Abstract
With the recent success in determining membrane protein structures, further detailed understanding of the identity and function of the bound lipidome is essential. Using a combined approach of High-energy native mass spectrometry (HE-nMS) and solution phase lipid profiling this protocol serves to determine the identity of the endogenous lipids that directly interact with the protein. Further, the method can identify systems where such lipid binding plays a key role towards regulating the oligomeric assembly of membrane proteins. The protocol begins with recording the native mass spectrum of the protein of interest, under successive delipidation conditions, to determine if delipidation leads to disruption of the oligomeric state. Subsequently, we propose employing a bi-pronged strategy where (i) using a HE-nMS platform that allows dissociation of detergent micelle at the front end of the instrument. This enables isolation of the protein-lipid complex at the quadrupole and successive fragmentation at the collision cell. This leads identification of the bound lipids masses. Simultaneously, coupling this with (ii) in solution LC-MS/MS based identification of extracted lipids reveals the complete identity of the interacting lipidome that co-purify with the proteins. Assimilation of these two sets of experiments divulges the complete identity of the set of lipids that directly interacts with the membrane protein of interests, and can further delineate its role in maintaining the oligomeric state of the protein. The entire procedure takes 2 days to be completed.
Keywords: Membrane protein, lipid, mass spectrometry, MS, MS/MS, LC-MS/MS, native mass spectrometry, native MS, structural biology, high-energy native mass spectrometry, high-energy native MS, HE-nMS, lipidome, protein-lipid interaction, delipidation, lipid extraction
Introduction
The study of membrane proteins by means of native MS (nMS) is gaining momentum with new discoveries and more research groups entering the field. 1–4 New insights reported include the stoichiometry of the beta-barrel assembly machinery 5 and evidence of the diverse and important roles lipids can play in maintaining membrane protein structure and function. 6 Recent work has also shown that lipids can play a key role in regulating the oligomeric states of membrane protein assemblies.7 Information about the oligomeric state and the identity of lipids at protein-protein interfaces can provide vital insights into their functional form. Integral membrane proteins are embedded in the lipid bilayer in vivo, and hence it seems natural that they may be co-purified with associated lipids. However, a long-standing problem has been the inability to identify and distinguish between different kinds of bound lipids. In particular, to differentiate those lipids that are in direct contact with the protein and play key functional and structural roles (specific and annular lipids), from co-purified “bulk lipids” that may be co-extracted as a result of the purification strategy and merely reflect the average composition of the membrane.8,9 This raises the question - how can we identify the key lipids that are likely to be implicated in mediating membrane protein function?
The current “gold standard” for visualising lipids in complex with membrane proteins is to observe the interaction directly through techniques such as cryo-electron microscopy and X-ray crystallography. This is exemplified by the recent visualisation of numerous phospholipid (PL) molecules surrounding the Ca2+-ATPase pump.10 However, optimising conditions for such experiments can be extremely challenging, and even when lipid molecules are present, they are often poorly resolved and cannot be differentiated from other molecules such as detergents. The low resolution also often prevents identification of both lipid class (head group) and chain length. A more routine approach to identifying the presence of lipid in complex with membrane proteins is through global size measurements such as analytical ultracentrifugation (AUC) 11, native polyacrylamide gel electrophoresis (PAGE) 12 or hyphenated approaches such as size exclusion chromatography (SEC) coupled to multi-angle light scattering (SEC-MALS).13 In these approaches, the relative proportions of protein, lipid and detergent can be estimated which in turn can provide evidence for co-purified lipid. However, the chemical identity of the lipid fraction cannot be determined. To achieve this, an “extraction-analysis” is often performed whereby all lipid components are removed by partitioning into organic solvent and analysed separately.
The extraction-analysis strategy can be extremely powerful if coupled with LC-MS based quantitative lipidomics. Using this approach both the number and molecular identity of the lipids that have co-purified with membrane proteins can be determined.14,15 In addition, comparison of the lipid profile that co-purifies with the protein of interest with that of the native membrane can provide indirect evidence of the protein of interest selective choses lipids in its vicinity, and by thus remodels the membrane.16 However, any extraction-analysis approach merely reports on lipids that have co-purified with the protein complex and is unable to differentiate specific from annular and bulk lipids. This remains a key issue in identifying regulatory lipids that are linked to function or stabilisation. In this protocol we describe the steps involved to identify and chemically characterize the lipidome that directly interacts and co-purifies with any membrane protein of interest and, within that, differentiate the tightly bound lipids that interact directly with the protein. The protocol also enables identification of systems where specific lipid binding may impart active regulation of the oligomeric assembly of membrane proteins.
Overview of the Procedure
This protocol aims to deconvolute the inventory of lipids directly interacting with proteins as opposed to those that are merely co-purified. Furthermore in specific membrane protein systems, where lipids play critical roles in maintaining the oligomeric assemblies, such lipids can be identified through successive delipidation followed by mass measurement at each stage. Assemblies can also be identified wherein removal of lipids leads to dissociation into lower order oligomers or monomeric subunits. The masses of bound lipids can be determined using the HE-nMS platform, which can remove the detergent micelle for subsequent isolation of protein lipid complexes. Correlating the masses of the bound lipid detected by native MS, with LC-MS/MS analysis of the lipids isolated from the same solution subjected to native MS, can reveal the classes of the individual lipid components.
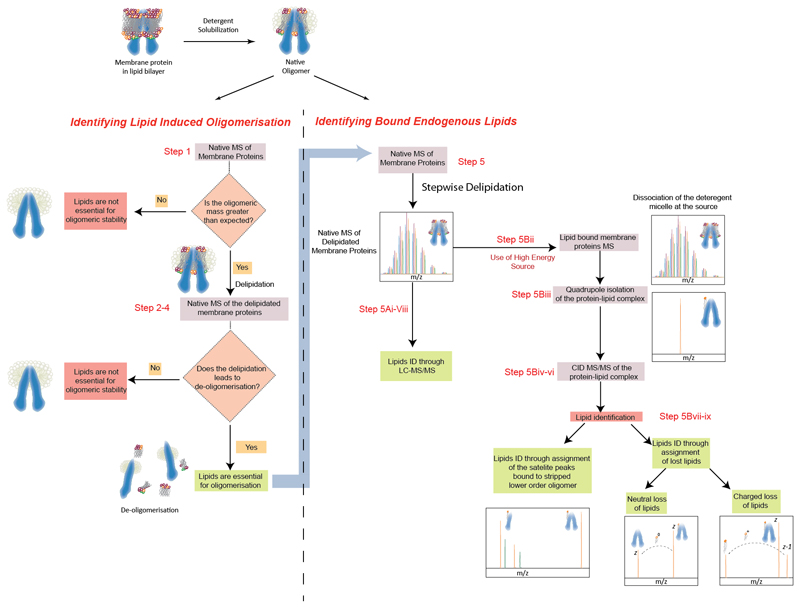
The stages developed to identify endogenous lipids that bind directly to membrane proteins involve a three-pronged strategy. In the first stage (Steps 1- 4), stepwise delipidation is combined with native MS, under native and delipidating detergent conditions, to refine conditions where key lipids remain bound (Figure 1). For oligomeric membrane proteins, this enables identification of systems where lipids play a critical role towards maintaining the oligomeric states. In the second stage (Step 5A, BOX 2), an extraction-analysis strategy is used to monitor stepwise gradual delipidation of the protein (either by varying the time, the amount of the detergent, or both) and determine the identity of the lipid cohort that co-purifies with the protein. The third stage (Steps 5B) employs the HE-nMS platform, at each point of delipidation, to perform multi step tandem MS on the protein-lipid complex to visualise, select and identify the lipids directly binding to the protein. Combining both sets of data determines the complete identity of lipids directly binding to the membrane protein of interest.
Figure 1. Schematic representation of the processes involving determination of bound lipids to membrane protein.
Overall workflow to identify membrane protein systems where lipid binding is critical for oligomeric stability. Following the native mass spectrum the HE-nMS platform is employed to identify the bound lipids through MS/MS of the isolated lipid-protein complex. In cases where the purpose of the experiment is to identify bound lipids, irrespective their role in maintaining the structural assembly, only the RHS of the diagram should be followed.
BOX 2. How to identify lipids from MS/MS experiments.
There are multiple ways of identifying lipids depending upon how the ions fragment
Scenario 1: MS/MS of the lipid-bound oligomer yields lower order oligomers. In such cases analyse the satellite peaks associated with the charge states corresponding to the lower order oligomers. Masses of these adducts will reveal the mass of the lipids bound to the oligomer
Scenario 2: MS/MS of the lipid bound membrane protein leads to neutral losses of lipid in the mass spectrum. In this case measure the mass loss from the parent ion, which corresponds to the mass of the lipid molecule.
Scenario 3: In certain cases, based on the head-group of the lipids, charged loss of the lipids may be observed. In such cases dissociated lipids may be present in the lower m/z region of the spectrum.
CRITICAL – It is suggested that the user adjusts the Quadrupole profile to maximise the transmission of the target/precursor ions. Typically the high collision gas pressures, that are used to detect protein oligomers, may deter the detection of lipid molecules. In the case of scenario 3 the Quadrupole profile and the collision gas phase may be further optimized for transmission over a wider m/z range.
