Abstract
Clostridium difficile is an anaerobic, toxin-producing pathogen that causes human infection possibly through the consumption of meat. Clostridium difficile was isolated from 45 of 415 (10.8%) various raw meat samples collected in nationwide markets in Korea between 2013 and 2014. Among the 45 isolated strains, the highest prevalence rate was found in September (28.6%) and detected in chicken (16.4%), pork (8.3%) and beef (6.8%). According to an antibiotic resistance test, resistance was found only for clindamycin (2.2%). The genetic similarity of ribotypes O78 and O27 and strains isolated from raw meats was determined using DiversiLab. Among the isolates studied, four different rep-PCR types were identified, genetically distinct from ribotypes O78 and O27. An ELISA reaction confirmed that the two strains have toxin A and toxin B and showed 89% genetic similarity. This study suggests that food animals could be potential routes of foodborne transmission in C. difficile-associated human infection.
Keywords: Clostridium difficile, Toxin, Ribotypes, Antimicrobial resistance, Raw meat
Introduction
Clostridium difficile is an anaerobic, spore-forming gastrointestinal pathogen that causes disease in susceptible humans, and the disease can be acquired by the consumption of meats from food animals via toxin production [1]. Clostridium difficile can be isolated from varying percentages of raw meats, such as pigs, cows, lambs and chickens [2, 3]. Food animals could be a potential source of C. difficile and one of the transmission routes from animals to humans [4]. Moreover, C. difficile strains found in food animals are often those that are implicated in C. difficile infections (CDI) [5–7]. In the last decade, many large outbreaks of CDI have been described worldwide, and the incidence has increased in the United States, Canada, Europe and Asia [7–14]. The major symptoms of a CDI range from asymptomatic colonization to mild diarrhea and severe life-threatening pseudomembranous colitis [1, 4]. Its major virulence factors are toxin A (TcdA) and toxin B (Tcd B), which are designated as an enterotoxin and a cytotoxin, respectively. In Canada, between 1991 and 2003, the incidence of C. difficile-associated diarrhea (CDAD) increased 4.5-fold from 35.6 to 156.36 per 100,000 people, and the death rate has increased fivefold from 4.5 to 22% [15]. In the USA, the prevalence of ribotype 027 has been reported to be as high as 14% [16]. Since 2003, mortality related to CDAD has been increasing due to the emergence of mutant BI/NAP1/027, which can produce the C. difficile toxin at approximately 10–20 times the rate of other strains. Recently, the mutant was shown to be a cause of the morbidity and mortality [15, 17]. In addition, recent studies have suggested that ribotype O78, which is a toxinotype V strain, may be associated with the disease. Interestingly, there is an overlap between the PCR ribotypes that are found in humans and animals [18], raising the possibility of interspecies transmission and suggesting possible routes of exposure to CDI in humans. Of particular interest, the incidence of CDI has increased during recent years in Korea [12, 19–21]. CDI has increased sixfold in domestic hospitals [20] and Hyper-virulent strains such as NAP1/027 have been detected [22]. Also, response rate to initial antibiotic is low and death can occur due to sepsis or complication [20]. However, there has been limited investigation into C. difficile contamination in food animals, and the source of human infection remains unclear. Therefore, epidemiological studies are needed to prevent recurrence through analysis of the characteristics of strains. The objective of this study is to analyze the characteristics of C. difficile isolated from raw meats distributed in Korea and to suggest that food animals can be a potential route for C. difficile-associated human infection.
Materials and methods
Samples
A total of 415 samples, including raw beef, pork and chicken meat, were purchased from different retail outlets (supermarkets) between April 2013 and March 2014 in Korea. One hundred and forty-nine samples were prepared from chicken, and 133 samples were prepared from beef and pork. The samples were transported and stored at a temperature between 1 and 4 °C and analyzed within 4 h from the time of sampling.
Detection method
The detection method used was based on the method described by Rodriguez-Palacios et al. [23] as follows: 5 g of each sample were transferred to 20 mL of C. difficile broth (CDB; Oxoid SR0048) (C. difficile Moxalactam Norfloxacin broth + 0.1% taurocholate) and incubated at 37 °C for 15 days under anaerobic conditions. 2 mL of the enrichment broth were added to 2 mL of 96% ethanol in a centrifuge tube and homogenized for 50 min on a shaker at room temperature. After centrifugation (3800×g for 10 min), a loopful of the sediment was streaked onto Clostridium difficile moxalactam norfloxacin (CDMN) agar (Merck, Darmstadt, Germany) and TSA with 5% blood agar (bioMérieux, Maecy L’Etoile, France). The plates were incubated for 24–48 h at 37 °C under anaerobic conditions, and up to 5 suspected colonies were subcultured on tryptone soya agar (Oxoid CM0131). Presumptive identification of suspected colonies was performed by testing for biochemical characterization using an API rapid ID 32A (20A) test kit (bioMérieux DR1107A, Maecy L’Etoile, France).
