Abstract
Lactic acid bacteria are known for their preservative effects on food products like meat and sausage. Since they are related to humans, these bacteria require proper characterization and identification among various other bacteria in the surroundings. For their identification, several typing methods have already been applied of which the genotyping methods provide reproducible and unambiguous results. In this study, PCR-based method called repetitive element PCR was used for typing 37 Leuconostoc mesenteroides with three primers, REP, ERIC, and (GTG)5, annealing to repetitive sequences present in the bacterial genome. Different fingerprints were obtained for the isolates showing distinguishing profiles. Further phylogenetic analysis was performed using UPGMA method of clustering which provided proper identification with genetic relatedness of all the isolates. It was finally observed that, out of the three primers used, (GTG)5 discriminated the strains precisely than the other two.
Keywords: Lactic acid bacteria, DNA profiling, rep-PCR, ERIC-PCR, (GTG)5-PCR
Introduction
Some classes of bacteria are seen as beneficial to and are being used on an industrial scale human [1]. One such example is the lactic acid bacteria (LAB) which include genera Lactobacillus, Leuconostoc, Lactococcus, Pediococcus, and Streptococcus that play a significant role in the dairy industry and in the production of various fermented foods and supplements [2, 3]. Research on LAB has become a major endeavor of the century and certain strains are gaining attention for their probiotic potential [4, 5].
Leuconostoc is a Gram-positive, heterofermentative bacteria, found in natural green habitats, and are known starter cultures in the food industry, but are also responsible for the spoilage of some foods [6, 7]. Twenty three species of this genus have been identified and among them, Leuconostoc mesenteroides having applications in the dairy industry are directly or indirectly related to humans and its surroundings and therefore, their identification has become a necessity [8, 9]. Traditionally, the phenotypic methods were known to have a good discriminatory ability, but, the genotyping methods like random amplified polymorphic DNA (RAPD-PCR), pulsed-field gel electrophoresis (PFGE), restriction fragment length polymorphism (RFLP), repetitive-element palindromic PCR (rep-PCR), and protein fingerprinting gained more popularity [6]. Owing to their strong discriminatory ability and better reliability, it is helpful for characterization of bacterial species at the subspecies and strain levels, than the traditional phenotypic methods that lack resolving power [10, 11].
A PCR technique, rep-PCR, is able to determine phylogeny among closely related species [12, 13]. In addition, rep-PCR typing is known to yield better and unambiguous results with a strong discriminatory ability [14] and is a practical tool that can differentiate the species by using short oligonucleotide primers [11, 15]. In the present study, three markers, REP (REP1R-I and REP2-I), ERIC (ERIC1R and ERIC2), and (GTG)5 were used for distinguishing LAB strains isolated from various fermented food materials in South Korea that have been earlier to analyze the genetic variability among certain strains [17, 18]. These primers target the highly conserved elements like repetitive extragenic palindromic elements and enterobacterial repetitive intragenic consensus elements, whose amplification provides a genetic fingerprinting pattern of the bacterial strains [16].
Materials and methods
The number of isolates and their growth
Various vegetables were used to extract 37 L. mesenteroides strains which have been identified using 16S rRNA gene sequencing and the sequences have been submitted in GenBank under the accession numbers KX289500 to KX289531. The sources included kimchi (11), radish (10), Chinese cabbage kimchi (4), radish kimchi (2), young radish (4), salted clam (2), salted fish (1), soya bean paste (1), onion (1), and salted small octopus (1) from different regions of South Korea. The strains were grown in MRS broth (Difco Laboratories, Detroit, MI, USA) and incubated overnight at 37 °C.
