Abstract
The budding yeast Saccharomyces cerevisiae reacts to increased external osmolarity by several cellular responses. Adaptive signaling largely relies on the high osmolarity glycerol (HOG) pathway, which is closely related to the mammalian p38 mitogen-activated protein kinase (MAPK) pathway in core architecture. To identify target proteins of the MAP kinase Hog1, we designed a mass spectrometry–based high-throughput analysis measuring the impact of Hog1 activation or inhibition on the S. cerevisiae phosphoproteome. In addition, we analyzed how deletion of RCK2, which encodes a known effector protein kinase target of Hog1, modulated stress-induced phosphorylation. Our results provide an overview of the diversity of cellular functions that are directly and indirectly affected by the activity of the HOG pathway and allow a clear assessment of Hog1-independent events during osmotic stress conditions. We extended the number of putative Hog1 direct targets by analyzing the modulation of motifs consisting of serine or threonine followed by a proline (S/T-P motif) and subsequently validated these by an in vivo interaction assay. Rck2 appears to act as a central hub for many Hog1-mediated secondary phosphorylation events. This study clarifies, to a large extent, speculations on the direct and indirect effects of HOG signaling and its stress-adaptive functions.
Introduction
Adaptive responses to fluctuations in extracellular parameters are generally controlled by complex signal transduction systems that transmit information on environmental cues to various effector molecules. These regulatory systems often constitute highly intertwined kinase and phosphatase networks rather than one single well-defined signal transduction pathway. To add further complexity, individual kinases and phosphatases can show different response kinetics depending on the stimulus, resulting in primary and complementary responses. Several high-throughput mass spectrometry (MS) shotgun studies have been undertaken to globally record cellular responses with the aim of determining the individual contribution of kinases to a given phosphoproteomic state (1–6). However, the collective frequency of phosphorylation and dephosphorylation events hampers the identification of specific kinase-substrate interactions (1, 7).
The hyperosmotic stress response of the budding yeast Saccharomyces cerevisiae is a paradigm for such responses and has been well-characterized using mRNA microarrays as well as MS-based approaches (4, 6, 8–10). Upon exposure to high osmolarity yeast cells experience rapid water loss and shrinkage. Reprogramming of gene expression patterns, a temporary cell-cycle arrest, and ultimately an increase in the intracellular concentration of the compatible osmolyte glycerol are the cornerstones of this response (11). Additionally, osmostress effects on glycolysis and cytoskeletal and mitotic spindle dynamics have been proposed (4, 6).
One of the main signaling cascades involved in the osmostress response is the high osmolarity glycerol (HOG) mitogen-activated protein kinase (MAPK) pathway (12–14), which is highly conserved across the fungal kingdom and homologous to the mammalian p38 stress-activated protein kinase (SAPK) pathway. Its central module essentially consists of the MAPK Hog1, the MAPK kinase (MAPKK) Pbs2, and the three MAPKK kinases (MAPKKK) Ssk2, Ssk22, and Ste11. Upon activation by extracellular hyperosmolarity, the MAPK undergoes dual phosphorylation at residues Thr174 and Tyr176. This dual phosphorylation correlates with Hog1 kinase activity, which peaks at 5 minutes after stress induction and returns to the original state within 20 to 30 minutes (11). The activated MAPK coordinates the osmostress response by phosphorylating motifs consisting of serine or threonine followed by a proline (S/T-P motifs) on several target proteins. Ultimately, the cascade leads to the activation of downstream kinases, such as Rck2 (15), which has been associated primarily with translation.
Despite the fact that many direct and indirect targets of Hog1 have already been described, several aspects of the hyperosmotic stress response are still not completely understood (11), including which cellular functions are directly controlled and which are indirectly controlled by Hog1. Soufi et al. and Kanshin et al. describe the scale and dynamics of the hyperosmotic stress response on a phosphoproteomic scale and find the response to be complex, involving many kinases and phosphatases (4, 6). Motif searches using sequences flanking dynamic phosphorylation sites reveal the involvement of basophilic protein kinase A (PKA) and p21-activating kinases (PAKs), proline-directed kinases [MAPKs, cyclin-dependent kinases (CDKs)], and others (4, 6). Although these datasets provide an excellent overview of osmostress-induced changes in the yeast proteome, the lack of experiments where Hog1 is specifically deactivated, hampers efforts to unambiguously define substrates of this MAPK. Moreover, identification of dynamically phosphorylated sites does not allow unequivocal assignment of phosphorylated targets to one specific kinase, which we demonstrated for S/T-P sites of Pan1, a protein involved in early endocytosis (7). To extract kinase-substrate interactions in a system-wide manner it is therefore necessary to use experimental means to resolve kinase dependencies of distinct phosphorylation sites.
Here we addressed this problem using a dual MS-shotgun approach based on stable isotope labeling with amino acids in cell culture (SILAC) (16, 17) with the aim of identifying substrates of Hog1. Integration of the MS datasets revealed more than 25 previously unidentified putative substrates and numerous indirect targets of Hog1. Identified target proteins were further validated by their ability to directly interact with Hog1 in vivo using a protein-protein proximity assay (18). In addition, we demonstrated the role of the kinase Rck2 as a master regulator of secondary responses downstream of Hog1.
Results
Experimental Setup to Identify Hog1 Targets
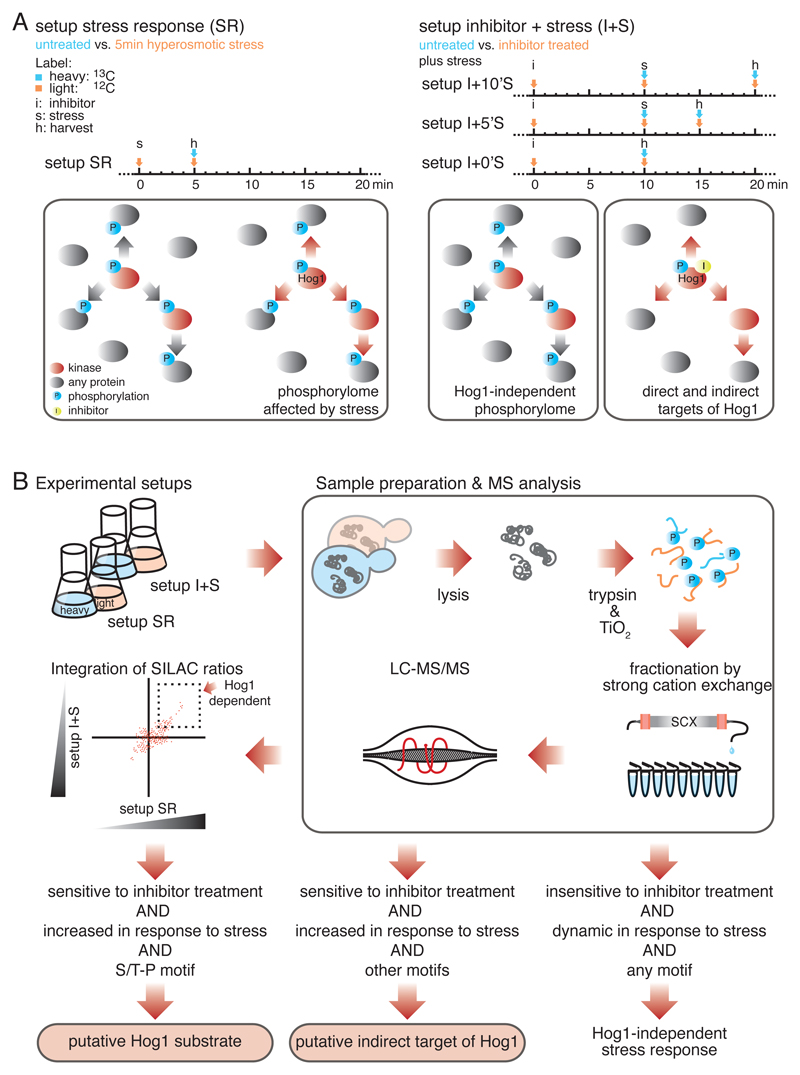
In yeast, extracellular hyperosmolarity activates the MAPK Hog1, which regulates various cellular processes important for stress adaptation and survival (11). Large-scale MS studies sought to predict specific Hog1-substrate interactions either by measuring osmostress induction or by using a hog1∆ strain (1, 4, 6). However, the overwhelming complexity of secondary effects impedes the discovery of specific substrates. We hypothesized that only the integration of both layers of information (induction and inactivation of Hog1, ideally by using a Hog1-specific inhibitor) would allow unambiguous assignment of phosphorylation events to Hog1 activity. Hence, to identify previously undescribed substrates of Hog1, we designed a two-step experimental setup using a quantitative proteomics strategy based on SILAC MS-labeling (Fig. 1A). As the first step we measured global changes in the phosphorylation pattern of the yeast proteome five minutes after exposure to increased extracellular salt concentrations, hereafter referred to as setup stress response (SR). As the second step we conducted a series of quantitative MS shotgun experiments, using a strain with a Hog1-allele sensitive to analogue-sensitive (as) inhibitor (19). After addition of the as-inhibitor, the kinase was specifically inactivated. Cells were subsequently exposed to hyperosmotic stress treatment for zero, five, or ten minutes, designated as setups I+0’S, I+5’S and I+10’S (Fig. 1A). Phosphorylated S/T-P motifs displaying more than a twofold increase in abundance in setup SR and sensitivity to inhibitor treatment in setups I+0’S, I+5’S or I+10’S were considered putative Hog1 substrates (Fig. 1B).
Fig. 1.
A two-step quantitative proteomics strategy to identify substrates of MAPK Hog1. A) Timing diagram (above) and schematic illustration (below) of the two experimental setups. In setup SR (left), global changes in the yeast phosphorylome in response to hyperosmotic stress were determined, n = 6 biological replicates. In setups I+0’S, I+5’S, and I+10’S (right), Hog1 was inactivated by an analogue sensitive (as) inhibitor followed by hyperosmotic stress treatment for zero, five or ten minutes, respectively. n = 2 biological replicates for each of the three experimental setups. Stress activated kinases are indicated in red and phosphorylation events in blue. The yellow dot indicates as-inhibitor. B) Schematic illustration of the applied workflow. Phosphorylation events displaying a more than twofold increase or decrease in abundance between SR and I+S setups were considered significant changes. Phosphorylated S/T-P motifs that increase in abundance in response to stress and show sensitivity to inhibitor treatment were considered putative substrates of Hog1.
Identification of Stress-Induced Changes in the S. cerevisiae Phosphorylome
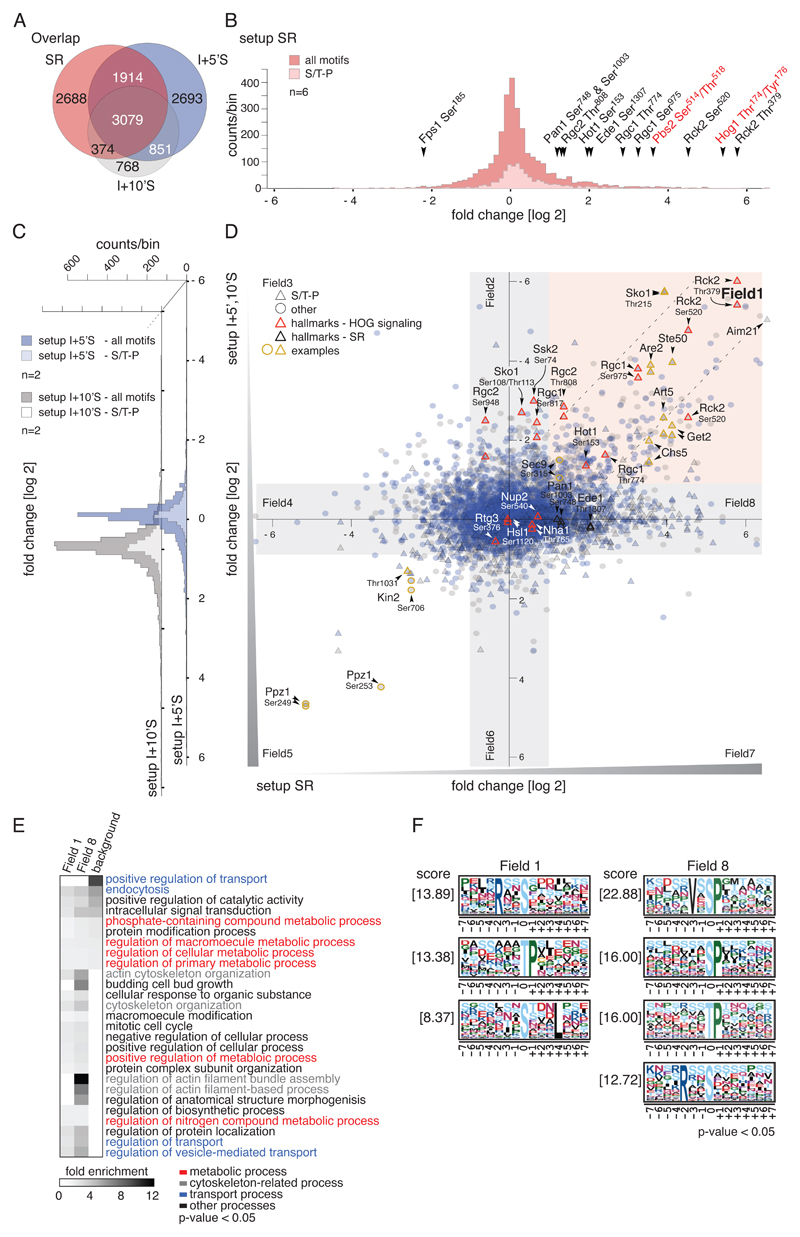
From our analysis of 283 liquid chromatography tandem MS runs, we identified 39055 peptides with a false positive rate below 1% for both peptides and proteins (table S1). On average we achieved a ~40% overlap for peptides between setups, allowing a systematic integrative analysis of the datasets (Fig. 2A). To facilitate phosphorylation site analysis we integrated all SILAC peptide ratios corresponding to a specific phosphorylation site. In this way we quantified 8055 phosphorylation sites in setup SR, 23% of which showed more than a twofold change in abundance (corresponding to 735 proteins), confirming the broadness of the hyperosmotic stress response in yeast. 77% of phosphorylation sites were static (≤ twofold change in abundance).
