Abstract
The development of multicellular organisms relies on a small set of construction techniques—assembly, sculpting, and folding—that are spatially and temporally regulated in a combinatorial manner to produce the diversity of tissues within the body. These basic processes are well conserved across tissue types and species at the level of both genes and mechanisms. Here we review the signaling, patterning, and biomechanical transformations that occur in two well-studied model systems of epithelial folding to illustrate both the complexity and modularity of tissue development. In particular, we discuss the possibility of a spatial code specifying morphogenesis. To decipher this code, engineers and scientists need to establish quantitative experimental systems and to develop models that address mechanisms at multiple levels of organization, from gene sequence to tissue biomechanics. In turn, quantitative models of embryogenesis can inspire novel methods for creating synthetic organs and treating degenerative tissue diseases.
Keywords: pattern formation, systems biology, quantitative, biomechanics, embryogenesis, organogenesis
INTRODUCTION
Most people think about embryonic development only when they are about to become parents. Developmental biologists think about it all the time as they explore the remarkable transformation of a single cell, a fertilized egg, into an adult organism containing multiple cell types, tissues, and organs. For centuries, embryology was a purely descriptive science. Embryologists examined and classified different stages of development, such as early cell divisions, blastula, and gastrula, but had no way to explore the underlying mechanisms. For example, in the 19th century Karl Ernst von Baer noticed that early embryos of all vertebrate animals are essentially indistinguishable (1). The origin of this similarity remained a mystery until the 1980s, when molecular geneticists discovered that all animals rely on the same genes, arranged in very similar regulatory networks, to first plan and then shape their bodies (2).
Today, as we learn more about the genetic and molecular basis of embryogenesis, we can begin to think about it within an increasingly quantitative and, eventually, engineering framework. A hallmark of such an approach is the ability to deconstruct a complex process into a small set of well-defined steps, similar to subroutines in a computer code or unit operations in chemical engineering. All structures in our bodies are constructed by reiterative application of three operations: assembly, sculpting, and folding (3) (Figure 1A–C). Just as a house can be built from materials delivered from different parts of the country, the muscular and skeletal structures of the face are constructed by different cells that are specified in the neural crest and then migrate to the location where a 3D nose, beak, or trunk is assembled (4–6). This is an example of assembly. Sculpting, or the removal of extra material, is used in shaping our limbs to separate fingers from each other (7, 8). Ducks have webbed feet owing to a lack of such programmed cell death (9).
Figure 1.
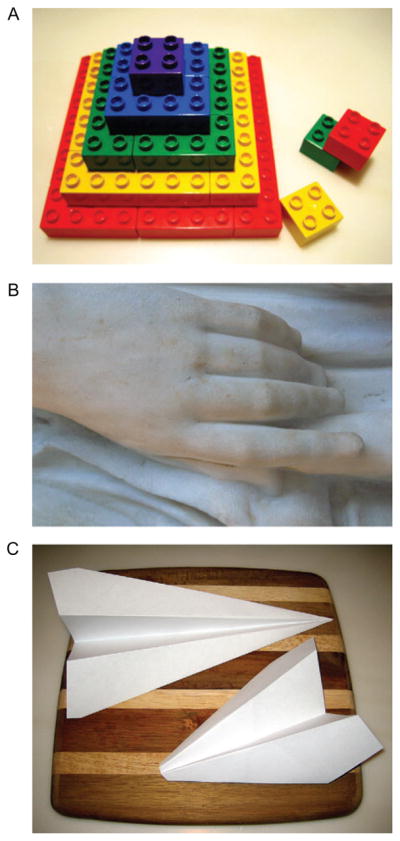
Unit operations of morphogenesis. Man-made structures can be built using several different strategies, including (A) assembly from basic building blocks, (B) sculpting from a block of material, (C) folding sheets of paper into 3D structures, or any combination of these three strategies.
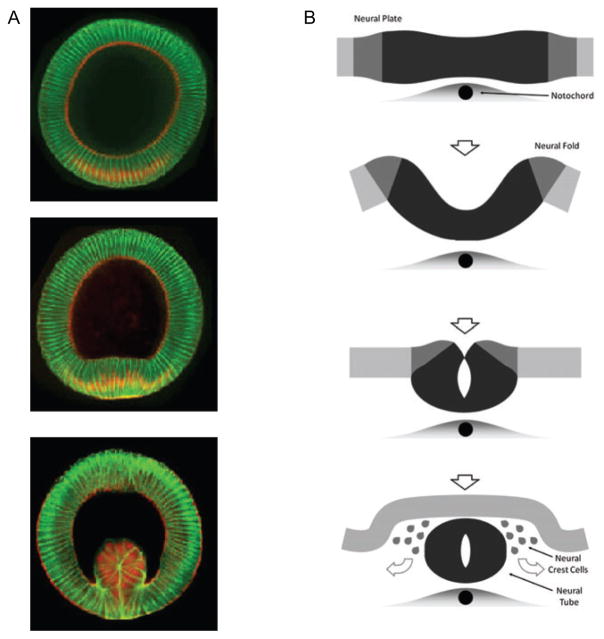
Three-dimensional structures can also form by folding 2D sheets composed of epithelial cells (Figure 2). Folding is probably the most basic of all tissue construction methods. Even simple multicellular organisms such as coral or invertebrates such as the fly undergo epithelial folding to form an additional mesodermal tissue layer (10) (Figure 2A). Higher organisms form more complex shapes by implementing spatial and temporal regulation in the patterning of epithelial bends and flexures to form complex shapes throughout all stages of embryonic development (11). One of the most important examples of epithelial folding is neurulation, the process in which a flat neural plate is folded into a neural tube (Figure 2B). Even the slightest imperfection in this process can lead to neural tube defects, some of the most common and disabling developmental disorders (12).
Figure 2.
Examples of epithelial folding. (A) Ventral furrow formation in Drosophila (images kindly provided by E. Wieschaus). (B) Bending of the neural plate to form the neural tube in vertebrates.
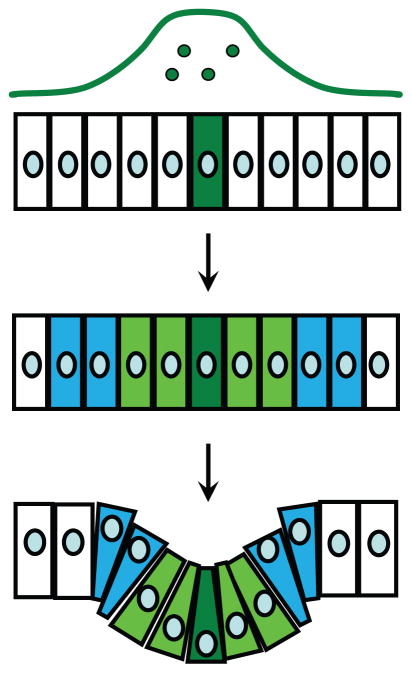
Epithelial folding is similar to origami, in which sequential folding of a 2D surface gives rise to a complex 3D structure. The process starts by specifying a pattern of future creases on a 2D surface. This pattern, which dictates the position of future folds and the sequence in which they are generated, is absolutely crucial, as one can quickly learn by trying to fold a simple box from an unpatterned piece of cardboard. In the case of epithelia, patterning is provided by chemical signals (morphogens), which are proteins secreted from localized sources (Figure 3). These molecules bind to cell surface receptors, activate them, and initiate a cascade of intracellular signals that regulate gene expression within the sheet (13). Patterned changes in gene expression then give rise to changes in cellular properties such as shape and motility, generating a localized force that deforms the cellular layer. This hierarchical framework is a working model, but at this point, there is not a single system in which we understand folding to the point that we can start with a 2D cell layer and controllably deform it into a target structure.
Figure 3.
Patterning of epithelial sheets by morphogen gradients precedes folding and other morphogenetic events in all animals. A concentration gradient of a patterning signal (a morphogen) provides spatial control of gene expression and cell shape changes within the sheet, leading to its controlled deformation.