DNA isolation
The genomic DNA from all the 37 isolates of L. mesenteroides was extracted by means of the AccuPrep Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea) [19].
rep-PCR
The PCR was performed using three primers synthesized by Bioneer, of which, two primers worked in pairs (forward and reverse), REP1R-I (5′-NNNNCGNCGNCATCNGGC-3′) as forward and REP2-I (5′-NCGNCTTATCNGGCCTAC-3′) as reverse, and ERIC1R (5′-ATGTAAGCTCCTGGGGATTCA-3′) as forward and ERIC2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′) as reverse primers, whereas (GTG)5 (5′-GTGGTGGTGGTGGTG-3′) [18] acted as both, forward and reverse primer. The reaction mixture from a PCR premix kit (Bioneer: 10 mM Tris–HCl, pH 9.0, 40 mM KCl, 1.5 mM of MgCl2, 250 µM dNTPs mix, 1 U of Taq polymerase) was used for amplification; 2 µL of each primer (10 pmol/µL) along with 2 µL of DNA template was added into the reaction mixture with subsequent addition of 16 µL of distilled water to make the final volume of 20 µL. The amplification was carried out in a thermal cycler (My cycler, Bio-Rad, CA, USA) at the following primer conditions:
REP1R-I and REP2-I
Pre-denaturation at 95 °C for 7 min followed by 35 cycles of denaturation at 95 °C for 1 min, annealing at 41 °C for 1 min, and extension at 65 °C for 3 min. Final extension was carried out at 65 °C for 16 min.
ERIC-PCR conditions
Pre-denaturation at 94 °C for 3 min, then 35 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 1 min 30 s, and extension at 68 °C for 8 min. Final extension was done at 68 °C for 8 min.
(GTG)5-PCR conditions
Pre-denaturation at 95 °C for 7 min, 4 cycles of PCR were carried out: denaturation at 95 °C for 2 min, annealing at 36 °C for 2 min, and extension at 72 °C for 2 min followed by 30 cycles of another PCR: denaturation at 95 °C for 1 min, annealing at 50 °C for 1 min, and extension at 72 °C for 1 min. The conclusive extension step was performed at 72 °C for 5 min. All the amplicons with each primer were stored at 4 °C [18].
Agarose gel electrophoresis
The PCR amplification was visualized with agarose gel electrophoresis in 1.5% agarose gel containing 0.05 µL/mL ethidium bromide and 1 × TAE buffer at a constant voltage of 70 V and a temperature of 4 °C for 6 h [3]. The band pattern was examined under UV light imaged using a gel documentation system (Bio-Rad Gel Doc XR+, Bio-Rad, CA, USA). A 100 bp extended range DNA ladder (Lonza, Rockland, NY, USA) and 1 kb DNA ladder (Takara Bio Inc., Shiga, Japan) were used as standard molecular markers.
Binary characters were applied to encode the bands, where 1 corresponded to the presence of a band and 0 corresponded to the absence of a band. The unweighted pair group method with an arithmetic mean (UPGMA) was used to analyze the correlation and similarity among the isolates in the NTSYS software.
Results and discussion
Evaluation of rep-PCR with primers
All three primer sets were initially standardized for the annealing temperature conditions. The amounts and ratios of the primers were also optimized using three isolates, 11,364, 11,392, and 11,436.
