Abstract
Bacteria are under constant predation from viruses, called bacteriophages (phages). This threat has driven the evolution of multiple defense systems, including the CRISPR-Cas immune pathway. Phages are not passive bystanders in their CRISPR-mediated demise however, as many have developed potent protein inhibitors of this bacterial adaptive immune system. Many CRISPR-Cas systems have been identified in nature, along with cognate “anti-CRISPR” proteins, the Type II CRISPR-Cas9 system has received much of the focus, after being co-opted as a revolutionary genome editing tool. Here, I review the work that led to the discovery of many distinct CRISPR-Cas inhibitor proteins. Furthermore, I outline how understanding their mechanisms of action has provided us with a suite of specific and high-affinity reagents to modulate and study CRISPR-Cas applications.
Introduction
Many powerful biotechnologies have been derived from the molecular arms race between bacteria and their viruses. Bacteriophages (phages) or “bacteria eaters” were discovered >100 years ago1, and are still shaping our understanding of molecular biology and providing new tools2. There are an estimated 1023 phage infections per second on the planet, driven by the massive numbers of phages and bacteria in the ocean3,4. Advances in high throughput sequencing technologies, extensive sampling, and microscopy have led to the realization that phages are a prominent member of nearly all ecological niches, including the human microbiome5. This appreciation of their abundance, but a poor understanding of their roles, in combination with a dire need for new mechanisms to combat antimicrobial resistance, has led phage biology into a renaissance in recent years. Historically, elucidating the mechanisms by which phages infect their host bacteria led to the identification of ligases, polymerases, recombinases, and restriction enzymes, among many other reagents6. More recently, efforts to identify new ways that bacteria protect themselves from phages led to the discovery of a novel and powerful new immune system, known as CRISPR-Cas7.
Clustered regularly interspaced short palindromic repeats (CRISPR) are arrays of repetitive DNA found in the genomes of bacteria and archaea. The spacing sequences between the direct repeats can possess sequence identity to phage genomes, representing a vaccination card or memory component of the first adaptive immune system identified in prokaryotes. Together with CRISPR-associated (cas) genes, this system harvests small sequences (~30 bp) from a phage genome, incorporates it into the CRISPR array, and subsequently transcribes, processes and packages these CRISPR RNAs (crRNAs) into Cas protein complexes that surveil the microbial cell for invasion. Detection of a foreign invader via complementarity between the crRNA sequence and the phage RNA or DNA, mediates recognition of the target, which is subsequently cleaved with remarkable specificity. Six distinct types of CRISPR-Cas system (Types I–VI) have been discovered to date8, divided broadly into two classes, those that utilize a multi-protein surveillance complex (Class 1, Types I, III, IV) and those that utilize a single protein effector nuclease (Class 2, Types II, V, VI).
The discovery that microbes program sequence-specific nucleases with RNA guides has been harnessed since 2012 to design and unleash precision double stranded breaks on genomes from many organisms, including humans, leading to the “CRISPR-Cas revolution” in genome editing9–12. While this technology initially focused on the Cas9 nuclease, other Class 2 effectors such as Cas12 (Cpf1) and Cas13 (C2c2) have recently been utilized due to the simplicity of single protein effectors guided by a single RNA13–15. As is the case with any formidable immune system, nature has developed powerful antagonists, and CRISPR-Cas systems are no exception. Here I describe the latest iteration in our understanding of CRISPR-Cas evolution, and yet another reagent borne out of the phage-bacteria arms race, anti-CRISPR proteins.
The phage counter attack
A recurrent theme in studying the molecular battle between phages and their hosts has been the emergence of counter-defence strategies deployed by phages. The ability of viruses to shut down immune pathways has also been well documented in eukaryotes16,17. Decades of work on the bacterial “innate” immune system, restriction-modification (R-M), has generated literature to inform searches for similar mechanisms of CRISPR-Cas evasion. The parallels between R-M and CRISPR-Cas extend much further, as the fundamental discovery of restriction enzymes from the phage-host battle enabled recombinant DNA construction, and now CRISPR-Cas has provided the in vivo equivalent for DNA manipulation. Phage-encoded inhibitors of R-M systems take many shapes and forms, largely following three themes: i) modifying the target of the immune system, ii) mimicking the target of the immune system iii) disabling the immune system18. These strategies have been paralleled by anti-CRISPR proteins, which function by either mimicking or occluding the target DNA, or directly disabling CRISPR nucleases, as described below.
