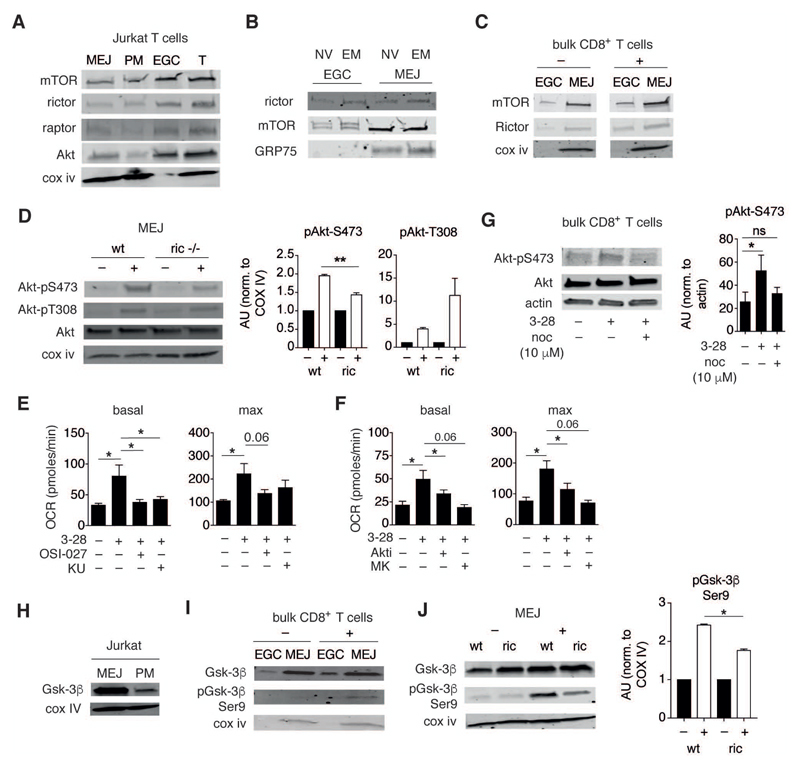
Figure 3. mTORC2, Akt and Gsk-3β signaling components are present in mitochondria–ER junctions.
(A) Immunoblots of MEJ, PM, and EGC fractions and total lysate (T) from Jurkat T cells. Blots were probed with mTOR, rictor, raptor, and Akt antibodies. Cox iv was used as loading and fraction-validation control (representative of n = 3 independent experiments).
(B) Immunoblots of EGC and MEJ fractions from NV and EM human CD8+ T cells probed with antibodies specific for rictor and mTOR. GRP75 was used to validate the respective fractions (representative of n = 2 independent samples, each sample consisted of 4x107 sorted cells, pooled from 2-4 donors).
(C) Immunoblot analysis of EGC and MEJ fractions from bulk CD8+ T cells treated with non-loaded control beads (–) or activated with α-CD3/α-CD28 mAb loaded beads (+) for 2 h. Blots were probed with mTOR and rictor antibodies. Cox iv was used as fraction validation control (representative of n = 3 independent experiments).
(D) Left, immunoblot analysis of MEJ fractions from wild type (wt) and rictor KO memory CD8+ T cells re-stimulated with OVA peptide for 1 h (+), or from non-activated counterparts (–). Blots were probed for total Akt, pAkt-S473, pAkt-T308, and Cox iv. Right, quantification was performed by normalization of targets to Cox iv. Bar graphs show fold change in Akt phosphorylation following activation relative to non-activated controls (n = 3 independent experiments).
(E,F) Summary of metabolic flux analysis on EM CD8+ T cells activated and assayed as in Figure 1A. Cells were treated with inhibitors of mTOR (OSI-027, 10 μM, and KU0063794, 10 μM) (E), or Akt (Akti, 10 μM, and MK2206, 10 μM) (F). Bar graphs show basal and maximal respiration (n = 4-8 donors).
(G) Immunoblot analysis of total cell lysates from EM CD8+ T cells activated for 2 h with α-CD3/α-CD28 mAb loaded beads only, or similarly activated in presence of nocodazole (10 μM). Bar graph shows Akt-Ser473 phosphorylation normalized to actin (n = 3 independent experiments).
(H) Immunoblots of MEJ and PM fractions from Jurkat T cells. Blots were probed with Gsk-3β and Cox iv antibodies (representative of n = 3 independent experiments).
(I) Immunoblots of MEJ and EGC fractions from bulk CD8+ T cells either left unstimulated (–) or activated (+) with α-CD3/α-CD28 mAb loaded beads for 2 h. Blots were probed for total Gsk-3β, pGsk-3β Ser9, and Cox iv (representative of n = 3 donors).
(J) Left, immunoblot analysis of wt and rictor KO memory OT-I cells activated as in Figure 3D. MEJ fractions were probed for Gsk-3β, pGsk-3β Ser9 and Cox iv. Right, bar graphs display fold change in phosphoprotein levels relative to non-activated controls (n = 3 independent experiments).
Data are presented as mean±SEM. Two-tailed paired Student’s t test (D,E,F,G,J) were used to compare groups. * P < 0.05, ** P < 0.01, ns = not significant. See also Figure S3.
We previously demonstrated that Akt signaling is required for stable upregulation of glycolysis in EM CD8+ T cells (Gubser et al., 2013). mTORC2 is the primary kinase that mediates Akt-Ser473 phosphorylation. In line with this notion, Akt was phosphorylated at Ser473 only in nascent activated EM, but not in similarly activated naïve CD8+ T cells (Fig. S3B). However, Akt-Ser473 is also a substrate of other kinases, such as integrin-linked kinase (ILK) (Lee et al., 2013). Thus, to firmly define the role of mTORC2 in the activation of Akt, we examined Akt-Ser473 phosphorylation in rictor deficient murine CD8+ T cells (Rictorflox/flox: CD4-Cre+) (Zhang et al., 2016). Lack of rictor protein in rictor KO OT-I cells was confirmed by Western blot (data not shown). We proceeded to determine whether Akt-Ser473 phosphorylation was subcellular compartment-specific in activated wild type (wt) and rictor KO CD8+ T cells. In vitro generated wt and rictor KO memory CD8+ T cells were re-stimulated with OVA peptide for 1 hour, and Akt, pAkt-Ser473, and pAkt-Thr308, a target of phosphoinositide-dependent kinase 1 (PDK1), were visualized by immunoblot analysis of mitochondria–ER fractions. Total Akt abundance was similar between wt and rictor KO memory cells (data not shown). There was, however, an approximately 50% reduction in Akt-Ser473 phosphorylation in MEJ fractions from activated rictor KO memory cells, compared to wt counterparts (Fig. 3D). Confirming specificity of this reduced phosphorylation, Akt-Thr308 phosphorylation was unaltered, and in some experiments even higher, in the MEJ fractions from activated rictor KO cells (Fig. 3D). These findings indicated that phosphorylation of Akt-Ser473 in these organellar contact sites, to a significant portion, was dependent on mTORC2 activity.
We then assessed the requirement of an intact mTORC2–Akt signaling axis for mitochondrial metabolic reprogramming. As in the experiments shown in Figure 1A, human EM CD8+ T cells were activated for 12 hours, yet now in the presence of inhibitors of mTOR (OSI-027 and KU-0063794) or Akt (Akti1/2 and MK-2206) –and mitochondrial respiration was examined by metabolic flux analysis. The increase in basal and maximal respiration following αCD3/αCD28 mAb stimulation was diminished in cells pre-treated with either mTOR or Akt inhibitors (Fig. 3E,F). This block in the capacity of memory CD8+ T cells to augment mitochondrial respiration indicated that the mTORC2–Akt axis was important for early metabolic reprogramming of mitochondria in EM CD8+ T cells. We then tested the impact of dissociating mitochondria–ER contacts on Akt activation. To that end, EM CD8+ T cells were stimulated for 2 hours with α-CD3/α-CD28 mAb in presence or absence of nocodazole, and whole cell lysates were assessed for Akt phosphorylation at Ser-473 (Fig. 3G). Dissociation of mitochondria–ER contacts largely prevented Akt phosphorylation, suggesting that intact mTORC2–Akt activity in this subcellular compartment was required for increased mitochondrial respiration of activated EM CD8+ T cells.
Akt has a number of known targets that can modulate mitochondrial metabolism (Manning and Cantley, 2007). One such target is Gsk-3β, a 47 kD kinase that was previously demonstrated in non-immune cells to impact glycolysis and glucose oxidation in mitochondria (Pastorino et al., 2005) (Hoshi et al., 1996). Akt-mediated phosphorylation of Ser9 inhibits the kinase activity of Gsk-3β (Cross et al., 1995). We therefore investigated the idea that inhibition of Gsk-3β by Akt is important for the remodeling of mitochondrial metabolism in activated EM CD8+ T cells. First, we examined the phosphorylation status of total Gsk-3β at Ser9 in activated, non-fractionated EM and NV CD8+ T cells. Intriguingly, Ser9 phosphorylation was significantly increased in activated EM cells but only to a much lesser extent in activated naïve cells (Fig. S3C). In addition, using a screen for phosphorylated Akt substrate motifs (RXRXXS*/T*), a band in the 40-50 kD region, which is the size range of Gsk-3β, was noticeably increased in EM CD8+ T cells activated for 1 hour but not in activated naïve cells (Fig. S3D). Together, these findings demonstrated that Gsk-3β activity was globally diminished, through inhibitory phosphorylation of Ser9, in newly activated EM CD8+ T cells. We thus proceeded to test whether Gsk-3β was present at mitochondria–ER contact sites of T cells. First, we assessed MEJ and PM fractions from Jurkat T cells. Similar to Akt (Fig. 3A), Gsk-3β was readily detected in MEJ fractions from Jurkat T cells, where it was more abundant than in pure mitochondria lysates (Fig. 3H). We then investigated whether Gsk-3β was also present at mitochondria–ER junctions of primary human CD8+ T cells. As shown in Figure 3I (upper lane), Gsk-3β was detected in MEJ fractions of both non-activated and activated CD8+ T cells. We further verified these findings by probing for Gsk-3β localization in activated EM CD8+ T cells using confocal microscopy. In agreement with the immunoblot analyses, Gsk-3β was detected in mitochondria/ER rich regions of these cells (Fig. S3E). Phosphorylation of the Akt target Ser9 in Gsk-3β was also increased in the MEJ fraction of activated human CD8+ T cells (Fig. 3I, middle lane). To investigate whether intact mTORC2 activity was important for phosphorylation of Gsk-3β in the mitochondria–ER subcellular compartment, we tested whether rictor KO memory CD8+ T cells exhibited deficits in Gsk-3β Ser9 phosphorylation in this compartment upon TCR stimulation. Indeed, Gsk-3β Ser9 phosphorylation in the fraction containing MEJs was diminished in rictor deficient cells by approximately 50% (Fig. 3J). These data demonstrated that mTORC2 activity at mitochondria–ER interaction sites was important for the inhibitory phosphorylation of Gsk-3β at Ser9. Combined with the observations above it is plausible that impaired Akt-Ser473 phosphorylation in rictor KO memory T cells led to deficits in Akt activity, and thus reduced phosphorylation of Gsk-3β at Ser9 in mitochondria–ER junctions. Moreover, these findings indicated that mitochondria–ER appositions function as a hub for mTORC2–Akt–Gsk-3β signaling during memory recall responses.