Fig. 2.
Effects of hyperosmotic stress and Hog1-inhibition on the S.cerevisiae phosphorylome. A) Venn diagram showing the overlap of unique phosphorylation sites between setups SR (n=6), I+5’S (n=2) and I+10’S (n=2). B) Histogram of SILAC ratios of quantified sites in setup SR. Light pink bins indicate the distribution of S/T-P motifs, and dark pink bins indicate other motifs. Ratios are log2-transformed. Hallmarks of osmostress and HOG signaling are highlighted with arrowheads; key residues of the MAPK Hog1 and MAPKK Pbs2 are indicated in red. C) Distribution of SILAC ratios of quantified sites in setups I+5’S (blue bins) and I+10’S (grey bins). S/T-P motifs: light blue and light grey bins, respectively; other motifs: dark blue and dark grey bins, respectively. Ratios are log2-transformed. D) Scatter plot overlaying SILAC ratios of setups SR, I+5’S, and I+10’S. X-axis: setup SR. Y-axis: setups I+5’S (blue) and I+10’S (grey), respectively. S/T-P motifs: triangles, other motifs: circles. Red outlines indicate hallmarks of HOG signaling; yellow outlines indicate examples discussed in the text. Ratios are log2-transformed. E) Heatmap of overrepresented GO terms in the individual fields defined in (C). Fold enrichment of biological processes, p-value < 0.05. Background: S. cerevisiae proteome. F) Overrepresented motifs in the individual fields were detected with MotifX, p-value < 0.05 (see also fig. S1C).
To test whether our setup SR adequately reflected the anticipated response of cells challenged with hyperosmolarity we looked at well-known phosphorylation events of osmostress signaling (Fig. 2B). First and foremost, we found phosphorylations at two key residues of Hog1 (Thr174 and Tyr176) to be increased approximately 70-fold, suggesting stress activation of the MAPK. In agreement with this, we also observed a more than 10-fold increase in phosphorylation at the regulatory amino acid residues of the MAPKK Pbs2 (Ser514, Thr518). Additionally, several well established Hog1 target sites showed increased phosphorylation, namely Thr379 and Ser520 of the Ca2+-or calmodulin-dependent protein kinase (CaMK)-like kinase Rck2 (15) (~50- and ~20-fold, respectively), Ser153 of transcription factor Hot1 (20) (~fourfold), Thr774 and Ser975 of Rgc1 (~10-fold), and Thr808 of Rgc2, also known as Ask10 (~threefold). Rgc1 and Rgc2 are regulators of the aquaglyceroporin Fps1 (21), which mediates the export of glycerol. We also confirmed a decrease in Ser185 phosphorylation of Fps1, which has been proposed to activate docking of Hog1 to this channel (4, 21).
Comparing setup SR with similar approaches (4) yielded a positive correlation (R = 0.53) for most of the dynamic phosphorylation sites (fig. S1 A and B). We thus recovered generally accepted hallmarks of the stress response and demonstrate full activation of Hog1 as well as data consistency with previous studies. We provide a detailed list of the integrated results of all shotgun experiments (table S2).
Hog1-Specific and General Responses to Hyperosmotic Stress
A previously unaddressed aspect of the HOG response concerns how many changes in the global phosphorylation pattern are mediated by Hog1 in a direct or indirect manner. To tackle this question we compared the corresponding SILAC ratios of datasets SR to I+5’S and I+10’S (Fig. 2, B–D). The distribution of SILAC fold changes, shown along the x-axis, represents phosphorylation changes in response to hyperosmotic stress. The dispersion along the y-axis indicates susceptibility to inhibitor treatment and therefore the degree of Hog1-dependency. By setting conservative ratio cutoffs (twofold) along both axes we defined eight data fields containing sites with distinct properties that can be characterized individually (Fig. 2D). Field 1 includes phosphorylation sites that increase in abundance in response to high osmolarity but remain unchanged if Hog1 is specifically inhibited. Therefore the phosphorylation status of these sites is directly or indirectly dependent on Hog1 activity. Field 2 includes phosphorylation sites that do not change in response to osmostress but are susceptible to inhibition of Hog1. Field 3 contains sites that decrease in abundance in response to stress and are also less abundant upon inhibitor treatment, and so forth. Finally, Field 8 includes all phosphorylation sites that increase in abundance in response to high osmolarity in a Hog1-independent manner. Although we observed a shift of data points towards higher values along the x-axis, suggesting an increase of phosphorylation events during stress in general, the distribution along the y-axis did not show any particular skewing. We expected changes along the y-axis to be minor given that the response was limited to inhibition of a very specific pathway. In total, 9% of phosphorylation sites covered in setup I+5’S (10% of setup I+10’S) were positively or negatively affected by inhibition of Hog1.
We evaluated the functional properties of the set of proteins clustering in Field 1 (Fig. 2E) and found enrichment for proteins involved in transport of solutes, endocytosis, and signal transduction, which is in line with previous reports (7, 9, 11, 15, 21–23). In addition, gene ontology (GO) terms related to the regulation of catalytic activity and diverse metabolic processes were also overrepresented, emphasizing the previously suggested role of Hog1 in metabolic fine-tuning (11). Though previously attributed to HOG signaling (4), actin- and cytoskeleton-related processes were enriched within the Hog1-independent set of proteins (Field 8).
We next tested for overrepresentation of peptide motifs surrounding dynamic phosphorylation sites using MotifX (24, 25) (Fig. 2F, fig. S1C) and first focused on phosphorylation at sites with prolines at position P+1 (S/T-P motifs), which is indicative of targeting by proline-directed kinases such as MAPKs and CDKs. Surprisingly, phosphorylation of most S/T-P motifs was independent of Hog1 (Field 8), suggesting that multiple proline-directed kinases are active during the stress response (Fig. 2, D and F, fig. S1D–F). In fact, only a minor fraction of S/T-P motifs was located in Field 1, the group of phosphorylation sites that increase in abundance in response to high osmolarity but remain unchanged if Hog1 is specifically inhibited. Twelve proteins contained both Hog1-dependently and Hog1-independently phosphorylated S/T-P motifs (in total, 542 proteins harbored sites from more than one field (table S3)) and might therefore represent hubs where multiple signaling cues are integrated (7). Inhibitor susceptibility of some S/T-P motifs might have been masked due to pathway crosstalk from Kss1, the MAPK of the filamentous growth pathway (26), which could result in a relocation of Hog1-substrates to Field 8. To determine whether the data were biased by Kss1-crosstalk, we determined stress-induced changes in the phosphorylome in cells in which both Hog1 and Kss1 were inactivated (fig. S2, A and B). A comparison of this dataset with setup SR revealed a linear correlation (R = 0.61) for almost the entire overlapping set (>90%) of Hog1-independent S/T-P motifs (63% of S/T-P motifs of Field 8 were covered), indicating that these sites were not affected by Kss1 crosstalk (fig. 2A). Hog1-dependent sites, on the other hand, did not show linear correlation except for Ser45 of Get2, which is probably targeted by both these MAPKs (fig. S2B). Furthermore, phosphorylation at Kss1 key residues Thr183 and Tyr185 was not altered by as-inhibitor treatment (fig.S 2C, table S2), and its four-fold increase in setup SR was negligible when compared to Hog1 (~70 fold) or Kss1 activity in a hog1∆ strain background (fig.S 2D).
We therefore conclude that the majority of stress-induced phosphorylation dynamics of S/T-P motifs are in fact phosphorylated not by Hog1 and Kss1 but by another proline-directed kinase (or kinases), and that the direct impact of Hog1 on the global phosphorylation pattern is probably smaller than anticipated.
Identification of Putative Hog1 Substrates
The 36 S/T-P motifs in Field 1 correspond to a total of 32 proteins that might represent direct targets of Hog1 (Table 1A, fig. S2E). In order to prevent the omission of targets due to incorrectly assigned phosphorylation sites, phosphorylations at S/T-S/T-P motifs were also considered. For completeness, setup I+0’S was also included in the analysis (Table 1A, table S2). We considered this set of phosphorylation sites, which included established hallmarks for Hog1-activity, such as Rck2 (Thr379, Ser520) (15), Hot1 (Thr153) (20), Rgc1 (Ser975, Thr774), and Rgc2 (Thr808) (21) as putative direct substrates of Hog1. Additionally, we found previously undescribed phosphorylation sites in well-established Hog1 targets such as the transcription factor Sko1 (Thr215) (27) and the adapter protein Ste50 (Thr244) (22, 23).
Table 1.
Putative direct target proteins of Hog1. (A) Candidates extracted from Field 1 that contain phosphorylated S/T-P and S/T-S/T-P motifs. SILAC ratios increasing ≥ 2-fold are highlighted in blue, and those increasing ≤ 0.5-fold in yellow. Phospho-accepting residues of S/T-P motifs are indicated in bold. “Up” indicates phosphorylation sites reported to increase in Kanshin et al. (4) as well. (B) Candidates extracted by indirect evidence (see text). SR n.a.: not assigned in setup SR. I+S n.a.: not assigned in inhibitor setups.
