Abstract
Aire mediates the expression of tissue-specific antigens in thymic epithelial cells to remove dangerous self-reactive T lymphocytes. However, the mechanism that allows expression of tissue-specific genes at levels that prevent harm is unknown. Here we show that Brg1 generates accessibility at tissue-specific loci to impose central tolerance. We found that Aire harbors an intrinsic repressive function that restricts chromatin accessibility and opposes Brg1 across the genome. Aire exerted this repressive influence within minutes upon recruitment to chromatin and restrained the amplitude of active transcription. Autoimmune mutations that impair Aire-induced activation also impair the its repression function, indicating dual roles for Aire. Together, Brg1 and Aire fine-tune the expression of tissue-specific genes at levels that prevent toxicity, yet promote immune tolerance.
The functional competence of the immune system requires development of T cells that promote defense against pathogens and tumors, while silencing or removing T cells that react against self-constituents. The resultant T cell repertoire is limited by the range of self-antigens presented in the thymus1. The ectopic expression of thousands of tissue-specific self-antigens (TSAs) encompassing all parenchymal organs leads to immune tolerance to these self-antigens2,3. This ectopic transcription is driven largely by Aire, which operates in medullary thymic epithelial cells (mTECs) and mutations in this transcription factor cause autoimmune polyendocrine syndrome type-1 (APS-1)4,5.
Tissue-specific expression programs are generally defined by the developmentally coordinated actions of positive and negative regulators that influence the recruitment and release of RNA polymerase II (Pol II) at enhancers and promoters of lineage-specific genes6. Aire releases paused Pol II for productive elongation7–10 but the mechanisms that lead to accessibility and Pol II binding are unclear. The determinants of this poising mechanism in mTECs are distinct from those in peripheral tissues, as the lineage-specific transcription factors essential in the latter contexts are unnecessary for their thymic expression11.
While Aire has a unique ability to activate a large spectrum of tissue-specific genes, mTECs must also limit the expression of these genes because many of them encode proteins that would perturb physiological processes if expressed at levels comparable to their later expression. Indeed, the expression of Aire-induced PTGs in mTECs is orders of magnitude lower than in their respective peripheral tissues10,12,13. The expression of PTG subsets within single mTECs is transient and shuttles between distinct gene subsets, reducing the number of mTECs necessary to present the entire PTG repertoire14. Elucidating how the duration and amplitude of tissue-specific expression is controlled is essential to understanding how Aire induces precisely quantified transcription.
Here, we report that Aire restricts chromatin accessibility through its multimerizing and histone-binding activities to restrain the amplitude of PTG transcription. We also found that Brg1 promotes accessibility at loci encoding tissue-specific antigens, thereby allowing Aire to function.
RESULTS
mTEChi differentiation promotes chromatin accessibility
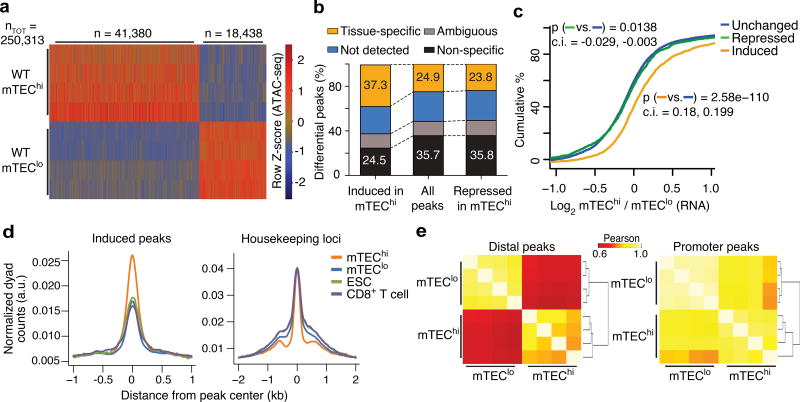
To elucidate the Aire-independent mechanisms that poise tissue-specific genes in mTECs, we determined when tissue-specific loci become accessible by examining the developmental control of chromatin accessibility during mTEC differentiation. Employing ATAC-seq15 and gene expression profiling, we defined accessibility landscapes and transcriptomes for progenitor mTECs (mTEClo) and Aire+ differentiated mTECs (mTEChi) distinguished by their surface expression of major histocompatibility class II (MHCII) (Supplementary Fig. 1a–e). We focused our ATAC-seq analyses on the main axis of variance from principal component analysis (PCA) that separates mTEChi and mTEClo samples (Supplementary Fig. 1f) to reduce the contribution of batch effects and other technical noise16. Using MAVRIC (Moskowitz and Greenleaf, manuscript submitted), a bioinformatic framework that attributes axes of variance to underlying sources (ATAC-seq peaks in this case), we identified 59,818 differentially accessible peaks that correlated with mTEChi maturation, accounting for ~ one-fourth of the total accessible genome detected (Fig. 1a). Nearly 70% of these peaks exhibited increases in accessibility during mTEChi differentiation and were enriched for their nearest gene being tissue-specific (Fig. 1a–d and Supplementary Fig. 1g). The mTEClo state exhibited low levels of accessibility at these regions, comparable to those in embryonic stem (ES) cells and naïve CD8+ T cells (Fig. 1d). Polycomb repressive complexes were enriched in ES cells (Supplementary Fig. 1h), suggesting that these tissue-specific sites were likely within facultative heterochromatin, consistent with the reported enrichment of lysine 27 trimethylation of histone H3 (H3K27me3) and depletion of Pol II binding and active histone marks at Aire-regulated tissue-specific genes17. Accessibility changes at the mTEChi-induced but not mTEChi-repressed peaks robustly correlated with the neighboring PTGs’ transcriptional changes (Fig. 1c). 90% of these peaks were > 1 kb away from the nearest transcriptional start site (TSS) (Fig. 1d,e). These results indicate a profound shift in the chromatin landscape during mTEChi differentiation that is driven by accessibility changes at distal cis-regulatory elements controlling transcription of tissue specific genes.
Figure 1.
mTEChi differentiation promotes chromatin accessibility at tissue-specific loci. (a) Heatmap of normalized ATAC-seq fragment density at differential peaks (rows). n = 4 independent experiments. (b) Tissue restrictions of indicated peak sets (filtered for peaks with accessibility fold-change > 2) assessed by tissue expression profile of the nearest gene. (c) Cumulative distribution function (CDF) plot of the transcriptional fold-change between mTEChi and mTEClo at tissue-specific peaks classified by differential accessibility changes (key). Representative of 4 independent experiments. Mann-Whitney U tests (two-tailed) performed for comparisons between classes of peaks. Confidence intervals (95%) are specified for each comparison. (d) Density plots of ATAC-seq fragment dyads from mTEChi, mTEClo, embryonic stem cells (ESC) and CD8+ T cells at induced peaks near tissue-specific genes upregulated in mTEChi (left) or near housekeeping genes (right). Representative of 4 independent experiments. (e) Clustered correlation of chromatin accessibility at ATAC-seq peaks distal (left) and proximal (right) to tissue-specifc genes with induced transcription in mTEChi vs. mTEClo. n = 4 independent experiments.
Aire and Brg1 have opposing influences on chromatin state in mTEChi
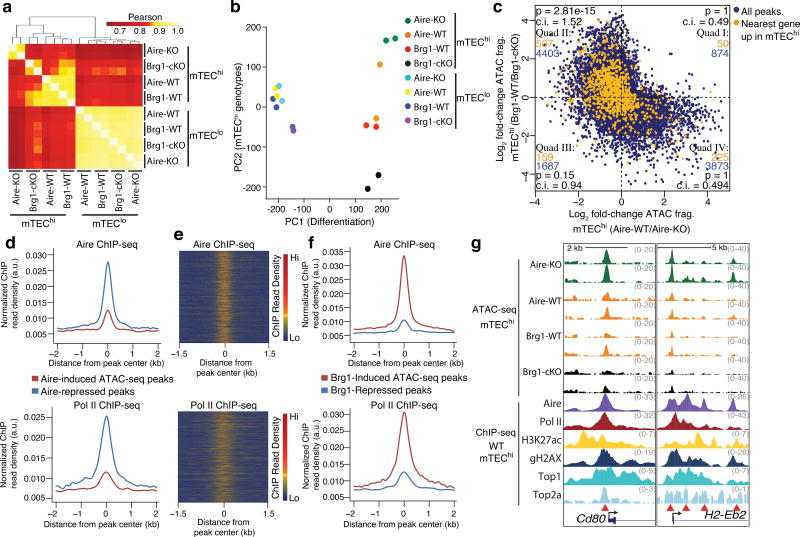
Subunits of the mSWI/SNF or BAF chromatin remodeling complex appeared in a hypothetical network for functional partners for Aire8 suggesting that BAF complexes specifically contribute to the poising of PTGs in mTECs. Thus, we profiled the accessibility landscapes in mTEChi and mTEClo from Foxn1ex9cre;Brg1F/F mice, which delete Smarca4, the catalytic subunit of Brg1 in TECs (Brg1-cKO hereafter). The frequencies of mTEC compartments and the punctate localization of Aire were largely normal in Brg1-cKO mice (Supplementary Fig. 2a–c). To compare the influence of Brg1 on the mTEChi chromatin state with that of Aire, we profiled the accessibility landscapes in mTECs from Aire-deficient (Aire-KO hereafter) mice. Hierarchical clustering based on the correlations of accessibility at mTEChi-induced peaks near PTGs separated the Brg1-cKO and Aire-KO mTEChi samples (Fig. 2a), indicating distinct influences of Brg1 and Aire on the mTEChi chromatin landscape. Aire-KO and Brg1-cKO mTEChi samples distributed at opposite poles of the principal component of variability attributable to the genotypes of mTEChi samples (PC2) (Fig. 2b), suggesting divergent influences of Aire and Brg1 on the mTEChi-specific accessibility state.