PCR with primers REP1R-I and REP2-I generated a profile of 296 in total having bands ranging from 100 to 3000 bp, corresponding to a maximum band pattern of 13 bands in isolate 11,289 and the minimum of 3 bands in isolate 11,460 (Fig. 1A). Except for three isolates 11,260, 11,392, and 11,416, a 300 bp band was found to be common to all the isolates. Similarly, a 1300 bp band was shared by all the isolates except for isolate 11,289 (from soybean paste). There were 268 bands with the (GTG)5 primer and they ranged from 200 to 1650 bp (Fig. 3A). The largest number of bands was yielded by three isolates namely, 11,259, 11,260, and 11,392, and the lowest number of bands was produced by a single isolate (11,426) with (GTG)5 primer. Unique bands of 1650 and 1500 bp were seen in isolate 11,259 extracted from soybean paste. Another band (1600 bp) was observed only in a single sample from onion namely, isolate 11,260. In addition, a band of 1500 bp was seen in isolate 11,392 (extracted from salted small octopus). A 200 bp band was observed only in 3 isolates that were extracted from soybean paste (isolate 11,259) and kimchi (isolates 11,438 and 11,439). Similar analysis by ERIC-PCR yielded a lesser number of bands than did the other primers, totaling 154 bands. The bands ranged between 200 and 2500 bp, showing a maximum number of 12 bands in isolate 11,260 and a minimum of 1 band in isolate 11,439 (Fig. 2A). A large number of bands with primers REP1R-I + REP2-I and (GTG)5 among the isolates helped to discriminate the strains more conveniently and precisely. The results produced differ with respect to the fingerprint patterns. According to the gel images with three primer sets, most of the strains sharing a common source seem to have similar arrangements of bands, e.g., isolates extracted from kimchi, 11,357, 11,359, 11,360, and 11,361. However, with REP1R-I and REP2-I, isolates 11,358 and 11,362 from kimchi share a common profile with the other isolates from kimchi, but, it did not seem similar for ERIC and (GTG)5 PCR profiles.
Fig. 1.
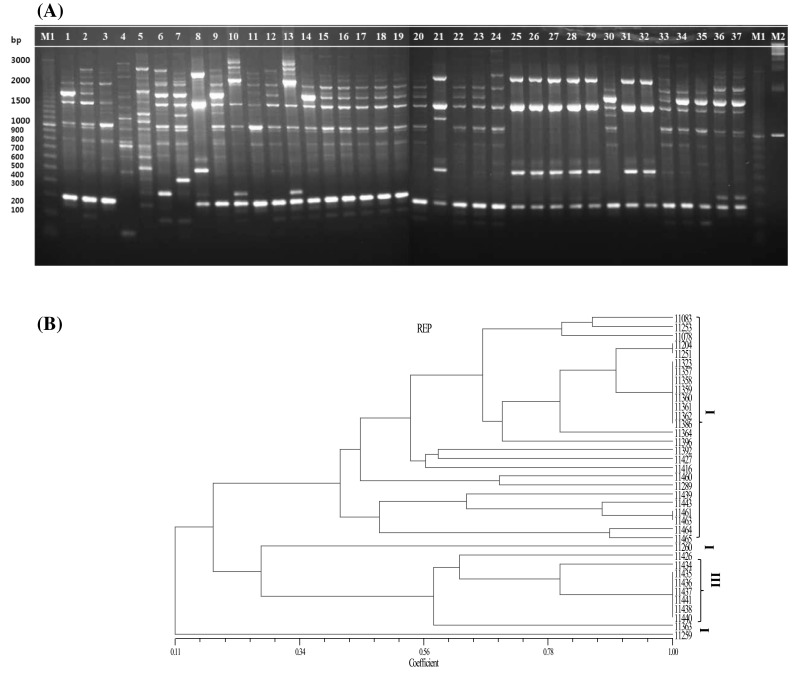
rep-PCR fingerprinting profiles of 37 isolates of Leuconostoc mesenteroides using REP1R-I + REP2-I primers. A Agarose gel electrophoresis. (Left to Right) M1: a 100 bp ladder of markers; 1-11,083; 2-11204; 3-11,253; 4-11,259; 5-11,260; 6-11,392; 7-11,416; 8-11,426; 9-11,427; 10-11,460;11-11,078; 12-11,251; 13-11,289; 14-11,323; 15-11,357; 16-11,358; 17-11,359; 18-11,360; 19-11,361; 20-11,362; 21-11,363; 22-11,364; 23-11,386; 24-11,396; 25-11,434; 26-11,435; 27-11,436; 28-11,437; 29-11,438; 30-11,439; 31-11,440; 32-11,441; 33-11,443; 34-11,461; 35-11,463; 36-11,464; 37-11,465; M1: 100 bp ladder of markers; M2: 1 kb ladder of markers. B UPGMA dendrogram of 37 Leuconostoc mesenteroides isolates
Fig. 3.