| Protein name | PhosphoSite | Field Assignment | setup SR | setup I+0'S | setup I+5'S | setup I+10'S | setup rck2∆ | motif | Kanshin et al. 2015 | known substrate | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Aim21 | 476(S) | Field 1 | 93.37 | 0.03 | 0.01 | PKA or STP | up | no | ||||
| Are2 | 175(S) | Field 1 | 12.03 | 0.08 | 0.07 | 0.07 | STP | no | |||||
| Art5 | 158(S) | Field 1 | 15.15 | 1.21 | 0.17 | 0.22 | 0.20 | CaMK or STP | up | no | |||
| Bck1 | 765(T) | Field 1 | 7.54 | 0.44 | 0.73 | STP | no | ||||||
| Bck2 | 258(S) | Field 1 | 22.67 | 0.50 | SL or STP | no | |||||||
| Boi1 | 647(T) | Field 1 | 7.55 | 1.08 | 0.48 | 0.51 | STP | no | |||||
| Bul1 | 107(T),111(T) | Field 1 | 25.96 | 0.67 | 0.37 | 0.17 | PKA or STP | no | |||||
| Chs5 | 373(T) | Field 1 | 11.70 | 0.95 | 0.36 | 0.25 | 0.91 | STP | no | ||||
| Ent3 | 320(T) | Field 1 | 11.14 | 0.38 | 1.18 | STP | no | ||||||
| Epo1 | 209(S) | Field 1 | 9.49 | 0.21 | STP | no | |||||||
| Erg11 | 458(S) | Field 1 | 4.88 | 0.95 | 0.46 | 0.40 | 0.93 | STP | no | ||||
| Ett1 | 33(S),34(T) | Field 1 | 2.19 | 0.46 | STP | no | |||||||
| Gal11 | 750(T) | Field 1 | 5.08 | 0.47 | 0.65 | 1.75 | STP | no | |||||
| Get2 | 45(S) | Field 1 | 17.56 | 0.23 | 0.19 | 0.58 | STP | no | |||||
| Gip3 | 260(S),264(T) | Field 1 | 2.06 | 0.47 | STP | no | |||||||
| Hal5 | 63(S),66(S) | Field 1 | 2.29 | 0.12 | STP | no | |||||||
| Hot1 | 153(S) | Field 1 | 3.88 | 0.77 | 0.39 | 1.02 | STP | static | yes | 20 | |||
| Ppz2 | 310(S) | Field 1 | 3.35 | 1.26 | 0.48 | 0.79 | STP | no | |||||
| Rck2 | 350(T) | Field 1 | 5.48 | 0.67 | 0.20 | STP | yes | 15 | |||||
| Rck2 | 379(T) | Field 1 | 55.24 | 0.02 | 0.02 | STP | up | yes | 15 | ||||
| Rck2 | 520(S) | Field 1 | 23.36 | 0.04 | 0.17 | STP | up | yes | 15 | ||||
| Reg1 | 898(S) | Field 1 | 3.18 | 0.05 | 1.09 | STP | no | ||||||
| Rgc1 | 774(T) | Field 1 | 5.42 | 0.32 | 0.56 | STP | yes | 21 | |||||
| Rgc1 | 975(S) | Field 1 | 9.67 | 0.07 | 0.08 | STP | up | yes | 21 | ||||
| Rgc2/Ask10 | 808(T) | Field 1 | 2.62 | 1.06 | 0.14 | 0.16 | 1.06 | STP | up | yes | 21 | ||
| Rod1 | 602(S),608(S) | Field 1 | 2.17 | 0.37 | STP or SL | no | |||||||
| Sfl1 | 556(S) | Field 1 | 7.43 | 1.00 | 0.35 | STP | no | ||||||
| Sko1 | 215(T) | Field 1 | 15.40 | 0.97 | 0.02 | 0.02 | 1.00 | STP | up | yes | 62 | ||
| Spt20 | 516(T) | Field 1 | 7.04 | 0.88 | 0.56 | 0.45 | 0.88 | STP | static | no | |||
| Ste50 | 244(T) | Field 1 | 17.64 | 0.06 | 0.91 | STP | yes | 22, 23 | |||||
| Tif4632 | 196(T) | Field 1 | 2.34 | 0.38 | 0.29 | 0.74 | STP | up | no | ||||
| Tod6 | 152(T) | Field 1 | 24.46 | 0.42 | STP | no | |||||||
| Vas1 | 294(S) | Field 1 | 2.74 | 0.56 | 0.40 | 1.25 | STP | no | |||||
| Vps9 | 375(S) | Field 1 | 3.95 | 0.80 | 0.37 | 0.26 | 0.76 | STP | up | no | |||
| Ylr257W | 8(T) | Field 1 | 2.49 | 0.81 | 0.61 | 0.50 | 0.64 | CaMK or STP | no | ||||
| Ylr257W | 7(S) | Field 1 | 2.53 | 0.95 | 0.50 | STP or SL | no | ||||||
| B | Rtg3 | 227(S) | SR n.a. | 0.94 | 0.46 | 0.70 | 0.87 | STP | up | yes | 29 | ||
| Ssk2 | 54(S),57(S) | SR n.a. | 0.22 | CaMK or STP | up | yes | 9 | ||||||
| Vps53 | 790(S) | SR n.a. | 0.15 | 0.17 | 0.07 | CaMK or STP | up | no | |||||
| Kic1 | 625(T) | I+S n.a. | 6.82 | STP | up | no | |||||||
| Tsl1 | 147(S) | I+S n.a. | 4.16 | 0.36 | CaMK or STP | up | no | ||||||
| Tsl1 | 161(S) | I+S n.a. | 16.97 | CaMK or STP | up | no | |||||||
| Ynl115C | 244(S) | I+S n.a. | 4.11 | STP | up | no | |||||||
| Ecm25 | 463(T),477(S) | SR n.a. | 0.18 | 0.18 | STP | no | |||||||
| Ppz1 | 49(S),67(T) | SR n.a. | 0.36 | 0.31 | STP | no | |||||||
Next, we aimed to extract leads for additional putative Hog1 targets from our datasets, focusing on S/T-P motifs that showed susceptibility to inhibitor treatment but were not covered in setup SR. This group of sites was compared with published data from Kanshin et al. (4) and browsed for increased phosphorylation in the early response to hyperosmotic stress. We found three phosphorylation sites that fitted these criteria: Ser790 of the Golgi-associated retrograde protein Vps53 (28), which has not previously been associated with the HOG pathway, Ser54 and Ser57 of the MAPKKK Ssk2 (9), and Ser227 of the retrograde and target of rapamycin (TOR) pathway transcription factor Rtg3 (29) (Table 1B).
In a second approach to scrutinize the dataset for potential targets, we grouped S/T-P motifs that get phosphorylated in setup SR but were not covered in at least one of the inhibitor datasets. Again we compared this group with the dataset of Kanshin et al., who hypothesized that conserved and putatively functional kinase-substrate interactions in the HOG response occur more rapidly than promiscuous interactions (4). This way we were able to extract three additional candidates, namely the trehalose synthase Tsl1 (Ser147 and Ser161), the p21 activated kinases (PAK)-Ste20 family protein kinase Kic1 (Thr625), and the uncharacterized protein Ynl115c (Ser244) (Table 1B). Of these four phosphorylation sites, Ser161 of Tsl1 showed the largest increase in stress-induced phosphorylation (~17 fold) and therefore strongly qualified for being a bona fide target of Hog1. Because trehalose functions as an osmolyte and protects the plasma membrane in E.coli (30), one could assume that it might exert a similar function in yeast in response to hyperosmotic stress (31, 32). Given the potential link to the HOG pathway, which had not yet been investigated, we performed some further in-depth analysis of the Tsl1 phosphorylation pattern in response to stress and Hog1 inhibition, using a validation strategy based on tandem affinity purification using a histidine biotin-tag (HTB) (7). Indeed, we confirmed Hog1-dependency for phosphorylation of two S/T-P motifs of Tsl1, Ser135 and Ser147 (fig. S3).
Finally, we also considered S/T-P motifs as potentially interesting if they were identified in at least two of the inhibitor setups, even if no other information on stress dependency was available. Four phosphorylation sites fulfilled these criteria: Ser176 of Are2 (33), Ser602 of Rod1 (34), Ser477 of Ecm25 (35) and Thr67 of Ppz1 (36). Of this group, Ecm25 and Ppz1 were not identified by any other selection criteria described above (Table 1B). To complete our search, we browsed the data from setups I+5’S and I+10’S for quantifications relying solely on one peptide identification and found as-inhibitor–susceptible S/T-P motifs from 22 proteins (table S4). It must be emphasized, however, that SILAC ratios based on a single identification must be considered as highly uncertain. This group contained some interesting candidates, for example the transcription factor Smp1, the protein kinase Ste50, and the F-Bar protein Syp1, all of which have been previously connected to HOG signaling (4, 22, 23, 37). The group also included protein kinases such as Akl1, Kin1, Rim15, and Ste20 (38–41).
In summary, our MS shotgun approach revealed a total of 40 putative target proteins of the MAPK Hog1: 32 from Field 1, six based on the integration of our data with previously published datasets, and two additional candidates that were selected based on coinciding ratios in two of the inhibitor datasets. In total, this set contains 32 putative Hog1-substrate interactions that have not been described previously (Table 1).
Identification of Hog1-Dependent Secondary Targets
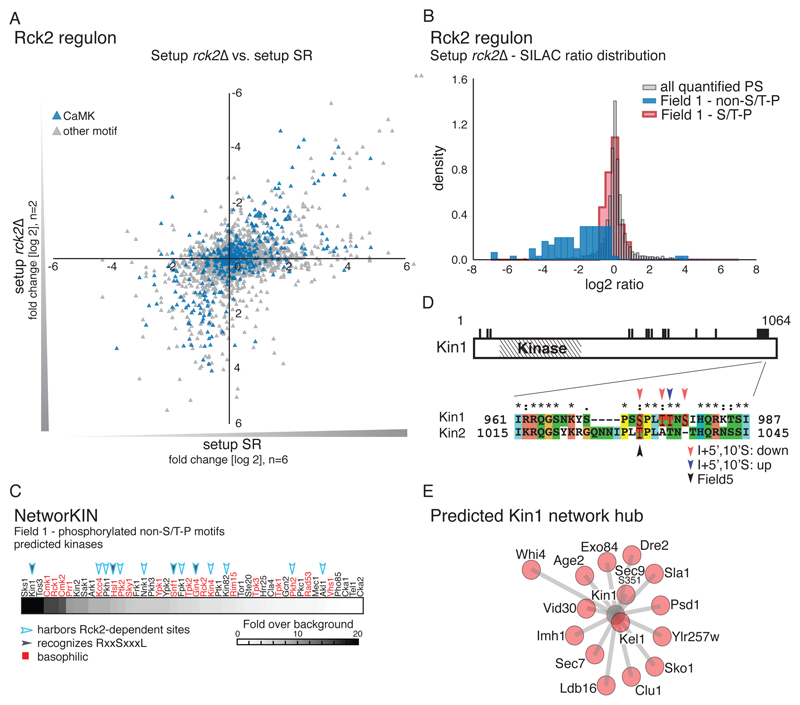
Because Hog1 activity influences the activity of other kinases, we also expected indirect effects of HOG signaling to be prominent in Field 1. Indeed, 82.4% of the phosphopeptides I Field 1were phosphorylated at non-S/T-P motif sequences. We considered this group to represent putative indirect targets of the Hog1 (table S5, fig.S 4A) and attempted to assign kinase dependencies for some of these phosphorylation sites. To do so, we initially searched for overrepresented peptide motifs and found R-X-X-S and S-X-X-X-L motifs to be significantly enriched (Fig. 2F). Motifs with an arginine at the P-3 position (R-X-X-S motifs) suggest the involvement of basophilic kinases, such as the members of the protein kinase A, G and C (AGC) and CaMK group or of the Casein kinase I family (42). Additional hydrophobic residues at the P+4 position (S-X-X-X-L motif) further suggest the involvement of a particular subfamily of the CaMK group (43, 44). Indeed, HOG signaling has previously been connected to CaMK-like kinases (15, 45, 46). For example, it is well known that the CaMK Rck2 is a target of the HOG pathway (15, 47) and its key residues are fully phosphorylated within 60 seconds of stress induction (4) (fig. S1B). Hence, it seemed likely that Rck2-mediated phosphorylation events also accumulated in Field 1. In order to systematically capture these, we performed an MS-shotgun experiment using a strain lacking RCK2 (setup rck2∆). We found the Rck2-dependent phosphoproteome to be complex, including positive or negative changes in 13.46% of quantified phosphorylation sites. At least 316 sites, including 108 basophilic kinase motifs, corresponding to 219 proteins were less phosphorylated in the rck2∆ strain than in wild-type cells in response to hyperosmotic stress (Fig. 3A and table S2). Rck2 seems to phosphorylate a large portion of the candidate indirect targets of Hog1 because most of the non-S/T-P motifs of Field 1 were affected by RCK2 deletion, whereas the impact of RCK2 deletion on S/T-P motifs of Field 1 was negligible (Fig. 3B, fig. S4, B–D). In total, we allocated 57 indirectly affected targets and an additional 48 stress- or inhibitor-responsive phosphorylation sites to Rck2 activity (table S2). Hence, we propose Rck2 as a major effector kinase of HOG signaling, influencing various cellular processes during osmostress.
Fig. 3.
The kinase Rck2 is a major effector of HOG signaling. A) Scatter plot comparing distribution of quantified phosphorylation sites between setup rck2∆ (SILAC ratio: rck2∆ (light): wild-type (heavy), both treated with NaCl (n=2)) and setup SR (n=6). B) Histogram showing the distribution of SILAC ratios of quantified sites in setup rck2∆. Red: Field 1 S/T-P motifs. Blue: Results for Field 1 non-S/T-P motifs. Grey: all quantified phosphorylation sites (PS). C) Prediction of kinases involved in the regulation of the indirect targets of Hog1. Basophilic kinases are indicated in red. Filled arrowheads: kinases recognizing S-X-X-X-L motifs. Open arrowheads: kinases containing Rck2-dependent phosphorylation sites. D) Diagram showing phosphorylated residues (black bars) of the kinase Kin1. Inset illustrates the conserved S/T-P motifs in Kin1 and Kin2, which include an as-inhibitor–sensitive serine (Ser973 in Kin1, black arrowhead). Phosphorylation at highlighted residues was either decreased (red arrowheads) or increased (blue arrowhead) upon inhibitor treatment or associated with Field 5 (black arrowhead). (E) Predicted interactions between Kin1 (grey) and substrates (red) interactions based on NetworKIN analysis. The relative length of each edge was calculated according to NetworkKIN scores: the closer to the kinase, the higher the score.