Distinguishing Specific Lipid Binding from Bulk and Annular Lipids
Native MS has proved an extremely powerful method for determining the oligomeric state of membrane protein assemblies and visualising the presence of ligands, lipids and drugs.7,16–19 To achieve this, conditions must be optimised in which the membrane protein complex is both stable in solution and able to be liberated intact, from its protective detergent micelle, inside the mass spectrometer. This involves selecting appropriate detergents for both membrane protein solubilisation, and also native MS, as described in our previous protocol.20 For identification of bound lipids, conditions are carefully selected to preserve the oligomeric state of the membrane protein and also to maintain the cohort of bound lipids. It should be noted that in certain cases non-specific gas phase association may lead to observation of non-physiological oligomeric states. Care should be taken to ensure the physiological viability of these associations. This requires both gas phase experimental validation as well as solution phase experiments that involve generating mutant proteins that lack probable lipid-protein/protein-protein association sites and expressing wild type proteins in specific lipid deficient environments 21,7. Once the presence of bound lipids is detected (by an increase in mass or distinct adducts present on the charge state series) the goal is to reduce the number of bound lipids so that only a key set of annular lipids remain. Two general strategies can be followed here.
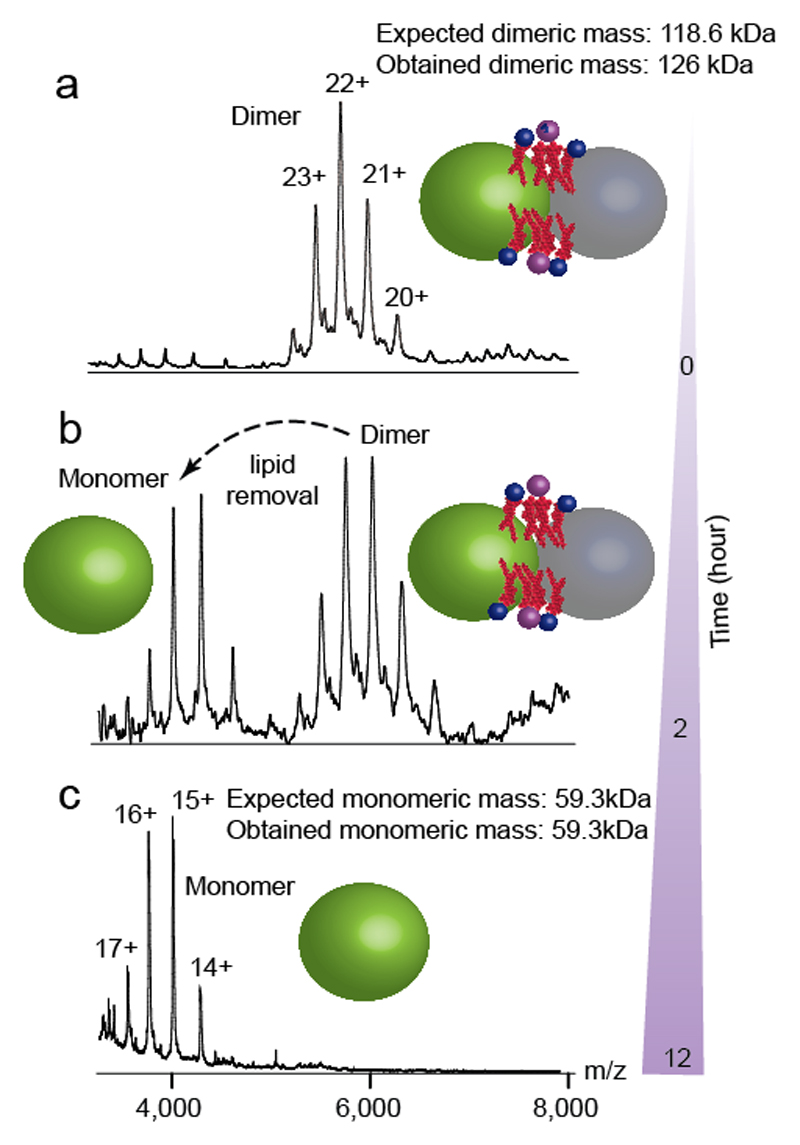
The first strategy (described in Steps 1-4) exploits the variable delipidating properties of different detergents.22 Detergents such as NG, Cymal-5, LDAO and the poly-ol class of detergents (such as C8E4 and C12E9) are known to be effective at delipidating membrane proteins. 20,22 Exchange into buffers containing these detergents may be sufficient to achieve substantial delipidation, which can then be monitored by native MS. The effect of exchange into a delipidating detergent is illustrated in Figure 2 for the bacterial leucine transporter LeuT from A. Aeolicus. In 1.0% wt/vol OG detergent (twice the amount of critical micelle concentration (CMC)) the mass spectrum of LeuT reveals a charge-state resolved dimer (Figure 2a). The mass measurement indicates an adduct of 7.4 kDa that persists even at high levels of activation.7 Upon delipidating the protein in NG detergent, 22 lipids are gradually removed from the dimer and a lipid-free monomer of LeuT is formed (Figure 2b,c). In this case, loss of lipid proceeds with concomitant dissociation of the complex. No lipid-free dimer was observed, implying that lipids may be essential for oligomerisation.
Figure 2. Delipidation of dimeric LeuT.
Progressive delipidation of LeuT dimer in 1% (wt/vol) delipidating detergent NG. a, Native MS of LeuT in OG shows dimer with 7.4 kDa lipid adduct (observed mass 126030 Da, expected mass of 119.8kDa). b, Progressive delipidation in NG detergent, monitored by native MS, indicates partial dissociation after 2h; however no apo dimer is observed and a 7.4kDa lipid adduct is retained with the dimer. Monomeric LeuT is devoid of lipid and the measured mass correlates well with the theoretical mass. c, Complete delipidation and dissociation of LeuT is achieved after 12h. Delipidation causes disintegration of the dimer into free monomers. Protein subunits are shown as grey and green spheres with lipids represented in red.The relative sizes of the protein and lipids are not to scale.
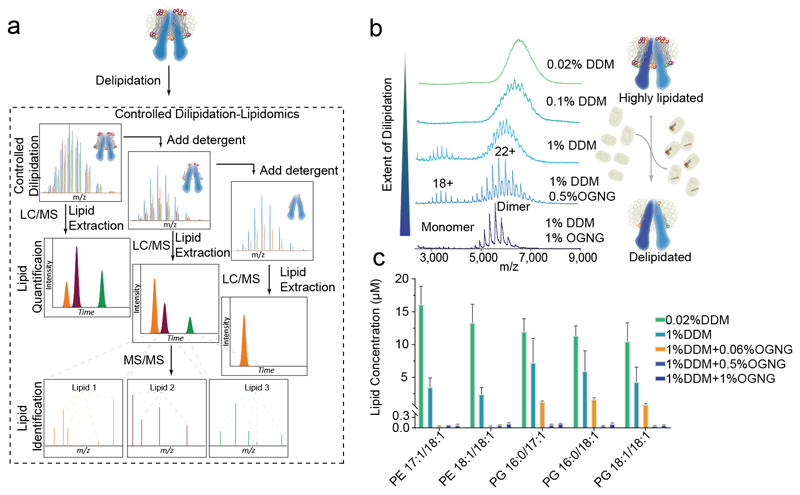
The second strategy (described in Steps 5A) is to gradually remove bulk lipids. Increasing the concentration of the detergents, or repeated exchange in the same detergent, can gradually remove the bulk lipid component surrounding the membrane protein complex.15 By varying the detergent concentration and class, stepwise delipidation can be achieved (Figure 3a). The most important lipids for mediating protein function are likely those that remain bound even after extensive delipidation. This is illustrated by the gradual delipidation of the ABC transporter MsbA (Figure 3b). When the protein is purified in 0.02% wt/vol DDM, a charge-state series cannot be discerned due to large amounts of bound lipid. Increasing the concentration of DDM to 0.10% wt/vol removes a proportion of the lipid and enables visualisation of a charge state series. Further delipidation occurs upon increasing the concentration to 1.0% wt/vol DDM. At this point, additional DDM had little effect. A mixed micelle containing 1% wt/vol OGNG was required to remove the majority of lipids present leaving only a low number of tightly bound lipids.
Figure 3. Schematic of performing successive delipidation of MsbA.
a, A stepwise protocol for carrying out progressive delipidation in-solution coupled with LC-MS to identify lipids. b, Native MS of MsbA upon progressive delipidation. Excess detergent was removed post-delipidation using SEC and the spectra were recorded from solution containing 0.02% (wt/vol) DDM. The native MS yields progressively well-resolved spectra with detection of apo-MsbA dimer following incubation in 1% (wt/vol) DDM + 1% (wt/vol) OGNG. Here the expected mass (134,547 Da) matches with that of the obtained mass (134,502Da). c, Corresponding in-solution quantitative lipid analysis of the delipidated proteins from the same protein sample. In the schematic lipids are represented as coloured spheres, whereas the detergents are in grey. The relative sizes of the protein and lipids are not scaled. Conventional lipid nomenclature has been followed where the first number stands for the number of carbons in the chain and the second number, separated by a colon, designates the number of unsaturations within the two aliphatic chains separated by /.
Combining stepwise delipidation with an extraction-analysis strategy (which involves extracting lipids present at each stage of extraction and subsequent identification by LC-MS/MS approaches, described in detail in BOX 1) can add an additional dimension to the analysis by defining the lipid cohort remaining at each stage. This approach is particularly powerful and has been used previously to determine the 1:1 binding stoichiometry of cardiolipin (CDL) to the K subunit in the K10 ring of V-type ATPases,14 and a key specific subset of negatively charged, functionally significant, annular phosphatidylglycerol (PG) lipids bound to the lipid flipase TmrAB. 15 In each case, native MS measurement provides evidence of lipids directly bound to the protein. The lipids are then extracted and LC-MS/MS of the lipid fraction is used to identify the entire set of lipids present in-solution, both in empty detergent micelles and in the proteo-micelles.
