Abstract
Various diseases have been linked to the human microbiota, but the underlying molecular mechanisms of the microbiota in disease pathogenesis are often poorly understood. Using acne as a disease model, we aim to understand the molecular response of the skin microbiota to host metabolite signaling in disease pathogenesis. By metatranscriptomic analysis, we found that the transcriptional profiles of the skin microbiota separated acne patients from healthy individuals. The vitamin B12 biosynthesis pathway in the skin bacterium, Propionibacterium acnes, was significantly down-regulated in acne patients. We hypothesized that host vitamin B12 modulates the activities of the skin microbiota and plays a role in acne pathogenesis. To test this hypothesis, we analyzed the skin microbiota in healthy subjects supplemented with vitamin B12. We found that the supplementation repressed the expression of vitamin B12 biosynthesis genes in P. acnes and altered the transcriptome of the skin microbiota. One of the ten subjects studied developed acne one week after vitamin B12 supplementation. To further understand the molecular mechanism, we revealed that vitamin B12 supplementation in P. acnes cultures promoted the production of porphyrins, which are known to induce inflammation in acne. Our findings suggest a new bacterial pathogenesis pathway in acne and provide one molecular explanation for the long-standing clinical observation that vitamin B12 supplementation leads to acne development. Our study discovered that vitamin B12, an essential nutrient in humans, modulates the transcriptional activities of the skin bacteria, and provided evidence that metabolite-mediated interactions between the host and the skin microbiota play essential roles in disease development.
Introduction
Microorganisms live symbiotically with humans and contribute to human health and disease. The taxonomic composition of the human microbiome has been characterized in both healthy and diseased populations (1–3), revealing that alterations in the microbiome composition are associated with certain diseases (4, 5). However, the underlying molecular mechanisms of the human microbiota in disease pathogenesis are not well understood. At many body sites, including skin, it has not been described whether host metabolites modulate the transcriptional activities of the microbiota. It is also unclear whether the microbiota responds to the host signals via altered metabolic activities, thus impacting on the host health/disease state. Understanding the molecular mechanisms of host-microbiota interactions may lead to more targeted therapy of many microbe-related human diseases.
Acne vulgaris (commonly called acne) provides a promising disease model to study the interactions between the host and the skin microbiota in disease pathogenesis, because the microbiota is less complex and the disease has been associated with a single dominant bacterium, Propionibacterium acnes (2, 6–8). As one of the most common skin diseases, acne affects more than 80% of adolescents and young adults globally (9, 10). Although not fatal, it can be extremely painful, disfiguring, and scarring, and in many patients can profoundly affect selfesteem and mental health (11, 12). Acne is a disease of the pilosebaceous unit (hair follicle), a unique skin compartment where resident bacteria interact with the host cells. Four main factors are believed to contribute to acne development: increased sebum production, follicular hyperkeratinization, proliferation of skin bacteria, and inflammation (13–16). The bacterial pathogenesis mechanism of acne remains to be defined. P. acnes has been long thought as a pathogenic factor for acne. However, it is a major skin commensal and dominates the skin microbiota in both acne patients and healthy individuals (2). Understanding whether the transcriptional activities of the skin microbiota are different between the two cohorts would provide key insights on the bacterial pathogenesis of acne.
Results
The transcriptional activities of the skin microbiota in acne patients were distinct from those in healthy individuals
To determine whether the transcriptional activities of the skin microbiota contribute to disease development, we compared the metatranscriptome of the skin microbiota in acne patients to the one in healthy individuals in a cross-sectional study. We collected the follicular contents of the nose skin from four acne patients and five healthy individuals (Supplementary Materials). We analyzed the microbial gene expression using RNA-Seq with a high sequencing depth of 44–182 million paired-end reads per sample (3.7G bp-18.1G bp) (table S1). We found that P. acnes was the most transcriptionally abundant bacterium (fig. S1). Additionally, Staphylococcus, Pseudomonas, and Shigella were detected in the metatranscriptome with much lower total transcriptional abundance. Given the predominance of P. acnes in all samples, we focused our further analysis on the gene expression profile of P. acnes.
We first quantified the P. acnes gene expression levels. Since each individual harbors various strains of P. acnes and the strain population structures of P. acnes are different among individuals (2), we created a non-redundant P. acnes gene set representative of all the genes encoded in various P. acnes strains. This enabled us to compare the gene expression level among different individuals even though they may harbor different strains. We binned the orthologous genes in 71 P. acnes genomes, which cover all the major lineages of P. acnes found on human skin (17), into 5,140 operational gene units (OGUs) as described in Materials and Methods. We then determined the expression level of each OGU in each sample. We identified a core set of 3,725 P. acnes OGUs (72.5% of all OGUs) expressed in all samples. This core set of OGUs covered most of the metabolic pathways encoded in P. acnes genomes, including carbohydrate metabolism, nucleic acid metabolism, amino acid metabolism, lipid metabolism, and the metabolism of cofactors and vitamins (fig. S2).
We next determined whether the gene expression profiles of P. acnes were distinct between acne patients and healthy individuals. We grouped the samples based on the expression profiles of P. acnes OGUs using an unsupervised hierarchical clustering algorithm. Microbiome samples of the acne patients formed a separate cluster from the samples of the healthy individuals (bootstrap value=99%) (Fig. 1A). We further showed that acne patients were significantly overrepresented in the cluster (P=0.0079, Fisher’s exact test). Other factors including gender, age, ethnicity, skin type, and hormone regulation were not significantly associated with the separation of the acne patients from the healthy individuals. These results demonstrate that P. acnes transcriptional activities in the skin microbiota of acne patients were distinct from those of healthy individuals. To our knowledge, this is a pioneering finding that the in vivo transcriptional activities of the microbiota clearly separate the healthy and disease states of the host. This finding also highlights the importance of studying the metatranscriptome of the human microbiota in addition to the microbial composition to better understand the role of the microbial community in human health and disease.
Fig. 1. The gene expression profiles of P. acnes in the skin microbiota were distinct between acne patients and healthy individuals.
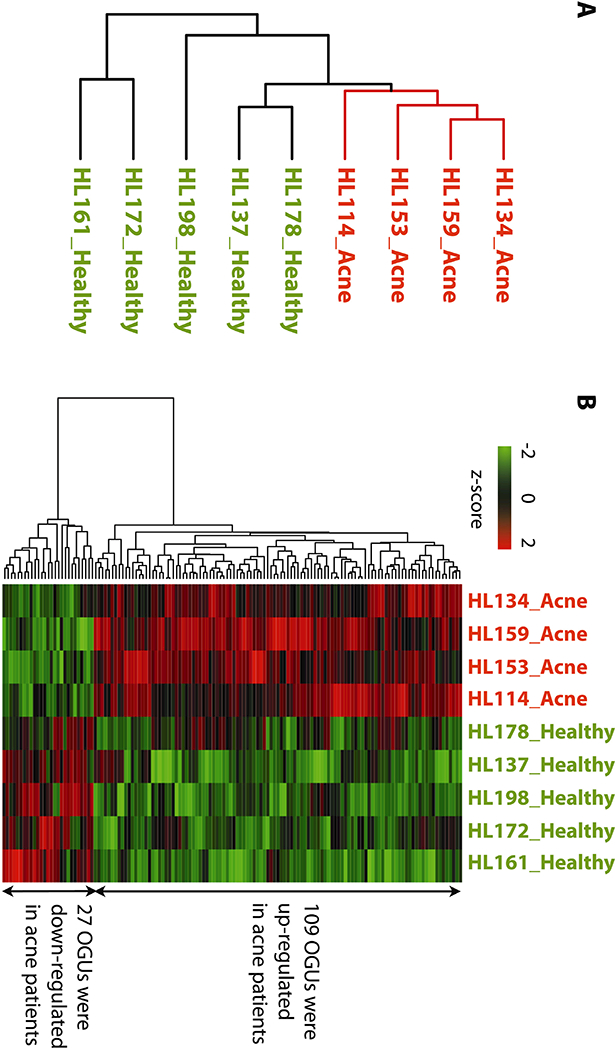
(A) Based on the gene expression of P. acnes in the skin microbiota, acne patients (labeled in red, “Acne”) formed a separate cluster from healthy individuals (labeled in green, “Healthy”) in an unsupervised hierarchical clustering analysis.
(B) 136 differentially expressed P. acnes OGUs were identified between acne patients and healthy individuals. Among them, 109 OGUs were up-regulated and 27 OGUs were down-regulated in acne patients. The OGU names are listed in table S2.