Among the affected proteins we found several suggested interactors of Rck2 (48–50) and 25 protein kinases from different families, including prominent basophilic kinases like Gin4, Hsl1, Kcc4, Sch9, and Snf1 (Fig. 3C, table S2). These kinases most likely contribute to secondary responses within the HOG- and Rck2 network, thus complicating the interpretation of kinase-substrate interactions. One such an example would be the regulation of the potassium transporter Trk1 (36, 51, 52) by the kinase Hal5 and the phosphatases Ppz1 and Ppz2, which have not previously been connected to HOG signaling. We found phosphorylation of Ser63 of Hal5 to be dependent on Rck2 (table S2), whereas Ppz1 and Ppz2 would, based on the MS-data, possibly constitute direct targets of Hog1 (Table 1). These regulatory events might explain changes in the phosphorylation pattern of Trk1, because we found Ser412 and Ser414, which are not present in S/T-P motifs, to increase in abundance in a stress- and HOG-dependent manner. Ser414 additionally showed susceptibility to deletion of RCK2, supporting a regulatory role for Rck2 in potassium transport (table S2).
To unravel further network hubs within the HOG-Rck2 network, we applied NetworKIN (53, 54), an algorithm designed to predict kinase-substrate interactions (Fig. 3C). The enrichment for both R-X-X-S and S-X-X-X-L motifs points towards a particular family of kinases of the CaMK group that may act downstream of Hog1 and Rck2 (42, 44, 55). NetworKIN analysis indeed showed that proteins with these motifs are likely targeted by Kin1, Kin2, Hsl1, Snf1or Gin4 (Fig. 3C, fig. S5). However, key residues of the activation loops of these kinases were either not identified in our MS-experiments or did not reveal HOG-Rck2–dependent activation, as in the case of Snf1 for example (table S2). Hence, we considered an alternative approach and hypothesized that, as previously suggested, functional phosphorylation sites would be more likely to be conserved than promiscuous sites (4). We therefore performed sequence alignments for the members of the individual kinase families and mapped affected phosphorylation sites obtained from our MS shotgun datasets. Thereby we identified a conserved cluster of multiple phosphorylation sites, including an S/T-P motif, at the C-terminal regulatory domain of Kin1 and Kin2 (39, 56, 57) (Fig. 3D). Even though the cluster could not be clearly assigned as a Hog1 or Rck2 substrate, the overall phosphorylation pattern hinted at dependency on HOG signaling. Furthermore, among the NetworKIN-predicted downstream targets of Kin1 or Kin2, we found Ser351 of Sec9, which is an in vitro substrate of these kinases (57) (Fig. 3D). This observation suggested that the kinases might indeed become activated in response to hyperosmotic stress and puts forward the hypothesis that Kin1 and Kin2 are activated in a manner that depends on HOG signaling.
Validation of Hog1-Substrate Interactions
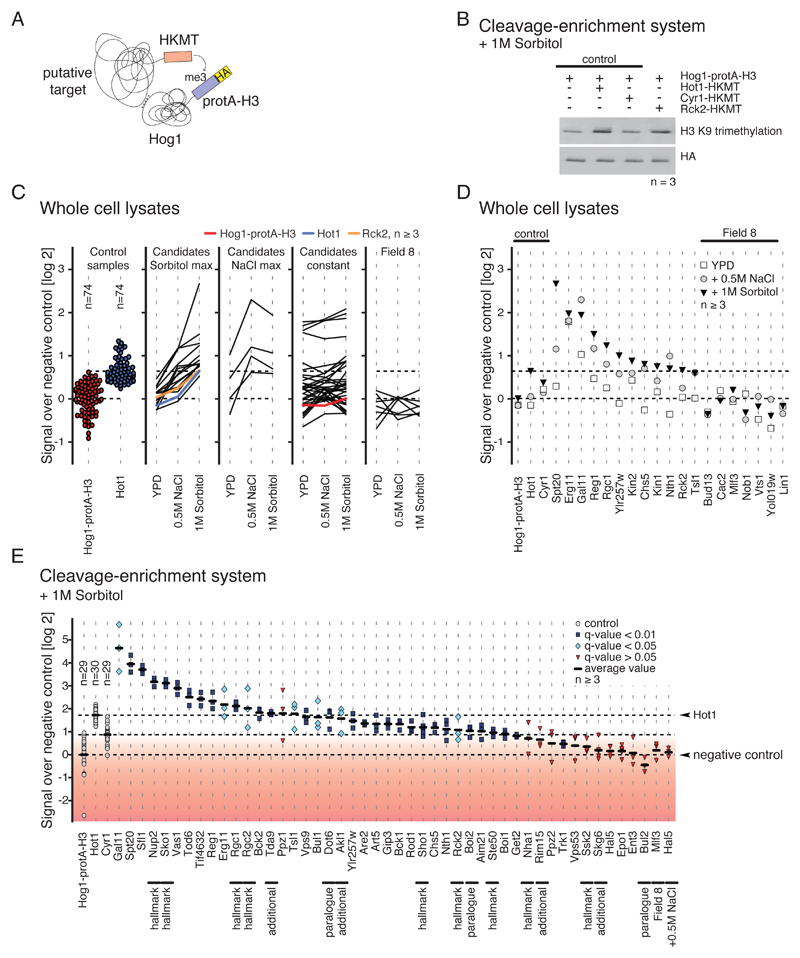
To confirm whether the newly identified putative target proteins (Table 1) directly interact with Hog1, we performed so-called M-track protein-protein proximity assays (18, 58), which is designed to capture transient interactions, such as those between kinases and their substrates. To do so, we tagged each of the candidate proteins with an active enzymatic domain from the SUV39 histone lysine methyltransferase (HKMT-myc (58)). Hog1 was fused with the prey sequence protA-H3 (58), which is a TEV-cleavable histone H3 tag fused to protein A- and hemagglutinin (HA)-tags that becomes permanently methylated upon coming into close proximity with the candidate protein bearing the HKMT-myc moiety (Fig. 4A). Each tagged candidate was coexpressed with Hog1-protA-H3 in yeast cells and the cells were subjected to various treatments. The proximity signal was detected by western blotting using an antibody directed against triple-methylated lysine 9 of histone H3 (me3K9H3) (Fig. 4B). We created functional HKMT-myc tag fusions for 35 of the 40 candidates described above (Table 1). Our analysis further included the following bait proteins and controls: i) the kinases Akl1 and Rim15 (38, 41), the transcription factor Tda9 (59), and membrane protein Skg6 (60) from table S4; ii) trehalase Nth1 (61), because we discovered a possible link between Hog1 signaling and trehalose metabolism, further supported by an S/T-P-containing peptide that was affected under stress conditions (table S2, fig. S6); iii) the potassium transporter Trk1, because three regulators of this transporter were identified as putative Hog1 substrates; iv) three known targets of Hog1, the osmosensor Sho1 (62), the Na+- and H+ antiporter Nha1(63), and the nucleoporin Nup2 (64); v) paralogues of three factors, Boi2, Bul2 and Dot6; and finally vi) Field 8-annotated proteins Bul13, Cac2, Mlf3, Nob1, Vts1 and Yol019w, and nuclear protein Lin1 as background controls. Background signal intensity was defined using a yeast strain expressing only Hog1-protA-H3.
Fig. 4.
Validation of kinase-substrate interactions. A) Cartoon illustrating the M-track protein-protein proximity assay. Hog1 (prey) is tagged with protA-H3 which gets permanently methylated when in close proximity to a putative target protein (bait) that is fused to an active domain of an histone lysine methyltransferase (HKMT). B) Representative Western blot showing M-track results obtained for Rck2 generated using cleavage-enrichment system. n≥3 replicates per sample. C) M-track proximity signals in whole cell extracts, comparing unstressed conditions to conditions of increased NaCl and sorbitol concentration. Each line corresponds to one protein. n ≥3 replicates per sample per condition. From left to right: loading controls, candidate proteins with maximum induction under sorbitol treatment, candidate proteins with maximum induction under NaCl treatment, candidate proteins that were not induced by sorbitol or NaCl, proteins derived from Field 8. Ratios are log2-transformed. D) Detailed proximity signals for tested targets described in the main text. n≥3 replicates per sample. Ratios are log2-transformed. E) Quantification of proximity signals. n ≥3 replicates per sample. Black lines indicate average proximity signal. Proximity signals that differ significantly from background are marked in light blue (q-value < 0.05, > 0.01) and blue (q-value < 0.01). The red gradient indicates low confidence based on q-values. One-tailed Welch’s t-test was applied and p-values were corrected for multiple testing by the Benjamini-Hochberg procedure. Proteins that were picked for validation based on less stringent criteria are marked with "additional".
We first analyzed whole-cell lysates in response to increased NaCl or sorbitol concentrations. Sorbitol treatment also activates Hog1 (65), but does not exert the same potentially toxic effects as sodium; thus, Hog1-substrate interactions captured under this condition additionally confirm the validity of our approach. To saturate proximity signals, we exposed cells for 40 minutes to the corresponding stimulus. 19 of our candidates indeed showed induction of the proximity signal after stress treatment, whereas the negative control (Hog1-protA-H3 alone) did not. Field 8-annotated proteins did not indicate proximity to Hog1 (Fig. 4C). Most of the strong proximity signals were observed with sorbitol treated samples (Fig., 4C and D, fig. S7). Some of the candidate factors, for example Erg11, showed high proximity signals even in the unstressed state, suggesting an interaction with Hog1 under isosmotic conditions. Finally, we also recovered candidate factors that were selected by less stringent search criteria such as for example Kin1 and Kin2, or Tsl1 and Nth1 (Fig. 4D).
However, due to the high variance in the signal intensities, the sensitivity of the applied statistical methods on the accumulated data was not optimal. To reduce variance and improve the signal-to-noise ratio, we performed additional M-track assays using a tag cleavage-enrichment system (58) and analyzed only the response to sorbitol (Fig. 4E). To identify significantly increased proximity signals, we applied Welch's t-test to compare signals to the negative control (Hog1-protA-H3 alone). 35 (74.5%) of the 47 kinase-substrate interactions we tested showed proximity signal intensities significantly above background intensity (based on a q-value of < 0.05). Within this group proximity signals, 28 (including 24 proteins that have not previously been associated with Hog1) showed a q-value of < 0.01, implying direct interaction with Hog1 after stress induction (Fig. 4E). This group of proteins included well-established hallmarks of HOG signaling such as Hot1, Nup2, Sko1, Rgc1, Sho1, and Ste50. Among the group of candidates with moderate proximity signals (q-values > 0.01 and < 0.05) we found three proteins containing both Hog1-dependent (S/T-P motifs) and Rck2-dependent sites, – Boi1, Gip3 and Rod1. This group also contained Akl1 and Bul1, both of which contain phosphorylation sites that were stress- and inhibitor responsive as well as Rck2-dependent (fig. S4D, table S2). This observation might indicate that either these sites are phosphorylated by both Hog1 and Rck2 or the corresponding proteins contain both Hog1-dependent and Rck2-dependent sites. In summary, our independent validation showed that most of the putative targets identified in our MS shotgun experiments indeed interact with the MAPK.
