Abstract
Global phosphoproteomic studies based on mass spectrometry have generated qualitative and quantitative data describing protein phosphorylation events in various biological systems. Since high throughput data for protein modifications are inherently incomplete we developed a strategy to extend and validate such primary data sets. We selected interesting protein candidates from a global screen in yeast and employed a modified histidine biotin tag that allows tandem affinity purifications of our targets under denaturing conditions. Products in question can be digested directly from affinity resins and phosphopeptides can be further enriched via TiO2 before MS analysis. Our robust protocol can be amended for SILAC as well as iTRAQ quantifications or label free approaches based on selective reaction monitoring, allowing completion of the phosphorylation pattern in a first step, followed by a detailed analysis of the phosphorylation kinetics. We exemplify the value of such a strategy by an in depth analysis of Pan1, a highly phosphorylated factor involved in early steps of endocytosis. The study of Pan1 under osmotic stress conditions in different mutant backgrounds allowed us to differentiate between mitogen-activated protein (MAP) kinase Hog1 driven and Hog1 independent stress responses.
Keywords: Endocytosis, Phosphorylation, Quantitative Proteomics, Signal Transduction
Introduction
The processing of environmental and intracellular signals involves highly intertwined networks of protein kinases and protein phosphatases that will alter a cell’s phosphorylome upon stimulation. Often different signaling cascades are integrated at the level of common target-proteins making the efforts to understand the complex nature of signaling responses extremely difficult. In addition, there are also cyclical phosphorylation/dephosphorylation events that are necessary for the execution of basic cellular mechanisms such as vesicular trafficking. Quantitative MS has become a powerful tool for the observation of global changes in the abundance as well as in the modification pattern of proteins during specific biological conditions. In the last decade high throughput MS studies have provided insight into the activity of signaling systems and have greatly helped to identify target proteins and sites modified by kinases and phosphatases [1–10]. However, due to the complexity of the samples, these shotgun studies provide incomplete modification patterns of a specific protein although the data help in selecting candidates that are best suited and interesting for subsequent functional and physiological studies. Also, apart from the incompleteness problem, high throughput experiments result in a certain fraction of false positive peptide identifications. Since they are further prone to quantitative variations, they generate a demand for procedures that allow the validation of such primary data.
A popular model for signal transduction processes is the cellular response of the budding yeast Saccharomyces cerevisiae to the challenges of osmotic stress (reviewed in [11]). S. cerevisiae reacts to increased external osmolarity by activation of the high-osmolarity glycerol (HOG) Mitogen-activated Protein Kinase (MAPK) pathway, a homologue to the mammalian p38 MAPK pathway. Upon activation the central MAPK Hog1 undergoes dual phosphorylation and coordinates the osmo-stress response by phosphorylating serine/threonine proline (S/T-P) motifs on several target proteins. Hog1 function has mainly been associated with the regulation of transcriptional events [11–15], however, several lines of evidence suggest that additional cellular roles of Hog1 must exist [16, 17]. With the help of SILAC based shotgun experiments we were able to identify several new candidate substrates of Hog1 including factors involved in early endocytosis such as Pan1.
Endocytosis is an evolutionarily conserved process by which cells absorb external or surface associated molecules. In the course of a coordinated series of events these molecules become selected and engulfed at specific sites on the cell surface, the so called coated pits. These cargo associated coated pits invaginate, leading to formation of a vesicle that ultimately becomes released into the cell and fused to the endosome (reviewed in [18]). A central factor of this process is the scaffold protein Pan1 [19–24]. Pan1 is first found at coated pits but remains at these sites throughout patch maturation and vesicle formation and thus is thought to coordinate the transition from early to late events [25]. The protein becomes modified at the N-terminus by cycles of phosphorylation and dephosphorylation mediated by kinases Ark1 and Prk1 [23, 26, 27] and the type 1 phosphatase Glc7 [28]. Our (and other [1–10]) shotgun studies provided several novel phosphorylation sites on Pan1, some of which appeared to become hyperphosphorylated in response to osmotic stress. To follow up this interesting observation we wanted to determine the number, the kinetics and stoichiometry of these phosphorylation events.
For this purpose, we developed a general strategy to validate and extend initial observations of shotgun screens. We applied a modified histidine biotin tandem affinity tag for a fast and robust affinity purification workflow and provide an in-depth classical MS analysis of purified Pan1 under different environmental conditions followed by quantitative MS measurements. First, we employed label based methods such as SILAC [3, 29] and iTRAQ [30] for a general phosphopattern characterization. Then we determined the phosphorylation kinetics of individual regulated sites with label free SRM [31]. Although several phosphorylation sites on the C-terminal half of Pan1 become up-regulated in response to osmotic stress our approach revealed that they differ in their response kinetics, stoichiometry and dependency on Hog1. Only one of these sites is directly dependent on the activity of Hog1 and thereby provides evidence for a direct involvement of this signal system in the endocytotic network in S. cerevisiae.
Materials and Methods
Yeast Strain and Plasmid construction
Yeast strains and plasmids (listed in Supplemental Table S13) were constructed as described in Supplemental Material and Methods.
One step and tandem affinity purification
The histidine biotin tandem affinity purification is based on methods described elsewhere [32] with modifications: cells were grown to an OD600 of 1, harvested by filtration and deep frozen in liquid N2. Cell breakage was performed using SPEX Freezer Mills (Mill 6770 or Mill 6870, SPEXSamplePrep, Metuchen, NJ, USA). Breakage conditions were chosen as follows: 10mins pre-cooling, then 7 cycles: 3mins breakage, 3mins cooling, 15 cycles per second. The cell powder was resuspended in denaturing buffer conditions (6 M Guanidine-HCl, 50 mM Tris, 5 mM NaF, 1 mM PMSF, 0.1% Tween, protease inhibitor cocktail (Roche, 11 873 580 001), pH 8) and cleared of debris by centrifugation 10000xg, 15 mins, 4°C. For one step purifications cell extracts were incubated for 4 hours with Streptavidine-Agarose (Thermo Scientific Pierce, Rockford, IL, USA, 20349). The beads were washed twice with urea buffer (8M urea, 50 mM Na-PO4, 300 mM NaCl, 0.01% Tween, pH 8), three times with urea buffer 6.3, three times with urea stringent wash buffer (8 M urea, 50 mM Na-PO4, 400 mM NaCl, 1 % SDS, 10 mM EDTA, 0.01% Tween, pH 8) and three times with urea wash buffer (8 M urea, 100 mM Tris, 200 mM NaCl, pH 8). Tandem purifications were performed as follows: cell extracts were incubated for 4 hours with Ni2+-Sepharose beads (GE Healthcare, Buckinghamshire, UK, 17-5318-06). The beads were washed twice with urea buffer and three times with urea buffer pH6.3. Proteins were eluted in three subsequent elution steps using urea buffer pH 4.3 containing 10 mM EDTA. The eluates were unified, incubated overnight with streptavidin-agarose beads and washed three times using urea wash buffer containing 1% SDS. SDS was removed by three washing steps with pure urea wash buffer.
MS-based analysis
A detailed description of labeling and MS procedures can be found in Supplemental Material and Methods.
Results
MAPK Hog1 is involved in the regulation of the early endocytotic pathway
S. cerevisiae reacts to increased external osmolarity by activation of the HOG MAPK pathway. To identify novel target proteins of the MAPK Hog1, we followed global changes in the phosphorylation pattern of the yeast proteome upon osmotic stress by employing shotgun experiments based on SILAC technology in combination with an enrichment protocol for phosphopeptides based on IMAC. Cells were stressed for 5 minutes and mixed with an equal amount of cells grown on 13C lysine/arginine medium, serving as unstressed control, prior to cell lysis. Proteins were extracted with TRIzol and subsequently digested with trypsin (Supplemental Table S1).