P. acnes vitamin B12 biosynthesis pathway was down-regulated in acne patients
To determine the molecular mechanism of the skin microbiota in acne pathogenesis, we identified the microbial genes and pathways that were differentially expressed between acne patients and healthy individuals. In acne patients, 109 P. acnes OGUs were up-regulated and 27 OGUs were down-regulated (Fig. 1B). Many of these differentially expressed OGUs encode cellular components that are involved in metabolite and protein transport, including metal transporters (iron, cobalt, and hemin), multi-drug transporters, protein export systems, and type II bacterial secretion systems. A few previously proposed bacterial factors involved in acne pathogenesis (18) were found to be up-regulated in acne patients, including a putative lipase (PPA1035), a putative adhesion protein (PPA1907), and a conserved hypothetical protein that may be involved in capsular polysaccharide biosynthesis (PPA0150).
Using a functional annotation clustering analysis, DAVID (19), we identified several differentially expressed metabolic pathways in acne. They include vitamin B12 biosynthesis, porphyrin metabolism, proteolysis, transport, arginine metabolic process, and glutamine family amino acid metabolism. In parallel, using the algorithm of ShotgunFunctionalizeR, we again identified that vitamin B12 biosynthesis pathway was significantly down-regulated (P=9.3E-51, ShotgunFunctionalizeR). We further mapped the differentially expressed OGUs to KEGG pathways (Fig. 2 and fig. S3). It indicated that in acne patients, P. acnes had increased transcriptional activities of the genes that encode carbohydrate uptake systems, enzymes that catalyze carbohydrate metabolism, and decreased transcriptional activities of the genes that encode enzymes in fatty acid biosynthesis and vitamin B12 biosynthesis.
Fig. 2. A schematic of the metabolic pathways in P. acnes illustrating the observed differentially expressed OGUs in acne patients compared to healthy individuals.
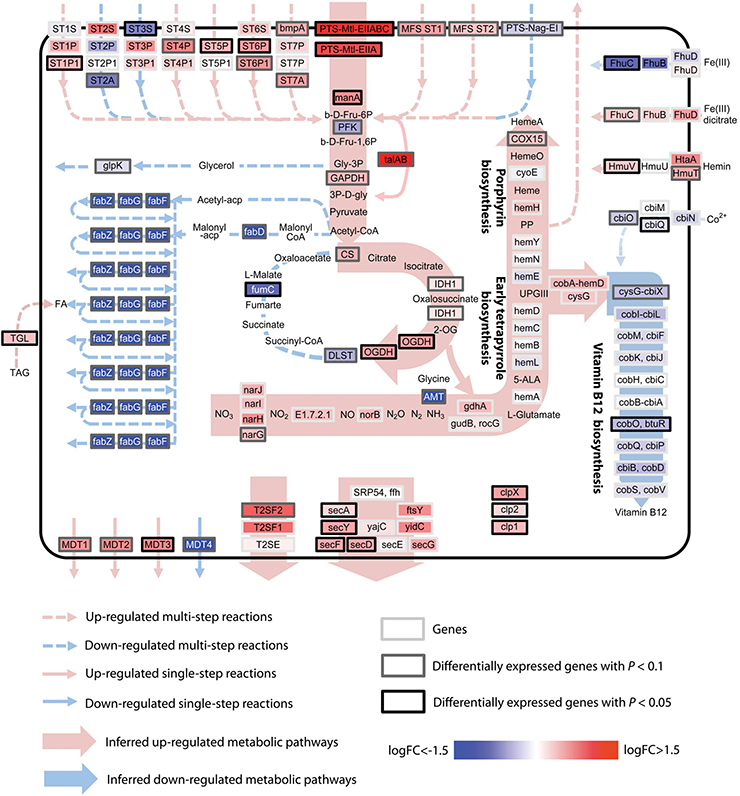
Genes encoding sugar transport and sugar metabolism pathways were up-regulated in acne patients. Genes in the vitamin B12 biosynthesis and fatty acid biosynthesis pathways were down-regulated. FC: fold change. The full gene names are listed in table S3.
Vitamin B12 has been implicated in acne pathogenesis. A number of clinical studies reported that supplementation of vitamin B12 induced acne in a subset of individuals (20–26). However, the molecular mechanism of vitamin B12 in acne development is not understood. To better understand the role of vitamin B12 in acne pathogenesis, we first verified the down-regulation of the vitamin B12 biosynthesis genes in the skin microbiota in a new cohort of subjects. We collected skin microbial samples from an additional nine acne patients and 15 healthy individuals. Using qRT-PCR, we quantified the expression levels of three genes in the vitamin B12 biosynthesis pathway. Among the three genes, cysG+cbiX (a fusion gene) encodes cobalamin synthesis protein, and cbiL encodes precorrin-2 C20-methyltransferase. The enzymes encoded by these two genes function at the initial steps of corrin ring synthesis controlling the metabolic flow entering the vitamin B12 biosynthesis pathway (27). The third gene btuR encodes cob(I)alamin adenosyltransferase, which adenosylates the synthesized corrin ring, and once inactivated, blocks the synthesis of vitamin B12 (27). Consistent with the RNA-Seq data obtained from our previous cohort, cysG+cbiX fusion gene and cbiL were significantly down-regulated in the nine acne patients (fold changes=3.34 and 1.94 and P=0.0023 and 0.0049, respectively; n=9 acne patients and 15 healthy individuals, Student’s t-test). Gene btuR also had lower expression levels in the acne patients compared to the healthy individuals, although the difference was not statistically significant (fold change=1.35, P=0.17, n=9 acne patients and 15 healthy individuals, Student’s t-test) (Fig. 3). In this new cohort of subjects, we verified that vitamin B12 biosynthesis gene expression in P. acnes was down-regulated in acne patients compared to healthy individuals.
Fig. 3. Vitamin B12 biosynthesis genes were down-regulated in the skin microbiota of acne patients compared to healthy individuals.
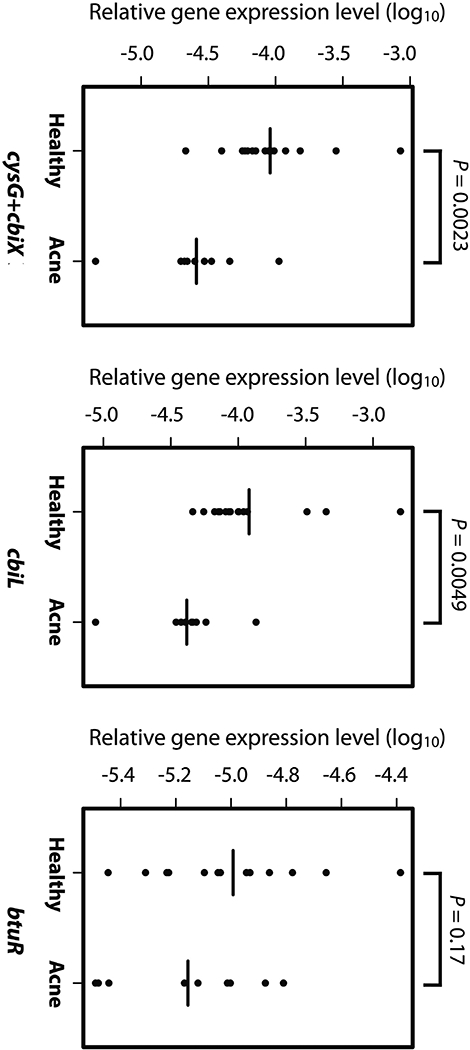
Down-regulation of the genes in P. acnes vitamin B12 biosynthesis pathway was validated in a separate cohort of acne patients (n=9) and healthy individuals (n=15). Consistent with the RNA-Seq data, cysG+cbiX and cbiL were significantly down-regulated, and btuR showed a lower average expression level in acne patients compared to healthy individuals. Significance was determined by Student’s t-test. The mean of the expression levels of each gene is indicated by a black bar. The gene expression data are listed in table S4.
Vitamin B12 supplementation in healthy subjects repressed vitamin B12 biosynthesis genes in P. acnes
It is known that vitamin B12 is a regulator of its own biosynthesis pathway. Previous studies in other bacterial species, including Salmonella typhimurium (28) and Escherichia coli (29), showed that vitamin B12 repressed the expression of the cob/cbi operons in vitamin B12 biosynthesis pathway through cobalamin riboswitches. Cobalamin riboswitches are conserved RNA structural elements that regulate the expression of vitamin B12 biosynthesis operons upon vitamin B12 binding (30). In P. acnes, the upstream regions of cob/cbi operons also encode cobalamin riboswitches (fig. S4).
