Abstract
Organ development is a sophisticated process of self-organization. However, despite growing understanding of the developmental mechanisms, little is known about how to reactivate them postnatally for regeneration. We found that treatment of adult non-hair fibroblasts with cell-free extract from embryonic skin conferred upon them the competency to regenerate hair follicles. Proteomics analysis identified three secreted proteins enriched in the embryonic skin, apolipoprotein-A1, galectin-1 and lumican that together were essential and sufficient to induce new hair follicles. These 3 proteins show a stage-specific co-enrichment in the perifolliculogenetic embryonic dermis. Mechanistically, exposure to embryonic skin extract or to the combination of the 3 proteins altered the gene expression to an inductive hair follicle dermal papilla fibroblast-like profile and activated Igf and Wnt signaling, which are crucial for the regeneration process. Therefore, a cocktail of organ-specific extracellular proteins from the embryonic environment can render adult cells competent to re-engage in developmental interactions for organ neogenesis. Identification of factors that recreate the extracellular context of respective developing tissues can become an important strategy to promote regeneration in adult organs.
1. Introduction
Compared with the profound spontaneous restoration of lost tissues and organs in lower vertebrates [[1], [2], [3]], mammals show limited regeneration after injury and damaged tissues are often replaced by morphologically and functionally debilitating scars [4]. To promote regeneration, attempts have been made by transplantation of stem cells, organoids or bioartificial organs [[5], [6], [7]], yet in most cases functional incorporation and maintenance of transplanted cells remains challenging [8]. In skin, it usually heals by fibrosis rather than regeneration after injury, leading to scar formation and permanent loss of skin appendages, prominently hair follicles (HFs) [9]. Although promotion of skin healing has been attempted by transplanting cultured keratinocytes or skin equivalents [10,11], regeneration of HFs is still an unmet clinical need.
The HF is a mini-organ composed of an epithelial cylinder and specialized dermal papilla (DP) fibroblasts [12]. Similar to other ectodermal mini-organs, HF development depends on well-choreographed epithelial-mesenchymal interactions initiated by the crosstalk between epithelial and dermal embryonic skin progenitors [12]. During HF morphogenesis, epithelial progenitors gradually adopt a follicular fate, whereas dermal progenitors differentiate into DP fibroblasts. In normal adult skin, new HFs generally can not form [12]. As an exception, HFs can regenerate spontaneously in the center of very large wounds and with low efficiency in humans via incompletely understood mechanisms [[13], [14], [15]]. Such examples indicate that, in principle, the potential for HF neogenesis is preserved and can possibly be unleashed.
To induce new HFs postnatally, Oliver et al. demonstrated that microdissected DP could induce new HFs from epidermis when transplanted into the subepidermal space [16]. Following this seminal work, HF neogenesis in adults has largely relied on culture-expanded DP fibroblasts, which are capable of reinitiating developmental epithelial-mesenchymal interactions with competent keratinocytes [[17], [18], [19], [20]]. The HF-inducing ability of cultured DP fibroblasts is easily lost during culture and this limits their use for large-scale HF regeneration [17]. In addition to adult DP cells, freshly isolated newborn murine dermal fibroblasts have also been demonstrated to be capable of inducing HF neogenesis [[7],[19],[21], [22], [23]], but such HF inductivity of dermal fibroblasts are quickly lost in postnatal life.
An alternative strategy for inducing new HFs is through the use of embryonic skin [24]. In contrast to adult skin, wounded embryonic skin heals without scarring and with extensive HF neogenesis [25]. When embryonic dermal tissues are combined with postnatal epithelium, they are able to induce new HFs [24]. What are the key factors that define the HF-inducing ability of embryonic skin? It was demonstrated that isolated embryonic dermal cells are able to induce new HFs when combined with keratinocytes [26]. On the other hand, it has also been suggested that embryonic tissues create unique extracellular environments conducive to regeneration [25,27]. The cell-free extracellular matrix from adult tissue has been shown to enhance tissue regeneration [6,[27], [28], [29], [30]]. Isolation of defined factors present in the extracellular matrix that can elicit neogenesis of an organ can open novel, therapeutically amenable organ-specific regeneration protocols. However, induction of the neogenesis of a specific organ by defined extracellular factors has not been achieved. Here, we demonstrate that a cocktail of 3 secreted protein factors enriched in the developmental skin can induce neogenesis of the HF miniorgan through reactivating the epithelial-mesenchymal crosstalk between adult skin cells.
2. Materials and methods
2.1. Animals
All animal experiments were approved by the Institutional Animal Care and Use Committee at National Taiwan University. Animals were housed in animal facilities of National Taiwan University. Wistar rats were purchased from BioLASCO Taiwan Co. and C57BL/6 mice and nude mice (BALB/cAnN-Foxn1nu/CrlNarl) were from Taiwan National Laboratory Animal Center. Z/AP mice expressing the lacZ reporter gene to overexpress β-galactosidase were from Jackson Laboratory [31]. For invasive experiments, animals were anesthetized by intramuscular injection of a mixture of zolazepam (Zoletil vet, Virbac Laboratories) and xylazine (Rompun, Bayer) (4:1, v/v).