Testing for Osmosensitive Phenotypes of the Putative Direct Targets of Hog1
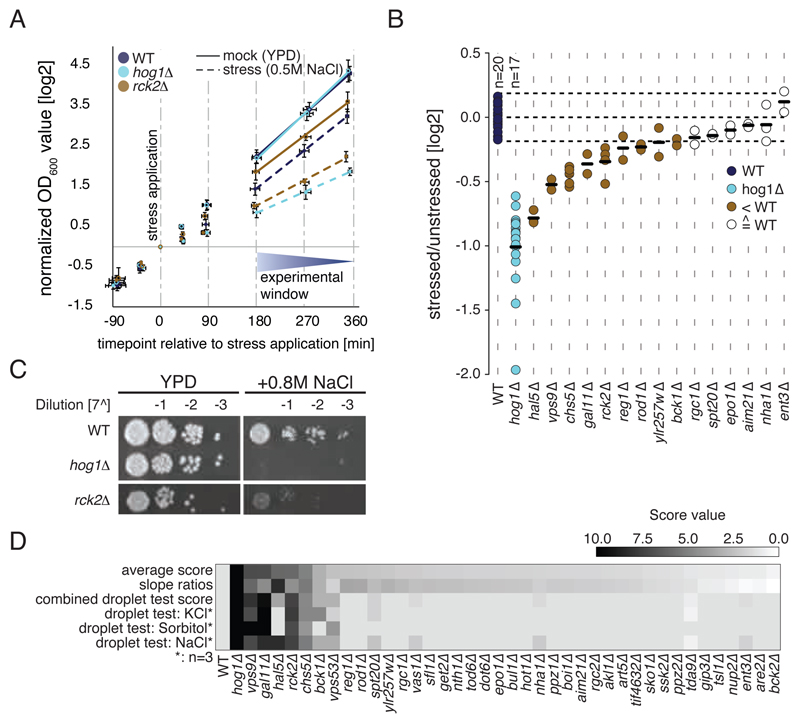
We next tested the newly identified Hog1 targets for potential functional relevance in osmostress adaptation and survival by performing growth assays. To capture short-term stress effects we compared the growth rates of hyper-osmotically challenged and unstressed deletion mutants for each of these proteins relative to those of wild-type and hog1∆ controls. 11 of the tested deletion mutants showed osmosensitive growth outside the fluctuation margin of the wild-type, and four of these had not previously been associated with osmo-sensitivity, namely reg1∆, rod1∆, vps9∆, and ylr275w∆ (Fig. 5A and 5B).
Fig. 5.
Characterization of osmosensitive phenotypes. A) Growth curves (log2-transformed) of wild-type, hog1∆, and rck2∆ cells in the absence of hyperosmotic stress and following induction of hyperosmotic stress. B) Stressed:unstressed (mock) ratios of log-transformed growth curve slopes depicted in (A), n ≥2 replicates per sample. (C) Serial dilution droplet test of the individual deletion mutants on YPD plates containing 0.8M NaCl. YPD: control. Only results obtained from rck2Δ cells are shown. (D) Heatmap illustrating scaled scores from dilution droplet tests under different conditions and growth curve analysis (slope score). Rows are sorted according to combined average score (0–10).
We determined the effects of long-term stress on growth with serial dilution droplet tests on complete medium (YPD) plates containing one of the following stressors: 0.5M NaCl, 0.8M NaCl, 1.2M NaCl, 0.8M KCl, 1.2M KCl, or 1.2M sorbitol. hog1Δ cells were most sensitive to osmotic shock, whereas wild-type and all the deletion mutants showed lower sensitivity to osmotic stress. Six of the tested candidates, including Rck2, were highly osmosensitive, indicating that they play an essential role in the response to increased extracellular osmolarity (Fig. 5C). Two additional deletion strains, spt20∆ and vps53∆, showed weak sensitivity to osmotic stress (fig. S8). Because these proteins are associated with processes other than increasing the intracellular concentration of osmolytes, we assume that Hog1 might affect additional functional modules in a post-translational manner. Results from growth curve analysis, as well as serial dilution droplet tests for different conditions (NaCl, sorbitol and KCl) are summarized in Fig. 5D.
Signaling networking revealing yet undiscovered functions of the MAPK Hog1
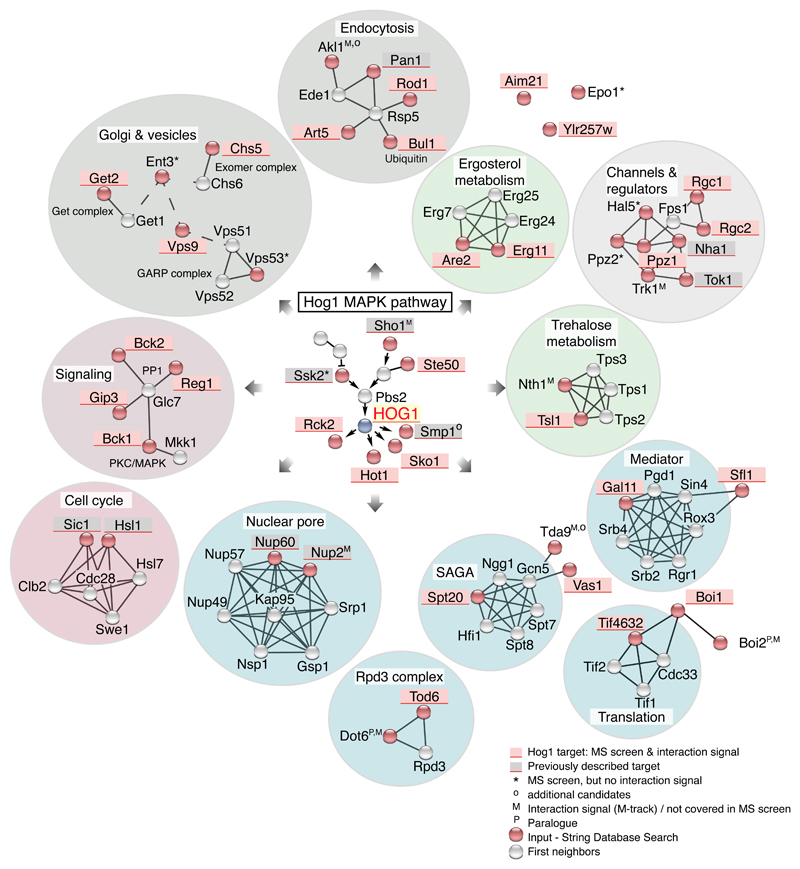
Our approach allowed the identification of several previously unidentified target proteins of Hog1; however, the total number is too small for a statistically solid GO-enrichment analysis. 7 of the protein candidates fall into the functional GO category of transcription and translation, 4 into the category of endocytosis, 3 into vesicle-related processes, and 4 into stress and starvation signaling (Fig. 6). For a better overview we created a protein network based on known protein-protein interactions from STRING database (66–68) using a list of 47 known and newly identified targets of Hog1 as input. Many of the cellular functions and pathways known to be directly affected by Hog1 signaling were captured, including cell cycle, signaling, nuclear pore complex, endocytosis, and glycerol channels plus their regulators. Moreover, the network included other molecular functions that have not been previously associated with HOG signaling, such as the Exomer and GET complexes, both of which are involved in vesicular transport to the Golgi; trehalose and ergosterol metabolism; and the SAGA, Mediator, and Rpd3 complex (Fig. 6). In summary, our analysis emphasizes that the Hog1-mediated response to hyperosmotic stress affects a broader set of functional units than previously anticipated.
Fig. 6.
Hog1 network based on STRING. Putative target proteins involved in the indicated cellular processes of Hog1 are shaded in red (Field 1 plus positive proximity signal). Previously described Hog1 substrates not included in our analysis are shaded in grey. Red circles indicate protein targets derived from the shotgun screen, with grey circles indicating their first neighbor according to STRING. Shaded circles enclosing proteins highlight functional groups, such as metabolism (green) and gene-expression related processes (blue).
Discussion
The study presented here provides a comprehensive MS-shotgun analysis designed to monitor the impact of Hog1 activity on the phosphoproteome of Saccharomyces cerevisiae. To ensure specificity of the screening, we integrated two experimental setups that monitor changes in phosphorylation patterns upon both exposure to hyperosmotic stress and inactivation of Hog1. Using a protein-protein proximity assay we further validated the candidates that we predicted to interact directly with Hog1 as substrates (18). In addition, we evaluated contributions made by the effector kinase Rck2. Our approach thus generated a comprehensive view of the Hog1 network, revealing at least 25 previously unidentified substrates as well as a broad set of indirect target proteins that extend our understanding of the HOG signaling system (Table 1).
Evaluating known Hallmarks of the Hog1 Response
Signaling through Hog1 has been previously associated with various cellular processes required for the adaptation to hyperosmotic stress, such as the regulation of intracellular glycerol concentration, gene expression, and cell cycle arrest. As a proof of principle for our experimental strategy, we expected to detect phosphosites of established Hog1 substrates that are involved in these processes, which was true for Hot (20), Rck2 (15), Rgc1, Rgc2 (21), Rtg3 (29), Sko1 (69), Ssk2 (9), and Ste50 (22, 23) (Fig. 2C and Table 1).
However, due to the inherent incompleteness of MS shotgun datasets, not all described hallmarks of Hog1 signaling were covered. Indeed, we did not identify or quantify the relevant phosphorylation sites of the osmosensor Sho1 (62), the CDK inhibitor Sic1 (70), or some other proteins previously implicated in Hog1 responses (37, 71–73). Notably, some known Hog1 target sites (29, 45, 63, 64) were influenced by neither stress nor inhibitor treatment (Fig. 2, table S2), which agrees with previous observations (4, 6). Given that Hog1 dependency of many of these sites has been established using in vitro kinase assays (29, 45, 63, 64), it is questionable whether they are genuine in vivo substrates of Hog1. Alternatively, such observations could be explained by promiscuous modifications due to a related kinase(s) such as Kss1. We addressed this possibility and provided evidence of Kss1 effectively having only a minor impact on our data (fig. S2). Another possible explanation could be that these sites are already saturated for phosphorylation by basal Hog1 activity. If the duration of inhibitor treatment is shorter than the turnover kinetics of phosphorylation, a decrease of phosphorylation events at these sites would not have been readily detected in our experiments.
Other well established Hog1 target sites showed more clear patterns of phosphorylation that were dependent on Hog1, but independent of osmotic stress (Field 2) - for example, Ser108 and Thr113 of Sko1 and Ser74 of Ssk2 (Fig. 2D, table S2). These S/T-P motifs are probably already modified to saturation by Hog1 under isosmotic conditions or undergo rapid dephosphorylation. These observations make it apparent that approaches to measure the turnover rates at individual sites are needed in the future to address such speculations.
Hog1-Dependent Phosphorylation Events Connected to Osmoadaptation
Our analysis provides insights into some open questions regarding the influence of Hog1 on the cellular metabolism (11) (Fig. 6), such as the previously underestimated interconnection of Hog1 signaling and trehalose metabolism and ergosterol biosynthesis. Although hyperosmotic stress tolerance in yeast has been primarily linked to glycerol production (13), it is also correlated with accumulation of trehalose (31, 32). This disaccharide is produced by the trehalose 6-phosphate synthase/phosphatase complex, which is composed of the catalytic subunit Tps1, the phosphatase Tps2, and the regulatory subunit Tsl1. When it is dispensable, trehalose is degraded by the neutral trehalases Nth1 and Nth2. In addition to being a candidate substrate of Hog1, Nth1 is also targeted by PKA (74–77), and we indeed recovered a phosphorylation event at Ser83 of Nth1, which lies in a predicted PKA target motif. The phosphorylation of this site decreased in response to stress (table S2), which is in line with previous observations on PKA-targeted motifs that are transiently affected under hyperosmotic stress conditions (78). We also recovered a phosphorylation event at Nth1 Ser66 (Fig. S6), which is targeted by Cdk1 to coordinate the cell cycle with central carbon metabolism (79, 80). Notably, a peptide including unphosphorylated Ser66 decreased five-fold upon hyperosmotic stress (table S2), suggesting stress-dependency of Nth1 as well. This observation, along with our detection of a significant Hog1-Nth1 proximity signal (Fig. 4E), leads us to propose that Nth1 is a direct target of Hog1, coinciding with an increase in Nth1-activity during recovery from stress (61). Moreover, we demonstrated Hog1-dependent changes in the phosphorylation pattern of Tsl1 (fig. S3) and prove proximity to Hog1 as well (Fig. 4E). Tps3, a known paralogue of Tsl1 (81), was not recovered as a Hog1 target in our analysis. Although the function of trehalose during cell stress is not fully understood and trehalose is usually connected to heat stress (31, 77, 82), it is a widespread assumption that trehalose is somehow involved in protecting and preserving membrane structure (83). Similarly, sterols, which are essential components of the eukaryotic plasma membrane that affect the fluidity and permeability of the membrane, have been implicated in osmostress adaptation in yeast, and transcription of ERG genes is negatively regulated by Hog1 (84). Here we provide evidence that Erg11, a lanosterol 14-alpha-demethylase, is directly targeted by Hog1, suggesting a transcription-independent and thereby faster mechanism for controlling ergosterol biosynthesis. These observations could thus provide novel insights into the mechanisms of plasma membrane protection during hyperosmotic stress.
