Abstract
Head and neck squamous cell carcinoma (HNSCC) is treated with cisplatin (CDDP) and radiotherapy (RT), and distinct results are observed among patients with similar clinicopathological aspects. This prospective study aimed to investigate whether MLH1 c.-93G>A (rs1800734), MSH2 c.211+9C>G (rs2303426), MSH3 c.3133G>A (rs26279), EXO1 c.1765G>A (rs1047840), and EXO1 c.2270C>T (rs9350) single nucleotide polymorphisms (SNPs) of the mismatch repair (MMR) pathway change side effects and response rate of 90 HNSCC patients treated with CDDP and RT. DNA from peripheral blood was analyzed by PCR-based methods to obtain genotypes. It was observed 4.27-fold and 4.69-fold increased risks of presenting pronounced nephrotoxicity with treatment in patients with MSH3 GG and EXO1 rs9350 CC genotypes compared with patients with GA or AA and CT or TT genotypes, respectively. MSH3 GG or GA and GT haplotype of EXO1 rs1047840 and rs9350 SNPs conferred to patients 10.29 and 4.00 more chances of presenting pronounced ototoxicity after treatment than MSH3 AA genotype and other EXO1 haplotypes, respectively. Patients with EXO1 rs1047840 GA or AA genotype and AC haplotype of EXO1 rs1047840 and rs9350 SNPs had both 9.55-fold increased risks of achieving partial response or stable disease instead of complete remission after treatment than patients with EXO1 GG genotype and other EXO1 haplotypes, respectively. For the first time, our data show preliminary indication that inherited alterations of DNA MMR pathway, related to MSH3 rs26279, EXO1 rs1047840 and EXO1 rs9350 SNPs, modify toxicity and response to chemoradiation in HNSCC, and may contribute to future personalized treatment of patients.
Keywords: head and neck squamous cell carcinoma, mismatch repair pathway, single nucleotide polymorphism, outcome
INTRODUCTION
The treatment of advanced unresectable head and neck squamous cell carcinoma (HNSCC) has been made with association between cisplatin (CDDP) and radiotherapy (RT) [1]. CDDP binds to DNA, forming adducts, and it also favors accumulation of intracellular free radicals [2]. RT induces lesion to DNA, direct and indirectly, by activity of photons and free radical generation, respectively [3]. DNA damages induced by CDDP and RT trigger apoptosis when not properly repaired by DNA repair pathways, such as the mismatch repair (MMR) [4, 5]. MutL homolog 1 (MLH1), MutS homolog 2 and 3 (MSH2 and MSH3) and exonuclease 1 (EXO1) proteins are crucial to identify CDDP-induced DNA lesion and enable its removal [4–7].
Variations in toxicity and response to therapy and/or in survival were seen in patients with lung [8–11], pancreatic [12], breast [13, 14], laryngeal [15], cervical [16], and ovarian [17] cancer treated with CDDP-based schemes and/or RT, which were attributed to abnormalities in production or function of proteins encoded by single nucleotide polymorphisms (SNPs) in genes of MMR pathway. In fact, variant “A”, “G”, “A”, and “A” alleles of MLH1 c.-93G>A (rs1800734) [18], MSH2 c.211+9C>G (rs2303426) [19], MSH3 c.3133G>A (rs26279) [20], and EXO1 c.1765G>A (rs1047840) [21], reduce levels of expressed protein compared with respective wild-type alleles, and have reduced DNA repair as consequence. It was also observed association of wild-type “C” allele of EXO1 c.2270C>T (rs9350) with decreased DNA repair, possibly due to its influence in preventing EXO1 involvement in the complex with MMR proteins [21].
Recently, we retrospectively analyzed MLH1 rs1800734, MSH2 rs2303426, MSH3 rs26279, EXO1 rs1047840, and EXO1 rs9350 SNPs in HNSCC subjects that received concurrent chemoradiotherapy with CDDP/carboplatin as neoadjuvant, adjuvant or definitive treatment, and observed that patients with wild-type “GG” genotypes of MSH3 rs26279 and EXO1 rs1047840 presented shorter relapse-free survival (RFS) and overall survival (OS) compared to others; however, side effects and response to therapy were not evaluated in study [20]. We investigated in this new prospective study the roles of the above mentioned SNPs in modulation of toxicity and response to therapy of HNSCC patients treated exclusively with CDDP chemoradiation.
RESULTS
HNSCC patients
The clinicopathological aspects of 90 patients enrolled in study are presented in Table 1.
Table 1. Clinical and tumor aspects of 90 patients with head and neck squamous cell carcinoma.
| Variable | Median (range) or N (%) |
|---|---|
| Age (years) | 56 (27–74) |
| Gender | |
| Male | 83 (92.2) |
| Female | 7 (7.8) |
| Body mass index (kg/m2) | 19 (13–31) |
| Tobacco consumption | |
| Smokers | 88 (97.8) |
| Nonsmokers | 2 (2.2) |
| Alcohol consumption | |
| Drinkers | 83 (92.2) |
| Abstainers | 7 (7.8) |
| Tumor location | |
| Oral cavity | 12 (13.3) |
| Pharynx | 55 (61.1) |
| Larynx | 23 (25.6) |
| Histological grade* | |
| Well + moderately | 60 (82.2) |
| Poorly + undifferentiated | 13 (17.8) |
| Tumor stage | |
| I + II | 6 (6.7) |
| III + IV | 84 (93.3) |
| Human papillomavirus type 16* | |
| Positive | 0 (0.0) |
| Negative | 57 (100.0) |
N: number of patients. *The number of patients differed from the total quoted in the study (N = 90) because it was not possible to obtain consistent information about histological grade and human papillomavirus type 16 status in some cases.
Sixty-eight patients were treated with three CDDP administrations, and 22 patients were treated with only two CDDP injections, because they presented hematologic or renal consistent toxicities. Median cumulative amount of administrated CDDP was 265 mg (range: 100–616). Adherence to antiemetics was medium or high in most of the patients (97.7%).