2.2. Preparation of cell-free extract
The dorsal skin containing the epidermis and dermis was isolated from Wistar rats (from E13.5 to postnatal stages) of either sex. The skin tissue was minced and mixed with phosphate-buffered solution (PBS) containing protease inhibitor (1 × Protease Arrest, G-Biosciences). The samples were repeatedly frozen at −80 °C and thawed on ice and then homogenized by repeated passages through a 25-G needle. Cell breakage was examined under a phase contrast microscope to ensure that >99% of cells were disrupted. Subsequent cell culture from the skin extract yielded no cell growth (data not shown). To prepare cell-free extract from epidermis or dermis alone, the whole skin was incubated in dispase (2 mg/ml in PBS, Sigma-Aldrich) at 37 °C for 1 h, and then the epidermis and dermis were separated with forceps before further extraction. The protein concentration of the cell-free extract was quantified by protein assay dye reagent (Protein Assay Dye Reagent Concentrate, Bio-Rad Laboratories). The protein concentration was ~1–2 μg/μl under our preparation procedure. The protein concentration was adjusted with PBS containing protease inhibitor to a concentration of 1 μg/μl for use in patch assays if not stated otherwise. If not used immediately, the extract solution was stored at −80 °C and was used within 1 month of preparation.
2.3. Dilution and pretreatment of skin extract
E16.5 skin extract was serially diluted in PBS or was pretreated with RNase A (2 μg/ml; Sigma-Aldrich), DNase I (100 U/ml; Sigma-Aldrich) or proteinase K (400 μg/ml; Sigma-Aldrich) for 10 min at 37 °C or was inactivated at 70 °C for 10 min before being tested in patch assays. Removal of apolipoprotein-A1, galectin-1 and lumican from the skin extract by immunodepletion was performed with specific antibodies followed by protein G–agarose beads (Roche) according to the manufacturer’s instructions. Antibodies used were anti-apolipoprotein-A1 (Novus, NB600-609), anti-galectin-1 (Abcam, ab138513), anti-lumican (Santa Cruz Biotechnology, sc-33785) and control rabbit IgG (RbIgG; GeneTex, GTX35035). The efficiency of immunodepletion for individual proteins was confirmed by western blotting.
2.4. Cells, culture conditions and mRNA transfection
Mouse DP cells were from vibrissae of 7- to 8-week-old C57BL/6 female mice. All fibroblasts and DP cells were expanded and cultured in DMEM (Gibco) with 10% FBS (Gibco) as described [32]. In the HF neogenesis assay, the contamination of preformed HFs in the isolated epidermal keratinocytes can lead to HF neogenesis without co-administration of inductive mesenchymal cells [21]. To avoid this confounding factor, we developed a protocol to eliminate preformed HFs from the isolated epidermal keratinocytes. Mouse P1 keratinocytes were isolated from the dorsal skin of C57BL/6 mice of either sex as described [21], and mouse adult keratinocytes were from the hairless areas of foot pads of 7- to 8-week-old C57BL/6 female mice. The isolated cells were transferred to a 5-cm plastic dish and examined under a phase contrast microscope to make sure that undissociated hair germ-like tissues were removed and the isolated keratinocytes alone did not grow new HFs in patch assays (Fig. S1b and S1b′). Keratinocytes were cultured with DKSFM with supplements (Invitrogen). To test the effect of E16.5 skin extract on cells, P1 keratinocytes and adult fibroblasts were cultured with medium containing E16.5 skin extract (2 μg/ml) for up to 3 days. For washout, fibroblasts cultured in E16.5 skin extract for 3 days were further cultured without E16.5 skin extract for 3 days. When fibroblasts were cultured with the indicated proteins, the concentration of each protein was 0.01 μg/ml. IGF1R inhibitor (NVP-AEW541, 10 nM; Selleck Chemicals) and Wnt inhibitor (XAV-939, 5 μM; Selleck Chemicals) were also used in fibroblast cultures. For mRNA transfection of cultured adult dermal fibroblasts, mRNAs were constructed from mouse Lgals (NM_008495.2), Apoa1(NM_009692.4) and Lum (NM_008524.2) genes respectively as we previously described [33]. Transfections of the three mRNAs were carried out by use of Stemfect™ (Stemgent, USA) mRNA transfection reagent in vitro [33]. Two hours after mRNA transfection, the fibroblasts were collected for patch assays.