To determine whether vitamin B12 represses P. acnes vitamin B12 biosynthesis pathway in vivo, we conducted a longitudinal study, in which the gene expression of the skin microbiota in healthy subjects with vitamin B12 supplementation was analyzed. We sampled ten healthy subjects with clear skin who were receiving intramuscular injection of vitamin B12 (1 mg hydroxocobalamin) for general well-being. It is documented that a single intramuscular injection of 1 mg hydroxocobalamin leads to a several fold increase of the serum vitamin B12 level to the range of 1,500–57,000 pg/mL, which lasts for at least two weeks (31). The serum vitamin B12 level usually returns to a normal range (200–900 pg/mL) in four weeks post injection (31–33). Stankler also showed that the skin vitamin B12 levels correlate with the serum vitamin B12 levels (34). We collected the follicular content of the nose skin from the ten healthy subjects at three time points: immediately before vitamin B12 injection as a baseline control, two days after the injection, and 14 days after the injection. We measured the gene expression levels of cysG+cbiX, cbiL, and btuR in vitamin B12 biosynthesis pathway using qRT-PCR (Fig. 4). Compared to the expression levels before or two days after vitamin B12 injection, these genes were significantly down-regulated on day 14 after the vitamin B12 injection (cysG+cbiX P values range from 1.6E-4 to 2.1E-4, cbiL P values range from 7.1E-4 to 7.7E-4, btuR P values range from 9.6E-5 to 0.0025, n=10, Student’s t-test). Moreover, the gene expression levels on day 14 were similar to those observed in the acne patients (Fig. 4). To control for effects not related to vitamin B12 supplementation, we re-sampled nine of the ten subjects with healthy skin three months later using the same sampling procedure without vitamin B12 supplementation. For the healthy subjects without vitamin B12 supplementation, there were no significant changes in the expression levels of vitamin B12 biosynthesis genes between day 0 and day 14. There were also no significant differences in the gene expression levels of the samples collected on day 0 between the original sampling period and the re-sampling period three months later (Fig. 4). Our study demonstrates that vitamin B12 supplementation in the host repressed the vitamin B12 biosynthesis genes in P. acnes.
Fig. 4. Vitamin B12 supplementation in healthy subjects repressed P. acnes cob/cbi operons.
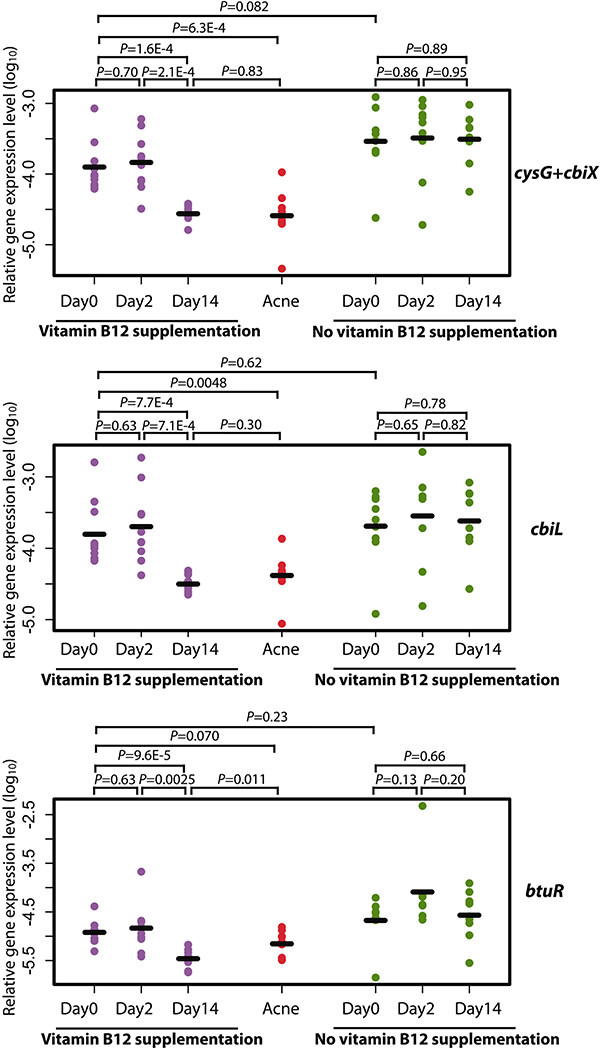
The gene expression levels of cbiL, cysG+cbiX, and btuR in P. acnes vitamin B12 biosynthesis pathway were significantly repressed in the healthy subjects (n=10) on day 14 after vitamin B12 supplementation, to a level similar to those observed in the acne patients (n=9). Without vitamin B12 supplementation, the expression levels of these genes did not change significantly on day 2 and day 14 compared to day 0 (n=9). The expression levels, quantified by qRT-PCR, for each gene on day 0, day 2, and day 14 are shown. Data from healthy subjects with vitamin B12 supplementation are shown in purple and without supplementation are shown in green. As a comparison, data from the acne patients are shown in red. The mean of the expression levels of each gene is indicated by a black bar. Significance was determined by Student’s t-test. The gene expression data are listed in tables S5A and S5B.
To determine whether vitamin B12 regulates the transcription of additional genes and pathways in skin bacteria, we analyzed the transcriptome of the skin microbiota in healthy subjects with vitamin B12 supplementation. We performed RNA-Seq to analyze P. acnes gene expression profiles of the samples collected on day 0 before supplementation and on day 14 after supplementation. We were able to construct sequencing libraries with good quality from seven samples collected on day 0 and ten samples on day 14. From the RNA-Seq data, we identified 93 OGUs that were differentially expressed between day 0 and day 14 (Fig. 5A). Although individual variations exist in some of these OGUs, the overall trend in up- or down-regulation of the OGUs was consistent among individuals. In addition to the vitamin B12 biosynthesis genes, the expression of other metabolic genes was also altered after vitamin B12 supplementation. PPA0750, which encodes S-adenosyl-methyltransferase MraW, was up-regulated. The activity of this enzyme depends on S-adenosylmethionine (SAM), which requires vitamin B12 for its regeneration. The changes in gene expression of the vitamin B12-dependent biological processes validated that our data reflected the regulatory effects of vitamin B12 on the transcriptional activities of P. acnes. Additionally, the gene expressions of a number of transcriptional regulatory components and DNA replication machinery components were altered after vitamin B12 supplementation. The expression of transporter genes was also affected, including up-regulated expression of iron compound, sugar and oligopeptide transporters and down-regulated expression of drug, proline/glycine betaine, and hypoxathine/guanine transporters. A sialidase gene (PPA1560), which has been suggested to function in sebocyte adherence and host tissue degradation (18), was up-regulated after vitamin B12 supplementation. Our results suggest that vitamin B12 supplementation to the host not only affects the expression of vitamin B12 biosynthesis genes in P. acnes, but also the gene expression of other metabolic enzymes and virulence factor.
Fig. 5. Vitamin B12 supplementation in the host altered the transcriptome of P. acnes in the skin microbiota.
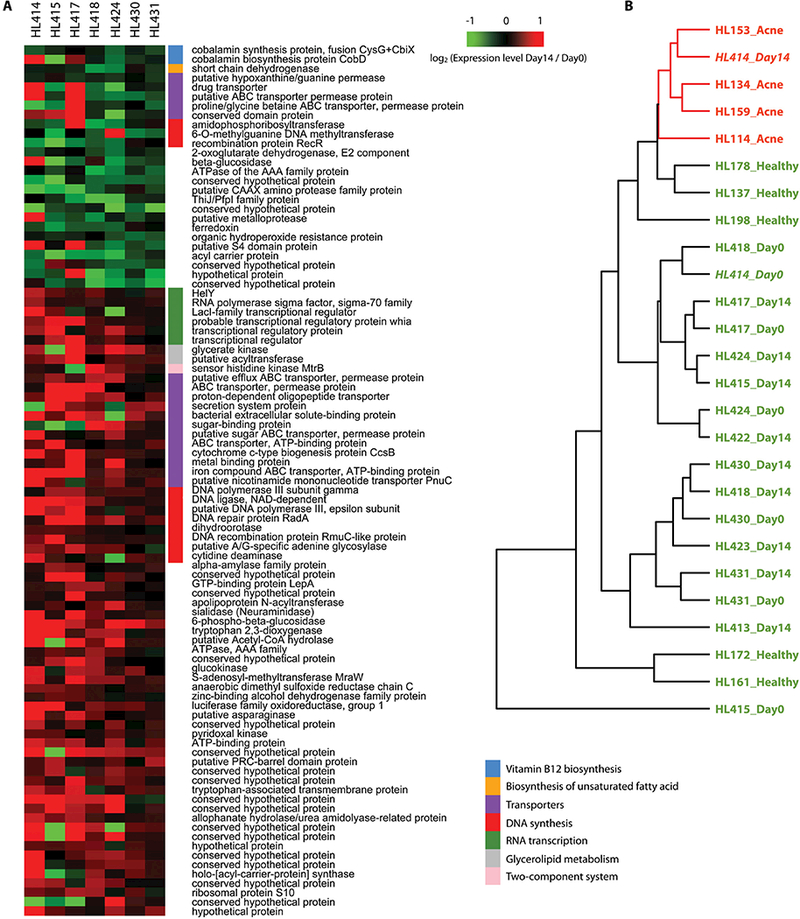
(A) The differentially expressed P. acnes OGUs between the Day0 samples and Day14 samples from the healthy subjects supplemented with vitamin B12. The red and green colors represent the fold change of the OGUs. Red indicates up-regulation after vitamin B12 supplementation and green indicates down-regulation. The functional categories of the OGUs are labeled by the color bars next to the OGUs’ names.