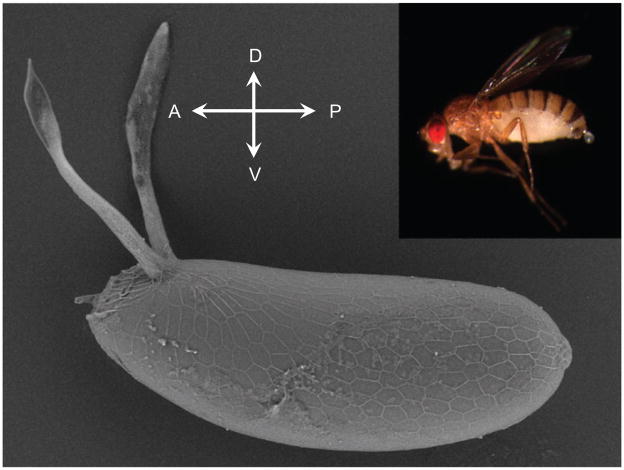
To control the folding of cellular sheets to the same level as paper folding in origami, we need to understand the underlying molecular and cellular processes. In particular, if we can learn the contextual syntax used by cells to interpret the extracellular signals that drive particular cellular responses, we can develop much more sophisticated tools for tissue engineering. Because these processes are highly conserved from coral to humans, we can study folding in model organisms. An ideal model will have simple anatomy, powerful genetics, and a sequenced genome. Few organisms fit this description better than the fruit fly, Drosophila (Figure 4, upper right panel). The fly has served as a useful model for many human diseases, such as obesity, diabetes, cancer, Huntington’s disease, Parkinson’s disease, and Alzheimer’s disease, to name a few (14–16).
Figure 4.
The fruit fly lays eggs that are decorated with tubular structures that project out of the main body of the egg to provide increased oxygen transport to the developing embryo (SEM image taken by Lily Cheung-Chang, Princeton University). The eggshell is a highly patterned 3D structure. Remarkably, the polarity of the eggshell predicts the polarity of the embryo and of the adult animal (see the inset with the image of the adult fruit fly; A, P, D, V denote the anterior, posterior, dorsal, and ventral sides of the structure). Proper patterning of the eggshell is essential for the correct specification of the body axes.
In this review we illustrate through two different examples how studying epithelial folding in the fly can teach us how organs are built. In the first example, we survey the mechanics and patterning of ventral furrow formation, which highlights the roles of apical constriction and the spatial regulation of myosin II in driving folding events (17). Next, we provide an overview of how the 2D spatial patterning of follicle cells results in the formation of tubular structures that form a part of the eggshell. Two recurring motifs can be derived from both systems: Spatially generated signals subdivide an epithelial cell layer into defined regions, and each region is defined by a set of expressed genes that specify the cellular activities and mechanical modifications necessary to generate epithelial folds. These motifs suggest that epithelial folding events in different contexts may share core modular subroutines, which can be altered to produce the diverse geometric transformations necessary for organ formation. We conclude with a discussion of how decoding this process could produce new methods for generating tissues de novo and lead to a more theoretical framework for tissue engineering.
VENTRAL FURROW FORMATION
Process Description and Insight from Models
At the onset of gastrulation—the initial process during embryogenesis that produces the three main cell types found in multicellular organisms—the Drosophila embryo consists of a single, largely uniform layer of epithelial cells that surround an incompressible fluid, the yolk. On the outside surface, the developing embryo is protected by an extracellular perivitelline membrane and eggshell consisting of chorion proteins. The first gastrulation event in the fly is ventral furrow formation, a process that results in a transient tubular structure (Figure 2A). Ventral furrow formation has been intensively studied using genetic as well as computational modeling approaches. As such, techniques that have been developed to study ventral furrow formation serve as a template for the engineering analysis of tissue development.
Gastrulation is preceded by patterning steps that subdivide the epithelial layer into two principal domains: ventral (or mesodermal) cells that form the tube, and more dorsally-located ectodermal cells that border the mesodermal cells. The formation of the ventral furrow occurs over the course of 20 min with a stereotypic progression of well-defined cellular shape changes (Figure 2A). First, cells within the ventral domain flatten their apical membranes (18, 19). Next, cells undergo a process of stochastic constriction leading to a shallow indentation on the ventral surface of the embryo (20). Cells within the furrow lengthen along the apical-basal axis to 1.7 times their original height (21). Afterward, the mesoderm cells shorten back to their original apico-basal length while still maintaining a constricted apical membrane surface, which results in the expansion of the basal membranes (21). Subsequent to invagination, the transiently-formed tube later disperses through an epithelial-to-mesenchyme transition (EMT) and migration of the mesoderm cells (21). The mesoderm cells undergo coordinated migration along the basal membrane of the outer ectodermal cells to form a layer basal to the ectoderm surface (21, 22).
Folding of the ventral cells depends on a combination of active force generation and passive deformations in both the ventral mesoderm and dorsal ectoderm, constrained by the geometry of the developing embryo. The relative structural simplicity and careful description of cellular changes that take place during ventral furrow formation have made it an attractive system to model the biomechanical mechanisms of epithelial folding. Such computational studies, which can be generalized to explain folding epithelia in other contexts, provide important insights into the role of biomechanics and geometry during animal development. Further, they provide predictions that can be verified by careful, quantitative analyses correlating the activity of proteins that modulate the cytoskeletal and adhesive properties of cells to the surface tensions and forces driving cell and tissue shape changes (23).
In an early computational study of ventral furrow formation and epithelial folding, Odell and colleagues posited that every cell is either constricted or unconstricted and that a constricted cell can pull on its neighbor and in this way switch it to the constricted state (20). In this model, the shapes of individual cells are described by damped springs (springs and dashpots) connecting the nodes of the cells, and the equations describing the length of the cellular edges result from a simple force balance. This mechanism can generate propagating waves of constriction within the epithelial layer, as demonstrated by a series of computational experiments that produced morphological changes similar to the ones observed experimentally. Subsequent experimental evidence, however, pointed to a more cell-autonomous model of cell constriction (24, 25).
Several other studies of epithelial folding have employed finite element modeling (FEM) approaches (20, 26–30). These models highlight the redundancy of possible mechanisms sufficient to drive epithelial folding, which lead to the testing of specific hypotheses about the biomechanics of folding. In particular, it is unclear which cell shape changes are active processes and which are passive deformations. Resolving this ambiguity has been a focus of several recent computational modeling efforts in 2D and 3D (28–30). In their FEM analyses, which also have been extended to 3D (29), Conte et al. employ a model that decomposes the finite deformations of the epithelial layer into active processes, such as apical constriction and apico-basal elongation, that can be associated with the known cellular transformations of ventral furrow formation. Their model does not include structural elements representing components of the cytoskeletal machinery, but rather relies on a kinematic description. This approach is advantageous because whereas it is very difficult to measure the material properties and forces in embryos, strains can easily be quantified.
Furthermore, Conte et al. assume that the epithelial cells can be approximated as a homogeneous hyperelastic material and that the deformation gradient tensor, which maps the current position of a material element to its initial position, can be decomposed into an active and a passive elastic component (30, 31). Munoz et al. found that only certain values of active cell shape changes (apical constriction and apical-basal elongation) can account for the folding patterns observed during ventral furrow formation (30). A 3D FEM model displays more robust invagination and also predicts the flow of yolk from the center of the embryo to the anterior or posterior regions of the egg chamber (29).
In their most recent study, Conte et al. took a systems approach to examine which combinations of active cell shape changes in ventral furrow formation are robust to mechanical and environmental perturbations (28). They found that a combination of apical constriction in the mesoderm and ectodermal pushing is most robust to ensuring complete invagination (closure of the ventral tube) as opposed to pure mesodermal apical constriction or cell lengthening, each of which alone does not robustly lead to tube formation. They conclude that ventral furrow formation is a result of different combinations of internally generated forces that are created by individual cells within the 2D tissue layer as well as passive deformations resulting from neighboring cells and extracellular surroundings imposing external forces on cells (19, 21, 32–34). As opposed to apical constriction, nothing is currently known about the genetic regulation and physical mechanisms of ectodermal cell shortening. Nor is it clear whether the cells that push are limited to a subset of the ventral mesoderm (19) or if all ectodermal cells are involved. Future efforts are needed to resolve these issues.
