Abstract
The structure of microbial communities in a typical Italian-style salami, including bacterial and fungal diversity, was investigated by high-throughput sequencing technology. A total of 6 phyla, 7 classes, 19 orders, 20 families and 28 genera were obtained from 16S rDNA sequences, and a total of 2 phyla, 4 classes, 4 orders, 5 families, 10 genera and 12 Species were obtained from 18S rDNA sequences. The core microbiota was composed of Staphylococcaceae, representing up to 97.52% of the total 16S rRNA, and Penicillium digitatum, accounting for 99.74% of the total classified 18S rRNA. Lactobacillales and Saccharomycetales were detected with a quite low proportion of 1.71 and 0.007%, respectively. This study contributes to the knowledge of the microbial diversity involved in salami and presents high-throughput sequencing as a useful tool to evaluate microbial diversity and monitor the food-borne pathogens in fermented sausage.
Keywords: High-throughput sequencing, Microbial diversity, Penicillium, Salami, Staphylococcaceae
Introduction
Salami is a ready-to-eat meat product and widely consumed in Italy, Spain and Germany, which is a typical dry sausages produced by microbial fermentation [1]. The ripening of salami is a complex biochemical process involving interactions between bacteria and fungi, and the microbial population play a fundamental role in the characterization of the product such as quality, safety and sensory properties [2]. Hence, it is essential to determine the composition and behavior of these microorganisms during fermentation to improve fermentation control and to obtain final products with the desired characteristics.
In the past several years, the microbial population in salami, mainly focusing on the presence of lactic acid bacteria and Gram-positive, catalase-positive cocci (GCC+), has been analyzed based on culture-dependent method that relied on the isolation of colonies for further characterization, and mostly Lactobacillus sakei, Lactobacillus curvatus, Lactobacillus plantarum, Staphylococcus xylosus, Staphylococcus saprophyticus and Staphylococcus equorum have been found in salami [3–5]. Although these basic microorganisms have been well described on a laboratory scale, the microbial communities in actual salami have yet to be fully elucidated because culture-dependent approach has the drawback of detecting only cultivable bacteria, potentially only a small portion of the true microbial population [6]. Hence, there is a high risk of misidentification of complex microbial ecosystems using culture-dependent methods.
Recently, several culture-independent methods based on the nucleic acids analysis without culture-step, such as PCR-denaturing gradient gel electrophoresis (PCR-DGGE) and PCR-temporal temperature gradient gel electrophoresis (PCR-TTGE) [7–9], have received increasing interests regarding the potential to overcome classical microbiology limitations, or more accurately, to complement traditional microbiology, and have been successfully applied to microbial analysis in food matrixes, which have brought new insights into microbial community and diversity in food matrixes. These newer techniques have identified a much higher microbial diversity than culture-dependent method [10]. Nevertheless, there are still some limitations regarding the resolution obtained by these molecular methods. It is difficult to distinguish less-common amplified sequences from the background by PCR-DGGE and PCR-TTGE, and those technologies have failed to determine the microbial community structure in complex matrixes completely because of their inherent limitation in terms of low-throughput and difficulties in extrapolating quantitative data by the analysis of band intensities [11]. Thereby, limitations in the conventional molecular methods still need to be overcome, especially for the analysis of matrices with diverse microbial communities [12].
Nowadays, with rapid development of molecular techniques, high-throughput sequencing (HTS) technology appeared leading to characterize more precisely microbial diversity of complex environmental ecosystems, and has become powerful tool for exploring natural diversity because it can generate thousands of sequences within a short time to cover the complex microbial communities as well as low-abundance microorganisms [13–15]. HTS technology has revolutionized the ecological microbial field. Thus far, this technology has been used to explore the bacterial composition in meat products such as fermented llama meat sausages and typical Italian salami [1, 2], indicating that these communities were richer than expected and that some of them might play a yet unsuspected role. However, those researches mainly focused on bacterial community especially Lactobacilli diversity based on 16S rRNA amplification, but the analysis of an eukaryotic ecosystem, target the 18S rRNA gene, have until recently been less reported in the literature. Yeast and filamentous fungi such as Dabaryomyces hansenula, Hansanula ciferrii and Pichia guilliermondii play a key role in fermented sausages just as bacteria do and contribute to the development of sensory properties of sausages, such as texture and flavor, due to the production of specific enzymes used in the metabolism of proteins and lipids [16, 17]. Therefore, both bacterial and eukaryotic ecosystem should be analyzed by HTS technology in order to grasp a deeper and more precise evaluation of microbial diversity and microbial quality in fermented sausages. Furthermore, considering that salami is a very popular ready-to-eat product and that the raw materials can be contaminated with pathogens, this product can put consumers at risk if a failure occurs during the manufacturing process [18]. However, many previous studies primarily focused on functional microorganisms analysis using HTS technology, but paid less attention to pathogens in fermented sausages [1, 2, 16, 19]. Thereby, the risk of contamination of salami by these pathogenic bacteria also should be paid meticulous attention.