Grade 2 or grade 3 nausea and grade 2 to grade 4 vomiting were seen in about two-thirds and one-third of cases, respectively. It was observed that one-third to half of cases presented grade 2 to grade 4 cytopenias, and half of cases had grade 2 to grade 5 nephrotoxicity or grade 2 to grade 4 ototoxicity. All available patients obtained complete response (CR) (N = 15), partial response (PR) (N = 53) or stable disease (SD) (N = 5) with treatment (Supplementary Table 1). The mean value (±SD) of CDDP found in urine was 237.0 ug/mL ± 116.2.
Cases were followed up for a median period of 21 months (range: 3.0–74). The assessed probabilities of 24-months event-free survival (EFS) and OS were 35.0% and 40.0%, respectively. In October 2017, 23 patients survived, 6 of them with disease and 17 without disease; and 67 patients died, 59 of them by tumor impacts and 8 by other causes.
Hardy-Weinberg equilibrium (HWE) was confirmed at MLH1 rs1800734 (χ2 = 2.56, P = 0.11), MSH2 rs2303426 (χ2 = 0.73, P = 0.39), MSH3 rs26279 (χ2 = 1.54, P = 0.21), EXO1 rs1047840 (χ2 = 0.42, P = 0.52), and rs9350 (χ2 = 2.80, P = 0.09) loci in patients’ samples. A linkage disequilibrium (LD) between EXO1 rs1047840 and EXO1 rs9350 (D’ = 19%) was found in our sample, and two EXO1 haplotypes (GT, AC) were included in further analyses due to possible clinical significance and frequency greater than 10%.
Genetic polymorphisms, side effects, response rate and prognosis
The genotypes of all SNPs and haplotypes of EXO1 rs1047840 and rs9350 in 90 studied patients stratified by toxicities and responses to therapy are presented in Table 2.
Table 2. MLH1 rs1800734, MSH2 rs2303426, MSH3 rs26279, EXO1 rs1047840 and EXO1 rs9350 single nucleotide polymorphism genotypes and EXO1 haplotypes in 90 patients with head and neck squamous cell carcinoma stratified by toxicity and response rate to concurrent chemoradiotherapy.
| Variable | Nephrotoxicity | Ototoxicity | Response rate | |||||
|---|---|---|---|---|---|---|---|---|
| G0 + G1 N (%) |
G2–G5 N (%) |
G0 + G1 N (%) |
G2–G4 N (%) |
CR + PR N (%) |
SD N (%) |
CR N (%) |
PR + SD N (%) |
|
| MLH1 rs1800734 | ||||||||
| GG + GA | 35 (52.2) | 32 (47.8) | 36 (52.2) | 33 (47.8) | 66 (93.0) | 5 (7.0) | 15 (21.1) | 56 (78.9) |
| AA | 1 (50.0) | 1 (50.0) | 0 (0.0) | 1 (100.0) | 2 (100.0) | 0 (0.0) | 0 (0.0) | 2 (100.0) |
| P-value | 0.99 | 0.99 | 0.99 | 0.99 | ||||
| OR (95% CI) | NE | NE | NE | NE | ||||
| GG | 20 (48.8) | 21 (51.2) | 20 (47.6) | 22 (52.4) | 42 (95.5) | 2 (4.5) | 9 (20.5) | 35 (79.5) |
| GA + AA | 16 (57.1) | 12 (42.9) | 16 (57.1) | 12 (42.9) | 26 (89.7) | 3 (10.3) | 6 (20.7) | 23 (79.3) |
| P-value | 0.29 | 0.46 | 0.10 | 0.47 | ||||
| OR (95% CI) | 1.76 (0.63–4.93) | 1.48 (0.53–4.13) | 0.13 (0.01–1.50) | 0.58 (0.13–2.55) | ||||
| MSH2 rs2303426 | ||||||||
| CC + CG | 29 (51.8) | 27 (48.2) | 27 (48.2) | 29 (51.8) | 54 (91.5) | 5 (8.5) | 10 (16.9) | 49 (83.1) |
| GG | 7 (53.8) | 6 (46.2) | 9 (64.3) | 5 (35.7) | 14 (100.0) | 0 (0.0) | 5 (35.7) | 9 (64.3) |
| P-value | 0.80 | 0.59 | 0.99 | 0.39 | ||||
| OR (95% CI) | 1.18 (0.33–4.24) | 1.43 (0.39–5.24) | NE | 2.01 (0.41–9.96) | ||||
| CC | 11 (61.1) | 7 (38.9) | 10 (55.6) | 8 (44.4) | 16 (94.1) | 1 (5.9) | 4 (23.5) | 13 (76.5) |
| CG + GG | 25 (49.0) | 26 (51.0) | 26 (50.0) | 26 (50.0) | 52 (92.9) | 4 (7.1) | 11 (19.6) | 45 (80.4) |
| P-value | 0.44 | 0.38 | 0.80 | 0.26 | ||||
| OR (95% CI) | 0.64 (0.21–1.98) | 0.59 (0.18–1.93) | 1.37 (0.12–15.71) | 0.42 (0.10–1.90) | ||||
| MSH3 rs26279 | ||||||||
| GG + GA | 31 (49.2) | 32 (50.8) | 31 (49.2) | 32 (50.8) | 65 (95.6) | 3 (4.4) | 14 (20.6) | 54 (79.4) |
| AA | 5 (83.3) | 1 (16.7) | 5 (71.4) | 2 (28.6) | 3 (60.0) | 2 (40.0) | 1 (20.0) | 4 (80.0) |
| P-value | 0.26 | 0.043 | 0.08 | 0.75 | ||||