Molecular Mechanisms of Cell Shape Changes
Of the three types of cellular deformations considered in the computational models of the previous section, the molecular mechanisms have been best defined for apical constriction, which requires nonmuscle myosin II to localize apically to form a ring tethered at the adherens junctions in the ventral cells (35, 36). As mentioned previously, analysis of ventral furrow formation reveals that apical constriction occurs in asynchronous pulses of a contracting actin-myosin network that is located in the apical medial cortex (36). Constriction of the actin-myosin network pulls adherens junctions inward, resulting in the incremental reduction of the cell’s apical membrane surface. Each contraction is followed by stabilization of the contracted network, ensuring that cell neighbors do not stretch the adherens junctions after each constriction pulse (36).
Each of these two steps, pulsed constriction and stabilization, is controlled by two transcription factors, Snail and Twist (34). Snail initiates the actin-myosin contraction, whereas Twist stabilizes the constriction (36). Downstream targets of Twist, known to mediate cell-shape changes, include Folded Gastrulation (Fog), a secreted peptide, and the transmembrane protein T48, which cooperatively regulate the activity of Rho guanosine 5′-triphosphate-exchange factor RhoGEF2 (35, 37). RhoGEF2 recruits myosin II from the basal side to the apical side of the invaginating cells. In addition to modulating the dynamics of the actin cytoskeleton, ventral furrow formation is also accompanied by changes in adhesion protein expression, in particular by downregulation of DE-cadherin and upregulation of DN-cadherin expression, a step that is important for the subsequent delamination and internalization of the presumptive mesoderm (38). The regulation of cell adhesion and actomyosin contractility in epithelial folding is a highly conserved process in all metazoa (39).
Genetic screens for proteins that are differentially expressed in the ventral versus lateral cells also reveal putative “difference-proteins” that are involved in a wide range of housekeeping processes including proteolysis, metabolism, and translation control (40), which suggests that the network regulating cell shape changes during ventral formation extends beyond the components of the adhesion and cytoskeletal machinery (40). However, a small set of regulators downstream of Twist and Snail that includes RhoGEF2 appears to be sufficient for specifying apical constriction (41). Essential regulators of morphogenesis such as RhoGEF2 have been identified through genetic screens, and their roles in ventral furrow formation were examined using genetic tools that characterize the power of fly genetics (41–43).
Patterning Mechanisms
The patterning of Twist and Snail, the two essential regulators of mesoderm invagination, depends on the inductive signaling of NF-κB homolog Dorsal, which specifies several domains of cells across a 1D dorsal-ventral gradient of nuclear accumulation (44–47). The nuclear accumulation gradient of Dorsal is controlled by the signaling of the Toll pathway through activation by the ligand Spätzle (reviewed in 44). Seventeen maternal factors are provided to ensure local activation of Toll signaling (44). Spätzle-Toll (SPZ-TL) induces signal transduction by the adaptor proteins Krapfen/dMyd88 (KRA/dMYD88) and Tube (TUB) and the kinase Pelle (PLL), which leads to the degradation of IκB homolog Cactus (CACT) (44). Degradation of CACT releases Dorsal to accumulate in the nucleus and regulate approximately 6070 target genes in multiple domains along the dorsal-ventral axis (reviewed in 45). Of these targets, >40 genes encode transcription factors or cell signaling components, which form >200 network connections (45). Interestingly, if the activity of Toll signaling expands laterally, the domain of ventral furrow formation can split into two furrows (44, 48). Consequently, Toll pathway signaling is modulated by multiple positive and negative feedback loops to ensure robust and precise patterning of the effectors of ventral furrow formation.
To summarize, the patterning required for ventral furrow formation depends on a gradient of Dorsal signaling, which regulates a small set of regulators (Twist and Snail) involved in the nonuniform patterning of forces and physiological changes required for driving apical constriction in a 2D field of cells. Furthermore, there is significant redundancy in the combinations of simple cellular shape changes that can lead to invagination. Different combinations of simple shape changes can also lead to a wide diversity of folding geometries, suggesting significant versatility. In this paradigm, signaling pathways, transcription factors, and enhancers are all components of a control network that regulates morphogenetic unit operations. Studies in ventral furrow formation have led the way in refining this paradigm and connecting protein activity to cell shape changes for a simple fold. In the next section we will describe the folding of a more complex structure.
PATTERNING AND MORPHOGENESIS IN OOGENESIS
Process Description
The second experimental model discussed in this review is oogenesis, the process of egg development, in Drosophila (49–54). The main product of oogenesis is the mature oocyte, which is ready to be fertilized and give rise to the embryo (52). In insects, including Drosophila, the oocyte and future embryo are covered by an eggshell, a 3D proteinaceous structure that protects the embryo and mediates its interaction with the environment (53, 55, 56). Figure 4 shows a scanning electron microscopy (SEM) micrograph of the Drosophila eggshell. One of the most striking features of this structure is its asymmetry, both front-to-back (anteroposterior) and top-to-bottom (dorsoventral). The two tubular structures located on the dorsal side of the eggshell are the so-called dorsal appendages, used for oxygen delivery to the embryo (57). The corrugated patch of chorionic material separating the two appendages is the operculum, a region of the eggshell from which the larva hatches when embryogenesis is complete.
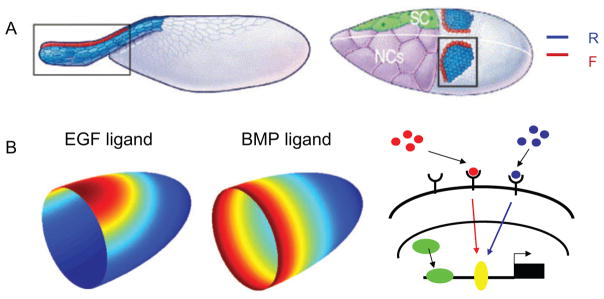
The eggshell is derived from the somatic follicle cells in the developing egg chamber, a precursor of the mature egg (Figure 5A). Most of the follicle cells are arranged in a columnar epithelial layer over the growing oocyte. During egg development, they are patterned by chemical signals from the oocyte and from the follicle cells themselves. The resulting 2D patterns of gene expression control the spatially regulated cell shape changes, migrations, and rearrangements that eventually transform a 2D region of the follicular epithelium into the 3D eggshell structures. Figure 5A (right) shows the follicular epithelium at a stage when it is still a 2D surface over the oocyte. Within 30 min, the two groups of cells at the dorsal anterior region of the follicular epithelium constrict, initiating the formation of the two tubular respiratory appendages shown in Figure 5A (left) (58, 59).
Figure 5.
Epithelial folding leading to tube formation. (A) The proteinaceous material of the eggshell is secreted by a layer of epithelial cells called the follicle cells. Patterning of the follicle cells results in the formation of groups of cells, including roof (R) and floor (F) cells, that undergo shape changes, including apical constriction, to fold into tubular structures. Also marked are the stretch cells (SCs) and nurse cells (NCs). Figure 5A is modified from Reference 58 and reprinted with permission. (B) The follicular epithelium is patterned by several chemical signals. The anterior-posterior (AP) patterning depends on the AP gradient of signaling through the bone morphogenetic protein (BMP) signaling pathway, and the dorsoventral (DV) patterning depends on the gradient of signaling through the epidermal growth factor (EGF) signaling pathway. Nonuniform activation of these signaling pathways provides spatial control of gene expression in the follicle cells.