In this study, the structure and diversity of microbial communities, including bacterial and eukaryotic ecosystem, in a typical Italian-style salami was investigated by an HTS approach conducted by Illumina Miseq™ analysis of the bacterial 16S hypervariable regions V4 and fungi 18S hypervariable regions V4, respectively, and the dominant populations that may affect the quality and flavor of salami were identified, which contribute to the knowledge of salami by characterizing its microbiota. Moreover, frequent pathogenic bacteria, namely Salmonella enterica, Escherichia coli, Staphylococcus aureus and Listeria monocytogenes, were also checked by HTS technology to evaluate the microbial safety of the salami.
Materials and methods
Samples
Salami with brand of Casa Tarradellas analyzed in this study were produced from ESPETEC (Gurb, Vich, Spain), which is a famous fermented Italian-style dry sausages and manufactured using traditional methods, without the addition of starter cultures. Salami samples from five different lot numbers were employed for community analysis by high-throughput sequencing technology, and each sample was sampled 10 g.
DNA extraction
The microbial DNA of the sample was extracted using the E.Z.N.A™ Mag-Bind Soil DNA Kit (OMEGA, USA) according to the manufacturer’s instructions, which is considered to be suitable for high throughput sequencing analysis [20]. The purity and integrity of extracted DNA was checked using the Nanodrop-2000 spectrophotometer (Thermo Fisher Scientific, USA) and agarose gel electrophoresis, respectively. Subsequently, the extracted DNA was quantified with a Qubit 2.0 fluorometer, using the Qubit™ dsDNA BR assay kit (Invitrogen, Paisley, UK), according to the manual.
Illumina high-throughput sequencing
The V4 region of the 16S and 18S rRNA gene was amplified using the universal prokaryotic primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 805R (5′-GACTGGAGTTCCTTGGCACCCGAGAATTCCAGGACTACHVGGGTATCTAATCC-3′), and universal fungal primers V43NDF (5′-GGCAAGTCTGGTGCCAG-3′) and Euk_V4_R(5′-GTGACTGGAGTTCCTTGGCACCCGAGAATTCCAACGGTATCTRATCRTCTTCG-3′), respectively, with the addition of a barcoded sequence and the required Illumina adapters.
The PCR reactions were carried out in 30 μL reactions with 15 μL of Phusion High-Fidelity PCR Master Mix (New England BioLabs Inc., Ipswich, MA), 0.2 μM primers, and approximately 10 ng template DNA. In order to reduce possible biases related to the primer extension, the two step-PCR approach was conducted with a first PCR step of 25 cycles using untagged primers, and a final step of 5 cycles with tagged primers and 1 μL of the first step products used as template. First PCR step consisted of initial denaturation at 94 °C for 3 min followed by 25 cycles of denaturation at 94 °C for 30 s, annealing at 45 °C for 20 s, elongation at 65 °C for 30 s, and a final extension at 72 °C for 5 min. The PCR final step consisted of initial denaturation at 95 °C for 3 min followed by 5 cycles of denaturation at 94 °C for 20 s, annealing at 55 °C for 20 s, elongation at 72 °C for 30 s, and a final extension at 72 °C for 5 min.
The PCR products were purified using Agencourt XP Ampure Beads (Beckman Coulter), and the quality of the final products was assessed with a Bioanalyzer 2100 (Agilent Technologies). The final PCR products were quantified using Quant-IT™ PicoGreen® kit (Invitrogen, Paisley, UK) and pooled in equal proportions for subsequent sequencing. Pair-end sequencing was conducted on the Illumina MiSeq sequencing platform (Illumina, USA) for 312 cycles at Sangon Biotech Co., Ltd (Shanghai, China) based on a standard protocol from the manufacturer. The raw gene sequence data were analyzed with the Quantitative Insights Into Microbial Ecology (QIIME) [21] software package and UPARSE pipeline [22]. Pairs of reads from the original DNA fragments were merged using FLASH [23], and filtered by QIIME quality filters.
