Abstract
The aims of this work were to investigate the effects of gamma irradiation on population, viability and aflatoxin B1 production of Aspergillus flavus strains isolated from nutmeg kernels. Thirteen strains of A. flavus were isolated and cultured in potato dextrose agar. Conidia were harvested, air-dried, and irradiated 0, 5, or 10 kGy with gamma rays using a Cobalt-60 source. Toxigenicity were determined using a coconut agar medium and thin layer chromatography. Genomes of strains were extracted from mycelia. Four sets of primers, regulatory (aflR) and structural genes (nor-1, ver-1, omt-1) were used to confirm the presence of the genes. Our results indicate that total fungal populations decreased significantly (P < 0.05) with increasing irradiation dose. However, doses of 5 and 10 kGy were insufficient to completely eliminate the viability of some A. flavus strains. Irradiation did not change toxigenicity and triggered surviving toxigenic strains to produce aflatoxin B1.
Keywords: Aspergillus flavus, Toxigenic, Non-toxigenic, Gamma irradiation, Aflatoxin B1
Introduction
Aflatoxins are secondary metabolites produced by some strains of Aspergillus section Flavi [1, 2]. These toxins are carcinogenic and extremely toxic. They often contaminate agricultural commodities including crops, herbs, and spices. The International Agency of Research on Cancer (IARC) has classified aflatoxins as Group I carcinogens [3]. Part of the Aspergillus section Flavi, Aspergillus flavus, is primarily found in tropical regions, but it is distributed worldwide. Regions with high temperatures and relative humidity favor fungal infections that cause spoilage and aflatoxin contamination [4, 5]. Fungal infection may occur during pre-harvest, harvest, and post-harvest handling (e.g., drying, storage, and transportation) [6]. There are many diverse strains of toxigenic and non-toxigenic A. flavus in nature [7, 8]. Each strain of A. flavus varies considerably in its ability to produce aflatoxin [9, 10]. Aflatoxin production by A. flavus is determined by fungal genes, substrate availability, geographic area, climate, and plant cultivation techniques [7, 11, 12].
Gamma irradiation is used to preserve agricultural commodities including crops, herbs, and spices. Application of gamma irradiation not only control fungal infection, but also degrades aflatoxin. The European Commission’s Scientific Committee on Food has determined that dried aromatic herbs and spices treated with gamma irradiation dose up to 10 kilogray (kGy) are safe [13]. Some reports have suggested that gamma irradiation reduces fungal population and aflatoxin concentration [14]. However, the sensitivity of fungi to irradiation depends on the fungal strain, number of fungi, moisture content of spores, and developmental state of fungi [15]. Exposing strains of A. flavus to gamma irradiation has the potential to make strains more toxigenic by increasing their aflatoxin production [16]. Currently, few studies have characterized A. flavus strains associated with nutmeg kernels and determined the effects of gamma irradiation on these strains. Our interest was (1) to enumerate different A. flavus strains after exposure to different doses of gamma irradiation, (2) to analyze toxigenicity of the surviving strains, and (3) to detect the presence of regulatory and structural genes involved in aflatoxin biosynthesis in these strains.
Materials and methods
Aspergillus flavus strains
Thirteen strains of A. flavus were isolated from nutmeg kernels obtained from the Kauditan sub-district, North Minahasa Regency, North Sulawesi Province, Indonesia. All strains were part of the culture collection at the Phytopathology Laboratory of the Southeast Asian Regional Centre for Tropical Biology (SEAMEO BIOTROP) in Bogor, Indonesia. Each isolate was sub-cultured for 7 days at 28 °C in petri dishes (9 cm diameter) containing potato dextrose agar (PDA, Difco Laboratories, Spark, MD, USA).