HK-I binding to VDAC controls mitochondrial metabolism in T cells
We went on to relate mTORC2–Akt–Gsk3β signaling at mitochondria–ER contact sites with the mitochondrial reprogramming uniquely observed in newly activated memory CD8+ T cells (Fig. 1A). Pyruvate dehydrogenase (PDH) is a known target of Gsk-3β whose activity is inhibited following phosphorylation by Gsk-3β (Hoshi et al., 1996). Hence, we first examined whether activation of EM CD8+ T cells resulted in increased PDH activity due to Gsk-3β inhibition by Akt. EM CD8+ T cells were activated for 1 hour, and PDH activity was examined in total cell lysates. There was no detectable difference in PDH activity between non-activated and activated cells (data not shown). Another Gsk-3β target localized to mitochondria and abundant at mitochondria–ER appositions is the voltage dependent anion channel (VDAC), which along with IP3R and GRP75 form a mitochondria–ER tethering complex (Szabadkai et al., 2006). Gsk-3β mediated phosphorylation of VDAC inhibits hexokinase (HK) binding in HeLa cells (Pastorino et al., 2005). As a functional readout of Gsk-3β inhibition mitochondria–ER junctions, we assessed HK recruitment to the mitochondria in activated CD8+ T cells by probing for HK abundance in the MEJ fraction from quiescent and activated human CD8+ T cells. HK-II was not detected in total lysates from bulk CD8+ T cells (data not shown). However, HK-I was enriched in MEJ fractions upon CD8+ T cell activation (Fig. 4A) We then interrogated whether destabilizing mitochondria–ER contacts attenuated HK-I recruitment to mitochondria in activated CD8+ T cells. HK-I abundance was decreased in mitochondrial lysates from nocodazole treated bulk CD8+ T cells, indicating that the recruitment of HK-I to mitochondria was indeed dependent on stable mitochondria–ER contacts (Fig. 4B). Combined, a molecular framework emerged from these findings, where stable mitochondria–ER contacts support mTORC2–Akt–Gsk-3β signaling, which in turn allows recruitment of HK-I to the mitochondria.
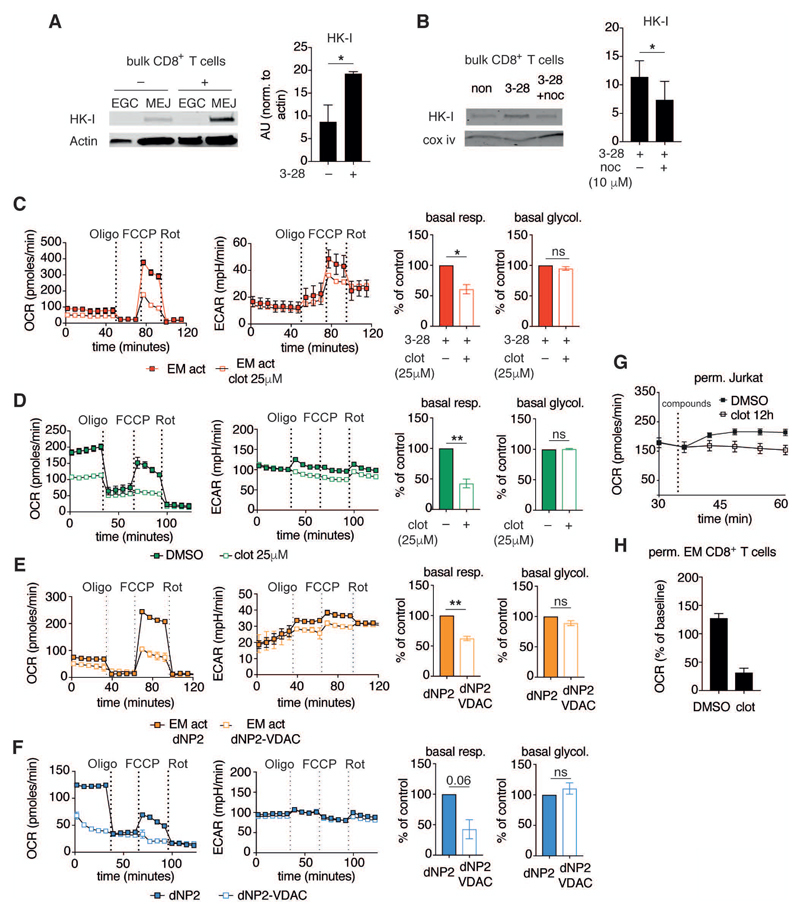
Figure 4. HK-I binding to VDAC regulates mitochondrial metabolism.
(A) Left, MEJ and EGC fractions from bulk human CD8+ T cells activated as in Figure 1A for 2 h. Immunoblots were probed for HK-I and actin. Right, summary bar graph of HK-I abundance in the mitochondria / mitochondria–ER junctions containing fraction normalized to actin (n = 3 donors).
(B) Left, Immunoblot of MEJ fractions from bulk human CD8+ T cells activated as in (A), or activated in the presence of nocodazole (10 μM). Right, summary bar graph of HK-I abundance in the mitochondria / mitochondria–ER junctions fraction normalized to Cox iv (n = 3 donors).
(C) Left, representative mitochondrial perturbation assay of human EM CD8+ T cells activated as in Figure 1A in presence of DMSO, or activated in the presence of clotrimazole for 12 h (25 μM). Right, summary bar graphs of basal respiration and glycolysis (n = 4 donors).
(D) Left, representative mitochondrial perturbation assay using Jurkat T cells treated with DMSO or clotrimazole (25 μM) for 12 h. Right, summary bar graphs of basal respiration and glycolysis (n = 3 independent experiments).
(E) Left, representative mitochondrial perturbation assay of human EM CD8+ T cells activated as in Figure 1A in presence of dNP2 peptide (5 µM), or activated in the presence of dNP2-VDAC peptide for 12 h (5 μM). Right, summary bar graphs of basal respiration and glycolysis (n = 3 donors).
(F) Left, representative mitochondrial perturbation assay using Jurkat T cells treated with dNP2 peptide (5 µM) or dNP2-VDAC peptide (5 μM) for 12 h. Right, summary bar graphs of basal respiration and glycolysis (n = 3 independent experiments).
(G) Oxygen consumption rates of permeabilized Jurkat T cells treated as in (D). Cells were permeabilized and respiration was assessed after addition of pyruvate, malate, and ADP (indicated as 'compounds' on the graph) (representative of n = 2 independent experiments).
(H) Oxygen consumption of permeabilized human EM CD8+ T cells activated as in Figure 1A for 12 h. Cells were then permeabilized and treated with malate, pyruvate, oligomycin, and DMSO/clotrimazole (25 μM), and changes in OCR were assessed relative to pre-injection rates (representative of n = 2 donors).
Data are presented as mean±SEM. Two-tailed paired Student’s t test (B,C,D,E,F) were used to compare groups. * P < 0.05, ** P < 0.01, ns = not significant. See also Figure S4.
To further elaborate on the impact of HK-I binding to VDAC on mitochondrial respiration, we examined metabolic remodeling in EM CD8+ T cells that were activated in the presence of clotrimazole or bifonazole, both drugs blocking HK binding to VDAC (Penso and Beitner, 1998). Clotrimazole treatment did not affect aerobic glycolysis in EM CD8+ T cells activated for 12 hours (Fig. 4C). However, mitochondrial respiration was clearly muted in the presence of this compound (Fig. 4C). Bifonazole induced a similar effect on mitochondrial respiration in nascent activated EM CD8+ T cells, while also leaving glycolysis unaltered (Fig. S4A). Moreover, clotrimazole pre-treatment did not attenuate the activation-induced early and stable glycolytic switch characteristic of EM CD8+ T cells (Fig. S4B), indicating that HK-I activity in nascent activated memory CD8+ T cells was independent of its subcellular localization and accessibility to mitochondrial ATP (van der Windt et al., 2013). Congruent with these experiments, clotrimazole also inhibited respiration in Jurkat T cells while leaving glycolysis largely unaffected (Fig. 4D). We then proceeded to specifically block HK-I binding to VDAC by using cell penetrating VDAC1 based peptides that were N-terminally linked to dNP2, a 22-residue long penetrating peptide from the human novel LZAP-binding protein (NLBP) (Arzoine et al., 2009; Lim et al., 2015; Lim et al., 2016). VDAC based peptides have previously been demonstrated to specifically prevent HK-VDAC interactions (Arzoine et al., 2009). In agreement with the above findings, hindering HK–VDAC interactions using dNP2-VDAC peptides attenuated mitochondrial respiration in activated EM CD8+ T cells (Fig. 4E) as well as in Jurkat T cells (Fig. 4F), whereas glycolysis remained largely unaffected (Fig. 4E and Fig. 4F). This reduction in mitochondrial metabolism was not due to increased cells death, since cellular viability was similar between dNP2 controls and dnP2-VDAC treated cells (Fig. S4C).