Dorsal appendage morphogenesis can be described as a sequence of steps that involves patterning of the follicle cells, their regulated migrations and rearrangements, and finally spatially regulated secretion of proteins that make the actual eggshell (56). Similar to the formation of the ventral furrow in the early embryo, the transformation of an epithelial sheet into a 3D structure is initiated by the spatially regulated shape changes within a predefined subpopulation of cells within the epithelium. In this case, however, the cells involved in morphogenesis are arranged not in a line, as in the ventral furrow, but in two patches. These cells are differentiated from their neighbors under the action of chemical signals that pattern the follicular epithelium.
Two-Dimensional Patterning
The patterning signals are provided by two highly conserved molecules related to human growth factors (Figure 5B). Both of them act as ligands that bind to cell surface receptors and lead to changes in gene expression. Both are distributed as concentration gradients that emerge from the combination of localized ligand production, extracellular diffusion, and degradation that is mediated by binding to receptors on the surfaces of the follicle cells. The first signal, Gurken (GRK), a molecule related to the mammalian epidermal growth factor (EGF), is secreted from the dorsal-anterior cortex of the oocyte and activates the EGF receptors (EGFRs) in the follicle cells (60). The second signal is Decapentaplegic (DPP), a molecule related to the bone morphogenetic proteins (BMPs) in vertebrates (61). DPP is secreted at the anterior border of the follicular epithelium and, similar to GRK, acts through receptors that are uniformly expressed in the follicle cells.
By binding to their cognate receptors, GRK and DPP activate highly conserved intracellular signaling pathways and in this way control gene expression in the follicle cells. EGFR acts through the mitogen activated protein kinase (MAPK) signaling pathway (62). Ligand-bound EGFR initiates a sequence of intracellular processes that lead to MAPK phosphorylation, which is essential for its catalytic activity. MAPK has multiple substrates inside the cell (63, 64). An important class of these targets are transcription factors that can change their activity in response to phosphorylation by MAPK. Similarly, DPP activates the serine-threonine catalytic activity of its cell surface receptor, which in turn phosphorylates the transcription factor SMAD (a name derived from combining the Drosophila homolog Mothers Against Decapentaplegic (MAD) and the C. elegans homolog SMA), which then translocates to the nucleus and can bind DNA to control gene expression (65).
The joint action of the GRK and DPP gradients is responsible for spatially nonuniform expression of at least 100 genes in the follicular epithelium (66). Abstractions of some of the resulting patterns, visualized by in situ hybridization experiments, are shown in Figure 6. Each of the images in this figure reports the expression of a messenger RNA for a single gene across the epithelial sheet that “prepares” for folding. Color reflects the level of expression: black means expressed, whereas white is not expressed. These genes are expressed in a wide range of patterns that can appear as spots, stripes, triangles, etc. The extent of molecular patterning revealed by these experiments is truly amazing. Compare it for a second with the patterning instructions in origami, in which a future 3D structure is specified by the locations of future folds and the order of folding. Is there a way to provide a comparable summary for the patterning that precedes eggshell morphogenesis?
Figure 6.
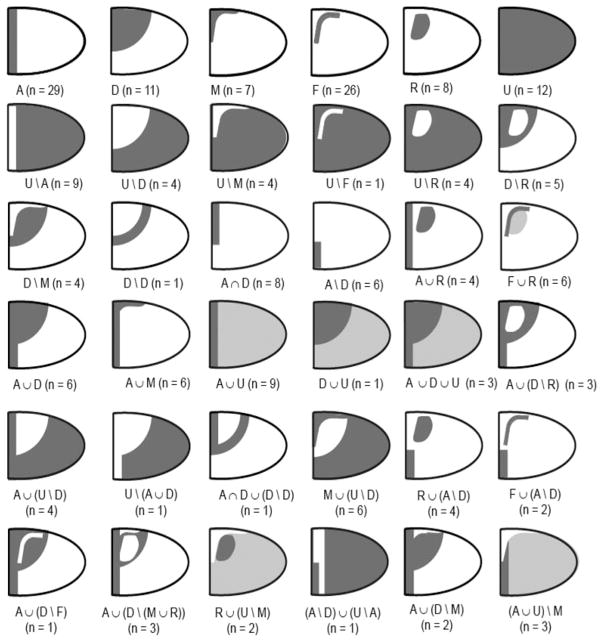
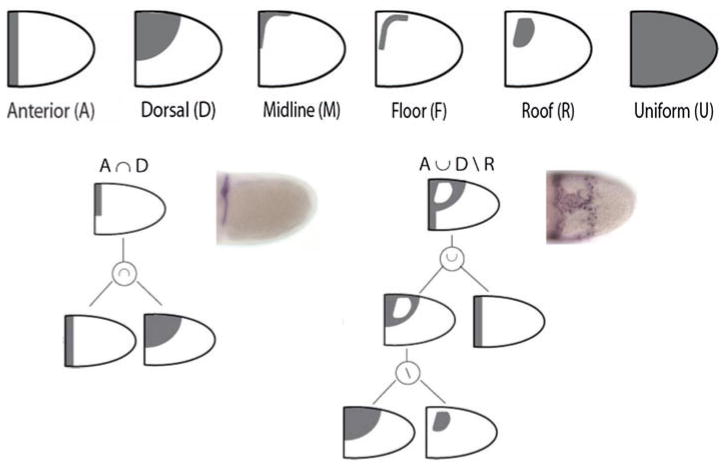
Representative sets of 2D patterns during oogenesis. Each abstraction represents the lateral view of oocyte-associated follicle cells. The Boolean expression is given below, as well as the number of observed gene expression patterns. The first panel shows the primitive shapes: A, D, M, F, R, U. Modified from Reference 66.
Epithelial Patterning Code
A systems-based approach to discovering genes downstream of the EGFR and DPP signaling pathways led to the observation that gene expression patterns follow a combinatorial code that is based on addition, intersection, and subtraction of just six building blocks (66) (Figure 7). One of these blocks, also called a primitive, is spatially uniform. The other primitives are related to the spatial distribution of the patterning signals. For example, the anterior stripe corresponds to the level sets, i.e., the lines of constant concentration, of the anteroposterior gradient of DPP signaling. Similarly, the dorsal semicircle can be viewed as a level set that corresponds to the intermediate level of the GRK gradient. The origins of the other three primitives, in the forms of a midline triangle (M), two dorsolateral patches (R), and two L-shaped stripes (F), are less understood. The working hypothesis is that their formation depends on feedback modulation of signals provided by GRK and DPP (65, 67–69). Importantly, the midline shape describes cells that contribute to the future operculum, whereas the roof and floor primitives describe the spatial arrangement of cells that contribute to the upper and lower parts of the tubes in the future dorsal appendages (58, 59).
Figure 7.
A patterning code for gene expression in the follicular epithelium. Each abstraction represents the lateral view of oocyte-associated follicle cells. The first panel shows the primitive shapes: anterior (A), dorsal (D), midline (M), floor (F), roof (R), and uniform (U). Two examples of Boolean expressions that yield more complex patterns are also presented. Modified from Reference 66.
Some of the gene expression patterns in the follicular epithelium can be described by just a single primitive. A majority of patterns, however, are more complex and their description requires several primitives, which are combined by the operations of intersection, union, addition, or subtraction. For example, the anterior segment that describes the expression of the gene argos (aos) is an intersection of a semicircle and vertical stripe. These geometric operations reflect the responses of the genes to patterning signals. The expression of aos, for example, requires high levels of both GRK and DPP signals. Cells exposed to high levels of GRK belong to the dorsal (D) primitive, whereas those exposed to high levels of DPP belong to the anterior (A) primitive. As a consequence, aos is expressed at the intersection of the anterior and dorsal primitives.