Detection of toxins by VIDAS
The C. difficile strains were examined for toxins with the VIDAS-CDAB Kit using mini-VIDAS (bioMérieux, Maecy L’Etoile, France) according to the manufacturer’s instructions. For VIDAS CDAB, an aliquot (200 μL) of well-mixed C. difficile strains was added to 1 mL of diluent and centrifuged for 5 min at 12,000×g. The supernatant (300 μL) was added to the sample well of the CDAB Kit. This assay is completed in approximately 75 min. The interpretation of each assay result can be positive, negative or equivocal according to the fluorescence intensity as described in the relevant package insert for each assay.
Antibiotic susceptibility testing (AST)
Antimicrobial susceptibility testing was performed following the Clinical and Laboratory Standards Institute (CLSI) guidelines [24]. The susceptibilities of the isolates to metronidazole, vancomycin, clindamycin and moxifloxacin were measured by the E-test (AB-BIOdisc, Solna, Sweden). The breakpoints were as follows: metronidazole, ≥ 32 mg/L; vancomycin, ≥ 8 mg/L; clindamycin, ≥ 8 mg/L; and moxifloxacin, ≥ 8 mg/L. The E-test was applied to the surface of an agar plate inoculated with the C. difficile strains, and the strip was incubated for 48 h in an anaerobic chamber. After the required incubation period, and only when an even lawn of growth was distinctly visible, the MIC value was read where the edge of the inhibition ellipse intersected the side of the strip. Clostridium difficile ATCC 700057 and ATCC (92,93) served as the quality control strains.
Rep-PCR DiversiLab microbial typing system
The C. difficile strains were cultured on CDMN agar for 24 h at 37 °C in an anaerobic chamber. Genomic DNA was quantified by Nanodrop (Thermo Scientific, Pierce division), and the working concentration was adjusted to 35 ng/μg. Rep-PCR was performed using the DiversiLab C. difficile Kit (Bacterial BarCodes, Inc, Houston, TX), which includes rep-PCR master mix 1, C. difficile primers, and kit-specific positive and negative controls, in accordance with the manufacturer’s product insert. PCR was performed on a T gradient using the following parameters: initial denaturation at 94 °C for 2 min and then 35 cycles of PCR (94 °C for 30 s, 45 °C for 30 s, and 70 °C for 90 s), with a final extension of 70 °C for 3 min. The analysis of the rep-PCR products was implemented using a DiversiLab system in which the amplified fragments of various sizes and intensities were separated Isolated strain 45 fingerprint patterns were automatically downloaded onto a secure laboratory designated DiversiLab website. The patterns were analyzed by DiversiLab software, which uses Pearson correlation coefficients to determine the distance matrices. The agreement between the methods was assessed at different rep-PCR SI cutoffs, including 80, 85, and 90%, as generated by the DiversiLab software, and the relatedness was determined by a cluster analysis according to the guidelines provided by the manufacturer.