2.5. Standard and modified patch assays
HF neogenesis was tested in patch assays in 7- to 10-week-old female nude mice [34,35]. For standard patch assays, 1 × 106 mesenchymal cells were mixed with 1 × 106 keratinocytes in 200 μl PBS and injected into the hypodermis of nude mice with an 18-G needle. Mice were sacrificed 2–3 weeks later, and the skin was removed for photography and further processed for histological examination or immunostaining. For the negative control, only keratinocytes were injected. We also modified patch assays to test HF inductivity of protein extracts or specific protein(s), and 1 × 106 keratinocytes were suspended in a solution containing cell-free skin extract or defined proteins to a final volume of 200 μl for injection. The cell-free skin extract was used at a protein concentration of 1 μg/μl or was serially diluted for testing. For defined proteins, each defined protein was initially serially diluted for testing. We ultimately used a concentration of 100 ng/μl for each protein because protein concentrations below this had lower or no inductive effects (data not shown). For fibroblasts transfected with mRNAs of Lgals (NM_008495.2), Apoa1(NM_009692.4) and Lum (NM_008524.2), 1 × 106 mRNA transfected fibroblasts cells were mixed with 1 × 106 keratinocytes in 200 μl PBS and injected into the hypodermis of nude mice. For comparison of HF neogenesis between different experimental groups, the rate of HF neogenesis was defined as (No. of patch assays with HF neogenesis)/(No. of total patch assays performed).
2.6. HF neogenesis in full-thickness wounds and preparation of dermal equivalents
A full-thickness wound (1 cm × 1 cm) was made by scissors on the backs of female nude mice and covered by Tegaderm™ (3 M) membranes. E16.5 skin extract (200 μl) containing 2 × 106 P1 keratinocytes was injected through the membrane into the wound.
For dermal equivalent preparation, adult mouse dermal fibroblasts were suspended in the culture medium (1:1 [v/v] DMEM/Ham’s F-12 supplemented with 10% FBS) containing a 1:10 (v/v) E16.5 skin extract solution/collagen I solution (BD Biosciences), with a final protein concentration of 1 μg/ml for the E16.5 skin extract, on ice and then transferred to an incubator for 3 days. The resulting dermal equivalent was transferred to a full-thickness wound and covered with Tegaderm™ (3 M) membranes. 2 × 106 P1 keratinocytes were injected through the membrane to the top of the dermal equivalent. Mice were sacrificed 4 weeks later, and the skin was removed for photography and was further processed for histological examination.
2.7. Real-time PCR analysis of gene expression
Total RNA was extracted from fibroblasts or DP cells using Trizol reagent (Invitrogen). cDNAs were synthesized with a RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific). Quantitative real-time PCR (qRT-PCR) was performed as described [36]. The sequences of the primers of mouse genes were as follows: Akp2 (NM_001287172.1, 5′-TTGGGCAGGCAAGACACAGA-3′ and 5′-TTGGCAACCCTGGGTAGACAG-3′), Bmp6 (NM_007556.2, 5′-CTGCGCACCAACCAAACTGAA-3′ and 5′-TTGGGGGAGGCGAACATTAGGTA-3′), Gapdh (GU214026.1, 5′-CGTAGACAAAATGGTGAAGGTCGG-3′ and 5′-AAGCAGTTGGTGGTGCAGGATG-3′), Hey1 (NM_010423.2, 5′-AGCGCCGACGAGACCGAATCAATA-3′ and 5′-TGGGGCAGCAGCAGAGGGTGTG-3′), Lef1 (NM_010703.4, 5′-CGCTAAAGGAGAGTGCAGCTA-3 and 5′-GCTGTCTCTCTTTCCGTGCT-3′), and versican (XM_011244471.1, 5′-GTGACTATGGCTGGCACAAATTCC-3′ and 5′-GGTTGGGTCTCCAATTCTCGTATTGC-3′).
2.8. RNA-sequencing and analysis
Total RNA was extracted from fibroblasts or DP cells with Trizol Reagent (Invitrogen) and RNeasy mini-columns (Qiagen). The cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen), and the resulting short DNA fragments were purified by QiaQuick PCR Extraction Kit (Qiagen). The cDNA libraries were sequenced from both the 5′ and 3′ ends on an Illumina GA II. The short reads with their adaptor sequences were trimmed and then aligned to UCSC mouse genome version 10 with STAR [37]. All the alignment parameters were set with default values. The process was performed on a server with 24 cores and 128 GB of memory. The average alignment rate was ~95%. Data analysis for expression changes was performed by Cufflinks pipeline [38], which includes cufflinks, cuffmerge and cuffdiff. The read counts of genes in samples were further normalized based on the reads per kilobase per million mapped reads (RPKM). Principal component analysis was performed with “prcomp” with the software package R [39], and was visualized with “ggplot” [40]. CummeRbund was also used to examine the RPKM data for each sample. The differentially expressed genes of fibroblasts exposed to selected protein(s) or to E16.5 skin extract were analyzed by the software SAS (Statistical Analysis System 9.1, SAS Institute Inc.) and Spearman’s rank correlation coefficient (rs) was calculated for comparison between groups. RNA-seq raw data have been submitted to http://www.ncbi.nlm.nih.gov/geo (GEO:GSE85303). More details are provided in Table S2. To identify the cellular processes/signaling pathways upregulated in fibroblasts after treatment with selected protein(s) or E16.5 skin extract, genes with greater than 1.2-fold increase were analyzed by DAVID Bioinformatics Resources 6.7 (https://david.ncifcrf.gov/). The significantly upregulated cellular processes/signaling pathways (p < 0.05) were selected and ranked. The top 20 cellular processes/signaling pathways were shown in Table S3.