Besides plasma membrane–related processes, several proteins related to vesicular transport as part of the endocytosis, exocytosis, and retrograde transport machineries were recovered as Hog1 substrates in our analysis. Our findings regarding the serine-threonine kinase Akl1 as a potential Hog1 target (38, 85) agrees with previous associations of the actin cytoskeleton regulatory complex member Pan1 with Hog1 (7). Both Akl1 and Pan1 are key constituents of the early endocytic machinery (86–90) that could be affected by Hog1-mediated phosphorylation. Regarding exocytosis, we identified Chs5, a scaffold protein of the exomer (91–93) complex, as a potential Hog1 substrate. Finally, Hog1-dependent phosphorylations of downstream kinases and phosphatases suggests the involvement of yet other signaling pathways, such as protein kinase C (PKC) (Bck1, (94, 95)) and protein phosphatase 1 (Reg1 and Gip3 (96)) signaling.
To explore additional putative Hog1-substrate interactions we also took into account fold changes of peptide variants containing potential target sites but carrying phosphogroups at different residues, as well as their respective unphosphorylated variants. If alternative —or even unphosphorylated—peptide variants showed dependency on Hog1, the corresponding protein might be somehow affected by Hog1 signaling. The corresponding peptides could then harbor phosphorylation sites that - whilst not necessarily covered in our MS-experiment - might be affected by Hog1 (for example see Ppz1in Fig. 2C, Field 5). These considerations pointed towards Kin1 and Kin2 (39, 56, 57), because their detected phosphopeptides resided in Field 5. The overall behavior of these sites suggested a Hog1-dependent phosphorylation at a conserved S/T-P motif at the C-terminus (Fig. 3D). Using the M-track assay we confirmed proximity of these proteins to Hog1 in response to sorbitol treatment (Fig. 4D). Both of these kinases belong to the Snf1 family of the CaMK group, which require structural rearrangements to unmask the kinase domain for activation. By analogy to Rck2 (15), Hog1 might affect exposure of the activation loop by phosphorylating C-terminal sites of these kinases. In fact, the only substrate of these kinases suggested so far, Ser351 of the t-SNARE protein Sec9 (57), was found amongst the group of proteins that we identified as indirect targets of Hog1.
The high number of indirectly affected targets of Hog1 we identified extends the diversity of putative cellular functions of Hog1. In a first attempt to dissect this signaling system into distinct kinase-target cascades, we focused on the proteins indirectly targeted by Hog1 through the CaMK Rck2, which reside in Field 1. We found the Rck2 network to be surprisingly complex, encompassing a large number of proteins of diverse cellular functions. In fact, the impact of rck2∆ under stress conditions on the sites of Field 1 was vast when compared to the set directly modified by Hog1. Rck2 seems to constitute a central hub of an underlying phosphorylation network, because phosphorylation sites of at least 16 kinases were affected by rck2∆. Some proteins also seemed to integrate Hog1 and Rck2 signaling, either by having separate Hog1 and Rck2 phosphorylation sites or having a phosphorylation site that could be targeted by either kinase. We conclude that the indirect targets of Hog1 are to a large extent controlled by Rck2 and therefore propose Rck2 as a major effector kinase of Hog1 signaling. Correspondingly, growth of a RCK2 deletion mutant was severely diminished in response to various hyperosmotic stress conditions. It will be interesting to see how future studies unravel the complexity of the Rck2 network, perhaps by applying a similar experimental strategy as presented here.
Hog1 Substrates Exerting Diverse Cellular Functions
Babazadeh et al. found that rewiring osmostress signaling through the Kss1 MAPK network reconstitutes osmoadaptation in hog1∆ cells (97). This reconstitution approach revealed that osmostress-induced up-regulation of intracellular glycerol concentration is sufficient for successful osmoadaptation. The authors further anticipated that the number of MAPK functions essential for the response might be smaller than estimated from knockout approaches and genome-wide analyses. However, if increasing intracellular glycerol concentration is sufficient for survival of and adaptation to hyperosmotic stress, it remains unclear why Hog1 would - directly and indirectly - affect such a variety of different cellular functions.
We challenged this hypothesis by analyzing growth phenotypes of deletion mutants of the identified candidate proteins and, for some, observed growth effects of differing severity from one another in response to hyperosmotic stress treatment. Although we cannot fully exclude that those targets could be bystander substrates that do not severely affect growth under hyperosmotic stress, the observed differences on growth under stress conditions indicate that some of the Hog1-modified processes might have more or less impact on overall cell physiology and growth rate. Proteins that appear to have only a subtle effect on cellular fitness during stress might be of more relevance when cells become exposed to continuous fluctuations in extracellular osmolarity or exhibit a cumulative phenotype when combined. It is intriguing to speculate that under constant fluctuation of environmental conditions such minor adjustments of phosphorylation patterns are important for competitive fitness.
Materials & Methods
Yeast strain and plasmid construction
Yeast strains and plasmids are listed in tables S6 and S7. Yeast strains WR557 (W303-1A SILAC Hog1as, Mat a) and WR564 (W303-1A SILAC Hog1as, Mat α) were obtained from a cross of WR210 with WR549 (7). Endogenous tagging (HTBeaq) and deletion mutations (kss1∆ and rck2∆) were obtained by methods described in (7, 98–100). Yeast strains used in M-track assays were generated as described in (58) using a strain-library available from Life Technologies (http://clones.lifetechnologies.com; (101)).
Growth conditions
Yeast cells were grown shaking (200rpm) at 30°C in synthetic medium (SD; 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose, amino acids as required) or rich medium (YPD; 1% yeast extract, 2% peptone, 2% glucose) for at least 7 generations until mid log phase (OD600nm ~ 1). For MS shotgun experiments (summarized in table S8) cells were subsequently treated, according to the experimental setup, with either 0.5M NaCl for 5 minutes (setups SR and rck2∆), or addition of the as-inhibitor (final concentration of 5µM) for 10 minutes (setup I+0'S) with subsequent salt treatment (0.5M NaCl) for 5 or 10 minutes, respectively (corresponding to setups HKi, I+5’S, I+10’S)). Conditions for growth and proximity assays are described below.
SILAC labeling
SILAC labeling was performed as described (3, 7, 17). For the experiments involving the analogue-sensitive inhibitor targeted at the Hog1 kinase, a yeast strain containing the Hog1as allele was used. With addition of the inhibitor to the cell culture (5µm final concentration) for 10 minutes, we ensure full inactivation of the Hog1 kinase. The inhibitor is designed to block the vicinity of the kinase that is usually reserved for ATP-binding (ATP-pocket) (19). Details on the experimental setups are provided in table S8.
Purification and Mass Spectrometric Analysis
Mass spectrometric shotgun experiments were performed as described (7). Briefly, cells were harvested by filtration and proteins were extracted using Trizol (Invitrogen) (7). Tryptic digests were subjected to TiO2 enrichment (102, 103). Phosphopeptides were fractionated offline by SCX chromatography and analyzed on a reverse-phase nano-HPLC-MS system coupled with an electrospray ionization interface (Proxeon Biosystems). MS-analysis was performed using a LTQ-Orbitrap Velos (CID mode) or a Q-Exactive (HCD mode) mass spectrometer (Thermo Scientific). Data analysis was performed using the SEQUEST algorithm (Proteome Discoverer 1.3 & 1.4). Parameter settings are described in the corresponding datasets in PRIDE (104) (see submissions PXD004294 - PXD004300). We performed both forward and reverse (decoy) searches and calculated protein and peptide FDR <1%. MS analysis of affinity-purified proteins was performed as described (7).
Computational Analysis of (Phospho)peptide Ratios
Normalization of data was performed as follows: Correction factors for the experiments were calculated as the geometric mean of the light-to-heavy (L/H)-ratios for all unphosphorylated peptides that contained either zero, one, two, etc. prolines (necessary correction for the signal loss due to arginine-proline conversion). The ratios measured for the phosphopeptides were divided by the correction factor.
To facilitate interpretation of phosphorylation sites, we grouped peptides together where the same residues are phosphorylated- regardless of potential missed cleavages, or additional modifications such as oxidation (corresponding to a so-called “phosphorylation-site group (PSG)”). The eventual ratio for the phosphorylation-site group is an average over all peptide ratios available in the group. Note that this kind of grouping could not be performed for the unphosphorylated peptide counterparts; however, we calculated the average and median of ratios corresponding to peptides of each protein, thereby estimating the impact experimental conditions had on the overall protein ratio.
For the analysis of the resulting peptide spectrum matches (PSMs) (FDR-calculation, normalization of ratios, extraction of quantified phosphorylation sites) we used in-house Python scripts (Python Software Foundation. Python Language Reference, version 2.7. Available at http://www.python.org). The scripts include a library of functions for extracting forward and reverse PSMs and the search summary for .msf-files allowing easier and much faster handling of the data. All scripts and functions are available on demand.
Gene Ontology Enrichment Analyses
Gene ontology (GO) enrichment analyses were performed with http://geneontology.org/page/go-enrichment-analysis, using the PANTHER classification system (105). Gene ontologies from the category “biological process” (GO-level 5) were extracted (corrected p-value of fold-enrichments <0.05); note that GO-levels for each term have been downloaded from http://supfam.cs.bris.ac.uk/SUPERFAMILY/ for proteins of each field designated in Figure 2.
Kinase Prediction Analyses
We used the MotifX algorithm (24, 25) to identify overrepresented linear motifs within our set of dynamic phosphorylation sites. Each Field was analyzed separately. The following settings were applied: min. occurrences: 10; background: SGD proteome. A binomial test was used to identify statistically enriched motifs, using a p-value of 0.05 as a significance cutoff. Further, we applied the NetworKIN algorithm (54) with default settings for kinase predictions for the dynamic phosphorylation sites. We determined the relative frequency for each kinase in individual Fields and divide the resulting values by kinase frequencies that occur in the group of static phosphorylation sites (cutoff for graph set at 0.3). Thereby we get an interpretable relative enrichment of kinases over a background represented by static phosphorylation sites.
Protein Network Analysis
Protein-protein interaction network for all putative targets of Hog1 was created using STRING database (version 10.0) (66). All factors listed in Figure 4F were used as search entries, with first neighbors automatically included in the network by the STRING database. All interaction predictions were based on physical, genetic and text mining evidence types with the minimal confidence score of > 0.7 (high) and a total of 40 white nodes and 0 interactors was allowed. We used the integrated MCL clustering algorithm according to STRING clustering instructions; clustering level was set to 3.
Growth tests
W303-1A wild-type and deletion mutant strains were grown until mid log phase, shifted to an OD600nm of 0.025 - 0.05, split, grown for 1-2 generations, then treated with either 0.5M NaCl (final concentration) or the corresponding volume of YPD (mock). OD600nm was monitored for a minimum of four generations after stress treatment. Slopes were obtained from log-transformed growth curves between generation 2 and 4 after stress application (experimental window, see Figure 5A) and a stressed (added 0.5M NaCl) versus unstressed (mock) slope ratio was calculated for every replicate (at least two per strain and condition, n=17 for hog1Δ, n=20 for wild-type). Slope ratios were log2-transformed (Figure 5B).
Protein-protein proximity assay (M-track)
M-Track protein-protein proximity assays were performed as described (18, 58). Cells were grown until mid log phase, treated with either 0.5M NaCl or 1M sorbitol (final concentration) for 40 minutes and harvested by filtration. Protein extraction was carried out under non-denaturing conditions by glass-bead lysis. Analysis of whole cell lysates (WCL) was performed as described (58) using buffer-1 (106). Immunoprecipitation and TEV cleavage (cleavage-enrichment) of prey proteins were performed as described (58). Trimethylation of histone 3 peptides was visualized by western blot using an antibody recognizing me3K9H3 (WCL: ab8898 (Abcam), IP: NBP 1-30141 (Novus Biochemicals). Loading was controlled using an antibody recognizing HA (12CA5). WCL loading was additionally controlled using an antibody targeting Pkg2 (Novex). Peak areas of signals were determined using ImageJ. Proximity signals were calculated as follows. Peak areas of histone H3 Lys9 trimethylation (me3K9H3) and hemagglutinin (HA) signals were determined using ImageJ, proximity signals were calculated as the log2-ratio of me3K9H3- over HA- signal of the individual candidates. Different western blot experiments were normalized by subtracting the average log2 signal intensity of the control samples Hot1 (positive control), Cyr1 (internal control) and Hog1-protA-H3 (negative control). We used a one-tailed Welch's t-test to identify the statistically significant candidates. In short, for each candidate gene the signal intensities of all replicates were compared against all signal intensities of the negative control. P-values were corrected for multiple testing by the Benjamini–Hochberg procedure with an alpha of 0.01 to generate q-values. Candidate genes with a mean signal intensity lower than the negative control were considered as not significant. Statistical analysis was performed independently for the whole cell extract and cleavage-enrichment datasets. Data analysis and figure generation was performed using Python scripts utilizing the libraries NumPy (107), matplotlib (108), seaborn(109), scipy (107) and statsmodels (110).