Results and discussion
Prevalence of C. difficile in meats
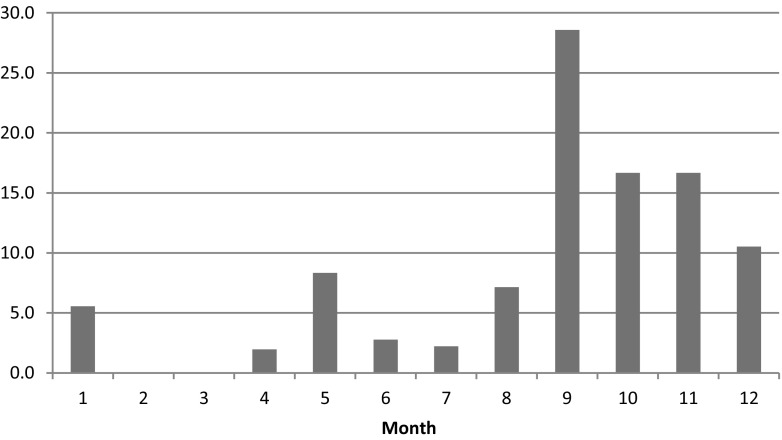
Between April 2013 and March 2014, a total of 415 raw meat samples were purchased from nationwide markets in Korea. Of these samples, 45 (10.8%) contained C. difficile. The highest prevalence of C. difficile was found in chicken at 16.4%, followed by pork at 8.3% and beef at 6.8% (Table 1). In other studies, the prevalence of C. difficile contamination in retail meat has varied widely, ranging from 0 to 42.4% [4–6, 23, 25, 26]. For example, one study found that 37 (42.0%) of 88 retail meat samples yielded C. difficile, including 42.4% in beef, 41.3% in pork, and 44.4% in turkey products [25]. However, C. difficile did not detect in meat samples other than beef [27]. Similar to results found in France [28]. An Austrian study reported a low prevalence of 3% for C. difficile in retail ground meat samples [29]. Contamination rates have varied from North America, where a high prevalence of C. difficile has been found, to Europe, where a lower prevalence has been reported [19, 28]. Such a large difference across countries in isolation frequencies of C. difficile from meat may reflect actual regional differences or may be caused by the use of different detection methods. Further improvements in the detection methods for C. difficile in foods is needed. Prevalence varied by season, with the highest prevalence (62%) observed between September and November (Fig. 1), September (29%), October (16%) and November (16%), Respectively. However, in Canada, the highest prevalence (11.5%) was observed between January and February [30]. In general, the prevalence patterns have differed across countries, and a seasonal difference has typically been observed. It is difficult to compare the detection rate because of various factors such as culture method and separation conditions. Therefore, standardized detection method is required for an accurate comparative analysis of the detection rate for across countries.
Table 1.
Prevalence of Clostridium difficile detected in beef, pork and chicken meat samples in Korea
| Meat type | No. of samples | No. of C. difficile positive samples |
|---|---|---|
| Beef | 133 | 9 (6.8%) |
| Pork | 133 | 11 (8.3%) |
| Chicken | 149 | 25 (16.8%) |
| Total | 415 | 45 (10.8%) |
Fig. 1.
A comparison between monthly isolation rate (%) of C. difficile in Korea
Investigation of the incidence of CDAD 027
In North America and Europe, increasing rates of Clostridium difficile-associated disease (CDAD) have been reported [5, 8, 16]. The hypervirulent strain in food is presumably associated with higher levels of toxin production by fluoroquinolone-resistant strains belonging to PCR ribotype 027, pulsed-field gel electrophoresis (PFGE) type NAP1, restriction endonuclease analysis (REA) type BI and toxinotype III [8]. The mutant BI/NAP1/027 can produce the C. difficile toxin A/B and binary toxin by altering the toxin-producing coordinator, making it approximately 10–20 times stronger than the regular strains [15, 17]. In Europe, outbreaks of CDAD caused by the new, highly virulent strain of C. difficile PCR ribotype 027, toxinotype III have been recognized in 75 hospitals in England, 16 hospitals in the Netherlands, 13 healthcare facilities in Belgium and nine healthcare facilities in France. Infections caused by BI/NAP1/027 seems to be strongly related to a very high mortality rate and severe clinical outcomes [8]. Active research is being conducted on C. difficile through North America and Europe. However, in Korea, diagnosis of CDAD associated diarrhea is conducted only in some large hospitals, and research related to this issue has been very rare. Therefore, investigation of toxin production, antimicrobial resistance profiles, and genetic diversity is necessary for better understanding C. difficile-associated disease (CDAD).