The observed difference in mitochondrial respiration between clotrimazole treated and control cells could be driven by differences in bioenergetic demands brought about by alterations in cellular activity. Therefore, we proceeded to assess mitochondrial respiration in permeabilized cells, which enabled us to control for this potential confounder. Mitochondria from permeabilized control Jurkat T cells were able to enhance respiration following addition of excess ADP, pyruvate and malate (State IIIADP respiration) (Fig. 4G). By contrast, cells treated with clotrimazole for 12 hours were unable to increase State IIIADP respiration. The unresponsiveness of clotrimazole-treated Jurkat T cells to state III metabolites suggested that blockade of HK binding to VDAC altered VDAC conductance for substrates either used in ATP turnover or substrate oxidation (Brand and Nicholls, 2011). Next, we evaluated the direct effect of clotrimazole on mitochondrial respiration in CD8+ T cells. For these assays, we examined oxygen consumption of permeabilized activated human EM CD8+ T cells in the presence of oligomycin, malate, and pyruvate. Control cells increased oxygen consumption upon addition of substrates, whereas in clotrimazole-treated cells oxygen consumption decreased (Fig. 4H). Together, these results suggested that HK-I binding to VDAC in memory CD8+ T cells alters VDAC conductance for substrates important in supporting mitochondrial respiration. In addition, the mTORC2–Akt–Gsk-3β dependent early increase in mitochondrial respiration, unique to EM CD8+ T cells, was mediated by this alteration in VDAC conductance.
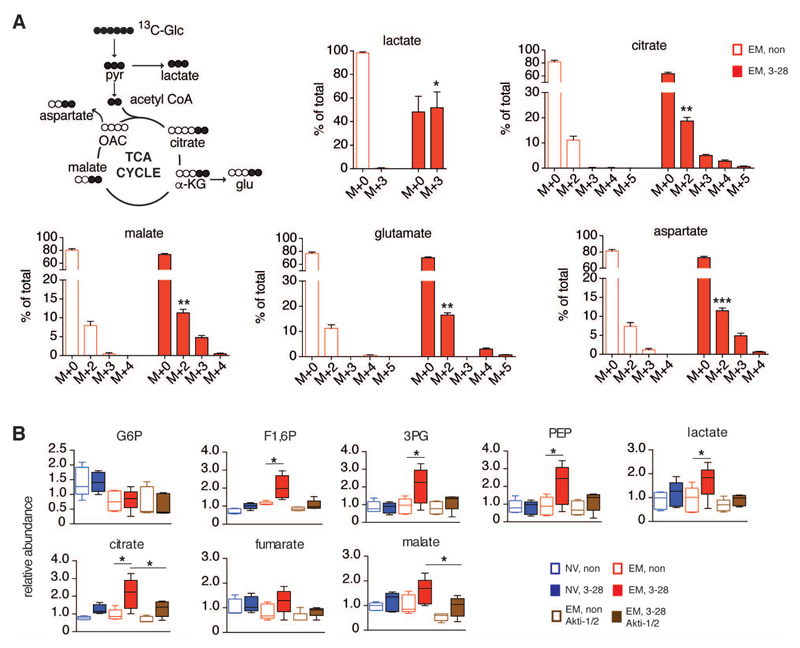
Glucose is metabolized in the mitochondria of activated EM CD8+ T cells
Aerobic glycolysis, or Warburg metabolism, is important for the early recall phase of memory CD8+ T cells (Gubser et al., 2013). This metabolic activity also generates pyruvate to fuel mitochondrial respiration. Given the observed mTORC2–Akt dependent increase in aerobic glycolysis and mitochondrial respiration following memory T cell activation, we examined how glucose is metabolized in memory CD8+ T cells immediately after activation. EM CD8+ T cells were incubated in uniformly labeled 13C6-glucose for 2 hours, and isotopologue distribution of glucose-derived metabolites was assessed under non-activating and activating conditions. Consistent with our previous findings (Gubser et al., 2013), glucose derived lactate (M+3) was significantly increased following activation (Fig. 5A). Yet, glucose derived isotopologues of citrate (M+2, M+3, M+4) and malate (M+2, M+3) were also increased (Fig. 5A). This established that tricarboxylic acid (TCA) cycle activity in EM CD8+ T cells was rapidly enhanced following activation. Lastly, glucose derived aspartate (M+2, M+3) was also more abundant in activated EM CD8+ T cells (Fig. 5A). These data suggested that a primary role of mTORC2–Akt activation at mitochondria–ER junctions in nascent stimulated EM CD8+ T cells could be to enhance pyruvate oxidation in mitochondria. Thus, to define in more detail the impact of Akt activation on mitochondrial metabolism, we utilized unbiased global metabolomic profiling to differentiate the metabolome of newly activated NV and EM CD8+ T cells from non-activated counterparts, and from EM cells activated in the presence of an Akt inhibitor (Akti). NV and EM cells were activated for 2 hours ±Akti, and metabolites were extracted. Congruent with the glucose tracing experiments shown in Figure 5A, most differentially abundant metabolites in activated NV vs. EM CD8+ T cells were part of the glycolytic pathway and the TCA cycle (Fig. 5B). Specifically, the glycolytic metabolites fructose 1,6 bisphosphate (F1,6P), 3-phosphoglycerate (3PG), phosphoenol pyruvate (PEP), and lactate were elevated in activated EM but not in naïve CD8+ T cells (Fig. 5B). In addition, TCA cycle metabolites (citrate, malate and, to some extent, fumarate) were also selectively increased in activated EM cells (Fig. 5B). Upon pre-treatment with Akti, all glycolytic and TCA cycle metabolites that were differentially increased in activated EM CD8+ T cells remained at baseline levels (Fig. 5B). Of note, the abundance of metabolites along other pathways was unaffected by Akt inhibition. This was true for metabolites from pathways that did not change activity upon T cell activation (e.g. ascorbate, glutathione), as well as for metabolites from pathways that altered activity in stimulated cells (e.g. carnitine, acetylcarnitine) (Fig. S5A). Akt is an established regulator of glucose uptake (Manning and Cantley, 2007). The observed reduction in glycolytic and TCA cycle metabolites following Akt inhibition could be due to an lack of glucose uptake. To probe for changes in glucose uptake following Akt inhibition, we incubated EM CD8+ T cells with 2-NBDG. Stimulation with α-CD3/α-CD28 mAb enhanced glucose uptake in EM T cells (Fig. S5B). As expected, glucose uptake following addition of Akti was reduced, yet remained clearly higher than non-stimulated counterparts. In agreement with these findings, we previously demonstrated that inhibition of the mTORC2–Akt axis does not globally impact EM T cell activation (Gubser et al., 2013). Against this background, the current findings suggested that a key role of the mTORC2–Akt–Gsk-3β signaling axis in nascent activated EM CD8+ T cell was enhancement of glucose oxidation in mitochondria, specifically by increasing conductance of metabolites across the outer mitochondrial membrane following HK-I binding to VDAC and thus providing for substrates essential in TCA cycle and electron transport chain (ETC) functionality.
Figure 5. Glucose is metabolized in the mitochondria of nascent activated EM CD8+ T cells.
(A) Glucose-derived carbon labeling of glycolysis and tricarboxylic acid (TCA) cycle metabolites. EM CD8+ T cells were cultured under basal and activating (α-CD3/α-CD28 mAb loaded beads) conditions for 6 h in 13C-glucose containing media. Bar graphs display mass isotopologue distribution of incorporated 13C. M+0 bars represent fractions of metabolites with no 13C-glucose incorporation (n = 4-5 donors).
(B) Glycolysis and TCA cycle metabolites in NV and EM CD8+ T cells stimulated with α-CD3/α-CD28 mAb loaded beads. Metabolites from EM CD8+ T cells (±activation) treated with Akti (10 μM) were further assessed. Box and whisker graphs reflect abundance of glucose-6-phosphate (G6P), fructose-1,6-bisphosphate (F1,6P), 3-phosphoglycerate (3PG), phosphoenolpyruvate (PEP), lactate, citrate, fumarate and malate in all samples tested (n = 5 donors).
Data are represented as mean±SEM (A) and median±min/max (B). Two-tailed paired Student’s t test (A), and Anova contrasts (B), were used to compare groups. * P < 0.05, ** P < 0.01, *** P < 0.001. See also Figure S5.