Figure 2.
Aire and Brg1 are determinants of an mTEChi-specific chromatin state with opposing influences on accessibility. (a) Correlations between indicated ATAC-seq samples at tissue-specific regions exhibiting induced accessibility during mTEChi differentiation. n = 4 independent experiments. (b) Principal component analysis of ATAC-seq data. (c) Comparison of the influences of Aire and Brg1 on mTEChi-specific accessibility state (ATAC-seq peaks correlated to PC2). Peaks whose nearest genes are upregulated in mTEChi vs. mTEClo are highlighted in orange and enumerated in each cartesian quadrant with total peaks (blue). P values indicate one-sided Fisher’s exact test with 95% confidence intervals (lower bound). (d) Aire (top) and Pol II (bottom) ChIP-seq fragment dyad density at Aire-induced or Aire-repressed ATAC-seq peaks. Representative of 2 independent experiments. (e) Heatmap of Aire (top) or Pol II (bottom) ChIP-seq fragment dyad density at Aire-repressed ATAC-seq peaks. Representative of 2 independent experiments. (f) Aire (top) and Pol II (bottom) ChIP-seq fragment dyad density at Brg1-induced or Brg1-repressed ATAC-seq peaks. Representative of 2 independent experiments. (g) Genomic signal tracks of ATAC-seq fragments at two loci from indicated mTEChi samples (top). ChIP-seq signal tracks (representative of 2 independent experiments) from WT mTEChi samples. Red arrowheads indicate differentially accessible regions.
Using MAVRIC, we identified peaks correlated to PC2 and found a negative correlation (r = −0.43) when comparing accessibility promoted by Aire versus that promoted by Brg1 (Fig. 2c). Using arbitrary fold-change cutoffs to classify induced and repressed peaks (FC >2, <0.5), cumulative distribution function (CDF) plots show that Brg1-induced peaks were predictive for Aire repression and Aire-induced peaks were predictive for Brg1 repression (Supplementary Fig. 2d), indicating that Aire and Brg1 exert opposing forces at the same sites. To assess the functional relevance of these regions for mTEChi differentiation, we assigned the nearest TSS to each ATAC-seq peak. mTEChi-activated genes were far more frequently near peaks whose accessibility was repressed by Aire and induced by Brg1 (Fig. 2c) than to peaks induced by Aire and repressed by Brg1 or those repressed by both Aire and Brg1 (Fig. 2c). To validate the negative influence of Aire on chromatin accessibility, we analyzed published ATAC-seq samples18 and found increases in fragment density in Aire-KO compared to wild-type mTEChi at peaks correlated to the primary axis of variance separating the genotypes (Supplementary Fig. 2h,i). Taken together, these data unveil that Aire has a repressive function at BAF-promoted accessible regions near genes upregulated during mTEChi differentiation.
To assess whether Aire is directly mediating these accessibility changes, we used published mTEChi Aire ChIP-seq data18 to assay Aire occupancy at differentially accessible peaks. Aire localization was robustly enriched at regions where chromatin accessibility was repressed by Aire compared to unaffected regions (Fig. 2d–g and Supplementary Fig. 2g,j). Furthermore, Aire bound to regions where accessibility was induced by Brg1 (Fig. 2f,g and Supplementary Fig. 2g), suggesting a direct role for Aire in repressing accessibility at Brg1-promoted sites. These Aire-repressed, Brg1-promoted regions were mostly gene-distal with only ~7% of peaks within promoters (Supplementary Fig. 2e,f). These regions were also enriched for Pol II, acetylation at lysine 27 of histone H3 (H3K27ac), topoisomerases I and IIa (Top1 and Top2a) and γ-H2AX deposition (Fig. 2d–g and Supplementary Fig. 2g,3), further supporting their enhancer potential. In contrast, regions where accessibility was increased by Aire were depleted of Aire, Pol II, H3K27ac, Top1, Top2a and γ-H2AX localization (Fig. 2d–g and Supplementary Fig. 3), suggesting an indirect influence of Aire on accessibility at these loci. These data suggest that the repressive function of Aire is executed after BAF-promotes accessibility.
Accessibility at PTGs is promoted by Brg1 and repressed by Aire
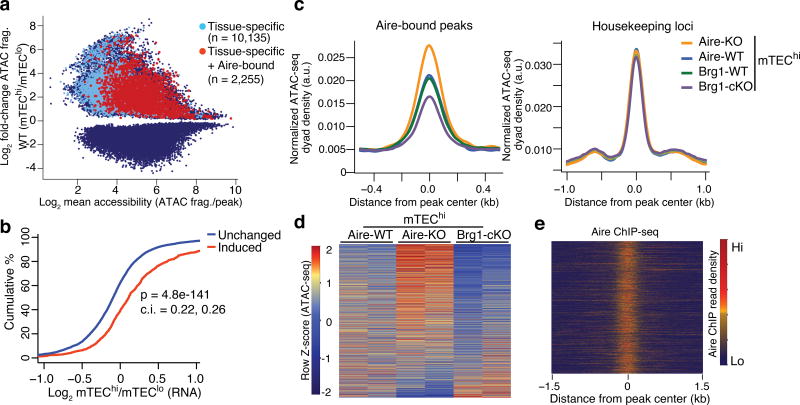
To gain insight into the mechanism of the ectopic expression of tissue-specific genes in mTECs, we focused our analyses on the regions whose accessibilities could serve to directly promote the expression of PTGs during mTEChi differentiation. We defined these regions as ATAC-seq peaks whose accessibility is specifically induced in mTEChi compared to mTEClo, is bound by Aire, and whose nearest gene is tissue-restricted. 2,255 peaks qualified for these criteria (Fig. 3a), which, considering the heterogeneity of individual mTECs expressing distinct subsets of the collective PTG repertoire10,11,13,14,19, is likely only a fraction of the total. These qualifying regions presumably reflect PTGs with higher probability of expression (up to 48% detected) in individual mTECs11. The accessibility changes at these peaks exhibited a strong correlation with the transcriptional changes during mTEChi maturation (Fig. 3b), indicating these peaks may serve as cis-regulatory elements that control PTG expression.
Figure 3.
Accessibility at tissue-specific loci are promoted by Brg1 and repressed by Aire. (a) MA plot of differential ATAC-seq peaks between mTEChi and mTEClo samples (blue), highlighted by those whose nearest gene is tissue-specific (light blue) and those that have Aire occupancy and neighbor tissue-specific genes (red). (b) CDF plot of the transcriptional fold-change between mTEChi and mTEClo at indicated loci classified by differential accessibility changes. P value from two-tailed Mann-Whitney U test with 95% confidence intervals. Representative of two independent experiments. (c) Density plots of ATAC-seq fragment dyads from mTEChi samples of indicated genotypes at Aire-bound, ATAC-seq peaks near tissue-specfic genes (left) or housekeeping genes (right). Representative of 2 independent experiments. (d) Heatmap of normalized ATAC-seq fragment dyad density between samples of indicated genotypes at mTEChi-induced, Aire-bound, tissue-specific loci. n = 2 independent independent experiments. (e) Heatmap of Aire ChIP-seq fragment dyad density at same regions as (d). Representative of 2 independent experiments.
To determine whether BAF promotes the poising of these tissue-specific regions by facilitating their accessibility, we quantified the ATAC-seq fragment density at these sites and found a substantial reduction of fragment density in Brg1-cKO mTEChi compared to wild-type (Fig. 3c,d). In contrast, the ATAC-seq fragment density at these sites in Aire-KO mTEChi increased (Fig. 3c–e), indicating that Aire repressed accessibility at these regions near transcriptionally upregulated PTGs. Furthermore, the increase in chromatin accessibility at these tissue-specific loci in mTEChi compared to mTEClo was also present in Aire-KO mTEChi cells (Supplementary Fig. 4a), indicating accessibility at these loci was induced in Aire-deficient mTECs. In contrast, the transcriptional activation of Aire-dependent tissue-specific genes was compromised in Aire-KO mTEChi compared to wild-type (Supplementary Fig. 4b). Similarly, the the transcriptional activation of PTGs during the mTEClo to mTEChi transition in wild-type mice was largely Aire-dependent (Supplementary Fig. 4d). In contrast, Aire was broadly dispensable for the accessibility induction at PTGs during normal mTEChi differentiation (Supplementary Fig. 4c). This is consistent with reports that the amount of DNA methylation and promoter-proximal transcription of Aire-induced PTGs does not change in Aire-KO mTECs compared to wild-type mTECs7,10. Taken together, these results suggested that Brg1 was essential for promoting accessibility near PTGs in mTEChi that Aire later targeted for transcriptional activation. Moreover, the negative influence of Aire on accessibility may restrain the transcriptional amplitude at PTGs.