Bioinformatics and data analysis
High-throughput sequencing data were sorted into different samples by matching the specific barcodes. Then, the primers, barcodes and adaptors were trimmed off using the collection command line tools of FASTX-Toolkits (http://hannonlab.cshl.edu/fastx_toolkit/). Then the sequence data were filtered, multiplexed and prepared for statistical analysis as previously described [1, 24, 25]. The remaining sequences were called effective sequence. The operational taxonomic units (OTUs) and the taxonomy based approaches were performed for the effective sequences analysis. The effective sequences were clustered into OTUs at the cut off level of 3% by using Mothur v1.30.0 [26], while the taxonomy matrices were obtained at a set confidence threshold of 80% using Rv 3.0.0 [27] with the Vegan package [28]. Alpha diversity was evaluated with QIIME to represent the microbial community diversity. Rarefaction curves, the Shannon diversity index, Chao 1 richness, and Good’s coverage were calculated to evaluate the alpha diversity.
Results and discussion
Characteristics of the sequencing data
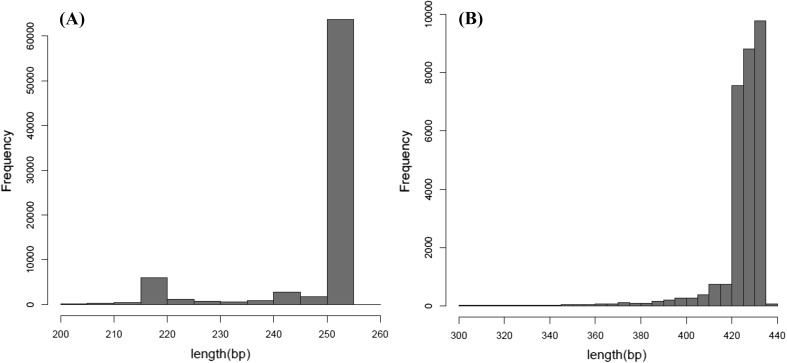
Based on the sequencing results, a total of 11,032 raw sequences were obtained. After the quality filtering, a total of 71,849 high-quality 16S rRNA gene sequences with an average length of 247 bp and 29,199 high-quality 18S rRNA gene sequences with an average length of 424 bp were recovered (Fig. 1), respectively, which were clustered into 2048 OTUs with the 97% identity level. To indicate the community diversity and sample coverage, the alpha-diversity indices, including the Shannon and Simpson diversity indices, were calculated and the results of the analysis of these indices are presented in Table 1. The final OTUs table contained 886 bacterial and 1162 eukaryotic distinct OTUs, and the Good’s coverage values were above 0.99 for bacterial and 0.96 for eukaryotic sequences, indicating that the majority of microbial phylotypes were detected.
Fig. 1.
Length distribution of reads for 16S rRNA gene sequences (A) and 18S rRNA gene sequences (B) in salami sample
Table 1.
Numbers of sequences analyzed, observed OTUs, Good’s coverage calculated, Chao1 diversity index and Shannon diversity index for the microbial communities of salami for high-throughput sequence reads determined at a 97% similarity
| Reads | Observed OTUs | Shannon index | ACE index | Chao1 index | Good’s coverage | Simpson | |
|---|---|---|---|---|---|---|---|
| 16S rRNA | 71,849 | 886 | 1.56 | 8102.49 | 3450.07 | 0.99 | 0.51 |
| 18S rRNA | 29,199 | 1162 | 1.18 | 118399.74 | 35975.33 | 0.96 | 0.46 |
Taxonomic complexity of bacterial community
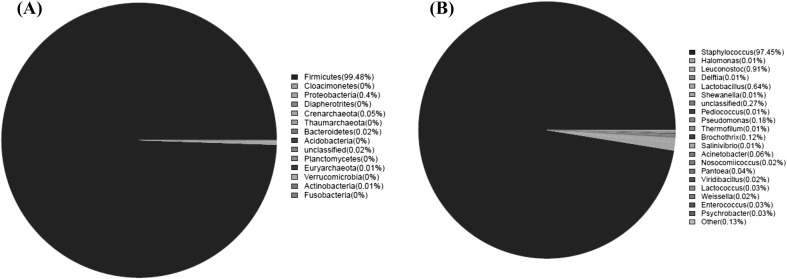
The effective sequences were blasted to different phylogenetic taxa in the Silva database. Within the classified reads, a total of 6 phyla, 7 classes, 19 orders, 20 families and 28 genera were obtained from 16S rDNA sequences. The bacterial population of the salami sample was investigated at both the phylum (Fig. 2A) and genus levels (Fig. 2B). In addition, a more precise percentage of the bacterial composition is reported in Table 2.
Fig. 2.
Relative abundance (%) of the bacteria of salami at the phyla (A) and genus (B) levels
Table 2.