| OR (95% CI) | 3.75 (0.38–37.03) | 10.29 (1.06–100.21) | 0.11 (0.01–1.31) | 1.49 (0.12–18.17) | ||||
| GG | 13 (35.1) | 24 (64.9) | 18 (46.2) | 21 (53.8) | 39 (95.1) | 2 (4.9) | 8 (19.5) | 33 (80.5) |
| GA + AA | 23 (71.9) | 9 (28.1) | 18 (58.1) | 13 (41.9) | 29 (90.6) | 3 (9.4) | 7 (21.9) | 25 (78.1) |
| P-value | 0.0071 | 0.13 | 0.61 | 0.56 | ||||
| OR (95% CI) | 4.27 (1.48–12.34) | 2.28 (0.79–6.58) | 0.60 (0.08–4.42) | 1.48 (0.39–5.59) | ||||
| EXO1 rs1047840 | ||||||||
| GG + GA | 34 (55.7) | 27 (44.3) | 33 (52.4) | 30 (47.6) | 60 (92.3) | 5 (7.7) | 14 (21.5) | 51 (78.5) |
| AA | 2 (25.0) | 6 (75.0) | 3 (42.9) | 4 (57.1) | 8 (100.0) | 0 (0.0) | 1 (12.5) | 7 (87.5) |
| P-value | 0.07 | 0.61 | 0.99 | 0.99 | ||||
| OR (95% CI) | 4.88 (0.86–27.55) | 0.64 (0.12–3.47) | NE | NE | ||||
| GG | 18 (56.3) | 14 (43.8) | 19 (61.3) | 12 (38.7) | 31 (93.9) | 2 (6.1) | 11 (33.3) | 22 (66.7) |
| GA + AA | 18 (48.6) | 19 (51.4) | 17 (43.6) | 22 (56.4) | 37 (92.5) | 3 (7.5) | 4 (10.0) | 36 (90.0) |
| P-value | 0.35 | 0.30 | 0.83 | 0.015 | ||||
| OR (95% CI) | 1.63 (0.59–4.51) | 0.58 (0.21–1.62) | 0.79 (0.10–6.56) | 9.55 (1.56–58.60) | ||||
| EXO1 rs9350 | ||||||||
| CC + CT | 36 (52.2) | 33 (47.8) | 36 (51.4) | 34 (48.6) | 68 (93.2) | 5 (6.8) | 15 (20.5) | 58 (79.5) |
| TT | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| P-value | 0.99 | 0.99 | 0.99 | 0.99 | ||||
| OR (95% CI) | NE | NE | NE | NE | ||||
| CC | 21 (42.0) | 29 (58.0) | 30 (57.7) | 22 (42.3) | 49 (94.2) | 3 (5.8) | 12 (23.1) | 40 (76.9) |
| CT | 15 (78.9) | 4 (21.1) | 6 (33.3) | 12 (66.7) | 19 (90.5) | 2 (9.5) | 3 (14.3) | 18 (85.7) |
| P-value | 0.022 | 0.06 | 0.60 | 0.27 | ||||
| OR (95% CI) | 4.69 (1.34–16.44) | 4.03 (1.17–13.93) | 0.59 (0.08–4.30) | 0.38 (0.07–2.15) | ||||
| EXO1 + EXO1 | ||||||||
| GT | 14 (87.5) | 2 (12.5) | 5 (31.3) | 11 (68.7) | 16 (88.9) | 2 (11.1) | 3 (16.7) | 15 (83.3) |
| Other haplotypes | 22 (41.5) | 31 (58.5) | 31 (57.4) | 23 (42.6) | 52 (94.5) | 3 (5.5) | 12 (21.8) | 43 (78.2) |
| P-value | 0.06 | 0.034 | 0.49 | 0.71 | ||||
| OR (95% CI) | 3.11 (1.85–44.75) | 4.00 (1.11–14.48) | 0.49 (0.07–3.72) | 0.71 (0.13–4.07) | ||||
| AC | 18 (48.6) | 19 (51.4) | 17 (43.6) | 22 (56.4) | 37 (92.5) | 3 (7.5) | 4 (10.0) | 36 (90.0) |
| Other haplotypes | 18 (56.3) | 14 (43.7) | 19 (61.3) | 12 (38.7) | 31 (93.9) | 2 (6.1) | 11 (33.3) | 22 (66.7) |
| P-value | 0.35 | 0.30 | 0.74 | 0.026 | ||||
| OR (95% CI) | 0.61 (0.22–1.70) | 0.58 (0.21–1.62) | 0.70 (0.08–5.90) | 9.55 (1.56–58.60) | ||||
G: grade of toxicity; N: number of patients; CR: complete response; PR: partial response; SD: stable disease; OR: odds ratio; CI: confidence interval; NE: not evaluated. ORs were adjusted by age, cumulative dose of cisplatin and body mass index in analyses of nephrotoxicity, by age, cumulative dose of cisplatin and tumor location in analyses of ototoxicity for age, cumulative dose of cisplatin, body mass index, tumor location and grade in analyses of response rate. The total number of patients differed from the total quoted in the study (N = 90) because it was not possible to obtain consistent information about response rate and toxicities in some cases; 1P bootstrap = 0.006, 2P bootstrap = 0.005, 3P bootstrap = 0.01, 4P bootstrap = 0.02, 5P bootstrap = 0.007, 6P bootstrap = 0.005.
Patients with MSH3 GG genotype had more frequently grade 2 to grade 5 nephrotoxicity than those with GA or AA genotype (64.9% vs. 28.1%); it was observed a 4.27-fold increased risk of substantial nephrotoxicity with treatment in patients with MSH3 GG genotype. The frequency of MSH3 GG or GA genotype was higher than AA in subjects with grade 2 to grade 4 ototoxicity (50.8% vs. 28.6%); after treatment, it was observed a 10.29-fold increased risk of substantial ototoxicity in patients with GG or GA genotype when compared to those with AA genotype.