Characteristics of C. difficile ribotype, antimicrobial susceptibility and toxin genes
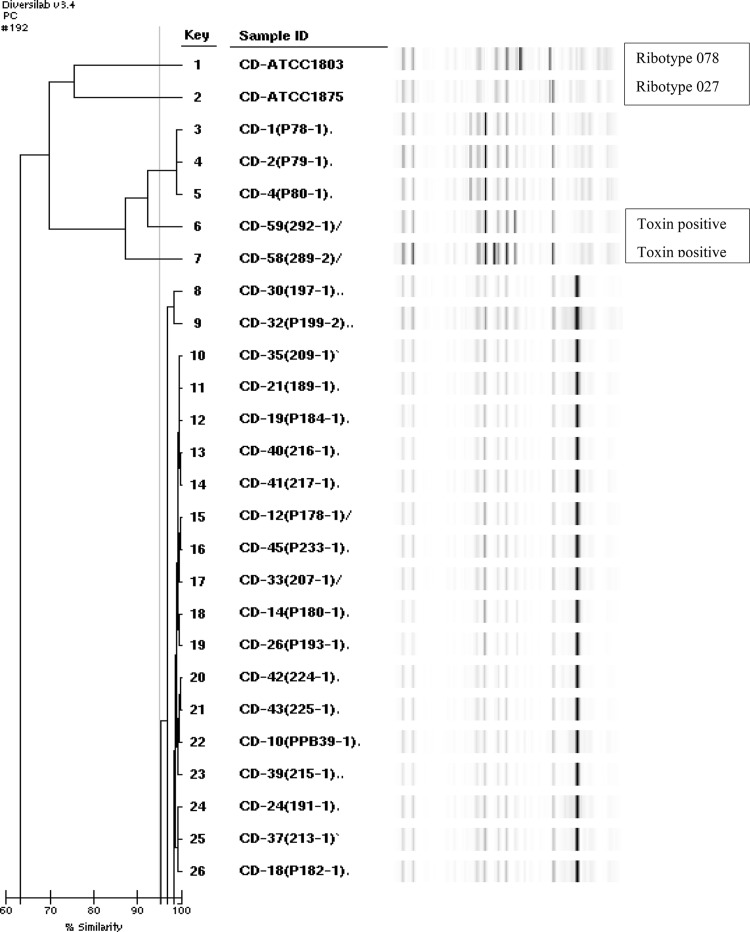
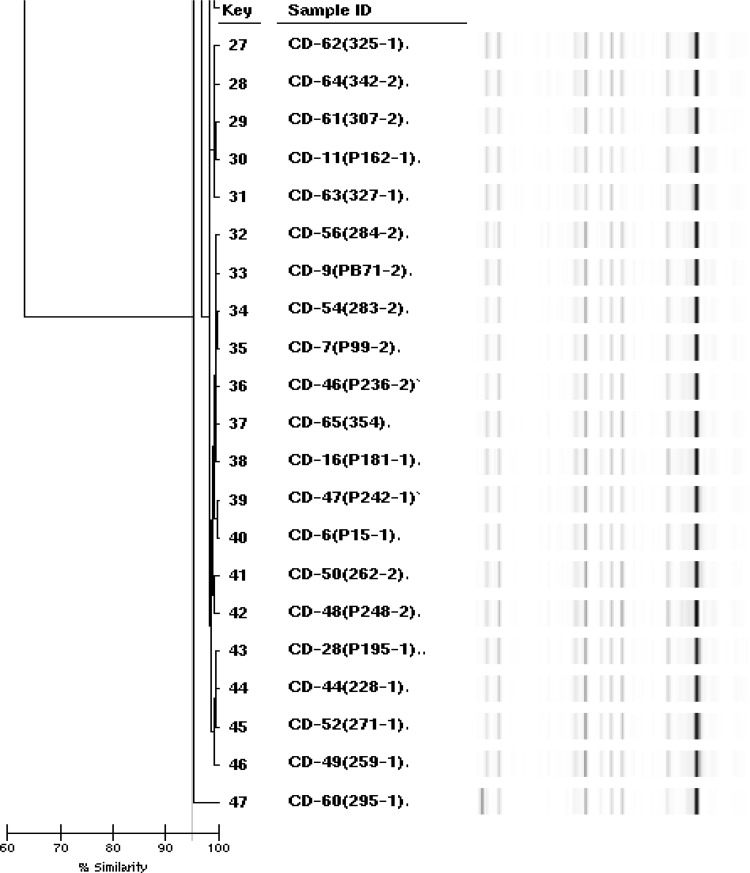
The genetic diversity of C. difficile was analyzed by rep-PCR method (DiversiLab) (Fig. 2). This method was useful in distinguishing several rep-PCR types of hypervirulent PCR ribotypes 027 and 078 [1]. Among the isolates studied, 4 different rep-PCR types, were identified as Type 1 (CD-1, 2, and 4), 2 (CD-59), 3 (CD-58) and 4 (40 strains). Type 4 was a dominant rep-PCR type, comprising 88.8% (40 isolates) of all isolates and Mean reproducibility gave a similarity index of 95.0% (Fig. 1). Ribotypes 027 and 078 were the genetically distinct strains isolated from meat products. But, the resistant 027 strain is increasing due to selective pressure from drug abuse [7] and the BI/NAP1/027 strain demonstrates a resistant pattern against fluoroquinolones such as moxifloxacin. Also, ribotype 027, which is highly susceptible to antimicrobials, has recently been emerging in Korea [12]. Thus warranting further study to prevent disease due to these strains. Clostridium difficile usually produces two toxins, toxin A (TcdA, an enterotoxin) and toxin B (TcdB, a cytotoxin) and is responsible for a range of diseases from mild diarrhea to pseudomembranous colitis [1, 31]. The VIDAS-CDAB test is a new ELFA that detects toxins A and B. Forty-five C. difficile isolates were isolated from 415 meats in culture. Among them, chicken-derived 2 strains had the genotype tcdA+/tcdB+ (4.4% prevalence) (Fig. 2), were susceptible to all antimicrobial agents. The strains were grouped into Type2 (sample ID cd-58) and Type3 (cd-59), respectively. In other study a total of 100 chicken fecal samples were collected from urban Zimbabwe and 29 (29%) contained C. difficile. Of these 29 strains, 26 (89.7%) contained toxin A + B + [32]. In addition C. difficile was isolated from twenty (17.4%) of 115 fecal samples in rural Zimbabwe. Of these 20 strains, 11 (55%) have toxin A + B+. Also, among C. difficile found in 22 soil samples around the bird, 95.5% had toxin A + B + [33]. Therefore, chicken can be a major reservoir of C. difficile. Detection of The toxins from symptomatic humans and food animals is important for diagnosis and prompt antimicrobial therapy [1].