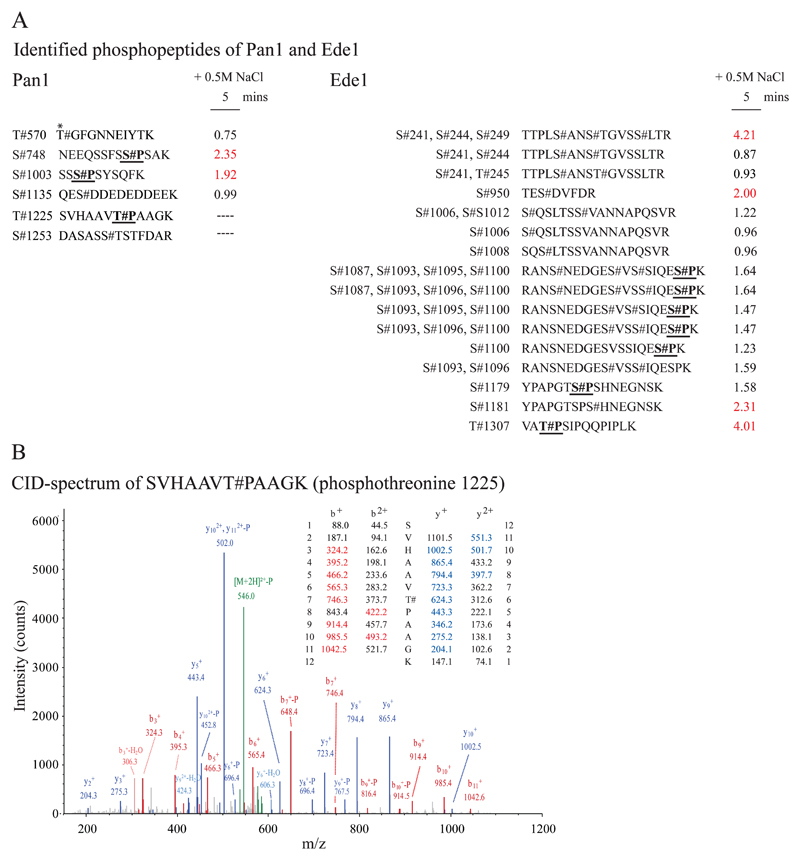
We identified Pan1 and Ede1, two factors annotated to the early endocytotic pathway, as putative targets of Hog1 based on an increase in phosphorylation at S/T-P MAPK motifs in response to hyperosmotic shock. In total, 6 phosphorylation sites of Pan1 have been identified in our shotgun approach (Fig 1 A), three of which (threonine 748 (covered by peptide sequence: NEEQSSFSS#PSAK), serine 1003 (SSS#PSYSQFK) and threonine 1225 (SVHAAVT#PAAGK)) are in accordance with the classical S/T-P consensus motif (Fig 2 A). Interestingly, we measured a 2.35 fold increase of phosphoserine 748 after 5 minutes exposure to hyperosmotic stress indicating a role of Pan1 in osmo-stress response (Fig 2 A). We also identified (amongst other phosphorylation sites) three S/T-P motifs to be phosphorylated on Ede1, one of which, namely phosphothreonine 1307, was up-regulated 4.01 fold in response to stress. Additionally, we could confirm Pan1 to be an in vitro substrates of kinase Hog1 (Supplemental Figure 3B). Similar observations were made with Ede1 (data not shown), hence our observations establish a direct interconnection of the osmotic stress response and the endocytotic machinery. Since Pan1 has previously been described as the central scaffolding factor of the endocytic process we have subsequently focused solely on this protein.
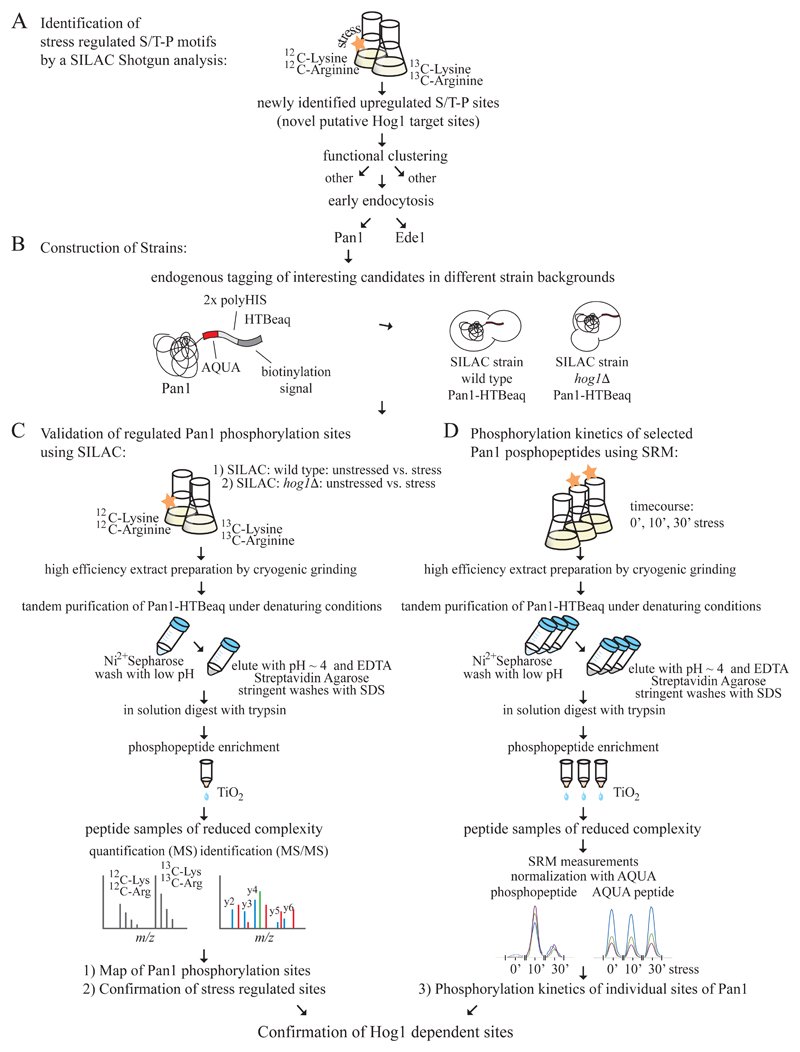
Figure 1. Representation of our methodological workflow for extending and validating high throughput data sets based on selective protein purification.
A: Primary shotgun experiments lead to the identification of candidate factors. In our case, Pan1 and Ede1, both factors of the early endocytosis machinery, popped up as putative novel Hog1 targets based on an increase in phosphorylation at S/T-P sites. B: Factors of interest become endogenously tagged with our modified histidine biotin tag (HTBeaq). Different strain backgrounds are used that allow determination of kinase dependency of phosphorylation sites. C: Completion of the phosphopattern and validation of regulated phosphorylation sites by SILAC or iTRAQ analysis of affinity purified proteins. Results obtained with wild type and mutant strains are compared. D: Determination of phosphorylation kinetics of individual sites using a targeted SRM approach with purified proteins.
Figure 2. The endocytotic factors Pan1 and Ede1 are a putative targets of MAPK Hog1.
A: Stressed/unstressed ratios of Pan1 and Ede1 phosphopeptides from our shotgun experiment. Phosphorylation sites are indicated with “#”. S/T-P MAPK consensus motifs are highlighted (underlined and bold). “*” indicates Prk1 consensus sites. Stress induced up-regulation is shown in red. B: Annotated collision induced dissociation (CID) spectrum of peptide SVHAAVT#PAAGK (phosphothreonine 1225). Singly and doubly charged N-terminal b ions and C-terminal y ions are indicated in red and blue respectively. Precursor is indicated in green.