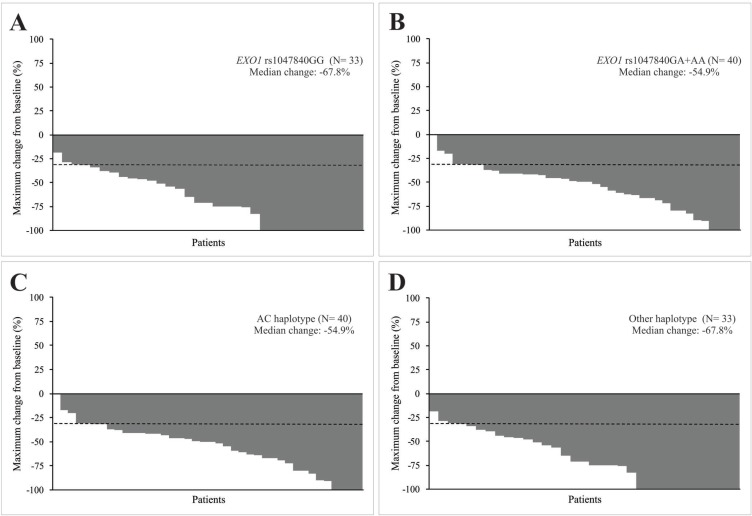
EXO1 rs1047840 GA or AA genotype was more common than GG genotype (90.0% vs. 66.7%) in patients with PR or SD; 9.55 more chances of presenting PR or SD instead of CR to chemoradiation were observed in patients with GA or AA genotype when compared to the ones with GG genotype. The median reduction in diameters of target lesions after chemoradiation was also less pronounced in patients with GA or AA genotype than in those with GG genotype (−54.9% vs. −67.8%) (Figure 1A, Figure 1B). EXO1 rs9350 CC genotype was more common than CT or TT genotype (58.0% vs. 21.1%) in patients with grade 2 to grade 5 nephrotoxicity; it was observed a 4.69-fold increased risk of substantial nephrotoxicity in subjects with CC genotype after treatment, when compared to those with the remaining genotypes. An excess of GT haplotype (wild-type and variant alleles of EXO1 rs1047840 and rs9350, respectively) was seen in subjects presenting grade 2 to grade 4 ototoxicity (68.7% vs. 42.6%) compared to those with other haplotypes; a 4.00-fold increased risk of substantial ototoxicity was observed in carriers of GT haplotype after chemoradiation, when compared to patients with other haplotypes. The AC haplotype (variant allele and wild-type allele of EXO1 rs1047840 and rs9350, respectively) was more frequent than other haplotypes in subjects who obtained PR or SD (90.0 vs. 66.7%); it was observed a 9.55-fold increased risk of achieving PR or SD instead of CR in carriers of AC haplotype after treatment, when compared to patients with other haplotypes. The median reduction in diameters of target lesions after chemoradiation was also less pronounced in patients with AC haplotype compared to others (−54.9% vs. −67.8%) (Figure 1C, Figure 1D).
Figure 1. Characteristics of response to concurrent cisplatin chemoradiotherapy of head and neck squamous cell carcinoma (HNSCC) patients.
Waterfall plots indicate the maximum change from baseline in the sum of reference diameters of target lesion in patients with EXO1 rs1047840 GG and GA or AA genotypes (A and B) an in patients with AC haplotype (variant allele of EXO1 rs1047840 and wild-type allele of EXO1 rs9350) and other haplotypes (C and D). The dashed lines indicate a 30% reduction in the tumor burden in the target lesion, as defined by Response Evaluation Criteria in Solid Tumors version 1.1.
No associations between SNPs and haplotypes were observed in subjects classified by nausea, vomiting, cytopenias and urinary CDDP (Supplementary Table 2).
At 24 months of follow-up, shorter EFS (32.7% vs. 83.3%, P = 0.01) and OS (35.7% vs. 100.0%, P = 0.009) were seen in patients with advanced tumors compared to those with localized tumors (Kaplan-Meier estimates). A shorter EFS and OS were observed in those patients in uni and multivariate Cox analyses. No associations of MLH1 rs1800734, MSH2 rs2303426, MSH3 rs26279, EXO1 rs1047840, EXO1 rs9350 SNPs, and EXO1 haplotypes were seen in survival analyses of HNSCC patients (Supplementary Table 3).
DISCUSSION
Clinical characteristics of patients, aspects of tumor, toxicity and response to chemoradiation, and survival rates found in the present study were similar to those previously described [1, 22, 23], indicating that our sample was satisfactory for investigations of new prognostic factors in HNSCC. None of our available patients had undergone HPV infection, as previously described in Brazilian HNSCC patients [24, 25]; this finding indicates that tobacco and alcohol were the most important factors related to tumor onset in our cases.
We found that MSH3 rs26279 GG and GG or GA genotypes were associated with pronounced nephrotoxicity and ototoxicity, respectively. MSH3 rs26279 altered radiosensitivity in breast cancer patients [13], response to platinum-based therapy and survival in lung cancer patients [9], response to CDDP chemoradiation and hematological toxicity in a unique case of laryngeal cancer [15] and survival in HNSCC patients treated by CDDP and RT [20], but its roles in nephrotoxicity and ototoxicity were not described in studies. We identified higher level of mRNA in MSH3 rs26279 GG individuals compared to those with GA or AA genotype in a previous study [20]. It was proposed that increased expression of MSH3 sequesters nuclear MSH2 and favors formation of MutSβ heterodimer, having a drastic change in MutSα and MutSβ proportion and reduction of efficiency in repairing base-base mismatches as consequence [26]. Thus, renal tubular cells and outer hair cells in carriers of “G” allele may be able to undergo to apoptosis in response to DNA damages due to CDDP and RT, with consequent nephrotoxicity and ototoxicity.
EXO1 rs1047840 GA or AA genotype and AC haplotype of EXO1 rs1047840 and rs9350 SNPs were associated with worst response to chemoradiation in this study. EXO1 rs1047840 GA genotype appeared to contribute to a CR and marked hematological toxicity seen in a patient with laryngeal cancer treated with CDDP and RT [15], and variant allele “A” of EXO1 rs1047840 was also associated with a better response rate in patients with cervical carcinoma [16]. Jin et al. (2008) [21] proposed that EXO1 rs1047840 variant “A” allele reduces the amount of mRNA of EXO1, and consequently decreases EXO1 protein levels and MMR activity, since it is fundamental for excision of DNA injuries initiated by CDDP, activating apoptosis [27, 28]. EXO1 rs9350 determines change of proline to leucine in codon 757 of EXO1 protein [29]. Proline tends to destabilize α-helices due to the lack of a backbone hydrogen bond and steric constraints [30], implying that the presence of this aminoacid could influence protein-protein interaction, especially with MSH2, resulting in incomplete repair of DNA lesions and apoptosis of damaged cells [29]. Thus, we expected to obtain a better response to therapy in carriers of variant “A” allele of EXO1 rs1047840 and wild-type “C” allele of rs9350 SNP, but the opposite result was found in this study. However, decrease of EXO1 expression in human fibroblasts or mouse embryonic fibroblasts caused a delay in DNA damage-induced apoptosis, and EXO1 may have a main role in caspase-3 cleavage, DNA fragmentation and cytochrome c release, participating in crucial phases of apoptosis; thus, SNPs in EXO1 may not only imply in DNA repair, but may also favor cell survival by apoptotic defects [31]. Since the roles of the mentioned SNPs in the treatment of HNSCC patients are still unknown, our data suggest that they are associated with a worst response to CDDP chemoradiation.