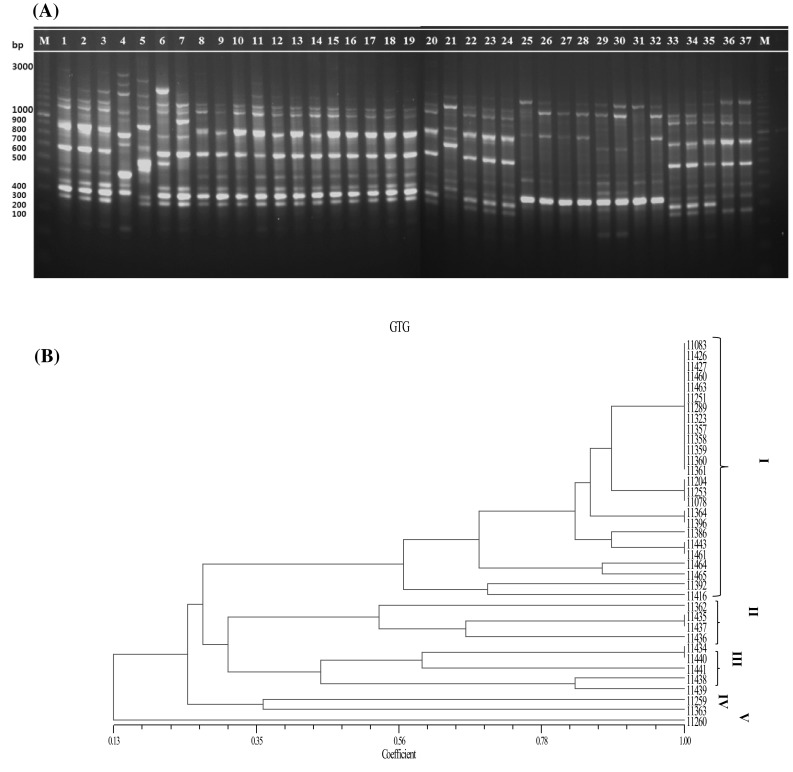
rep-PCR fingerprinting profiles of 37 isolates of Leuconostoc mesenteroides using (GTG)5 primers. A (Left to Right) M: a 100 bp ladder of markers; 1-11,083; 2-11,204; 3-11,253; 4-11,259; 5-11,260; 6-11,392; 7-11,416; 8-11,426; 9-11,427; 10-11,460; 11-11,078; 12-11,251; 13-11,289; 14-11,323; 15-11,357; 16-11,358; 17-11,359; 18-11,360; 19-11,361; 20-11,362; 21-11,363; 22-11,364; 23-11,386; 24-11,396; 25-11,434; 26-11,435; 27-11,436; 28-11,437; 29-11,438; 30-11,439; 31-11,440; 32-11,441; 33-11,443; 34-11,461; 35-11,463; 36-11,464; 37-11,465; M: a 100 bp ladder of markers. B UPGMA dendrogram of 37 Leuconostoc mesenteroides isolates
Fig. 2.