2.9. In situ hybridization
Paraffin sections were prepared for in situ hybridization as we described [41]. Digoxigenin-labeled probes against rat apolipoprotein-A1 (nucleotides 44–700; NM_012738.1), galectin-1 (nucleotides 72–479; NM_008495.2) and lumican (nucleotides 417–1727; NM_008524.2) were synthesized with a DIG RNA Labeling Kit (Roche) [41]. The sections were counterstained with eosin.
2.10. Western blot
Western blotting was performed by using antibodies against the following proteins: Akt (Cell Signaling, 9272; 1:1000), phospho-Akt (Cell Signaling, 9271S; 1:1000), apolipoprotein-A1 (Novus, NB600-609; 1:1000), β-actin (Santa Cruz Biotechnology, sc-47778; 1:1000), β-catenin (BD Biosciences, 610153; 1:1000), Erk (Cell Signaling, 9102S; 1:1000), phospho-Erk (Cell Signaling, 4370S; 1:1000), GAPDH (Millipore, MAB374; 1:1000), galectin-1 (Abcam, ab138513; 1:1000), IRS2 (LifeSpan BioSciences, LS-C33930; 1:1000), phospho-IRS2 (Novus, NBP1-72215; 1:1000), lumican (Santa Cruz Biotechnology, sc-33785; 1:1000) and TATA binding protein (TBP) (Cell Signaling, 8515S; 1:1000). Fractionation of cells into nuclear and cytoplasmic content was performed with the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific) supplemented with Protease Inhibitor Cocktail (Thermo Scientific) as we described [42].
2.11. Recombinant proteins
The recombinant proteins used for patch assays and cell culture were as follows: apolipoprotein-A1 (PeproTech, 350–11), galectin-1 (Novoprotein, C285), lumican (Novoprotein, C372), annexin A2 (PROSPEC, PRO-777), gelsolin (Abnova, H00002934-P01), fibrinogen beta-chain fragment (American Peptide Company, AP42-1-33A), fibronectin (Innovative Research, Inc. IHFBN) and sulfated glycoprotein 1(Psap) (Abnova, H00005660-P01).
2.12. Histology, immunofluorescence, β-galactosidase activity assay and alkaline phosphatase activity assay
Skin samples were fixed in 4% paraformaldehyde overnight at 4 °C, serially dehydrated and embedded in paraffin. The specimens were sectioned and stained with hematoxylin and eosin (H&E). Immunofluorescence staining was performed with routine antigen retrieval as suggested by the antibody manufacturer [41]. The slides were examined under a confocal microscope (Meta 510, Carl Zeiss). The primary antibodies for immunostaining used in this study were as follows: fibrinogen beta-chain fragment (Abcam, EPR3083; 1:200), sulfated glycoprotein 1 (Abcam, ab68466; 1:200), galectin-1 (Abcam, ab138513; 1:200), lumican (Abcam, ab168348; 1:200), annexin A2 (Genetex, GTX101902, 1:200), gelsolin (Genetex, GTX101185; 1:200), apolipoprotein-A1 (Novus, NBP1-77008; 1:200) and fibronectin (Genetex, GTX61207; 1:200). Secondary antibodies were Alexa Fluor 594 donkey anti–mouse IgG (H+L) (Jackson ImmunoResearch, 715-585-151; 1:500), Cy3-conjugated AffiniPure donkey anti–rabbit IgG (Jackson ImmunoResearch, 711-165-152; 1:500) and Alexa Fluor 594-AffiniPure F (ab′)2 Fragment rabbit anti–chicken IgY (IgG) (H+L) (Jackson ImmunoResearch, 303-586-003; 1:500). β-galactosidase activity and alkaline phosphatase activity were detected as described [43,44].