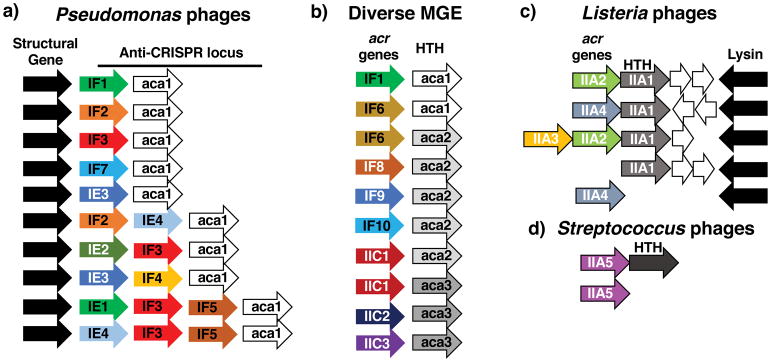
The first report of proteins inhibiting CRISPR-Cas function emerged in 2013, encoded by phages that infect the opportunistic human pathogen, Pseudomonas aeruginosa19. This discovery was enabled by the observation that integrated phage genomes were inactivating the endogenous CRISPR-Cas system, suggesting that they expressed inhibitor proteins. These anti-CRISPR proteins inactivate the Class 1, Type I CRISPR-Cas system, which is comprised of a multi-subunit ribonucleoprotein effector complex and a trans-acting nuclease/helicase, Cas320–22. The most remarkable aspect of the first set of five “anti-CRISPR” proteins discovered, was their diversity. The only commonality between these genes was that they were small (~150–450 base pairs) and encoded by closely related phages at the same genomic position, or “anti-CRISPR locus” (Fig. 1a). By expressing each of these proteins in P. aeruginosa, it was revealed that they each possessed anti-CRISPR activity against either the type I-F or type I-E CRISPR-Cas system19,23, both of which are found in P. aeruginosa isolates24,25. Anti-CRISPR genes are named in the order with which they were discovered and describe the system they inhibit, for example acrIF1 (anti-CRISPR for type I-F). Note that this has been updated from earlier nomenclature, acrF1.
Figure 1.
a) The Pseudomonas aeruginosa phage anti-CRISPR locus. A syntenic region from ten different phage genomes is shown to highlight the varied assortment of Type I-F (IF1–IF5, IF7) and I-E (IE1–IE4) anti-CRISPR genes that are found in related phage genomes, anchored adjacent to a conserved structural gene (black). The presence of anti-CRISPR associated gene 1 (aca1) is highly conserved in these anti-CRISPR loci, b) The helix-turn-helix (HTH) encoding aca1 gene enabled identification of new anti-CRISPR genes, found next to novel HTH proteins (aca2, aca3), leading to the identification of Type II-C anti-CRISPRs (IIC1–3), c) Type II-A anti-CRISPR genes (IIA1–4) are shown with their associated HTH gene, which is also an anti-CRISPR (IIA1), d) AcrIIA5, found in Streptococcus phages, associated with an HTH in certain phages. The gene size and spacing are not to scale.
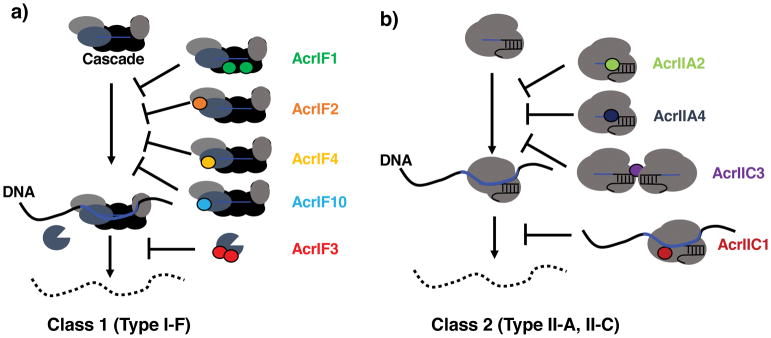
While the anti-CRISPR genes themselves did not possess informative homologs or domain structures, biochemical experiments revealed that Type I-F anti-CRISPR proteins interact directly with Cas proteins (Fig. 2a). For example, two distinct anti-CRISPR proteins, AcrIF1 and AcrIF2 interact with the Cascade surveillance complex members Cas7 and Cas8, respectively, which blocks DNA binding26. A third anti-CRISPR protein, AcrIF3, prevents the recruitment of the Cas3 nuclease via a direct interaction26. Subsequent structures of anti-CRISPRs both bound and unbound to their targets confirmed the truly distinct nature of these proteins, as they possessed no common fold27,28. However, thorough analysis of structures of the I-F Cascade complex bound to various anti-CRISPRs (i.e. AcrIF1, AcrIF2, AcrIF10) has revealed common strategies including steric occlusion of DNA binding and DNA mimicry29,30. Anti-CRISPR diversity across even closely related phages underscores the strong selective pressures placed on an immune/anti-immune effector battle and emphasizes how this conflict is a “hot spot” for the generation of novel genes31. With 14 Type I anti-CRISPR proteins having been identified19,23,32,33, the only commonality that unites them is often a shared genomic locus within a mobile element such as a prophage, and an association with a putative regulatory helix-turn-helix (HTH) protein.