Enhancement of mitochondrial metabolism driven by mitochondria–ER junctions is important for the rapid response of memory CD8+ T cells
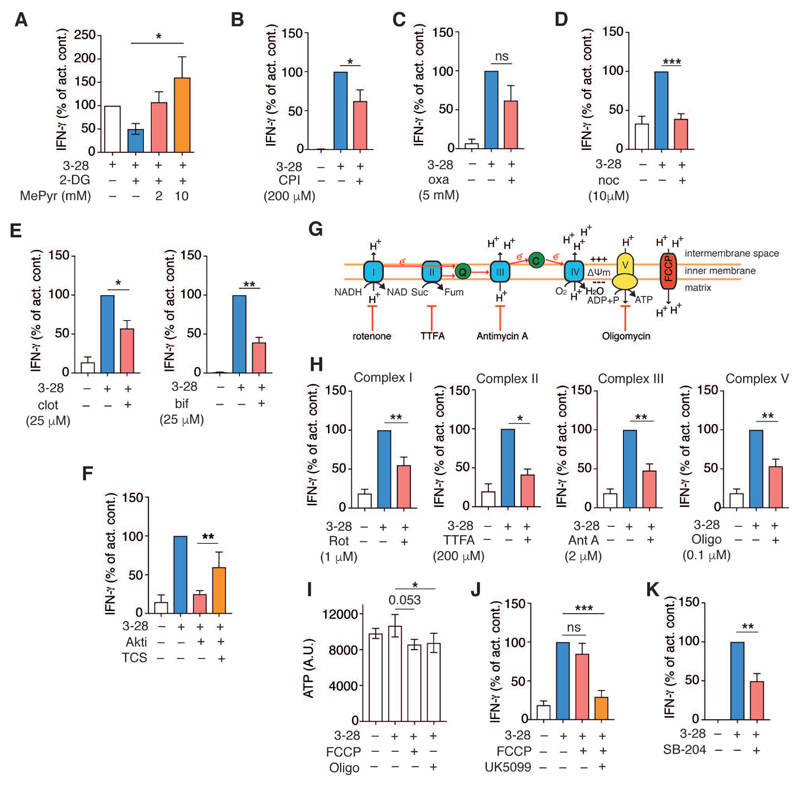
Given the selective enhancement of mitochondrial respiration in newly activated memory CD8+ T cells, IFN-γ production during the immediate-early recall response could be linked to pyruvate oxidation in the mitochondria. To test this idea, we assessed whether the membrane permeable form of pyruvate (methyl pyruvate, MePyr) could rescue IFN-γ production in cells treated with the glycolysis inhibitor, 2-deoxyglucose (2-DG). As expected, treatment with 2-DG decreased IFN-γ production by activated EM CD8+ T cells (Fig. 6A). However, addition of increasing doses of MePyr fully circumvented the inhibitory effect of 2-DG on IFN-γ production (Fig. 6A). Pyruvate sits at a metabolic branch point, since it is either metabolized in the TCA cycle or converted to lactate. Thus, we examined the impact of PDH or lactate dehydrogenase (LDH) inhibition on IFN-γ production. CPI-613, a PDH inhibitor, reduced IFN-γ production in activated EM CD8+ T cells (Fig. 6B). Inhibition of lactate dehydrogenase with oxamate did not consistently and not significantly affect IFN-γ production (Fig. 6C). These results established that metabolism of glucose-derived pyruvate in the mitochondria was required to optimally support the rapid recall response of EM CD8+ T cells.
Figure 6. Intact mitochondria–ER contacts and ETC function, but not mitochondrial ATP production, are required to support the early response of memory CD8+ T cells.
(A) IFN-γ production by EM CD8+ T cells 12 h post-stimulation. The bar graph displays fold change in IFN-γ production relative to activation with α-CD3/α-CD28 mAb loaded beads alone, or in cells co-treated with 2-DG (1 mM) or 2-DG + MePyr (2 or 10 mM) (n = 3 donors).
(B-E) IFN-γ production by EM CD8+ T cells following activation with α-CD3/α-CD28 mAb loaded beads for 12 h or activation in the presence of CPI-613 (PDH inhibitor, 200 μM) (B), (n = 6 donors); oxamate (Oxa, lactate dehydrogenase inhibitor, 5mM) (C), (n = 5 donors); nocodazole (noc) (10 μM) (D), (n = 9 donors); left, clotrimazole (clot) (25 μM), right, bifonazole (bif) (25 µM) (E), (clot, n = 6 donors; bif, n = 4 donors).
(F) IFN-γ production by human EM CD8+ T cells activated as in (B). Cells were treated with Akti (10 μM) alone, or in combination with TCS-2002 (Gsk-3β inhibitor, 10 μM), (n = 9 donors).
(G) Schematic representation of mitochondrial electron transport chain (ETC) complexes along with their respective inhibitors.
(H) IFN-γ production by EM CD8+ T cells activated with α-CD3/α-CD28 mAb loaded beads for 12 h ± ETC inhibitors (n = 7-9 donors).
(I) ATP measurement in EM CD8+ T cells with no stimulation or activated with α-CD3/α-CD28 mAb loaded beads ± FCCP (5 μM) or oligomycin (Oligo, 0.1 μM) for 12 h, (n = 5 donors).
(J) IFN-γ production by EM CD8+ T cells activated as in (B) in the presence of the uncoupling agent FCCP (5 μM) ± UK5099 (10 µM) (n = 5-7 donors).
(K) IFN-γ production by EM CD8+ T cells activated as in (B) in the presence of the ACLY inhibitor SB-204990 (3 µM) (n = 5 donors).
Data are presented as mean±SEM. Two-tailed paired Student’s t tests were used throughout to compare groups. * P < 0.05, ** P < 0.01, *** P < 0.001, ns = not significant. See also Figure S6.
We then interrogated the role of mitochondria–ER interactions in potentiating the immediate-early EM CD8+ T cell recall response. Rapid IFN-γ production was prevented in nocodazole treated EM CD8+ T cells (Fig. 6D). In addition, inhibiting recruitment of HK-I to the mitochondria by clotrimazole and bifonazole also reduced IFN-γ production (Fig. 6E). IFNG transcripts were likewise reduced in both nocodazole and clotrimazole treated cells (Fig. S6A). Viability of EM CD8+ T cells was not affected by either nocodazole or clotrimazole (Fig. S6B). Lastly, blockade of HK-I and VDAC interaction using the cell penetrating peptide dNP2-VDAC also reduced IFN-γ production (Fig. S6C).
These data indicated that, by providing a signaling platform for the mTORC2–Akt–GSK-3β pathway, intact mitochondria–ER contacts were required for reprogramming of mitochondrial respiration and enhancement of IFN-γ production. Given these findings, we postulated that reduced IFN-γ production in activated EM CD8+ T cells with inhibited Akt function (thus prevented from phosphorylating and hence inhibition of Gsk-3β) should be rescued by pharmacologic inhibition of Gsk-3β. In agreement with our previous experiments, Akt inhibition reduced IFN-γ gene transcription and protein production by EM T cells (Fig. S6D and Fig. 6F). Combined pre-activation treatment of cells with both Akti and the GSK-3β inhibitor, TCS-2002, completely rescued IFN-γ production at the transcriptional, and partially at the protein level (Fig. S6D and Fig. 6F). Together these results established a mitochondria–ER junction localized mTORC2–Akt–Gsk3β signaling axis that significantly contributed to the rapid recall response of memory CD8+ T cells by promoting HK-I binding to VDAC, and enhancing VDAC conductance for cytoplasmic metabolites that drive mitochondrial respiration.
In addition to their key role in producing ATP, mitochondria are also important for the generation reactive oxygen species (ROS); calcium buffering; regulation of apoptosis, and for biomolecular synthesis (Weinberg and Chandel, 2015). Since fueling of the TCA cycle by pyruvate was a key function of early glycolytic remodeling in EM CD8+ T cells, we asked which aspect of mitochondrial function was required to support rapid IFN-γ production. To begin to address this question, EM CD8+ T cells were activated in the presence of a series of mitochondrial respiration inhibitors (Fig. 6G). Under each of the indicated conditions we then probed for effects on proliferation (Fig. S6E), activation (by assessing CD69 surface expression) (Fig. S6F), and production of IFN-γ (Fig. 6H). Inhibitors that targeted mitochondrial electron transport chain (ETC) complexes I (rotenone), II (TTFA), III (antimycin-A), and V (oligomycin) all blocked cell proliferation (Fig. S6E), but had little or no effect on CD69 cell surface expression (Fig. S6F). This indicated that inhibition of mitochondrial respiration had a limited impact on early activation as reflected by upregulation of CD69, but it efficiently prevented cell proliferation, potentially due to the reduction of ATP availability (Chang et al., 2013). Intriguingly, all ETC complex-inhibitors also reduced IFN-γ production, which showed that maintaining electron transport function was an important aspect of the rapid recall response of EM CD8+ T cells (Fig. 6H). In addition, IFNG transcripts were also reduced in the presence ETC inhibitors (Fig. S6G). Of note, none of the ETC inhibitors used in these experiments affected cell viability (Fig. S6B). Next, we assessed the impact of dissipating the mitochondrial proton gradient on IFN-γ production by using the mitochondrial uncoupler, FCCP. Similar to the effect of ATP synthase inhibition with oligomycin, chemical uncoupling of the proton gradient diminished ATP production (Fig. 6I). We hypothesized that, if deficits in ATP synthesis following inhibition of the ETC led to a decrease in IFN-γ production, then chemical uncoupling of the proton gradient should mimic this effect. Analogous to the inhibition of ETC complexes, chemical uncoupling of mitochondrial respiration had little impact on CD69 expression yet it blocked EM CD8+ T cell proliferation (data not shown). However, in clear contrast to ETC inhibition, disruption of the electrochemical gradient did not inhibit IFN-γ production (Fig. 6J). However, addition of the mitochondrial pyruvate carrier inhibitor, UK5099, significantly reduced their capacity to produce IFN-γ (Fig. 6J). This demonstrated that decreased ATP synthesis by mitochondria was not essential for immediate-early IFN-γ production and that pyruvate oxidation in the mitochondria is important for the recall response. We previously demonstrated that acetylation of H3K9 at the IFNG promoter is dependent on glycolysis (Gubser et al., 2013). It is plausible that glucose oxidation in the mitochondria fuels the TCA cycle to drive the synthesis of intermediates that are important for epigenetic regulation of gene expression in memory CD8+ T cells. Conversion of glucose-derived citrate into acetyl-CoA –which then can be used for histone acetylation– is mediated by the enzyme ATP citrate-lyase (ACLY) (Balmer et al., 2016; Wellen et al., 2009). Indeed, treatment of activated EM CD8+ T cells with SB-204990, an ACLY inhibitor, attenuated IFN-γ production (Fig. 6K). In line with this finding, exposure to histone acetyltransferase inhibitors also reduced IFN-γ production in these cells (Fig. S6H).
Mitochondrial ROS regulates IL-2 production in T cells (Sena et al., 2013). We thus also examined the impact of mitochondrial ROS on IFN-γ production by treating activated EM CD8+ T cells with a mitochondria targeted anti-oxidant (mitotempo, MTP). Targeting mitochondrial ROS did not alter IFN-γ production (Fig. S6I). Lastly, blockade of mitochondrial calcium buffering, by using inhibitors that target the mitochondrial calcium uniporter (MCU), Ru360 and ruthenium red (RR), also did not affect IFN-γ production (Fig. S6J).