2.13. Mass spectrometry for proteomic analysis
To compare the proteomes of E16.5 and P1 cell-free extracts, each extract was lysed in lysis buffer: 12 mM sodium deoxycholate (SDC), 12 mM sodium lauryl sulfate (SLS), protease inhibitor cocktail (Roche) and phosphatase inhibitors (Sigma-Aldrich) in 100 mM triethylammonium bicarbonate (TEABC). Lysates were heated at 95 °C for 5 min, sonicated for 15 min and then reduced with 10 mM dithiothreitol (DTT) for 30 min and alkylated with 50 mM iodoacetamide for 30 min. The alkylated protein solution was diluted five-fold with 50 mM TEABC before Lys-C and trypsin digestion overnight [45]. After digestion, SDC and SLS were removed by acidified ethyl acetate for phase transfer [46]. Finally, the peptides were desalted by using reversed-phase Stage Tip [47]. For proteome analysis, a TripleTOF 5600 system (AB SCIEX) was equipped with a nanoACQUITY UPLC (Waters Corporation). The 3-μm ReproSil-Pur C18-AQ particles (Dr. Maisch HPLC GmbH) were packed into a 15-cm self-pulled column with a 100-μm inner diameter. The LC system consisted of 0.1% formic acid in water (buffer A) and 0.1% formic acid in acetonitrile (buffer B). Peptides were separated through a gradient of up to 80% buffer B over 120 min at a flow rate of 500 nl/min. Data were acquired using an ion spray voltage of 2.6 kV and an interface heater temperature of 150 °C. For information-dependent acquisition, the MS survey scan range was m/z 300–1,500, and data were acquired for 200 ms. The top 15 precursor ions were selected based on a threshold of 150 counts/s in each MS survey scan, and each acquisition scan was performed for 150 ms. The collision energy was automatically adjusted by the rolling CID function of Analyst TF 1.5. To minimize repeated scans, dynamic exclusion was set for 6 s, and the precursor was then removed from the exclusion list. For proteome data analysis, the raw MS/MS data were processed as described [48]. Searches were performed against the Swiss-Prot database (version 57.8, Rattus norvegicus) by using Mascot (Matrix Science, London, UK; version 2.3). To allow for tryptic peptides with up to two missed cleavage sites, a fragment ion mass tolerance of 0.1 Da and a parent ion tolerance of 20 p.p.m. were selected. Carbamidomethyl (C) was selected as the fixed modification and oxidation (M) was selected as the variable modification. The FDR was evaluated by a search against a randomized decoy database at the peptide-spectrum matches (PSMs) level. Only PSMs with p < 0.01 were accepted and the FDR at the PSMs level was 0.02.
Label-free quantitative analysis was performed by IDEAL-Q software as described [49,50]. Alignment of elution times was then performed based on the peptide information list using linear regression across different LC-MS/MS runs followed by the correction of aberrational chromatographic shift across fragmental elution-time domains. To increase the correct assignment, the detected peptide peaks were validated by (a) a signal-to-noise ratio > 3 and (b) an accurate charge state. Only proteins with at least two identified peptides were selected for quantification. Bioinformatics analysis was performed by Perseus (version 1.5.0.15) [51]. Two-sample t-test (Volcano plot in Fig. 3a; FDR = 0.05, S0 = 0.6) was applied to compare across all data sets.
2.14. Statistics
Data are expressed as the mean ± standard deviation of the mean (S.D.). Comparisons between groups were performed with a two-tailed Student’s t-test using GraphPad Prism version 5.00 (GraphPad Software) software. The rate of HF neogenesis between different groups was compared using Fisher’s exact test by GraphPad Prism version 5.00 (GraphPad Software). Differences were considered statistically significant at p < 0.05.
3. Results
3.1. Induction of HF neogenesis by cell-free extract from E15.5-E17.5 embryonic rat skin
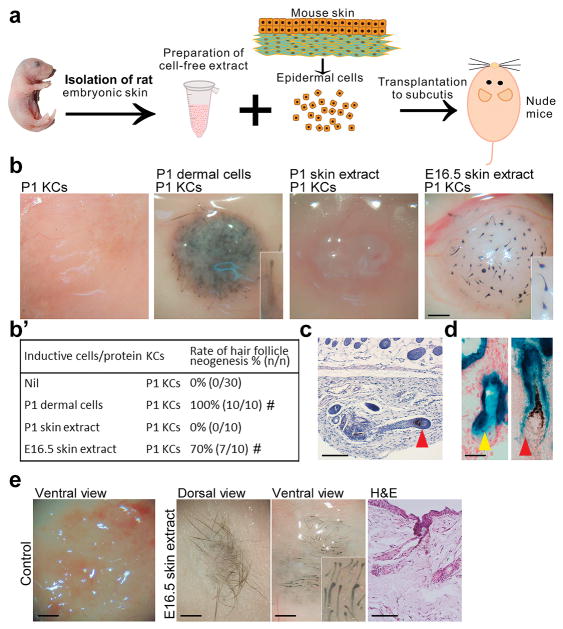
We first determined whether cell-free extracts from embryonic skin can induce HF neogenesis. In the HF neogenesis assay, the contamination of preformed HFs in keratinocytes can lead to HF formation without co-administration of inductive mesenchymal cells [21]. We carefully removed preformed HFs from epidermal keratinocytes to avoid this and the isolated keratinocytes alone did not grow any new HFs in patch assays for HF neogenesis (Fig. S1b, and S1b′). Formation of new HFs in the HF neogenesis assay indicated the presence of mesenchymal inductivity. Whole-skin cell-free extracts from embryonic day 13.5 (E13.5) to postnatal day 1 (P1) rat embryos were tested in a modified patch assay (Fig. 1a). In the conventional patch assay [34], keratinocytes are combined with inductive mesenchymal cells, such as newborn skin dermal progenitors or adult DP fibroblasts (Fig. S1b and S1b′) and are co-injected subcutaneously into nude mice. We modified this assay by replacing the inductive mesenchymal cells with cell-free skin extracts and collected the skin specimens for examination at 2–3 weeks after cell implantation. Skin extracts from E15.5–17.5, the time period when HF morphogenesis is initiated (Fig. S1a), induced new HFs from P1 mouse keratinocytes (Fig. 1b and b′; Fig. S1b and S1b′), with highest inductivity at E16.5 (70%). With serial dilution, the E16.5 extract gradually lost its HF inductivity (Fig. S1c and S1c′). The E16.5 extract also induced HF neogenesis from adult mouse keratinocytes, albeit at a lower rate (20%) as compared with P1 keratinocytes (Fig. S1b and S1b′). When embryonic skin epidermis and dermis were separated, only dermal extract induced new HFs (Fig. S1b and S1b′), suggesting that factors present in the dermis are sufficient for the observed inductive effect.