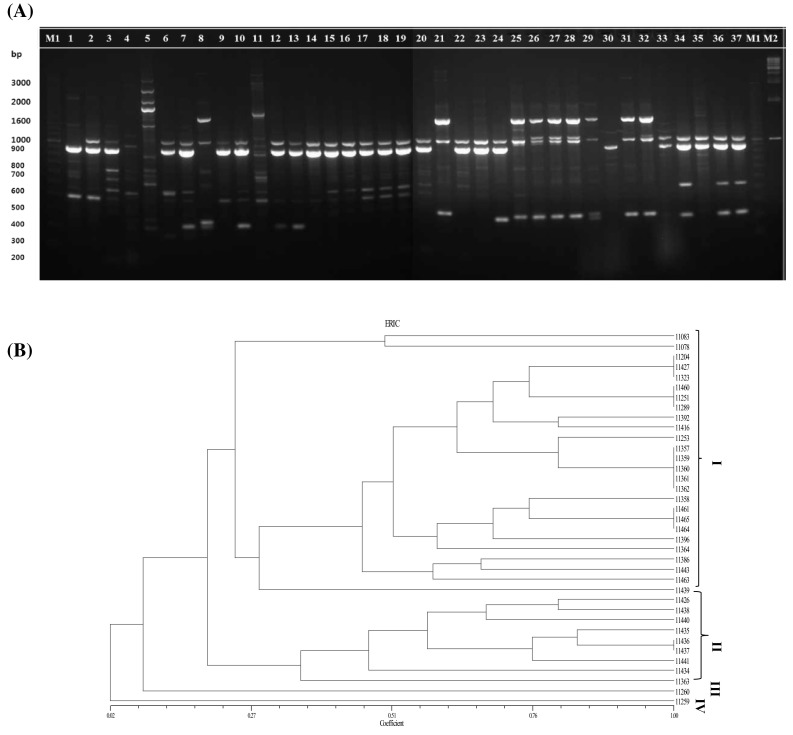
rep-PCR fingerprinting profiles of 37 isolates of Leuconostoc mesenteroides using ERIC1R + ERIC2 primers. A Agarose gel electrophoresis. (Left to Right) M: a 100 bp ladder of markers; 1-11,083; 2-11,204; 3-11,253; 4-11,259; 5-11,260; 6-11,392; 7-11,416; 8-11,426; 9-11,427; 10-11,460;11-11,078; 12-11,251; 13-11,289; 14-11,323; 15-11,357; 16-11,358; 17-11,359; 18-11,360; 19-11,361; 20-11,362; 21-11,363; 22-11,364; 23-11,386; 24-11,396; 25-11,434; 26-11,435; 27-11,436; 28-11,437; 29-11,438; 30-11,439; 31-11,440; 32-11,441; 33-11,443; 34-11,461; 35-11,463; 36-11,464; 37-11,465; M1: a 100 bp ladder of markers; M2: a 1 kb ladder of markers. B UPGMA dendrogram of 37 Leuconostoc mesenteroides isolates
These results were further analyzed by the phylogenetic details of the strains. Using the Unweighted pair group method with arithmetic mean (UPGMA) algorithm, four clusters were formed with REP1R-I + REP2-I and ERIC primers (Figs. 1B, 2B) in contrast to five clusters with (GTG)5 primer (Fig. 3B). The observations were similar with those obtained from the banding pattern and the maximum number of strains were linked to cluster I in all the rep-PCR profiles. For REP1R-I + REP2-I and ERIC profiles, the same 26 strains were grouped into cluster I, whereas 25 strains found in (GTG)5 cluster I were different from the ones found in the other two. It could be inferred that all the strains that were highly similar were isolated mostly from the same source. The strains in other clusters were also discriminated well. Other studies have shown that with REP1R-I + REP2-I and (GTG)5 primers, various LAB strains are typed, but only Lactobacillus strains yield a good profile [17, 20]. In the present analysis, primers REP1R-I + REP2-I and (GTG)5 yielded higher band intensity than ERIC primer and produced reliable results with the strains of L. mesenteroides as well.
Contrary to the REP1R-I + REP2-I and ERIC primers, (GTG)5 was able to discriminate between some of the strains that the other two primer sets could not. Moreover, ERIC and REP1R-I + REP2-I profiles showed similar discriminative abilities, whereas the (GTG)5 profiles formed 5 clusters thus differentiating the strains more precisely at the molecular level. Genotyping with the (GTG)5 primer is known to have a strong intraspecies discriminatory ability that can be used for a large number of LAB strains [3]. Many other researchers have obtained reproducible results differentiating LAB isolates at the strain level with ERIC-PCR [17, 20, 21], in contrast to the present study.