NF-κB target sites are hypersensitive to regulation by Aire and Brg1
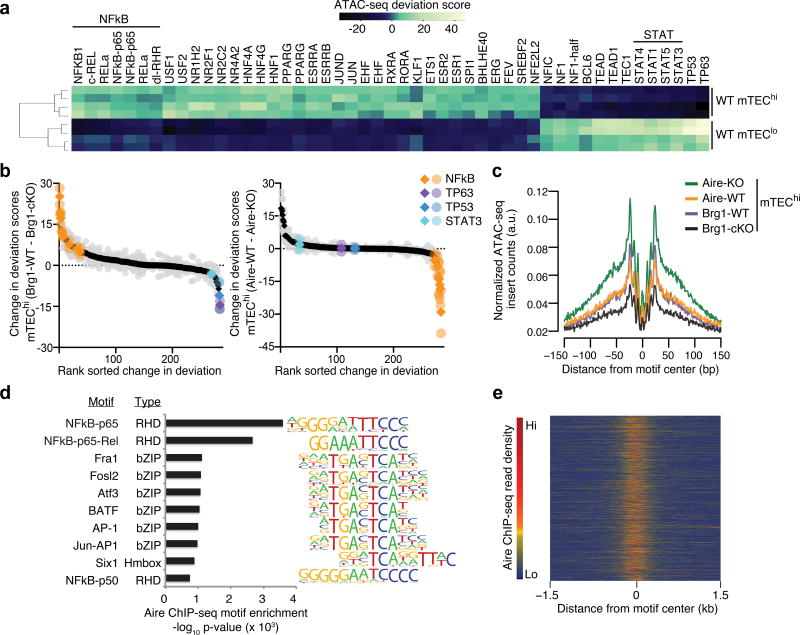
Because the 8 DNA-binding domains of the BAF subunits have only limited sequence specificity20 and Aire does not exhibit sequence-specific DNA-binding21,22, we sought to identify transcription factors potentially working in tandem with either complex that promote the mTEChi-specific chromatin state. Steric hindrance between transcription factors and the Tn5 transposase can be inferred from ATAC-seq data, allowing enrichment analyses for transcription factor footprinting15. Using an established framework to quantify the changes in accessibility across ATAC-seq peaks sharing known transcription factor motifs23, we identified a battery of transcription factors that were differentially enriched in wild-type mTEChi compared to mTEClo. Peaks with motifs for the transcription factors cRel, RelA and NF-κB1 exhibited the most prominent increases in accessibility (Fig. 4a and Supplementary Fig. 5a), consistent with the essential roles of NF-κB signaling in mTEChi maturation24,25. We also inferred the differential increase in binding of NF-κB complexes in mTEChi vs. mTEClo using transcription factor footprinting (Supplementary Fig. 5e). We also noticed significant decreases in accessibility at sites containing STAT, P53 and P63 motifs in mTEChi compared to mTEClo (Fig. 4a and Supplementary Fig. 5a), along with the putative loss in binding of these factors (Supplementary Fig. 5f,k,l,n–p), consistent with the reported role of Stat3 and p63 in the expansion and survival of the progenitor mTEClo compartment26–28.
Figure 4.
Regions containing NF-kB motifs are highly sensitive to opposing regulation by Aire and Brg1. (a) Heatmap of deviations from expected ATAC-seq signal from mTEChi or mTEClo samples at ATAC-seq peaks containing known trans-factor motifs. n = 4 independent experiments (b) Change in deviation from expected accessibility signal at ATAC-seq peaks containing known trans-factor motifs (key) between indicated samples. Mean (diamond) and replicates (circles) from n = 2 independent experiments. (c) Accessibility footprints at NF-kB motifs within ATAC-seq peaks from indicated mTEChi samples. (d) Top known motif enrichment within Aire ChIP-seq peaks. (e) Heatmap of Aire ChIP-seq fragment dyad density at ATAC-seq peaks with NF-kB motifs. Representative of 2 independent experiments.
Because NF-κB depends on pre-existing accessibility for binding29, the substantial increase in accessibility at sites with NF-κB motifs during mTEChi maturation suggested NF-κB-binding requires chromatin remodeling to impose the mTEChi state. Indeed, Brg1 promoted chromatin accessibility most robustly at peaks containing NF-κB motifs (Fig. 4b and Supplementary Fig. 5b,d). Furthermore, the NF-κB footprinting along with the flanking accessibility was substantially reduced in Brg1-cKO mTEChi at these sites compared to wild-type (Fig. 4c). These data indicate that BAF prepared the chromatin landscape to allow NF-κB-binding at target loci and drive mTEChi differentiation.
Activation of NF-κB signaling in mTECs precedes the expression of Aire25. To determine the influence of Aire on the NF-κB and Brg1-induced chromatin accessibility, we quantified the changes in accessibility between Aire-KO mTEChi and wild-type mTEChi across peaks containing all known motifs. The largest changes were observed at peaks with NF-κB motifs, whose accessibilities were repressed by Aire (Fig. 4b and Supplementary Fig. 5c,d). Moreover, the flanking accessibility and magnitude of the NF-κB footprinting was increased in Aire-KO mTEChi vs. wild-type controls (Fig. 4c). Robust Aire occupancy at NF-κB motif-containing ATAC-seq peaks indicated direct repression by Aire at Brg1-promoted sites (Fig. 4d,e). Considering the central role of NF-κB activation in mTEChi differentiation and the negative regulation of mTEChi cellularity by Aire5,13,30,31, these results suggest Aire directly curtails fundamental components of the mTEChi differentiation program by limiting chromatin accessibility. Indeed, Aire bound to and diminished chromatin accessibility near genes encoding factors essential for mTEChi differentiation, e.g. Cd80, H2-Eb2 and Fam18a (Fig. 2g and Supplementary Fig. 2g).
Brg1 imposes immunological tolerance
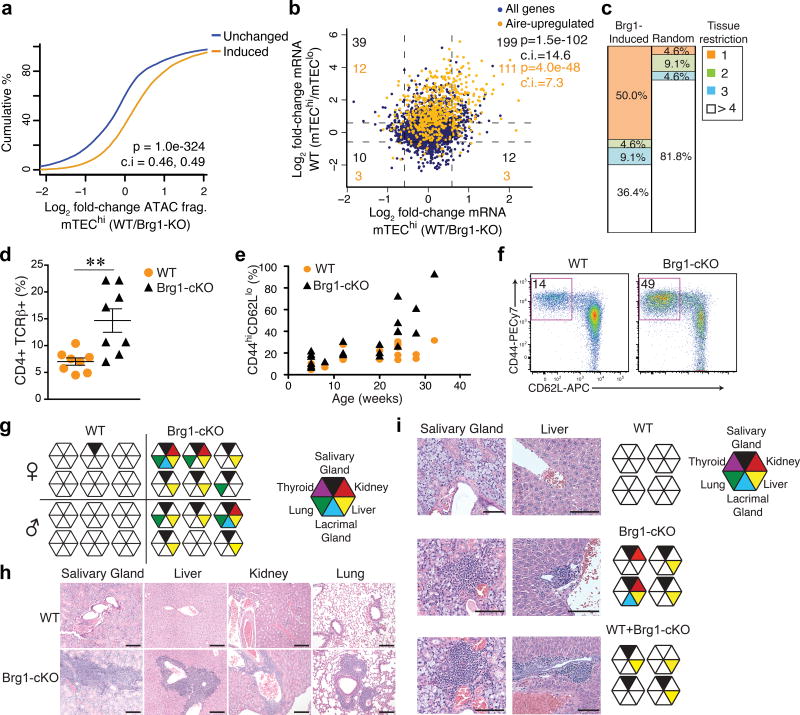
Because Aire acts on a preexisting chromatin landscape, we asked whether the induced accessibility at tissue-specific loci was important for ectopic transcription of PTGs and central tolerance induction. Accessibility changes at tissue-specific loci during the mTEClo to mTEChi transition were robustly predictive of the accessibility changes between Brg1-cKO and wild-type mTEChi (Fig. 5a). Gene-expression profiling of Brg1-cKO mTEChi showed that the activation of hundreds of PTGs was diminished in Brg1-cKO mTEChi compared to wild-type, especially at Aire-regulated genes (Fig. 5b). In addition, the top decile of Brg1-upregulated genes was highly enriched for PTGs expressed in a single peripheral tissue (Fig. 5c). Taken together, these results suggested that BAF bolstered PTG expression by promoting accessibility at distal regulatory elements during mTEC differentiation. To determine how the compromised activation of PTGs in Brg1-cKO mice affected the self-reactivity of the T cell repertoire, we first assayed the frequency of activated T cells in the periphery. While the frequency of thymocyte compartments were largely normal (Supplementary Fig. 6a,b), the Brg1-cKO mice exhibited a doubling of the frequency of CD4+ CD44hi CD62Llo T effector cells compared to wild-type, with nearly half of the splenic T cell compartment consisting of activated CD44hi CD62Llo T effector cells at 6 months of age (Fig. 5d–f). However, the number of total splenic T cells in Brg1-cKO mice was approximately half of that in wild-type mice (Supplementary Fig. 6e), likely due to the compromised function of Brg1-deficient cTECs that promote early thymocyte differentiation (Supplementary Fig. 6c,d). We next asked whether the activated effector T cells in Brg1-cKO mice could provoke autoreactive tissue damage. We found substantial lymphocytic infiltration in the kidney, liver, lung, lacrimal and salivary glands at 3 – 6 months old Brg1-cKO mice (Fig. 5g,h). Immunohistochemistry showed strong CD3 staining within these tissue infiltrates (Supplementary Fig. 6h).
Figure 5.