Figure 2.
Summary of known mechanisms of CRISPR-Cas inhibition. a) In multi-protein subunit Class 1 CRISPR-Cas system, Type I-F inhibitors function by binding to the RNA-guided Cascade complex and preventing DNA binding (AcrIF1, AcrIF2, AcrIF4), or by interacting with the trans-acting nuclease Cas3 and preventing recruitment to the DNA target (AcrIF3). b) In the single protein effector Class 2 CRISPR-Cas system, Type II-A and II-C inhibitors bind directly to Cas9 and block DNA binding (AcrIIA2, AcrIIA4, AcrIIC3), or block DNA cutting by limiting the movement of the HNH nuclease domain upon DNA binding (AcrIIC1). In the absence of these inhibitor proteins, the DNA is degraded (dashed lines)
CRISPR-Cas9 inhibitors
The predictive power and importance of anti-CRISPR loci was clearly demonstrated by the discovery of the first inhibitors of the Type II CRISPR-Cas9 system. The locus where Type I anti-CRISPRs (acrIF1-F5, acrIF7, and acrIE1-4) were found always possessed a conserved, predicted helix-turn-helix protein (HTH, anti-CRISPR associated protein 1, aca1) adjacent to anti-CRISPR genes. While the function of this gene is still unknown, it served as a powerful indicator gene for discovery of new anti-CRISPRs acrIF6, and acrIF8-F10 (Fig. 1b). While identifying new Type I anti-CRISPRs, Pawluk and colleagues identified a new HTH anti-CRISPR associated gene, aca232. Homologs of this gene were identified next to new anti-CRISPRs, which in turn, led to a new gene encoding a predicted HTH-protein, aca3. Remarkably, this bioinformatic “guilt-by-association” based search led to anti-CRISPR loci in Neisseria meningitidis mobile elements, an organism with a well characterized Type II-C CRISPR-Cas9 system34,35. Three candidates identified in this way possessed inhibitory activity against NmCas9 in natural Neisseria hosts, in vitro, and in human cells36. Subsequent structural and biochemical work has revealed that AcrIIC1 is a broad spectrum Type II-C Cas9 inhibitor, which prevents movement of the HNH nuclease domain toward its DNA substrate, while AcrIIC3 inhibits Cas9 through a mechanism involving a forced dimerization of Cas937 (Fig. 2b).
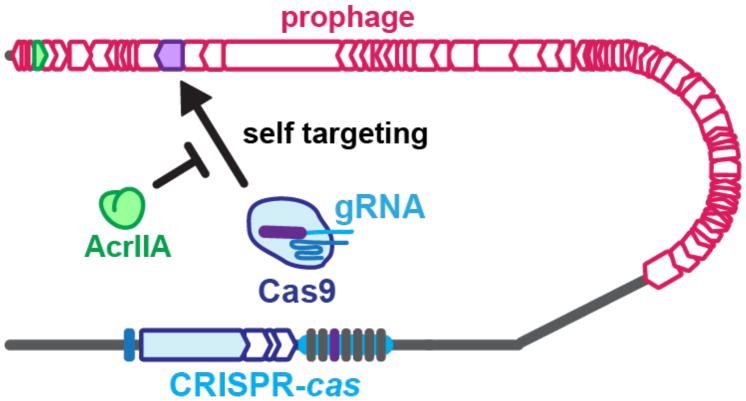
Cas9 has been the most widely utilized CRISPR-Cas enzyme for biotechnological applications. Specifically, the Type II-A orthologue from Streptococcus pyogenes (SpyCas9) has been deployed for gene editing in numerous organisms, as well as many DNA-binding applications derived from catalytically dead Cas9 (dCas9)12. To identify anti-CRISPRs that function against the Type II-A system, a bioinformatics approach was developed, which surveyed genomes for “self-targeting” (Fig. 3). This describes a situation where one or more endogenous CRISPR spacers encode for the production of a crRNA complex that would mediate the cleavage of a target in the same genome. Cleavage of a bacterial genome in this manner would be lethal and therefore, broadly speaking, the self-targeting CRISPR system must be disabled in some manner for the continued survival of the cell. There are many possible ways to inactivate CRISPR systems, including mutation, transcriptional repression, or anti-CRISPR inhibition. In the bacterium Listeria monocytogenes, however, self-targeting was revealed to be directly stabilized by one of four anti-CRISPR proteins (AcrIIA1-4), present in numerous related prophages (Fig. 1c). Again, a characteristic anti-CRISPR locus was observed where conserved phage genes flanked a diverse locus with genes possessing anti-CRISPR function38.