BOX 1. LC-MS/MS extraction analysis for identification of lipids present in solution.
Timing: 2 Days
1. Digest 0.2 - 1 mg of delipidated protein sample from Step 5A(vii) in the detergent) by incubating it with 1:50 molar ratio of trypsin for overnight, at 37°C.
Δ CRITICAL STEP It is important to note that in principle, the lipid isolation could also be performed by denaturing and precipitating the protein with hydrophobic solvents alone. But often it results in closely bound lipids being precipitated out with the proteins.
2. Dry down the digest by using a vacuum concentrator (SpeedVac) and re-suspend it in a minimal volume (20 to 40 μL) of 30% LC buffer B/70% LC buffer A. Alternatively, samples can also be lyophilized and resuspended as stated.
3. Sonicate the sample for at least 30 min in a sonicator bath (without heating, at room temperature). Subsequently centrifuge the sample at 5000g for 15 minutes (4°C) and collect the supernatant.
4. Dilute the sample to a final concentration of 10 pg/ul to 1 ng/ul, depending on the complexity of co-purified lipids, in 30% LC buffer B/70% LC buffer A.
Δ CRITICAL STEP Low concentrations of lipids must be used at this stage to minimize lipid aggregation in nanospray. For lipid quantification it is important to measure linear dynamic ranges of the LC-MS system using lipid standards to estimate a proper lipid concentration range for quantification.
5. Perform LC-MS/MS analysis using a reversed-phase column at 40 °C. Less hydrophobic columns (such as C8) should be selected for the more hydrophobic lipids such as CDL. Load the total lipid extract into a 1 μL loop using the autosampler and loading pump, then transfer onto a C18 column (Acclaim PepMap 100, C18, 75 μm × 15 cm, Thermo Fisher Scientific) at a flow rate of 300 nL/min, using 30% Solution B.
6. Reveal the lipid classes and their structural identity by comparing with the published databases or using lipid identification tools. 29,30 Subsequently, quantify the lipids following the protocols reported in Reference 15.
In the current work, we applied this approach to MsbA. Different concentrations of detergent were used during the membrane solubilisation step and the quantities of predominant phosphatidylethanolamine (PE) and PG species were monitored at different stages of delipidation (Figure 3c). Increasing concentrations of DDM from 0.02% wt/vol to 1% wt/vol reduces the amount of both PG and PE lipid present. A further small addition of 0.06% wt/vol OGNG greatly reduces the total concentration of bound lipid. This illustrates the importance of selecting appropriate detergents for delipidation. Most interestingly, the fraction of lipids that remains bound after delipidation with DDM/OGNG mixed micelles is heavily enriched in negatively charged PG lipids. This suggests that the annular belt around MsbA is primarily composed of PG. Further increasing the OGNG concentration to 1% wt/vol reduces this cohort of PG lipids to those presumably in direct contact with the MsbA dimer.
It is important to note that this extraction-analysis approach merely identifies lipids that are present in solution with the protein of interest. If delipidation can be refined sufficiently to leave only the level of the annular belt of lipids directly interacting with the protein, then this approach alone can identify binding partners. In practice it is often not possible to refine delipidation conditions to such a degree. Further experiments performed on the protein-lipid complex itself are required to definitively identify lipids that are bound directly (see below).
Determining the Identity of Lipids Directly Bound to Membrane Protein Complexes
The major limitation of the extraction-analysis approach is that it reports on the entire set of lipids present in the solution, both bound to membrane proteins and present in bulk, either in empty or proteo micelles. Hence, only a subset of these lipids may be in contact with the membrane protein. To identify endogenous binding partners, explicit evidence of binding is required. A solution to this problem would be to detect and interrogate individual protein-lipid complexes, directly, inside the mass spectrometer (Steps 5B). This approach has been successful for soluble protein complexes in so-called ‘catch and release’ type experiments.23 Here, protein-ligand complexes are transferred from solution into the mass spectrometer, selected in the quadrupole and then dissociated in a collision cell, leading to identification of the ligand by mass measurement. These experiments are not routinely possible for membrane protein-lipid complexes however, as the protective detergent micelle must first be removed. For common MS platforms (e.g. Q-ToF, hybrid Orbitrap and FTICR systems 24) this is achieved by transmitting the proteo-micelle complex through the source region to the mass analyser (quadrupole). Subsequently the detergent micelle is removed via collisions with an inert gas in the collision/HCD cell.16,20,25 In this configuration, further dissociation experiments are not possible as there is no additional mass analyser between the collision cell and detector to isolate individual lipid-bound species for further activation.
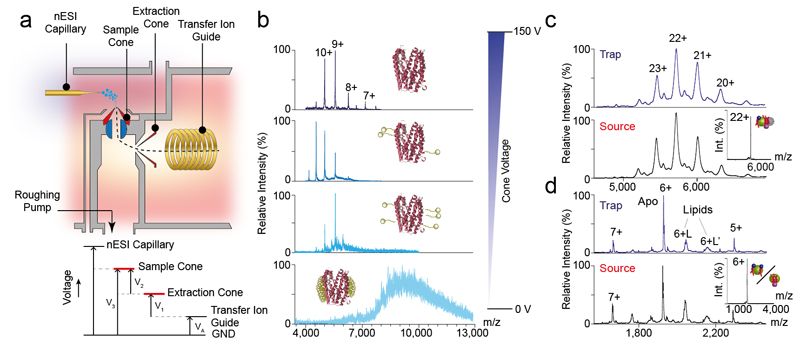
To overcome this limitation, we developed a platform that enables isolation of protein-lipid complexes, earlier in their transmission, removing detergent micelles in the source region of the mass spectrometer (Figure 4a). Elevated voltages, applied to the sample and extractor cones in the source region of our modified Q-ToF instrument, allow increased activation of ions in this region and removal of the detergent micelle. Using this approach, gradual liberation of the monomeric transporter MATE from C8E4 micelles was achieved (Figure 4b). In-source activation also allows liberation of LeuT (Figure 4c) and semiSWEET (Figure 4d) dimers from OG and C8E4 micelles respectively, producing results comparable to those obtained when detergent is removed in the collision cell. Most importantly this process releases protein-lipid complexes that can be subsequently selected with the quadruple for dissection in the collision cell (Inset to Figure 4c-d).
Figure 4. Schematic of the high energy platform and its application on various membrane proteins.
a, Schematic of the modified high-energy source and voltage diagram of key components. To achieve additional activation in the source region the extraction cone voltage (V1) can be adjusted from 0-200 V. The sample cone voltage (V3) is controlled by an external power supply. Total activation energy = V3-VA ≡ V1+V2 b) Release of monomeric MATE (PDB id: 3VVN) from C8E4 detergent micelle. Increasing cone voltage in the source region from 0 - 150 V allows MATE to be liberated gradually from the detergent micelle (observed mass 50016 Da). Similar results obtained for c, lipid-bound LeuT and d, semiSWEET. In both these cases the spectra generated through liberation of the protein at the trap is identical to source dissociation. The Insets show isolation spectra of lipid-bound LeuT and SemiSWEET, isolated in the quadrupole following fragmentation of the proteo-micelle in the source. The PLs are represented blue head-groups and red chain, whereas the head-group of CDL is depicted in violet. The relative sizes of the protein and lipids are not scaled. The L and L’ in panel d correspond to different lipid bound 6+ charge states. All the spectra were acquired on a Q-TOF instrument.
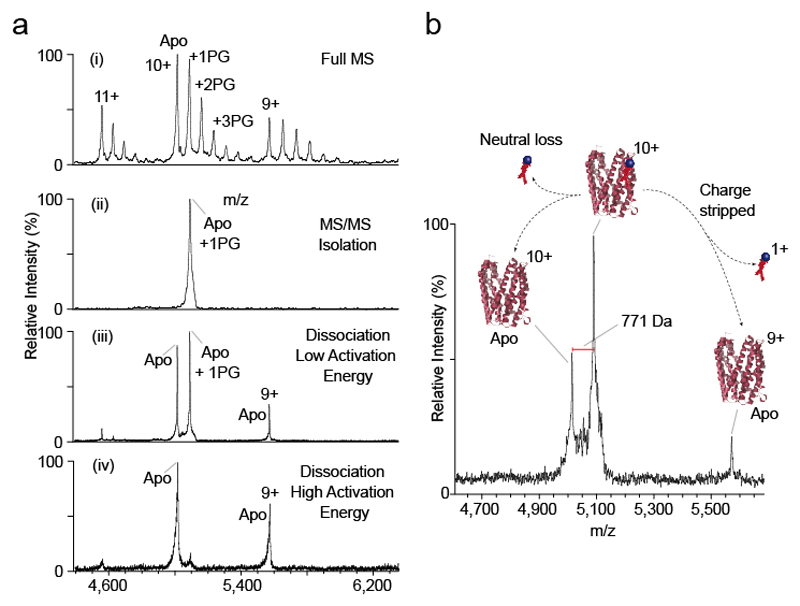
A straightforward example of this multi-step native MS workflow is shown in Figure 5. A complex of MATE and POPG lipid is present in the gas phase after detergent removal in the high-energy source. Up to 3 POPG lipids remain bound to the protein (Figure 5a(i)). The MATE10+ charge state bound to one POPG bound is then selected in the quadrupole (Figure 5a(ii)). Low energy activation in the collision cell results in dissociation of the lipid, which is lost either as a charged or neutral species (Figure 5a(iii)). Increasing the collision energy results in complete loss of the lipid (Figure 5a(iv)). The mass of the bound lipid can then be calculated from the difference between the bound and apo peaks (Figure 5b).