Brg1 imposes immunological tolerance. (a) CDF plot of accessibility fold-changes at tissue-specific peaks classified as differentially accessible in WT mTEChi vs. mTEClo. P value (two-tailed Mann-Whitney U-test with 95% confidence intervals). Representative of 2 independent experiments. (b) Role of Brg1 on the ectopic induction of tissue-specific genes during mTEChi differentiation. Dotted lines indicate 1.5- and 0.66 fold-change for each comparison. Corner numbers indicate total (black) and Aire-upregulated (orange) genes in each quadrant. P values correspond to genes in upper right quadrant (one-tailed Fisher’s exact test with 95% confidence intervals). (c) Fraction of genes from top decile of Brg1-activated genes in mTEChi in each indicated tissue-restriction group (number of peripheral tissues expressing gene) compared to random group of genes. (d) Frequencies of activated splenic CD4+ T cells in 4 wk-old mice (n = 8 independent experiments). Mean +/− s.e.m. **P = 0.002 (two-tailed t-test). (e) Comparison of the frequencies of activated splenic T cells as a function of age from indicated genotypes. (f) Representative cytometry plot from 4 independent experiments of the frequency of activated CD4+ T cells in spleen from 6 month-old mice of indicated genotypes. (g) Histological analyses of indicated tissues (left) from 3–6 month-old WT or Brg1cKO mice via H&E staining for infiltrating lymphocytes. Each octagon represents individual mouse. n = 6 independent experiments. (h) Representative H&E stainings (n = 6 independent experiments) of the histopathology in diseased tissues of Brg1cKO mice. Scale bars = 200um. (i) Histological analyses of tissues via H&E staining for infiltrating lymphocytes from nude mice, 14 wk post-thymus transplants from indicated donors (right). Representative H&E stainings (left) from 2 independent experiments. Scale bars = 100um.
We next addressed whether the autoimmunity in Brg1-cKO mice was due to failure in the negative selection of self-reactive T cells and/or the positive selection of Foxp3+CD4+25+ regulatory T cells (Treg cells) or to the diminished expression of tissue-specific self-antigens (TSAs) in mTECs. Brg1-cKO mice had normal frequency, number and function of splenic Foxp3+ Treg cells compared to those in wild-type mice (Supplementary Fig. 6f,g), suggesting a functional Treg cell compartment. To test whether the autoimmunity in Brg1-cKO mice was driven by the reduced thymic output, which might extend the neonatal window of physiologic lymphopenia, allowing aberrant homeostatic proliferation of self-reactive T cells32, or by deficiencies in the selection or function of the Treg cell compartment, we grafted thymic stroma depleted of hematopoietic cells by treatment with 2-deoxyguanosine from wild-type mice, Brg1-cKO mice or both under the kidney capsules of wild-type athymic nude mice. This experimental setting allowed us to test whether the normalized thymic output and the provision of Treg cells from the wild-type co-transplanted thymus could rescue the autoimmune pathology driven by the self-reactive T cells escaping negative selection in the Brg1-cKO thymus. Mice transplanted with both wild-type and Brg1-cKO thymic stroma exhibited lymphocytic infiltration in multiple organs (Fig. 5i), indicating the self-reactive T cells from the Brg1-cKO thymus had a dominant activity. However, the range of organs affected in these co-transplanted chimeric hosts was reduced to the liver and salivary gland compared to those transplanted with only Brg1-cKO thymic stroma that exhibited infiltrations in the kidney and lacrimal glands (Fig. 5i), suggesting a potential role for defective positive selection of Treg cells and/or lymphopenia-induced proliferation to the autoimmunity in Brg1-cKO mice. Taken together, these data underscore the importance of Brg1 in promoting central tolerance induction by facilitating the accessibility at tissue-specific loci, an essential component for TSA expression and selection of a functional T cell repertoire.
Aire rapidly represses accessibility upon recruitment to chromatin
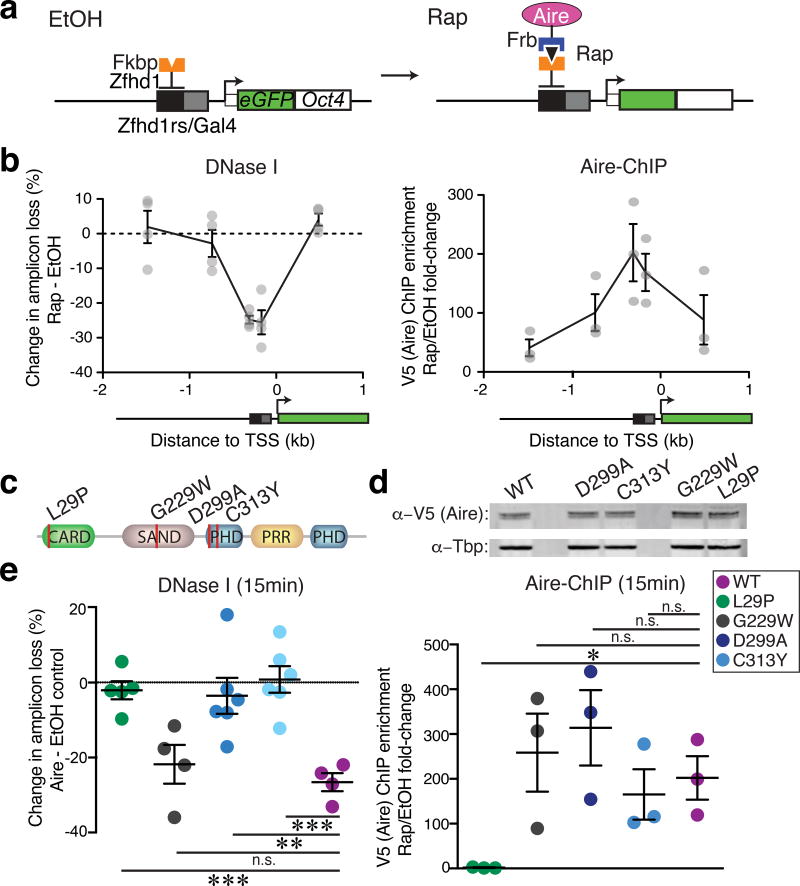
To elucidate the mechanism of the repressive function of Aire, we used the chromatin in vivo assay (CiA) system33–35, which allows precise temporal control of the recruitment of chromatin regulators to specific genetic loci via chemically-induced proximity within living cells. The minute-by-minute assessment of effects of recruitment of a chromatin regulator allows one to distinguish between rapid direct biochemical actions of Aire from slow secondary and indirect effects, e.g. influences on cell cycle or the actions of factors encoded by Aire-target genes. Mouse embryonic fibroblasts (MEFs) from CiA mice have two arrays of binding sites for transcription factors and GFP inserted into one allele of the Oct4 gene (CiA:Oct4)33, which is an Aire-regulated, tissue-specific locus in mTEChi 31(Supplementary Fig. 7a). This locus is faithfully integrated into the endogenous chromatin context with appropriate long-range, topological conformations34. Furthermore, this locus is transcriptionally silent and allows assessment of effects independent of transcription. We generated stable cell lines from these mice expressing the ZFHD1 DNA-binding domain fused to FKBP12 (component of mTOR signaling pathway) and Aire fused to the FRB domain of mTOR and V5 epitope serving as the anchor and recruitment partner, respectively (Fig. 6a). Addition of rapamycin elicited the rapid association of FKBP12 and FRB causing the robust recruitment of Aire (> 200-fold measured by V5 ChIP) to the CiA:Oct4 locus within minutes (Fig. 6b). We assessed the impact of Aire recruitment to the CiA:Oct4 locus and found an extensive loss of chromatin accessibility at the recruitment site (measured by DNase I sensitivity) within 15 minutes of rapamycin addition compared to control ethanol treatment (Fig. 6b). The magnitude of the accessibility loss at the modified Oct4 locus appeared comparable to that seen after 1 hour or after 5 days of Aire recruitment (Supplementary Fig. 7b), suggesting Aire induces its maximum repressive effect within minutes of recruitment. These observations indicated that Aire has a direct repressive activity. This rapid repression was in contrast to the quick gain in accessibility upon BAF complex recruitment to the CiA:Oct4 locus (Supplementary Fig. 7c,d)34,35. Furthermore, the transcriptionally silent state of the CiA:Oct4 locus indicated that the repressive activity of Aire is uncoupled to transcription (Supplementary Fig. 7e). In a distinct approach, we employed an orthogonal recruitment strategy using dCas9 and MS2-binding aptamers36 in the human cortical thymic epithelium cell line 4D637 (Supplementary Fig. 7f,g). We chose an open tissue-specific locus upstream of PSMB11 (encoding the β5t subunit of the thymoproteosome38) to test the influence of Aire recruitment on accessibility. Aire recruitment upstream of PSMB11 induced loss of accessibility compared to the recruitment of dCas9 alone (Supplementary Fig. 7h), confirming the results at the CiA:Oct4 locus in MEFs. Taken together, Aire exerted its repressive influence within minutes of recruitment to an Aire-regulated locus independent of transcription, indicating a direct biochemical function.
Figure 6.