Previously, rep-PCR has proven to be an easy and reliable fingerprinting technique, with the (GTG)5 primer yielding reproducible results for various food-related bacterial strains [3]. In the present study, three rep-PCR primers have been compared and analyzed for 37 isolates of L. mesenteroides from food samples which have been precisely differentiated, providing data on the fingerprinting pattern and their genetic relatedness. It is seen that rep-PCR is feasible, reproducible and reliable, where the results can be obtained within a day [12, 13, 15]. Previously, L. mesenteroides strains have been identified and distinguished with respect to other LAB [20, 22, 23], but such a work on investigating only the L. mesenteroides strains rep-PCR has not been published, specifically in South Korea.
Acknowledgements
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through the High Value-added Food Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA; Grant Number: 314073-03-2-HD040).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Stiles ME, Holzapfel WH. Lactic acid bacteria of foods and their current taxonomy. Int. J. Food Microbiol. 1997;36:1–29. doi: 10.1016/S0168-1605(96)01233-0. [DOI] [PubMed] [Google Scholar]
- 2.Rattanachaikunsopon P, Phumkhachorn P. Lactic acid bacteria: their antimicrobial compounds and their uses in food production. Ann. Biol. Res. 2010;1:218–228. [Google Scholar]
- 3.Gevers D, Huys G, Swings J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 2001;205:31–36. doi: 10.1111/j.1574-6968.2001.tb10921.x. [DOI] [PubMed] [Google Scholar]
- 4.Chagnaud P, Machinis K, Coutte LA, Marecat A, Mercenier A. Rapid PCR-based procedure to identify lactic acid bacteria: application to six common Lactobacillus species. J. Microbiol. Methods. 2001;44:139–148. doi: 10.1016/S0167-7012(00)00244-X. [DOI] [PubMed] [Google Scholar]
- 5.Muñoz MCC, Benomar N, Lerma LL, Gálvez A, Abriouel H. Antibiotic resistance of Lactobacillus pentosus and Leuconostoc pseudomesenteroides isolated from naturally-fermented Aloreña table olives throughout fermentation process. Int. J. Food Microbiol. 2014;172:110–118. doi: 10.1016/j.ijfoodmicro.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 6.Dan T, Liu W, Sun Z, Lv Q, Xu H, Song Y, Zhang H. A novel multi-locus sequence typing (MLST) protocol for Leuconostoc lactis isolates from traditional dairy products in China and Mongolia. BMC Microbiol. 2014;14:150. doi: 10.1186/1471-2180-14-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hemme D, Foucaud-Scheunemann C. Leuconostoc, characteristics, use in dairy technology and prospects in functional foods. Int. Dairy J. 2004;14:467–494. doi: 10.1016/j.idairyj.2003.10.005. [DOI] [Google Scholar]
- 8.Kot W, Neve H, Heller KJ, Vogensen FK. Bacteriophages of Leuconostoc, Oenococcus, and Weissella. Front. Microbiol. 2014;5:186. doi: 10.3389/fmicb.2014.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohania D, Nagpal R, Kumar M, Bhardwaj A, Yadav M, Jain S, Marotta F, Singh V, Parkash O, Yadav H. Molecular approaches for identification and characterization of lactic acid bacteria. J. Dig. Dis. 2008;9:190–198. doi: 10.1111/j.1751-2980.2008.00345.x. [DOI] [PubMed] [Google Scholar]
- 10.Amor KB, Vaughan EE, Vos WM. Advanced molecular tools for the identification of lactic acid bacteria. J. Nutr. 2007;137:741S–747S. doi: 10.1093/jn/137.3.741S. [DOI] [PubMed] [Google Scholar]
