Abstract
Recent studies have revealed that relationships between plant pathogens and their vectors differ depending on species, strains and associated host plants. Turnip mosaic virus (TuMV) is one of the most important plant viruses worldwide and is transmitted by at least 89 aphid species in a non-persistent manner. TuMV is fundamentally divided into six phylogenetic groups; among which Asian-BR, basal-BR and world-B groups are known to occur in Japan. In Kyushu Japan, basal-BR has invaded approximately 2000 and immediately replaced the predominant world-B virus group. To clarify the relationships between TuMV and vector aphids, we examined the effects of the TuMV phylogenetic group on the population growth of aphid vectors in turnip plants. The population growth of a generalist aphid, Myzus persicae, was not significantly different between non-infected and TuMV-infected treatments. The population growth of a specialist aphid, Lipaphis erysimi, was higher in TuMV-infected plants than non-infected ones. Similar results were obtained in experiments using world-B and basal-BR groups of TuMV. Therefore, we conclude that L. erysimi is more mutualistic with TuMV than M. persicae, and differences in TuMV phylogenetic groups do not affect the growth of aphid vectors on turnip plants.
Introduction
Many plant pathogenic viruses expand their infection range with the aid of sucking insect vectors such as aphids, whiteflies and leafhoppers [1, 2]. Transmission by these insects is divided into persistent and non-persistent manners. In persistent viruses, vectors retain viruses inside their bodies for a relatively long time and continue the infective state through molting. Therefore, mutualistic relationships evolve easily between the virus and the vector [3]. In non-persistent viruses, vectors acquire and retain viruses only in their stylets for a relatively short period of time, and easily lose viruses after molting or feeding on an uninfected plant [4].
Recent studies have revealed that relationships between plant pathogens and their vectors differ depending on species, strains and associated host plants [5–10]. For example, a study [9] showed that tomato yellow leaf curl virus infecting tomato plants positively affected the growth performance of the vector, biotype Q of Bemisia tabaci, but negatively affected biotype B. This difference may be the cause for changes in dominant whitefly biotypes by Q in various localities in China. Although the majority of agricultural crop viruses causing severe economic damage are transmitted in a non-persistent manner especially by aphids [11], relationships between vectors and non-persistent viruses have seldom been investigated because of the extremely short contact time between the virus and the vector [3].
Turnip mosaic virus (TuMV) is one of the most important plant viruses worldwide and is transmitted by at least 89 aphid species in a non-persistent manner [12]. TuMV is fundamentally divided into six phylogenetic groups [13–14]. Of these groups, the Asian-BR, basal-BR and world-B groups are known to occur in Japan. In Japan, basal-BR invaded the Kyushu Region in approximately the year 2000 and immediately overcame the predominant world-B group, but the cause of this replacement remains unknown [15]. A previous study [16] reported that the reproductive performance of the generalist aphid vector Myzus persicae (Sulzer) increased in Nicotiana benthamiana Domin (Solanaceae) and Arabidopsis thaliana L. (Heyn.) (Brassicaceae) infected with TuMV. However, the effects of TuMV infection on the reproductive performance of vector aphids has not been investigated in brassicaceous crops, which suffer severe economic damages from TuMV in Japan [17–19]. In addition, these vegetables are infested by both generalist aphid vectors including M. persicae and specialist aphid vectors [20–21]. Therefore, clarification of interactions among brassicaceous crops, phylogenetic groups of TuMV and vector aphids is important for establishing management measures for these agricultural pests and pathogens.
The purpose of this study was to clarify whether the relationships between TuMV and vector aphids are different among phylogenetic groups of TuMV and vector aphid species. To assess these relationships, we surveyed the population growth of two aphid vectors, generalist aphid M. persicae and specialist aphid Lipaphis erysimi (Kaltenbach), on TuMV-infected and non-infected Japanese turnip plants using isolates of world-B and basal-BR phylogenetic groups of TuMV.