(B) The P. acnes gene expression profile of subject HL414 on day 14 after vitamin B12 supplementation (HL414-Day14) was similar to those from the acne patients and clustered together. The Day0 sample from subject HL414 (“HL414_Day0”, healthy skin, before vitamin B12 supplementation) was similar to the samples from other healthy subjects. Samples from the acne patients were labeled in red (“Acne”), and samples from the healthy subjects were labeled in green (“Healthy”, “Day0”, “Day14”), except for HL414-Day14.
One of the ten subjects developed acne one week after vitamin B12 supplementation
Consistent with multiple clinical studies previously reporting that vitamin B12 supplementation induces acne in subsets of individuals (20–26), in the current study, one of the ten healthy subjects, HL414, had multiple erythematous papules and comedones developed on the face one week after vitamin B12 supplementation (Supplementary Materials). It is not understood how vitamin B12 supplementation leads to acne in some individuals and whether the vitamin B12-associated acne has a separate pathogenesis mechanism. To determine whether vitamin B12 supplementation in subject HL414 altered the gene expression of the skin microbiota with a profile similar to those observed in the acne patients, we clustered the P. acnes gene expression profiles of the samples collected from HL414 and the other healthy subjects with vitamin B12 supplementation together with the samples from our earlier cross-sectional study (Fig. 5B). On day 0, the P. acnes gene expression pattern of HL414 was similar to other healthy subjects, but on day 14 it resembled the expression pattern seen in the acne patients and was clustered within the acne group (bootstrap value=100%). This suggests that the transcriptional changes in the skin microbiota of HL414 after vitamin B12 supplementation are not different from those in acne patients. This result also strongly supports the hypothesis that the host vitamin B12 level modulates the transcriptional activities of the skin bacteria, which in turn affect the health or disease state of the host skin. The Day14 samples from the nine subjects who did not develop acne after supplementation were clustered outside of the acne group. Some of these samples, including HL417_Day14, HL431_Day14, and HL430_Day14, were clustered with their corresponding Day0 samples. This suggests that the effects of the vitamin B12 supplementation on regulating the transcriptional activities of the skin microbiota vary among individuals, possibly due to both host factors and microbial factors. These differences may partly explain why only a subset of individuals develops acne after vitamin B12 supplementation.
The transcriptional profile of the skin microbiota in HL414 was different from other healthy subjects after vitamin B12 supplementation
To understand why subject HL414, not others in our healthy cohort, developed acne after vitamin B12 supplementation, we further investigated the specific bacterial transcriptional changes in HL414 compared to the other subjects 14 days after vitamin B12 supplementation. Based on the RNA-Seq data, we found 397 OGUs differentially expressed in HL414-Day14 compared to the other nine Day14 samples. We compared the list of these genes (HL414 vs. others on day 14) with the genes that were significantly altered after vitamin B12 supplementation in all healthy subjects (Day0 vs. Day14 samples). This comparison is based on the reasoning that bacterial factors involved in vitamin B12-associated acne should be regulated by vitamin B12. We found that 11 OGUs that were differentially expressed between HL414-Day14 and the other Day14 samples were also regulated by vitamin B12 with differential expression between Day0 and Day14 samples (table S6). Among them, PPA0693 encodes the E2 component of the 2-oxoglutarate dehydrogenase complex, which functions in the tricarboxylic acid (TCA) cycle. It was down-regulated in all the healthy subjects after vitamin B12 supplementation (fold change=0.86, P=0.044, n=7, Student’s t-test). Among all the Day14 samples, HL414-Day14 had the lowest expression. This gene was also down-regulated when we compared the acne patients to the healthy individuals in our cross-sectional study (fold change=0.76, P=0.075, n=4 acne patients and 5 healthy individuals, Student’s t-test) (fig. S5). This suggests that the gene down-regulation of the E2 component of the 2-oxoglutarate dehydrogenase complex is regulated by vitamin B12 and is involved in acne pathogenesis.
We further investigated the potential role of PPA0693 in acne pathogenesis. Based on the metabolic pathways in P. acnes (Fig. 2), 2-oxoglutarate dehydrogenase complex converts 2-oxoglutarate to succinyl-CoA. 2-oxoglutarate is a substrate for the biosynthesis of L-glutamate, which is a precursor for both vitamin B12 biosynthesis and porphyrin biosynthesis (35). It has been shown that the biosynthesis of porphyrins in Propionibacteria is inversely correlated with the biosynthesis of vitamin B12 (36, 37). Bykhovskii et al. and Zaitseva et al. found that inhibiting vitamin B12 biosynthesis in Propionibacterium shermanii, either by enzyme inhibitor or by depleting cobalt, led to an increase in porphyrin biosynthesis (36, 38). Porphyrins produced by P. acnes are known to induce inflammation in acne (39–41). They interact with molecular oxygen, generate free radicals to damage adjacent keratinocytes, and stimulate the production of inflammatory mediators in keratinocytes (39–41).
Based on the above data, we propose a molecular mechanism for the bacterial pathogenesis of acne through vitamin B12 regulation. We hypothesize that elevated vitamin B12 level in the host modulates the transcriptional activities of the skin microbiota. Among the alterations in P. acnes, PPA0693 is down-regulated, resulting in an increase in 2-oxoglutarate and L-glutamate. The vitamin B12 biosynthesis pathway is also down-regulated, resulting in the increased metabolic flow of L-glutamate being shunted to the porphyrin biosynthesis pathway. These result in an over-production of porphyrins by P. acnes, which can induce an inflammatory response in the host cells leading to acne. This hypothesis can potentially explain the molecular mechanism of vitamin B12 in acne pathogenesis. In a subset of individuals, similar to HL414, the expression level of PPA0693 after vitamin B12 supplementation is lower than in other individuals, resulting in further increased level of porphyrins produced and leading to acne development.
Porphyrin production is promoted in P. acnes with vitamin B12 supplementation
To test the above hypothesis, we investigated whether porphyrin production is promoted in P. acnes by vitamin B12 supplementation. We supplemented 10 μg/mL vitamin B12 to P. acnes cultures, and found that vitamin B12 persistently repressed the expression of cob/cbi operons. The previously mentioned three genes in the vitamin B12 biosynthesis pathway, cysG+cbiX, cbiL, and btuR, were significantly down-regulated by the addition of vitamin B12 from day 2 to day 14 (P values range from 0.057 to 0.0031, n=2 to 3, Student’s t-test) in cultures (Fig. 6A). In the meantime, we compared the amounts of porphyrins produced in the P. acnes cultures with or without vitamin B12 supplementation. We found that the addition of vitamin B12 significantly increased porphyrin production by 39% during the exponential phase on day 8 and nearly 19% at the stationary phase on day 14 (P=0.0043 and 6.8E-4 respectively; n=3 to 4, Student’s t-test) (Fig. 6B). Our results demonstrated that vitamin B12 supplementation repressed the gene expression of vitamin B12 biosynthesis in P. acnes and increased the biosynthesis of porphyrins.
Fig. 6. Vitamin B12 supplementation repressed the expression of cob/cbi operons in the vitamin B12 biosynthesis pathway and promoted porphyrin production in P. acnes cultures.
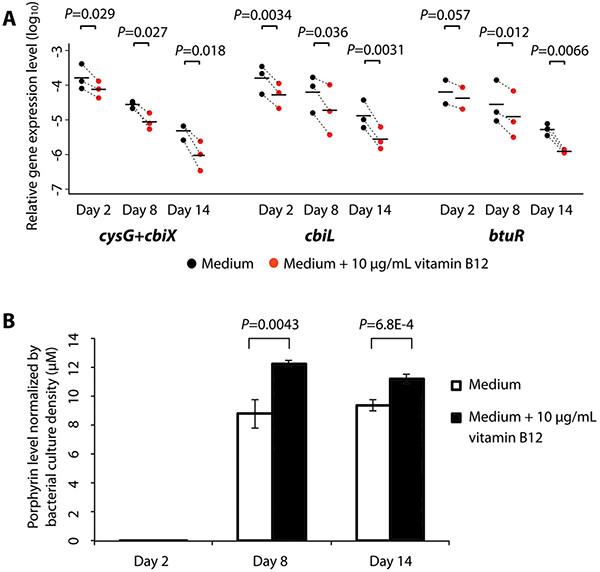
(A) The expression levels of cbiL, cysG+cbiX, and btuR were significantly repressed on day 2, day 8, and day 14 after vitamin B12 supplementation in P. acnes cultures. The experiments were repeated three times independently. Each dot represents the average gene expression level of two to three technical replicates in each independent experiment. The dotted lines indicate the changes between the P. acnes cultures in medium only (black) and the cultures in medium with vitamin B12 supplementation (red). The mean of the gene expression levels from the three independent experiments is shown as a black bar. Significance was determined by Student’s t-test. The gene expression data are listed in table S7.