Serial dilution droplet test
W303-1A wild-type and deletion mutant strains were grown mid log phase. OD600nm values of cultures were equalized to 0.1 and serial dilution steps of 1:7 dilutions were prepared. Droplets of 2.5µl were transferred onto hyperosmotic stress plates (YPD +0.5M NaCl, +0.8M NaCl, +1.2M NaCl, +0.8M KCl, +1.2M KCl, and +1.2M sorbitol) and onto a YPD plate with no additives as a control. Plates were incubated at 30°C, and growth was monitored for four days.
Scoring System in Serial Dilution Droplet Test
Growth was monitored for 1-4 days and compared to wild-type and hog1∆. Long-term growth effects (Figure 5C) were assayed using an additive scoring system, endowing a score of two for a strong phenotype, a score of one for a weak phenotype and score of zero in case no phenotype was observed. Scores were further weighted according the molarity of the stressor (multiplied with a weight of three for 0.5M, two for 0.8M and one for 1.2M), resulting in a maximum score of 40 (applied for hog1Δ). Strains with scores corresponding to ≥ 13.5 (threefold of the wild-type) were regarded as highly osmosensitive candidates, scores between < 13 and ≥ 5 as moderate or weakly sensitive. Mutants with a score similar to wild-type (lower than 5) were regarded as insensitive to hyperosmotic stress.
Finally, to calculate a final score integrating both droplet-test scores and slope ratios, we scaled scores from these assays from 0 to 10 and computed the average score (see Figure 5D).
Supplementary Material
Acknowledgements
We thank Jillian Augustine for comments; Markus Hartl and Verena Unterwurzacher for help with mass spectrometry analyses; Jillian Augustine, Morton Grøtli, Andrea Brezovich, and Claudine Kraft for providing reagents; and Bernd Klaus and Matt Rogon for advice on statistical and network analysis.
Funding: This work was supported by grants from the 7th EU framework project (UNICELLSYS) and the Austrian Science Fund (Project F34) and by the foundation “Verein zur Förderung der Genomforschung”.
Footnotes
Author contributions: G.A. and W.R. designed the project. N.R, D.H., M.J., D.A., and W.R. performed experiments and analyzed data. W.R. wrote the manuscript. All authors commented and edited the text.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium through the PRIDE partner repository (104) with the dataset identifiers PXD004294 - 004300. The plasmids require a material transfer agreement from AAAS, U.S.A.
References
- 1.Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O, Brusniak MY, et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, Ahn J, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Kanshin E, Bergeron-Sandoval LP, Isik SS, Thibault P, Michnick SW. A cell-signaling network temporally resolves specific versus promiscuous phosphorylation. Cell Rep. 2015;10:1202–1214. doi: 10.1016/j.celrep.2015.01.052. [DOI] [PubMed] [Google Scholar]
- 5.Mascaraque V, Hernaez ML, Jimenez-Sanchez M, Hansen R, Gil C, Martin H, Cid VJ, Molina M. Phosphoproteomic analysis of protein kinase C signaling in Saccharomyces cerevisiae reveals Slt2 mitogen-activated protein kinase (MAPK)-dependent phosphorylation of eisosome core components. Mol Cell Proteomics. 2013;12:557–574. doi: 10.1074/mcp.M112.020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soufi B, Kelstrup CD, Stoehr G, Frohlich F, Walther TC, Olsen JV. Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol Biosyst. 2009;5:1337–1346. doi: 10.1039/b902256b. [DOI] [PubMed] [Google Scholar]
- 7.Reiter W, Anrather D, Dohnal I, Pichler P, Veis J, Grotli M, Posas F, Ammerer G. Validation of regulated protein phosphorylation events in yeast by quantitative mass spectrometry analysis of purified proteins. Proteomics. 2012;12:3030–3043. doi: 10.1002/pmic.201200185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chasman D, Ho YH, Berry DB, Nemec CM, MacGilvray ME, Hose J, Merrill AE, Lee MV, Will JL, Coon JJ, Ansari AZ, et al. Pathway connectivity and signaling coordination in the yeast stress-activated signaling network. Mol Syst Biol. 2014;10:759. doi: 10.15252/msb.20145120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharifian H, Lampert F, Stojanovski K, Regot S, Vaga S, Buser R, Lee SS, Koeppl H, Posas F, Pelet S, Peter M. Parallel feedback loops control the basal activity of the HOG MAPK signaling cascade. Integr Biol (Camb) 2015;7:412–422. doi: 10.1039/c4ib00299g. [DOI] [PubMed] [Google Scholar]
- 10.Vaga S, Bernardo-Faura M, Cokelaer T, Maiolica A, Barnes CA, Gillet LC, Hegemann B, van Drogen F, Sharifian H, Klipp E, Peter M, et al. Phosphoproteomic analyses reveal novel cross-modulation mechanisms between two signaling pathways in yeast. Mol Syst Biol. 2014;10:767. doi: 10.15252/msb.20145112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saito H, Posas F. Response to hyperosmotic stress. Genetics. 2012;192:289–318. doi: 10.1534/genetics.112.140863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Nadal E, Ammerer G, Posas F. Controlling gene expression in response to stress. Nat Rev Genet. 2011;12:833–845. doi: 10.1038/nrg3055. [DOI] [PubMed] [Google Scholar]
- 13.Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Posas F, Chambers JR, Heyman JA, Hoeffler JP, de Nadal E, Arino J. The transcriptional response of yeast to saline stress. J Biol Chem. 2000;275:17249–17255. doi: 10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- 15.Teige M, Scheikl E, Reiser V, Ruis H, Ammerer G. Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc Natl Acad Sci U S A. 2001;98:5625–5630. doi: 10.1073/pnas.091610798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 17.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 18.Zuzuarregui A, Kupka T, Bhatt B, Dohnal I, Mudrak I, Friedmann C, Schuchner S, Frohner IE, Ammerer G, Ogris E. M-Track: detecting short-lived protein-protein interactions in vivo. Nat Methods. 2012;9:594–596. doi: 10.1038/nmeth.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein M, Morillas M, Vendrell A, Brive L, Gebbia M, Wallace IM, Giaever G, Nislow C, Posas F, Grøtli M. Design, synthesis and characterization of a highly effective inhibitor for analog-sensitive (as) kinases. PLoS One. 2011;6:e20789. doi: 10.1371/journal.pone.0020789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alepuz PM, de Nadal E, Zapater M, Ammerer G, Posas F. Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. EMBO J. 2003;22:2433–2442. doi: 10.1093/emboj/cdg243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J, Reiter W, Dohnal I, Gregori C, Beese-Sims S, Kuchler K, Ammerer G, Levin DE. MAPK Hog1 closes the S. cerevisiae glycerol channel Fps1 by phosphorylating and displacing its positive regulators. Genes Dev. 2013;27:2590–2601. doi: 10.1101/gad.229310.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao N, Zeng Y, Elston TC, Dohlman HG. Control of MAPK specificity by feedback phosphorylation of shared adaptor protein Ste50. J Biol Chem. 2008;283:33798–33802. doi: 10.1074/jbc.C800179200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto K, Tatebayashi K, Tanaka K, Saito H. Dynamic control of yeast MAP kinase network by induced association and dissociation between the Ste50 scaffold and the Opy2 membrane anchor. Mol Cell. 2010;40:87–98. doi: 10.1016/j.molcel.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Chou MF, Schwartz D. Biological sequence motif discovery using motif-x. Curr Protoc Bioinformatics. 2011;Chapter 13:Unit 13 15–24. doi: 10.1002/0471250953.bi1315s35. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 26.O'Rourke SM, Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Proft M, Pascual-Ahuir A, de Nadal E, Arino J, Serrano R, Posas F. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 2001;20:1123–1133. doi: 10.1093/emboj/20.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz-Roig C, Noriega N, Duch A, Posas F, de Nadal E. The Hog1 SAPK controls the Rtg1/Rtg3 transcriptional complex activity by multiple regulatory mechanisms. Mol Biol Cell. 2012;23:4286–4296. doi: 10.1091/mbc.E12-04-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zahringer H, Thevelein JM, Nwaka S. Induction of neutral trehalase Nth1 by heat and osmotic stress is controlled by STRE elements and Msn2/Msn4 transcription factors: variations of PKA effect during stress and growth. Mol Microbiol. 2000;35:397–406. doi: 10.1046/j.1365-2958.2000.01706.x. [DOI] [PubMed] [Google Scholar]
- 31.Neves MJ, Francois J. On the mechanism by which a heat shock induces trehalose accumulation in Saccharomyces cerevisiae. Biochem J. 1992;288:859–864. doi: 10.1042/bj2880859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parrou JL, Teste MA, Francois J. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology. 1997;143:1891–1900. doi: 10.1099/00221287-143-6-1891. [DOI] [PubMed] [Google Scholar]
- 33.Zweytick D, Leitner E, Kohlwein SD, Yu C, Rothblatt J, Daum G. Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur J Biochem. 2000;267:1075–1082. doi: 10.1046/j.1432-1327.2000.01103.x. [DOI] [PubMed] [Google Scholar]
- 34.Becuwe M, Vieira N, Lara D, Gomes-Rezende J, Soares-Cunha C, Casal M, Haguenauer-Tsapis R, Vincent O, Paiva S, Leon S. A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J Cell Biol. 2012;196:247–259. doi: 10.1083/jcb.201109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lussier M, White AM, Sheraton J, di Paolo T, Treadwell J, Southard SB, Horenstein CI, Chen-Weiner J, Ram AF, Kapteyn JC, Roemer TW, et al. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yenush L, Mulet JM, Arino J, Serrano R. The Ppz protein phosphatases are key regulators of K+ and pH homeostasis: implications for salt tolerance, cell wall integrity and cell cycle progression. EMBO J. 2002;21:920–929. doi: 10.1093/emboj/21.5.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Nadal E, Casadome L, Posas F. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol Cell Biol. 2003;23:229–237. doi: 10.1128/MCB.23.1.229-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cope MJ, Yang S, Shang C, Drubin DG. Novel protein kinases Ark1p and Prk1p associate with and regulate the cortical actin cytoskeleton in budding yeast. J Cell Biol. 1999;144:1203–1218. doi: 10.1083/jcb.144.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamb A, Tibbetts M, Hammond CI. The product of the KIN1 locus in Saccharomyces cerevisiae is a serine/threonine-specific protein kinase. Yeast. 1991;7:219–228. doi: 10.1002/yea.320070304. [DOI] [PubMed] [Google Scholar]
- 40.Leberer E, Dignard D, Harcus D, Thomas DY, Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidan S, Mitchell AP. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol Cell Biol. 1997;17:2688–2697. doi: 10.1128/mcb.17.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunter T, Plowman GD. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 43.Dale S, Wilson WA, Edelman AM, Hardie DG. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 1995;361:191–195. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- 44.Mok J, Kim PM, Lam HY, Piccirillo S, Zhou X, Jeschke GR, Sheridan DL, Parker SA, Desai V, Jwa M, Cameroni E, et al. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci Signal. 2010;3:ra12. doi: 10.1126/scisignal.2000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clotet J, Escote X, Adrover MA, Yaakov G, Gari E, Aldea M, de Nadal E, Posas F. Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J. 2006;25:2338–2346. doi: 10.1038/sj.emboj.7601095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizuno T, Masuda Y, Irie K. The Saccharomyces cerevisiae AMPK, Snf1, Negatively Regulates the Hog1 MAPK Pathway in ER Stress Response. PLoS Genet. 2015;11:e1005491. doi: 10.1371/journal.pgen.1005491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bilsland-Marchesan E, Arino J, Saito H, Sunnerhagen P, Posas F. Rck2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase Hog1. Mol Cell Biol. 2000;20:3887–3895. doi: 10.1128/mcb.20.11.3887-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 49.Sharifpoor S, van Dyk D, Costanzo M, Baryshnikova A, Friesen H, Douglas AC, Youn JY, VanderSluis B, Myers CL, Papp B, Boone C, et al. Functional wiring of the yeast kinome revealed by global analysis of genetic network motifs. Genome Res. 2012;22:791–801. doi: 10.1101/gr.129213.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, Wilson WA, Fujino MA, Roach PJ. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol Cell Biol. 2001;21:5742–5752. doi: 10.1128/MCB.21.17.5742-5752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mulet JM, Leube MP, Kron SJ, Rios G, Fink GRR. Serrano, A novel mechanism of ion homeostasis and salt tolerance in yeast: the Hal4 and Hal5 protein kinases modulate the Trk1-Trk2 potassium transporter. Mol Cell Biol. 1999;19:3328–3337. doi: 10.1128/mcb.19.5.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yenush L, Merchan S, Holmes J, Serrano R. pH-Responsive, posttranslational regulation of the Trk1 potassium transporter by the type 1-related Ppz1 phosphatase. Mol Cell Biol. 2005;25:8683–8692. doi: 10.1128/MCB.25.19.8683-8692.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horn H, Schoof EM, Kim J, Robin X, Miller ML, Diella F, Palma A, Cesareni G, Jensen LJ, Linding R. KinomeXplorer: an integrated platform for kinome biology studies. Nat Methods. 2014;11:603–604. doi: 10.1038/nmeth.2968. [DOI] [PubMed] [Google Scholar]