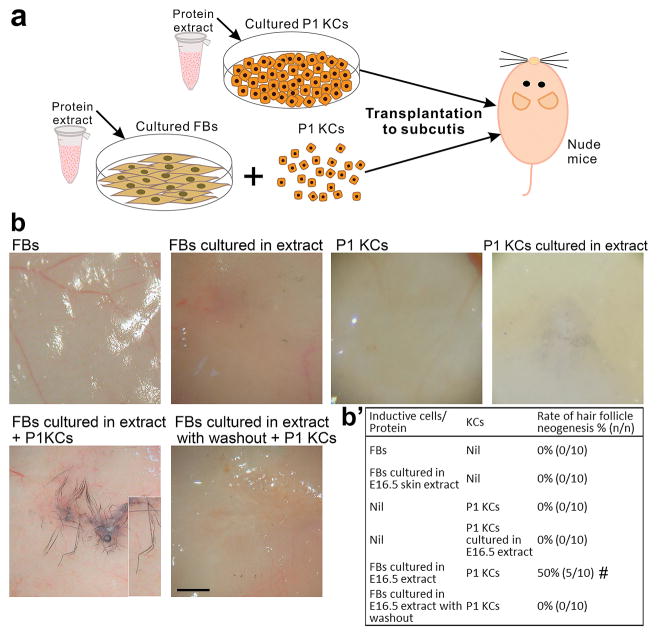
Fig. 1. Cell-free extract from stage-specific embryonic skin induces HF neogenesis.
(a) Preparation of skin extract and HF neogenesis assay. (b, b′) Effect of skin extract on HF induction. Regenerated HFs are pigmented because the P1 keratinocyte preparation also contains melanocyte progenitors. #p < 0.05 with Fisher’s test, compared with P1 KCs only (n = 10). (c) E16.5 skin extract induced new HFs with new DPs (red arrowhead). (d) New HFs formed from lacZ+ keratinocytes exhibited β-galactosidase activity in the epithelium, including the sebaceous gland (yellow arrowhead), but not in the DP (red arrowhead). (e) In full-thickness wounds of nude mice, incorporation of E16.5 extract into dermal equivalents induced HF neogenesis from transplanted C57BL/6 mouse keratinocytes. #p < 0.05 with Fisher’s test, compared with nascent E16.5 skin extract (n = 10). All insets show enlarged images of the regenerated HFs. KC: keratinocyte. Bar: histology, 100 μm; gross images, 500 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Regenerated HFs showed a normal morphology, complete with alkaline phosphatase+ DPs and hair shafts with cuticles (Fig. 1c; Fig. S2a and S2b). Longer hair shafts could be observed when the skin specimens were collected later for examination at about nearly 3 weeks after cell implantation. To determine the origin of epithelial and mesenchymal cells of the neogenic HFs, we performed modified patch assays using keratinocytes from Z/AP mice carrying the lacZ reporter gene to overexpress β-galactosidase. β-galactosidase activity was only detected in the epithelial portion of the neogenic HFs (Fig. 1d), indicating that new DPs formed from the host mesenchyme, likely from the local fibroblasts.
We further tested HF inductivity of the E16.5 extract in the full-thickness wound and in the dermal equivalent models. Treatment with keratinocytes plus E16.5 extract and treatment with keratinocytes plus dermal equivalents containing E16.5 extract both induced new HFs (n = 3) whereas no HF neogenesis was observed in the control groups without addition of E16.5 extract (Fig. 1e; Fig. S1d).
3.2. Protein component from embryonic skin extract is required for HF inductivity
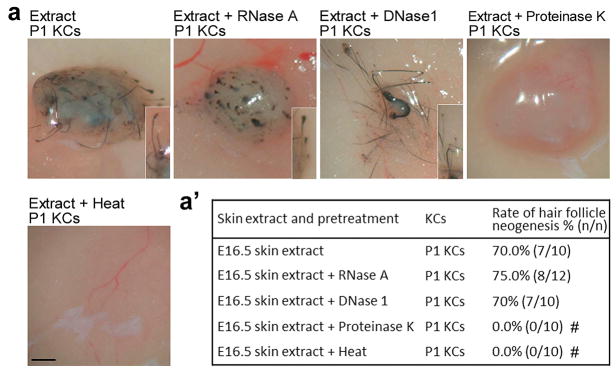
Because crude tissue extracts might contain molecules other than proteins, we pretreated the extracts with RNase, DNase, proteinase, or heat denaturation and found that HF inductivity was abolished when proteins were digested or heat denatured (Fig. 2a and a′). These findings suggest that protein factors present in embryonic skin at the time of early HF morphogenesis are able to induce HF neogenesis from adult cells.