- 11.Rademaker JLW, Hoste B, Louws FJ, Kersters K, Swings J, Vauterin L, Vauterin P, Bruijn FJ. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA–DNA homology studies: Xanthomonas as a model system. Int. J. Syst. Evol. Microbiol. 2000;50:665–677. doi: 10.1099/00207713-50-2-665. [DOI] [PubMed] [Google Scholar]
- 12.Tafvizi F, Ebrahimi MT. Application of repetitive extragenic palindromic elements based on PCR in detection of genetic relationship of lactic acid bacteria species isolated from traditional fermented food products. J. Agric. Sci. Technol. 2015;17:87–98. [Google Scholar]
- 13.Manzano M, Giusto C, Iacumin L, Cantoni C, Comi G. Molecular methods to evaluate biodiversity in Bacillus cereus and Bacillus thuringiensis strains from different origins. Food Microbiol. 2009;26:259–264. doi: 10.1016/j.fm.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Olive DM, Bean P. Principles and applications of methods for DNA based typing of microbial organisms. J. Clin. Microbiol. 1999;37:1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee CM, Sieo CC, Cheah YK, Abdullah N, Ho YW. Discrimination of probiotic Lactobacillus strains for poultry by repetitive sequenced based PCR fingerprinting. J. Sci. Food Agric. 2012;92:660–666. doi: 10.1002/jsfa.4627. [DOI] [PubMed] [Google Scholar]
- 16.Vuyst LD, Camu N, Winter TD, Vandemeulebroecke K, Perre VV, Vancanneyt M, Vos PD, Cleenwerck I. Validation of the (GTG)5-rep-PCR fingerprinting technique for rapid classification and identification of acetic acid bacteria, with a focus on isolates from Ghanaian fermented cocoa beans. Int. J. Food Microbiol. 2008;125:79–90. doi: 10.1016/j.ijfoodmicro.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 17.Stephenson DP, Moore RJ, Allison GE. Comparison and utilization of repetitive-element PCR techniques for typing Lactobacillus isolates from the chicken gastrointestinal tract. Appl. Environ. Microbiol. 2009;75:6764–6776. doi: 10.1128/AEM.01150-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Versalvoic J, Schneider M, Bruijn FJ, Lupski JR. Genomic fingerprinting of bacteria using repetitive sequence based polymerase chain reaction. Methods Mol. Biol. 1994;5:25–40. [Google Scholar]
- 19.Kaur J, Lee S, Park Y-S, Sharma A. RAPD analysis of Leuconostoc mesenteroides strains associated with vegetables and food products from Korea. LWT Food Sci. Technol. 2017;77:383–388. doi: 10.1016/j.lwt.2016.11.078. [DOI] [Google Scholar]
- 20.Ruiz P, Sesena S, Palop ML. A comparative study of different PCR-based DNA fingerprinting techniques for typing of lactic acid bacteria. Eur. Food Res. Technol. 2014;239:87–98. doi: 10.1007/s00217-014-2197-9. [DOI] [Google Scholar]
- 21.Saito S, Kobayashi M, Nira HK, Aoki R, Mizumachi K, Miyata S, Yamamoto K, Kitagawa Y, Suzuki C. Intraspecies discrimination of Lactobacillus paraplantarum by PCR. FEMS Microbiol. Lett. 2011;316:70–76. doi: 10.1111/j.1574-6968.2010.02193.x. [DOI] [PubMed] [Google Scholar]
- 22.Terzic-Vidojevic A, Mihajlovic S, Uzelac G, Veljovic K, Tolinacki M, Nikolic M, Topisirovic L, Kojic M. Characterization of lactic acid bacteria isolated fromm artisanal Travnik young cheeses, sweet creams and sweet kajmaks over four seasons. Food Micrbiol. 2014;39:27–38. doi: 10.1016/j.fm.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Ouadghiri M, Vancanneyt M, Vandamme P, Naser S, Gevers D, Lefebvre K, Swings J, Amar M. Identification of lactic acid bacteria in Moroccan raw milk and traditionally fermented skimmed milk ‘Iben’. J. Appl. Microbiol. 2009;106:486–495. doi: 10.1111/j.1365-2672.2008.04016.x. [DOI] [PubMed] [Google Scholar]