Figure 5. HE-nMS experiment on MATE.
(a) Detection, isolation and fragmentation of POPG-bound MATE through (i) in-source dissociation of the proteo-micelle that leads to the MS detection of lipid-bound MATE, (ii) subsequent quadrupole isolation of the lipid-bound state and fragmentation (iii) low and (iv) high CID energy. Mass spectra show that with increasing collision energy higher quantities of delipidated protein are formed. (b) The difference between the m/z values of the isolated peak and the naked protein correspond to the mass of the bound lipid. (MATE PDB id: 3VVN)
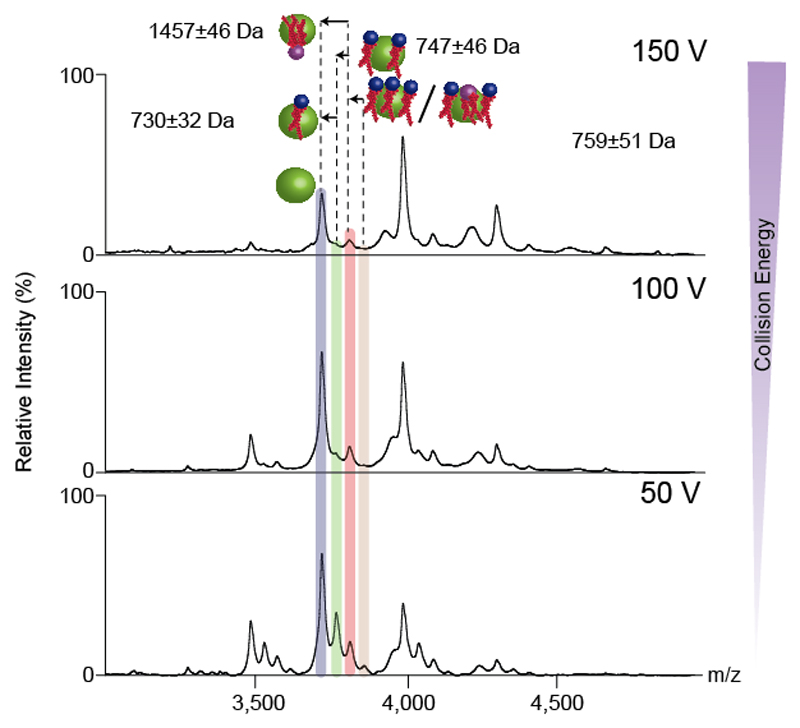
Alternatively, for some multimeric assemblies, activation of the lipid-bound species may lead directly to disintegration of the complex into lower order oligomers. This behaviour is a strong indication that lipids may be involved in maintaining the oligomeric state. Here, lipid identification cannot simply be achieved by considering dissociation of lipids. In this case examining the sub-complexes formed after activation, for lipid adducts that are retained, can be used to identify a subset of lipids that were bound to the original complex. The LeuT dimer exhibits this behaviour. Figure 6 shows the MS/MS spectrum of the dimeric LeuT with 7.4kDa lipid plug. Upon CID activation the dimer dissociates into monomer, with satellite peaks corresponding to bound lipids that were part of the 7.4kDa lipid plug. Hence, by determining the masses of the satellite peaks one can reveal the identity of the bound lipids.
Figure 6. HE-nMS of dimeric lipid bound LeuT.
Lipid identification of LeuT through identification of the satellite peaks bound to the stripped monomer species generated through MS/MS of the lipid bound dimeric state (as shown in Figure 4c). The satellite peaks correspond to three PL and one CDL bound to each monomer. The masses of the lipids are calculated by subtracting the masses of the apo and lipid bound states. The errors in the masses of lipids determined represent the sum of the errors incurred in measuring masses of the individual peaks. The spectra were obtained for the dimeric LeuT in OG. Phospholipids and cardiolipins are represented with blue and purple spheres respectively. The relative sizes of the protein and lipids are not scaled.
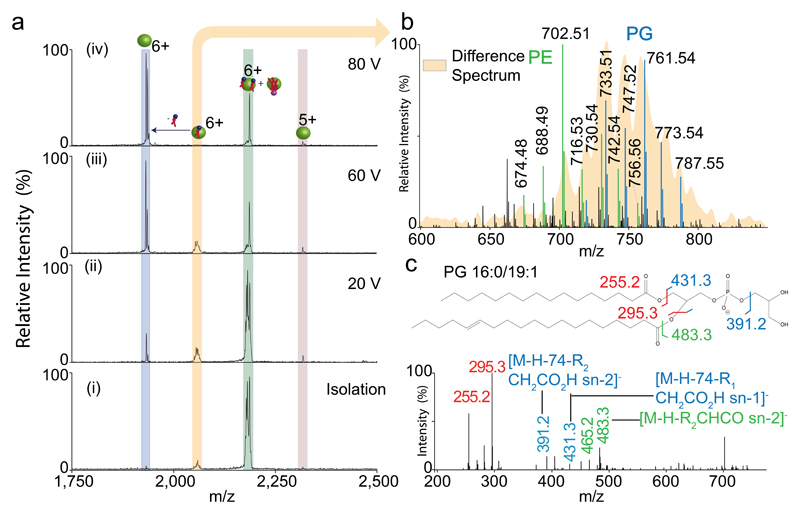
In specific cases, when heterogeneous endogenous lipids are present, differentiation between individual lipid-bound species is often difficult. For example, it may not be possible to differentiate two glycerol phospholipids from one cardiolipin, when bound to protein, due to their similar masses. In these cases selection of the mixed lipid adduct, followed by sequential activation in the collision cell, can be used to separate the different classes of lipids. An example of this is presented in detail in the ANTICIPATED RESULTS section below (Figure 7) where increasing the level of collisional activation allows the 1,300-1,400 Da adduct bound to semiSWEET to be dissociated sequentially, and a mixture of PG, PE and CDL lipids identified.
Figure 7. HE-nMS and in solution lipid identification of semiSWEET.
Comprehensive lipid identification of semiSWEET through the unison of high energy and in-solution based lipid identification. Fragmentation of the (i) isolated 6+ charge state (green) of semiSWEET at increasingly high CID energy (ii)-(iv) leads to generation of a species assigned to a bound PL (yellow) and apo protein (blue). The fine structure and broad nature of the peak assigned to one bound PL is characteristic of the heterogeneity in PL. The identity of these PL species was determined through in-solution lipidomics using the same protein preparation subjected to native MS. The heterogeneity observed in the apo-spectrum corresponds to different Na+ bound species. (b) the spectrum shows the m/z values of the PL species identified through in-solution lipidomics overlaid with the spectrum of the PL-bound protein peak (yellow - generated by subtracting the mass of the apo protein). As seen, the bound lipids correspond primarily to PGs. (c) MS/MS spectra of PG lipids show the presence of peaks characteristic of the head-group. The phospholipids and cardiolipins attached to the protein are represented with blue and purple spheres respectively. The relative sizes of the protein and lipids are not scaled.
The examples presented in Figures 5-7 show how our delipidation strategy, followed by HE-nMS, can be used to identify key lipid binding partners for membrane protein complexes where lipids play important functional roles. As such our protocol can be employed to distinguish lipid classes with distinctly different mass ranges and binding energies, for example between PG and CDL. When combined with lipid profiling it can discriminate between lipid classes that are much closer in mass, such as PG and PE. In cases where further molecular detail is required combining with a lipid extraction step and analysis by LC MS/MS will enable definition of the lipidome directly bound to the membrane protein of interest.
Advantages and Limitations
Lipid extraction followed by LC-MS/MS based analysis has thus far remained the method of choice for identifying the lipidome interacting with proteins of interest. Nevertheless this falls short of identifying the subset of lipids, among all lipids that co-purify with the protein, that bind directly to the protein of interest. This protocol combines direct identification of bound lipids through MS/MS analysis with in solution LC-MS/MS analysis to give a complete view of the entire lipidome that co-purifies with the protein and further identifies the subset of lipids bound directly to the protein. Coupling this protocol with stepwise delipidation, and measuring the structural and functional perturbation delipidation causes to the protein, can further determine the role of the lipid towards maintaining the structural and/or functional integrity of the protein of interest. In our hands, using HE-nMS, we have been successful in liberating membrane protein-lipid complexes from various classes of commonly used detergents. However, it is possible that some detergents may not be readily removed even at very high activation energies. In such cases alternative detergents should be trialled. It must be stated that the target proteins are often expressed in convenient hosts, such as E. coli, rather than the native organisms, which may have different lipid compositions. Hence, before making any biological conclusion on the effects of lipid binding, it is important to note whether the specific, or chemically equivalent lipid, is also present in the native membrane environment. A comparison of the lipid profile of the host organism with that of the native organism will be essential in this regard.
Experimental Design
Here, using specific examples of LeuT, MATE, SemiSWEET and MsbA we illustrate how different strategies can be employed to directly determine bound lipids. We show that while stepwise delipidation followed by LC-MS/MS analysis of the extracted lipid can reveal the lipidome that is present at different stages of delipidation, HE-nMS analysis of the protein-lipid complex determines directly the bound lipids. The unison of these two strategies reveals the complete identity of the bound lipids.
It is important to remember a few points before starting experiments
At various stages of the protocol, affinity purification is used as a method of exchanging the membrane protein from one detergent to another, or to adjust the concentration of the detergent. Hence, the protein of interest should be expressed with the desirable affinity tags to perform these steps. We have used His-tagged protein for this protocol, nevertheless, the protocol is compatible with other affinity tags. Also if the protein cannot be expressed with an affinity tag, size exclusion chromatography can be employed.