- 54.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith FC, Davies SP, Wilson WA, Carling D, Hardie DG. The SNF1 kinase complex from Saccharomyces cerevisiae phosphorylates the transcriptional repressor protein Mig1p in vitro at four sites within or near regulatory domain 1. FEBS Lett. 1999;453:219–223. doi: 10.1016/s0014-5793(99)00725-5. [DOI] [PubMed] [Google Scholar]
- 56.Donovan M, Romano P, Tibbetts M, Hammond CI. Characterization of the KIN2 gene product in Saccharomyces cerevisiae and comparison between the kinase activities of p145KIN1 and p145KIN2. Yeast. 1994;10:113–124. doi: 10.1002/yea.320100111. [DOI] [PubMed] [Google Scholar]
- 57.Elbert M, Rossi G, Brennwald P. The yeast par-1 homologs kin1 and kin2 show genetic and physical interactions with components of the exocytic machinery. Mol Biol Cell. 2005;16:532–549. doi: 10.1091/mbc.E04-07-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brezovich A, Schuschnig M, Ammerer G, Kraft C. An in vivo detection system for transient and low-abundant protein interactions and their kinetics in budding yeast. Yeast. 2015;32:355–365. doi: 10.1002/yea.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walkey CJ, Luo Z, Madilao LL, van Vuuren HJ. The fermentation stress response protein Aaf1p/Yml081Wp regulates acetate production in Saccharomyces cerevisiae. PLoS One. 2012;7:e51551. doi: 10.1371/journal.pone.0051551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomishige N, Noda Y, Adachi H, Yoda K. SKG6, a suppressor gene of synthetic lethality of kex2Delta gas1Delta mutations, encodes a novel membrane protein showing polarized intracellular localization. J Gen Appl Microbiol. 2005;51:323–326. doi: 10.2323/jgam.51.323. [DOI] [PubMed] [Google Scholar]
- 61.Nwaka S, Holzer H. Molecular biology of trehalose and the trehalases in the yeast Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1998;58:197–237. doi: 10.1016/s0079-6603(08)60037-9. [DOI] [PubMed] [Google Scholar]
- 62.Hao N, Behar M, Parnell SC, Torres MP, Borchers CH, Elston TC, Dohlman HG. A systems-biology analysis of feedback inhibition in the Sho1 osmotic-stress-response pathway. Curr Biol. 2007;17:659–667. doi: 10.1016/j.cub.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 63.Proft M, Struhl K. MAP kinase-mediated stress relief that precedes and regulates the timing of transcriptional induction. Cell. 2004;118:351–361. doi: 10.1016/j.cell.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 64.Regot S, de Nadal E, Rodriguez-Navarro S, Gonzalez-Novo A, Perez-Fernandez J, Gadal O, Seisenbacher G, Ammerer G, Posas F. The Hog1 stress-activated protein kinase targets nucleoporins to control mRNA export upon stress. J Biol Chem. 2013;288:17384–17398. doi: 10.1074/jbc.M112.444042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Westfall PJ, Thorner J. Analysis of mitogen-activated protein kinase signaling specificity in response to hyperosmotic stress: use of an analog-sensitive HOG1 allele. Eukaryot Cell. 2006;5:1215–1228. doi: 10.1128/EC.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Proft M, Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell. 2002;9:1307–1317. doi: 10.1016/s1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- 70.Escote X, Zapater M, Clotet J, Posas F. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat Cell Biol. 2004;6:997–1002. doi: 10.1038/ncb1174. [DOI] [PubMed] [Google Scholar]
- 71.Dihazi H, Kessler R, Eschrich K. High osmolarity glycerol (HOG) pathway-induced phosphorylation and activation of 6-phosphofructo-2-kinase are essential for glycerol accumulation and yeast cell proliferation under hyperosmotic stress. J Biol Chem. 2004;279:23961–23968. doi: 10.1074/jbc.M312974200. [DOI] [PubMed] [Google Scholar]
- 72.Mollapour M, Piper PW. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol Cell Biol. 2007;27:6446–6456. doi: 10.1128/MCB.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thorsen M, Di Y, Tangemo C, Morillas M, Ahmadpour D, Van der Does C, Wagner A, Johansson E, Boman J, Posas F, Wysocki R, et al. The MAPK Hog1p modulates Fps1p-dependent arsenite uptake and tolerance in yeast. Mol Biol Cell. 2006;17:4400–4410. doi: 10.1091/mbc.E06-04-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.App H, Holzer H. Purification and characterization of neutral trehalase from the yeast ABYS1 mutant. J Biol Chem. 1989;264:17583–17588. [PubMed] [Google Scholar]
- 75.Kopp M, Nwaka S, Holzer H. Corrected sequence of the yeast neutral trehalase-encoding gene (NTH1): biological implications. Gene. 1994;150:403–404. doi: 10.1016/0378-1119(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 76.Thevelein JM, Beullens M. Cyclic AMP and the stimulation of trehalase activity in the yeast Saccharomyces cerevisiae by carbon sources, nitrogen sources and inhibitors of protein synthesis. J Gen Microbiol. 1985;131:3199–3209. doi: 10.1099/00221287-131-12-3199. [DOI] [PubMed] [Google Scholar]
- 77.Zahringer H, Holzer H, Nwaka S. Stability of neutral trehalase during heat stress in Saccharomyces cerevisiae is dependent on the activity of the catalytic subunits of cAMP-dependent protein kinase, Tpk1 and Tpk2. Eur J Biochem. 1998;255:544–551. doi: 10.1046/j.1432-1327.1998.2550544.x. [DOI] [PubMed] [Google Scholar]
- 78.Reiter W, Klopf E, De Wever V, Anrather D, Petryshyn A, Roetzer A, Niederacher G, Roitinger E, Dohnal I, Gorner W, Mechtler K, et al. Yeast protein phosphatase 2A-Cdc55 regulates the transcriptional response to hyperosmolarity stress by regulating Msn2 and Msn4 chromatin recruitment. Mol Cell Biol. 2013;33:1057–1072. doi: 10.1128/MCB.00834-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ewald JC, Kuehne A, Zamboni N, Skotheim JM. The Yeast Cyclin-Dependent Kinase Routes Carbon Fluxes to Fuel Cell Cycle Progression. Mol Cell. 2016;62:532–545. doi: 10.1016/j.molcel.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao G, Chen Y, Carey L, Futcher B. Cyclin-Dependent Kinase Co-Ordinates Carbohydrate Metabolism and Cell Cycle in S. cerevisiae. Mol Cell. 2016;62:546–557. doi: 10.1016/j.molcel.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vuorio OE, Kalkkinen N, Londesborough J. Cloning of two related genes encoding the 56-kDa and 123-kDa subunits of trehalose synthase from the yeast Saccharomyces cerevisiae. Eur J Biochem. 1993;216:849–861. doi: 10.1111/j.1432-1033.1993.tb18207.x. [DOI] [PubMed] [Google Scholar]
- 82.Diamant S, Eliahu N, Rosenthal D, Goloubinoff P. Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stresses. J Biol Chem. 2001;276:39586–39591. doi: 10.1074/jbc.M103081200. [DOI] [PubMed] [Google Scholar]
- 83.Conlin LK, Nelson HC. The natural osmolyte trehalose is a positive regulator of the heat-induced activity of yeast heat shock transcription factor. Mol Cell Biol. 2007;27:1505–1515. doi: 10.1128/MCB.01158-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Montanes FM, Pascual-Ahuir A, Proft M. Repression of ergosterol biosynthesis is essential for stress resistance and is mediated by the Hog1 MAP kinase and the Mot3 and Rox1 transcription factors. Mol Microbiol. 2011;79:1008–1023. doi: 10.1111/j.1365-2958.2010.07502.x. [DOI] [PubMed] [Google Scholar]
- 85.Henry KR, D'Hondt K, Chang JS, Nix DA, Cope MJ, Chan CS, Drubin DG, Lemmon SK. The actin-regulating kinase Prk1p negatively regulates Scd5p, a suppressor of clathrin deficiency, in actin organization and endocytosis. Curr Biol. 2003;13:1564–1569. doi: 10.1016/s0960-9822(03)00579-7. [DOI] [PubMed] [Google Scholar]
- 86.Duncan MC, Cope MJ, Goode BL, Wendland B, Drubin DG. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- 87.Tang HY, Xu J, Cai M. Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol Cell Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toshima J, Toshima JY, Duncan MC, Cope MJ, Sun Y, Martin AC, Anderson S, Yates JR, 3rd, Mizuno K, Drubin DG. Negative regulation of yeast Eps15-like Arp2/3 complex activator, Pan1p, by the Hip1R-related protein, Sla2p, during endocytosis. Mol Biol Cell. 2007;18:658–668. doi: 10.1091/mbc.E06-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wendland B, Emr SD. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wendland B, Steece KE, Emr SD. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huranova M, Muruganandam G, Weiss M, Spang A. Dynamic assembly of the exomer secretory vesicle cargo adaptor subunits. EMBO Rep. 2016;17:202–219. doi: 10.15252/embr.201540795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Trautwein M, Schindler C, Gauss R, Dengjel J, Hartmann E, Spang A. Arf1p, Chs5p and the ChAPs are required for export of specialized cargo from the Golgi. EMBO J. 2006;25:943–954. doi: 10.1038/sj.emboj.7601007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang CW, Hamamoto S, Orci L, Schekman R. Exomer: A coat complex for transport of select membrane proteins from the trans-Golgi network to the plasma membrane in yeast. J Cell Biol. 2006;174:973–983. doi: 10.1083/jcb.200605106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Irie K, Takase M, Lee KS, Levin DE, Araki H, Matsumoto K, Oshima Y. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol Cell Biol. 1993;13:3076–3083. doi: 10.1128/mcb.13.5.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee KS, Levin DE. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol Cell Biol. 1992;12:172–182. doi: 10.1128/mcb.12.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pinsky BA, Kotwaliwale CV, Tatsutani SY, Breed CA, Biggins S. Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol Cell Biol. 2006;26:2648–2660. doi: 10.1128/MCB.26.7.2648-2660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Babazadeh R, Furukawa T, Hohmann S, Furukawa K. Rewiring yeast osmostress signalling through the MAPK network reveals essential and non-essential roles of Hog1 in osmoadaptation. Sci Rep. 2014;4:4697. doi: 10.1038/srep04697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 99.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 100.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 101.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 102.Mazanek M, Mituloviae G, Herzog F, Stingl C, Hutchins JR, Peters JM, Mechtler K. Titanium dioxide as a chemo-affinity solid phase in offline phosphopeptide chromatography prior to HPLC-MS/MS analysis. Nat Protoc. 2007;2:1059–1069. doi: 10.1038/nprot.2006.280. [DOI] [PubMed] [Google Scholar]
- 103.Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protoc. 2006;1:1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 104.Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O'Kelly G, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol Cell Proteomics. 2006;5:737–748. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- 107.VanDerWalt S, Colbert SC, Varoquaux G. The NumPy array: A structure for efficient numerical computation. Computing in Science & Engineering. 2011;13 [Google Scholar]
- 108.Hunter JD. Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering. 2007;9 [Google Scholar]
- 109.Waskom M, Evans C, Warmenhoven J, Yarkoni T, Meyer KK, Rocher L, Hobson P, Halchenko Y, Koskinen M. Seaborn: v0.6.0. Zenodo. 2015 doi: 10.5281/zenodo.19108. [DOI] [Google Scholar]
- 110.Seabold S, Perktold J. paper presented at the Proceedings of the 9th Python in Science Conference; Austin, Texas, USA. June 28 - July 3 2010. [Google Scholar]
- 111.Chong YT, Koh JL, Friesen H, Duffy SK, Cox MJ, Moses A, Moffat J, Boone C, Andrews BJ. Yeast Proteome Dynamics from Single Cell Imaging and Automated Analysis. Cell. 2015;161:1413–1424. doi: 10.1016/j.cell.2015.04.051. [DOI] [PubMed] [Google Scholar]
- 112.Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.