Aire rapidly represses accessibility upon recruitment to chromatin. (a) Schematic of inducible CiA system: Recruitment to modified Oct4 locus via rapamycin (Rap)-induced dimerizing of Aire-Frb and Fkbp-Zfhd1 fusion proteins. (b) Changes in DNase I hypersensitivity (left, n = 4 independent experiments - circles) at CiA:Oct4 locus upon Rap-induced (15 min) Aire recruitment measured by V5 ChIP (right, n = 3 independent experiments). Mean +/− s.e.m. (c) Depiction of APS-1 patient mutations in CARD, SAND and PHD1 domains. (d) Western blot of Aire-Frb (and mutants) transgenic expression in CiA system. Representative of two independent experiments. (e) Changes in DNase I hypersensitivity (left, n = 4 independent experiments) at CiA:Oct4 locus upon Rap-induced (15 min) WT Aire or variant (key) recruitment measured by V5 ChIP (right, n = 3 independent experiments). Mean +/− s.e.m. Statistical significance for each mutant relative to WT by two-tailed t-test. P values < 0.05 (*), < 0.01 (**), < 0.001 (***).
Aire repressive function requires the PHD1 and multimerization domains
Aire directly interacts with chromatin through its PHD1, a histone-binding module whose recognition of unmodified H3-tail is necessary for tolerance induction22,39,40. Because the CiA system uses a ZFHD1-FKBP anchor, the recruitment of Aire to the CiA:Oct4 locus does not depend on its own domains, and as such can be used to uncouple the targeting and repressive functions of Aire. The Aire C311Y mutation in humans and C313Y in mice (Fig. 6c) disrupt the zinc coordination of PHD141, preventing Aire interaction with chromatin22. This mutation gives rise to the autoimmune polyendocrine syndrome type-1 (APS-1)4. When we recruited a Aire-C313Y variant to the CiA:Oct4 locus via rapamycin, the repressive activity of Aire was abolished compared to the recruitment of wild-type Aire, despite the comparable protein expression and locus recruitment between cells expressing Aire-C313Y and Aire-WT (Fig. 6d,e). Because the unfolding of the PHD1 finger in the Aire-C313Y mutant might affect other domains, we next recruited an Aire variant with a designed mutation (Aire-D299A) that mitigates the histone-binding activity of Aire, but maintains the integrity of the PHD finger22,41. Aire-D299A was unable to repress accessibility at the CiA:Oct4 locus compared to that of wild-type Aire (Fig. 6e), indicating the importance of the histone-binding module of Aire for both its repressive and activating functions. The G228W APS-1 mutation in the SAND domain of Aire (G229W in mice) exhibits dominant negative activity to cripple Aire localization42. In contrast to the PHD1 mutants Aire-C313Y and Aire-D299A, recruitment of Aire-G229W to the CiA:Oct4 locus decreased chromatin accessibility similar to that of Aire-WT (Fig. 6e), indicating that the targeting defects of Aire-G229W were rescued by the ZFHD1-FKBP anchor.
Aire multimerizes through its caspase recruitment domain (CARD) to form a multi-protein complex of ~750 kDa18,43. The L28P APS-1 mutation (L29P in mice) disrupts the ability of Aire to multimerize, causing defects in subnuclear localization and activation of target genes43. The recruitment of Aire-L28P to the CiA:Oct4 locus was ~50-fold less than the recruitment of Aire-WT, despite comparable protein expression in the respective CiA MEFs (Fig. 6d,e), and the repressive activity of Aire-L28P was largely abolished compared to Aire-WT (Fig. 6e), suggesting that the multivalent conformation of Aire within an oligomerized complex is essential to its repressive function. Taken together, the repressive function of Aire is keyed by the same multimerization and histone-binding activities that are necessary for promoting ectopic transcription, indicating a unique duality in the functional domains of Aire.
Aire recruitment to an active locus restrains transcriptional amplitude
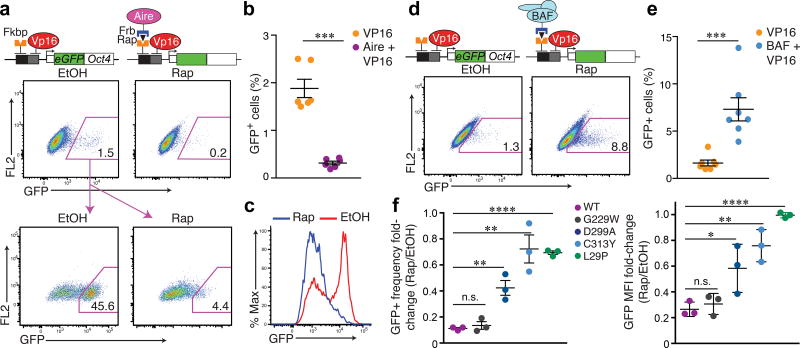
If the repressive influence of Aire on chromatin accessibility serves to constrain the transcription of tissue-specific genes, directed Aire recruitment to an active locus should reduce gene expression. To directly test the impact of Aire on active transcription, we constitutively recruited the transcriptional activator VP16 to the CiA:Oct4 locus and sorted for cells that activated the GFP reporter encoded in the CiA:Oct4 locus before a 3 day treatment with rapamycin to co-recruit Aire (Fig. 7a). Both the intensity of GFP expression and the frequency of GFP+ cells were reduced upon Aire and VP16 locus co-recruitment compared to cells not given rapamycin (Fig. 7a–c). In contrast, co-recruitment of VP16 and the BAF complex to the CiA:Oct4 locus increased the frequency of GFP+ cells (Fig. 7d,e). The repressive influence of Aire was dependent in part on the histone-binding module and multimerization potential of Aire (Fig. 7f). These observations suggest that multimerized Aire and the recognition of unmodified H3-tails through the PHD1 fingers contribute to fortifying a barrier for active transcription. Taken together, these results indicate that during mTEC development, the initial poising by Brg1 and the subsequent transcriptional triggering by Aire was restrained by the repressive influence of Aire on chromatin accessibility, akin to a rheostat in an electrical circuit (Supplementary Fig. 7i).
Figure 7.
Aire recruitment to an active locus restrains transcriptional amplitude. (a) Top: Sequential recruitment of Aire (Rap) to CiA:Oct4 locus for 3 days post VP16 recruitment, frequency of GFP+ cells over ethanol control (EtOH) assessedvia flow cytometry. Representative cytometry plots of 6 independent experiments. Bottom: GFP+ cells post-VP16 recruitment were sorted, then treated +/− Rap for 3 days. Representative cytometry of 5 independent experiments. (b) GFP+ frequency quantification via VP16 +/− Aire co-recruitment (n = 6 independent experiments). Mean +/− s.e.m. ***P <0.001 (two-tailed t-test). (c) Representative histogram of Rap- vs. EtOH-treated cells described in (a-bottom) for GFP expression. Representative of 5 independent experiments. (d,e) Sequential recruitment of BAF via Ss18 (Rap) for 3 days post VP16 recruitment, frequency of GFP+ cells quantified (n = 7 independent experiments), Mean +/− s.e.m. ***P <0.001 (two-tailed t-test). (f) Quantification of fold-change in frequency (left) and mean flourescence intensity (right) of GFP+ cells between EtOH- and Rap-treated conditions after sorting as in (a). n = 3 independent experiments. Mean +/− s.e.m. Two-tailed t-tests between each mutant and WT Aire recruitment: P < 0.05 (*), < 0.01 (**), < 0.001 (***), < 0.0001 (****).
DISCUSSION
Here we provide new insight on the underlying mechanisms that promote activation of thousands of tissue-specific genes in the thymic epithelium, at levels that are sufficient for immune tolerance, but below those that could be harmful. We find that Aire, the transcriptional regulator necessary for much of the mTEC expression of tissue-specific genes, has an intrinsic repressive function that restricts chromatin accessibility. Our analyses indicate that Aire bound Brg1-induced, mTEChi-specific regulatory sites and repressed their accessibility to curtail mTEChi differentiation. Taken together with the repressive actions of Aire at tissue-specific loci, it appears that Aire limits the duration and amplitude of transcription of tissue-specific genes, as well as the number of cells that could become competent for this ectopic expression. The physiological necessity for this negative regulation can be explained by the negative consequences of overexpressing tissue-specific factors such as insulin, calcitonin, and blood coagulation factors in mTECs44–46.
Our recruitment studies using chemically-induced proximity at an Aire-regulated locus indicates that Aire exerts its repressive influence within minutes of recruitment to chromatin, indicating a direct biochemical mechanism. Because this locus is transcriptionally silent, the repressive influence of Aire on accessibility precludes any dependence on active transcription. This repressive function required key residues that facilitate the multimerization and histone-binding activities of Aire. Importantly, the same domains and residues are essential for expression of tissue-specific genes and prevention of autoimmunity45,40. The histone-binding activity of Aire stabilizes its interaction with target loci, contributing to its targeting function39,40. However, the CARD-mediated multivalent state allows the simultaneous recognition of multiple histone tails that likely favor nucleosomal occupancy and perhaps repression of transcription initiation. To our knowledge, this intrinsic functional duality has not been described for a single transcription regulatory factor.
We find that the Brg1 catalytic subunit of the mSWI/SNF remodeling complex is essential for promoting the mTEChi-specific chromatin state by facilitating accessibility at tissue-specific loci prior to their expression. The mSWI/SNF or BAF complex represent one of about 30 non-redundant ATP dependent chromatin remodeling complexes with often instructive roles in developmental lineage specification and tumor suppression. BAF antagonizes Polycomb occupancy at target sites via direct physical interaction and ATP-dependent eviction34,47. Considering the enrichment of Polycomb subunits and H3K27me3 at Aire-regulated loci in peripheral tissues and ES cells17, BAF likely employs similar mechanisms in mTECs to facilitate the poising of tissue-specific genes. In addition, BAF complexes associate with topoisomerase II (Top2) for the resolution of facultative heterochromatin35,48,49. These observations are consistent with our studies, as the mTEChi-specific regulatory regions whose accessibilities are promoted by Brg1 showed enrichment of Top2a. Importantly, Aire inhibits topoisomerase II18,50 suggesting a mechanism by which Aire could reduce accessibility and provide a repressive function.