In all these data established that mitochondria–ER contact-driven enhancement of mitochondrial respiration was important for the rapid CD8+ T cell recall response, and that mitochondria dependent immediate-early IFN-γ production was not linked to ATP synthesis, mitochondrial ROS signaling, or calcium buffering.
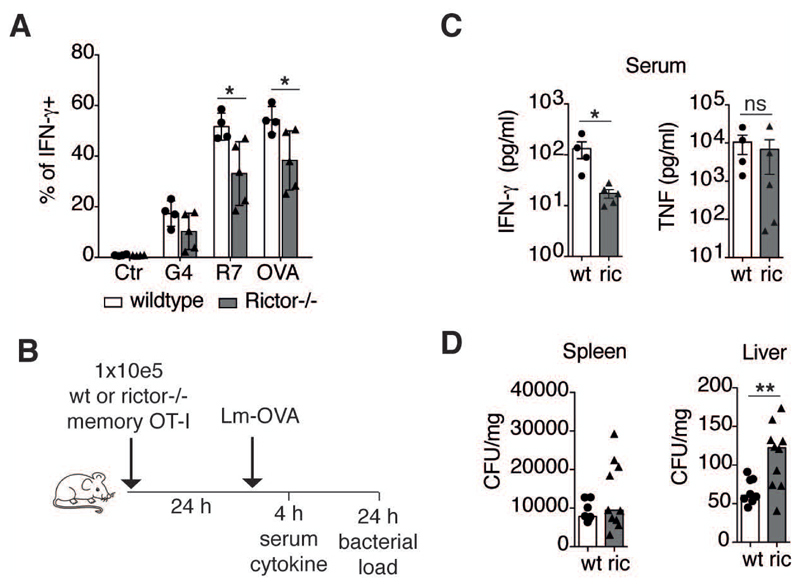
Rictor deficiency attenuates the rapid recall response of memory CD8+ T cells in vitro and in vivo
Rapid IFN-γ production by human EM CD8+ T cells requires mTORC2 activation (Gubser et al., 2013). Having observed increased abundance of mitochondria–ER contacts in EM CD8+ T cells and mTORC2 being localized to this subcellular structure, we next assessed whether IFN-γ production by mouse memory OT-I cells was similarly dependent on mTORC2 following re-stimulation with its cognate peptide (OVA), or with altered peptide variants possessing lower TCR affinities (OVA>R7>G4). Rapid IFN-γ production of in vitro differentiated rictor KO memory OT-I cells was lower than in wild type counterparts across all TCR:peptide affinities tested (Fig. 7A). These differences in immune functionality were not due to the altered differentiation or cellular activation of rictor deficient memory cells, since both differentiation and activation markers (CD44, CD127, CXCR3, Ly6C, CD62L, KLRG1, CD27, CD25, CD69, CX3CR1, and CCR7) were all similarly expressed by wt and rictor KO memory cells (Fig. S7). We further examined whether the reduction in IFN-γ production by rictor deficient memory cells in vitro also translated to reduced immune function in vivo. To that end, rictor KO and wt memory OT-I cells were adoptively transferred into mice, which were infected with LmOVA at 24 hours post-transfer (Fig. 7B). These experiments were designed to probe the rapid recall functionality of adoptively transferred memory OT-I cells. At 4 hours post-infection we evaluated serum cytokine concentrations (IFN-γ, TNF), and at 24 hours post-infection bacterial loads were assessed in the spleen and liver. IFN-γ serum concentrations were markedly reduced in infected mice that received rictor deficient memory cells (Fig. 7C). Additionally, the bacterial burden in the liver was significantly elevated in mice that received rictor KO memory OT-I cells (Fig. 7D). These findings confirmed the importance of mTORC2 activation in memory CD8+ T cells during the very early phase of the recall response.
Figure 7. Rictor deficiency attenuates memory recall response, in vivo.
(A) Frequency of IFN-γ producing cells from in vitro derived wt and rictor KO memory OT-I cells re-stimulated for 4 h with OVA, R7, or G4 peptide (n = 4-5 independent experiments).
(B) Schematic diagram of adoptive transfer and infection strategy.
(C) Serum IFN-γ and TNF levels 4 h after infection of mice with LmOVA. (n = 4-5 independent experiments).
(D) Listeria monocytogenes CFU in spleen and liver at 24 h post-infection (n = 8-10 independent experiments).
Each dot represents data obtained from cells isolated from one mouse, bars indicate mean±SEM. Two-tailed paired Student’s t test were used to compare groups. * P < 0.05, ** P < 0.01, ns = not significant. See also Figure S7.
In summary, our data identified compartmentalized metabolic reprogramming in memory CD8+ T cells. Specifically, we demonstrated selective abundance of mitochondria–ER junctions in memory CD8+ T cells. This subcellular compartment was a hub that, by default, brought together signaling molecules necessary for rapid glycolytic remodeling and enabled recruitment HK-I to VDAC. Binding of HK-I to VDAC supported mitochondrial import of metabolites needed for respiration, and oxidation of glucose in the mitochondria was the metabolic requirement for rapid production of IFN-γ–potentially by providing substrate for histone acetylation.
Discussion
It was recently demonstrated that exhausted CD8+ T cells from mice with chronic virus infection, as well as functionally suppressed tumor infiltrating CD8+ T cells, exhibit deficits in mitochondrial biogenesis and function (Bengsch et al., 2016; Scharping et al., 2016). These findings align with our observation that augmentation of mitochondrial metabolism in CD8+ T cells is critical for sustaining effector capacity (Balmer and Hess, 2016). Memory CD8+ T cells typically possess greater mitochondrial mass than naïve and effector counterparts (van der Windt et al., 2012), and mitochondria of memory T cells are morphologically and functionally distinct from those of effector cells (Buck et al., 2016). Yet, how and when mitochondrial function is reprogrammed to provide the advantage that memory CD8+ T cells display during the recall response is not known. Likewise, how upregulation of glycolysis –a metabolic hallmark of activated memory CD8+ T cells– relates to altered mitochondrial activity has not been explored in any detail. Using a combination of imaging; biochemical; metabolic and immunologic methods, our study demonstrates that mitochondrial contact with the ER is a key driver of rapid metabolic and functional reprogramming in memory CD8+ T cells (Fig. S8). We show that mitochondria–ER contacts are enriched in memory CD8+ T cells and required for enhancement of mitochondrial respiration in newly activated cells. In a preformed manner mTORC2, Akt and Gsk-3β were present in this subcellular compartment of non-activated memory CD8+ T cells, and activation-induced signaling activity of this pathway in mitochondria–ER junctions was essential for recruitment of HK-I to the mitochondria via binding to VDAC. HK-I binding was important for enhancing mitochondrial respiration in newly activated memory T cells, which we further identified as a requirement for rapid IFN-γ production.
We previously demonstrated that histone 3, lysine 9 (H3K9) acetylation in the IFNG promoter is increased following activation of EM CD8+ T cells (Gubser et al., 2013). Acetyl-CoA used for acetylation of histones is primarily derived from glucose oxidized in the mitochondria (Martinez-Reyes et al., 2016; Wellen et al., 2009). We now show that glucose-derived citrate, the primary metabolite exported from mitochondria and converted to acetyl-CoA by ATP citrate lyase, was abundant in activated EM CD8+ T cells, but not in Akt inhibited counterparts. Preventing acetyl-CoA formation from citrate, as well as blocking histone acetyl-transferase activity both reduced the capacity of nascent activated EM CD8+ T cells to produce IFN-γ. Moreover, dissociation of mitochondria–ER contacts; inhibition of mTORC2–Akt signaling, and dissociation of HK-I from mitochondria all negatively impacted the selective enhancement of mitochondrial respiration and IFN-γ production in nascent activated EM CD8+ T cells. Therefore, these findings point to mitochondria–ER junctions as a hub bringing together signaling and metabolic components that potentiate glucose oxidation, mitochondrial respiration and citrate production in newly activated EM CD8+ T cells to enable the rapid recall response.
Increased calcium in the mitochondrial matrix is known to potentiate respiration by enhancing the function of TCA cycle dehydrogenases (Denton, 2009). Mitochondria–ER interactions can directly influence mitochondrial metabolism by regulating calcium flux via the VDAC–mitochondrial calcium uniporter complex (Raturi and Simmen, 2013). Of note, this subcellular compartment was observed to steer constitutive inositol-3-phosphophate receptor (IP3R) mediated calcium transfer from the ER to the mitochondria in transformed chicken B cell lines, which was important for maintenance of mitochondrial respiration (Cardenas et al., 2010). By contrast, insulin was shown to promote the mTORC2–Akt dependent inhibition of IP3R activity in fibroblasts, which limited ER–mitochondria calcium flux and decreased mitochondrial membrane potential (Betz et al., 2013). Here, i.e. in nascent activated memory CD8+ T cells, blocking calcium buffering did not influence rapid production of IFN-γ, suggesting that mitochondria–ER junctions in T cells were not essential to this process.
HK–VDAC interaction can modulate mitochondrial outer membrane (OMM) conductance to small molecules (organic phosphate) and ions (Ca2+) (Shoshan-Barmatz and Mizrachi, 2012). However, the impact of this interaction on conductance of metabolites, such as ADP, pyruvate and malate, is not well established. Our results show that dissociation of HK-I from the mitochondria did not affect cell-wide aerobic glycolysis early after activation, but reduced mitochondrial respiration. We further show that oxidation of glucose in the mitochondria was important in newly activated EM CD8+ T cells to support rapid cytokine production. By hindering HK-I binding to VDAC in permeabilized cells, we additionally provide evidence that HK-I binding to VDAC indeed augmented metabolite flux through the OMM, thereby promoting TCA activity and hence respiration. Thus, in this scenario, the recruitment of HK-I to VDAC linked the initial step of glycolysis to glucose oxidation (i.e. pyruvate oxidation) in the mitochondria.