Harvesting conidia and gamma irradiation
Asexual spores (conidia) were harvested from the sub-cultured strains by adding 10 mL of 0.05% sterile Tween 80 to the surface the petri dish containing fungi. Colonies were scrapped off the plate with a small sterilized brush. Spores in the suspension were counted using a hemocytometer and diluted to a final concentration of 107 spores per mL. Then, 1 mL of the diluted conidial suspension was transferred to a 1.5 mL microcentrifuge tube. Tubes were centrifuged at 6000×g for 15 min. Supernatants were discarded and conidia were air-dried for 24 h at 28 °C. Dried conidia were irradiated at dose either 5 or 10 kGy in a 60Cobalt source that emits gamma rays with an activity of 878.339 kilocurie (kCi). Irradiation was performed at the Center for Application of Isotope and Radiation Technology of the National Nuclear Energy Agency in Jakarta, Indonesia. Experiments were done in triplicate for each A. flavus strain and irradiation dose. Conidia with no irradiation were used as control treatment. After irradiation, all conidia were re-suspended immediately in 1 mL of sterile distilled water. Serial dilutions (10−1–10−4 conidia mL−1) were made and plated onto aspergillus flavus and parasiticus agar (AFPA) (10 g L−1 peptone, 20 g L−1 yeast extract, 0.5 g L−1 ferric ammonium citrate, 100 mg L−1 chloramphenicol, 18 g L−1 bacto agar, and 2 mg L−1 dichloran) [17]. After incubating the plates for 6 days at 28 °C, CFUs of irradiated and non-irradiated conidia were counted and the percentage of surviving conidia were determined. Conidial viability also was observed microscopically based on the presence of conidial germtube.
Qualitatively determining toxin production using fluorescence
To determine if strains produced aflatoxin B1 (AFB1), all fungal strains were inoculated on coconut agar medium (CAM) (36 g L−1 bacto agar and 100 mL L−1 of coconut cream extracted from freshly shredded coconut endosperm) [18]. The medium was adjusted to pH 7.0 using 2 N NaOH and sterilized for 20 min at 120 °C. A small fragment of a colony was inoculated onto the center of a petri dish (9 cm in diameter) containing CAM and incubated for 5 days, 28 °C. A yellow pigment was observed when the plate was examined from the top and from the bottom. The presence or absence of blue fluorescence on each plate was determined using long-wave (365 nm) ultra violet (UV) light. An uninoculated plate was used as a reference.
Measuring AFB1 using thin layer chromatography (TLC)
Quantitative assessment of AFB1 production was done using TLC according to Bainton et al. [19]. A colony of A. flavus (from a CAM plate incubated at 28 °C for 5 days showing blue fluorescence was mixed with 50 mL of ethanol in a waring blender; the suspension was extracted for 30 min and filtered using filter paper (Whatman #1). The filtrate then was transferred to a 250 mL separating funnel and extracted twice with 50 mL of n-hexane and cleaned with 50 mL of chloroform. The extract was dehydrated in a vial and filtered using anhydrate sodium sulfate (Na2SO4). Using a microsyringe, 10 µL of the residue was spotted onto a TLC plate (MERCK #1.05554, Silica gel 60, F254) and run for 20 min. The developing solvent was chloroform:acetone (9:1). Commercially available aflatoxins were used as standards (Sigma-Aldrich Chemical Company, USA).
Deoxyribonucleic acid (DNA) extraction
We also used PCR to detect toxigenic and non-toxigenic strains using a molecular method. Each A. flavus strains was sub-cultured in a 125 mL flask containing 75 mL of potato dextrose broth (PDB) and grown for 7 days at 28 °C. Mycelia were filtered through a Buchner funnel with sterile filter paper (Whatman #1). Mycelia were rinsed twice with sterile distilled water using centrifugation. After the final centrifugation, supernatants were removed and samples were frozen at − 80 °C. Fungal mycelia (approximately 100 mg wet weight) were transferred to a mortar and pestle containing liquid nitrogen and ground to a fine powder. DNA was extracted from homogenized mycelia using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Briefly, as much as 400 µL of lysis buffer [50 mM Tris–HCl (pH 8.0), 10 mM ethylenediamine tetraacetic acid (EDTA) (pH 8.0), 0.1% sodium dodecyl sulfate (SDS), 0.1 M NaCl], and 4 µL RNase were added. The suspension was incubated at 65 °C for 10 min. One hundred and thirty µL of P3 buffer [3.0 M potassium acetate (pH 5.5)] were added and the mixture was incubated for 5 min on ice. Then, the lysate was centrifuged at 20,000×g for 5 min. The supernatant was filtered using a Q1 shredder minispin column that was placed in a 2 mL collection tube and centrifuged 20,000×g for 2 min using a DNA binding column. As much as 1.5 volumes of AW1 buffer was added to the supernatant. The mixture was transferred to a DNeasy minispin column, which was placed in a 2 mL collection tube. The sample was incubated for 5 min at room temperature and centrifuged at 6000×g for 1 min. Columns were washed twice with 500 µL of AW2 buffer by centrifugation at 20,000×g for 2 min; 100 µL of AE buffer were used for the elution. After adding elution buffer, the sample was incubated for 5 min at room temperature (15–25 °C) and then centrifuged at 6000×g for 1 min. Subsequently, DNA was examined by agarose gel (1%) electrophoresis.