Fig. 2.
rep PCR-ribotypes, VIDAS-CDAB A/B toxin gene profiles of C. difficile isolated from beef, pork, and chicken meat samples
Antibiotic resistance of all the isolates was measured for metronidazole, vancomycin, clindamycin and moxifloxacin by E-test method. Metronidazole and vancomycin are the primary therapeutic antibiotics of C. difficile. especially metronidazole has a high activity against C. difficile and the resistance rate is low [13]. No resistance was found for metronidazole (0%) and vancomycin (0%) among isolated tested. All strains were sensitive to metronidazole, but only one isolate (strain 7) was resistant to clindamycin. The resistance rate of clindamycin was 2.2% (Table 2) and derived from chicken. Also, intermediate is 28.9%, which means that the probability of going to resistance is high.
Table 2.
Antimicrobial resistance of 45 C. difficile strains isolated from beef, pork, and chicken meat samples in Korea
| Antimicrobial agent | Sensitive | Intermediate | Resistant |
|---|---|---|---|
| Metronidazole | 45 (100%) | 0 (0%) | 0 (0%) |
| Vancomycin | 43 (95.55%) | 2 (4.44%) | 0 (0%) |
| Clindamycin | 31 (68.88%) | 13 (28.88%) | 1 (2.22%) |
| Moxifloxacin | 43 (95.55%) | 2 (4.44%) | 0 (0%) |
Although toxin-producing strains of C. difficile turned out to be uncommon in Korea in our study, additional investigation is necessary because genetically diverse ribotypes among C. difficile strains from meat were found, suggesting the potential presence of other virulent strains. This study has been conducted to isolate C. difficile from raw meat, examine the molecular similarity, characterize the toxin and antimicrobial resistance profiles. Clostridium difficile is detected at relatively high prevalence in meat products of food animals. This suggests that the food animal is a potential route to human transmission. Therefore, further studies on C. difficile detected from food animals are needed to prevent C. difficile-associated disease (CDAD).
Acknowledgements
This research was supported by Main Research Program (E0156500-03) of the Korea Food Research Institute (KFRI) funded by the Ministry of Science, ICT.