Materials and methods
Aphid populations
Colonies of M. persicae and L. erysimi were collected from Brassica napus L. in Saga City, Saga, Japan in 2014 (Saga strains: 33°14’12.2”N, 130°17’47.2”E). Because the Saga strain of L. erysimi expired during the course of this study, another colony of L. erysimi was collected from Japanese radish in Ogi City, Saga, Japan in 2014 (Ogi strain: 33°16’40.6”N, 130°13’06.6”E). Saga strains were acquired from Honjo Park and did not require specific permission to collect. Ogi strain was acquired from a private land and we obtained the owner’s permission to conduct the study on the site. Aphids were any protected or endangered species present in these fields. These colonies were continuously reared on turnip plants (Brassica rapa cv. “Hakatasuwari”) in an insect chamber at 25°C and 16L:8D photoperiodic conditions.
Virus isolates and inoculation of source plants
The TuMV isolates of C42J [22] from B. rapa and OGI11X2 from Raphanus sativus L. were used as representatives of world-B and basal-BR phylogenetic groups, respectively [23]. Both isolates were inoculated onto Chenopodium quinoa Wild. and serially cloned one time through single lesions. Each virus was propagated in B. rapa cv. Hakatasuwari or N. benthamiana plants. To prepare the virus source plants, each TuMV isolate was separately inoculated onto young seedlings of N. benthamiana at the 1-true leaf developmental stage by mechanical sap inoculation [22]. Then, seedlings were kept for at least ten days in a glasshouse at 25°C until use as inoculation source plants.
Reproductive performance experiments on TuMV-infected turnips
Virus source plants infected with world-B and basal-BR isolates were homogenized in 0.01 M potassium phosphate buffer (pH 7.0) and mechanically inoculated onto B. rapa cv. Hakatasuwari plants at the 1-true leaf stage. Healthy plants were treated with the same buffer solution at the same growth stage and used as non-infected controls. Inoculated plants were kept for ten days in a glasshouse at 25°C, and then used for experiments.
To assess the effects of TuMV infection on aphid reproduction, five first instar nymphs of M. persicae or L. erysimi were placed on world B-infected and non-infected turnip plants. Similarly, one first instar nymph (due to low numbers of first instars) of M. persicae or L. erysimi was placed on basal-BR-infected and non-infected turnip plants. All aphids on each plant were counted after six days. Nine or ten replications were performed for each treatment and control. The Saga strain of L. erysimi was used in the experiments with the world-B group and the Ogi strain in the experiments with the basal-BR group. The numbers of aphids between in infected and non-infected turnip plants were analyzed using a Student’s t-test.
Results
Reproductive performance experiments on TuMV-infected turnips
The number of M. persicae on turnip plants was not significantly different between non-infected and world-B infected treatments (t = 0.71, p = 0.49, Student’s t-test; Fig 1A). Meanwhile, the number of L. erysimi was significantly higher on world-B infected turnip plants than non-infected ones (t = -2.45, p < 0.05, Student’s t-test; Fig 1B).
Fig 1.
Mean number of (a) M. persicae and (b) L. erysimi on non-infected and world-B infected turnip plants. Vertical bars represent ±1 standard error of the mean. Asterisk and “NS” indicate significant and non-significant differences, respectively, between non-infected and world-B infected turnip plants (p < 0.05).