(B) Porphyrin production in P. acnes cultures on day 8 and day 14 after vitamin B12 supplementation was significantly increased. Significance was determined by Student’s t-test. Data are presented as the mean ± standard deviation. The experiments were repeated three times independently with three to four biological replicates each time. The porphyrin levels are listed in table S8.
To further determine that the increased porphyrin production is directly linked to the reduction in vitamin B12 biosynthesis, we measured the porphyrin level in P. acnes under the condition that vitamin B12 biosynthesis was repressed without the regulation by vitamin B12. We repressed vitamin B12 biosynthesis by depleting cobalt (Co2+ ion), which is an essential component required for vitamin B12 biosynthesis (38). We observed that the porphyrin production in P. acnes cultures was significantly increased after Co2+ depletion (fig. S6). This is consistent with previous studies in other Propionibacteria (36, 37). Our result demonstrates that vitamin B12 biosynthesis pathway and porphyrin biosynthesis pathway share the same pool of precursor substrates and are inversely correlated at the metabolic levels. It further supports that the down-regulation of vitamin B12 pathway promotes porphyrin production.
Discussion
Originally reported more than a half century ago, it is observed that vitamin B12 leads to acne development in a subset of population. However, the underlying mechanism of this clinical observation was not understood. In this study, by investigating the transcriptional activities and regulations of the skin microbiota, we revealed a molecular link between vitamin B12 and acne pathogenesis. We found that the vitamin B12 biosynthesis pathway was down-regulated in acne patients and in healthy subjects who were supplemented with vitamin B12 (Figs. 3 and 4). Several lines of evidence suggest that a high level of serum vitamin B12 is associated with acne phenotype. Goldblatt et al. (42) found that the serum level of vitamin B12 was significantly higher in acne patients than in healthy individuals. Moreover, they and Karadag et al. reported that in acne patients, the serum level of vitamin B12 was significantly decreased after treatment (42, 43). Based on our findings and the evidence mentioned above, we propose a vitamin B12-mediated bacterial mechanism for acne pathogenesis (Fig. 7). In healthy skin, when the vitamin B12 level is normal, the vitamin B12 biosynthesis pathway in the skin bacterium P. acnes is expressed and porphyrins are produced at a low level. When the host vitamin B12 level is elevated, it signals transcriptional changes in P. acnes. The levels of 2-oxoglutarate and L-glutamate are increased, and the vitamin B12 biosynthesis in P. acnes is repressed. The metabolic flow of L-glutamate is shunted toward the porphyrin biosynthesis pathway, leading to an over-production of porphyrins by P. acnes in the follicle. The over-produced porphyrins, secreted by P. acnes, induce an inflammatory response in the host cells leading to acne development. Supporting this proposed mechanism, clinical improvement after acne treatment has been associated with a lower level of porphyrins produced in follicles (44–47). Borelli et al. (46) also reported that in patients with a poor treatment outcome the porphyrin level was not decreased. Our proposed mechanism of vitamin B12-mediated interactions between the host and the skin microbiota provides one potential molecular explanation for the bacterial pathogenesis of acne. It also emphasizes the importance of metabolite-mediated interactions between the host and the microbiota in human health and disease.
Fig. 7. A model of vitamin B12 modulating the transcriptional and metabolic activities of the skin bacterium P. acnes in acne pathogenesis.
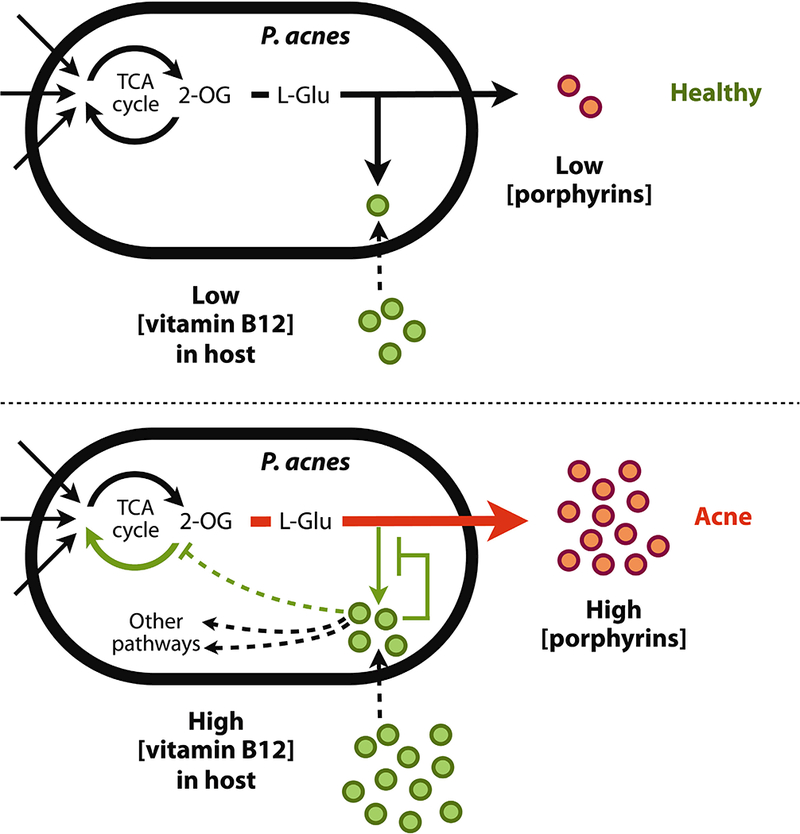
In healthy skin, when the host vitamin B12 level is normal, the vitamin B12 biosynthesis pathway in P. acnes is expressed and porphyrins are produced at a low level. When the host vitamin B12 level is elevated, it signals transcriptional changes in P. acnes. The levels of 2-oxoglutarate and L-glutamate are increased, and the vitamin B12 biosynthesis in P. acnes is repressed. The metabolic flow of L-glutamate is shunted toward the porphyrin biosynthesis pathway, leading to an over-production of porphyrins by P. acnes in the follicle. The overproduced porphyrins, secreted by P. acnes, induce an inflammatory response in the host cells leading to acne development in a subset of individuals. 2-OG: 2-oxoglutarate. L-Glu: L-glutamate.
In our study, acne developed in subject HL414 in a short period of time after vitamin B12 supplementation, but not in other healthy subjects. This observation is consistent with previous clinical findings that not all individuals developed acne after vitamin B12 supplementation (20–26). We also found that after vitamin B12 supplementation, HL414 had a significantly lower gene expression level in the E2 component of the 2-oxoglutarate dehydrogenase complex compared to the other subjects. The low expression level of this gene may contribute to a higher level of 2-oxoglutarate and L-glutamate in P. acnes, leading to a greater amount of porphyrins produced. This may partially explain why HL414 developed acne while others did not during the 14-day period. In addition to PPA0693, we found several other bacterial factors that are associated with acne development after vitamin B12 supplementation (table S6). Among them, PPA2220 was up-regulated in all the acne patients compared to the healthy individuals and also in the healthy subjects after vitamin B12 supplementation (fig. S5). Moreover, subject HL414 had a significantly higher expression level of PPA2220 than other healthy subjects after vitamin B12 supplementation. The function of PPA2220 is not yet known. It encodes a protein containing a photosynthetic reaction center (PRC) barrel domain and a domain of unknown function (DUF2382). Its orthologues in other organisms are often located next to the genes that encode electron transfer chain components, acyl-CoA transferase, and/or dehydrogenases. PPA2220 was also the second most highly expressed genes in the P. acnes transcriptome in our samples. These findings suggest that PPA2220 may play an important role in acne pathogenesis and needs to be further studied. Other pathogenic factors, from the host and/or the bacteria, regulated by vitamin B12 or not, may also be important in the disease development and need to be investigated in future studies.
To our knowledge, the metatranscriptome of the human skin microbiota in health and disease states has not been described prior to this study. While some other human body sites, such as oral cavity, gut, urogenital tract, and upper respiratory tract, can have sufficient biomass for microbiota characterization (48–51), the skin presents a challenging site with low microbial biomass density (6, 52). Studies of the skin microbiome have been limited to mostly taxonomic composition analysis (53). It is important to understand the transcriptional changes of the skin microbiota during diseases in addition to its taxonomic composition and metagenomic content. In this study, we applied RNA-Seq to quantitatively measure the metatranscriptome of the skin microbiota in acne patients and in healthy individuals. We revealed microbial differences at the transcriptional level separating acne patients and healthy individuals. By vitamin B12 supplementation in healthy individuals and in vitro experiments, we revealed that host vitamin B12 modulates the transcriptional and metabolic activities of the skin bacteria, leading to increased production of porphyrins, which induce inflammation in acne. Our findings suggest a new metabolite-mediated bacterial mechanism for acne pathogenesis.