Our studies indicate that Aire is unique in that it has, in the same protein, mediated by the same domains, both activating and repressive functions. These bifunctional domains may have evolved to cooperate with both repressors and activators to allow ectopic expression of tissue-specific antigens at levels generally several orders of magnitude lower than their expression in their peripheral tissues. Although thousands of tissue-specific genes become accessible to the transcription factors present in mTECs, these potential regulatory regions appear to be held in check by Aire’s ability to oppose accessibility. This opposition potentially guards tissue-specific genes from the hundreds of transcription factors (like NF-κB) expressed in mTECs to yield safe but immunologically effective levels of tissue-specific antigens in the thymus.
ONLINE METHODS
Mice
B6. Aire+/− mice5 obtained from Jackson Laboratory were bred to generate Aire−/− and Aire+/+ littermates. Brg1F/F and Foxn1ex9cre mice51,52 on C57BL/6 background were bred to generate Brg1F/F and Foxn1ex9cre;Brg1F/F littermates. All experiments used female and male littermate controls. All mice were maintained in accordance with Stanford University’s Animal Care and Use Committee guidelines. Animal protocol is annually approved by the Administrative Panel on Laboratory Animal Care of the Research Compliance Office of Stanford University.
Cell Lines
Mouse embryonic fibroblasts (MEFs) were acquired from mice with the CiA:Oct4 allele at embryonic day 14.5 as previously described33. MEFs were transformed with simian virus 40 large T antigen and single cell sorted after transfection with LGmCreER (self-deleting) plasmid53 (Addgene #33340) to enrich for cells with excised neo cassette. Clones were screened for growth rate, VP16-mediated eGFP activation and DNase accessibility at the CiA:Oct4 locus. A single clone was used for all CiA recruitment experiments. MEFs were grown in high-glucose DMEM (Life Technologies, 11960) supplemented with 10% FBS (Omega Scientific, FB-11), 10 mM HEPES pH 7.5, Minimal AA, glutaMAX, Na Pyruvate, Pen/Strep, 2-Mercaptoethanol at 37° C., 5% CO2. The 4D6 human thymic epithelial cell line37 was a gift from Maria Toribio and Diane Mathis. The 4D6 cells were grown in RPMI 1640 (Life Technologies, 21870092) supplemented with 10% FBS (Omega Scientific, FB-11) and Pen/Strep at 37° C., 5% CO2.
Flow cytometry
mTEC subsets were isolated as previously described54,55. Thymi were dissected, capsules incised and triturated with glass pipettes to release thymocytes from stromal fragments. Stroma were digested with Liberase TM (Roche) and DNase I (Roche), and TECs were enriched using α-CD45 MACS microbeads (Miltenyi) or centrifugation on a Percoll PLUS gradient (GE Healthcare). Enriched TECs were stained with flurochrome-conjugated antibodies (BioLegend) against CD45 (30-F11), Ly-51 (6C3), MHC-II I-A/I-E (M5/114.15.2) and EpCAM (G8.8) along with fluorescein labeled UEA-I (Vector Labs) and DAPI (Life Tech). Thymocytes were stained with antibodies against TCRβ (H57–597), CD4 (RM4–5), CD8 (53-6.7), CD25 (PC61) and CD69 (H1.2F3). Spleen and lymph nodes were isolated, minced and stained with antibodies against TCRβ, CD4, CD8, CD25, CD44 (IM7) and CD62L (MEL-14). Intracellular staining for Aire (5H12) and Foxp3 (FJK-16s) were performed using the eBiosciences Foxp3 staining kit. All antibody stainings were preceded by FcγR block (2.4G2). Cell sorting was performed on FACS Aria II (BD), data collected using LSR II flow cytometer (BD) and analyzed using FACS Diva (BD) and FlowJo (Tree Star).
Gene Expression Profiling
mTECs were sorted into Trizol LS (Life Technologies), RNA was extracted according to manufacturer’s instructions, then purified using the RNeasy Micro Kit (Qiagen). Purified RNA was processed, amplified, labeled and hybridized to Affymetrix GeneChip MoGene 2.0 ST microarrays as previously described56. Expression signals were normalized using the Signal Space Transformation-Robust Multi-Chip Average (SST-RMA) algorithm on the Affymetrix Expression Console software. Normalized signals were analyzed using the Transcriptome Analysis Console software (Affymetrix). Tissue restriction for each gene was determined using assignments from the previously reported dynamic step method17. Briefly, expression values for 64 non-thymic physiological samples from the GNF Mouse GeneAtlas V357,58 were calculated and hierarchically clustered into 35 groups. Guided by thresholds typically chosen for microarray data analysis, we define tissue restricted probes as those with a minimum normalized expression value of 50 that showed a moderated exponential step-up in expression, such that expression was substantially higher in 1–5 tissue groups than in the 6th highest tissue group. Only genes with unanimously tissue-restricted probe sets were designated as tissue-restricted.
ATAC-seq sample preparation
Sixty thousand cells were sorted into a V-bottom 2ml tube (E&K Scientific) and 1 ml of RSB buffer without detergent (10mM Tris 7.4, 10mM NaCl, 3mM MgCl2) was added prior to pelleting by a swinging bucket centrifuge. Cells were resuspended in 200ul RSB buffer with 0.1% Tween-20 (Sigma-Aldrich) and incubated on ice for 5 min. Cells were pelleted, resuspended in 50ul TD buffer with 2.5ul Tn5 transposase (Illumina) and incubated at 37° C. for 30 min. Transposition reactions were cleaned up with MinElute columns (Qiagen) and libraries were constructed as previously described59. Libraries were sequenced using paired-end, dual-index sequencing on an Illumina HiSeq2000 instrument with 51 × 8 × 8 × 51 cycle reads.
ATAC-seq data analysis
ATAC-seq data were processed as previously described23,60. Reads were trimmed with a custom script23,60, aligned using Bowtie2 and filtered for unique reads with alignment quality > q30. Reads mapping to mitochondrial, ChrY and unmapped contigs were removed. Peaks were called using MACS2 and filtered with a custom blacklist23. Peak summits were extended +/− 250-bp and filtered for non-overlapping, maximally significant 500-bp peaks (n = 250,313). ATAC-seq fragment counts for each sample were calculated across all 250,313 peaks, quantile normalized and GC-content normalized as previously described23,60. The nearest gene to each peak was annotated using HOMER. PCA was performed on normalized data using the MAVRIC R package (Moskowitz and Greenleaf, 2017, manuscript submitted) for the top 100,000 peaks with highest variance (ntop = 100000, alpha =0.01, effsize = 0.05, corMethod = “pearson”). Peaks with variance significantly correlated to a given PC (α < 0.05) were identified using MAVRIC [dimdesc($pcaobj)]. Hierarchical clustering was performed using Pearson correlation as the distance metric on normalized data. Normalized bedGraph files were created for each sample using bedtools genomecov to visualize insertion tracks. The sum of reads within peaks associated with “housekeeping” genes (median microarray signal > 75, coefficient of variation < 0.15, fold-change WT mTEChi vs. mTEClo > 0.9 and < 1.1) was used as the scaling factor for each sample as previously described61. Transcription factor (TF) deviation scores were calculated as described previously23,60. TF footprinting was assessed by plotting the normalized distribution of the 5’ ends of fragments (replicates pooled) spanning a 300bp window relative to the motif center. Similarly, the normalized distribution of fragment dyads (midpoint) were plotted spanning a 4 kb window relative to indicated peak center or transcription start-site (TSS).
ChIP-seq data analysis
Fastq files for Aire, Pol II, Top1, Top2a, γ-H2AX and H3K27ac ChIP-seq18 from mTEChi; Lsd1, CoREST, Hdac262, Suz1263, Ring1b64, LaminB65 in ES cells were aligned to the mouse genome (mm9) with Bowtie using default settings except filtering reads with multiple alignments (command line parameter –m 1). Bam files were processed by removing duplicates, unaligned reads and those aligned to ChrM. Fragment sizes were estimated and reads were extended accordingly for each replicate. MACS2 was used to call peaks using -callpeak function with IgG ChIP as controls. Signal tracks were generated using bedtools genomecov and normalized by sequence depth. These signal track were cross-validated for significance using MACS2 with commands bdgcmp –m logLR –p 0.00001 to generate log10-likelihood bedGraph files. Enriched TF motifs for peak sets were identified using HOMER (findMotifsGenome.pl –size given –mask).