Different membrane proteins are stable in a variety of detergent conditions. Hence, before initiating the experiment it is imperative to understand the stability of the protein in the detergents selected.
Check all the connections related to the HE platform before the experiment to ensure it is correctly assembled.
Materials
Reagents
Ammonium acetate (7.5M solution, Sigma, cat. No. A2706)
Ammonium hydroxide (extra pure, 25% (wt/vol) solution in water, Acros organics, cat. No. 10730751)
Anapoe-C12E8 (Affymetrix, cat. no. APO128)
Anapoe-C12E9 (Affymetrix, cat. no. APO129)
Cesium iodide (CsI; 99.999%; Sigma-Aldrich, cat. no. 203033)
Chloroform (ACS spectroscopic grade, >99.8%; Sigma-Aldrich, cat. no. 366919)
C8E4 (Tetraethylene glycol monooctyl ether, Anagrade, ≥99%; Affymetrix, cat. no. T350)
C8E5 (Pentaethylene glycol monooctyl ether, Anagrade, ≥99%; Affymetrix, cat. no. P350)
Decyl maltose neopentyl glycol (Affymetrix, cat. no. NG322)
Dodecyl-β-D-maltopyranoside (DDM, Sol-grade, Anatrace, cat. No. D310S)
Imidazole (>99%; Fisher Scientific/Acros Organics, cat. no. I/0010/53)
Lauryldimethylamine-N-Oxide (LDAO, Anatrace, cat. no. D360)
Methanol (CHROMASOLV, for HPLC, >99.9%; Sigma-Aldrich, cat. no. 34860)
n-Octyl-β-D-glucopyranoside (OG, Sol-Grade, ≥97%; Affymetrix, cat. no. O311S)
n-Nonyl-β-D-glucopyranoside (NG, Sol-Grade, ≥97%; Affymetrix, cat. no. N324S)
Octyl Glucose Neopentyl Glycol (OGNG, Anatrace, cat. No. NG311)
PE 12:0/13:0, LM-1100 (Avanti Polar lipids)
PE 17:0/14:1, LM-1104 (Avanti Polar lipids)
PG 12:0/13:0, LM-1200 (Avanti Polar lipids)
PG 17:0/14:1, LM-1204 (Avanti Polar lipids)
Protein of interest. We demonstrate our Protocol using the following example proteins: MsbA, LeuT, SemiSWEET and MATE (see Reagent Setup).
Sodium chloride (>99.5%; Sigma-Aldrich, cat. no. S7653)
Tris (Trizma base, 99.9%; Sigma-Aldrich, cat. no. T1503)
Trypsin Gold, Mass Spectrometry Grade V5280, Promega
Equipment
Boroscilicate glass capillaries (Harvard Apparatus); 1.0 mm outer diameter (o.d.) × 0.5 mm inner diameter (i.d.) with filament (cat. no. GC100FS-10) and 1.0 mm o.d. × 0.58 mm i.d. (cat. no. GC100-10)
Superdex 200 Increase 5/150 GL (GE Healthcare 28990945)
Acclaim PepMap 100, C18, 75 μm × 15 cm, Thermo Fisher Scientific
Bio-Rad Micro Bio-Spin chromatography columns (Empty; Bio-Rad, cat. no. 732-6204)
Buffer-exchange devices, Micro Bio-Spin 6 chromatography columns, MW exclusion limit 6 kDa (Tris buffer, Bio-Rad, cat. no. 732-6221)
Centrifugal concentrators (MWCO 50/100 kDa, Amicon Ultra-15 Centrifugal Filter Unit with Ultracel-30 K membrane, Millipore, cat. No. UFC906096; NMWL Amicon Ultra-0.5 Centrifugal Filter Unit with Ultracel-50 K, ultracel-100 K membranes)
Centrifuge tubes with screw caps (Ultra High Performance 15 mL, VWR, cat. No. 21008-214 and Ultra High Performance 50 mL, VWR, 21008-240)
AKTA Purifier (GE Healthcare)
HisTrap™ HP column (1 mL, GE Healthcare, cat. No. 17-5247-01)
SpeedVac
Sonicator
UHPLC: We have used Dionex-3000 UHPLC for performing the LC-MS/MS analysis of the lipids.
Mass Spectrometer: We have used Synapt G1(Waters) mass spectrometer for High-energy nMS and LTQ Orbitrap XL™ Hybrid Ion Trap-Orbitrap Mass Spectrometer (Thermo Fisher Scientific corp.) for lipidomics
MassLynx software™ version 4.1 (Waters corp.) was used to analyse MS data from Waters instrument and Xcalibur™ (Thermo Fisher Scientific corp.) was used to analysed the lipidomics data LC-MS/MS data. These were obtained along with the instrument, or can be purchased as stand-alone from the respective vendors.
Reagent setup
Membrane protein expression and purification
Depending on the target membrane protein complex we suggest the following previously published protocols for expression and purification strategies. 7, 26 We used these strategies for the purification of the following proteins as examples for the Procedures described in this Protocol: MsbA, LeuT, SemiSWEET and MATE.7, 26 The plasmids of these proteins are also available upon request.
CsI calibration solution
CsI calibration solution contains 100 mg ml–1 CsI dissolved in water; the solution can be stored at room temperature (23 °C) for up to 3–4 weeks. It is advisable to perform instrument calibration before the performing the native MS experiments to ensure data calibration.
Detergents
Stocks of detergent are usually made up at 10% (wt/vol) (except C8E4 which is available as 50% (wt/vol) stock) in water according to variable detergent solubility and stored at -20°C for up to six months.
Immobilised-metal affinity chromatography (IMAC) delipidation buffer
Mix 20 mM Tris, 50 mM sodium chloride, 50 mM imidazole and detergents of choice, and adjust the pH to 8.0 with hydrochloric acid at room temperature, store at 4 °C for up to a month.
IMAC elution buffer
Mix 20 mM Tris, 50 mM NaCl, 500mM imidazole and 2X CMC detergent (depending on protein), and adjust the pH to 8.0 with hydrochloric acid at room temperature, store at 4 °C for up to a month.
Imidazole stock solution (pH 8)
Stock solution is made up 0.5 M imidazole dissolved in water, and the pH is adjusted to 8 with hydrochloric acid at room temperature, store at 4 °C up to a month.
SEC buffer
Mix 20 mM Tris, 50 mM sodium chloride and 2X CMC (depending on protein), and adjust the pH to 8.0 with hydrochloric acid at room temperature, store at 4 °C up to a month.
Membrane MS buffer
Mix 200 mM ammonium acetate and 2× CMC detergent of choice, and adjust the pH to 7.2 or 8.0 with ammonium hydroxide. See note below regarding buffers containing detergents. This can be stored at 4 °C up to a month.
Lipidomics LC/MS Buffer A
ACN:H2O 60:40, 10 mM ammonium formate, and 0.1% (vol/vol) formic acid. This can be stored at 4 °C up to a month.
Lipidomics LC/MS Buffer B
IPA:ACN 90:10, 10 mM ammonium formate, and 0.1% (vol/vol) formic acid. This can be stored at 4 °C up to a month.
Equipment setup
Nano-electrospray capillaries
These were prepared in-house from borosilicate capillaries as previously described. 27
Mass spectrometer
In this study, we used a Synapt G2 HDMS instrument (Waters corp.) with a Z-spray source including modifications previously described19. In brief, modifications include a valve to adjust the source pressure and a modified radio frequency (RF) generator enabling quadrupole selection of ions up to m/z of 32,000. To allow access to higher voltages on the extraction cone in the source region of the Synapt G2, the configuration file entry for the extraction cone was modified to increase the maximum voltage setting from 10 V to 200 V. Altering this setting alone would restrict the maximum sample cone voltage that could be accessed, owing to the limits imposed by the power supplies. To overcome this limitation, the sample cone voltage was supplied from an external powered supply (Spellman’s Bertan 230 Series benchtop high voltage power supply). This was accomplished by introducing a patch cable between the instrument lens control PCB and the source ion block. Similar advancements can also be achieved in the next generation Waters instruments, such G2Si, without any additional changes and maintaining equivalent values for other parameters, such as source pressure. Prior to introducing these modifications it is recommended that users contact the instrument manufacturer for advice.
If applying up to 0-10V at the extractor cone and 0-200V at the sample cone the internal power supplies are sufficient. The acceleration voltage in the source region is calculated from V1+V2 and is read directly from the software interface. When the sample cone is driven by the external power supply the acceleration in the source region is calculated as V3-VA (see Figure 4a). As the acceleration occurs in two stages, first between the sample and extractor cones, then between the extractor and transfer optic, it is important to record both cone voltages.