Gene amplification by quadruplex polymerase chain reaction (PCR)
The GeneAmp PCR System 9700 (Roche Molecular Systems Inc., USA) was used to amplify the regulatory gene, aflR1, and structural genes ver-1, nor-1, and omt-1. Primer sets (Integrated DNA Technologies, Singapore) used were: aflR-F 5′-TATCTCCCCCCGGGC ATCTCCCGG-3′ and aflR-R 5′-CCGTCAGACAGCCACTGGACACGG-3′, Ver-1- F 5′-ATGTCGGATAATCACCGTTTAGATGGC-3′ and Ver-1- R 5′-CGAAAAGCGCCACCA TCCACCCCAATG-3′, Nor-1-F 5′-ACCGCTACGCCGGCACTCTCGGCAC-3′ and Nor-1-R 5′-GTTGGCCGCCAGCTTCGACACTCCG-3′, Omt-1-F 5′-GGCCCGGTTCCTTGG CTCCTAAGC-3′ and Omt-1-R 5′-CGCCCCAGTGAGACCCTTCCTCG-3′. The 1032, 895, 400, 1232 bp fragments were amplified, respectively. These primers have been used previously by Criseo et al. [27]. The amplification mix consisted of 12.5 PCR mix, 2.5 µL of 10 µM F and R each primer, 2.5 µL H2O, 5 µL DNA template were added and the final reaction volume was 25 µL. PCR was performed as follows: a preincubation step at 94 °C for 10 min followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 65 °C for 2 min, extension at 72 °C for 2 min, and a final extension for 5–7 min at 72 °C. Tm for all primers was 72 °C. The experiment was completed in approximately 3 h and 20 min. PCR products were analyzed by electrophoresis using 1.5% agarose gels in 1 × TAE [40 mM Tris–acetate, 1 mM EDTA (pH 8)]. Gels were stained with 0.5 µg µL−1 ethidium bromide, visualized under UV light (260 nm), and photographed using Kodak Gel Logic 200 (Eastman Kodak Company, NY). A 1 kb plus DNA ladder (Promega) was used as a standard to determine the size of the.
Statistical analysis
We employed a factorial completely randomized design. The observed data were analyzed using analysis of variance (ANOVA) for statistically significant differences, followed by Duncan’s multiple range test at the 5% probability level. Statistical analysis SPSS software, version 22 (IBM Inc. New York, USA) was used.
Results and discussion
Effects of gamma irradiation on viability toxigenic and non-toxigenic A. flavus strains
All of the 13 strains of A. flavus tested produced conidia (log 7.1–7.4 CFU mL−1) with no significantly different and most of them were non-toxigenic. Only two strains, AF5 and AF12, were toxigenic before irradiation. The effect of 5 and 10 kGy of gamma irradiation on strain viability varied. All 13 strains were viable before radiation (Table 1). When 7 day old, dried conidia were exposed to either 5 or 10 kGy of gamma irradiation, viability of all strains was drastically impacted. Irradiated fungi were either non-viable or their viability was reduced significantly by approximately 6 logs.
Table 1.