Chromatin in vivo Assay (CiA)
Mouse embryonic fibroblasts (MEFs) were acquired from mice with the CiA:Oct4 allele at embryonic day 14.5 as previously described33. MEFs were transformed with simian virus 40 large T antigen and single cell sorted after transfection with LGmCreER (self-deleting) plasmid53 (Addgene #33340) to enrich for cells with excised neo cassette. Clones were screened for growth rate, VP16-mediated eGFP activation and DNase accessibility at the CiA:Oct4 locus. A single clone was used for all CiA recruitment experiments. Embryonic stem cells (ESCs) from mice with CiA:Oct4 allele were established as previously described33. Murine Aire with C-terminal Frb tandem repeat (Aire-Frb2x- V5) or Gal4-binding domain (Aire-Gal4BD-V5) were cloned into a previously described lentiviral backbone33 for rapamycin-induced- or constitutive recruitment, respectively. BAF was recruited via the Ss18 subunit, using previously described constructs (Frb2x-V5-Ss18, ZFHD1-Ss18)34,35. CiA anchor (ZFHD1-FKBP12) and VP16 (Gal4BD-VP16) constructs were described previously33. Lentivirus was generated as previously described66. MEFs and ESCs were infected and selected with puromycin (2ug/ml), blasticidin (10ug/ml) and/or hygromycin (200ug/ml). For experiments using chemical-induced proximity (CIP), rapamycin (Selleckchem) was added at 3nM. Media containing rapamycin (Rap) was changed daily for experiments spanning longer than 24h. For early time points (< 20 min), Rap at 12nM was added to media as well as to the fresh trypsin used to dissociate cells.
CRISPR-Cas9 guided recruitment
Catalytic-dead Cas9 (dCas9) mediated recruitment was performed as previously described36. Three small guide RNAs (sgRNAs) targeting a DNase hypersensitivity site (DHS) 200 bp upstream of the PSMB11 TSS were selected based on scores for off-target matches as previously described67. These sgRNAs were cloned into the sgRNA(MS2)_zeo lentiviral backbone (Addgene #61427) containing MS2-specific hairpin aptamers. Murine Aire with C-terminal MS2 (Aire-MS2-V5) was cloned into a previously described lentiviral backbone33. 4D6 human thymic epithelial cell line was sequentially transduced with lentiviruses encoding the sgRNAs and dCas9 or the sgRNAs, dCas9 and Aire-MS2.
Chromatin Immunoprecipitation (ChIP)
ChIPs were essentially performed as previously described34,35,47. Cells were dissociated, washed and 30 million cells were formaldehyde crosslinked (1%) at 37° C. for 12 min. For experiments using CIP (< 20 min), Rap at 3nM was added to media quenching trypsin, PBS wash and fix buffer. Nuclei were sonicated using Covaris E220 ultrasonicator at 5% duty cycle, 4 intensity, 140W PIP and 200 cycles per burst for 13 min. Insoluble chromatin was pelleted, supernatant diluted 1:1 in 2× ChIP buffer (100 mM HEPES pH 7.5, 600mM NaCl, 2 mM EDTA pH 8.0, 2% Triton X-100, 0.2% sodium deoxycholate, 0.1% SDS) and divided into four ChIP reactions (4–5 ug antibody/reaction, α-V5, clone R960-25, Life Technologies) which incubated overnight at 4° C. 25 ul of Protein G Dynabeads (Thermo Fisher) slurry washed in ChIP buffer were added to ChIP reactions and rotated at 4° C. for 1h. Beads were washed 3× in ChIP buffer, 1× in DOC buffer and 1× in TE before consecutive elutions (2×) in 150 ul of 0.1M NaHCO3, 1% SDS. Eluates were subjected to RNase A and proteinase K before reverse cross-linking at 65° C. overnight. ChIP DNA was precipitated using phenol/chloroform extraction and reconstituted in TE buffer for qPCR reactions. Amplicon detection for each target region was normalized to that at respective control regions for all samples. Enrichment was calculated to be the fold-change between the normalized bound/input values of Rap- treated vs. ethanol treated samples. Primers used for ChIP studies at CiA:Oct4 locus are summarized in Supplementary Table 133,34. V5 ChIP at CiA:Oct4 locus was normalized to that at the housekeeping Rps29 promoter (c).
DNase Accessibility Assay
DNase I sensitivity assays were performed as previously described68. Cells were lysed in hypotonic Buffer A, washed in Buffer A and 5 million nuclei were pelleted for each DNase I reaction. For experiments using CIP (< 20 min), Rap at 3nM was added to media quenching trypsin, PBS wash and Buffer A. Nuclei were subjected to varying concentrations of DNase I (Sigma) for 3 min at 37° C. Digestions were terminated with Stop buffer and subjected to proteinase K for 1h at 55° C. Samples were treated with RNase A and DNA was purified over MinElute columns (Qiagen) for qPCR reactions. Amplicon detection at CiA:Oct4 locus (Supplementary Table 1) for all DNase I conditions was normalized to that at a DNase I-insensitive region at the Rho locus (Supplementary Table 1). Amplicon detection at promoter of PSMB11 locus for all DNase I conditions was normalized to that at DNase I-insensitive region at the RHO locus (Supplementary Table 1).
Histopathology and Immunohistochemistry
Histopathology was performed as previously described40. Briefly, tissues were fixed in buffered 10% formalin and paraffin-embedded. Hematoxylin and eosin stainings were performed using standard methods. Immunohistochemistry was performed on 4-um sections with the ABC Vectastain kits (Vector Laboratories) using α-CD3 antibody (DakoCytomation, cat#A045229), and developed with DAB.
Thymus Transplants
Kidney capsule thymus transplants were performed as previously described69. Briefly, thymi from newborn Brg1cKO and WT littermates were cultured in 1.35 mM 2-deoxyguanosine for 7 days to deplete hematopoietic compartments. Thymic stroma were washed and transplanted under the kidney capsule of 6–8 week-old female nude mice. Thymopoiesis was monitored via cytofluorimetric analysis of blood at 5 and 10 weeks post-transplantation. Animals were examined 13 – 15 weeks after transplantation for T cell reconstitution and peripheral organs were collected for histopathology.
Statistics
Fisher’s exact tests for over-representation in fold-change vs. fold-change plots were one-sided tests with 95% confidence intervals for the lower bound of odd-ratios (Fig. 2c, 5b). For cumulative distribution function plots, Mann-Whitney-U tests were used for significance between comparisons with 95% confidence intervals at medians of pairwise comparisons (Fig. 1c,3b,5a and Supplementary Fig. 2d,4). Correlations between different peak sets were performed using Pearson correlation test (Fig. 1e, 2a). Student t-tests were performed as two-tailed tests with n specified for independent experiments in individual figure legends (Fig. 5,6,7, and Supplementary 6).
Data and Code Availability
ATAC-seq and microarray datasets reported in this manuscript, including raw reads and fully processed count matrices can be accessed in GEO with accession codes GSE102526 and GSE102525, respectively. Publicly available ChIP-seq and ATAC-seq datasets referenced in this study are as follows: GSE92597 (ChIP-seq) for Aire, Pol II, Top1, Top2a, γ-H2AX, H3K27ac in mTECs; GSE39513 (ChIP-seq) for Suz12 in ES cells; GSE42466 (ChIP-seq) for Ring1b in ES cells; GSE28247 (ChIP-seq) for LaminB in ES cells; GSE27841 (ChIP-seq) for Lsd1, CoREST, Hdac2 in ES cells; GSE94041 (ATAC-seq) for ES cells. Any custom code will be made available upon request.
Supplementary Material
Acknowledgments
We are grateful to N. Manley for Foxn1ex9cre mice, N. Hathaway, C. Kadoch, S. Braun and E. Chory for CiA constructs, M. Toribio and D. Mathis for 4D6 TEC line, D. Mathis, M. Anderson and S. Denny for insightful comments, Y. Chien, C. Weber, J. Kirkland, L. Wagar and J. Ronan for critical reading of the manuscript, and J. Gardner, P. Chu and R. Li for technical assistance. We thank the Stanford Shared FACS facility and S. Kim for flow cytometry and cell sorting. Stanford BioX3 cluster was used for computational analyses (NIH S10 grant 1S10RR02664701). This work was supported by the Howard Hughes Medical Institute (to G.R.C.) and NIH grants CA163915 and NS046789 (to G.R.C.), P50-HG007735 (to H.Y.C. and W.J.G.), T32HG000044 (to J.D.B.), T32 GM007790 (to E.L.M.). A.S.K. was supported by the Lymphoma and Leukemia Society and D.M.M. was supported by Stanford Biomedical Informatics Training Grant from the National Library of Medicine (LM-07033).
Footnotes
Author Contributions: A.S.K. and G.R.C. conceived of the study and wrote the paper. A.S.K. planned and performed all experiments and data analysis. A.S.K., E.L.M., J.D.B. and D.M.M. performed ATAC-seq data analysis. J.W. performed kidney capsule transplants. W.J.G. and H.Y.C. provided conceptual insights and advised on data analysis and experimental design.
Competing Financial Interests: W.J.G. and H.Y.C. declare competing interests as co-founders of Epinomics, Inc. Stanford University has filed a patent on ATAC-seq, on which W.J.G. and H.Y.C. are inventors.