The instrument parameters, such as Capillary Voltage, Source Pressure, Trap Pressure, Bias, Wave Height and Velocities in trap and transfer were tuned to maximise transmission for the protein complex of interest. 20
LC-MS/MS for lipidomics
In this study, we used a Dionex UltiMate 3000 UHPLC coupled with LTQ Orbitrap XL (Thermo Scientific) via a dynamic nanospray source. The total lipid extract was loaded firstly into a 5 µL sample loop using auto-sampler by full-loop method with an overfill factor of 1.4, then transferred to a 1 µL injection loop using a loading-pump at a flow rate of 5 µL/min 30% Solution B with an overfill factor of 3. The lipids in the injection loop were injected to a C18 column (Acclaim PepMap 100, C18, 75 μm × 15 cm, Thermo Fisher Scientific) using a nano-pump at a flow rate of 300 nL/min, 30% Solution B. The lipids are separated on the C18 column at 40 °C by a gradient starting from 30% LC buffer B. After 10 min buffer B was ramped to 65% over 1 min, then 80% over 6 min, before being held at 80% for 10 min, then ramped to 99% over 6 min and held for 7 min (adapted from Reference 28). Typical MS conditions were a spray voltage of 1.8 kV and capillary temperature of 175 °C in negative ion mode. MS was operated in a data-dependent acquisition mode with one MS scan in the Orbitrap (m/z 350-2000, resolution of 60000) followed with five MS/MS scans for the top 5 most intense ions in the linear ion trap (a normalized collision energy of 38% at an activation of q=0.25 and an activation time of 30 ms).15,16
Data analysis
Data acquired from mass spectrometry experiments was processed with MassLynx software™ version 4.1 (Waters corp.). Quantitative lipidomics data were processed as described previously.15 For generating the difference spectrum in Figure 7b, the native MS should first be transformed. This can be achieved readily by copying the raw spectral data into an appropriate data processing package, such as Microsoft Excel, and transforming the x axis of the native spectrum by subtracting the mass (or m/z for non-deconvoluted spectra) of the apo protein along with the correct number of protons. This yields a difference spectrum which can be displayed behind a summed LC/MS profile for direct comparison. An example of this is shown in (Fig 7b) where the distributions of PE and PG identified from lipidomics can now be matched to adducts on the native MS spectra of semiSWEET.
Procedure
Identifying cases where lipids may play a critical role in maintaining oligomeric state
TIMING (Step 1: 30-60 min, Step 2: 4-12 h, Step 3: 1-2 h, Step 4: 30-60 min)
CRITICAL Steps 1-4 can be performed on any purified membrane protein-lipid complex to establish whether a cohort of bound lipids may be important for maintaining the oligomeric state of the membrane protein.
1| Native MS of membrane protein-lipid complex: Establish native MS conditions to obtain the best possible mass spectrum for the membrane protein lipid complex. Please refer to the Equipment Setup and our previously published protocol (Ref. 20) for details.
Δ CRITICAL STEP: Proteins should be expressed in suitable vectors and expression systems. This protocol is designed for proteins that have been modified with a cleavable histidine tag but can be readily adapted for proteins with other affinity tags or separation techniques used for the protein of interest.
Δ CRITICAL STEP: It is important to screen suitable detergent conditions that stabilize a specific membrane protein as well as yield a good native spectrum see our previously published protocol. 20
2| Exchange of membrane protein-lipid complexes into delipidating detergents: Start with a 50 µL aliquot of the purified membrane protein-lipid complex of interest, with a protein concentration ranging from 2-10 µM. To this sample, add an excess of the desired delipidating detergent powder (C8E4 (liquid stock) (or other higher chain polyol classes of detrgents), LDAO powder, OGNG, NG or Cymal 5) to a final concentration of 2-20 x CMC. Keep the sample at 4°C for 4-12 hours. Considering the efficiency of removing lipids and ease of performing nMS experiments, we generally recommend following the order, as described above. We also recommend for each detergent start from a low concentration, such as 2 xCMC, for 4 hours. Subsequently, depending on the results wither increase the time or amount or both.
Δ CRITICAL STEP The process of delipidation occurs either because the detergent has higher affinity for the lipid than the protein or by mass-action due to the presence of large excess of the detergent. Hence, the time and the concentration of detergent required for delipidation can vary depending on the protein and detergent. Nevertheless, the concentration should always be more than 2 x CMC.
3| Removal of excess delipidating detergent by size exclusion chromatography: Filter the delipidated membrane protein-lipid complex using a 0.22 µm spin filter. Inject the filtered sample onto a 3 mL SEC column (Superdex 200 10/150) pre-equilibrated in with SEC buffer containing 2 × CMC of the chosen delipidating detergent. Elute with 3 mL SEC buffer and monitor the sample elution at 280nm. Collect the protein-containing fractions, pool them together and concentrate to 2-10 µM using a spin concentrator with an appropriate MWCO (molecular weight cut off) at 4 °C, following the speed recommended for the volume and rotor type.
Δ CRITICAL STEP Care should be taken not to concentrate the detergent while concentrating the protein. Choose a spin concentrator with MWCO higher than the detergent micelle size.
TROUBLESHOOTING
4| Verification of oligomerisation state after exchange into delipidating detergent: Acquire a native mass spectrum of the delipidated protein using the optimised conditions determined in Step 1. If delipidation leads to de-oligomerisation, as observed by the recorded mass, then the protein system is a likely candidate where lipids may mediate oligomerisation (see Figure 2 for an example). In cases where delipidation does not lead to any change in the measured mass of the complex, compared to that recorded prior to delipidation (Step 1), then exchange to another delipidating detergent should be performed (see Step 2). If delipidation leads to detection of expected mass of the bare oligomer, then oligomerisation for that system is not lipid dependent.
5| Identifying interacting lipids: We propose using a bi-pronged strategy proposed below to identify lipids that interact with the membrane protein of interest. Follow the progressive delipidation strategy (Path A) to gradually remove the lipid shell co-purified with the protein. At each stage LC MS/MS analysis is performed to reveal the identity, class and chain composition of the entire set of lipids that co-purify with the protein of interest. Simultaneously, at each stage follow Path B to perofmr HE-nMS analysis to directly identify the masses of the bound lipids. Now by combining both sets of data, we can determine the complete identity of subsets of co-purified lipids that directly interact with the protein of interest.
Path A: Progressive delipidation, TIMING 3 hours
i| To screen a range of detergents equilibrate a 1mL IMAC column (HisTrap™ HP column), coupled to an GE AKTA Purifier ™ instrument operated at 4 °C, with 25 mL of the desired IMAC delipidation buffer (see Reagent Setup) containing the detergent of choice, depending on the results of Step 1 -4. We suggest initiating with 2xCMC of detergent, in the order described in Step 2.
Δ CRITICAL STEP In case the protein of interest cannot be tagged with poly-His, other affinity purification techniques may be followed and SEC maybe used to replace IMAC. IMAC was used here due to the time scale and ease of the method.
Δ CRITICAL STEP At no point should the detergent concentration fall below 2× CMC of the detergent.
ii| Add an imidazole stock solution (pH 8) to the 2-10 µM protein sample to a final imidazole concentration of 0.05 M and filter the sample using a 0.22 µm spin filter. This will reduce nonspecific binding to Ni resin during loading.
iii| Divide the protein sample into aliquots of 100 µL each (the number depends on the number of conditions to be screened) and load one aliquot onto this pre-equilibrated IMAC column. Wash with IMAC delipidation buffer (containing the same detergent) at 2.5 mL/min for 1 h. Elute using 10 mL IMAC elution buffer and collect the eluate, monitor absorption at 280 nm and pool the protein-containing fractions.
iv|(optional step) If cleavage of the affinity tag is required, perform cleavage in an appropriate buffer containing the same detergent composition as used for IMAC or for SEC.
v| Concentrate the IMAC eluate with a spin concentrator, with an appropriate MWCO, at 4 °C down to approximately 100 µL. Filter the concentrated eluate with 0.22 μm filter
ΔCRITICAL STEP Care should be taken not to concentrate the detergent while concentrating the protein. Choose a spin concentrator with MWCO higher than the detergent micelle size.
vi| Inject 50 µL of the delipidated membrane protein complex onto a size-exclusion column (Superdex-200 5/150, 3 mL) that has been pre-equilibrated in 10 mL SEC buffer (with the same amount of the detergent used in Step 5A(i)). Elute with 3 mL SEC buffer and monitor the elution profile at 280 nm. Collect fractions containing delipidated membrane proteins and concentrate to 2-10 µM using a spin concentrator with an appropriate MWCO (molecular weight cut off).
□ PAUSE POINT Often samples can be stable up to 24 h at 4°C following SEC. However, if MS data is not acquired immediately, it is strongly suggested that the sample buffer must contain 10% (vol/vol) glycerol and the sample can be snap frozen using liquid nitrogen and kept at -80 °C for longer storage.
vii| Perform native MS analysis on the delipidated sample (see Step 1 for how to perform native MS of membrane proteins) or subject the protein preparation to lipid extraction followed LC-MS/MS analysis (BOX 1) for in-solution lipid identification.
TROUBLESHOOTING
viii| Repeat Steps i-vii with the next aliquot, with a different detergent content. We suggest to increase the detergent concentration from 2× CMC to upto 10× CMC before progressing to more delipidating detergents. This will yield a set of fractions with varying degrees of delipidation.
Path B: Identifying Lipids Bound Directly to Membrane Protein Complexes
TIMING 2 hours
i| Preparing the samples and instrument for native MS: Using spin columns, or SEC (see Step 5A(vi) for details), buffer exchange the protein sample (50 μL at a concentration approximately 2-10 μM) into membrane MS buffer containing 2× CMC of the desired detergent, as found to be optimal in Step 4, either in terms of keeping the oligomeric state with bond lipids (for lipid dependent oligomers), or maintaining endogenous lipids (for cases where the goal is to identify bound endogenous lipids). Optimise the instrument conditions for transmission of the protein-lipid complex by keeping the extractor cone voltage low (<10 V) and increasing the collision energy (trap or transfer voltages, from 10-200 eV) in the trap to release the protein from the detergent micelle. For further details on how to obtain native mass spectra of membrane proteins see Step 1.
ii| Removing detergent in the source region of the mass spectrometer: Once a well resolved mass spectrum has been obtained using standard conditions (activation and micelle removal in the trap) then decrease the trap collision energy (to approximately 10V) and increase the sample and extractor cone voltages (from 10-250 V) and fine tune these parameters to yield sufficient activation of the proteo-micelle complex at the source region. Depending on the systems, comprising of both protein and the detergent micelle, while certain proteins prefer higher ejection energies from the extractor, some are improved by a higher potential difference between the sample and the extractor cone.