Population, viability and toxigenicity (indicated by fluorecence on CAM medium) A. flavus strains after gamma irradiation at doses 5 and 10 kGy
| A. flavus strains | Gamma irradiation doses (kGy)/population (log CFU mL−1) | |||||||
|---|---|---|---|---|---|---|---|---|
| No irradiation | 5 | 10 | ||||||
| Population | Fluorescence | Population | Viability (%) | Fluorescence | Population | Viability (%) | Fluorescence | |
| AF1 | 7.2a | − | 0d | 0 | − | 0d | 0 | − |
| AF2 | 7.1a | − | 1.0bcd | 1.1 | − | 0d | 0 | − |
| AF3 | 7.2a | − | 1.6b | 1.3 | − | 1.0bcd | 1.1 | + |
| AF4 | 7.1a | − | 1.0bcd | 1.1 | − | 1.3bc | 1.3 | − |
| AF5 | 7.1a | + | 0d | 0 | − | 0d | 0 | − |
| AF6 | 7.2a | − | 1.7b | 1.4 | ++ | 1.3bc | 1.3 | − |
| AF7 | 7.2a | − | 1.0bcd | 1.1 | − | 0d | 0 | − |
| AF8 | 7.3a | − | 0d | 0 | − | 0d | 0 | − |
| AF9 | 7.3a | − | 1.3bc | 1.3 | − | 0d | 0 | − |
| AF10 | 7.4a | − | 0d | 0 | − | 0d | 0 | − |
| AF11 | 7.3a | − | 1.0bcd | 1.1 | + | 1.0bcd | 1.1 | − |
| AF12 | 7.1a | + | 1.0bcd | 1.1 | ++ | 1.0bcd | 1.1 | − |
| AF13 | 7.3a | − | 1.6b | 1.3 | − | 1.0bcd | 1.1 | − |
+ Slightly fluorescence, ++ more fluorescence, and − no fluorescence under UV (365 nm) in CAM medium
Numbers followed by same letters not significantly different (P < 0.05) according to Duncan’s multiple range test (DMRT)
Strains AF1, AF5, AF8, and AF10 were non-viable after receiving either 5 or 10 kGy, other strains showed more than one surviving colonies were found. Further, gamma irradiation of AF6 with 5 kGy affected both toxigenicity and viability. After this strain was irradiated, 10 of 50 CFUs mL−1 were toxigenic. For AF11 and AF12, only 10 CFUs mL−1 were aflatoxin producers. Also, irradiation with 10 kGy resulted in 10 CFUs mL−1 of toxigenic AF3. After irradiation with 5 kGy, strains AF6, AF3, and AF13 had the most viable colonies (50, 30, and 30 CFUs mL−1, respectively).
All toxigenic strains were characterized by the production of a yellow pigment on the reverse side of a colony after 5 days of incubation on CAM plates. The yellow pigment also can be seen in the marginal area of the upper side of the colony. Production of the yellow pigment by toxigenic A. flavus is one method of determining aflatoxin production without the use of UV light [20]. We observed slight yellow pigmentation in the marginal area of the upper side of some non-toxigenic colonies, however, no yellow pigment was present on the reverse side of their colonies. Presence of a beige ring around the colony and its fluorescence under UV light are indications of aflatoxin production. The amount of aflatoxin corresponds to fluorescence intensity. Our results are consistent with Davis et al. [18] and Sudini et al. [21]. These previous studies differentiated toxigenic and non-toxigenic A. flavus and A. parasiticus strains by culturing fungi on coconut agar medium and exposing the colonies to UV (365 nm).
High diversity of toxigenic and non-toxigenic strains of A. flavus was described by Ehrlich [8]. Our data agree with that earlier study, because high variability in the viability rate, radiosensitivity and toxin production was observed in tested strains after gamma irradiation exposure. Exposure to 5 and 10 kGy of gamma irradiation reduced significantly (P < 0.05) the number of colony forming unit per mL (CFU mL−1) for each strain when compared to non-irradiated samples (Table 1). Treating with 10 kGy of irradiation reduced viability by > 99.9% for all strains. In addition, the effect of irradiation on each strain between the 5 and 10 kGy treatment groups varied significantly. Further, irradiation up to 10 kGy was insufficient to completely destroy population some strains. It was assumed that the high initial conidial density (log 7.1–7.3 CFU mL−1) and dry 7 days old spores (conidia) contributed to the viability and sensitivity of the conidia during irradiation. Previous studies show that A. flavus is an Aspergilli that is relatively resistant to gamma irradiation [22]. Total elimination of mold on natural ingredients and poultry feed by gamma irradiation was observed using 8 kGy. A. flavus and A. parasiticus were the most resistant to irradiation [23]. In our results, 9 and 6 strains had some viable strains remain after treatment with 5 and 10 kGy, respectively. The dryness of samples exposed to irradiation also contributed to their viability [15]. This result is consistent with Farkas [24], who stated that the absence of water molecules during ionizing irradiation reduces the sensitivity of microorganisms.