We observed that EXO1 rs9350 CC genotype was associated with pronounced nephrotoxicity and GT haplotype of EXO1 rs1047840 and rs9350 SNPs was associated with pronounced ototoxicity. Decrease in DNA repair was previously attributed to wild-type “C” allele of EXO1 rs9350, possibly due to its influence in protein-protein interaction, preventing EXO1 involvement in the complex with MMR proteins [21]. Thus, renal tubular cells in patients with CC genotype may undergo to apoptosis in response to DNA damages induced by CDDP and RT, with consequent nephrotoxicity. However, the association of wild-type “G” and variant “T” alleles of EXO1 rs1047840 and rs9350 SNPs, associated with increased DNA repair [21], with ototoxicity was not expected in this study; these apparent controversial findings could be attributed to different roles of EXO1 alleles, especially rs9350 “C”, which may be specific in distinct tissues/organs [32].
No association of analyzed genotypes and haplotypes with survival of 90 HNSCC was seen in this study. MSH3 rs26279 and EXO1 rs9350 altered survival of 180 lung cancer [9] and 602 lung cancer [10] patients treated with platinum-based chemotherapy. RFS and OS of 397 patients with HNSCC were also altered by MSH3 rs26279 and EXO1 rs1047840 SNPs in a previous study conducted by our group [20]. Differences of results could be related to different tumor types, sample sizes and median times of follow-up, which was about 2.0 times higher in our previous study than in the present one.
In conclusion, for the first time, this present study shows preliminary indication that inherited variations promoted by MSH3 rs26279, EXO1 rs1047840 and EXO1 rs9350, involved in DNA MMR pathway, may alter side effects and response CDDP and RT in HNSCC patients. We are aware that further analysis in a larger number of patients and functional analyses of relevant SNPs will be required to confirm results and elucidate their roles in disease. We believe that these results may contribute to the future personalized treatment of HNSCC patients, possibly with the use of varying CDDP doses and protective agents against CDDP-induced nephro- and ototoxicity [33, 34].
MATERIALS AND METHODS
Ninety HNSCC patients at diagnosis, seen at the University of Campinas from June 2011 to February 2014, were enrolled in this prospective study. Patients were chosen to CDDP chemoradiation as definitive treatment according to the following inclusion criteria: did not accepted surgical resolution facing expected anatomic or functional sequels, locoregional unresectable tumor, or a strategy of organ preservation. Declaration of Helsinki was conducted and the institutional Ethics Committee approved this study (n. 274/2011), and all patients provided written informed consent.
Patients were separated as smokers vs. nonsmokers and drinkers vs. abstainers [35]. HNSCC was diagnosed and staged based on conventional criteria [36, 37]. P16 immunohistochemistry and in situ hybridization were performed in tumor fragments, aiming to test the presence of human papillomavirus type 16 (HPV 16) [38, 39].
Patients were treated with a starting dose of 80–100 mg/m2 of “in bolus” intravenous injection of CDDP on 1st, 22th and 43th days concomitant with RT (70 Gy; 35 sessions); lower dose of CDDP (50–75 mg/m2) was delivered in second and/or third infusion in patients who presented toxicity with the first administration of the agent [1, 40]. As hydration and antiemetic protocols, they received intravenous saline solution, mannitol, ondansetron and dexamethasone before CDDP administration, as well as intravenous physiological saline and oral dexamethasone and metoclopramide during three days after each CDDP infusion [41, 42]. Antiemetic adherence was analyzed [43].
Nausea and vomiting were assessed immediately after each CDDP infusion and in the four following days. Cytopenias were evaluated with complete blood counts performed after each CDDP administration. Nephrotoxicity was analyzed using glomerular filtration rate measured by 51Cr-EDTA and ototoxicity was assessed by audiometric exams, both performed pre and post chemoradiation, respectively [44]. The worst grades of toxicities seen during the entire treatment were considered for analyses.
Response to therapy was categorized as CR, PR, SD or progressive disease (PD) [45]. Immediately after each CDDP administration, 48-hours urine collection was performed for estimation of CDDP by high-performance liquid chromatography assay [46]; the sum of cumulative measurement of urinary CDDP estimates acquired after each CDDP infusion was considered the final concentration.
Subjects with PR after treatment or tumor relapse and good clinical condition were selected for surgical tumor resection; palliative intravenous methotrexate was indicated to patients with unfavorable clinical performance [47].
Genotyping was performed by polymerase chain reaction (PCR) and enzymatic digestion or PCR real-time assay, using DNA from patients’ peripheral blood samples as reported for MLH1 rs1800734 [48], MSH2 rs2303426 [49], MSH3 rs26279 [20], EXO1 rs1047840 and rs9350 [27]. Total of 15% of genotypes were carried out by two independent experiments with total concordance.
For goodness-of-fit test was used chi-square (χ2) statistics to evaluate HWE. The Haploview 4.2 software was used to perform pairwise LD analyses of EXO1 haplotypes. To analyze differences between groups, χ2 or Fisher’s exact test were used. To obtain adjusted odds ratio (OR) values and to assess associations among SNPs, toxicity and response to treatment, and urinary CDDP, model of logistic regression was used. To ensure the stability of model was used the bootstrapping (N = 1,000) based on repeatedly random sampling, applying the bias-corrected and accelerated method [50].
EFS and OS were computed from the date of diagnosis to the first relapse, death from disease or last follow-up, and from the date of diagnosis until the death, resulting from any cause, or last follow-up, respectively. Kaplan-Meier method was used to analyze EFS and OS. Multivariate Cox regression was used to estimate hazard ratios (HRs) adjusted for possible discrepancies in clinical aspects (P-values ≤ 0.10 in univariate Cox regression).
All statistical tests were done using the SPSS 21.0 software (SPSS Incorporation, IL, USA), and two-sided significance was achieved when P-values were < 0.05.
SUPPLEMENTARY MATERIALS TABLES
Acknowledgments
We would like to thank all patients and their families, and all nurses of the Clinical Oncology Service.