Figure 3.
The spacers in a CRISPR array dictate the identity of the guide RNAs (gRNAs) that will be generated by that strain. In this example, a ‘purple’ gRNA possesses perfect complementarity to a sequence in the bacterial genome, specifically in a prophage. This will result in the destruction of the genome and cellular death, if not for an “AcrIIA” protein that neutralizes the threat.
The recent discovery of a fifth Type II-A Cas9 inhibitor (AcrIIA5) was made by immunizing strains of Streptococcus thermophilus with spacers that target various phages and identifying a phage that resists targeting39 (Fig. 1d). Iterative cloning of phage open reading frames led to the identification of acrIIA5, which possessed Cas9-inhibitory activity when expressed in isolation. Notably, acrIIA5 is encoded both by lytic and temperate phages, a property which is also shared by acrIIA4. AcrIIA5 was also functional against SpyCas9, much like AcrIIA4, suggesting broad spectrum inhibition.
Proteins AcrIIA2 and A4 were shown to block Cas9-DNA-binding using CRISPRi assays in E. coli. This mechanism was later supported by direct biochemical evidence that these proteins interact tightly with Cas9 after sgRNA loading (Kd= 4 nM) and prevent DNA binding in vitro40 (Fig. 2b). Structural studies revealed that AcrIIA4 mimics DNA and binds to the Cas9 residues that are required for binding to the PAM nucleotides in the DNA template40–42. No structural or biochemical mechanisms have been demonstrated for the other Type II-A Cas9 inhibitors, but it can be expected that diverse routes to Cas9 inhibition will be advantageous for downstream applications.
Applications for phage inhibitor proteins
Phage enzymes such as T4 DNA ligase, T7 polymerase, and Cre recombinase have been applied broadly as in vitro and in vivo biotechnologies for decades. Phages and phage proteins are also being widely studied and utilized in the fight against antimicrobial resistant pathogens. Some well-studied phages such as M13 and T7 have even been co-opted for numerous phage display-based technologies, which enable screens for protein-protein interactions. However, it is less common for non-enzymatic phage-derived inhibitors (such as anti-CRISPRs) to have been co-opted for biotechnology. One powerful example, however, is the use of the uracil glycosylase inhibitor (UGI) from Bacillus subtilis phage PBS1, which displays remarkably strong inhibition of the human enzyme. This has recently enabled precise base editing technologies with fused domains to dCas943,44. Given the strong binding affinity that many phage-encoded inhibitors display for their target (e.g. UGI IC50 = 12 pM), uses in eukaryotic systems may be plentiful.
All anti-CRISPR proteins are defined by their ability to block CRISPR-Cas-mediated DNA cleavage in vivo. Moreover, all characterized anti-CRISPRs function through direct interactions with a target Cas protein26,37,40. Therefore, Cas9 inhibitors have utility as a specific and direct “off-switch” for Cas9 gene editing in heterologous hosts such as human cells. One of the largest hurdles currently faced by gene editing applications is off-target mutations that can occur during editing45,46. While on-target events occur with faster kinetics than off-targeting, the long-term stability of Cas9 in the host nucleus also leads to off-target mutations. The extent of this problem can be mitigated by thoughtful sgRNA design47, and varies depending on many factors including cell type, Cas9 concentration, and Cas9 delivery modality48,49. For therapeutic applications, a limitation arises with the need to target a particular site during allele-specific targeting, where investigators are limited to a single target site and sgRNA sequence. In these cases, anti-CRISPRs may represent a particularly useful reagent to limit off-target editing. As a proof-of-principle, the specific anti-Cas9 activity of AcrIIA4 (SpyCas9 inhibitor), has been shown to reduce off-target editing in cell culture when deployed via protein or plasmid delivery41. Many other strategies are also being used to limit off-target editing such as mutagenesis of Cas9 to enhance accuracy, leading to the generation of eSpyCas9(1.1), SpyCas9-HF1, and most recently, HypaCas950–52. Other strategies to enable enhanced control in cells include the addition of ligand binding domains to generate an allosteric switch53 and small molecule-mediated degradation54. While not yet experimentally tested, inhibitor proteins are likely compatible with these engineering efforts and could be applied in concert to enable enhanced precision and safety for future applications where preventing off-target editing is absolutely critical.