Integration of our dataset with previous high throughput studies [1–10] revealed one previously unknown phosphorylation site of Pan1: phosphothreonine 1225 (Fig 2 A and B). Even though several phosphopeptides of Pan1 have independently been identified in these studies, changes in phosphorylation in response to environmental conditions have not been quantified. We therefore decided to validate modification events by directly analyzing purified Pan1 (Fig 1 B - D).
Pan1 is highly phosphorylated
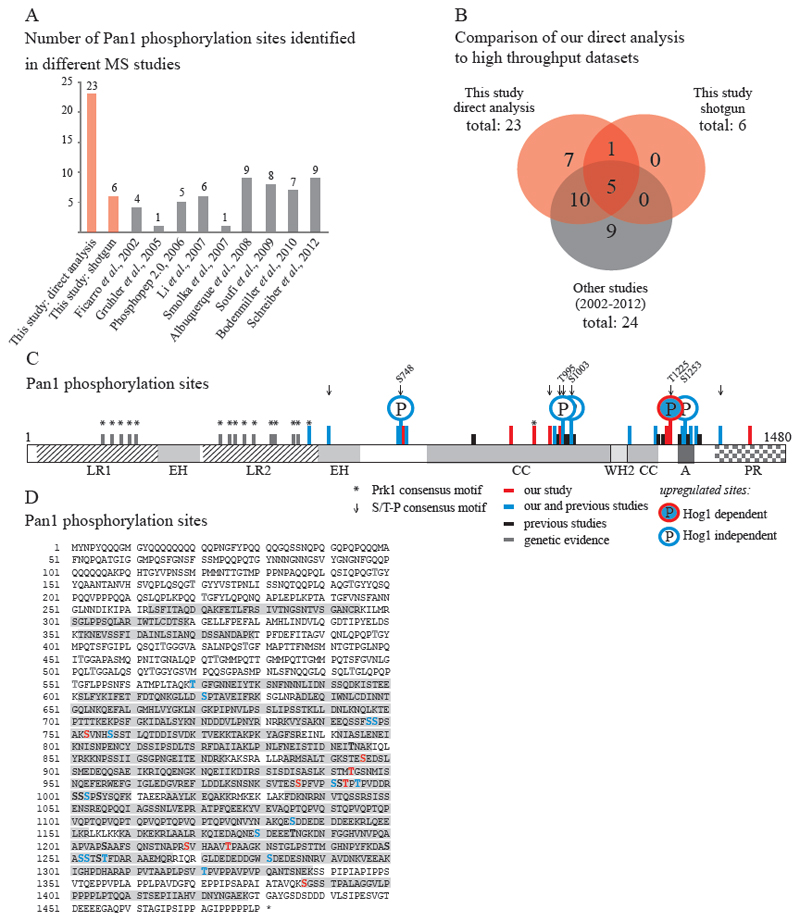
To follow up modification events on Pan1 in vivo, we chose a tandem affinity approach, that works under denaturing conditions ensuring that post-translational modifications are not altered during extract preparation and sample processing such as provided by the HB-tag (poly histidine, biotinylation signal) family [33]. We reconstructed and modified the original HB-tag to improve efficiency and versatility of the purification protocol. We named the new tag HTBeaq (HTB-effective cleavage plus absolute quantification) (Fig 1 B, for details see Supplemental Results and Supplemental Figure 1). MS/MS and MS/MS/MS spectra of tandem purified Pan1-HTBeaq samples were obtained on an LTQ-FT or a Velos Orbitrap mass spectrometer. In total, we covered 57.4 percent of the Pan1 peptide sequence with tryptic digests, including most of the C-terminal half of Pan1. The N-terminal part of Pan1 was less covered due to peptide stretches absent of arginine and lysine, preventing the generation of suitable tryptic peptides. Up to 23 phosphorylation sites with a site probability > 75% [34] have been identified in our in-depth MS analysis of purified Pan1 (Fig 3 A and B, Table 1 and Supplemental Table S2), including all 6 phosphorylation sites that have been described above. When comparing our data to previous studies [1–10] 15 sites have independently been covered (Fig 3 B). The 8 novel unambiguously identified phosphorylation sites from our study are serines 753, 896, 986, 1219 and 1387, and threonines 944, 993 and 1225. Assignments with a site probability below 75% were considered as ambiguous and are listed in Supplemental Table S3.
Figure 3. Pan1 is highly phosphorylated.
A and B: Comparison of the number of Pan1 phosphorylation sites identified by us (red) to the number identified in independent high throughput MS studies [1–10] (grey). C: Scheme of protein Pan1. The two long repeat regions (LR), two Eps15 homology domains (EH), the coiled-coil domain (coil), the acidic domain (A) and the proline-rich domain (PRD) are indicated. Phosphorylation sites identified by us are indicated by red and blue (also identified by others [1–10]) bars. Black bars: Phosphorylation sites identified in independent MS studies [1–10]. Grey bars: Phosphorylation sites identified by genetic studies [23]. Up-regulated phosphorylation sites are highlighted by circles and indicated amino acid positions. S/T-P MAPK motifs (arrow) and Prk1 (*) motifs are indicated. D: Pan1 sequence with highlighted phosphorylation sites identified by us (red), identified also by others (blue) and identified by independent MS-studies (black) and genetic studies (dark grey). Sequence coverage is highlighted in light grey.
Table 1.
Summary of high confidence PAN1-phosphorylation sites from all LC-MS/MS runs. Site probability was determined by phosphoRS 2.0. Phosphopeptides are split for Site Probability >95 % and 95%> x >75%.