In LPS+ATP stimulated macrophages, mTORC1 dependent expression of HK-I is essential for NLRP3 inflammasome activation. In this setting, HK-I was found to localize to mitochondria, however, its effects on mitochondrial metabolism were not investigated (Moon et al., 2015). By contrast, in CD8+ T cells, HK-I is constitutively expressed and translocation to the mitochondria was dependent on mTORC2–Akt–Gsk-3β signaling. More importantly, in our system HK-I binding to VDAC modulated mitochondrial respiration. NLRP3 inflammasome components in THP1 cells were previously shown to co-localize with mitochondria and ER (Zhou et al., 2011) Moreover, HK is a pattern recognition receptor that mediates NLRP3 activation (Wolf et al., 2016). Together, these findings suggest that mitochondria–ER interactions may also serve as a subcellular hub for immune cell inflammasome activation, potentially via the initial recruitment of HK-I to the mitochondria. In addition, since endogenous NLRP3 inflammasome activation was recently shown to control human Th1 responses following TCR/CD46 stimulation (Arbore et al., 2016), it would be interesting to investigate whether this subcellular compartment is involved in T cell inflammasome activation.
In summary, our data uncovered that integrating ultrastructural aspects of the cell –such as the ER–mitochondria network– is vital to fully appreciate how the intersection of signaling and metabolic pathway usage impacts immune cell functionality.
Experimental Procedures
Human CD8+ T cell isolation and sorting
Blood samples were obtained from healthy blood donors as buffy coats after written informed consent (Blood donor center, University Hospital Basel). Peripheral blood mononuclear cells (PBMC) were isolated by standard density-gradient centrifugation protocols (Lymphoprep Fresenius Kabi, Norway). CD8+ T cells were enriched by positive selection using magnetic CD8+ beads (Miltenyi Biotec, Germany). Cells were rested overnight prior to cell sorting in RPMI-1640 medium (Gibco, USA) containing 10% fetal bovine serum (FBS, Gibco, USA), 50U/ml penicillin and 50 μg/ml streptomycin (Gibco, USA) (R10FBS). For sorting of naïve and EM CD8+ T cells, CD8+ were stained with anti-CD62L (ImmunoTools, Germany) and anti-CD45RA (Beckman Coulter, USA) antibodies. NV and EM CD8+ T cells were identified as CD62L+ CD45RA+ and CD62L– CD45RA– populations, respectively. Cell sorting was performed with a BD influx cell sorter (BD Bioscience, USA). Cells were rested in R10FBS for 2-4 hours at 37°C prior to further experiments.
Animals
T cell specific rictor ko mice were generated by crossing CD4 promoter controlled Cre recombinase transgenic mice with rictor KO mice. T cell specific rictor KO mice were further crossed with OT-I TCR transgenic mice. All mouse strains were backcrossed to the C57Bl/6J background. Mice were maintained at the specific pathogen free animal facilities at the University of Lausanne and at the University of Basel. All animal experiments were performed in accordance with local and Swiss federal guidelines.
Mouse T cell isolation and culture
Spleen and lymph nodes were removed from OT-I wt and rictor ko mice. Single cell suspensions from both tissues were activated with OVA peptide for 3 d and then cultured for an additional 3 d in IL-15 (10 ng/ml). Mouse OT-I cells were cultured in RPMI 1640 containing 10% FCS, 100 U/mL penicillin, 100 μg streptomycin, 0.29 mg/mL L-glutamine, 50 μM β-Mercaptoethanol. In vivo derived, OT-I memory T cells were isolated from L. monocytogenes infected mice (> 40 days post-infection) that were transferred with naïve OT-I cells 24 hours before infection. T cells were isolated from splenocytes, and memory OT-I cells sorted using a BD influx cell sorter (BD Bioscience, USA).
Cells and reagents
Jurkat T cells were cultured in R10FBS supplemented with 2 mM glutamine. Chemicals and inhibitors used in this study were as follows: 2-DG (10 mM), Akti1/2 (10 μM), Osi-027 (10 μM), KU-0063794 (10 μM), Rotenone (1 μM), TTFA (200 μM), Antimycin A (2 μM), Oligomycin (0.1 or 1 μM) or 4-(trifluoromethoxy) phenylhydrazone (FCCP, 2 μM), Clotrimazol (25 μM), nocodazole (10 μM), oxamate (5 mM), methyl pyruvate (1 or 10 mM), bifonazole (25 µM), MB-3 (1, 10 µM), 2-NBDG (0.5 mM) and mitotempo (20 μM) (all from Sigma-Aldrich). TCS-2002 (10 µM), SB 204990 (3 µM) and garcinol (1, 10 µM) (Tocris) was used at 10 μM. VDAC peptides corresponding to loop 4 (LP4 - KKLETAVNLAWTAGNSN) flanked N-, and C-terminally by the tryptophan zipper motif for increased stability (SWTWE and KWTWK, respectively) were linked to tandem repeats of the DNP2 fusion peptide (KIKKVKKKGRKKIKKVKKKGRK). The complete sequence for the peptides used are: dNP2 (KIKKVKKKGRKKIKKVKKKGRKSWTWEKWTWK) and dNP2-VDAC (KIKKVKKKGRKKIKKVKKKGRKSWTWEKKLETAVNLAWTAGNSNKWTWK). Both peptides were synthesized by Biomatik.
T cell activation
Human CD8+ T cell activation was performed using in house generated anti-CD3/anti-CD28 coated microbeads unless indicated otherwise. Polybead microspheres (4.5 μm, Polyscience Eppenheim) were incubated with 10 μg anti-human CD28 IgG1 antibody (CD28.2 Biolegend, USA) and 1.5 μg anti-human CD3 IgG2a antibody (HIT3a, Biolegend, USA). Antibody coupling was tested using 2nd antibodies against IgG1 or IgG2a (SouthernBiotech, USA), respectively. Quality control was performed by measuring the amount of antibody molecules per bead and using Quantum MESF microspheres (Bangslab, USA) for reference. All measurements were performed using a BD AccuriC6 flow cytometer (Becton Dickinson, USA) and data analyzed with FlowJo 10.0.8 software (Tree Star, USA). All bead based T cell activations were performed using a 2:1 bead to cell ratio. For 12 hour activation of human T cells used in metabolic flux assays, cells were activated with plate bound anti-CD3 (5 μg/ml, HIT3A) and soluble anti-CD28 (10 μg/ml) antibodies. For direct measurement of activation induced glycolytic switch by metabolic flux analysis, activation beads (5:1 bead to cell ratio) were directly added through the injection port. For in vitro reactivation of mouse T cells, memory OT-I cells were re-stimulated with 10 μM peptide ligands (OVA, R7, G4) for 1 hour (cell fractionation for immunoblots), or 4 hours (intracellular cytokine staining).
Adoptive transfer and bacterial infection
1x105 in vitro generated memory OT-I cells from wt or rictor KO mice were adoptively transferred intravenously (i.v.) into C57Bl/6 mice. Recipient mice were then infected with 5x105 CFU virulent LmOVA-N4 at 24 hours post-transfer. Mice were bled at 4 hours post-infection to measure serum cytokines and sacrificed at 24 hours post-infection. Spleens and livers were removed and both organs were homogenized in 0.5% Tergitol/PBS with a Tissuelyser (QIAGEN). Organ suspensions were plated on brain-heart-infusion (BHI) and bacterial colonies counted at 24 hours after plating.
Metabolic assays
A Seahorse XF-96e extracellular flux analyzer (Seahorse Bioscience, Agilent) was used to determine the metabolic profile of cells. NV and EM CD8+ T cells were plated (3x105 cells/well) onto Celltak (Corning, USA) coated cell plates. Mitochondrial perturbation experiments were carried out by sequential addition of oligomycin (1 μM, Sigma), FCCP (2 μM, Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone, Sigma), and rotenone (1 μM, Sigma). Oxygen consumption rates (OCR, pmol/min) and extracellular acidification rates (ECAR, mpH/min) were monitored in real time after injection of each compound. For monitoring oxygen consumption in permeabilized cells, Jurkat and CD8+ T cells were resuspended in MAS buffer (70 mM sucrose, 220 mM mannitol, 10 mM KH2PO4, 5 mM MgCl2, 2 mM Hepes, and 1 mM EGTA), then treated with the XF plasma membrane permeabilizer (Seahorse, Agilent), which was followed by treatment with pyruvate (5 mM)/malate (2.5 mM)/ADP (4 mM), or pyruvate/malate/oligomycin (2 μM) for monitoring State IIIADP respiration and non-ATP synthase dependent respiration, respectively. ATP quantitation (Abcam) was performed on 2.5x104 non-activated and activated T cells according to the manufacturer’s instructions.
IFN-γ measurement
EM CD8+ T cells were plated on flat-bottom 96 well plates (3x105 cells/well) using R10FBS. In select experiments, cells were pre-incubated for 30 min. with inhibitors and then activated using anti-CD3/anti-CD28 loaded beads. Cell supernatants were harvested 12 hours after activation and IFN-γ was measured using either an IFN-γ ELISA kit (eBioscience, USA) or a human Th1 cytokine bead-based immunoassay (Legendplex, Biolegend) according to manufacterer’s instructions. For cytokine measurements in mouse sera, a bead-based immunoassay was also used.
Metabolomics
NV and EM CD8+ T cells were cultured for 2 hours in a 24 well plate (3x106 cells/well) under non-activating and activating conditions. For Akt inhibition, cells were treated with Akti1/2 (10 μM, Sigma-Aldrich). Cell pellets were washed twice with cold PBS, snap frozen in EtOH containing dry ice and stored at -80°C. Metabolomic assays and analysis were performed by Metabolon Inc. (Durham, USA).