Δ CRITICAL STEP: Remember that the minimum sample cone voltage should be VA+V1 when using the HE-nMS platform (See Figure 4a).
Δ CRITICAL STEP: Beware that an excessively high sample cone voltage may eventually lead to denaturation of the protein in the source.
Δ CRITICAL STEP: It is to be noted that the collision energy used to dissociate micelle may also result in loss of bound lipids. Various detergent micelles require different energies (for detail listing see Ref 20), hence the residual bound lipids may differ. Hence, before inferring the structural significance of any bound lipid, their presence in different micelles should be checked.
CAUTION!: Sample cone voltages of more than approximately 350 V may lead to breakdown in the source and electrical discharge inside the mass spectrometer. Similarly, a large potential difference between the sample and the extractor cone can also lead to breakdown between the cones. Hence, raising the sample cone to very high voltages should be accompanied by increasing the extractor cone voltage. Also, using an external power supply to drive the sample cone circumvents the instrument safety interlocks. Care must be taken while dealing with high voltages.
TROUBLESHOOTING
iii| Isolation of lipid bound species (Steps iii-iv): Identify peaks in the native MS corresponding to lipid bound adducts by comparison with the expected mass of the protein complex and note their m/z values (see Figure 5a for an example).
iv| Switch the instrument into MS/MS mode and isolate the m/z of interest in the quadrupole, corresponding to the lipid bound species. Start with high m/z ranges for the isolation and then iteratively narrow the window whilst monitoring the total ion current for the isolated species. Attempt to achieve the narrowest isolation window possible such that it selects the species of interest with workable ion intensity.
Δ CRITICAL STEP Ensure the trap voltage is low (1-10 V) during these experiments to limit complex dissociation.
TROUBLESHOOTING
v| Dissociation of lipid bound species (Steps v-vi): Once a suitable lipid bound species has been isolated, gradually increase the trap collision energy recording spectra at 5-10 V steps. Monitor the spectrum for the appearance of new peaks or a decrease in intensity of the parent ion.
vi| Keep increasing the trap energy until product ions are present with satisfactory intensity, along with the parent ion (see Figure 5a for an example). At this point fine-tune the gas pressure in the trap to maximise ion intensity in the MS/MS spectra.
Δ CRITICAL STEP: Quickly scanning through a large range of trap energies and monitoring the spectrum can help in quickly finding the energy ranges at which important dissociation events occur. Voltage should then be systematically ramped in small increments focusing on these ranges.
TROUBLESHOOTING
vii| Identifying the classes of bound phospholipids (Steps vii-ix) To determine the classes of bound lipids directly from native MS/MS spectra perform HE-nMS/MS analysis as directed in Box 2.
viii| In cases where classes of lipids could not be determined by mass measurement alone, separate lipids in solution. Perform LC-MS/MS analysis on the extracted lipids, from the proteo-micelle solution that was subjected to HE-nMS/MS analysis, and identify the lipids based on their fragmentation pattern (as explained in BOX-1). This will determine the entire set of lipid masses and their classification.
ix| Once both lipidomics data and native mass spectra have been obtained the two can be compared. This can aid in identification of the lipid adducts observed in the native MS, especially when baseline resolution of the protein-lipid complex cannot be achieved. To allow direct comparison of this data the native MS should first be transformed. This can be achieved simply by transforming the x axis of the native spectrum by subtracting the mass (or m/z for non-deconvoluted spectra) of the apo protein along with the correct number of protons. This yields a difference spectrum which can overlaid with a summed LC/MS profile. An example of this is shown in (Fig 7b) where the distributions of PE and PG identified from lipidomics can now be matched to the adducts on the native MS spectra of semiSWEET.
TROUBLESHOOTING
Troubleshooting advice can be found in Table 1
Table 1. Troubleshooting Table.
| Step | Problem | Possible reason | Possible solution |
|---|---|---|---|
| 3 | The protein is unstable, as detected by monitoring the SEC profile or observing precipitates. | Certain proteins are stable only in specific detergents. | Screen different detergent to determine a list of detergent for the protein of interest to achieve the correct balance between stability and delipidation. |
| 5Avii | The used detergents are incompatible with Native MS. This often results into use of excessive collision energy to ablate the micelle. Beside the presence of highly charged species, ion-mobility based unfolding analysis can reveal the denatured state of the protein. 31 | Often certain membrane proteins, in spite of being stable in certain detergents, do not yield good spectra under the nMS conditions. This is largely due to the stronger affinity of the specific protein for the detergent and/or large micelle size. | In these cases charge reducing agents (such as imidazole) can be added to the nMS electrospray buffer 32 or a different MS ion polarity can be used in an effort to stabilise the membrane protein complex.26 If this is not successful an additional screening process should be performed in an appropriate detergent for native MS, following our previous protocol. 20 |
| 5Bii | The HE platform is inefficient in removing the detergent micelle | It is feasible that the additional high energy available is still not sufficient to ablate the detergent micelle. | In our hands, using the high-energy activation method, we have been successful in liberating membrane protein-lipid complexes from various classes of commonly used detergents. However it is possible that some detergents may not be able to be removed even at very high activation energies. In such cases alternative detergents should be trialled. |
| 5Biv | Lipid bound peak of interest could not be isolated for fragmentation | Due to presence of multiple peaks within close proximity at the m/z scale exclusive isolation of a single peak of interest may become difficult. | Check if the isolated spectrum contains only the lipid bound species, if more species are present reduce the quadrupole isolation window. A balance must be sought while selecting the window range. A large isolation window increases the intensity of the isolated peak but at the same time may include the non-lipid bound state or higher order lipid bound states. |
| 5Bvi | Absence of signal at the lower m/z | Since the generated fragment ions of lipids or lower order oligomers are of lower m/z values than the parent ions, the instrumental condition set for the isolation of the parent ion may not optimum. | a. The trap pressure may need adjustment to ensure optimal transmission and detection of lower mass species. b. When increasing the trap energy to dissociate isolated protein-lipid complexes care must be taken to avoid increasing the energy so as to induce further dissociation and/or fragmentation of the product ions. This can lead to complex spectra that are difficult to interpret |
| 5Bvi | The parent ion is recalcitrant to fragmentation | Due to greater Coulombic repulsion and the availability of larger numbers of mobile protons, higher charge states typically yield richer fragmentation spectra following CID. Nevertheless, a balance must be sought while choosing between the highest charge state and the parent ion intensity. | We suggest that experiments first be performed on the charge state with maximum intensity and if the resulting fragmentation pattern is not satisfactory, perform the isolation and fragmentation experiments on progressively higher charge states. |
TIMING
Step 1 -Native MS of membrane protein-lipid complex: 30-60 minutes
Step 2 - Exchange of membrane protein-lipid complexes into delipidating detergents: 4-12 hours
Step 3 -Removal of excess delipidating detergent by size exclusion chromatography: 60-120 minutes
Step 4 -Verification of oligomerisation state after exchange into delipidating detergent : 30-60 minutes
Step 5A - Progressive delipidation: 2 hours
Step 5B - Identifying Lipids Bound Directly to Membrane Protein Complexes: 3 hours
BOX 1: 2 Days
Anticipated Results
We subjected three proteins, LeuT, SemiSWEET and MATE to tandem MS experiments using the HE-nMS/MS platform described here. The protocol described shows that we can determine the entire cohort of lipids that co-purify with the protein of interest and further identify the subset of lipids that directly bind to the protein of interest (Figures 2 – 6). For all of these proteins an intact protein -lipid complex could be liberated from the detergent micelle using the HE-nMS platform in the source region of the mass spectrometer. So far we have been successful in ablating all the membrane protein we have used thus far from their detergent cargoes. Moreover these examples highlight the various approaches that can be taken to identify bound lipids.
In the case of MATE, upon collisional activation, the lipid-bound peak readily yields the 10+ charge state of the naked protein (Figure 5a). The mass difference between this and the isolated peak corresponds to the mass of the bound lipid, providing a direct means of determining the mass of the bound lipids (Figure 5b). With increase in collision energy, the intensity of the isolated peak decreases yielding a higher abundance of the daughter ions. The spectra also reveal the presence of the apo 9+ charge state of the protein, plausibly due to loss of positively charged POPG. This presents an example where by inspecting the neutral losses yields the masses of the bound lipids.
Often intact mass measurements of protein-lipid complexes cannot distinguish between different lipid classes, semiSWEET presents once such example. The MS/MS of the lipid-bound charge state of semiSWEET leads to generation of two sets of peaks, formed by neutral losses, assigned to one lipid-bound and apo-protein. (Figures 4d and 7a). This ‘non-sequential’ loss is indicative of the binding of two lipid species corresponding to a protein with both two PLs and one CDL bound. Furthermore, fine structure observed in the peak assigned to binding of one PL, suggests that various chain lengths are present. In-solution LC-MS/MS identification of the lipids extracted from this sample, subjected to native MS, deconvolutes the heterogeneity and can be directly compared to the native mass spectrum (Figure 7b). A superimposition of the lipid masses corresponding to PLs obtained through native MS and our in-solution approach indicates the identity of the bound lipids to be largely PG. The identities of the lipids were obtained through characteristic fragment ions (Figure 7c). In this case a combined native MS and identification of lipids in solution reveals the cohort of lipids bound to SemiSWEET.