| Protein Site | Sequence | pRS Site Probabilities >95% | Modifications | Activation Type | XCorr | # MC | Charge | m/z [Da] | MH+ [Da] | ΔM [ppm] |
|---|---|---|---|---|---|---|---|---|---|---|
| T570 | tGFGNNEIYTK | T(1): 100.0 | T1(Phospho) | CID | 3.77 | 0 | 2 | 662.284 | 1323.562 | -0.10 |
| S621 | GLLDsPTAVEIFR | S(5): 100.0 | S5(Phospho) | CID | 4.17 | 0 | 2 | 749.371 | 1497.734 | -0.34 |
| S747 | NEEQSSFsSPSAK | S(8): 99.4 | N-Term(iTRAQ4plex) S9(Phospho) K13(iTRAQ4plex) | HCD | 3.43 | 0 | 2 | 883.398 | 1765.789 | 0.24 |
| S748 | NEEQSSFSsPSAK | S(9): 99.6 | S9(Phospho) | CID | 3.81 | 0 | 2 | 739.296 | 1477.585 | 0.78 |
| S753 | sVNHSSSTLQTDDISVDK | S(1): 100.0 | S1(Phospho) | CID | 6.36 | 0 | 2 | 1006.945 | 2012.884 | 1.33 |
| S986 | sVTESsPFVPSSTPTPVDDR | S(6): 96.9 | N-Term(iTRAQ4plex) S6(Phospho) | CID | 2.25 | 0 | 2 | 1165.042 | 2329.077 | 2.10 |
| T995 | SVTESSPFVPSSTPtPVDDR | T(15): 100.0 | T15(Phospho) | CID | 4.86 | 0 | 2 | 1092.987 | 2184.967 | -1.16 |
| S991 T995 | SVTESSPFVPsSTPtPVDDR | S(11): 99.5 T(15): 99.5 | S11(Phospho) T15(Phospho) | CID | 5.46 | 0 | 2 | 1132.973 | 2264.938 | 1.04 |
| S1003 | SSsPSYSQFK | S(3): 99.8 | S3(Phospho) | CID | 3.42 | 0 | 2 | 599.245 | 1197.482 | -0.11 |
| S1135 | QEsDDEDEDDEEk | S(3): 100.0 | S3(Phospho); K13(Label:13C(6)) | CID | 4.52 | 0 | 2 | 834.772 | 1668.536 | -0.87 |
| S1180 | KQIEDAQNEsDEEETNGK | S(10): 100.0 | S10(Phospho) | CID | 6.53 | 1 | 2 | 1072.438 | 2143.869 | 1.25 |
| S1219 | sVHAAVTPAAGk | S(1): 100.0 | S1(Phospho) K12(Label:13C(6)) | CID | 2.99 | 0 | 2 | 597.802 | 1194.597 | -0.26 |
| T1225 | SVHAAVtPAAGK | T(7): 100.0 | T7(Phospho) | CID | 3.41 | 0 | 2 | 594.792 | 1188.577 | 0.13 |
| S1252 | dASAsSTSTFDAR | S(5): 99.6 | N-Term(iTRAQ4plex); S5(Phospho) | HCD | 2.85 | 0 | 2 | 770.326 | 1539.645 | 0.12 |
| S1253 | DASASsTSTFDAr | S(6): 95.9 | S6(Phospho); R13(Label:13C(6)) | CID | 3.11 | 0 | 2 | 701.285 | 1401.563 | 0.40 |
| S1253 T1256 | DASASsTStFDAR | S(6): 96.1 T(9): 99.8 | S6(Phospho); T9(Phospho) | CID | 3.94 | 0 | 2 | 738.258 | 1475.509 | 0.37 |
| S1281 | GLDEDEDDGWsDEDESNNR | S(11): 100.0 | S11(Phospho) | CID | 6.53 | 0 | 2 | 1138.892 | 2276.776 | 1.11 |
| T1321 | aPPVTAAPLPSVtPVPPAVPVPQANTSNEk | T(13): 100.0 | N-Term(iTRAQ4plex) T13(Phospho) K30(iTRAQ4plex) | HCD | 5.95 | 0 | 4 | 829.446 | 3314.763 | -0.31 |
| S1387 | sGSSTPALAGGVLPPPPPLPTQQASTSEPIIAHVDNYnGAEk | S(1): 97.8 | N-Term(iTRAQ4plex) S1(Phospho) N38(Deamidated) K42(iTRAQ4plex) | HCD | 6.91 | 0 | 3 | 1512.428 | 4535.268 | 2.11 |
| Protein Site | Sequence | pRS Site Probabilities >75% and <95% | Modifications | Activation Type | XCorr | # MC | Charge | m/z [Da] | MH+ [Da] | ΔM [ppm] |
|---|---|---|---|---|---|---|---|---|---|---|
| S757 | SVNHsSSTLQTDDISVDKTVEK | S(5): 93.3 | S5(Phospho) | CID | 5.33 | 1 | 3 | 824.050 | 2470.135 | 0.32 |
| S896 | STEsEDSLSMEDEQQSAEIKR | S(4): 76.9 | S4(Phospho) | CID | 4.24 | 1 | 3 | 827.011 | 2479.018 | -0.03 |
| T944 | STMtGSNMISNQEFER | T(4): 92.3 | T4(Phospho) | CID | 5.60 | 0 | 2 | 956.385 | 1911.762 | 0.26 |
| T993 | SVTESSPFVPSStPTPVDDR | T(13): 93.8 | T13(Phospho) | CID | 4.27 | 0 | 2 | 1092.988 | 2184.969 | -0.33 |
A comparison of the Pan1 phosphorylation sites with known kinase motifs showed that 8 sites, namely serines 621, 748, 986, 1003 and threonines 993, 995, 1225 and 1321 lie within a S/T-P MAPK consensus motif. Threonine 570 is a target site of kinase Prk1.
Pan1 becomes hyperphosphorylated in response to hyperosmotic stress
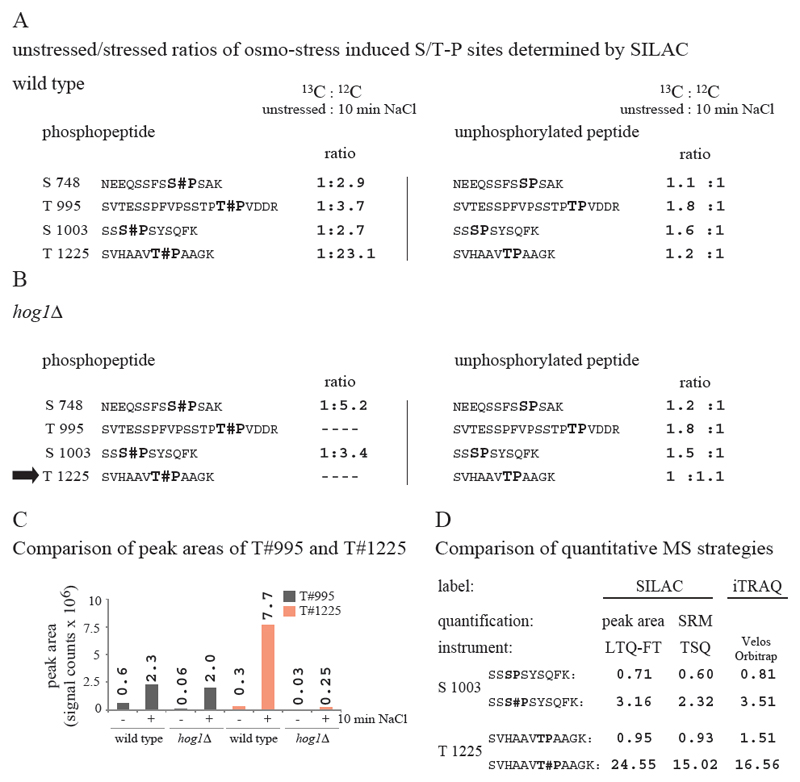
We assessed which of the phosphorylation sites of Pan1 become up or down-regulated in response to hyperosmotic stress. We first made use of SILAC labeling because this method allows the simultaneous quantification of all phosphorylation events within the covered protein sequence and also provided a dataset directly comparable to our shotgun approach. Furthermore, we wanted to identify suitable peptide-ions for SRM measurements to study phosphorylation kinetics (see below). Our analysis confirmed that five phosphorylation sites, namely phosphoserines 748 (covered by peptide sequence: NEEQSSFSS#PSAK), 1003 (SSS#PSYSQFK), 1253 (DASASS#TSTFDAR) and phosphothreonines 995 (SVTESSPFVPSSTPT#PVDDR) and 1225 (SVHAAVT#PAAGK), became significantly up-regulated after a 10 minutes exposure to osmotic stress (Fig 4 A and Supplemental Table S4). Interestingly, with the exception of phosphoserine 1253 all of these sites lie within a S/T-P MAPK consensus motif. No dephosphorylation events have been detected by our analysis.
Figure 4. Regulated S/T-P phosphorylation sites of Pan1.
A and B: Pan1-HTBeaq was purified from either wt (A) or hog1Δ (B) strain backgrounds. Untreated (13C arginine and 13C lysine) to osmo-stressed (12C arginine and 12C lysine) ratios are shown. Phosphothreonine 1225 is marked with an arrow. C: Peak areas of phosphopeptides SVTESSPFVPSSTPT#PVDDR (T995) and SVHAAVT#PAAGK (T1225). Note: No 13C peaks were detected in a hog1Δ strain background for both phosphopeptides. However, 12C peak areas of phosphothreonine 995 were comparable between wild type and hog1Δ mutant indicating a Hog1 independent phosphorylation of the site. Data from a representative experiment are shown. D: Comparison of different strategies for quantification. Ratios (stressed/unstressed) obtained for phosphopeptides SSS#PSYSQFK (S1003) or SVHAAVT#PAAGK (T1225) and the corresponding unphosphorylated peptides of Pan1 are indicated. SRM measurements have been corrected for proline conversation (correction value was obtained from SILAC quantification Supplemental Table S12).