Fig. 2. Protein fraction of embryonic skin extract is required for HF induction.
(a, a′) Effect of various pretreatments on the HF inductivity of E16.5 extract. HF inductivity of E16.5 extract was lost after inactivation of proteins by heat and proteinase K treatment. #p < 0.05 with Fisher’s test, compared with nascent E16.5 skin extract (n = 10). All insets show enlarged images of the regenerated HFs. KC: keratinocyte. Bar: gross images, 500 μm.
3.3. Three secreted proteins, Apoa1, Lgals1 and Lum, that together induce HF neogenesis
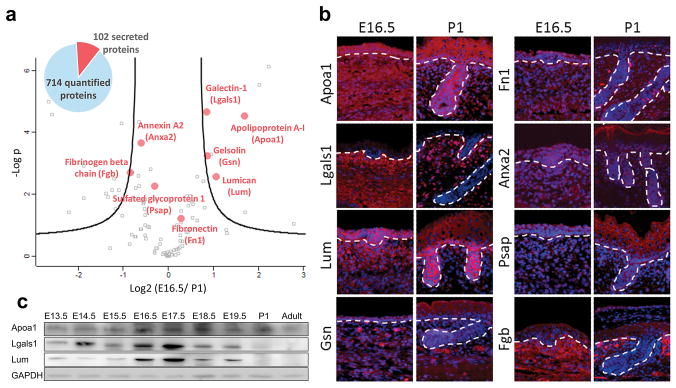
To identify the specific proteins capable of inducing HF neogenesis, we quantitatively compared the proteomes of E16.5 and P1 whole-skin extracts by liquid chromatography–tandem mass spectrometry (LC-MS/MS) (Table S1). Based on a Volcano plot of 714 quantifiable proteins (false discovery rate [FDR] = 5%; Fig. 3a), we selected secreted proteins that were significantly enriched in the E16.5 extract and were abundant in the dermis. Immunostaining verified that eight candidate proteins, apolipoprotein-A1 (Apoa1), galectin-1 (Lgals1), lumican (Lum), gelsolin (Gsn), fibronectin (Fn1), annexin A2 (Anxa2), sulfated glycoprotein 1 (Psap) and fibrinogen beta chain (Fgb), were expressed in the dermis of E16.5 and E17.5 skin (Fig. 3b; Fig. S3a).
Fig. 3. Proteomic analysis identifies secreted proteins enriched in the embryonic skin.
(a) LC-MS/MS analysis of the E16.5 and P1 skin extract proteomes and identification of key secreted proteins. Volcano plot shows the distribution of quantified proteins according to a t-test (p < 0.05) and fold change. Black curved lines indicate the significance levels. The data points located to the left of the left curve and to the right of the right curve indicate significantly down- and up-regulated proteins, respectively. The proteins marked by red dots were selected for follow-up biological validation. (b) Immunostaining of the eight selected proteins in the E16.5 and P1 skin. Red, specific protein; blue, nuclear DAPI staining; dashed line, basement membrane. (c) Western blotting for the expression of Apoa1, Lgals1 and Lum in skin extract from different ontogenetic stages. Bar: 100 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
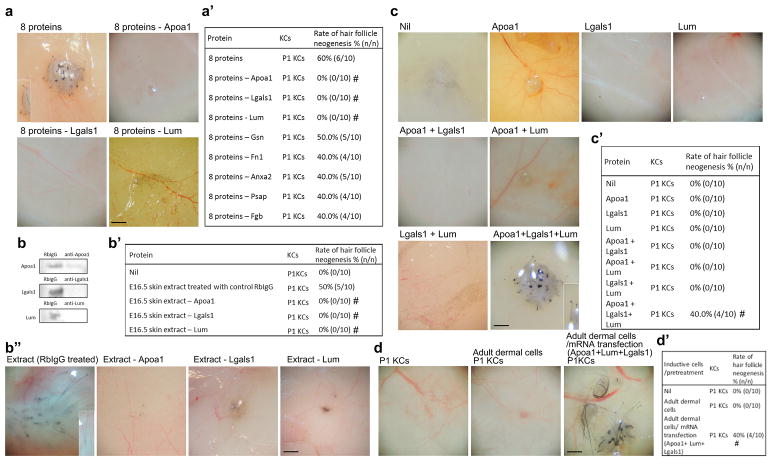
To determine whether defined factors can induce HF neogenesis, the inductivity of these eight proteins was tested in patch assays. The eight proteins together induced new HFs (Fig. 4a and a′; Fig. S4a and S4b). When individual proteins were removed, we found differential effects on HF neogenesis efficiency; complete suppression of neogenesis was observed when Apoa1, Lgals1 or Lum was individually removed (Fig. 4a and a′; Fig. S4a and S4b). Depletion of the E16.5 whole-skin extract of either one of these three proteins also eliminated HF inductivity (Fig. 4b, b′ and b″). These results indicated that Apoa1, Lgals1 and Lum are essential for the inductive effect of whole-skin extract. We further tested whether these three proteins together are sufficient to induce new HFs. Each protein alone or combinations of two proteins did not induce HF neogenesis (Fig. 4c and c′), whereas the mixture of all three proteins did (Fig. 4c and c′), albeit with lower efficiency compared with either the eight-protein mix or the whole-skin extract.