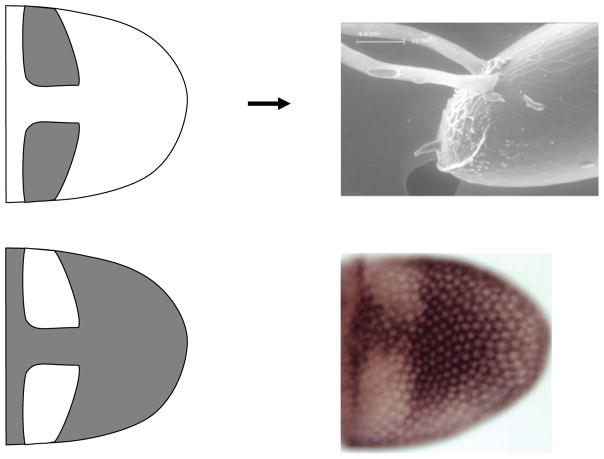
With this code, it is possible to summarize hundreds of 2D expression patterns for hundreds of different genes in terms of the primitives and the geometric operations through which they are combined into more complex patterns (Figure 6, 7). Significantly, the proposed code provides a guide for future mechanistic studies of both gene regulation and subsequent morphogenesis. This point is illustrated in a study of the expression, regulation, and function of Cad74A, a gene that encodes a cadherin protein (70). Cadherins are key regulators of epithelial morphogenesis in all multicellular animals and have multiple functions, most notably control of cell-cell adhesion (71–74). The pattern of Cad74A expression can be best characterized as the difference between the uniform and roof primitives, suggesting that Cad74A is repressed in the roof cells (Figure 8). Indeed, Broad (BR), a transcription factor expressed in the roof cells and controlled by GRK and DPP gradients, represses Cad74A expression (70, 75, 76). Furthermore, work in our lab has demonstrated that the pattern of Cad74A is functionally significant for eggshell morphogenesis. In particular, filling in the two holes in the wild-type Cad74A pattern is sufficient to disrupt the normal morphogenesis of the dorsal appendages (70).
Figure 8.
Cad74A is required for robust eggshell morphogenesis. The roof cells form the top (roof) of the projecting dorsal appendages and are initially specified as two domains of cells (gray patches). Cad74A is expressed in most of the follicle cells contacting the oocyte, but is specifically repressed in the roof cells. The cartoon image (lower left) summarizes the in-situ hybridization pattern of Cad74A (lower right), which is marked by cells stained a dark brown color.
These studies illustrate a systematic approach to multiple steps in epithelial morphogenesis: from patterning signals to gene regulation to the formation of 3D structures. Cad74A is certainly not the only gene that must be patterned to generate the 3D eggshell structures (77). A current focus of investigation is the regulation of the functions of other candidate effectors of morphogenesis. The main goals are to understand how the joint action of cytoskeletal, adhesion, and motor proteins leads to cell shape changes and how these shape changes then lead to the emergence of 3D multicellular structures. Given the highly conserved nature of the molecules and processes involved in epithelial morphogenesis, the results of these studies have implications that go well beyond the formation of the two tubes on an insect eggshell (78). In fact, they might reveal the general principles of morphogenetic programs that convert 2D cellular layers into complex 3D structures.
A GENERAL CODE FOR EPITHELIAL ORIGAMI
A small set of strategies exists for constructing complicated organs: assembly, sculpting, and folding. We have examined the steps of patterning and morphogenesis for epithelial folding in two separate contexts to illustrate the common themes found in building 3D structures from 2D sheets. Ventral furrow and dorsal appendage formation are two examples of epithelial sheets transforming into 3D structures under the action of chemical signals that control gene expression and in this way affect the force generation and mechanical transformations of epithelial cells. This basic picture suggests that different structures can be generated from the same sheet by perturbing the spatial patterning of genes (79). The small set of spatial primitives, together with a modified Boolean syntax, describes a much larger set of gene expression possibilities and suggests the potential for a morphogenetic code.
A true indication that a morphogenetic code governs the formation of structures would be reiterative patterns of gene expression in different tissues with similar organization. Modifications in expression patterns would necessarily reflect differences in tissue function and organization. Further, a morphogenetic code will share similar traits with other codes, such as the genetic code, computer program codes, and human language:
Codes necessarily consist of a small set of units (modules) that form the building blocks for a defined vocabulary. In the case of the proposed morphogenetic code, the small number of spatial primitives in the ovary can be used to describe the much more complicated patterns of gene expression.
Only a fraction of the possible sets of symbols that comprise the alphabet form the vocabulary of the code. For example, it is clear that the vocabulary of human languages consists of only a small fraction of the possible combinations of letters. Similarly, only a small fraction of the total possible number of spatial patterns that can be constructed from the primitives in oogenesis are observed in screens of gene expression patterns (66, 77).
A morphogenetic code will carry a certain level of redundancy. Misspelled words are often understandable to an audience, and putative effector genes, such as cadherins in the ovary and protocadherins in the central nervous system, often play redundant roles and are not essential to any given morphogenetic process. Nonetheless, they play a cumulative role in robust organ development.
Within this framework, conserved mechanical regulators are controlled by different signals in different contexts to define the geometric primitives of the code. Epithelial structures that share a similarity in folding events would be expected in at least some cases to reflect conserved spatial relationships of gene expression. One indication that this may in fact be the case is the similar patterns of overlapping gene expression found in the formation of the larval posterior spiracles during embryogenesis and in defining the follicle cell domains (77, 80). The posterior spiracles are openings for the larval trachea and are also formed by invagination. Genes such as Cad86C and Cad74A are expressed in nonoverlapping spatial patterns in the posterior spiracles (80), similar to in the follicle cells.
The set of known molecular effectors that are involved in implementing the required cellular shape changes has grown significantly, and in the case of the wiring of the nervous system, the morphogenetic code is extremely complex (81, 82). As a starting point, deciphering the code will require systems-based approaches to summarize and classify the expression data. Although spatial expression can provide strong hints to the functions of and interactions between gene sets, functional and quantitative analysis of gene perturbations will also be necessary for decoding the epithelial code. An important first step to connecting signaling pathways to epithelial properties is quantitative studies of multiple pathways in several different epithelial contexts (83, 84). Understanding how the morphogenetic code works to segregate and define subpopulations of cells within organs may prove revolutionary for advancing the state of tissue engineering and our knowledge of how to build tissues de novo.
EMBRYONIC DEVELOPMENT AS A THEORETICAL FRAMEWORK FOR TISSUE ENGINEERING
Traditionally, building organs in the laboratory requires the integration of cells with synthetic parts such as polymer-based scaffolds (85). Progress in tissue engineering has been incremental and has relied mostly on experimental tinkering and inventing because of the lack of a comprehensive theory or framework (86). Ultimately, it is hoped that by reverse engineering tissue morphogenesis in model organisms, theoretical frameworks for tissue engineering and regenerative medicine can be developed. For example, understanding the mechanisms of epithelial folding can guide advances in the treatment of neural tube defects.
An engineering analysis of animal development can provide the basis for constructing a theory for tissue engineering. Decoding pattern formation entails elucidating the transcriptional regulation of gene expression as well as connecting information about gene expression to biological processes such as cell proliferation, locomotion, apoptosis, and differentiation. In this analogy, embryogenesis is described by a block flow diagram of morphogenetic unit operations (87). Signaling pathways, transcription factors, and enhancers are all components of the control network that encodes the spatial properties of cells within a 2D tissue. Engineers can contribute by building and analyzing models, developing new experimental techniques, designing tissue mimetics for understanding normal morphogenesis, and exploring morphogenesis in tissue engineering designs.
Both computational modeling and experiments are necessary for a completely quantitative understanding of tissue formation. Experiments provide the raw data needed for discovering the components of the gene regulatory network and the fitting parameters for computational models. Hypothesis formation and prioritization of future experiments are driven by predictions from computational models (88). Engineers have much to offer in both spheres, as the same tools that are used to analyze bridge construction and plant design can be modified and applied to analyze both the chemical patterning steps and mechanical transformations that occur during tissue formation.
An open question still to be answered is whether there exists a general code of master regulators that specify and control the processes of morphogenesis in a modular fashion that transcends context. A comparative systems-level analysis of multiple organs built from similar unit operations will be required to answer this question. Based on the high degree of similarity at the levels of single genes, signaling pathways, and developmental modules (i.e., Hox genes), it would not be a complete surprise if conserved morphogenetic unit operations can be identified that are used in a combinatorial manner to specify the building of all organs. The ability to rearrange and recapitulate such morphogenetic processes de novo represents a future vision for tissue engineering.