Metabolite-mediated interactions between the host and the microbiota play essential roles in human health and disease. This has been recognized in the gut microbiota (54). Host nutritional states or interventions, such as diets and use of antibiotics and other small-molecule drugs, have been shown to modulate the composition and transcriptional activities of the gut microbiota (55–58). In response, the gut microbiota produces metabolites that are linked to diseases (54, 59–61). Vitamin B12 is an essential nutrient to humans. Its microbial biosynthesis and human absorption mainly take place in the gut. It has been shown that vitamin B12 regulates gut microbial gene expression and affects the selection and competition among microbial species (62). Our study presented here showed that vitamin B12 supplementation to the host modulates the transcriptome of the skin microbiota and leads to acne development in a subset of the individuals. Our study not only provides an explanation for the long-standing clinical observations of the correlation between increased vitamin B12 level and acne development, but also suggests a molecular mechanism for acne pathogenesis. Future follow-up studies may potentially lead to the development of new therapeutics for this medically important disease.
There are a few limitations of our study. First, we had a limited number of subjects investigated in our longitudinal study. Among the ten healthy subjects, one individual developed acne after vitamin B12 supplementation. Although it is consistent with the previous reports that vitamin B12 leads to acne development in only a subset of individuals (20–26), increasing the cohort size would render more power in statistical analysis and would allow us to further establish the molecular link between vitamin B12 and acne pathogenesis. Second, the number of samples in our RNA-Seq metatranscriptomic analysis was limited. This was mainly due to the fact that skin samples have low bacterial biomass and it is difficult to extract sufficient amount of RNAs from each sample. Some of our samples failed during sample processing, RNA extraction, or sequencing library construction. Future improvements of sample collection and bacterial RNA extraction from samples with limited biomass will empower us to better understand the microbial transcriptional activities and their regulations on the human skin.
Materials and Methods
Study design
A cross-sectional study to compare the transcriptional activities of the skin microbiota between acne patients and healthy individuals was performed. Among the nine subjects recruited, four were acne patients (two males and two females) and five were healthy individuals (two males and three females). The average age of the acne patients was 24.8 (19 – 38) and the average age of the healthy subjects was 22.8 (13 – 32). Among the acne patients, two are African American and two are Hispanic. Among the healthy subjects, one is African American, one is Hispanic, two are Caucasians, and one is Asian. There were no significant differences in gender, age, and ethnicity between the acne patients and the healthy subjects. The average face acne score of the four acne patients was 2.5 and the average nose acne score was 0.25 (Supplementary Materials). Twenty-four additional subjects were recruited for the gene expression analysis of the vitamin B12 biosynthesis pathway. Among them, nine were acne patients (three males and six females) and 15 were healthy subjects (seven males and eight females). The average age of the acne patients was 23.8 (15 – 42) and the average age of the healthy subjects was 33.8 (21 – 44). Among the acne patients, four are Hispanic, two are Caucasian, two are Asian, and one has more than one race. Their average face acne score was 2.8 and the average nose acne score was 1.6 (Supplementary Materials). Among the healthy subjects, three are Hispanic, three are Caucasian, eight are Asian, and one has more than one race. There were no significant differences in gender and ethnicity, but age, between the acne patients and the healthy subjects.
Among the 15 healthy subjects, ten were receiving intramuscular injection of vitamin B12 (1 mL of 1,000mcg/mL hydroxocobalamin) for general well-being and were included in the longitudinal study. The longitudinal study was to investigate the effects of vitamin B12 supplementation on the transcriptional activity of the skin microbiota in healthy individuals.
None of the healthy subjects reported any current or past acne treatment. None of the acne patients were being treated with antibiotics at the time of sampling. Four acne patients were being treated with other topical acne therapies at the time of sampling, including tazarotene, tretinoin, salicylic acid, and benzoyl peroxide. Seven acne patients had been treated in the past. None of the subjects had cosmetic procedures.
All subjects were recruited in Southern California. The diagnosis of acne was made by board-certified dermatologists. The presence of acne was graded on a scale of 0 to 5 relating closely to the Global Acne Severity Scale (63). Subjects with healthy skin were determined by board-certified dermatologists and were defined as people who had no acneiform lesions on the face, chest, or back. All subjects provided written informed consent. All protocols and consent forms were approved by both the UCLA and Los Angeles Biomedical Research Institute IRBs. The study was not blinded.
Sample collection
The follicular contents of the nose skin were sampled from the subjects using Bioré Deep Cleansing Pore Strips (Kao Brands Company) following the instruction of the manufacturer (2). This sampling method is different from the conventional tape stripping method. It samples mostly the follicular content of the pilosebaceous unit, while the conventional tape stripping method samples the stratum corneum of the epidermis. A comparison of the samples collected from opposite sides of the nose of the same individual using these two different methods is illustrated in fig. S7. Clean gloves were used for each sampling. After removal from the nose, the strip was placed into a 50 mL sterile tube and labeled with a coded sample name.
For the ten healthy subjects with vitamin B12 supplementation, the first samples (Day0) were taken from one side of the nose before they received vitamin B12 injection. The second samples (Day2) were taken from the other side of the nose two days after the injection. The third samples (Day14) were taken from the entire nose 14 days after the injection. As a control, three months later, nine of the ten healthy subjects were sampled again using the same sampling protocol over a period of 14 days without vitamin B12 supplementation.
Sequence Analysis
Sequence reads mapped to the human genome were removed following the procedure used in the Human Microbiome Project (64). Low quality reads were trimmed or filtered out (details in Supplementary Materials). The rRNA reads were then removed by mapping against P. acnes 16S, 23S, and 5S rRNA sequences and against the SILVA database (release 108) (65). Sequence mapping was performed using Bowtie (version 0.12.7) allowing up to three mismatches per read (66). The remaining reads were aligned to 71 P. acnes genomes (17). For paired-end mapping, we required that the read pairs were aligned to the same reference genome on different strands with the distance between the two reads shorter than 1 Kb. The remaining reads were then mapped against the Human Microbiome Project reference genomes (http://www.hmpdacc.org/HMREFG/). The resulting unaligned reads were further searched against NCBI non-redundant nucleotide database (RefSeq release 48), including bacterial, fungal, and viral genomes.
OGU construction
P. acnes OGUs were constructed in a similar way as previously described (67) with minor modification. Genes in all 71 P. acnes genomes were binned based on their protein sequences using CD-HIT version 4.3 (68) and its default parameters (≥ 90% protein sequence identity).
Quantification of OGU expression level
We assigned a coverage score of 1 to each reference nucleotide aligned by a read with unique alignment. The total coverage score of each reference nucleotide was calculated by summing the scores of the nucleotide from all the aligned reads.
The reads with multiple alignment hits were analyzed as previously described (69). The coverage score of each nucleotide within the aligned region in the reference genome was averaged by the total number of hits (M).
The expression level of a gene was calculated as the total coverage score for all the nucleotides within the gene region normalized by the gene length. To adjust for sequencing depth differences among different samples, the expression level of each P. acnes gene was further normalized by the total number of base pairs aligned to P. acnes genomes. Reads aligned to P. acnes rRNAs and tmRNAs were removed prior to normalization. OGU expression level was calculated by summing the normalized expression level of each gene member of the OGU.