Flow cytometry
For cell proliferation assays, EM CD8+ T cells were loaded prior to activation with the cell-proliferation dye carboxyfluorescein succinimidyl ester (1 μM CFSE, Molecular probes, USA) and seeded on 48-well plates (5x105 cells/well). Plated cells were pretreated with inhibitors as indicated, and cultured with activation beads and IL-2 (1000 U/ml) (ImmunoTools, Germany) for 3 days. For surface staining of CD8+ T cells, human T cells were stained with antibodies targeting CD8, CD3, CD45RA, CD62L, CD69, and mouse OT-I cells were stained with antibodies targeting CD69, CD44, CD127, CXCR3, Ly6C, CD62L, KLRG1, CD27, CD25, CD69, CX3CR1, and CCR7. For intracellular IFN-γ measurements, cells were treated with Brefeldin A after 2 hours of activation and stained using a standard intracellular cytokine staining protocol (BD Bioscience). For measurement of mitochondrial mass and membrane potential, cells were stained with mitotracker green (MTG, 25 nM) or mitotracker red (MTR, 25 nM), respectively. Data was acquired using a BD AccuriC6 flow cytometer and analyzed with Flowjo 10.0.8 (Tree Star, USA).
Total cell lysate preparation and cell fractionation
To prepare total cell lysates, cells were activated for 1 hour (beads or peptide). Cell pellets were washed with cold PBS and lysed using RIPA buffer (Thermo Scientific, Rockford IL, USA) containing protease and phosphatase inhibitors (Roche). To prepare mitochondrial fractions that are enriched in mitochondria–ER junctions, cells were washed and resuspended in homogenization buffer (225 mM mannitol, 75 mM sucrose, 0.1 mM EGTA and 30 mM Tris–HCl pH 7.4), and homogenized with 20 strokes (CD8+ T cells) or 50 strokes (Jurkat T cells) using a teflon homogenizer. Cell homogenates were centrifuged twice at 600xg for 5 min. (4°C) and pellets were discarded after each spin. Supernatants were then centrifuged at 10'300xg, 10 min. (4°C). Supernatants (containing ER, golgi and cytoplasm) were collected, and pellets (containing mitochondria–ER junction-enriched mitochondria) were resuspended in RIPA buffer (Thermo Scientific, Rockford IL, USA). A digitonin-based protocol using glass homogenizers was used to prepare pure mitochondrial fractions.
Immunoblots
Protein concentrations were determined by BCA protein assay kit (Thermo Scientific). Total cell lysates, cytoplasmic, mitochondria–ER junction-enriched mitochondrial fractions or pure mitochondrial fractions were separated using 4-15% Mini Protean TGX Gel (Bio-Rad, Hercules CA, USA), and transferred to nitrocellulose membranes using Trans-Blot Turbo Transfer (Bio-Rad, Hercules CA, USA). Membranes were probed with antibodies against ACC, rictor, mTOR, HK-I, HK-II, Sin1, Cox iv, Akt, pAkt Thr308, pAkt Ser473, p-(S/T) Akt substrate motif, pGsk-3β Ser9 (all from cell signaling), GRP75, VDAC1, Gsk-3β (from Abcam), actin (Sigma-Aldrich), or raptor (Bethyl). Blots were stained with appropriate secondary antibodies and the odyssey imaging system (LICOR, Lincoln NE, USA) was used for visualization.
Electron microscopy
Transmission electron microscopy was performed at the Biozentrum (University of Basel). Cells were sequentially fixed in 3% paraformaldehyde, 0.5% glutaraldehyde and 1% osmium tetraoxide, embedded and then cut into 60 nm sections. Micrographs (27’000x magnification) were obtained with a Morgagni 268 (FEI, Hillsboro OR, USA) transmission electron microscope operated at 80 kV. Image J software (NIH, Bethesda, USA) was used for measuring mitochondrial length (major axis) and width (minor axis). To quantify the morphology of mitochondria, the average aspect ratio was calculated (major axis to minor axis), an aspect ratio of 1 indicates a circular mitochondrial section. To quantify mitochondria–ER contacts for each cell, total mitochondria and mitochondria in contact with ER were counted and the frequency of mitochondria–ER contacts calculated. Mitochondria–ER contacts were defined as mitochondria juxtaposed to ER at a distance of < 100 nM.
Proximity Ligation Assay
Anti-VDAC1 and anti-IP3R1 antibodies were used to visualize mitochondria–ER junctions. T cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton-X 100, and blocked with 5% goat serum. PLA was performed according to the manufacturers instructions (Duolink, Sigma). Fluorescent spots were detected by using an inverted spinning disk confocal microscope, Visiscope CSU-W1 (Visitron). Quantification of detected spots per nucleus was performed by using the Image J software.
RNA mediated interference
Jurkat T cells were transfected with pools of Grp75 siRNA or control scrambled siRNA (Qiagen) for 72 hours using the AMAXA T cell nucleofection kit (Lonza). Knockdown efficiency was verified by immunoblot analysis.
Confocal imaging
Experiments with fixed cells were performed on a Nikon A1 confocal microscope (Microscopy core facility, DBM) with a 40x oil-immersion objective. Cells were allowed to attach by gravity on culture slides (BD Bioscience) coated with poly-d-lysine for 20 min. Cells were pre-stained with mitotracker deep red and then fixed with 4% paraformaldehyde and permeabilized with 0.1% TritonX-100. Cells were stained with antibodies against rictor (Atlas antibodies), KDEL motif (Abcam) or Gsk-3β (Abcam), labeled with the appropriate secondary antibodies, and counterstained with DAPI. All images were processed and analyzed using ImageJ (NIH). Experiments with live cells were performed using the 3I spinning disk confocal microscope (Intelligent Imaging Innovations, Denver CO, USA) with a 100x oil-immersion objective at 5% CO2 and 37°C (IMCF Facility, Biozentum, University of Basel). Cells were loaded with 50 nM MTG (Invitrogen) and 25 nM tetramethylrhodamine-ethyl-ester-perchlorate (TMRE, Invitrogen). Cells were incubated for 30 min. with both dyes, washed and resuspended in fresh media. Since CD8+ T cells are non-adherent, dye-loaded T cells were allowed to settle onto culture slides (Ibidi, Munich, Germany) coated with poly-d-lysine for 20-30 min. before imaging. Stacked images were then acquired at 0.5 μm intervals throughout the body of each cell. Quantification of mitochondrial structure was carried out on cells labeled with MTG. Extraction of mitochondrial structural parameters was performed using a multi-step image-processing algorithm. Briefly, MTG intensities in raw images were enhanced by linear contrast stretch, spatially process using a 7x7 top hat filter to isolate mitochondrial objects and converted into a binary image by a threshold operation. Mitochondrial circularity (4π area/perimeter2), length, and form factor (1/circularity) were then calculated using ImageJ (NIH, Bethesda MD). Mitochondrial membrane potential (∆ψm) was calculated as the ratio of TMRE to MTG fluorescence intensities. MTG was included to normalize for mitochondrial mass differences between cell populations.
13C6 glucose labeling
2x106 EM CD8+ T cells were plated onto 48 well plates in RPMI10-FBS with 4 mM U-13C glucose and left non-activated or activated with anti-CD3/anti-CD28 loaded beads for 6 hours. Metabolites were extracted with 50% MeOH/30% Acetonitrile. Isotopologue distribution was assessed by LC-MS.
Statistical analysis
Statistical significance was analyzed using Prism 6.0 (GraphPad Software, USA). P values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgements
The authors would like to thank the flow sorting team of the Department of Biomedicine at Basel University. Funding: C.H. SNSF 310030_153059 and 31003A_172848; M.F. SNSF 323530_139181; M.L.B., S.D. and P.M.G. Basel University Young Investigator grants; M.R. SNSF PP00P3_144863.