Percentages of the most abundant taxonomical groups of bacteria in the salami sample
| Phylum | Class | Order | Family | Genus |
|---|---|---|---|---|
| Firmicutes 99.48% | Bacilli 99.47% | Bacillales 97.73% | Staphylococcaceae 97.52% | Staphylococcus 97.45% |
| Nosocomiicoccus 0.07% | ||||
| Listeriaceae 0.12% | Brochothrix 0.12% | |||
| Planococcaceae 0.03% | Viridibacillus 0.03% | |||
| Bacillaceae 0.03% | Geobacillus 0.03% | |||
| Lactobacillales 1.71% | Leuconostocaceae 0.93% | Leuconostoc 0.91% | ||
| Weissella 0.02% | ||||
| Lactobacillaceae 0.65% | Lactobacillus 0.64% | |||
| Pediococcus 0.91% | ||||
| Lactococcus 0.01% | ||||
| Streptococcaceae 0.04% | Streptococcus (0.03%) | |||
| Enterococcaceae 0.03% | Enterococcus 0.03% |
In total, 6 phyla, namely Firmicutes, Proteobacteria, Crenarchaeota, Bacteroidetes, Actinobacteria, and Euryarchaeota, were found and the most abundant bacterial taxonomic group was the Firmicutes, representing up to 99.48% of the total sequences. Within Firmicutes, the Bacilli was the most dominant class representing up to 99.47% of the total OTUs, and Clostridia represented in a small portion (0.01%). Bacillales was the most dominant order within Bacilli accounting for 97.73% of all the sequences, followed by Lactobacillales with a quite low proportion of 1.71%. Bacterial families were largely dominated by Staphylococcaceae (97.52%), followed by Lactobacillaceae (0.65%) and rare families in lower percentages. Sequence assignment at genus level confirmed Staphylococcus as the most representative genera in the salami sample, with a minor presence of Lactobacillus and Lactococcus. Previous studies had found that Staphylococcaceae. spp. is the most dominant bacterial, followed by Lactobacillaceae. spp. using culture-dependent method or culture-independent methods [1]. This observation is consistent with a previous report that indicated Staphylococcaceae. spp. to be the predominant microorganism in salami.
Staphylococcaceae xylosus is a frequent species isolated from salami samples [29–31], which has been developed as commercial starter cultures for the manufacture of fermented sausages. Moreover, in a recent study on typical Italian dry fermented sausages [1], the presence of 32 different Staphylococcaceae, including Staphylococcaceae. succinus, Staphylococcaceae. equorum, Staphylococcaceae. saprophyticu and so on by using DNA-based HTS coupled with DGGE techniques was reported, which may be metabolically active and can really contribute to determining the final characteristics of the products. Apart from Staphylococcus species, the diversity of Lactobacilli in salami samples was also found to be very high [31, 32], and 18 major Lactobacilli species, including L. sakei, Lactobacillus acidipiscis, Lactobacillus casei and so on, were determined from typical Italian dry fermented sausages by Połka and others [1]. Interestingly, the data obtained from our studies showed that Staphylococcaceae was the dominant bacterial and its relative abundance reached up to 97.52%, while the relative abundance of Lactobacillales was only 1.72%, which was far fewer abundant than previous results obtained by culture-dependent method. Moreover, Pediococcus was detected with a low abundance, which is most often involved in sausage fermentations [33].
Taxonomic complexity of fungal community
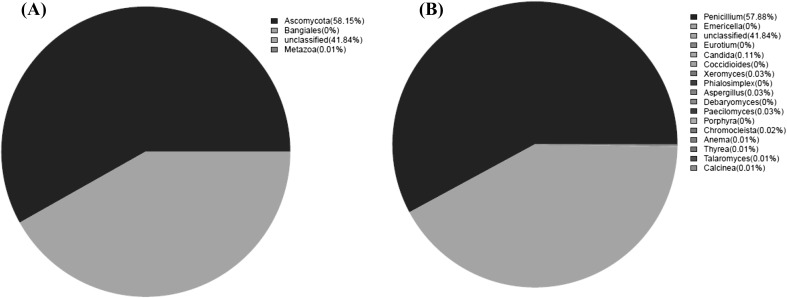
The eukaryotic ecosystem of salami was investigated at both the phylum and genus levels (Fig. 3). In addition, a more precise percentage of the fungal composition is presented in Table 3.
Fig. 3.
Relative abundance (%) of the fungi of salami at the phyla (A) and genus (B) levels
Table 3.