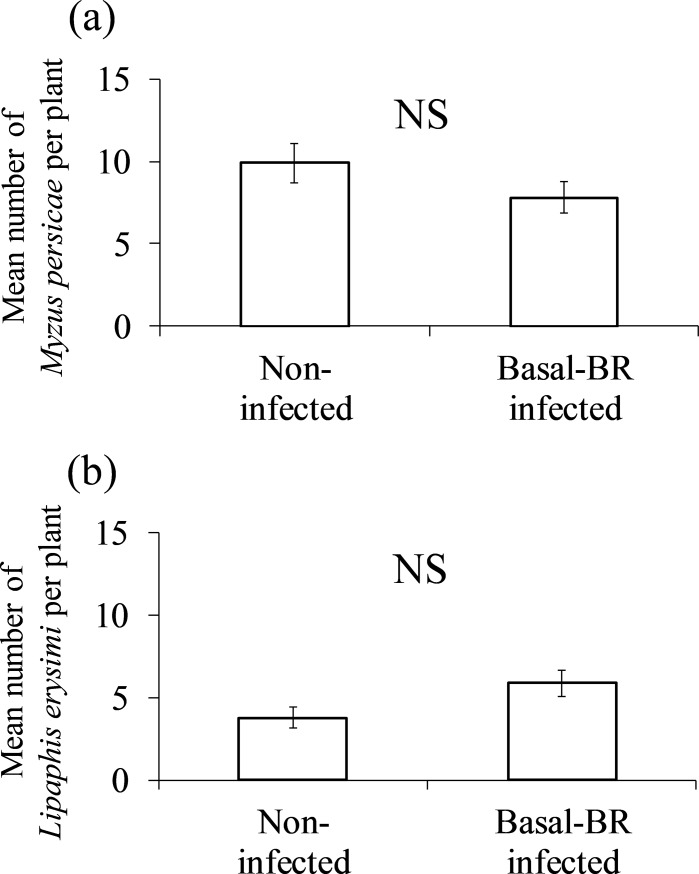
Similarly, no significant differences were detected in the number of M. persicae on turnip plants between non-infected and basal-BR infected treatments (t = 1.41, p = 0.18, Student’s t-test; Fig 2A), while the number of L. erysimi tended to be higher on basal-BR infected turnip plants than on non-infected ones (t = -2.10, p = 0.052, Student’s t-test; Fig 2B).
Fig 2.
Mean number of (a) M. persicae and (b) L. erysimi on non-infected and basal-BR infected turnip plants. Vertical bars represent ±1 standard error of the mean. “NS” indicates no significant difference between non-infected and basal-BR infected turnip plants (p < 0.05).
Discussion
In this study, we examined the population growth of two aphid vectors of TuMV on turnip plants infected by world-B and basal-BR phylogenetic groups of TuMV. The population growth of M. persicae was not promoted by TuMV infection, regardless of phylogenetic group. This is consistent with results shown by a previous study [24], in which the reproductive performance of M. persicae did not change on TuMV-infected N. tabacum. However, another previous study reported that the performance of M. persicae on N. benthamiana and A. thaliana increased with TuMV infection [16]. The previous report [24] demonstrated that, under the presence of M. persicae, a viral protein, Nla-Pro, localized to a vacuole in N. benthamiana but not in N. tabacum, and concluded that this phenomenon critically affects the increase of M. persicae reproductive performance in N. benthamiana. Further studies are needed to clarify whether the localization of Nla-Pro is observed or not in our system.
In contrast to M. persicae, the population growth of L. erysimi significantly increased on world-B infected plants and tended to increase on basal-BR infected plants, when compared to non-infected treatments. As mentioned earlier, L. erysimi is a specialist aphid associated only with Brassicaceae, while M. persicae is a generalist with a broad host range of over 40 plant families [25]. Therefore, the former species may be better adapted to Brassicaceae plants with TuMV. In addition, our results suggest that not only differences in plant species but also combinations of vectors and host plants are important factors affecting the performance of vector aphids.
In the present study, effects of TuMV infection on aphid reproductive performance were similar between world-B and basal-BR groups, suggesting that other factors are related to the replacement of the TuMV phylogenetic group world-B with the basal-BR in Kyushu. A previous study [26] showed that host range and infection efficiency are different among TuMV phylogenetic groups. Therefore, infection capacities related to Japanese crops and wild host plants may have caused the replacement in Kyushu. Another possibility is co-infection with other plant viruses may have affected the dominance of TuMV phylogenetic groups because mixed infections of TuMV and other viruses are often confirmed in field situations [12, 17–18]. Further studies are needed to clarify the factors causing the replacement of TuMV phylogenetic groups in Kyushu.
In summary, this study shows the relationships between TuMV and its aphid vectors differ between L. erysimi and M. persicae, but do not appear to differ between the two phylogenetic groups of TuMV, world-B and basal-BR.