However, while phosphoserines 748 and 1003 and phosphothreonine 995 of the S/T-P motifs became only moderately induced between 2- to 4- fold, we observed a 23-fold increase in abundance of phosphothreonine 1225 (Fig 4 A and Table 2). We also quantified the corresponding unphosphorylated peptides to get information on the stoichiometry of the phosphopeptides (Fig 4 A). We observed that the levels of unphosphorylated peptides covering threonine 995 and serine 1003 become reduced in the same order of magnitude as the corresponding phosphopeptides become increased. Furthermore, at basal conditions the signals of the unphosphorylated and of the phosphorylated precursors were of similar intensities. Therefore, we conclude that a stoichiometric fraction of Pan1 molecules is already phosphorylated at these sites before stress application. Similar observations were made for the S/T-P independent phosphoserine 1253 (Supplemental Table S4). In contrast, the unphosphorylated peptide of threonine 1225 did not change in abundance although we observed a significant increase of the phosphopeptide signal. Therefore we predict that only a very minor percentage of Pan1 is phosphorylated at this site before stress exposure, and that probably most of Pan1 remains unphosphorylated at threonine 1225 even after osmo-stress induction. Hence this site, while strikingly up-regulated, is phosphorylated only at sub-stoichiometric levels (Fig 4 A).
Table 2.
Fold changes of PAN1 phosphorylation sites upon salt stress in wild type yeast (SILAC and iTRAQ), and in the mutants hog1Δ and hog1Δkss1Δ (SILAC). Ratios were manually normalized and corrected for Arg-Pro conversion and averaged over biological replicates. Quantification of site-specific phosphorylation events as well as unambiguous site assignment require that the phosphopeptide isoforms are chromatographically resolved which (according to the XICs) is the case for the phosphopeptides covering S1252-S1253 but not for those covering S747-S748.
| SILAC | iTRAQ | ||||
|---|---|---|---|---|---|
| Protein Site | Peptide | wt | hog1Δ | hog1Δkss1Δ | wt |
| T570 | T#GFGNNEIYTK | 0.84 | 1.07 | 1.14 | 1.28 |
| S621 | GLLDS#PTAVEIFR | noID | noID | 0.99 | noID |
| S747-S748 | NEEQSSFSS#PSAK | 2.94 | 4.66 | 3.67 | 2.30 |
| S747-S748 | KVYSAKNEEQSSFS#SPSAK, KVYSAKNEEQSSFSS#PSAK |
3.27 | 5.16 | noID | noID |
| S753 | S#VNHSSSTLQTDDISVDK | noID | noQuan | noID | noID |
| S753 | S#VNHSSSTLQTDDISVDKTVEK | noQuan | noID | noID | noID |
| S757 | SVNHS#SSTLQTDDISVDKTVEKK | 8.26 | noID | noID | noID |
| S896 | STES#EDSLSMEDEQQSAEIKR | 4.38 | noID | noID | noID |
| T944 | STMT#GSNMISNQEFER | 2.56 | noID | noID | noID |
| S986 | SVTESS#PFVPSSTPTPVDDR | noID | noID | 1.56 | noQuan |
| T993 | SVTESSPFVPSST#PTPVDDr | noQuan | noID | noQuan | noQuan |
| T995 | SVTESSPFVPSSTPT#PVDDR | 3.73 | noQuan | noQuan | 2.91 |
| S991 & T995 | SVTESSPFVPS#STPT#PVDDR | 7.86 | noQuan | noQuan | 1.24 |
| S1003 | SSS#PSYSQFK | 2.74 | 3.09 | 4.76 | 3.51 |
| S1135 | QES#DDEDEDDEEk | noID | 0.72 | noID | 1.11 |
| S1135 | QES#DDEDEDDEEKR | 1.17 | 1.04 | 1.02 | noID |
| S1180 | QIEDAQNES#DEEETNGK | noID | 0.89 | 1.15 | 1.24 |
| S1180 | kQIEDAQNES#DEEETNGk | noID | 0.86 | 0.98 | noID |
| S1219 | S#VHAAVTPAAGK | 1.89 | noQuan | noQuan | noID |
| T1225 | SVHAAVT#PAAGK | 23.10 | noQuan | noQuan | 16.56 |
| S1252 | DASAS#STSTFDAR | noID | noID | noQuan | 1.96 |
| S1253 | DASASS#TSTFDAR | 3.90 | 2.52 | 2.57 | 3.22 |
| S1253 & T1256 | DASASS#TST#FDAR | 6.97 | noID | noID | noID |
| S1281 | GLDEDEDDGWS#DEDESNNR | 1.06 | 0.99 | 1.07 | 1.16 |
| S1281 | GLDEDEDDGWS#DEDESNNRVAVDNKVEEAK | 1.00 | 1.05 | 1.04 | noID |
| T1321 | APPVTAAPLPSVT#PVPPAVPVPQANTSNEK | noQuan | noID | noQuan | 2.33 |
| S1387 | S#GSSTPALAGGVLPPPPPLPTQQASTSEPIIAHVDNYNGAEK | noID | noID | noID | 2.02 |
| S1387 & T1391 | S#GSST#PALAGGVLPPPPPLPTQQASTSEPIIAHVDNYNGAEK | noID | noID | 0.88 | noID |
To investigate whether the up-regulation of phosphorylation in response to osmotic stress depends on the MAPK Hog1 we repeated our analysis using a W303 SILAC hog1Δ strain (Fig 4 B and Supplemental Table S5). Interestingly, unphosphorylated and phosphopeptides of serine 748 and serine 1003 gave similar fold change like in the wild type. Regarding phosphothreonine 995, no induction ratio was determined due to the absence of the 13C labeled variant of the peptide. However, a comparison of peak areas of phosphopeptide SVTESSPFVPSSTPT#PVDDR (phosphothreonine 995) revealed similar intensities for the stressed samples (12C) in the wild type and hog1Δ mutant (Fig 4 C). Furthermore, Kss1, the MAPK of the filamentous growth pathway of budding yeast, can partly complement the effects of a hog1Δ deletion due to pathway crosstalk. To test whether the phosphorylation sites are affected by pathway crosstalk of Kss1 [35], we performed an additional series of experiments using a hog1Δkss1Δ double deletion mutant (Supplemental Table S6). Regarding serine 748 and serine 1003 we obtained similar phosphorylation patterns in wild type, the hog1Δ single deletion (Fig 4 A and B) and the hog1Δkss1Δ double deletion mutant (Table 2). Peak areas of phosphothreonine 995 again revealed similar intensities for the stressed samples in the wild type and double mutant (data not shown). This strongly indicates that phosphorylation at these sites is independent of Hog1 and that therefore they become phosphorylated by another, unknown kinase in response to osmotic stress. In contrast, the signal for the stressed, 12C-labeled phosphothreonine 1225 was almost abolished in a hog1Δ deletion strain compared to wild type (Fig 4 C) strongly indicating that this site indeed becomes directly phosphorylated by the MAPK. Thus, our targeted approach revealed that the stress regulated phosphorylation events which initially made Pan1 an interesting novel putative Hog1 target are in fact independent of the MAPK. Nevertheless, our observations on phosphothreonine 1225 (a site not quantified in our primary shotgun experiment) strongly suggests that Pan1 is a direct target of Hog1. This illustrates the significance of our validation approach, which allowed us to differentiate kinase dependencies of individual S/T-P sites.