Abbreviations
- 51Cr-EDTA
chromium-51 labeled ethylenediamine tetraacetic acid
- 95% CI
95% confidence interval
- CDDP
cisplatin
- CR
complete response
- CreCl
creatinine clearance
- EFS
event-free survival
- D’
disequilibrium coefficient
- DNA
deoxyribonucleic acid
- EXO1
exonuclease 1
- FAPESP
São Paulo Research Foundation
- G
grade of toxicity
- Gy
gray
- HNSCC
head and neck squamous cell carcinoma
- HPV 16
human papillomavirus type 16
- HR
hazard ratio
- HWE
Hardy-Weinberg equilibrium
- LD
linkage disequilibrium
- MDR
Dimensionality Reduction Model
- MLH1
MutL homolog 1
- MMR
mismatch repair
- MSH2
MutS homolog 2
- MSH3
MutS homolog 3
- N
number of patients
- NCI
National Cancer Institute
- NE
not evaluated
- OR
odds ratio
- OS
overall survival
- PCR
polymerase chain reaction
- PD
progressive disease
- PR
partial response
- RECIST 1.1
Response Evaluation Criteria in Solid Tumors 1.1
- RT
radiotherapy
- SD
stable disease
- SNP
single nucleotide polymorphism
- χ2
chi-square
Author contributions
GASN, EFDC, LLA, TRPL, MBV, ECP, PM, and CSPL made relevant collaboration to conception of work, treatment of patient, genotyping, and data interpretation. MBV, ECP, and PM performed the identification and control of side effects, and the urinary CDDP measurements. GJL made statistical analysis. LC and CTC performed and analyzed audiometric tests. FVM and AMAMA analyzed histological grade and human papillomavirus type 16 status. JMCA analyzed the images of computed tomography of neck of patients. CDR analyzed glomerular filtration rate measured by 51Cr-EDTA. GASN and CSPL drafted the manuscript. All authors approved the final manuscript.
CONFLICTS OF INTEREST
Nothing to report.
FUNDING
This study was supported by State of São Paulo Research Foundation (FAPESP) (grant numbers 2012/01807-2, 2012/01418-6).
REFERENCES
- 1.Adelstein DJ, Li Y, Adams GL, Wagner H, Jr, Kish JA, Ensley JF, Schuller DE, Forastiere AA. An intergroup phase III comparison of standard radiation therapy and two schedules of concurrent chemoradiotherapy in patients with unresectable squamous cell head and neck cancer. J Clin Oncol. 2003;1:92–98. doi: 10.1200/JCO.2003.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Deavall DG, Martin EA, Horner JM, Roberts R. Drug-induced oxidative stress and toxicity. J Toxicol. 2012;2012:645460. doi: 10.1155/2012/645460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lomax ME, Folkes LK, O’Neill P. Biological consequences of radiation-induced DNA damage: relevance to radiotherapy. Clin Oncol. 2013;25:578–585. doi: 10.1016/j.clon.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Mello JA, Acharya S, Fishel R, Essigmann JM. The mismatch-repair protein hMSH2 binds selectively to DNA adducts of the anticancer drug cisplatin. Chem Biol. 1996;3:579–589. doi: 10.1016/s1074-5521(96)90149-0. [DOI] [PubMed] [Google Scholar]
- 5.Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clin Cancer Res. 2008;14:1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 6.Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34:359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 7.Berwick M, Vineis P. Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J Natl Cancer Inst. 2000;92:874–897. doi: 10.1093/jnci/92.11.874. [DOI] [PubMed] [Google Scholar]
- 8.Matakidou A, el Galta R, Webb EL, Rudd MF, Bridle H, GELCAPS Consortium. Eisen T, Houlston RS. Genetic variation in the DNA repair genes is predictive of outcome in lung cancer. Hum Mol Genet. 2007;16:2333–2340. doi: 10.1093/hmg/ddm190. [DOI] [PubMed] [Google Scholar]
- 9.Xu XL, Yao YL, Xu WZ, Feng JG, Mao WM. Correlation of MSH3 polymorphisms with response and survival in advanced non-small cell lung cancer patients treated with first-line platinum-based chemotherapy. Genet Mol Res. 2015;14:3525–3533. doi: 10.4238/2015.April.15.16. [DOI] [PubMed] [Google Scholar]
- 10.Li R, Gu J, Heymach JV, Shu X, Zhao L, Han B, Ye Y, Roth J, Wu X. Hypoxia pathway genetic variants predict survival of non-small-cell lung cancer patients receiving platinum-based chemotherapy. Carcinogenesis. 2017;38:419–424. doi: 10.1093/carcin/bgx014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Y, Deng Z, Yin J, Wang S, Lu D, Wen X, Li X, Xiao D, Hu C, Chen X, Zhang W, Zhou H, Liu Z. The association of genetic variations in DNA repair pathways with severe toxicities in NSCLC patients undergoing platinum-based chemotherapy. Int J Cancer. 2017;141:2336–2347. doi: 10.1002/ijc.30921. [DOI] [PubMed] [Google Scholar]
- 12.Dong X, Jiao L, Li Y, Evans DB, Wang H, Hess KR, Abbruzzese JL, Li D. Significant associations of mismatch repair gene polymorphisms with clinical outcome of pancreatic cancer. J Clin Oncol. 2009;27:1592–1599. doi: 10.1200/JCO.2008.20.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangoni M, Bisanzi S, Carozzi F, Sani C, Biti G, Livi L, Barletta E, Costantini AS, Gorini G. Association between genetic polymorphisms in the XRCC1, XRCC3, XPD, GSTM1, GSTT1, MSH2, MLH1, MSH3, and MGMT genes and radiosensitivity in breast cancer patients. Int J Radiat Oncol Biol Phys. 2011;81:52–58. doi: 10.1016/j.ijrobp.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 14.De Langhe S, Mulliez T, Veldeman L, Remouchamps V, van Greveling A, Gilsoul M, De Schepper E, De Ruyck K, De Neve W, Thierens H. Factors modifying the risk for developing acute skin toxicity after whole-breast intensity modulated radiotherapy. BMC Cancer. 