Acknowledgments
We are grateful to Drs. Y. Hayakawa, H. Tatsuta, Y. Sakamaki, D. Taylor and K. Fujiwara for their valuable comments on the manuscript.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Andret-Link P. and Fuchs M. Transmission specificity of plant viruses by vectors. J Plant Pathol. 2005; 87: 153–165. http://www.jstor.org/stable/41998234?seq=1#page_scan_tab_contents [Google Scholar]
- 2.Hogenhout SA, Ammar ED, Whitfield AE, Redinbaugh MG. Insect vector interactions with persistently transmitted viruses. Annu Rev Phytopathol. 2008; 46: 327–359. http://www.annualreviews.org/doi/abs/10.1146/annurev.phyto.022508.092135 [DOI] [PubMed] [Google Scholar]
- 3.Mauck KE, Consuelo MDM, Mark CM. Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proc Natl Acad Sci USA. 2010; 107: 3600–3605. http://www.pnas.org/content/107/8/3600.short 10.1073/pnas.0907191107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng JCK, Perry KL. Transmission of plant viruses by aphid vectors. Mol Plant Pathol. 2004; 5: 505–511. http://onlinelibrary.wiley.com/doi/10.1111/j.1364-3703.2004.00240.x/full [DOI] [PubMed] [Google Scholar]
- 5.Kersch-Becker MF, Thaler JS. Virus strains differentially induce plant susceptibility to aphid vectors and chewing herbivores. Oecologia. 2014; 174: 883–892. https://link.springer.com/article/10.1007/s00442-013-2812-7 [DOI] [PubMed] [Google Scholar]
- 6.Kluth S, Kruess A, Tscharntke T. Insects as vectors of plant pathogens: mutualistic and antagonistic interactions. Oecologia. 2002; 133: 193–199. https://link.springer.com/article/10.1007/s00442-002-1016-3 [DOI] [PubMed] [Google Scholar]
- 7.Li M, Liu J, Liu SS. Tomato yellow leaf curl virus infection of tomato does not affect the performance of the Q and ZHJ2 biotypes of the viral vector Bemisia tabaci. Insect Sci. 2011; 18: 40–49. http://onlinelibrary.wiley.com/doi/10.1111/j.1744-7917.2010.01354.x/full [Google Scholar]
- 8.Liu J, Li M, Li JM, Huang CJ, Zhou XP, Xu FC, et al. Viral infection of tobacco plants improves performance of Bemisia tabaci but more so for an invasive than for an indigenous biotype of the whitefly. J. Zhejiang Univ-Sci B (Biomed & Biotechnol). 2010; 11: 30–40. https://link.springer.com/article/10.1631/jzus.B0900213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan H, Chu D, Liu B, Shi X, Guo L, Xie W, et al. Differential effects of an exotic plant virus on its two closely related vectors. Sci Rep. 2013; 3: 2230 https://www.nature.com/articles/srep02230?sa=X&ved=0CC0Q9QEwC2oVChMIxJrlpvL6xgIV5CxyCh1_dAxG 10.1038/srep02230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinheiro PV, Ghanim M, Alexander M, Rebelo AR, Santos RS, Orsburn BC, et al. Host plants indirectly influence plant virus transmission by altering gut cysteine protease activity of aphid vectors. Mol Cell Proteomics. 2017; S230–S243. http://www.mcponline.org/content/16/4_suppl_1/S230.short 10.1074/mcp.M116.063495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomlinson JA. Epidemiology and control of virus diseases of vegetables. Ann Appl Biol. 1987; 110: 661–681. http://onlinelibrary.wiley.com/doi/10.1111/j.1744-7348.1987.tb04187.x/full [Google Scholar]
- 12.Shattuck VI. The biology, epidemiology and control of turnip mosaic virus. Plant Breed Rev. 1992; 14: 199–238. [Google Scholar]
- 13.Ohshima K. Plant potyvirus evolution: the survey of the genetic structure of populations. Virus. 2012; 62: 151–160. (In Japanese with English summary.) [DOI] [PubMed] [Google Scholar]