Comparison of different strategies for quantification
Both, our shotgun analysis and our validation approach with purified Pan1, were based on SILAC labeling as a quantitative method for MS. To investigate whether our experimental settings are generally applicable for different quantitative MS methods we analyzed the stress dependent increase in phosphorylation of Pan1 by three independent quantitative methods for MS: SILAC, iTRAQ and SRM. We focused on two phosphopeptides of Pan1 which become phosphorylated with a different stoichiometry and compared ratios of unstressed to osmo-stressed samples. We analyzed phosphopeptides SSS#PSYSQFK (phosphoserine 1003) and SVHAAVT#PAAGK (phosphothreonine 1225) and their corresponding unmodified counterparts. Pan1 was isolated by HTBeaq tandem affinity purification from SILAC labeled cultures and the sample was split to allow an analysis by two independent methods. Ratios were either determined by peak area integration of extracted ion chromatograms measured on an LTQ-FT or by quantifying SRM transitions on a triple quadrupole instrument (TSQ). Interestingly, we obtained highly consistent ratios for all measured peptides (Fig 4 D and Supplemental Table S8). Additionally, Pan1 peptides obtained by tandem purification and digestion with trypsin were labeled with 4-plex iTRAQ reagents (Supplemental Table S7). Again we obtained highly consistent unstressed to osmo-stressed ratios by iTRAQ measurements, even for the low abundance peptide SVHAAVT#PAAGK (Fig 4 D). Hence, our purification workflow leads to peptide samples of sufficiently reduced complexity to allow a general application for all current types of quantitative mass spectrometry. Importantly, known limitation due to co-eluting peptides in the isolation window, such as the iTRAQ ratio compression [36] or the background interference on SRM transitions, are minimized or even eliminated when HTBeaq purified samples are analyzed.
Phosphorylation on threonine 1225 is regulated by MAPK Hog1
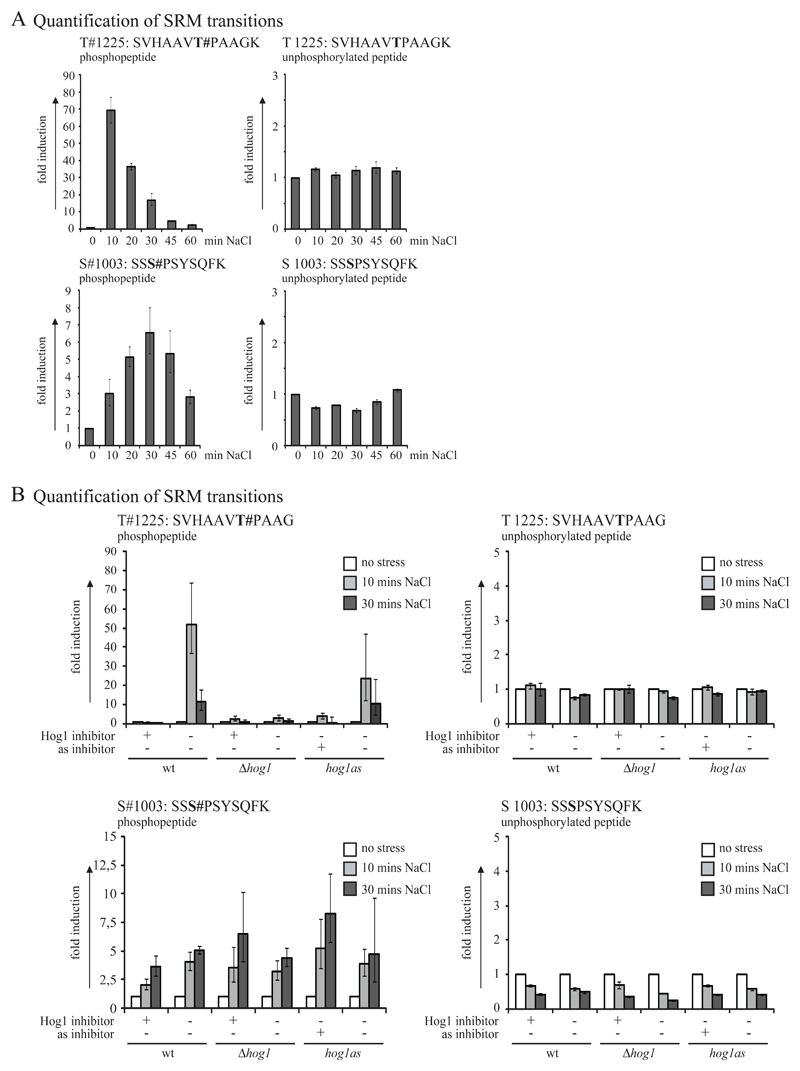
To verify threonine 1225 of Pan1 as a direct substrate of the MAPK Hog1 we studied the phosphorylation kinetics of this site during the response to osmotic stress. We prepared Pan1-HTBeaq purifications (Supplemental Figure 1E shows SRM sample preparation conditions) from cultures at logarithmic growth and after 10, 20, 30, 45 and 60 minutes induction with 0.5 M NaCl and analyzed the abundance of peptides of interest with SRM. Our analysis showed that phosphorylation at threonine 1225 reaches a maximum (70 fold induction) after 10 minutes and subsequently decreases back to basal levels within 60 minutes (Fig 5 A and Supplemental Table S10). Again, no change in the abundance of the unphosphorylated peptide was detected, indicating that most of the Pan1 proteins of a cell remain unphosphorylated at this site throughout the osmotic stress response. This is an indication for a very transient phosphorylation at threonine 1225. The observed phosphorylation kinetics reflects those of kinase Hog1 which becomes activated by a dual phosphorylation at residues threonine 174 and tyrosine 176 within a similar timeframe (see [14]).
Figure 5. Phosphorylation kinetics of S/T-P sites of Pan1.
A: Time course of phosphorylation of threonine 1225 and serine 1003 of Pan1 using SRM analysis. Pan1-HTBeaq has been purified from cells exposed to 0.5M NaCl for 0, 10, 20, 30, 45 and 60 minutes. Shown are geometric means of 4 technical replicates. Error bars denote the geometric standard deviation. B: Time course of phosphorylation of threonine 1225 and serine 1003 of Pan1 using Hog1 specific inhibitors. A specific Hog1 inhibitor was used for wild type and hog1Δ strains, for as-inhibitor treatment a hog1as strain was used. Cells have been exposed to 0.5M NaCl for 0, 10 and 30 minutes in presence (+) or absence (-) of the inhibitors. Shown are geometric means of 3 technical replicates. Error bars denote the geometric standard deviation.
Interestingly, similar experiments covering serine 1003 showed a different kinetic of phosphorylation: The abundance of phosphoserine 1003 continuously rises until a maximum (~ 6.5 fold) is reached after 30 minutes of stress induction. Thereafter the abundance of this phosphopeptide drops continuously, however levels were still not completely back to basal after 60 minutes. Again we could observe a reduction in the abundance of the corresponding unphosphorylated peptide, reaching a minimum at 30 minutes. Taken together the results of the phosphorylation kinetics measurements further supported our observation that threonine 1225 of Pan1 is a Hog1 phosphorylation site.