Percentages of the most abundant taxonomical groups of fungi in the salami sample
| Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|
| Ascomycota 58.15% | Eurotiomycetes 58.01% | Eurotiales 58.01% | Trichocomaceae 57.97% | Penicillium 57.88% | Penicillium digitatum Pd1 57.73% |
| Penicillium roqueforti 0.15% | |||||
| Peniillium charles 0.01% | |||||
| Paecilomyces 0.03% | Byssochlamys spectabilis 0.03% | ||||
| Aspergillus 0.03% | Penicillium malachiteum 0.03% | ||||
| Chromocleista 0.02% | Talaromyces wortmannii 0.02% | ||||
| Talaromyces 0.01% | Aspergillus clavatus NRRL1 0.01% | ||||
| Monascaceae 0.03% | Xeromyce 0.03% | Xeromes bisporus 0.03% | |||
| Saccharomycetes 0.12% | Saccharomycetales 0.12% | Saccharomycetales 0.12% | Candida 0.12% | Candidazey lanoides 0.12% | |
| Lichinomycetes 0.02% | Lichinales 0.02% | Lichinaeae 0.02% | Thyrea 0.01% | Thyreaconfusa 0.01% | |
| Anema 0.01% | Clathrina Wistariensis 0.01% |
Just two phyla, namely Ascomycota and Metazoa, were detected in the salami as Fig. 3(A) shows. Ascomycota was the most abundant phyla, accounting for 58.15% of the total sequences, while Metazoa represented in a small portion (0.01%). Moreover, 41.84% of the total sequences were unclassified. Within Ascomycota, Eurotiomycetes was the most dominant classes, representing 58.01% of the total OTUs, whereas Saccharomycetes and Lichinomycetes occupied a quite low proportion of 0.12 and 0.02%, respectively. At the family level, 5 different families were acquired by blasting, including Trichocomaceae, Saccharomycetales, Monascaceae, Lichinaeae and Clostridia, in which Trichocomaceae accounted for 57.97% of all the sequences, and other families occupied lower than 1% of all the sequences. Saccharomycetales was detected with a quite low proportion of 0.12%, which is far fewer abundant than previous results obtained by culture-dependent method. In terms of species, 11 species were determined in the salami as Table 3 shows, and were largely dominated by Penicillium digitatum Pd1 with a quite high relative abundance of 57.73% of all the sequences.
Fungal development on the surface of dry sausages has an important role, in terms of quality, especially during the seasoning period of both industrially and artisanal products. Although the surface mycoflora is very heterogeneous, the predominant genus is Penicillium. In the present study, the Penicillium was the most dominant genus, accounting for 99.53% of the total classified sequences, based on the data obtained by HTS method. The proteolytic, lipolytic and antioxidant activities have been detected in atoxigenic Penicillium species, such as Penicillium nalgiovense, P. digitatum, Penicillium chrysogenum and Penicillium salamii isolated from dry fermented sausages as starter culture to standardize and improve the quality of dry cured sausages production, giving rise to amino acids and fatty acids [33]. The further transformation of these substances into volatile compounds, such as 2-and 3-methyl-1-butanol, 2-and 3-methylbutanal and 2-methylpropanal, and various aroma compounds including methylketones are essential for the development of the characteristic flavor of dry fermented sausages. In the present study, P. digitatum Pd1 was the most dominant species, representing 99.74% of the total Penicillium species. The environmental conditions of manufacturing rooms for salami production are also suitable for the growth of undesirable ochratoxin A (OTA) producing species, including Penicillium verrucosum and Penicillium nordicum, which are the most serious concern in dry fermented sausages [34]. The sequence reads of P. verrucosum and P. nordicum were not found in the salami sample, which suggests microbial safety of the salami is satisfactory.
Food-borne pathogens risk assessment
It has been reported that the most frequent pathogenic bacterium in spices is S. enterica, followed by E. coli, S. aureus and L. monocytogenes, in terms of frequency and seriousness of the diseases [18, 35]. Considering that salami is a very popular ready-to-eat product and that the raw materials can be contaminated with pathogens, which can put consumers at risk if a failure occurs during the manufacturing process. Consequently, the risk of contamination of salami by these pathogenic bacteria should be paid meticulous attention. The food-borne pathogens risk, namely S. enterica, E. coli, S. aureus and L. monocytogenes, in salami was assessed by HTS technology at genus levels. Enterococcus with a quite low relative abundance of 0.03% was observed in the salami sample (Table 2), which were previously observed in raw meat [36]. The small amount of Enterococcus present in the salami sample maybe derive from raw meat. Apart from Enterococcus, the sequence reads of S. enterica, S. aureus and L. monocytogenes present in the salami sample were scarce. These observations strongly suggest that microbial safety of the final products is satisfactory. The classical culture-dependent method has long been used to trace the sources of contamination for those pathogenic bacteria in food products. However, some drawbacks have been identified, namely tedious operation, low sensitivity and poor specificity, resulting in inaccurate detection of trace pathogenic bacteria in food matrixes. In the present study, a HTS method, which can rapidly, simply, and cheaply perform MLST analysis using a next-generation sequencer (NGS) that can analyze a large volume of base sequences at once was developed.