References
- 1.Lyon SA. Clostridium difficile in healthy food animals and development of a PCR assay for detection in enriched food and fecal samples. PhD thesis, University of Georgia, Athens (2009)
- 2.Houser BA, Soehnlen MK, Wolfgang DR, Lysczek HR, Burns CM, Jayarao BM. Prevalence of Clostridium difficile toxin genes in the feces of veal calves and incidence of ground veal contamination. Foodborne Pathog. Dis. 2012;9:32–36. doi: 10.1089/fpd.2011.0955. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Palacios A, Stämpfli HR, Duffield T, Peregrine AS, Trotz-Williams LA, Arroyo LG, Brazier JS, Weese JS. Clostridium difficile PCR ribotypes in calves, Canada. Emerg retail ground meat, Canada. Emerg. Infect Dis. 2006;13:485–487. doi: 10.3201/eid1303.060988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Boer E, Zwartkruis-Nahuis A, Heuvelink AE, Harmanus C, Kuijper EJ. Prevalence of Clostridium difficile in retailed meat in the Netherlands. Int. J. Food Microbiol. 2011;144:561–564. doi: 10.1016/j.ijfoodmicro.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Harvey RB, Norman KN, Andrews K, Hume ME, Scanlan CM, Callaway TR, Anderson RC, Nisbet DJ. Clostridium difficile in poultry and poultry meat. Foodborne Pathog. Dis. 2011;8:1321–1323. doi: 10.1089/fpd.2011.0936. [DOI] [PubMed] [Google Scholar]
- 6.Hoffer E, Haechler H, Frei R, Stephan R. Low occurrence of Clostridium difficile in fecal samples of healthy calves and pigs at slaughter and in minced meat in Switzerland. J. Food Protect. 2010;73:973–975. doi: 10.4315/0362-028X-73.5.973. [DOI] [PubMed] [Google Scholar]
- 7.McDonald LC, Killgore GE, Thompson A, Owens RC, Jr, Kazakova SV, Sambol SP, Johnson S, Gerding DN. An epidemic, toxin gene–variant strain of Clostridium difficile. New Engl. J. Med. 2005;353:2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 8.Kuijper E, Coignard B, Tüll P. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin. Microbiol. Infec. 2006;12:2–18. doi: 10.1111/j.1469-0691.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 9.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M. A predominantly clonal multi-institutional outbreak of Clostridium difficile–associated diarrhea with high morbidity and mortality. New Engl. J. Med. 2005;353:2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 10.Thomas C, Stevenson M, Riley TV. Antibiotics and hospital-acquired Clostridium difficile-associated diarrhoea: a systematic review. J. Antimicrob. Chemoth. 2003;51:1339–1350. doi: 10.1093/jac/dkg254. [DOI] [PubMed] [Google Scholar]
- 11.Yan Q, Zhang J, Chen C, Zhou H, Du P, Cui Z, Cen R, Liu L, Li W, Cao B. Multilocus sequence typing (MLST) analysis of 104 Clostridium difficile strains isolated from China. Epidemiol. Infect. 2013;141:195–199. doi: 10.1017/S0950268812000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H, Lee Y, Moon HW, Lim CS, Lee K, Chong Y. Emergence of Clostridium difficile ribotype 027 in Korea. Kor. J. Lab. Med. 2011;31:191–196. doi: 10.3343/kjlm.2011.31.3.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang H, Weintraub A, Fang H, Wu S, Zhang Y, Nord CE. Antimicrobial susceptibility and heteroresistance in Chinese Clostridium difficile strains. Anaerobe. 2010;16:633–635. doi: 10.1016/j.anaerobe.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 14.George W, Rolfe R, Finegold S. Clostridium difficile and its cytotoxin in feces of patients with antimicrobial agent-associated diarrhea and miscellaneous conditions. J. Clin. Microbiol. 1982;15:1049–1053. doi: 10.1128/jcm.15.6.1049-1053.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pépin J, Valiquette L, Alary ME, Villemure P, Pelletier A, Forget K, Pépin K, Chouinard D. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. Can. Med. Assoc. J. 2004;171:466–472. doi: 10.1503/cmaj.1041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walk ST, Micic D, Jain R, Lo ES, Trivedi I, Liu EW. Almassalha LM, Ewing SA, Ring C, Galecki AT. Clostridium difficile ribotype does not predict severe infection. Clin. Infect. Dis. 2012;55:1661–1668. doi: 10.1093/cid/cis786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faris B, Blackmore A, Haboubi N. Review of medical and surgical management of Clostridium difficile infection. Tech. Coloproctol. 2010;14:97–105. doi: 10.1007/s10151-010-0574-3. [DOI] [PubMed] [Google Scholar]