Porphyrin quantification in P. acnes cultures
P. acnes was cultured in reinforced clostridium broth anaerobically without exposure to light at 37°C. The culture medium was supplemented with 10 μg/mL vitamin B12 (Sigma), a concentration similar to the one used in a previous study in E. coli (70). The controls were cultured without vitamin B12 supplementation. After 14 days, 200 μL bacterial culture was used to measure optical density at 595 nm, and 500 μL bacterial culture was used to extract porphyrins using the method described previously (71) with minor modifications. Briefly, bacterial culture was mixed with 250 μL ethyl acetate/acetic acid (4:1) for 10 seconds by vortexing, and then centrifuged for 5 minutes at 12,000 rpm. The upper phase was transferred to a new tube and mixed with 250 μL 1.5M HCl for 10 seconds. After centrifuging for 2 minutes at 12,000 rpm, 200 μL extracted porphyrins in HCl lower phase was taken and quantified by measuring its absorbance at 405 nm. All the optical absorbance was quantified on GENios Microplate Reader (TECAN). The absorbance at 405 nm was then converted to concentration based on a standard curve determined using coproporphyrin III.
cob/cbi gene expression analysis
Total RNA was extracted from P. acnes cell cultures or skin microbial samples collected from the subjects using the protocol described in Supplementary Materials. The total RNA was converted to single-stranded cDNA using Superscript III first-strand synthesis supermix (Life Technologies). Quantitative RT-PCR was performed on LightCycler 480 (Roche) using the LightCycler 480 High Resolution Melting Master Mix (Roche) and the following primers: cbiL-forward: 5’-GCGCGAGGCAGACGTGATCC-3’, cbiL-reverse: 5’-GACACCGGACCTCTCCCGCA-3’, cysG+cbiX-forward: 5’-TGTATTCCGCCCCGCTGTTGC-3’, cysG+cbiX-reverse: 5’-GAGCACTGCCGACGTGTCCC-3’, btuR-forward: 5’-GGAAGATGCTCTTCGGGCGCT-3’, btuR-reverse: 5’ -GCCTCAGGGTTCTCCGCAGC-3’, 16S-forward: 5’-GGGGCTTAACCCTGAGCGTGC-3’, 16S-reverse: 5’-TTCGCTCCCCACGCTTTCGC-3’. The qRT-PCR protocol was set as the following: initial denaturation at 95°C for 5 minutes, followed by 50 cycles of 95°C for 10 seconds, 62°C for 30 seconds, and 72°C for 30 seconds. The expression level of each gene was expressed as the logarithm of its relative expression level to 16S rRNA transcript level.
Statistics
The differentially expressed OGUs in the RNA-Seq data were identified using Student’s t-test (details in Supplementary Materials). The threshold of statistical significance was set at P < 0.05. The differentially expressed OGUs were further confirmed by ShotgunFunctionalizeR (72), with a cutoff of Akaike’s information criterion < 5,000 and adjusted P < 0.05. The differentially expressed KEGG pathways in acne patients were determined by ShotgunFunctionalizeR, with a cutoff of Akaike’s information criterion < 5,000 and adjusted P < 0.05. DAVID (19) was used to identify the enriched metabolic pathways and functional annotation clusters. Student’s t-test was used to determine the statistical significance of the differences in vitamin B12 gene expression quantified by qRT-PCR and in porphyrin production between the culture conditions.
Unsupervised hierarchical clustering was generated based on the correlation metric of OGU expression levels and the average linkage clustering method was used. The robustness of the clustering was evaluated by bootstrapping method using R package pvclust (73). The significance of the association of the skin clinical status, as well as other subject information, with the clustering was determined by Fisher’s exact test.
Supplementary Material
One Sentence Summary: Vitamin B12 in the host modulates the transcriptional activities of the skin microbiota, leading to increased bacterial porphyrin biosynthesis and acne development.
Acknowledgments:
We thank S. Tomida for suggestions on experimental design and A.C. Maretti-Mira for assistance in sample collection. We thank E. Barnard, T. Johnson, B. Chiu, J. Torres, and L. Altieri for assistance in performing experiments and data analysis. B. Tan provided the photographic comparison of the two different skin sampling methods. We also thank E. Barnard, D. Eisenberg, Z. Guo, M. Liu, M. Pellegrini, and B. Tan for suggestions on improving the manuscript. UC Davis Sequencing Center and UCLA BSCRC Sequencing Center provided sequencing support.
Funding: This study was funded by the NIH grants R01GM099530 from NIGMS and UH2AR057503 from NIAMS.
Footnotes
Author contributions: D.K. performed the experiments, analyzed the data, and wrote the manuscript. B.S. analyzed the data and revised the manuscript. M.C.E. collected clinical samples. N.C. conceived and directed the clinical study, recruited subjects, collected samples, interpreted clinical correlations and data, and revised the manuscript. H.L. conceived and directed the project, analyzed the data, and wrote the manuscript.
Competing interests: The Regents of the University of California is the owner of a patent application (US application No. 62/078,275, titled “New acne treatment targets”) related to the role of vitamin B12 in acne, which names D.K., B.S., N.C., and H.L. as inventors. The other author declares no competing interests.
Data and materials availability: The RNA-Seq reads have been deposited in the NCBI Sequence Read Archive (BioProject number PRJNA260091).
The link to the final version of the manuscript published in Science Translational Medicine is: http://stm.sciencemag.org/content/7/293/293ra103
References and Notes:
- 1.The Human Microbiome Project Consortium, Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitz-Gibbon S, Tomida S, Chiu BH, Nguyen L, Du C, Liu M, Elashoff D, Erfe MC, Loncaric A, Kim J, Modlin RL, Miller JF, Sodergren E, Craft N, Weinstock GM, Li H, Propionibacterium acnes Strain Populations in the Human Skin Microbiome Associated with Acne. J Invest Dermatol 133, 2152–2160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA, Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho I, Blaser MJ, The human microbiome: at the interface of health and disease. Nat Rev Genet 13, 260–270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pflughoeft KJ, Versalovic J, Human microbiome in health and disease. Annu Rev Pathol 7, 99–122 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Marples RR, Leyden JJ, Stewart RN, Mills OH Jr., Kligman AM, The skin microflora in acne vulgaris. J Invest Dermatol 62, 37–41 (1974). [DOI] [PubMed] [Google Scholar]
- 7.Leyden JJ, McGinley KJ, Mills OH, Kligman AM, Propionibacterium levels in patients with and without acne vulgaris. J Invest Dermatol 65, 382–384 (1975). [DOI] [PubMed] [Google Scholar]
- 8.Holland KT, Cunliffe WJ, Roberts CD, The role of bacteria in acne vulgaris: a new approach. Clin Exp Dermatol 3, 253–257 (1978). [DOI] [PubMed] [Google Scholar]
- 9.Cunliffe WJ, Gould DJ, Prevalence of facial acne vulgaris in late adolescence and in adults. Br Med J 1, 1109–1110 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghodsi SZ, Orawa H, Zouboulis CC, Prevalence, severity, and severity risk factors of acne in high school pupils: a community-based study. J Invest Dermatol 129, 2136–2141 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Gupta MA, Gupta AK, Depression and suicidal ideation in dermatology patients with acne, alopecia areata, atopic dermatitis and psoriasis. Br J Dermatol 139, 846–850 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Halvorsen JA, Stern RS, Dalgard F, Thoresen M, Bjertness E, Lien L, Suicidal ideation, mental health problems, and social impairment are increased in adolescents with acne: a population-based study. J Invest Dermatol 131, 363–370 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Gollnick H, Current concepts of the pathogenesis of acne: implications for drug treatment. Drugs 63, 1579–1596 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Jappe U, Pathological mechanisms of acne with special emphasis on Propionibacterium acnes and related therapy. Acta Derm Venereol 83, 241–248 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Webster GF, Inflammation in acne vulgaris. J Am Acad Dermatol 33, 247–253 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Williams HC, Dellavalle RP, Garner S, Acne vulgaris. Lancet 379, 361–372 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Tomida S, Nguyen L, Chiu BH, Liu J, Sodergren E, Weinstock GM, Li H, Pan-genome and comparative genome analyses of propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. MBio 4, e00003–00013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruggemann H, Henne A, Hoster F, Liesegang H, Wiezer A, Strittmatter A, Hujer S, Durre P, Gottschalk G, The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 305, 671–673 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA, DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4, P3 (2003). [PubMed] [Google Scholar]
- 20.Jadassohn W, Paillard R, Hofer R, Golaz M, Vitamine B12 et poussée aenéïforme. Dermatologica 116, 349 (1958). [Google Scholar]
- 21.Puissant A, Vanbremeersch F, Monfort J, Lamberton JN, [A new iatrogenic dermatosis: acne caused by vitamin B 12]. Bull Soc Fr Dermatol Syphiligr 74, 813–815 (1967). [PubMed] [Google Scholar]
- 22.Dugois P, Amblard P, Imbert R, de Bignicourt B, [Acne caused by vitamin B 12]. Lyon medical 221, 1165 (1969). [PubMed] [Google Scholar]
- 23.Braun-Falco O, Lincke H, [The problem of vitamin B6/B12 acne. A contribution on acne medicamentosa (author’s transl)]. MMW Munch Med Wochenschr 118, 155–160 (1976). [PubMed] [Google Scholar]
- 24.Dupre A, Albarel N, Bonafe J, Christol B, Lassere J, Vitamin B-12 induced acnes. Cutis; cutaneous medicine for the practitioner 24, 210 (1979). [PubMed] [Google Scholar]
- 25.Sherertz E, Acneiform eruption due to “megadose” vitamins B6 and B12. Cutis; cutaneous medicine for the practitioner 48, 119 (1991). [PubMed] [Google Scholar]