Many anti-CRISPRs function by inhibiting DNA-binding by CRISPR-Cas complexes (AcrIF1, IF2, IF4, IIA2, IIA4, IIC1, IIC3)26,36,38. This inhibits various CRISPR-Cas applications such as gene editing itself36,38,41, CRISPRi38, and CRISPR-based imaging36. For applications that do not result in a change to the DNA sequence (i.e. CRISPRi, imaging), inhibitors are also likely of use due to their ability to impede a dCas9 based application such as transcriptional repression. Although AcrIIA4 is unable to displace Cas9 from bound DNA, cellular factors that likely contribute to Cas9 displacement could provide free Cas9 for AcrIIA4 to bind, ultimately titrating dCas9 away from its DNA target. At a fundamental level, stoichiometric inhibitor proteins are also useful to understand the dynamics of dCas9 based activities, prevent leaky CRISPRi activity, and build circuits that cycle dCas9 based modalities between tightly controlled on and off states.
Not all anti-CRISPR proteins are complete off-switches for CRISPR-Cas function. Remarkably, some specifically disable DNA cleavage, while still permitting DNA-binding. In both the Type I systems (AcrIF3 inhibits Cas3 recruitment) and in Type II systems, (AcrIIC1 inhibits Cas9 HNH domain activation), cleavage inhibition has been reported, which serves to enable CRISPRi (Fig. 2)26,28,37. While the physiological importance of this partial inactivation of CRISPR-Cas in bacteria is not yet clear, these proteins clearly enable CRISPRi transcriptional repression with CRISPR-Cas systems that are otherwise genetically wild-type26. AcrIIC1 performing this function was only recently reported37, and thus applications have not yet been fully explored, however the ability to toggle Cas9 between catalytically active and dead states in this manner could enable new applications.
Future Considerations
Future work to explore applications will focus, like CRISPR-Cas9 technology itself, on delivery. Inhibitors have been successfully provided on plasmids or as protein41 and will need to be continually evaluated for their impact on the ratio of on- to off-target editing in any particular setting. Of particular note is the advantage of a genetically encoded Cas9 off-switch as CRISPR-Cas9 technology is applied to living organisms. For example, provision of an inhibitor gene in the same viral vector as a CRISPR effector nuclease would ensure that every cell in a given organism receives both the nuclease and the off-switch. Additionally, there are valid concerns that gene drive technology (which forces non-Mendelian inheritance of a mutant allele using CRISPR-Cas955,56) could become uncontrolled or used for nefarious purposes. In theory, Cas9 inhibitor expression would be able to halt a gene drive in its tracks.
As demonstrated by Figure 1, the identification of an anti-CRISPR locus for any CRISPR-Cas subtype is a remarkable finding, as it can function as a key to discovering numerous independent proteins that inhibit CRISPR-Cas function through diverse mechanisms. Once a locus is identified, there is massive potential for novel gene discovery, limited only by the extent to which the relevant elements have been sequenced. As researchers continue to discover and mine these mysterious loci, which are seemingly assembled as a “grab bag” of anti-CRISPRs, these proteins provide novel reagents for mechanistic and structural studies as well as applications described here. For more consideration of the natural biology and evolutionary trajectory of anti-CRISPRs, their loci and how bacteria might fight back, I direct the reader to recently published reviews33,57. Although only a small number of such anti-CRISPR loci have been identified thus far, discovering more will continue to bring large sums of phage genomic “dark matter” constituents into the light.
Table 1.