2014;14:711. doi: 10.1186/1471-2407-14-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopes-Aguiar L, Visacri MB, Nourani CM, Costa EF, Nogueira GA, Lima TR, Pincinato EC, Moriel P, Altemani JM, Lima CS. Do genetic polymorphisms modulate response rate and toxicity of cisplatin associated with radiotherapy in laryngeal squamous cell carcinoma: a case report. Medicine (Baltimore) 2015;94:e578. doi: 10.1097/MD.0000000000000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu JH, Xi P, Chai YL, Wang J, Wang T, Liu Z, Dai PG. Association of DNA repair gene polymorphisms with response to cisplatin-based concurrent chemoradiotherapy in patients with cervical carcinoma. DNA Repair (Amst) 2016;41:69–72. doi: 10.1016/j.dnarep.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 17.Shi T, Jiang R, Wang P, Xu Y, Yin S, Cheng X, Zang R. Significant association of the EXO1 rs851797 polymorphism with clinical outcome of ovarian cancer. Onco Targets Ther. 2017;10:4841–4851. doi: 10.2147/OTT.S141668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perera S, Mrkonjic M, Rawson JB, Bapat B. Functional effects of the MLH1-93G>A polymorphism on MLH1/EPM2AIP1 promoter activity. Oncol Rep. 2011;25:809–815. doi: 10.3892/or.2010.1129. [DOI] [PubMed] [Google Scholar]
- 19.Marra G, D’Atri S, Corti C, Bonmassar L, Cattaruzza MS, Schweizer P, Heinimann K, Bartosova Z, Nyström-Lahti M, Jiricny J. Tolerance of human MSH2 ± lymphoblastoid cells to the methylating agent temozolomide. Proc Natl Acad Sci U S A. 2001;98:7164–7169. doi: 10.1073/pnas.121136498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nogueira GA, Lourenço GJ, Oliveira CB, Marson FA, Lopes-Aguiar L, Costa EF, Lima TR, Liutti VT, Leal F, Santos VC, Rinck-Junior JA, Lima CS. Association between genetic polymorphisms in DNA mismatch repair-related genes with risk and prognosis of head and neck squamous cell carcinoma. Int J Cancer. 2015;137:810–818. doi: 10.1002/ijc.29435. [DOI] [PubMed] [Google Scholar]
- 21.Jin G, Wang H, Hu Z, Liu H, Sun W, Ma H, Chen D, Miao R, Tian T, Jin L, Wei Q, Huang W, Lu D, et al. Potentially functional polymorphisms of EXO1 and risk of lung cancer in a Chinese population: a case-control analysis. Lung Cancer. 2008;60:340–346. doi: 10.1016/j.lungcan.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 22.de Castro G, Jr, Snitcovsky IM, Gebrim EM, Leitão GM, Nadalin W, Ferraz AR, Federico MH. High-dose cisplatin concurrent to conventionally delivered radiotherapy is associated with unacceptable toxicity in unresectable, non-metastatic stage IV head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2007;264:1475–1482. doi: 10.1007/s00405-007-0395-9. [DOI] [PubMed] [Google Scholar]
- 23.Martins RG, Parvathaneni U, Bauman JE, Sharma AK, Raez LE, Papagikos MA, Yunus F, Kurland BF, Eaton KD, Liao JJ, Mendez E, Futran N, Wang DX, et al. Cisplatin and radiotherapy with or without erlotinib in locally advanced squamous cell carcinoma of the head and neck: a randomized phase II trial. J Clin Oncol. 2013;31:1415–1421. doi: 10.1200/JCO.2012.46.3299. [DOI] [PubMed] [Google Scholar]
- 24.Ribeiro KB, Levi JE, Pawlita M, Koifman S, Matos E, Eluf-Neto J, Wunsch-Filho V, Curado MP, Shangina O, Zaridze D, Szeszenia-Dabrowska N, Lissowska J, Daudt A, et al. Low human papillomavirus prevalence in head and neck cancer: results from two large case-control studies in high-incidence regions. Int J Epidemiol. 2011;40:489–502. doi: 10.1093/ije/dyq249. [DOI] [PubMed] [Google Scholar]
- 25.López RV, Levi JE, Eluf-Neto J, Koifman RJ, Koifman S, Curado MP, Michaluart-Junior P, Figueiredo DL, Saggioro FP, de Carvalho MB, Kowalski LP, Abrahão M, de Góis-Filho F, et al. Human papillomavirus (HPV) 16 and the prognosis of head and neck cancer in a geographical region with a low prevalence of HPV infection. Cancer Causes Control. 2014;25:461–471. doi: 10.1007/s10552-014-0348-8. [DOI] [PubMed] [Google Scholar]
- 26.Drummond JT, Genschel J, Wolf E, Modrich P. DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSalpha/hMutSbeta ratio and reduces the efficiency of base-base mismatch repair. Proc Natl Acad Sci U S A. 1997;94:10144–10149. doi: 10.1073/pnas.94.19.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai MH, Tseng HC, Liu CS, Chang CL, Tsai CW, Tsou YA, Wang RF, Lin CC, Wang HC, Chiu CF, Bau DT. Interaction of Exo1 genotypes and smoking habit in oral cancer in Taiwan. Oral Oncol. 2009;45:e90–94. doi: 10.1016/j.oraloncology.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Jagmohan-Changur S, Poikonen T, Vilkki S, Launonen V, Wikman F, Orntoft TF, Møller P, Vasen H, Tops C, Kolodner RD, Mecklin JP, Järvinen H, Bevan S, et al. EXO1 variants occur commonly in normal population: evidence against a role in hereditary nonpolyposis colorectal cancer. Cancer Res. 2003;63:154–158. [PubMed] [Google Scholar]
- 29.Yamamoto H, Hanafusa H, Ouchida M, Yano M, Suzuki H, Murakami M, Aoe M, Shimizu N, Nakachi K, Shimizu K. Single nucleotide polymorphisms in the EXO1 gene and risk of colorectal cancer in a Japanese population. Carcinogenesis. 2005;26:411–416. doi: 10.1093/carcin/bgh335. [DOI] [PubMed] [Google Scholar]
- 30.Chang DK, Cheng SF, Trivedi VD, Lin KL. Proline affects oligomerization of a coiled coil by inducing a kink in a long helix. J Struct Biol. 1999;128:270–279. doi: 10.1006/jsbi.1999.4182. [DOI] [PubMed] [Google Scholar]