Effect gamma irradiation on aflatoxin B1 production
Irradiation of some A. flavus strains with 5 or 10 kGy resulted in non-viable fungi and some surviving strains began to produce toxin (Table 2). Previous studies demonstrate that gamma irradiation of wheat grains stimulates toxigenic fungi to produce more aflatoxin [25]. Other studies have shown that A. flavus and A. parasiticus strains inoculated into corn grains after being treated with 2 kGy of gamma irradiation produce twice as much AFB1 compared to non-irradiated strains [16]. The presence of toxigenic colonies in AF6, AF11, and AF3 samples after irradiation with 5 and 10 kGy at doses were found. These toxigenic colonies were not isolated from a single conidium. Among the strains tested, toxigenic AF12 expressed more toxin after irradiation with 5 kGy. Environmental stress caused by irradiation triggers toxigenic strains to produce aflatoxin. Previous studies, such as Kebede et al. [26], report that the increase in aflatoxin contamination in corn by A. flavus was associated with stressful environmental changes.
Table 2.
Aflatoxin B1 production by A. flavus strains before and after irradiation with 5 and 10 kGy
| A. flavus strain | Irradiation doses (kGy) and AFB1 production (ppb) | ||
|---|---|---|---|
| No irradiation | 5 kGy | 10 kGy | |
| AF3 | < 1 | < 1 | 40.52 |
| AF4 | < 1 | < 1 | < 1 |
| AF5 | 22.8 | 0 | 0 |
| AF6 | < 1 | 101.31 | 0 |
| AF11 | < 1 | 86.19 | < 1 |
| AF12 | 57.2 | 93.46 | 0 |
Detection of genes involved in aflatoxin biosynthesis
Codes of A. flavus strains for genes detection before and after irradiation with 5 and 10 kGy in aflatoxin biosynthesis were shown on Table 3.
Table 3.
Codes of A. flavus strains before and after irradiation with 5 and 10 kGy
| Non-irradiated strains (control) | Toxigenicity | Irradiated strains at 5 and 10 kGy | Toxigenicity |
|---|---|---|---|
| AF3 | Non-toxigenic | 10AF3 | Toxigenic |
| AF4 | Non-toxigenic | 5AF4 | Non-toxigenic |
| AF6 | Non-toxigenic | 5AF6 | Toxigenic |
| AF11 | Non-toxigenic | 5AF11 | Toxigenic |
| AF12 | Toxigenic | 5AF12 | Toxigenic |
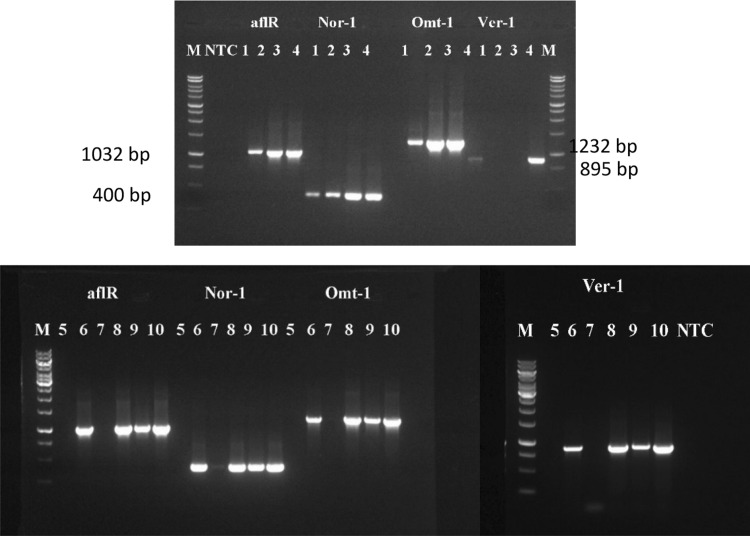
PCR products of regulatory (aflR) and structural genes (nor-1, ver-1, and omt-1) were amplified using genomic DNA isolated from select non-irradiated strains (AF3, AF4, AF6, AF11, and AF12), strains irradiated at 5 kGy (5AF4, 5AF6, 5AF11, and 5AF12) and strains irradiated at 10 kGy (10AF3). These data are presented in Fig. 1.