Anti-CRISPR Proteins
| Name | Type | Source | Inhibits (Organism) | Accession |
|---|---|---|---|---|
| AcrIF1 | I-F | Pseudomonas aeruginosa phage | Cascade (Pae, Pec) | YP_007392342.1 |
| AcrIF2 | I-F | P. aeruginosa phage | Cascade (Pae, Pec) | YP_002332454.1 |
| AcrIF3 | I-F | P. aeruginosa phage | Cas3 (Pae) | YP_007392440.1 |
| AcrIF4 | I-F | P. aeruginosa phage | Cascade (Pae) | WP_016068584.1 |
| AcrIF5 | I-F | P. aeruginosa phage | Target unknown (Pae) | YP_007392740.1 |
| AcrIF6 | I-E/I-F | P. aeruginosa prophage | Target unknown (Pae, Pec) | WP_043884810.1 |
| AcrIF7 | I-F | P. aeruginosa phage | Target unknown (Pae, Pec) | YP_009146150.1 |
| AcrIF8 | I-F | Pectobacterium atrosepticum phage | Target unknown (Pae, Pec) | YP_007006940.1 |
| AcrIF9 | I-F | Vibrio parahaemolyticus prophage | Target unknown (Pae, Pec) | WP_031500045 |
| AcrIF10 | I-F | Shewanella xiamenensis prophage | Target unknown (Pae, Pec) | WP_037415910.1 |
| | ||||
| AcrIE1 | I-E | P. aeruginosa phage | Target unknown (Pae) | YP_007392738.1 |
| AcrIE2 | I-E | P. aeruginosa phage | Target unknown (Pae) | YP_007392439.1 |
| AcrIE3 | I-E | P. aeruginosa phage | Target unknown (Pae) | YP_950454.1 |
| AcrIE4 | I-E | P. aeruginosa phage | Target unknown (Pae) | NP_938238.1 |
| | ||||
| AcrIIA1 | II-A | L. monocytogenes prophage | Target unknown (Lmo) | WP_003722518.1 |
| AcrIIA2 | II-A | L. monocytogenes prophage | Cas9 (Lmo, Spy) | WP_003722517.1 |
| AcrIIA3 | II-A | L. monocytogenes prophage | Target unknown (Lmo) | WP_014930691.1 |
| AcrIIA4 | II-A | L. monocytogenes prophage | Cas9 (Lmo, Spy) | WP_003723290.1 |
| AcrIIA5 | II-A | Streptococcus thermophilus phage | Target unknown (Sth, Spy) | |
| | ||||
| AcrIIC1 | II-C | Neisseria meningitidis mobile element | Cas9 (Nme, Cje, Geo) | WP_049360089.1 |
| AcrIIC2 | II-C | N. meningitidis prophage | Cas9 (Nme) | WP_042743678.1 |
| AcrIIC3 | II-C | N. meningitidis prophage | Cas9 (Nme) | WP_042743676.1 |
Note: Anti-CRISPR nomenclature has been updated to reflect the type and the subtype. For example, what was formerly AcrF1 is now AcrIF1.
Pae: Pseudomonas aeruginosa, Lmo: Listeria monocytogenes, Spy: Streptococcus pyogenes, Sth: Streptococcus thermophilus, Nme: Neisseria meningitidis, Cje: Campylobacter jejuni, Geo: Geobacillus stearothermophilus
Acknowledgments
I thank A. Borges for critical reading of this manuscript, and A. Borges, B. Rauch, and F. Jiang (J. Doudna lab) for contributing to the figures. CRISPR-Cas work in the Bondy-Denomy lab is supported by the University of California San Francisco Program for Breakthrough in Biomedical Research, funded in part by the Sandler Foundation, and an NIH Office of the Director Early Independence Award (DP5-OD021344).