- 31.Bolderson E, Richard DJ, Edelmann W, Khanna KK. Involvement of Exo1b in DNA damage-induced apoptosis. Nucleic Acids Res. 2009;37:3452–3463. doi: 10.1093/nar/gkp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Y, Hayes RB, Huang WY, Caporaso NE, Burdette L, Yeager M, Chanock SJ, Berndt SI. DNA repair gene polymorphisms and tobacco smoking in the risk for colorectal adenomas. Carcinogenesis. 2011;32:882–887. doi: 10.1093/carcin/bgr071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bulacio RP, Anzai N, Ouchi M, Torres AM. Organic anion transporter 5 (Oat5) urinary excretion is a specific biomarker of kidney injury: evaluation of urinary excretion of exosomal Oat5 after N-acetylcysteine prevention of cisplatin induced nephrotoxicity. Chem Res Toxicol. 2015;28:1595–1602. doi: 10.1021/acs.chemrestox.5b00176. [DOI] [PubMed] [Google Scholar]
- 34.Doğan M, Polat H, Yaşar M, Kaya A, Bayram A, Şenel F, Özcan İ. Protective role of misoprostol against cisplatin-induced ototoxicity. Eur Arch Otorhinolaryngol. 2016;273:3685–3692. doi: 10.1007/s00405-016-4031-4. [DOI] [PubMed] [Google Scholar]
- 35.Hayes RB, Bravo-Otero E, Kleinman DV, Brown LM, Fraumeni JF, Jr, Harty LC, Winn DM. Tobacco and alcohol use and oral cancer in Puerto Rico. Cancer Causes Control. 1999;10:27–33. doi: 10.1023/a:1008876115797. [DOI] [PubMed] [Google Scholar]
- 36.Cardesa A, Gale N, Nadal A, Zidar N. Squamous cell carcinoma. In: Barnes L, Eveson JW, Reichart P, Sidransky D, editors. World Health Organization classification of tumours, pathology & genetics, head and neck tumours. 2nd edn. IARC Press; Lyon: 2005. pp. 118–121. [Google Scholar]
- 37.American Joint Committee on Cancer . Cancer Staging Handbook From the AJCC Cancer Staging Mannual. 7th ed. Springer; Philadelphia: 2010. pp. 21–97. [Google Scholar]
- 38.Begum S, Gillison ML, Ansari-Lari MA, Shah K, Westra WH. Detection of human papillomavirus in cervical lymph nodes: a highly effective strategy for localizing site of tumor origin. Clin Cancer Res. 2003;9:6469–6475. [PubMed] [Google Scholar]
- 39.Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer. 2010;116:2166–2173. doi: 10.1002/cncr.25033. [DOI] [PubMed] [Google Scholar]
- 40.Mendenhall WM, Werning JW, Pfister DG. Treatment of head and neck cancers. In: De Vita VT, Lawrence TS, Rosemberg SA, editors. Cancer: Principles & Practice of Oncology. 9 ed. Philadelphia: Lippincott Williams & Wilkins; 2011. pp. 729–780. [Google Scholar]
- 41.Cunningham D, Dicato M, Verweij J, Crombez R, de Mulder P, du Bois A, Stewart A, Smyth J, Selby P, van Straelen D, Parideans R, McQuade B, McRae J. Optimum anti-emetic therapy for cisplatin induced emesis over repeat courses: ondansetron plus dexamethasone compared with metoclopramide, dexamethasone plus lorazepam. Ann Oncol. 1996;7:277–282. doi: 10.1093/oxfordjournals.annonc.a010572. [DOI] [PubMed] [Google Scholar]
- 42.Kris MG, Tonato M, Bria E, Ballatori E, Espersen B, Herrstedt J, Rittenberg C, Einhorn LH, Grunberg S, Saito M, Morrow G, Hesketh P. Consensus recommendations for the prevention of vomiting and nausea following high-emetic-risk chemotherapy. Support Care Cancer. 2011;19:S25–32. doi: 10.1007/s00520-010-0976-9. [DOI] [PubMed] [Google Scholar]
- 43.Morisky DE, Green LW, Levine DM. Concurrent and predictive validity of a self-reported measure of medication adherence. Med Care. 1986;24:67–74. doi: 10.1097/00005650-198601000-00007. [DOI] [PubMed] [Google Scholar]
- 44.National Cancer Institute Cancer therapy evolution program. Common terminology criteria for adverse events (CTCAE) v 4.0 (2009). Available at: http://ctep.cancer.gov. Accessed August 28, 2016.
- 45.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 46.Lopez-Flores A, Jurado R, Garcia-Lopez P. A high-performance liquid chromatographic assay for determination of cisplatin in plasma, cancer, cell and tumor samples. J Pharmacol Toxicol Methods. 2005;52:366–372. doi: 10.1016/j.vascn.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Forastiere AA, Metch B, Schuller DE, Ensley JF, Hutchins LF, Triozzi P, Kish JA, McClure S, VonFeldt E, Williamson SK, Von Hoff DD. Randomized comparison of cisplatin plus fluorouracil and carboplatin plus fluorouracil versus methotrexate in advanced squamous cell carcinoma of the head and neck: a Southwest Oncology Group study. J Clin Oncol. 1992;10:1245–1251. doi: 10.1200/JCO.1992.10.8.1245. [DOI] [PubMed] [Google Scholar]
- 48.Park SH, Lee GY, Jeon HS, Lee SJ, Kim KM, Jang SS, Kim CH, Lee WK, Kam S, Park RW, Kim IS, Jung TH, Park JY. -93G>A polymorphism of hMLH1 and risk of primary lung cancer. Int J Cancer. 2004;112:678–682. doi: 10.1002/ijc.20359. [DOI] [PubMed] [Google Scholar]
- 49.Jung CY, Choi JE, Park JM, Chae MH, Kang HG, Kim KM, Lee SJ, Lee WK, Kam S, Cha SI, Kim CH, Han SB, Jung TH, et al. Polymorphisms in the hMSH2 gene and the risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:762–768. doi: 10.1158/1055-9965.EPI-05-0834. [DOI] [PubMed] [Google Scholar]
- 50.Austin PC, Steyerberg EW. Events per variable (EPV) and the relative performance of different strategies for estimating the out-of-sample validity of logistic regression models. Stat Methods Med Res. 2017;26:796–808. doi: 10.1177/0962280214558972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.