How gene expression is related to cell mechanics remains largely a mystery. Several models have been used to describe the mechanics of ventral furrow formation, but quantitative studies of morphogenesis and the mechanical modeling of dorsal appendages have not yet been addressed. In the future, we need to develop multiscale, systems-level models that incorporate the regulation of genes that affect the mechanical and physiological properties of cells in describing how complicated organ structures form. Because context is important, several model systems need to be investigated at a quantitative level to provide a basis for comparison. Several challenges must be overcome to accomplish this ambitious goal, including taking quantitative measurements of inductive signaling, gene and protein expression, and the forces that are generated by cells during morphogenesis. Ultimately, connecting signaling pathways (the input) to the forces that shape tissues (the output) will provide a more unified theory of tissue morphogenesis by integrating genetics with mechanics.
SUMMARY POINTS.
Organisms such as Drosophila are useful as models for many human diseases, including cancer, as well as developmental defects. The small size, relative genetic simplicity, and genetic and imaging tools available for Drosophila research make it a powerful system for studying the biophysics of epithelial morphogenesis.
Organs are built through the spatial control of a small number of morphogenetic processes such as epithelial folding, programmed cell death, and cell migration. The effectors and signaling pathways involved in regulating these unit operations are conserved from flies to humans.
Examples of epithelial folding that serve as paradigms and points of comparison to human development include ventral furrow formation and dorsal appendage formation.
Computational models of morphogenesis lead to predictions and insights into the mechanics of epithelial folding.
A systems-level analysis of gene expression patterns suggests the possibility of a hierarchical epithelial code that directs the pattern of epithelial folds.
Engineering approaches are needed to provide systematic, quantitative analysis of the cell signaling and biomechanics of each of the unit operations of tissue development.
Acknowledgments
The authors thank N. Yakoby, T. Schupbach, A. Martin, G.W. Brodland, J. Veldhuis, and all members of the Shvartsman lab for many stimulating discussions. The authors also thank L.S. Cheung, R.A. Shvartsman, and D.J. Shvartsman for help in preparing Figures 1 and 2. J.J.Z. was funded in part by the Fannie and John Hertz Foundation and the Princeton Wu Fellowship program. S.Y.S. was supported by the R01 GM078079 NIH grant.
Glossary
- Morphogens
secreted molecules, usually small peptides that diffuse across tissues and control gene expression in a concentration-dependent manner
- Gastrulation
the first morphogenesis events that occur during embryogenesis to form the body
- Hyperelastic material
an ideal elastic material, of which the work done is independent of the deformation path, stress relaxation, or heat dissipation
- Cadherins
calcium-dependent adhesion molecules involved in cell-cell adhesion, organization of the cytoskeleton, and cell signaling
Footnotes
RELATED RESOURCES
Flymove: http://flymove.uni-muenster.de/
An online resource for exploring the stages of Drosophila embryogenesis; see Reference 89.
The Interactive Fly: http://www.sdbonline.org/fly/aimain/1aahome.htm
A online guide to Drosophila development with multiple links to movies and images of Drosophila gastrulation and organogenesis.
Flybase: http://flybase.org
A comprehensive database of fly genetics and genomics, with searchable information on each of the genes discussed in this review.
Berg Laboratory: http://berglab.gs.washington.edu/tmp/Research.Movies.html
Online time lapse videos illustrating dorsal appendage formation (58).
Center for Cell Dynamics Morphogenesis Tutorial: http://celldynamics.org/celldynamics/tutorials/morph/index.html
A downloadable java program that allows the user to simulate 2D morphogenesis.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Cretekos CJ, Rasweiler JJ, IV, Behringer RR. Comparative studies on limb morphogenesis in mice and bats: a functional genetic approach towards a molecular understanding of diversity in organ formation. Reprod Fertil Dev. 2001;13:691–95. doi: 10.1071/rd01115. [DOI] [PubMed] [Google Scholar]
- 2.Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301:331–33. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- 3.Montell DJ. Morphogenetic cell movements: diversity from modular mechanical properties. Science. 2008;322:1502–5. doi: 10.1126/science.1164073. [DOI] [PubMed] [Google Scholar]
- 4.Kurosaka S, Kashina A. Cell biology of embryonic migration. Birth Defects Res C Embryo Today. 2008;84:102–22. doi: 10.1002/bdrc.20125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tucker RP. Neural crest cells: a model for invasive behavior. Int J Biochem Cell Biol. 2004;36:173–77. doi: 10.1016/s1357-2725(03)00243-7. [DOI] [PubMed] [Google Scholar]
- 6.Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–68. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- 7.Merino R, Gañán Y, Macias D, Rodríguez-León J, Hurle JM. Bone morphogenetic proteins regulate interdigital cell death in the avian embryo. Ann N Y Acad Sci. 1999;887:120–32. doi: 10.1111/j.1749-6632.1999.tb07927.x. [DOI] [PubMed] [Google Scholar]
- 8.Zuzarte-Luis V, Hurle JM. Programmed cell death in the embryonic vertebrate limb. Semin Cell Dev Biol. 2005;16:261–69. doi: 10.1016/j.semcdb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Merino R, Rodriguez-Leon J, Macias D, Ganan Y, Economides AN, Hurle JM. The BMP antagonist Gremlin regulates outgrowth, chondrogenesis and programmed cell death in the developing limb. Development. 1999;126:5515–22. doi: 10.1242/dev.126.23.5515. [DOI] [PubMed] [Google Scholar]
- 10.Hayward DC, Miller DJ, Ball EE. snail expression during embryonic development of the coral Acropora: blurring the diploblast/triploblast divide? Dev Genes Evol. 2004;214:257–60. doi: 10.1007/s00427-004-0398-0. [DOI] [PubMed] [Google Scholar]
- 11.Pilot F, Lecuit T. Compartmentalized morphogenesis in epithelia: from cell to tissue shape. Dev Dyn. 2005;232:685–94. doi: 10.1002/dvdy.20334. [DOI] [PubMed] [Google Scholar]
- 12.Nicholas DEG, Andrew JC. Development of the vertebrate central nervous system: formation of the neural tube. Prenat Diagn. 2009;29:303–11. doi: 10.1002/pd.2206. [DOI] [PubMed] [Google Scholar]
- 13.Lewis J. From signals to patterns: space, time, and mathematics in developmental biology. Science. 2008;322:399–403. doi: 10.1126/science.1166154. [DOI] [PubMed] [Google Scholar]
- 14.Schutt C, Froesch B, Hafen E. Drosophila—a model system for targets and lead identification in cancer and metabolic disorders. In: Carroll PM, Fitzgerald KJ, editors. Model Organisms in Drug Discovery. Chichester, West Sussex, UK/Hoboken, NJ: Wiley; 2003. pp. 119–51. [Google Scholar]
- 15.Li H, Garza D. Drosophila as a tool for drug discovery. In: Carroll PM, Fitzgerald KJ, editors. Model Organisms in Drug Discovery. Chichester, West Sussex, UK/Hoboken, NJ: Wiley; 2003. pp. 83–117. [Google Scholar]
- 16.Kango-Singh M, Singh A. Regulation of organ size: insights from the Drosophila Hippo signaling pathway. Dev Dyn. 2009;238:1627–37. doi: 10.1002/dvdy.21996. [DOI] [PubMed] [Google Scholar]