Based on these observations, we reasoned that threonine 1225 should be the only site of Pan1 susceptible to inhibitors specifically affecting the Hog1 kinase. Therefore, we tested the effect of two novel kinase inhibitors on the phosphorylation pattern of Pan1 (Fig 5 B, Supplemental Figure 2 and Supplemental Table S11). The inhibitors were a newly designed, Hog1-specific inhibitor [37] and a novel analogue sensitive (as) inhibitor [38]. Pan1-HTBeaq was purified from yeast treated for 10 minutes with 5µM of inhibitor prior the harvest or prior an additional 10 or 30 minutes exposure of 0.5 M NaCl. For as-inhibitor studies a W303 SILAC hog1as strain was used (in absence of an as-inhibitor the hog1as allele is fully functional [38]). Additionally, a hog1Δ deletion strain was included in the study. Incubation with DMSO was used for mock samples and induction of Hog1 signaling was controlled by western blots (data not shown). Unphosphorylated peptides and phosphopeptides of serine 1003 and threonine 1225 were quantified by SRM analysis (Fig 5 B). The abundance of phosphoserine 1003 and the corresponding unphosphorylated peptide was not affected by the inhibitors, whereas phosphothreonine 1225 could not be detected. However, we were able to monitor standard phosphorylation kinetics for the latter site in mock samples (Fig 5 B). These results clearly indicate that phosphorylation of threonine 1225 is regulated by the MAPK Hog1 in response to osmotic stress.
Discussion
The study presented in this manuscript provides a general strategy for validation and expansion of primary high throughput datasets based on selective protein purification under denaturing conditions. We demonstrate the value of our strategy by an in-depth phosphorylation analysis of Pan1, an essential factor of early endocytosis in Saccharomyces cerevisiae. Our analysis revealed different kinase dependencies of the protein.
A two-step approach for the validation of regulated protein modifications
Phosphorylation is one of the most common modes for regulation of protein function. Identification of novel kinase (and phosphatase) substrates is of high relevance in signaling research. Nowadays high throughput MS screens provide large datasets on putative target sites. However, besides reliable allocation of phosphorylation sites, an in-depth (ideally quantitative) characterization of the protein of interest provides the groundwork for functional and physiological studies. Here we presented a methodological workflow (Fig 1) for extending and validating high throughput data sets based on selective protein purification and showed that it reliably yielded data on kinase-dependency, stoichiometry and kinetics of individual regulated phosphorylation sites.
Modification studies of purified protein by MS are not necessarily new and individual steps of our workflow have been previously described in detail. However, here we provide an outline how to combine these steps into an experimental pipeline. A key element of our strategy is the analysis of peptide samples of reduced complexity provided by a robust and efficient protocol for protein purification. In our view, the choice of a modified histidine biotin tag is justified because it allows enrichment of individual proteins under continuous denaturing conditions [33]. Samples become significantly reduced in their complexity while posttranslational modifications are preserved. Moreover, tandem as well as one step purifications proofed to be scalable and allowed optimal preparative conditions for very low to medium abundant protein targets. Most importantly, peptide samples can be amended for all current types of quantitative MS including label based methods such as SILAC as well as iTRAQ, or label free methods like SRM.
This universal applicability was important for our study on the endocytotic scaffold protein Pan1, because it provided flexibility in the choice of quantification methods. According to our study Pan1 becomes phosphorylated by at least three independent kinases: Prk1, Hog1 and a yet unidentified kinase. Identification of phosphorylation sites alone was not sufficient for confident allocation to the responsible kinase. Only additional information such as the kinetics and stoichiometry of phosphorylation obtained by different quantitative MS methods allowed us to differentiate between individual signaling inputs.
As many proteins are targeted by multiple regulators our validation strategy of using purified proteins (as depicted in Fig 1 C and D) might be optimal for unraveling complex networks. First, SILAC or iTRAQ labeling can be used for the discovery of the phosphorylation pattern including the signal induced changes. Second, label free and cost effective SRM can be set up to determine phosphorylation kinetics in time-course experiments. Studies in mutants and/or treatment with specific kinase inhibitors are useful for revealing the underlying regulatory mechanisms. We believe that the first step provides the necessary characteristics of peptide-ion species for a targeted SRM setup. For example, information on the correct phosphorylation site localization, retention time, missed cleavage, ionization efficiency, charge state, additional PTMs, oxidation, and to some extent suitable transitions should be obtained beforehand. Furthermore, stoichiometry of phosphorylation can only be described by comparing the signals of the corresponding precursors (of the unphosphorylated and the phosphopeptide). Nevertheless information about stoichiometry is helpful in interpreting SRM data. The analysis of the unphosphorylated counter-peptide can yield supportive information and is, in some cases of stoichiometric modifications levels, even sufficient for description of temporal changes. For instance, the reversible decrease of peptide SSSPSYSQFK over time also reflects the phosphorylation kinetics of phosphoserine 1003. However, for phosphorylation changes on threonine 1225 the levels of the unphosphorylated peptide are not informative except for indicating the low stoichiometry (Fig 5 A).
Similar performance of quantitative MS methods due to robust protein purification
Considerable effort has been undertaken to improve quantitative MS methods such as SILAC, iTRAQ or SRM. These techniques are based on different methodical principles (reviewed in [39]) and to test whether they deliver similar results is of high significance. Here we present a direct comparison of the three methods by analyzing the stress dependent increase in phosphorylation of two phosphopeptides of Pan1 which become phosphorylated with a different stoichiometry. Samples used for the direct comparison were obtained by equal culture and protein purification conditions. Interestingly, we obtained highly consistent unstressed to osmo-stressed ratios for both phosphopeptides by integration of extracted ion chromatograms of SILAC labeled samples measured on a LTQ-FT and by comparison of the reporter ion signals in iTRAQ samples measured on a Velos Orbitrap and by quantifying SRM transitions on a triple quadrupole instrument. In general, SILAC and iTRAQ datasets were highly consistent regarding quantification, number of identified phosphorylation sites and sequence coverage, indicating that iTRAQ ratio compression [36] is minimized when samples of low complexity are analyzed (Fig 4 D). Therefore MS analysis of purified proteins is not limited by the choice of labels, which is important for studies that are restricted in the options of quantitative MS methods.
The endocytotic factor Pan1 is a target of kinase Hog1
We have been able to pinpoint threonine 1225 of Pan1 to be a substrate of Hog1. To our knowledge for the first time, the involvement of the HOG MAPK pathway in the endocytotic machinery is shown. Further observations provide additional proof for this interconnection. First, Pan1 is not the only factor of the endocytic machinery that becomes hyperphosphorylated in response to osmotic stress. We also detected an up-regulation of one S/T-P motif of the endocytotic factor Ede1 (Fig 2 A). Second, Pan1 is also an in vitro substrate of kinase Hog1 (Supplemental Figure 3B). Third, deletion mutants of interaction partners of Pan1 (ede1Δ or sla1Δ) are sensitive to high osmolarity conditions (data not shown) suggesting that a return to functional endocytosis is required for survival under osmotic stress and that the Hog1 site might be important for this process.
At the moment we can only speculate about the biological relevance of the different phosphorylation events on Pan1 during normal growth as well as stress. Pan1 lies at the core of a multi protein complex promoting assembly of the coat complex at the endocytic sites and is involved in the coupling of the vesicle coat to the actin network (for review see [40]). Actin polymerization provides a mechanical force to drive membrane invagination leading to the formation of endocytotic vesicles. Complex assembly and dissociation has been shown to be regulated by cycles of Prk1 dependent phosphorylation and PP1 dependent dephosphorylation of the N-terminal domain of Pan1 [23]. Here we present evidence that additional Pan1 domains might be the target of cyclical phosphorylation/dephosphorylation patterns as indicated by the accumulation of Hog1 independent sites. We could exclude Prk1 as a candidate kinase because all these sites lack the consensus (L/I/V/M)xx(Q/N/T/S)xTG motif [26]. Furthermore, the only Prk1 phosphorylation site of Pan1 covered in our quantitative analysis, serine 570, does not change its phosphorylation level upon osmotic stress (Fig 2 A and Supplemental Figure 3A).
Finally, the complexity of Pan1 phosphorylation patterns might serve as a good example for how important and useful a detailed phosphorylation analysis could be before any genetic and physiological approaches are initiated.