References
- 1.Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see) Nat Rev Immunol. 2014;14:377–391. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahama Y, Ohigashi I, Baik S, Anderson G. Generation of diversity in thymic epithelial cells. Nat Rev Immunol. 2017;17:295–305. doi: 10.1038/nri.2017.12. [DOI] [PubMed] [Google Scholar]
- 3.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 4.Bruserud O, Oftedal BE, Wolff AB, Husebye ES. AIRE-mutations and autoimmune disease. Curr Opin Immunol. 2016;43:8–15. doi: 10.1016/j.coi.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 6.Long HK, Prescott SL, Wysocka J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell. 2016;167:1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giraud M, et al. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci U S A. 2012;109:535–540. doi: 10.1073/pnas.1119351109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giraud M, et al. An RNAi screen for Aire cofactors reveals a role for Hnrnpl in polymerase release and Aire-activated ectopic transcription. Proc Natl Acad Sci U S A. 2014;111:1491–1496. doi: 10.1073/pnas.1323535111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida H, et al. Brd4 bridges the transcriptional regulators, Aire and P-TEFb, to promote elongation of peripheral-tissue antigen transcripts in thymic stromal cells. Proc Natl Acad Sci U S A. 2015;112:E4448–4457. doi: 10.1073/pnas.1512081112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meredith M, Zemmour D, Mathis D, Benoist C. Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat Immunol. 2015;16:942–949. doi: 10.1038/ni.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villasenor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc Natl Acad Sci U S A. 2008;105:15854–15859. doi: 10.1073/pnas.0808069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jolicoeur C, Hanahan D, Smith KM. T-cell tolerance toward a transgenic beta-cell antigen and transcription of endogenous pancreatic genes in thymus. Proc Natl Acad Sci U S A. 1994;91:6707–6711. doi: 10.1073/pnas.91.14.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derbinski J, Pinto S, Rosch S, Hexel K, Kyewski B. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl Acad Sci U S A. 2008;105:657–662. doi: 10.1073/pnas.0707486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinto S, et al. Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self-antigen diversity. Proc Natl Acad Sci U S A. 2013;110:E3497–3505. doi: 10.1073/pnas.1308311110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner A, Regev A, Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nature biotechnology. 2016;34:1145–1160. doi: 10.1038/nbt.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sansom SN, et al. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome research. 2014;24:1918–1931. doi: 10.1101/gr.171645.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bansal K, Yoshida H, Benoist C, Mathis D. The transcriptional regulator Aire binds to and activates super-enhancers. Nat Immunol. 2017 doi: 10.1038/ni.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennecke P, et al. Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat Immunol. 2015;16:933–941. doi: 10.1038/ni.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, et al. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc Natl Acad Sci U S A. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bottomley MJ, et al. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nature structural biology. 2001;8:626–633. doi: 10.1038/89675. [DOI] [PubMed] [Google Scholar]
- 22.Koh AS, et al. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc Natl Acad Sci U S A. 2008;105:15878–15883. doi: 10.1073/pnas.0808470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manley NR, Condie BG. Transcriptional regulation of thymus organogenesis and thymic epithelial cell differentiation. Progress in molecular biology and translational science. 2010;92:103–120. doi: 10.1016/s1877-1173(10)92005-x. [DOI] [PubMed] [Google Scholar]
- 25.Rossi SW, et al. RANK signals from CD4(+)3(−) inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J Exp Med. 2007;204:1267–1272. doi: 10.1084/jem.20062497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 27.Satoh R, et al. Requirement of Stat3 Signaling in the Postnatal Development of Thymic Medullary Epithelial Cells. PLoS genetics. 2016;12:e1005776. doi: 10.1371/journal.pgen.1005776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomada D, et al. Stat3 Signaling Promotes Survival And Maintenance Of Medullary Thymic Epithelial Cells. PLoS genetics. 2016;12:e1005777. doi: 10.1371/journal.pgen.1005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Natoli G. Control of NF-kappaB-dependent transcriptional responses by chromatin organization. Cold Spring Harbor perspectives in biology. 2009;1:a000224. doi: 10.1101/cshperspect.a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J, et al. Increased generation of Foxp3(+) regulatory T cells by manipulating antigen presentation in the thymus. Nature communications. 2016;7:10562. doi: 10.1038/ncomms10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillard GO, Dooley J, Erickson M, Peltonen L, Farr AG. Aire-dependent alterations in medullary thymic epithelium indicate a role for Aire in thymic epithelial differentiation. J Immunol. 2007;178:3007–3015. doi: 10.4049/jimmunol.178.5.3007. [DOI] [PubMed] [Google Scholar]
- 32.Asano M, Toda M, Sakaguchi N, Sakaguchi S. Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J Exp Med. 1996;184:387–396. doi: 10.1084/jem.184.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hathaway NA, et al. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadoch C, et al. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet. 2016 doi: 10.1038/ng.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller EL, et al. TOP2 synergizes with BAF chromatin remodeling for both resolution and formation of facultative heterochromatin. Nat Struct Mol Biol. 2017 doi: 10.1038/nsmb.3384. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandez E, et al. Establishment and characterization of cloned human thymic epithelial cell lines. Analysis of adhesion molecule expression and cytokine production. Blood. 1994;83:3245–3254. [PubMed] [Google Scholar]
- 38.Murata S, et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- 39.Org T, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO reports. 2008;9:370–376. doi: 10.1038/sj.embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koh AS, Kingston RE, Benoist C, Mathis D. Global relevance of Aire binding to hypomethylated lysine-4 of histone-3. Proc Natl Acad Sci U S A. 2010;107:13016–13021. doi: 10.1073/pnas.1004436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chakravarty S, Zeng L, Zhou MM. Structure and site-specific recognition of histone H3 by the PHD finger of human autoimmune regulator. Structure (London, England : 1993) 2009;17:670–679. doi: 10.1016/j.str.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su MA, et al. Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. The Journal of clinical investigation. 2008;118:1712–1726. doi: 10.1172/jci34523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pitkanen J, et al. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. The Journal of biological chemistry. 2000;275:16802–16809. doi: 10.1074/jbc.M908944199. [DOI] [PubMed] [Google Scholar]
- 44.Iglesias P, Diez JJ. Management of endocrine disease: a clinical update on tumor-induced hypoglycemia. European journal of endocrinology. 2014;170:R147–157. doi: 10.1530/eje-13-1012. [DOI] [PubMed] [Google Scholar]
- 45.Schneider R, Heverhagen AE, Moll R, Bartsch DK, Schlosser K. Differentiation between thyroidal and ectopic calcitonin secretion in patients with coincidental thyroid nodules and pancreatic tumors - a report of two cases. Experimental and clinical endocrinology & diabetes : official journal, German Society of Endocrinology [and] German Diabetes Association. 2010;118:520–523. doi: 10.1055/s-0029-1231083. [DOI] [PubMed] [Google Scholar]
- 46.Falanga A, Schieppati F, Russo D. Cancer Tissue Procoagulant Mechanisms and the Hypercoagulable State of Patients with Cancer. Seminars in thrombosis and hemostasis. 2015;41:756–764. doi: 10.1055/s-0035-1564040. [DOI] [PubMed] [Google Scholar]
- 47.Stanton BZ, et al. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet. 2016 doi: 10.1038/ng.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dykhuizen EC, et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIalpha. Nature. 2013;497:624–627. doi: 10.1038/nature12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trotter KW, King HA, Archer TK. Glucocorticoid Receptor Transcriptional Activation via the BRG1-Dependent Recruitment of TOP2beta and Ku70/86. Molecular and cellular biology. 2015;35:2799–2817. doi: 10.1128/mcb.00230-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abramson J, Giraud M, Benoist C, Mathis D. Aire's partners in the molecular control of immunological tolerance. Cell. 2010;140:123–135. doi: 10.1016/j.cell.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 51.Chi TH, et al. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19:169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- 52.Gordon J, et al. Specific expression of lacZ and cre recombinase in fetal thymic epithelial cells by multiplex gene targeting at the Foxn1 locus. BMC developmental biology. 2007;7:69. doi: 10.1186/1471-213x-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Z, et al. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes & development. 2010;24:1389–1402. doi: 10.1101/gad.1940710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gardner JM, et al. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science. 2008;321:843–847. doi: 10.1126/science.1159407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jain R, Gray DH. Isolation of thymic epithelial cells and analysis by flow cytometry. Current protocols in immunology. 2014;107:3.26.21–15. doi: 10.1002/0471142735.im0326s107. [DOI] [PubMed] [Google Scholar]
- 56.Cipolletta D, et al. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lattin JE, et al. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome research. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Current protocols in molecular biology. 2015;109:21.29.21–29. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Denny SK, et al. Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell. 2016;166:328–342. doi: 10.1016/j.cell.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whyte WA, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jia J, et al. Regulation of pluripotency and self-renewal of ESCs through epigenetic-threshold modulation and mRNA pruning. Cell. 2012;151:576–589. doi: 10.1016/j.cell.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morey L, Aloia L, Cozzuto L, Benitah SA, Di Croce L. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell Rep. 2013;3:60–69. doi: 10.1016/j.celrep.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 65.Handoko L, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nature protocols. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 67.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.John S, et al. Genome-scale mapping of DNase I hypersensitivity. Current protocols in molecular biology. 2013 doi: 10.1002/0471142727.mb2127s103. Chapter 27, Unit 21.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson MS, et al. The cellular mechanism of Aire control of T cell tolerance. Immunity. 2005;23:227–239. doi: 10.1016/j.immuni.2005.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ATAC-seq and microarray datasets reported in this manuscript, including raw reads and fully processed count matrices can be accessed in GEO with accession codes GSE102526 and GSE102525, respectively. Publicly available ChIP-seq and ATAC-seq datasets referenced in this study are as follows: GSE92597 (ChIP-seq) for Aire, Pol II, Top1, Top2a, γ-H2AX, H3K27ac in mTECs; GSE39513 (ChIP-seq) for Suz12 in ES cells; GSE42466 (ChIP-seq) for Ring1b in ES cells; GSE28247 (ChIP-seq) for LaminB in ES cells; GSE27841 (ChIP-seq) for Lsd1, CoREST, Hdac2 in ES cells; GSE94041 (ATAC-seq) for ES cells. Any custom code will be made available upon request.