Fig. 1.
Detection of genes involved in toxin production using PCR and gel electrophoresis. Bands represent amplified fragments of fungal DNA (aflR = 1032 bp, nor-1 = 400 bp, omt-1 = 1232 bp, and ver-1 = 895 bp). DNA was extracted from A. flavus AF4 (line 1), 5AF4 (line 2), AF6 (line 3), 5AF6 (line 4), AF3 (line 5), 10AF3 (line 6), AF11 (line 7), 5AF11 (line 8), AF12 (line 9), and 15AF12(line 10). M = marker ladder and NTC (no primer or template control)
Bands corresponding to the aflR, nor-1, omt-1, and ver-1 genes can be seen at 1032, 400, 1232, and 895 bp, respectively. AF12, 5AF6, 5AF11, 5AF12, and 10AF3 expressed all of the genes, indicating that these strains are aflatoxin producers. The other five strains (AF4, 5AF4, AF6, AF3, and AF11) showed varying patterns and did not amplify at least one of the genes, indicating that these strains were non-toxigenic.
The presence of regulatory and structural genes suggests that 5AF6 is an aflatoxin producer. Criseo et al. [27] examined toxigenicity of A. flavus strains by combining sets of primers for aflR, nor-1, ver-1, and omt-1, which are genes in the aflatoxin biosynthetic pathway. They found that amplification of all four genes indicated toxigenic strains, while non-aflatoxin producers had only some of the genes in involved in this pathway. Erami et al. [28] reported the presence of structural (nor-1, ver-1, and omt-1) and regulatory (aflR) genes differentiated toxigenic A. flavus from non-toxigenic strains. Our positive results show the presence of structural and regulatory genes in toxigenic and non-toxigenic A. flavus strain before and after irradiation. Strains reported to produce aflatoxin using culture-based and analytical methods (i.e., florescence on CAM plate and TLC, respectively) also had all of the regulatory and structural genes needed to produce toxin (Table 4).
Table 4.
Comparison of A. flavus toxigenicity by molecular, culture-based, and analytical methods
| A. flavus strain | Gene detected by PCR | Fluorescence on a CAM plate | Aflatoxin B1 detected by TLC | |||
|---|---|---|---|---|---|---|
| AflR | Nor-1 | Omt-1 | Ver-1 | |||
| AF4 | − | + | − | + | − | − |
| 5AF4 | + | + | + | − | − | − |
| AF6 | + | + | + | − | − | − |
| 5AF6 | + | + | + | + | + | + |
| AF3 | − | − | − | − | − | − |
| 10AF3 | + | + | + | + | + | + |
| AF11 | − | + | − | − | − | − |
| 5AF11 | + | + | + | + | + | + |
| AF12 | + | + | + | + | + | + |
| 5AF12 | + | + | + | + | + | + |
+ Positive for an amplicon and − Negative for an amplicon
Each strain of A. flavus isolated from nutmeg kernels had a different toxigenicity. Just the use of gamma irradiation, therefore, is insufficient to reduce A. flavus present at conidial densities (log 7.1–7.3 CFU mL−1). Increasing the dose of ionizing irradiation is not possible because the Food and Agriculture Organization, International Atomic Energy Agency, World Health Organization [29] and European Commission (EC) [13] concluded in their reports that irradiation of commodities such as dried aromatic herbs, vegetable seasonings, and spices can be treated with a maximum overall average dose of 10 kGy. Combining irradiation with good management practices prior to harvest, during harvesting, and post-harvest to minimize fungal infection and aflatoxin contamination in dried nutmeg kernels is required to ensure the safe consumption of this agricultural commodity.