References
- Arbore G, West EE, Spolski R, Robertson AA, Klos A, Rheinheimer C, Dutow P, Woodruff TM, Yu ZX, O'Neill LA, et al. T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4(+) T cells. Science. 2016;352 doi: 10.1126/science.aad1210. aad1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzoine L, Zilberberg N, Ben-Romano R, Shoshan-Barmatz V. Voltage-dependent anion channel 1-based peptides interact with hexokinase to prevent its anti-apoptotic activity. The Journal of biological chemistry. 2009;284:3946–3955. doi: 10.1074/jbc.M803614200. [DOI] [PubMed] [Google Scholar]
- Bagchi S, Fredriksson R, Wallen-Mackenzie A. In Situ Proximity Ligation Assay (PLA) Methods Mol Biol. 2015;1318:149–159. doi: 10.1007/978-1-4939-2742-5_15. [DOI] [PubMed] [Google Scholar]
- Balmer ML, Hess C. Feeling Worn Out? PGC1alpha to the Rescue for Dysfunctional Mitochondria in T Cell Exhaustion. Immunity. 2016;45:233–235. doi: 10.1016/j.immuni.2016.07.024. [DOI] [PubMed] [Google Scholar]
- Balmer ML, Ma EH, Bantug GR, Grahlert J, Pfister S, Glatter T, Jauch A, Dimeloe S, Slack E, Dehio P, et al. Memory CD8(+) T Cells Require Increased Concentrations of Acetate Induced by Stress for Optimal Function. Immunity. 2016;44:1312–1324. doi: 10.1016/j.immuni.2016.03.016. [DOI] [PubMed] [Google Scholar]
- Bantug GR, Galluzzi L, Kroemer G, Hess C. The spectrum of T cell metabolism in health and disease. Nature reviews. Immunology. 2017 doi: 10.1038/nri.2017.99. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Kartashov AV, Liu C, Imamichi H, Yang W, Peng W, Lane HC, Zhao K. Rapid Recall Ability of Memory T cells is Encoded in their Epigenome. Sci Rep. 2017;7:39785. doi: 10.1038/srep39785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengsch B, Johnson AL, Kurachi M, Odorizzi PM, Pauken KE, Attanasio J, Stelekati E, McLane LM, Paley MA, Delgoffe GM, Wherry EJ. Bioenergetic Insufficiencies Due to Metabolic Alterations Regulated by the Inhibitory Receptor PD-1 Are an Early Driver of CD8(+) T Cell Exhaustion. Immunity. 2016;45:358–373. doi: 10.1016/j.immuni.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, Hall MN. Where is mTOR and what is it doing there? The Journal of cell biology. 2013;203:563–574. doi: 10.1083/jcb.201306041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, Stracka D, Prescianotto-Baschong C, Frieden M, Demaurex N, Hall MN. Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12526–12534. doi: 10.1073/pnas.1302455110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. The Biochemical journal. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R, Vicencio JM, Parra V, Troncoso R, Munoz JP, Bui M, Quiroga C, Rodriguez AE, Verdejo HE, Ferreira J, et al. Increased ER-mitochondrial coupling promotes mitochondrial respiration and bioenergetics during early phases of ER stress. Journal of cell science. 2011;124:2143–2152. doi: 10.1242/jcs.080762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, O'Sullivan D, Klein Geltink RI, Curtis JD, Chang CH, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJ, et al. Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell. 2016;166:63–76. doi: 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas C, Miller RA, Smith I, Bui T, Molgo J, Muller M, Vais H, Cheung KH, Yang J, Parker I, et al. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell. 2010;142:270–283. doi: 10.1016/j.cell.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O'Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Denton RM. Regulation of mitochondrial dehydrogenases by calcium ions. Biochimica et biophysica acta. 2009;1787:1309–1316. doi: 10.1016/j.bbabio.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Farber DL. Biochemical signaling pathways for memory T cell recall. Semin Immunol. 2009;21:84–91. doi: 10.1016/j.smim.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomello M, Pellegrini L. The coming of age of the mitochondria-ER contact: a matter of thickness. Cell death and differentiation. 2016;23:1417–1427. doi: 10.1038/cdd.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubser PM, Bantug GR, Razik L, Fischer M, Dimeloe S, Hoenger G, Durovic B, Jauch A, Hess C. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nature immunology. 2013;14:1064–1072. doi: 10.1038/ni.2687. [DOI] [PubMed] [Google Scholar]
- Harty JT, Badovinac VP. Shaping and reshaping CD8+ T-cell memory. Nature reviews. Immunology. 2008;8:107–119. doi: 10.1038/nri2251. [DOI] [PubMed] [Google Scholar]
- Hoshi M, Takashima A, Noguchi K, Murayama M, Sato M, Kondo S, Saitoh Y, Ishiguro K, Hoshino T, Imahori K. Regulation of mitochondrial pyruvate dehydrogenase activity by tau protein kinase I/glycogen synthase kinase 3beta in brain. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:2719–2723. doi: 10.1073/pnas.93.7.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan A, Huang W, Huang F, August A. Signal transduction via the T cell antigen receptor in naive and effector/memory T cells. Int J Biochem Cell Biol. 2012;44:2129–2134. doi: 10.1016/j.biocel.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Geltink RI, O'Sullivan D, Corrado M, Bremser A, Buck MD, Buescher JM, Firat E, Zhu X, Niedermann G, Caputa G, et al. Mitochondrial Priming by CD28. Cell. 2017;171:385–397 e311. doi: 10.1016/j.cell.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krippner A, Matsuno-Yagi A, Gottlieb RA, Babior BM. Loss of function of cytochrome c in Jurkat cells undergoing fas-mediated apoptosis. The Journal of biological chemistry. 1996;271:21629–21636. doi: 10.1074/jbc.271.35.21629. [DOI] [PubMed] [Google Scholar]
- Lee SL, Chou CC, Chuang HC, Hsu EC, Chiu PC, Kulp SK, Byrd JC, Chen CS. Functional Role of mTORC2 versus Integrin-Linked Kinase in Mediating Ser473-Akt Phosphorylation in PTEN-Negative Prostate and Breast Cancer Cell Lines. PloS one. 2013;8:e67149. doi: 10.1371/journal.pone.0067149. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lim S, Kim WJ, Kim YH, Lee S, Koo JH, Lee JA, Yoon H, Kim DH, Park HJ, Kim HM, et al. dNP2 is a blood-brain barrier-permeable peptide enabling ctCTLA-4 protein delivery to ameliorate experimental autoimmune encephalomyelitis. Nature communications. 2015;6:8244. doi: 10.1038/ncomms9244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Lee JA, Koo JH, Kang TG, Ha SJ, Choi JM. Cell Type Preference of a Novel Human Derived Cell-Permeable Peptide dNP2 and TAT in Murine Splenic Immune Cells. PloS one. 2016;11:e0155689. doi: 10.1371/journal.pone.0155689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annual review of immunology. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Reyes I, Diebold LP, Kong H, Schieber M, Huang H, Hensley CT, Mehta MM, Wang T, Santos JH, Woychik R, et al. TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Molecular cell. 2016;61:199–209. doi: 10.1016/j.molcel.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JS, Hisata S, Park MA, DeNicola GM, Ryter SW, Nakahira K, Choi AMK. mTORC1-Induced HK1-Dependent Glycolysis Regulates NLRP3 Inflammasome Activation. Cell reports. 2015;12:102–115. doi: 10.1016/j.celrep.2015.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Murley A, Nunnari J. The Emerging Network of Mitochondria-Organelle Contacts. Molecular cell. 2016;61:648–653. doi: 10.1016/j.molcel.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhill N, Lynes EM, Nanji JA, Blagoveshchenskaya AD, Fei H, Carmine Simmen K, Cooper TJ, Thomas G, Simmen T. The subcellular distribution of calnexin is mediated by PACS-2. Molecular biology of the cell. 2008;19:2777–2788. doi: 10.1091/mbc.E07-10-0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, Qiu J, Smith AM, Lam WY, DiPlato LM, et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olenchock BA, Rathmell JC, Vander Heiden MG. Biochemical Underpinnings of Immune Cell Metabolic Phenotypes. Immunity. 2017;46:703–713. doi: 10.1016/j.immuni.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino JG, Hoek JB, Shulga N. Activation of glycogen synthase kinase 3beta disrupts the binding of hexokinase II to mitochondria by phosphorylating voltage-dependent anion channel and potentiates chemotherapy-induced cytotoxicity. Cancer research. 2005;65:10545–10554. doi: 10.1158/0008-5472.CAN-05-1925. [DOI] [PubMed] [Google Scholar]
- Peng M, Yin N, Chhangawala S, Xu K, Leslie CS, Li MO. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science. 2016;354:481–484. doi: 10.1126/science.aaf6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penso J, Beitner R. Clotrimazole and bifonazole detach hexokinase from mitochondria of melanoma cells. European journal of pharmacology. 1998;342:113–117. doi: 10.1016/s0014-2999(97)01507-0. [DOI] [PubMed] [Google Scholar]
- Prasad M, Walker AN, Kaur J, Thomas JL, Powell SA, Pandey AV, Whittal RM, Burak WE, Petruzzelli G, Bose HS. Endoplasmic Reticulum Stress Enhances Mitochondrial Metabolic Activity in Mammalian Adrenals and Gonads. Molecular and cellular biology. 2016;36:3058–3074. doi: 10.1128/MCB.00411-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raturi A, Simmen T. Where the endoplasmic reticulum and the mitochondrion tie the knot: the mitochondria-associated membrane (MAM) Biochimica et biophysica acta. 2013;1833:213–224. doi: 10.1016/j.bbamcr.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annual review of immunology. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, Ferris RL, Delgoffe GM. The Tumor Microenvironment Represses T Cell Mitochondrial Biogenesis to Drive Intratumoral T Cell Metabolic Insufficiency and Dysfunction. Immunity. 2016;45:374–388. doi: 10.1016/j.immuni.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang CR, Schumacker PT, Licht JD, Perlman H, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoshan-Barmatz V, Mizrachi D. VDAC1: from structure to cancer therapy. Frontiers in oncology. 2012;2:164. doi: 10.3389/fonc.2012.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadkai G, Bianchi K, Varnai P, De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzuto R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. The Journal of cell biology. 2006;175:901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurey P, Rieusset J. Mitochondria-Associated Membranes Response to Nutrient Availability and Role in Metabolic Diseases. Trends in endocrinology and metabolism: TEM. 2017;28:32–45. doi: 10.1016/j.tem.2016.09.002. [DOI] [PubMed] [Google Scholar]
- Thome JJ, Yudanin N, Ohmura Y, Kubota M, Grinshpun B, Sathaliyawala T, Kato T, Lerner H, Shen Y, Farber DL. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 2014;159:814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs E, Theurey P, Vial G, Bendridi N, Bravard A, Chauvin MA, Ji-Cao J, Zoulim F, Bartosch B, Ovize M, et al. Mitochondria-associated endoplasmic reticulum membrane (MAM) integrity is required for insulin signaling and is implicated in hepatic insulin resistance. Diabetes. 2014;63:3279–3294. doi: 10.2337/db13-1751. [DOI] [PubMed] [Google Scholar]
- van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Windt GJ, O'Sullivan D, Everts B, Huang SC, Buck MD, Curtis JD, Chang CH, Smith AM, Ai T, Faubert B, et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14336–14341. doi: 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nature chemical biology. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng NP, Araki Y, Subedi K. The molecular basis of the memory T cell response: differential gene expression and its epigenetic regulation. Nature reviews. Immunology. 2012;12:306–315. doi: 10.1038/nri3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nature protocols. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- Wolf AJ, Reyes CN, Liang W, Becker C, Shimada K, Wheeler ML, Cho HC, Popescu NI, Coggeshall KM, Arditi M, Underhill DM. Hexokinase Is an Innate Immune Receptor for the Detection of Bacterial Peptidoglycan. Cell. 2016;166:624–636. doi: 10.1016/j.cell.2016.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tschumi BO, Lopez-Mejia IC, Oberle SG, Meyer M, Samson G, Ruegg MA, Hall MN, Fajas L, Zehn D, et al. Mammalian Target of Rapamycin Complex 2 Controls CD8 T Cell Memory Differentiation in a Foxo1-Dependent Manner. Cell reports. 2016;14:1206–1217. doi: 10.1016/j.celrep.2015.12.095. [DOI] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.