References
- 1.d’Herelle F. An invisible microbe that is antagonistic to the dysentery bacillus. Comptes rendus Acad Sciences. 1917;165:373–375. [Google Scholar]
- 2.Salmond GPC, Fineran PC. A century of the phage: past, present and future. Nat Rev Micro. 2015;13:777–786. doi: 10.1038/nrmicro3564. [DOI] [PubMed] [Google Scholar]
- 3.Suttle CA. Marine viruses--major players in the global ecosystem. Nat Rev Micro. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 4.Suttle CA. Viruses in the sea. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 5.Waller AS, Yamada T, Kristensen DM, Kultima JR, Sunagawa S, Koonin EV, Bork P. Classification and quantification of bacteriophage taxa in human gut metagenomes. The ISME Journal. 2014;8:1391–1402. doi: 10.1038/ismej.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snyder L. Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents? Molecular Microbiology. 1995;15:415–420. doi: 10.1111/j.1365-2958.1995.tb02255.x. [DOI] [PubMed] [Google Scholar]
- 7.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 8.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nature Biotechnology. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 13.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:eaam9321–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. Antiviral RNA Interference in Mammalian Cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Mierlo JT, Overheul GJ, Obadia B, van Cleef KWR, Webster CL, Saleh MC, Obbard DJ, van Rij RP. Novel Drosophila Viruses Encode Host-Specific Suppressors of RNAi. PLoS Pathog. 2014;10:e1004256. doi: 10.1371/journal.ppat.1004256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samson JE, Magadán AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nat Rev Micro. 2013;11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 19.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westra ER, van Erp PBG, Künne T, Wong SP, Staals RHJ, Seegers CLC, Bollen S, Jore MM, Semenova E, Severinov K, de Vos WM, Dame RT, de Vries R, Brouns SJJ, van der Oost J. CRISPR Immunity Relies on the Consecutive Binding and Degradation of Negatively Supercoiled Invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiedenheft B, van Duijn E, Bultema JB, Bultema J, Waghmare SP, Waghmare S, Zhou K, Barendregt A, Westphal W, Heck AJR, Heck A, Boekema EJ, Boekema E, Dickman MJ, Dickman M, Doudna JA. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proceedings of the National Academy of Sciences. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pawluk A, Bondy-Denomy J, Cheung VHW, Maxwell KL, Davidson AR. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. mBio. 2014;5:e00896–14. doi: 10.1128/mBio.00896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cady KC, White AS, Hammond JH, Abendroth MD, Karthikeyan RSG, Lalitha P, Zegans ME, O’toole GA. Prevalence, conservation and functional analysis of Yersinia and Escherichia CRISPR regions in clinical Pseudomonas aeruginosa isolates. Microbiology. 2011;157:430–437. doi: 10.1099/mic.0.045732-0. [DOI] [PubMed] [Google Scholar]
- 25.van Belkum A, Soriaga LB, LaFave MC, Akella S, Veyrieras JB, Barbu EM, Shortridge D, Blanc B, Hannum G, Zambardi G, Miller K, Enright MC, Mugnier N, Brami D, Schicklin S, Felderman M, Schwartz AS, Richardson TH, Peterson TC, Hubby B, Cady KC. Phylogenetic Distribution of CRISPR-Cas Systems in Antibiotic-Resistant Pseudomonas aeruginosa. mBio. 2015;6:e01796–15. doi: 10.1128/mBio.01796-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, Wiedenheft B, Maxwell KL, Davidson AR. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature. 2015;526:136–139. doi: 10.1038/nature15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maxwell KL, Garcia B, Bondy-Denomy J, Bona D, Hidalgo-Reyes Y, Davidson AR. The solution structure of an anti-CRISPR protein. Nature Communications. 2016;7:13134. doi: 10.1038/ncomms13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Yao D, Xu J-G, Li A-R, Xu J, Fu P, Zhou Y, Zhu Y. Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3. Nat Struct Mol Biol. 2016;23:868–870. doi: 10.1038/nsmb.3269. [DOI] [PubMed] [Google Scholar]
- 29.Chowdhury S, Carter J, Rollins MF, Golden SM, Jackson RN, Hoffmann C, Nosaka L, Bondy-Denomy J, Maxwell KL, Davidson AR, Fischer ER, Lander GC, Wiedenheft B. Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex. Cell. 2017;169:47–57 e11. doi: 10.1016/j.cell.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo TW, Bartesaghi A, Yang H, Falconieri V, Rao P, Merk A, Eng ET, Raczkowski AM, Fox T, Earl LA, Patel DJ, Subramaniam S. Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex. Cell. 2017;171:414–426 e12. doi: 10.1016/j.cell.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carter J, Hoffman C, Wiedenheft B. The Interfaces of Genetic Conflict Are Hot Spots for Innovation. Cell. 2017;168:9–11. doi: 10.1016/j.cell.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pawluk A, Staals RHJ, Taylor C, Watson BNJ, Saha S, Fineran PC, Maxwell KL, Davidson AR. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nature Microbiology. 2016;1:1–6. doi: 10.1038/nmicrobiol.2016.85. [DOI] [PubMed] [Google Scholar]