Acknowledgements
Authors acknowledge Directorate General of Higher Education and the Ministry of Research and Technology of the Republic of Indonesia for their financial support (Grant 017/SP2H/LT/DRPM/II/2016). We are grateful to the Plant Pathology and Biotechnology Laboratories of the Southeast Asian Regional Centre for Tropical Biology (SEAMEO BIOTROP) in Bogor, Indonesia for use research facilities.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Varga J, Frisvad JC, Samson RA. A reappraisal of fungi producing aflatoxins. World Mycotoxin J. 2009;2(3):263–277. doi: 10.3920/WMJ2008.1094. [DOI] [Google Scholar]
- 2.Baranyi N, Kocsubé S, Vágvölgyi C, Varga J. Current trends in aflatoxin research (Review) Acta Biologica Szegediensis. 2013;57(2):95–107. [Google Scholar]
- 3.IARC. International Agency of Research on Cancer. Monographs on the evaluation of carcinogenic risk to humans. Some traditional herbal medicines, some mycotoxins, naphthalene and styrene. IARC Monoger Eval. Carcinog. Risk Hum. 82: 169–366 (2002) [PMC free article] [PubMed]
- 4.Sauer DB, Meronuck RA, Christensen CM. Microflora, pp. 313–340. In: Storage of Cereal Grains and Their Product. Sauer DB (ed). 4th ed, American Association of Cereal Chemist, Minnesota, USA (1992)
- 5.Godet M, Munaut F. Molecular strategy for identification in Aspergillus section Flavi [research letter] FEMS Microbiol. Lett. 2010;304:157–168. doi: 10.1111/j.1574-6968.2009.01890.x. [DOI] [PubMed] [Google Scholar]
- 6.Dharmaputra OS, Ambarwati S, Retnowati I, Nurfadila N. Fungal infection and aflatoxin contamination in stored nutmeg (Myristica fragrans Houtt.) at various stages of the delivery chain in North Sulawesi province. Biotropia. 2015;22(2):129–139. [Google Scholar]
- 7.Vaamonde G, Patriarca A, Pinto VF, Comerio R, Degrossi C. Variability of aflatoxin and cyclopiazonic acid production by Aspergillus flavus section flavi from different substrates in Argentina. Int. J. Food Microbiol. 2003;88:79–84. doi: 10.1016/S0168-1605(03)00101-6. [DOI] [PubMed] [Google Scholar]
- 8.Ehrlich KC. Non-aflatoxigenic Aspergillus flavus to prevent aflatoxin contamination in crops: advantages and limitations (Review article) Frontiers in Microbiol. 2014;5(50):1–9. doi: 10.3389/fmicb.2014.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egel DS, Cotty PJ, Elias KS. Relationships among isolates of Aspergillus sec. flavi that vary in aflatoxin production. Phytopathol. 84: 906–912 (1994)
- 10.Mellon JE, Cotty PJ. Expression of pectinase activity among Aspergillus flavus isolates from southwestern and southwestern United States. Mycopathologia. 2004;157:333–338. doi: 10.1023/B:MYCO.0000024181.36900.15. [DOI] [PubMed] [Google Scholar]
- 11.Horn BW, Grene RL, Sobolev VS, Dorner JW, Powell JH, Layton RC. Association of morphology and mycotoxin production with vegetative compatibility groups in Aspergillus flavus, A. parasiticus, and A. tamarii. Mycologia. 1996;88:574–587. doi: 10.2307/3761151. [DOI] [Google Scholar]
- 12.Perrone G, Gallo A, Logrieco AF. Biodiversity of Aspergillus section Flavi in Europe in relation to the management of aflatoxin risk [review] Frontiers Microbiol. 2014;5:1–5. doi: 10.3389/fmicb.2014.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Commission. Revision of the opinion of the Scientific Committee on Food on the irradiation of food. European Commission, Brussels. Available from: http://europa.eu.int/comm/food/fs/sc/scf/out135_en.pdf. Accessed Feb. 5, 2003