Functional profiling of prokaryotic reads
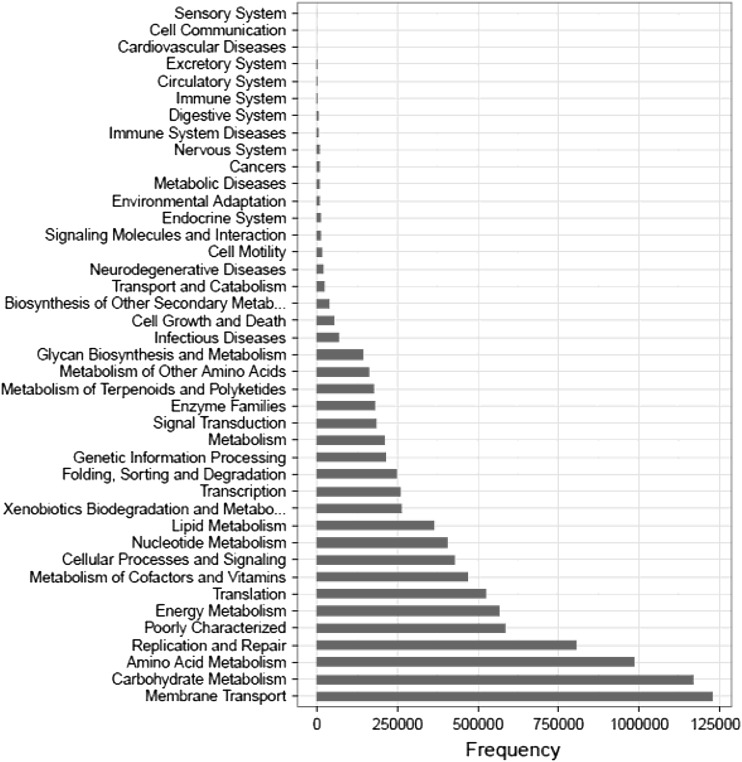
To assess the functional capacities present in bacteria metagenome, the reads were searched against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Fig. 4). Approximately 42% of the reads corresponded to metabolism-related genes involved in carbohydrate, amino acid and energy metabolism, and were largely dominated by carbohydrate metabolism, followed by acid metabolism and energy metabolism. Moreover, the number of sequences classified into the functional categories of membrane transport represented in a large portion. On the other hand, the number of genes involved in biosynthesis of polyketides and glycan were far fewer.
Fig. 4.
Functional classifications of prokaryotic reads derived from HTS approach. The functional genes were determined using a BLASTP search according to the KEGG database
Conclusions
The high-throughput sequencing method was applied to investigate the structure of microbial communities, including prokaryotic and eukaryotic population, in a typical Italian-style salami. A total of 71,849 effective 16S rRNA gene sequences with an average length of 247 bp and 29,199 effective 18S rRNA gene sequences with an average length of 424 bp were obtained. The Firmicutes were the most abundant prokaryotic phyla, accounting for 99.48% of the 16S rRNA gene total sequences, and bacterial families were largely dominated by Staphylococcaceae, representing up to 97.52%. However, Lactobacillales was detected with a quite low proportion of 1.71%, which was far fewer abundant than previous results obtained by culture-dependent method. Penicillium was the most dominant eukaryotic genus, accounting for 99.53% of the total classified 18S rRNA sequences, while Saccharomycetales was far fewer abundant than previous results obtained by culture-dependent method, just accounting for 0.007% of the total classified 18S rRNA sequences. Furthermore, the food-borne pathogens risk, namely S. enterica, E. coli, S. aureus and L. monocytogenes, in salami was assessed by HTS technology at genus levels. Enterococcus was observed in the salami sample with a quite low relative abundance of 0.03%. Apart from Enterococcus, the sequence reads of S. enterica, S. aureus and L. monocytogenes present in the salami sample were not detected, which suggests microbial safety of the final products is satisfactory. The present study provides a broad characterization of the microbial composition of salami, including bacterial and fungal diversity, allowing a description of microorganisms not previously detected in this product. Further studies are needed to better understand the relationship between the volatile flavor, organic acid and amino acid components and the microbial composition.