Alternatively, MS/MS of a lipid bound oligomer can lead to generation of lower order oligomers. For example the CID MS/MS spectra of dimeric LeuT with a 7.4k Da lipid mass (Figure 4c and 6). The lipid-bound dimer readily yields monomer, with satellite peaks corresponding to lipid bound states. Since all of these species are generated from the precursor, lipids that remain bound must be part of the cohort comprising total lipid mass of 7.4 kDa. Analysis of these peaks leads to the identification of three PLs with masses 730 (±32), 747 (±46) and 759 (±51) Da. The mass addition for the second satellite peak also corresponds to that of CDL (mass 1,457 (±46) Da). Whether this peak corresponds to binding combinations of PL, or a mixture of PL and CDL, can be further elucidated by performing a collision energy ramp. With increase in collision energy only the second satellite peak persists. Had the second peak been assigned to a PL-bound species, increase in collision energy would have resulted in a sequential disappearance of the three satellite peaks. Three PL and one CDL correspond to an additional mass of 3.7kDa bound to each subunit, constituting the 7.4kDa lipids addition to the dimer.
In summary we have illustrated how using the high-energy MS platform described here, in conjunction with in-solution lipidomics, we can deconvolute the lipodome that interacts directly with the membrane protein of interest. For specific cases, such as LeuT, lipids may be essential for maintaining the oligomeric state. Alternatively binding of lipids may be promiscuous, or without obvious functional significance. The protocol described here can be applied in all of these scenarios.
Editorial Summary.
This protocol describes a native mass spectrometry-based approach for identifying the key lipids that interact with membrane proteins.
Acknowledgments
The Robinson group is funded by a Wellcome Trust Investigator Award (104633/Z/14/Z), an ERC Advanced Grant ENABLE (695511) and an MRC programme grant (MR/N020413/1). K.G. is a Junior Research Fellow at St Catherine’s College, Oxford and supported by the Royal Commission for the Exhibition of 1851. J.G. is a Junior Research Fellow of The Queen’s College. Joseph Donlan, Waters Corporation and OMass Technologies are thanked for their support.
Footnotes
Please indicate up to four primary research articles where the protocol has been used and/or developed.
1. The role of interfacial lipids in stabilizing membrane protein oligomers. Gupta K, Donlan JAC, Hopper JTS, Uzdavinys P, Landreh M, Struwe WB, Drew D, Baldwin AJ, Stansfeld PJ, Robinson CV. Nature. 2017 Jan 19;541(7637):421-424.
2. Different modes of lipid binding to membrane proteins probed by mass spectrometry. Bechara C, Robinson CV. J Am Chem Soc. 2015 Apr 29;137(16):5240-7
3. A subset of annular lipids is linked to the flippase activity of an ABC transporter. Bechara C, Nöll A, Morgner N, Degiacomi MT, Tampé R, Robinson CV. Nat Chem. 2015 Mar;7(3):255-62
Author contributions
K.G. expressed and purified LeuT and SemiSWEET and performed the high energy experiments with the help of J.T.S.H. C.B. developed the successive delipidation protocol and J.L. expressed and purified MsbA and performed the successive delipidation experiment. C.B. and J.G. performed the lipidomics experiments and analysed the data. D.W. set up the lipidomics platform. I.L. expressed, purified and performed the MS experiments on MATE. J.L.P.B., J.T.S.H and K.G. (Kevin Giles) designed the high energy source. K.G. and C.V.R. wrote the manuscript with assistance from J.G. and I.L. and inputs from all the authors.
Competing financial interests
The authors declare that they have no competing financial interests.
References
- 1.Konijnenberg A, et al. Global structural changes of an ion channel during its gating are followed by ion mobility mass spectrometry. Proc Natl Acad Sci U S A. 2014;111:17170–17175. doi: 10.1073/pnas.1413118111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiffrin B, et al. Skp is a multivalent chaperone of outer-membrane proteins. Nat Comm. 2016;23:786–793. doi: 10.1038/nsmb.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvey SR, Liu Y, Liu W, Wysocki VH, Laganowsky A. Surface induced dissociation as a tool to study membrane protein complexes. Chem Comm. 2017;53:3106–3109. doi: 10.1039/c6cc09606a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, et al. Native Mass Spectrometry Characterizes the Photosynthetic Reaction Center Complex from the Purple Bacterium Rhodobacter sphaeroides. J Am Soc Mass Spectrom. 2017;28:87–95. doi: 10.1007/s13361-016-1451-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iadanza MG, et al. Lateral opening in the intact beta-barrel assembly machinery captured by cryo-EM. Nat Comm. 2016;7:12865. doi: 10.1038/ncomms12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landreh M, Marklund EG, Uzdavinys P, Degiacomi MT. Integrating mass spectrometry with MD simulations reveals the role of lipids in Na+/H+ antiporters. Nat Comm. 2017;8:13993. doi: 10.1038/ncomms13993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta K, et al. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017;541:421–424. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hedger G, Sansom MS. Lipid interaction sites on channels, transporters and receptors: Recent insights from molecular dynamics simulations. Biochimica et biophysica acta. 2016;1858:2390–2400. doi: 10.1016/j.bbamem.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landreh M, Marty MT, Gault J, Robinson CV. A sliding selectivity scale for lipid binding to membrane proteins. Curr Opin Struct Biol. 2016;39:54–60. doi: 10.1016/j.sbi.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norimatsu Y, Hasegawa K, Shimizu N, Toyoshima C. Protein-phospholipid interplay revealed with crystals of a calcium pump. Nature. 2017;545:193–198. doi: 10.1038/nature22357. [DOI] [PubMed] [Google Scholar]
- 11.Le Roy A, et al. AUC and Small-Angle Scattering for Membrane Proteins. Meth in Enz. 2015;562:257–286. doi: 10.1016/bs.mie.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Maekawa S, Kobayashi Y, Morita M, Suzaki T. Tight binding of NAP-22 with acidic membrane lipids. Neuroscience Lets. 2015;600:244–248. doi: 10.1016/j.neulet.2015.06.025. [DOI] [PubMed] [Google Scholar]
- 13.Deme JC, et al. Purification and interaction analyses of two human lysosomal vitamin B12 transporters: LMBD1 and ABCD4. Mol Membrane Biol. 2014;31:250–261. doi: 10.3109/09687688.2014.990998. [DOI] [PubMed] [Google Scholar]
- 14.Zhou M, et al. Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science. 2011;334:380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bechara C, et al. A subset of annular lipids is linked to the flippase activity of an ABC transporter. Nature Chem. 2015;7:255–262. doi: 10.1038/nchem.2172. [DOI] [PubMed] [Google Scholar]
- 16.Gault J, et al. High-resolution mass spectrometry of small molecules bound to membrane proteins. Nature Methods. 2016 doi: 10.1038/nmeth.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcoux J, et al. Mass spectrometry reveals synergistic effects of nucleotides, lipids, and drugs binding to a multidrug resistance efflux pump. Proc Natl Acad Sci U S A. 2013;110:9704–9709. doi: 10.1073/pnas.1303888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liko I, et al. Dimer interface of bovine cytochrome c oxidase is influenced by local posttranslational modifications and lipid binding. Proc Natl Acad Sci U S A. 2016;113:8230–8235. doi: 10.1073/pnas.1600354113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehmood S, et al. Mass spectrometry captures off-target drug binding and provides mechanistic insights into the human metalloprotease ZMPSTE24. Nat Chem. 2016;8:1152–1158. doi: 10.1038/nchem.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laganowsky A, Reading E, Hopper JT, Robinson CV. Mass spectrometry of intact membrane protein complexes. Nat Protoc. 2013;8:639–651. doi: 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun J, Kitova EN, Sun N, Klassen JS. Method for identifying nonspecific protein-protein interactions in nanoelectrospray ionization mass spectrometry. Anal Chem. 2007;79:8301–8311. doi: 10.1021/ac0709347. [DOI] [PubMed] [Google Scholar]
- 22.Ilgu H, et al. Variation of the detergent-binding capacity and phospholipid content of membrane proteins when purified in different detergents. Biophys J. 2014;106:1660–1670. doi: 10.1016/j.bpj.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J, Kitova EN, Klassen JS. Method for stabilizing protein-ligand complexes in nanoelectrospray ionization mass spectrometry. Anal Chem. 2007;79:416–425. doi: 10.1021/ac061109d. [DOI] [PubMed] [Google Scholar]
- 24.Campuzano ID, et al. Native MS Analysis of Bacteriorhodopsin and an Empty Nanodisc by Orthogonal Acceleration Time-of-Flight, Orbitrap and Ion Cyclotron Resonance. Anall Chem. 2016;88:12427–12436. doi: 10.1021/acs.analchem.6b03762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles protect membrane complexes from solution to vacuum. Science. 2008;321:243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 26.Liko I, Hopper JT, Allison TM, Benesch JL, Robinson CV. Negative Ions Enhance Survival of Membrane Protein Complexes. J Am Soc Mass Spectrom. 2016;27:1099–1104. doi: 10.1007/s13361-016-1381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 28.Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Lipidomics profiling by high-resolution LC-MS and high-energy collisional dissociation fragmentation: focus on characterization of mitochondrial cardiolipins and monolysocardiolipins. Analytical chemistry. 2011;83:940–949. doi: 10.1021/ac102598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.http://www.lipidmaps.org/data/structure/index.php.
- 30.Tsugawa H, et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature Methods. 2015;12:523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allison TM, et al. Quantifying the stabilizing effects of protein-ligand interactions in the gas phase. Nat Commun. 2015;6 doi: 10.1038/ncomms9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehmood S, et al. Charge reduction stabilizes intact membrane protein complexes for mass spectrometry. J Am Chem Soc. 2014;136:17010–17012. doi: 10.1021/ja510283g. [DOI] [PMC free article] [PubMed] [Google Scholar]