- 14.Aquino S, Ferreira F, Riberio DHB, Corrêa B, Greiner R, Villavincencio ALCH. Evaluation of viability of Aspergillus flavus and aflatoxin degradation in irradiated samples of maize. Braz. J. Microbiol. 2005;36:352–356. doi: 10.1590/S1517-83822005000400009. [DOI] [Google Scholar]
- 15.Calado T, Venâncio A, Abrunhosa L. Irradiation for mold and mycotoxin control (Review) Comprehensive Review in Food Sci. Food Safety. 2014;13:1049–1061. doi: 10.1111/1541-4337.12095. [DOI] [Google Scholar]
- 16.Ribeiro J, Cavaglieri L, Vital H, Cristofolini A, Merkis C, Astoreca A, Orlando J, Carú M, Dalcero A, Rosa CAR. Effect of gamma radiation on Aspergillus flavus and Aspergillus ochraceus ultrastructure and mycotoxin production. Rad. Phys. Chem. 2011;80:658–663. doi: 10.1016/j.radphyschem.2010.12.017. [DOI] [Google Scholar]
- 17.Pitt JI, Hocking AD, Glen DR. An improve medium for the detection of Aspergillus flavus and A. parasiticus. J. Appl. Bacteriol. 1983;52:109–114. doi: 10.1111/j.1365-2672.1983.tb01307.x. [DOI] [PubMed] [Google Scholar]
- 18.Davis ND, Iyer SK, Diener UL. Improved method of screening for aflatoxin with a coconut agar medium. Appl. Environ. Microbiol. 1987;53(7):1593–1595. doi: 10.1128/aem.53.7.1593-1595.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bainton SJ, Coker RD, Jones BD, Morley EM, Naglar MJ, Turner RL. Mycotoxin Training Manual. London: Tropical Product Institute; 1980. pp. 1–176. [Google Scholar]
- 20.Lin MT, Dianese JC. A coconut-agar medium for rapid detection of aflatoxin production by Aspergillus spp. Phytopathol. 1976;66:1466–1469. doi: 10.1094/Phyto-66-1466. [DOI] [Google Scholar]
- 21.Sudini H, Srilakshmi P, Kumar VK, Njoroge SMC, Osiru M, Seetha A, Waliyar F. Detection of aflatoxigenic Aspergillus strains by cultural and molecular methods: A critical Review. Afr. J. Microbiol. Res. 2015;9(8):484–491. doi: 10.5897/AJMR2014.7309. [DOI] [Google Scholar]
- 22.Saleh YG, Mayo MS, Ahearn DG. Resintance of some common fungi to gamma irradiation. Appl. Environ. Microbiol. 1988;54(8):2134–2135. doi: 10.1128/aem.54.8.2134-2135.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribeiro J, Cavaglieri RL, Vital HD, Kruger CD, Rosa CAD. Gamma radiation on the mycoflora of poultry feed and Aspergillus species. Ciência Rural. 2009;39:1452–1458. doi: 10.1590/S0103-84782009005000076. [DOI] [Google Scholar]
- 24.Farkas J. Irradiation for better foods. Trends Food Sci. Technol. 2006;17:148–152. doi: 10.1016/j.tifs.2005.12.003. [DOI] [Google Scholar]
- 25.Paster N, Bullerman LB. Mould spoilage and mycotoxin formation in grains as controlled by physical means. Int. J. Food Microbiol. 1988;7:257–265. doi: 10.1016/0168-1605(88)90044-X. [DOI] [PubMed] [Google Scholar]
- 26.Kebede H, Abbas HK, Fisher DK, Bellaloui N. Relationship between aflatoxin contamination and physiological responses of corn plants under drought and heat stress. Toxins. 2012;4:385–1403. doi: 10.3390/toxins4111385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Criseo G, Bagnara A, Bisignano G. Differentiation of aflatoxin-producing and non-producing strains of Aspergillus flavus group. Lett. Appl. Microbiol. 2001;33:291–295. doi: 10.1046/j.1472-765X.2001.00998.x. [DOI] [PubMed] [Google Scholar]
- 28.Erami M, Hashemi SJ, Pourbakhsh SA, Shahsavandi S, Mohammadi S, Shooshtari AH, Jahanshiri Z. Application on PCR on detection of aflatoxigenic fungi (Short Communication) Arch, Razi Inst. 2007;2(62):95–100. [Google Scholar]
- 29.FAO/IAEA/WHO. Food and Agriculture Organization/International Atomic Energy Agency/World Health Organization. The wholesomeness of irradiated food. Report of a Joint FAO/IAEA/WHO Expert Committee. Technical Report Series No. 659, World Health Organization, Geneva, Switzerland (1981)