- 26.Balta I, Ozuguz P, Vitamin B12-induced acneiform eruption. Cutan Ocul Toxicol 33, 94–95 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC, The biosynthesis of adenosylcobalamin (vitamin B12). Nat Prod Rep 19, 390–412 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Ravnum S, Andersson DI, Vitamin B12 repression of the btuB gene in Salmonella typhimurium is mediated via a translational control which requires leader and coding sequences. Mol Microbiol 23, 35–42 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Nou X, Kadner RJ, Coupled changes in translation and transcription during cobalamin-dependent regulation of btuB expression in Escherichia coli. J Bacteriol 180, 6719–6728 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS, Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA 9, 1084–1097 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glass GB, Skeggs HR, Lee DH, Jones EL, Hardy WW, Hydroxocobalamin. I. Blood levels and urinary excretion of vitamin B12 in man after a single parenteral dose of aqueous hydroxocobalamin, aqueous cyanocobalamin and cyanocobalamin zinc-tannate complex. Blood 18, 511–521 (1961). [PubMed] [Google Scholar]
- 32.Tauber SA, Skeggs HR, Itkin A, Stanley JL, Parenteral studies with vitamin B12 complexes. Am J Clin Nutr 10, 480–483 (1962). [DOI] [PubMed] [Google Scholar]
- 33.Skouby AP, Retention and distribution of B12 activity, and requirement for B12, following parenteral administration of hydroxocobalamin (Vibeden). Acta medica Scandinavica 180, 95–105 (1966). [DOI] [PubMed] [Google Scholar]
- 34.Stankler L, The vitamin B12 level in psoriatic skin and serum. Br J Dermatol 81, 911–918 (1969). [DOI] [PubMed] [Google Scholar]
- 35.Murakami K, Hashimoto Y, Murooka Y, Cloning and characterization of the gene encoding glutamate 1-semialdehyde 2,1-aminomutase, which is involved in delta-aminolevulinic acid synthesis in Propionibacterium freudenreichii. Appl Environ Microbiol 59, 347–350 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bykhovskii V, Zaitseva NI, Bukin VN, [Possible competition in the use of delta-aminolevulinic acid for the biosynthesis of vitamin B 12 and porphyrin by resting suspensions of Propionibacterium shermanii]. Dokl Akad Nauk SSSR 180, 232–234 (1968). [PubMed] [Google Scholar]
- 37.Friedmann HC, Cagen LM, Microbial biosynthesis of B12-like compounds. Annu Rev Microbiol 24, 159–208 (1970). [DOI] [PubMed] [Google Scholar]
- 38.Zaitseva NI, Bykhovskii V, Bukin VN, [Regulation of vitamin B-12 and prophyrin biosynthesis in Propionibacterium shermanii]. Dokl Akad Nauk SSSR 190, 1476–1479 (1970). [PubMed] [Google Scholar]
- 39.Saint-Leger D, Bague A, Cohen E, Chivot M, A possible role for squalene in the pathogenesis of acne. I. In vitro study of squalene oxidation. Br J Dermatol 114, 535–542 (1986). [DOI] [PubMed] [Google Scholar]
- 40.Gribbon EM, Shoesmith JG, Cunliffe WJ, Holland KT, The microaerophily and photosensitivity of Propionibacterium acnes. J Appl Bacteriol 77, 583–590 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Schaller M, Loewenstein M, Borelli C, Jacob K, Vogeser M, Burgdorf WH, Plewig G, Induction of a chemoattractive proinflammatory cytokine response after stimulation of keratinocytes with Propionibacterium acnes and coproporphyrin III. Br J Dermatol 153, 66–71 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Goldblatt S, [Vitamin B-12 levels in blood serum in acne-like skin diseases]. Hautarzt 17, 106–108 (1966). [PubMed] [Google Scholar]
- 43.Karadag AS, Tutal E, Ertugrul DT, Akin KO, Effect of isotretinoin treatment on plasma holotranscobalamin, vitamin B12, folic acid, and homocysteine levels: noncontrolled study. Int J Dermatol 50, 1564–1569 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Martin RJ, Kahn G, Gooding JW, Brown G, Cutaneous porphyrin fluorescence as an indicator of antibiotic absorption and effectiveness. Cutis 12, 758–764 (1973). [Google Scholar]
- 45.Mills OH Jr., Kligman AM, Pochi P, Comite H, Comparing 2.5%, 5%, and 10% benzoyl peroxide on inflammatory acne vulgaris. Int J Dermatol 25, 664–667 (1986). [DOI] [PubMed] [Google Scholar]
- 46.Borelli C, Merk K, Schaller M, Jacob K, Vogeser M, Weindl G, Berger U, Plewig G, In vivo porphyrin production by P. acnes in untreated acne patients and its modulation by acne treatment. Acta Derm Venereol 86, 316–319 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Mayr-Kanhauser S, Kranke B, Aberer W, Efficacy of octenidine dihydrochloride and 2-phenoxyethanol in the topical treatment of inflammatory acne. Acta Dermatovenerol Alp Panonica Adriat 17, 139–143 (2008). [PubMed] [Google Scholar]
- 48.Simon GL, Gorbach SL, Intestinal flora in health and disease. Gastroenterology 86, 174–193 (1984). [PubMed] [Google Scholar]
- 49.Washio J, Sato T, Koseki T, Takahashi N, Hydrogen sulfide-producing bacteria in tongue biofilm and their relationship with oral malodour. J Med Microbiol 54, 889–895 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG, Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 184, 957–963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ling Z, Liu X, Chen X, Zhu H, Nelson KE, Xia Y, Li L, Xiang C, Diversity of cervicovaginal microbiota associated with female lower genital tract infections. Microb Ecol 61, 704–714 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Leyden JJ, McGinley KJ, Nordstrom KM, Webster GF, Skin microflora. J Invest Dermatol 88, 65s–72s (1987). [DOI] [PubMed] [Google Scholar]
- 53.Grice EA, Segre JA, The skin microbiome. Nat Rev Microbiol 9, 244–253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tremaroli V, Backhed F, Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, Li H, Alekseyenko AV, Blaser MJ, Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488, 621–626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ, Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ, Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD, Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H, Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt E, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL, Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N, Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI, Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6, 279–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dreno B, Poli F, Pawin H, Beylot C, Faure M, Chivot M, Auffret N, Moyse D, Ballanger F, Revuz J, Development and evaluation of a Global Acne Severity Scale (GEA Scale) suitable for France and Europe. J Eur Acad Dermatol Venereol 25, 43–48 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Consortium THMP, A framework for human microbiome research. Nature 486, 215–221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO, The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41, D590–596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langmead B, Trapnell C, Pop M, Salzberg SL, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, Heath AC, Knight R, Gordon JI, Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci U S A 108 Suppl 1, 4599–4606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li W, Jaroszewski L, Godzik A, Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 17, 282–283 (2001). [DOI] [PubMed] [Google Scholar]
- 69.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J, The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250–255 (2010). [DOI] [PubMed] [Google Scholar]
- 70.Lundrigan MD, Kadner RJ, Altered cobalamin metabolism in Escherichia coli btuR mutants affects btuB gene regulation. J Bacteriol 171, 154–161 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Piomelli S, Davidow B, Guinee VF, Young P, Gay G, The FEP (free erythrocyte porphyrins) test: a screening micromethod for lead poisoning. Pediatrics 51, 254–259 (1973). [PubMed] [Google Scholar]
- 72.Kristiansson E, Hugenholtz P, Dalevi D, ShotgunFunctionalizeR: an R-package for functional comparison of metagenomes. Bioinformatics 25, 2737–2738 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Suzuki R, Shimodaira H, Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, Delong EF, Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci U S A 105, 3805–3810 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Poretsky RS, Bano N, Buchan A, LeCleir G, Kleikemper J, Pickering M, Pate WM, Moran MA, Hollibaugh JT, Analysis of microbial gene transcripts in environmental samples. Appl Environ Microbiol 71, 4121–4126 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA, The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4, 41 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M, KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40, D109–114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oksanen J, Blanchet FG, Kindt R, Legendre P, O’Hara RG, Simpson GL, Solymos P, Henry M, Stevens H, Wagner H, vegan: Community Ecology Package. R package version 1.17–0. 2010. [Google Scholar]
- 79.Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B, gplots: Various R programming tools for plotting data. R package version 2.7.4. 2009. [Google Scholar]
- 80.Cristino AS, Tanaka ED, Rubio M, Piulachs MD, Belles X, Deep sequencing of organ- and stage-specific microRNAs in the evolutionarily basal insect Blattella germanica (L.) (Dictyoptera, Blattellidae). PLoS One 6, e19350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P, iPath2.0: interactive pathway explorer. Nucleic Acids Res 39, W412–415 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kasimatis G, Fitz-Gibbon S, Tomida S, Wong M, Li H, Analysis of complete genomes of Propionibacterium acnes reveals a novel plasmid and increased pseudogenes in an acne associated strain. BioMed research international 2013, 918320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