Supplementary Material
Acknowledgements
We thank C. Kraft, A. Schmidt, S. K. Yelamanchi for reading of the manuscript. K. Mechtler (IMP Vienna) provided synthetic peptide standards. P. Kaiser provided HB-tagging plasmids. We thank A. Vendrell for the W303 hog1as strain and L. Subirana for the in vitro kinase assay. W.R. would like to dedicate this work to his father. W.R., I.D. and P.P. are supported by the Christian Doppler Society and the 7th EU framework project UNICELLSYS. The project was also funded by the Austrian Science Fund Project F3402-B03.
Abbreviations
- AGC
automatic gain control
- AQUA
absolute quantification
- ETD
electron-transfer dissociation
- FDR
false discovery rate
- HB
poly histidine, biotinylation signal
- HCD
higher energy collision dissociation
- HTB
poly histidine, TEV-protease cleavage site, biotinylation signal
- HTBeaq
HTB-effective cleavage plus absolute quantification
- HOG
high-osmolarity glycerol
- IAA
iodoacetamide
- MAPK
mitogen-activated protein kinase
- SC
synthetic complete medium
- SGD
Saccharomyces cerevisiae genome database
- TEAB
triethylammonium bicarbonate
- YPD
yeast extract, peptone, D-glucose medium
References
- [1].Albuquerque CP, Smolka MB, Payne SH, Bafna V, et al. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bodenmiller B, Wanka S, Kraft C, Urban J, et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].de Godoy LM, Olsen JV, Cox J, Nielsen ML, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- [4].Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- [5].Ghaemmaghami S, Huh WK, Bower K, Howson RW, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- [6].Gruhler A, Olsen JV, Mohammed S, Mortensen P, et al. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- [7].Li X, Gerber SA, Rudner AD, Beausoleil SA, et al. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- [8].Schreiber TB, Mausbacher N, Soroka J, Wandinger SK, et al. Global Analysis of Phosphoproteome Regulation by the Ser/Thr Phosphatase Ppt1 in Saccharomyces cerevisiae. J Proteome Res. 2012;11:2397–2408. doi: 10.1021/pr201134p. [DOI] [PubMed] [Google Scholar]
- [9].Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Soufi B, Kelstrup CD, Stoehr G, Frohlich F, et al. Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol Biosyst. 2009;5:1337–1346. doi: 10.1039/b902256b. [DOI] [PubMed] [Google Scholar]
- [11].Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].de Nadal E, Ammerer G, Posas F. Controlling gene expression in response to stress. Nat Rev Genet. 2011;12:833–845. doi: 10.1038/nrg3055. [DOI] [PubMed] [Google Scholar]
- [13].de Nadal E, Posas F. Multilayered control of gene expression by stress-activated protein kinases. EMBO J. 2010;29:4–13. doi: 10.1038/emboj.2009.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Reiser V, Ruis H, Ammerer G. Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:1147–1161. doi: 10.1091/mbc.10.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rep M, Reiser V, Gartner U, Thevelein JM, et al. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol. 1999;19:5474–5485. doi: 10.1128/mcb.19.8.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mollapour M, Piper PW. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol Cell Biol. 2007;27:6446–6456. doi: 10.1128/MCB.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Westfall PJ, Patterson JC, Chen RE, Thorner J. Stress resistance and signal fidelity independent of nuclear MAPK function. Proc Natl Acad Sci U S A. 2008;105:12212–12217. doi: 10.1073/pnas.0805797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Toret CP, Drubin DG. The budding yeast endocytic pathway. J Cell Sci. 2006;119:4585–4587. doi: 10.1242/jcs.03251. [DOI] [PubMed] [Google Scholar]
- [19].Barker SL, Lee L, Pierce BD, Maldonado-Baez L, et al. Interaction of the endocytic scaffold protein Pan1 with the type I myosins contributes to the late stages of endocytosis. Mol Biol Cell. 2007;18:2893–2903. doi: 10.1091/mbc.E07-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Duncan MC, Cope MJ, Goode BL, Wendland B, Drubin DG. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- [21].Miliaras NB, Park JH, Wendland B. The function of the endocytic scaffold protein Pan1p depends on multiple domains. Traffic. 2004;5:963–978. doi: 10.1111/j.1600-0854.2004.00238.x. [DOI] [PubMed] [Google Scholar]
- [22].Toshima J, Toshima JY, Duncan MC, Cope MJ, et al. Negative regulation of yeast Eps15-like Arp2/3 complex activator, Pan1p, by the Hip1R-related protein, Sla2p, during endocytosis. Mol Biol Cell. 2007;18:658–668. doi: 10.1091/mbc.E06-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Toshima J, Toshima JY, Martin AC, Drubin DG. Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat Cell Biol. 2005;7:246–254. doi: 10.1038/ncb1229. [DOI] [PubMed] [Google Scholar]
- [24].Zeng G, Yu X, Cai M. Regulation of yeast actin cytoskeleton-regulatory complex Pan1p/Sla1p/End3p by serine/threonine kinase Prk1p. Mol Biol Cell. 2001;12:3759–3772. doi: 10.1091/mbc.12.12.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- [26].Huang B, Zeng G, Ng AY, Cai M. Identification of novel recognition motifs and regulatory targets for the yeast actin-regulating kinase Prk1p. Mol Biol Cell. 2003;14:4871–4884. doi: 10.1091/mbc.E03-06-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zeng G, Cai M. Regulation of the actin cytoskeleton organization in yeast by a novel serine/threonine kinase Prk1p. J Cell Biol. 1999;144:71–82. doi: 10.1083/jcb.144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zeng G, Huang B, Neo SP, Wang J, Cai M. Scd5p mediates phosphoregulation of actin and endocytosis by the type 1 phosphatase Glc7p in yeast. Mol Biol Cell. 2007;18:4885–4898. doi: 10.1091/mbc.E07-06-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- [30].Ross PL, Huang YN, Marchese JN, Williamson B, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- [31].Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci U S A. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tagwerker C, Flick K, Cui M, Guerrero C, et al. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol Cell Proteomics. 2006;5:737–748. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- [33].Tagwerker C, Zhang H, Wang X, Larsen LS, et al. HB tag modules for PCR-based gene tagging and tandem affinity purification in Saccharomyces cerevisiae. Yeast. 2006;23:623–632. doi: 10.1002/yea.1380. [DOI] [PubMed] [Google Scholar]
- [34].Taus T, Kocher T, Pichler P, Paschke C, et al. Universal and Confident Phosphorylation Site Localization Using phosphoRS. J Proteome Res. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- [35].O'Rourke SM, Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ow SY, Salim M, Noirel J, Evans C, et al. iTRAQ underestimation in simple and complex mixtures: "the good, the bad and the ugly". J Proteome Res. 2009;8:5347–5355. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- [37].Diner P, Veide Vilg J, Kjellen J, Migdal I, et al. Design, synthesis, and characterization of a highly effective Hog1 inhibitor: a powerful tool for analyzing MAP kinase signaling in yeast. PLoS One. 2011;6:e20012. doi: 10.1371/journal.pone.0020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Klein M, Morillas M, Vendrell A, Brive L, et al. Design, synthesis and characterization of a highly effective inhibitor for analog-sensitive (as) kinases. PLoS One. 2011;6:e20789. doi: 10.1371/journal.pone.0020789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Eyrich B, Sickmann A, Zahedi RP. Catch me if you can: mass spectrometry-based phosphoproteomics and quantification strategies. Proteomics. 2011;11:554–570. doi: 10.1002/pmic.201000489. [DOI] [PubMed] [Google Scholar]
- [40].Huang B, Cai M. Pan1p: an actin director of endocytosis in yeast. Int J Biochem Cell Biol. 2007;39:1760–1764. doi: 10.1016/j.biocel.2006.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.