Acknowledgements
The research was financially supported by Science and Technology Department of Sichuan Province (2017JY0088) and the Open Subject of Key Laboratory of Xihua University (szjj2016-085). The authors thank Sangon Biotech for technical assistance in carrying out the analyses.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.Połka J, Rebecchi A, Pisacane V, Morelli L, Puglisi E. Bacterial diversity in typical Italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiol. 2015;46:342–356. doi: 10.1016/j.fm.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 2.Rebecchi A, Pisacane V, Miragoli F, Polka J, Falasconi I, Morelli L, Puglisi E. High-throughput assessment of bacterial ecology in hog, cow and ovine casings used in sausages production. Int. J. Food Microbiol. 2015;212:49–59. doi: 10.1016/j.ijfoodmicro.2015.04.047. [DOI] [PubMed] [Google Scholar]
- 3.Ammor MS, Mayo B. Selection criteria for lactic acid bacteria to be used as functional starter cultures in dry sausage production: an update. Meat Sci. 2007;76(1):138–146. doi: 10.1016/j.meatsci.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 4.Albano H, van Reenen CA, Todorov SD, Cruz D, Fraga L, Hogg T, Dicks LMT, Teixeira P. Phenotypic and genetic heterogeneity of lactic acid bacteria isolated from “Alheira”, a traditional fermented sausage produced in Portugal. Meat Sci. 2009;82:389–398. doi: 10.1016/j.meatsci.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Holko I, Hrab J, Salakov A, Rada V. The substitution of a traditional starter culture in mutton fermented sausages by Lactobacillus acidophilus and Bifidobacterium animalis. Meat Sci. 2013;94:275–279. doi: 10.1016/j.meatsci.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Giraffa G, Neviani E. DNA-based, culture-independent strategies for evaluating microbial communities in food-associated ecosystems. Int. J. Food Microbiol. 2001;67(2):19–34. doi: 10.1016/S0168-1605(01)00445-7. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez A, Hierro N, Poblet M, Mas A, Guillamon JM. Application of molecular methods to demonstrate species and strain evolution of acetic acid bacteria population during wine production. Int. J. Food Microbiol. 2005;102:295–304. doi: 10.1016/j.ijfoodmicro.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 8.Hierro N, Esteve-Zarzoso B, Mas A, Guillamon JM. Monitoring of Saccharomyces and Hanseniaspora populations during alcoholic fermentation by real-time quantitative PCR. FEMS Yeast Res. 2007;7:1340–1349. doi: 10.1111/j.1567-1364.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 9.Prakitchaiwattana CJ, Fleet GH, Heard GM. Application and evaluation of denaturing gradient gel electrophoresis to analyse the yeast ecology of wine grapes. FEMS Yeast Res. 2004;4:865–877. doi: 10.1016/j.femsyr.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Chodak M, Golebiewski M, Morawska-Ploskonka J, Kuduk K, Niklińska M. Diversity of microorganisms from forest soils differently polluted with heavy metals. Appl. Soil Ecol. 2013;64:7–14. doi: 10.1016/j.apsoil.2012.11.004. [DOI] [Google Scholar]
- 11.Dolci P, Alessandria V, Rantsiou K, Bertolino M, Cocolin L. Microbial diversity, dynamics and activity throughout manufacturing and ripening of Castelmagno PDO cheese. Int. J. Food Microbiol. 2010;143:71–75. doi: 10.1016/j.ijfoodmicro.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Ogier JC, Son O, Gruss A, Tailliez P, Delacroixbuchet A. Identification of the Bacterial Microflora in Dairy Products by Temporal Temperature Gradient Gel Electrophoresis. Appl. Environ. Microbiol. 2002;68(8):3691–3701. doi: 10.1128/AEM.68.8.3691-3701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quigley L, Sullivan O, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl. Enviro Microbiol. 2012;78:5717–5723. doi: 10.1128/AEM.00918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalmasso A, Maria DSR, Civera T, Pattono D, Cardazzo B, Bottero MT. Characterization of microbiota in Plaisentif cheese by high-throughput sequencing. LWT-Food Sci. Technol. 2016;69:490–496. doi: 10.1016/j.lwt.2016.02.004. [DOI] [Google Scholar]
- 15.Carmen PM, Mas A. Analysis of microbial diversity and dynamics during wine fermentation of Grenache grape variety by high-throughput barcoding sequencing. LWT-Food Sci. Technol. 2016;72:317–321. doi: 10.1016/j.lwt.2016.05.009. [DOI] [Google Scholar]
- 16.Wang XH, Ren HY, Wang W, Bai T, Li JX, Zhu WY. Effects of inoculation of commercial starter cultures on the quality and histamine accumulation in fermented sausages. J. Food Sci. 2015;80(2):377–383. doi: 10.1111/1750-3841.12765. [DOI] [PubMed] [Google Scholar]