- 33.Borges AL, Davidson AR, Bondy-Denomy J. The Discovery, Mechanisms, and Evolutionary Impact of Anti-CRISPRs. Annual Review of Virology. 2017;4 doi: 10.1146/annurev-virology-101416-041616. annurev–virology–101416–041616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, Vogel J, Sontheimer EJ. Processing-Independent CRISPR RNAs Limit Natural Transformation in Neisseria meningitidis. Mol Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Rajan R, Seifert HS, Mondragón A, Sontheimer EJ. DNase H Activity of Neisseria meningitidis Cas9. Mol Cell. 2015;60:242–255. doi: 10.1016/j.molcel.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pawluk A, Amrani N, Zhang Y, Garcia B, Hidalgo-Reyes Y, Lee J, Edraki A, Shah M, Sontheimer EJ, Maxwell KL, Davidson AR. Naturally Occurring Off-Switches for CRISPR-Cas9. Cell. 2016;167:1829–1838 e9. doi: 10.1016/j.cell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrington LB, Doxzen KW, Ma E, Liu J-J, Knott GJ, Edraki A, Garcia B, Amrani N, Chen JS, Cofsky JC, Kranzusch PJ, Sontheimer EJ, Davidson AR, Maxwell KL, Doudna JA. A Broad-Spectrum Inhibitor of CRISPR-Cas9. Cell. 2017 doi: 10.1016/j.cell.2017.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rauch BJ, Silvis MR, Hultquist JF, Waters CS, McGregor MJ, Krogan NJ, Bondy-Denomy J. Inhibition of CRISPR-Cas9 with Bacteriophage Proteins. Cell. 2017;168:150–158 e10. doi: 10.1016/j.cell.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hynes AP, Rousseau GM, Lemay ML, Horvath P, Romero DA, Fremaux C, Moineau S. An anti-CRISPR from a virulent streptococcal phage inhibits Streptococcus pyogenes Cas9. Nature Microbiology. 2017;315:1. doi: 10.1038/s41564-017-0004-7. [DOI] [PubMed] [Google Scholar]
- 40.Dong D, Guo M, Wang S, Zhu Y, Wang S, Xiong Z, Yang J, Xu Z, Huang Z. Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein. Nature. 2017 doi: 10.1038/nature22377. [DOI] [PubMed] [Google Scholar]
- 41.Shin J, Jiang F, Liu JJ, Bray NL, Rauch BJ, Baik SH, Nogales E, Bondy-Denomy J, Corn JE, Doudna JA. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci Adv. 2017;3:e1701620. doi: 10.1126/sciadv.1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang H, Patel DJ. Inhibition Mechanism of an Anti-CRISPR Suppressor AcrIIA4 Targeting SpyCas9. Mol Cell. 2017:1–17. doi: 10.1016/j.molcel.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, Mosbaugh DW, Tainer JA. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell. 1995;82:701–708. doi: 10.1016/0092-8674(95)90467-0. [DOI] [PubMed] [Google Scholar]
- 44.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnology. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nature Biotechnology. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature Biotechnology. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cameron P, Fuller CK, Donohoue PD, Jones BN, Thompson MS, Carter MM, Gradia S, Vidal B, Garner E, Slorach EM, Lau E, Banh LM, Lied AM, Edwards LS, Settle AH, Capurso D, Llaca V, Deschamps S, Cigan M, Young JK, May AP. Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat Methods. 2017;14:600–606. doi: 10.1038/nmeth.4284. [DOI] [PubMed] [Google Scholar]
- 49.Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, Joung JK. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods. 2017;14:607–614. doi: 10.1038/nmeth.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017 doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oakes BL, Nadler DC, Flamholz A, Fellmann C, Staahl BT, Doudna JA, Savage DF. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nature Biotechnology. 2016;34:646–651. doi: 10.1038/nbt.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maji B, Moore CL, Zetsche B, Volz SE, Zhang F, Shoulders MD, Choudhary A. Multidimensional chemical control of CRISPR-Cas9. Nat Chem Biol. 2017;13:9–11. doi: 10.1038/nchembio.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, James AA. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proceedings of the National Academy of Sciences. 2015;112:E6736–43. doi: 10.1073/pnas.1521077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gantz VM, Bier E. Genome editing. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science. 2015;348:442–444. doi: 10.1126/science.aaa5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pawluk A, Davidson AR, Maxwell KL. Anti-CRISPR: discovery, mechanism and function. Nat Rev Micro. 2017;1 doi: 10.1038/nrmicro.2017.120. nrmicro.2017.120. [DOI] [PubMed] [Google Scholar]