- 17.Martin AC. Pulsation and stabilization. Contractile forces that underlie morphogenesis. Dev Biol. 2010;341:114–25. doi: 10.1016/j.ydbio.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 18.Sweeton D, Parks S, Costa M, Wieschaus E. Gastrulation in Drosophila: the formation of the ventral furrow and posterior midgut invaginations. Development. 1991;112:775–89. doi: 10.1242/dev.112.3.775. [DOI] [PubMed] [Google Scholar]
- 19.Leptin M, Grunewald B. Cell shape changes during gastrulation in Drosophila. Development. 1990;110:73–84. doi: 10.1242/dev.110.1.73. [DOI] [PubMed] [Google Scholar]
- 20.Odell GM, Oster G, Alberch P, Burnside B. The mechanical basis of morphogenesis. I Epithelial folding and invagination. Dev Biol. 1981;85:446–62. doi: 10.1016/0012-1606(81)90276-1. [DOI] [PubMed] [Google Scholar]
- 21.Costa M, Sweeton D, Wieschaus E. Gastrulation in Drosophila: cellular mechanisms of morphogenetic movements. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Plainview, NY: Cold Spring Harbor Lab; 1993. pp. 425–65. [Google Scholar]
- 22.McMahon A, Supatto W, Fraser SE, Stathopoulos A. Dynamic analyses of Drosophila gastrulation provide insights into collective cell migration. Science. 2008;322:1546–50. doi: 10.1126/science.1167094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidson LA, Oster GF, Keller RE, Koehl MAR. Measurements of mechanical properties of the blastula wall reveal which hypothesized mechanisms of primary invagination are physically plausible in the sea urchin Strongylocentrotus purpuratus. Dev Biol. 1999;209:221–38. doi: 10.1006/dbio.1999.9249. [DOI] [PubMed] [Google Scholar]
- 24.Leptin M. Gastrulation movements: the logic and the nuts and bolts. Dev Cell. 2005;8:305–20. doi: 10.1016/j.devcel.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 25.Leptin M, Roth S. Autonomy and nonautonomy in Drosophila mesoderm determination and morphogenesis. Development. 1994;120:853–59. doi: 10.1242/dev.120.4.853. [DOI] [PubMed] [Google Scholar]
- 26.Brodland GW, Clausi DA. Cytoskeletal mechanics of neurulation: insights obtained from computer simulations. Biochem Cell Biol. 1995;73:545–53. doi: 10.1139/o95-060. [DOI] [PubMed] [Google Scholar]
- 27.Brodland GW, Clausi DA. Embryonic tissue morphogenesis modeled by FEM. J Biomech Eng. 1994;116:146–55. doi: 10.1115/1.2895713. [DOI] [PubMed] [Google Scholar]
- 28.Conte V, Munoz JJ, Baum B, Miodownik M. Robust mechanisms of ventral furrow invagination require the combination of cellular shape changes. Phys Biol. 2009;6:16010. doi: 10.1088/1478-3975/6/1/016010. [DOI] [PubMed] [Google Scholar]
- 29.Conte V, Munoz JJ, Miodownik M. A 3D finite element model of ventral furrow invagination in the Drosophila melanogaster embryo. J Mech Behav Biomed Mater. 2008;1:188–98. doi: 10.1016/j.jmbbm.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Munoz JJ, Barrett K, Miodownik M. A deformation gradient decomposition method for the analysis of the mechanics of morphogenesis. J Biomech. 2007;40:1372–80. doi: 10.1016/j.jbiomech.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez EK, Hoger A, McCulloch AD. Stress-dependent finite growth in soft elastic tissues. J Biomech. 1994;27:455–67. doi: 10.1016/0021-9290(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 32.Keller R, Davidson LA, Shook DR. How we are shaped: the biomechanics of gastrulation. Differentiation. 2003;71:171–205. doi: 10.1046/j.1432-0436.2003.710301.x. [DOI] [PubMed] [Google Scholar]
- 33.Leptin M. Gastrulation in Drosophila: the logic and the cellular mechanisms. EMBO J. 1999;18:3187–92. doi: 10.1093/emboj/18.12.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leptin M. Drosophila gastrulation: from pattern formation to morphogenesis. Annu Rev Cell Dev Biol. 1995;11:189–212. doi: 10.1146/annurev.cb.11.110195.001201. [DOI] [PubMed] [Google Scholar]
- 35.Dawes-Hoang RE, Parmar KM, Christiansen AE, Phelps CB, Brand AH, Wieschaus EF. Folded gastrulation, cell shape change and the control of myosin localization. Development. 2005;132:4165–78. doi: 10.1242/dev.01938. [DOI] [PubMed] [Google Scholar]
- 36.Martin AC, Kaschube M, Wieschaus EF. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 2009;457:495–99. doi: 10.1038/nature07522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolsch V, Seher T, Fernandez-Ballester GJ, Serrano L, Leptin M. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science. 2007;315:384–86. doi: 10.1126/science.1134833. [DOI] [PubMed] [Google Scholar]
- 38.Oda H, Tsukita S, Takeichi M. Dynamic behavior of the cadherin-based cell-cell adhesion system during Drosophila gastrulation. Dev Biol. 1998;203:435–50. doi: 10.1006/dbio.1998.9047. [DOI] [PubMed] [Google Scholar]
- 39.Lecuit T, Lenne PF. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol. 2007;8:633–44. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- 40.Puri M, Goyal A, Senutovich N, Dowd SR, Minden JS. Building proteomic pathways using Drosophila ventral furrow formation as a model. Mol Biosyst. 2008;4:1126–35. doi: 10.1039/b812153b. [DOI] [PubMed] [Google Scholar]
- 41.Barrett K, Leptin M, Settleman J. The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell. 1997;91:905–15. doi: 10.1016/s0092-8674(00)80482-1. [DOI] [PubMed] [Google Scholar]
- 42.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–37. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 43.Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–69. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- 44.Moussian B, Roth S. Dorsoventral axis formation in the Drosophila embryo–shaping and transducing a morphogen gradient. Curr Biol. 2005;15:R887–99. doi: 10.1016/j.cub.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 45.Hong J-W, Hendrix DA, Papatsenko D, Levine MS. How the Dorsal gradient works: insights from postgenome technologies. Proc Natl Acad Sci. 2008;105:20072–76. doi: 10.1073/pnas.0806476105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanodia JS, Rikhy R, Kim Y, Lund VK, DeLotto R, Lippincott-Schwartz J, Shvartsman SY. Dynamics of the Dorsal morphogen gradient. Proc Natl Acad Sci USA. 2009;106(51):21707–12. doi: 10.1073/pnas.0912395106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liberman LM, Reeves GT, Stathopoulos A. Quantitative imaging of the Dorsal nuclear gradient reveals limitations to threshold-dependent patterning in Drosophila. Proc Natl Acad Sci USA. 2009;106(52):22317–22. doi: 10.1073/pnas.0906227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roth S. The origin of dorsoventral polarity in Drosophila. Philos Trans R Soc Lond Ser B. 2003;358:1317–29. doi: 10.1098/rstb.2003.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng WM, Bownes M. Patterning and morphogenesis of the follicle cell epithelium during Drosophila oogenesis. Int J Dev Biol. 1998;42:541–52. [PubMed] [Google Scholar]
- 50.Dobens LL, Raftery LA. Integration of epithelial patterning and morphogenesis in Drosophila ovarian follicle cells. Dev Dyn. 2000;218:80–93. doi: 10.1002/(SICI)1097-0177(200005)218:1<80::AID-DVDY7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 51.Horne-Badovinac S, Bilder D. Mass transit: epithelial morphogenesis in the Drosophila egg chamber. Dev Dyn. 2005;232:559–74. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- 52.Spradling AC. Developmental genetics of oogenesis. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Plainview, NY: Cold Spring Harbor Lab; 1993. pp. 1–70. [Google Scholar]
- 53.Waring GL. Morphogenesis of the eggshell in Drosophila. Int Rev Cytol. 2000;198:67–108. doi: 10.1016/s0074-7696(00)98003-3. [DOI] [PubMed] [Google Scholar]