- 17.Casaburi A, Aristoy MC, Cavella S, Monaco RD, Ercolini D, Toldrá F, Villani F. Biochemical and sensory characteristics of traditional fermented sausages of Vallo di Diano (Southern Italy) as affected by the use of starter cultures. Meat Sci. 2007;76(2):295–307. doi: 10.1016/j.meatsci.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Polese P, Torre M, Lucia SM. Prediction of the impact of processing critical conditions for Listeria monocytogenes growth in artisanal dry-fermented sausages (salami) through a growth/no growth model applicable to time-dependent conditions. Food Control. 2017;75:167–180. doi: 10.1016/j.foodcont.2016.12.002. [DOI] [Google Scholar]
- 19.Fontana C, Bassi D, Lopez C, Pisacane V, Otero MC, Puglisi E, Rebecchi A, Cocconcelli PS, Vignolo G. Microbial ecology involved in the ripening of naturally fermented llama meat sausages A focus on lactobacilli diversity. Int. J. Food Microbiol. 2016;23:617–625. doi: 10.1016/j.ijfoodmicro.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Di BJ, Bao Y, Gloor GB, Burton JP, Reid G. High throughput sequencing methods and analysis for microbiome research. J. Microb. Meth. 2013;95:401–414. doi: 10.1016/j.mimet.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD. QIIME allows analysis of high-throughput community sequencing data. Nature Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 23.Mago R, Simkova H, Brown-Guedira G, Dreisigacker S, Breen J, Jin Y, Singh R, Appels R, Lagudah ES, Ellis J, Dolezel J, Spielmeyer W. An accurate DNA marker assay for stem rust resistance gene Sr2 in wheat. Theor. Appl. Genet. 2011;122(4):735–744. doi: 10.1007/s00122-010-1482-7. [DOI] [PubMed] [Google Scholar]
- 24.Bassi D, Puglisi E, Cocconcelli PS. Understanding the bacterial communities of hard cheese with blowing defect. Food Microbiol. 2015;52:106–118. doi: 10.1016/j.fm.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Vasileiadis S, Puglisi E, Trevisan M, Scheckel KG, Langdon KA, McLaughin MJ, Lombi E, Donner E. Changes in soil bacterial communities and diversity in response to long-term silver exposure. FEMS Microbiol. Ecol. 2015;91:108–114. doi: 10.1093/femsec/fiv114. [DOI] [PubMed] [Google Scholar]
- 26.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Webber CF. Introducing mothur: open-source, platform in dependent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Development Core Team. R: a Language and Environment for Statistical Computing, Reference Index Version 2.14.1. R Foundation for Statistical Computing (2011).
- 28.Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 29.Iacumin L, Comi G, Cantoni C, Cocolin L. Molecular and technological characterization of Staphylococcus xylosus isolated from naturally fermented Italian sausages by RAPD. Rep-PCR and Sau-PCR analysis. Meat Sci. 2006;74:281–288. doi: 10.1016/j.meatsci.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 30.Janssens M, Van derMijnsbrugge A, Sánchez-Mainar M, Balzarini T, De Vuyst L, Leroy F. The use of nucleosides and arginine as alternative energy sources by coagulase-negative staphylococci in view of meat fermentation. Food Microbiol. 2014;39:53–60. doi: 10.1016/j.fm.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Greppi A, Ferrocino I, La Storia A, Rantsiou K, Ercolini D, Cocolin L. Monitoring of the microbiota of fermented sausages by culture independent rRNA-based approaches. Int. J. Food Microbiol. 2015;212:67–75. doi: 10.1016/j.ijfoodmicro.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Wang XH, Ren HY, Liu DY, Zhu WY, Wang W. Effects of inoculating Lactobacillus sakei starter cultures on the microbiological quality and nitrite depletion of Chinese fermented sausages. Food Control. 2013;32:591–596. doi: 10.1016/j.foodcont.2013.01.050. [DOI] [Google Scholar]
- 33.Castellari C, Quadrelli AM, Laich F. Surface mycobiota on Argentinean dry fermented sausages. Int. J. Food Microbiol. 2010;142:149–155. doi: 10.1016/j.ijfoodmicro.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 34.Battilani P, Pietri VA, Giorni P, Formenti S, Bertuzzi T, Toscani T, Virgili R, Kozakiewicz Z. Penicillium populations in dry-cured ham manufacturing plants. J. Food Protect. 2007;70:975–980. doi: 10.4315/0362-028X-70.4.975. [DOI] [PubMed] [Google Scholar]
- 35.European Food Safety Authority (EFSA). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA Journal 13:3991–4156 (2015). [DOI] [PMC free article] [PubMed]
- 36.Yu Q, Zhai LG, Bie XM, Lu ZX, Zhang C, Tao TT, Li JJ, Lv FX, Zhao HZ. Survey of five food-borne pathogens in commercial cold food dishes and their detection by multiplex PCR. Food Control. 2016;59:862–869. doi: 10.1016/j.foodcont.2015.06.027. [DOI] [Google Scholar]