- 14.Yasaka R, Fukagawa H, Ikematsu M, Soda H, Korkmaz S, Golnaraghi A, et al. The timescale of emergence and spread of turnip mosaic potyvirus. Sci Rep. 2017; 7: 4240 https://www.nature.com/articles/s41598-017-01934-7 10.1038/s41598-017-01934-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomitaka Y, Ohshima K. A phylogeographical study of the Turnip mosaic virus population in East Asia reveals an ‘emergent’ lineage in Japan. Mol Ecol. 2006; 15: 4437–4457. http://onlinelibrary.wiley.com/doi/10.1111/j.1365-294X.2006.03094.x/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casteel CL, Alwis MD, Bak A, Dong H, Whitham SA, Jander G. Disruption of ethylene responses by turnip mosaic virus mediates suppression of plant defense against the green peach aphid vector. Plant Physiol. 2014; 169: 209–218. http://www.plantphysiol.org/content/early/2015/06/19/pp.15.00332 10.1104/pp.15.00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sano Y, Kojima M. Increase in cucumber mosaic virus concentration in Japanese radish plants co-infected with turnip mosaic virus. Ann Phytopath Soc Japan. 1989; 55: 296–302. https://www.jstage.jst.go.jp/article/jjphytopath1918/55/3/55_3_296/_article [Google Scholar]
- 18.Tsuchizaki T, Goto T, Fujisawa I, Yoshida K. Virus disease occurring legumes and vegetables in Hokkaido. Res Bull Hokkaido Natl Agric Exp Stn. 1981; 131: 71–93. [In Japanese with English summary]. [Google Scholar]
- 19.Walsh JA, Jenner CE. Turnip mosaic virus and the quest for durable resistance. Mol Plant Pathol. 2002; 3: 289–300. 10.1046/j.1364-3703.2002.00132.x [DOI] [PubMed] [Google Scholar]
- 20.Moritsu M. Aphids of Japan in Colors. Zenkoku Noson Kyoiku Kyokai, Tokyo, Japan. 1983. [In Japanese]. [Google Scholar]
- 21.Nishi Y. Study on the transmission of plant viruses by aphids. Bull Kyushu Agric Exp Sta. 1963; 8: 355–356. [In Japanese]. [Google Scholar]
- 22.Ohshima K, Yamaguchi Y, Hirota R, Hamamoto T, Tomimura K, Tan Z, et al. Molecular evolution of Turnip mosaic virus: evidence of host adaptation, genetic recombination and geographical spread. J Gen Virol. 2002; 83: 1511–1521. http://jgv.microbiologyresearch.org/content/journal/jgv/10.1099/0022-1317-83-6-1511 [DOI] [PubMed] [Google Scholar]
- 23.Ohshima K, Tomitaka Y, Wood JT, Minematsu Y, Kajiyama H, Tomimura K, et al. Patterns of recombination in turnip mosaic virus genomic sequences indicate hotspots of recombination. J Gen Virol. 2007; 88: 298–315. http://jgv.microbiologyresearch.org/content/journal/jgv/10.1099/vir.0.82335-0#tab1 [DOI] [PubMed] [Google Scholar]
- 24.Bak A, Cheung AL, Yang C, Whitham SA, Casteel CL. A viral protease relocalizes in the presence of the vector to promote vector performance. Nat Comm. 2017; 8: 14493 https://www.nature.com/articles/ncomms14493 10.1038/ncomms14493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blackman RL, Eastop VF. Aphids on the world’s plants. 2017; http://www.aphidsonworldsplants.info/ (Accessed on 23 November 2017.)
- 26.Tomimura K, Špak J, Katis N, Jenner CE, Walsh JA, Gibbs AJ, et al. Comparisons of the genetic structure of populations of Turnip mosaic virus in West and East Eurasia. Virology. 2004; 330: 408–423. https://www.sciencedirect.com/science/article/pii/S004268220400652X 10.1016/j.virol.2004.09.040 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.