- 54.Yakoby N, Bristow CA, Gouzman I, Rossi MP, Gogotsi Y, et al. Systems-level questions in Drosophila oogenesis. IEE Proc Syst Biol. 2005;152:276–84. doi: 10.1049/ip-syb:20050039. [DOI] [PubMed] [Google Scholar]
- 55.Berg CA. The Drosophila shell game: patterning genes and morphological change. Trends Genet. 2005;21:346–55. doi: 10.1016/j.tig.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 56.Cavaliere V, Bernardi F, Romani P, Duchi S, Gargiulo G. Building up the Drosophila eggshell: first of all the eggshell genes must be transcribed. Dev Dyn. 2008;237:2061–72. doi: 10.1002/dvdy.21625. [DOI] [PubMed] [Google Scholar]
- 57.Hinton HE. Respiratory systems of insect egg shells. Annu Rev Entomol. 1969;14:343–68. doi: 10.1146/annurev.en.14.010169.002015. [DOI] [PubMed] [Google Scholar]
- 58.Dorman JB, James KE, Fraser SE, Kiehart DP, Berg CA. bullwinkle is required for epithelial morphogenesis during Drosophila oogenesis. Dev Biol. 2004;267:320–41. doi: 10.1016/j.ydbio.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 59.Ward EJ, Berg CA. Juxtaposition between two cell types is necessary for dorsal appendage tube formation. Mech Dev. 2005;122:241–55. doi: 10.1016/j.mod.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 60.Neuman-Silberberg FS, Schupbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGFα-like protein. Cell. 1993;75:165–74. [PubMed] [Google Scholar]
- 61.Twombly V, Blackman RK, Jin H, Graff JM, Padgett RW, Gelbart WM. The TGF-beta signaling pathway is essential for Drosophila oogenesis. Development. 1996;122:1555–65. doi: 10.1242/dev.122.5.1555. [DOI] [PubMed] [Google Scholar]
- 62.Nilson LA, Schupbach T. EGF receptor signaling in Drosophila oogenesis. Curr Top Dev Biol. 1999;44:203–43. doi: 10.1016/s0070-2153(08)60471-8. [DOI] [PubMed] [Google Scholar]
- 63.Astigarraga S, Grossman R, Diaz-Delfin J, Caelles C, Paroush Z, Jimenez G. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 2007;26:668–77. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mantrova E, Hsu T. Down-regulation of transcription factor CF2 by Drosophila Ras/MAP kinase signaling in oogenesis: cytoplasmic retention and degradation. Genes Dev. 1998;12:1166–75. doi: 10.1101/gad.12.8.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shravage BV, Altmann G, Technau M, Roth S. The role of Dpp and its inhibitors during eggshell patterning in Drosophila. Development. 2007;134:2261–71. doi: 10.1242/dev.02856. [DOI] [PubMed] [Google Scholar]
- 66.Yakoby N, Bristow CA, Gong D, Schafer X, Lembong J, et al. A combinatorial code for pattern formation in Drosophila oogenesis. Dev Cell. 2008;15:725–37. doi: 10.1016/j.devcel.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yakoby N, Lembong J, Schupbach T, Shvartsman SY. Drosophila eggshell is patterned by sequential action of feedforward and feedback loops. Development. 2008;135:343–51. doi: 10.1242/dev.008920. [DOI] [PubMed] [Google Scholar]
- 68.Zartman JJ, Kanodia JS, Cheung LS, Shvartsman SY. Feedback control of the EGFR signaling gradient: superposition of domain-splitting events in Drosophila oogenesis. Development. 2009;136:2903–11. doi: 10.1242/dev.039545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lachance JF, Lomas MF, Eleiche A, Kerr PB, Nilson LA. Graded Egfr activity patterns the Drosophila eggshell independently of autocrine feedback. Development. 2009;136:2893–902. doi: 10.1242/dev.036103. [DOI] [PubMed] [Google Scholar]
- 70.Zartman JJ, Yakoby N, Bristow CA, Zhou X, Schlichting K, et al. Cad74A is regulated by BR and is required for robust dorsal appendage formation in Drosophila oogenesis. Dev Biol. 2008;322:289–301. doi: 10.1016/j.ydbio.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 72.Takeichi M. Cadherins: a molecular family important in selective cell-cell adhesion. Annu Rev Biochem. 1990;59:237–52. doi: 10.1146/annurev.bi.59.070190.001321. [DOI] [PubMed] [Google Scholar]
- 73.Tepass U, Truong K, Godt D, Ikura M, Peifer M. Cadherins in embryonic and neural morphogenesis. Nat Rev Mol Cell Biol. 2000;1:91–100. doi: 10.1038/35040042. [DOI] [PubMed] [Google Scholar]
- 74.Wheelock MJ, Johnson KR. Cadherins as modulators of cellular phenotype. Annu Rev Cell Dev Biol. 2003;19:207–35. doi: 10.1146/annurev.cellbio.19.011102.111135. [DOI] [PubMed] [Google Scholar]
- 75.Deng WM, Bownes M. Two signaling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Development. 1997;124:4639–47. doi: 10.1242/dev.124.22.4639. [DOI] [PubMed] [Google Scholar]
- 76.Tzolovsky G, Deng W-M, Schlitt T, Bownes M. The function of the Broad-Complex during Drosophila melanogaster oogenesis. Genetics. 1999;153:1371–83. doi: 10.1093/genetics/153.3.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zartman JJ, Kanodia JS, Yakoby N, Schafer X, Watson C, et al. Expression patterns of cadherin genes in Drosophila oogenesis. Gene Expr Patterns. 2009;9:31–36. doi: 10.1016/j.gep.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berg CA. Tube formation in Drosophila egg chambers. Tissue Eng A. 2008;14:1479–88. doi: 10.1089/ten.tea.2008.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Newman SA, Bhat R. Dynamical patterning modules: a “pattern language” for development and evolution of multicellular form. Int J Dev Biol. 2009;53:693–705. doi: 10.1387/ijdb.072481sn. [DOI] [PubMed] [Google Scholar]
- 80.Lovegrove B, Simões S, Rivas ML, Sotillos S, Johnson K, et al. Coordinated control of cell adhesion, polarity, and cytoskeleton underlies Hox-induced organogenesis in Drosophila. Curr Biol. 2006;16:2206–16. doi: 10.1016/j.cub.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 81.Dreyer WJ. The area code hypothesis revisited: olfactory receptors and other related transmembrane receptors may function as the last digits in a cell surface code for assembling embryos. Proc Natl Acad Sci USA. 1998;95:9072–77. doi: 10.1073/pnas.95.16.9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Redies C, Takeichi M. Cadherins in the developing central nervous system: an adhesive code for segmental and functional subdivisions. Dev Biol. 1996;180:413–23. doi: 10.1006/dbio.1996.0315. [DOI] [PubMed] [Google Scholar]
- 83.Widmann TJ, Dahmann C. Wingless signaling and the control of cell shape in Drosophila wing imaginal discs. Dev Biol. 2009;334:161–73. doi: 10.1016/j.ydbio.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 84.Widmann TJ, Dahmann C. Dpp signaling promotes the cuboidal-to-columnar shape transition of Drosophila wing disc epithelia by regulating Rho1. J Cell Sci. 2009;122:1362–73. doi: 10.1242/jcs.044271. [DOI] [PubMed] [Google Scholar]
- 85.Shvartsman SY. Quantitative analysis of developing tissues. AIChE J. 2005;51:1312–18. [Google Scholar]
- 86.Viola J, Lal B, Grad O. The emergence of tissue engineering as a research field. 2003 http://www.nsf.gov/pubs/2004/nsf0450/
- 87.Davies JA. Synthetic morphology: prospects for engineered, self-constructing anatomies. J Anat. 2008;212:707–19. doi: 10.1111/j.1469-7580.2008.00896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lembong J, Yakoby N, Shvartsman SY. Pattern formation by dynamically interacting network motifs. Proc Natl Acad Sci USA. 2009;106:3213–18. doi: 10.1073/pnas.0810728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weigmann K, Klapper R, Strasser T, Rickert C, Technau G, et al. FlyMove—a new way to look at development of Drosophila. Trends Genet. 2003;19:310–11. doi: 10.1016/S0168-9525(03)00050-7. [DOI] [PubMed] [Google Scholar]