- 18.Debast SB, Van Leengoed LA, Goorhuis A, Harmanus C, Kuijper EJ, Bergwerff AA. Clostridium difficile PCR ribotype 078 toxinotype V found in diarrhoeal pigs identical to isolates from affected humans. Environ. Microbial. 2009;11:505–511. doi: 10.1111/j.1462-2920.2008.01790.x. [DOI] [PubMed] [Google Scholar]
- 19.Hensgens MP, Keessen EC, Squire MM, Riley TV, Koene MG, de Boer E, Lipman LJ, Kuijper EJ. Clostridium difficile infection in the community: a zoonotic disease? Clin. Microbiol. Infec. 2012;18:635–645. doi: 10.1111/j.1469-0691.2012.03853.x. [DOI] [PubMed] [Google Scholar]
- 20.Lee YJ, Choi MG, Lim CH, Jung WR, Cho HS, Sung HY, Nam KW, Chang JH, Cho YK, Park JM. Change of Clostridium difficile Colitis during Recent 10 Years in Korea. Korean J. Gastroenterology. 2010;55:169–174. doi: 10.4166/kjg.2010.55.3.169. [DOI] [PubMed] [Google Scholar]
- 21.Lee JH, Lee SY, Kim YS, Park SW, Park SW, Jo SY, Ryu SH, Lee JH, Moon JS, Whang DH. The incidence and clinical features of Clostridium difficile infection; single center study. Korean J. Gastroenterology. 2010;55:175–182. doi: 10.4166/kjg.2010.55.3.175. [DOI] [PubMed] [Google Scholar]
- 22.Chung HT, Jung SA, Song HJ, Kim SE. First Case of Antibiotic-associated Colitis by Clostridium difficile PCR Ribotype 027 in Korea. J. Korean Med. Sci. 2009;24:520–524. doi: 10.3346/jkms.2009.24.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Palacios A, Staempfli HR, Duffield T, Weese JS. Clostridium difficile in retail ground meat, Canada. Emerg. Infect. dis. 2007;13:485–487. doi: 10.3201/eid1303.060988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hecht DW, Citron DM, Cox M. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard. Clin. Lab. Stand. Inst. 27: NO. 2 (2007)
- 25.Songer JG, Trinh HT, Killgore GE, Thompson AD, McDonald LC, Limbago BM. Clostridium difficile in retail meat products, USA, 2007. Emerg. Infect. Dis. 15: No. 5 (2009) [DOI] [PMC free article] [PubMed]
- 26.Weese J, Reid-Smith R, Avery B, Rousseau J. Detection and characterization of Clostridium difficile in retail chicken. Lett. Appl. Microbiol. 2010;50:362–365. doi: 10.1111/j.1472-765X.2010.02802.x. [DOI] [PubMed] [Google Scholar]
- 27.Martirani Von Abercron SM, Karlsson F, Trowald Wigh G, Wierup M, Krovacek K. Low occurrence of Clostridium difficile in retail ground meat in Sweden. J. Food Protect. 2009;72:1732–1734. doi: 10.4315/0362-028X-72.8.1732. [DOI] [PubMed] [Google Scholar]
- 28.Eckert C, Burghoffer B, Barbut F. Contamination of ready-to-eat raw vegetables with Clostridium difficile in France. J. Medi. Microbiol. 2013;62:1435–1438. doi: 10.1099/jmm.0.056358-0. [DOI] [PubMed] [Google Scholar]
- 29.Jöbstl M, Heuberger S, Indra A, Nepf R, Köfer J, Wagner M. Clostridium difficile in raw products of animal origin. Int. J. food microbiol. 2010;138:172–175. doi: 10.1016/j.ijfoodmicro.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 30.Gould LH, Limbago B. Clostridium difficile in food and domestic animals: a new foodborne pathogen? Clin. Infect. Dis. 2010;51:577–582. doi: 10.1086/655692. [DOI] [PubMed] [Google Scholar]
- 31.Ed J. Kuijper MD, Jaap T. van Dissel MD. Spectrum of Clostridium difficile infections outside health care facilities. Can. Medi. Assoc. 179: 747–748 (2008) [DOI] [PMC free article] [PubMed]
- 32.Simango C, Mwakurudza S. Clostridium difficile in broiler chickens sold at market places in Zimbabwe and their antimicrobial susceptibility. Int. J. Food Microbiol. 2008;124:268–270. doi: 10.1016/j.ijfoodmicro.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 33.Simango C. Prevalence of Clostridium difficile in the environment in a rural community in Zimbabwe. T. Roy. Soc. Trop. Med. H. 2006;100:1146–1150. doi: 10.1016/j.trstmh.2006.01.009. [DOI] [PubMed] [Google Scholar]