Fig. 4. Induction of HF neogenesis by defined proteins and mRNAs.
P1 keratinocytes were used for all patch assays. (a, a′) Effect of eight- and seven-protein mixtures on HF neogenesis. #p < 0.05 with Fisher’s test, compared with the eight-protein mixture (n = 10). (b, b′, b″) Removal of Apoa1, Lgals1 or Lum from the E16.5 skin extract with specific antibodies inhibited HF neogenesis. Removal of each protein was confirmed by western blotting. #p < 0.05 with Fisher’s test, compared with 16.5 extract treated with RbIgG (n = 10). (c, c′) The effect of single proteins and of two- or three-protein mixtures on HF neogenesis. #p < 0.05 with Fisher’s test, compared with P1 KCs only (n = 10). (d, d′) The effect of mRNA transfection of the three genes Apoa/Lgals1/Lum to adult dermal fibroblasts on HF neogenesis. Adult fibroblasts were transfected with mRNA of the three proteins before being tested in patch assays. #p < 0.05 with Fisher’s test, compared with P1 KCs only (n = 10). Insets show enlarged images of the regenerated HFs. KC: keratinocyte; RbIgG: control rabbit IgG. Bar: 500 μm.
3.4. Fibroblasts become competent for hair regeneration after exposure to embryonic skin proteins
To explore the mechanism of extract-induced HF neogenesis, P1 keratinocytes and adult dermal fibroblasts were cultured in the presence of E16.5 extract for 3 days before being tested in patch assays (Fig. 5a). Only fibroblasts could acquire hair-forming competence in response to this culture condition (Fig. 5b and b′), suggesting that fibroblasts are the target cells for these proteins. Importantly, the hair-forming ability of extract-treated fibroblasts was lost after further culture in extract-free conditions for three additional days (Fig. 5b and b′).
Fig. 5. E16.5 skin extract confers hair-forming ability on adult fibroblasts.
(a) P1 KCs and fibroblasts were cultured with E16.5 skin extract before HF neogenesis assays. (b, b′) Effect of E16.5 skin extract on keratinocytes and adult fibroblasts. Short-term exposure of fibroblasts to skin extract conferred on them a transient hair-forming ability. #p < 0.05 with Fisher’s test, compared with P1 KCs only (n = 10).
We then induced transient expression of these three proteins Apoa1, Lgals1 and Lum with mRNA delivery to adult fibroblasts in culture before they were combined with keratinocytes for patch assay analysis [33]. We found that transfection with these 3 mRNAs of Apoa/Lgals1/Lum to adult fibroblasts also rendered them competent to induce HF neogenesis (Fig. 4d and d′).
3.5. Three secreted proteins, Apoa1, Lgals1 and Lum, are co-enriched in the dermis of perifolliculogenetic stage of embryonic skin
Since the 3 proteins together are able to confer the ability to initiate HF neogenesis upon fibroblasts, we explored whether these three proteins are preferentially expressed in the dermis of embryonic skin. Through western blotting (Fig. 3c), in situ hybridization and immunostaining (Fig. 3b; Fig. S3a and S3b), we confirmed increased expression of Apoa1, Lgals1 and Lum was co-enriched in the E16.5–17.5 embryonic skin.
3.6. Exposure to embryonic skin extract or a combination of the 3 proteins, Apoa1, Lgals1 and Lum, alters the gene expression to a HF dermal papilla fibroblast-like profile
Next, using RNA sequencing, we determined how the exposure of adult fibroblasts to E16.5 extract, the Apoa1/Lgals1/Lum mix or each of the 3 proteins alters their transcriptome and also compared the resultant transcriptomes with those of adult DP and embryonic dermal fibroblasts (Table S2). Gene expression of adult fibroblasts exposed to E16.5 extract and gene expression of adult fibroblasts exposed to the Apoa1/Lgals1/Lum mix changed notably and, compared with gene expression of fibroblasts exposed to each of the three proteins alone, both exhibited a pattern more similar to that of DP fibroblasts (Fig. 6a). After exposure to E16.5 extract, key DP signature genes became upregulated (Fig. 6b), and alkaline phosphatase activity, which is characteristic of inductive DP fibroblasts in vitro [52], was present (Fig. 6c). The results above show that E16.5 protein extract and the Apoa1/Lgals1/Lum mix alter the gene expression of adult dermal fibroblasts toward a DP-like profile and render them competent to regenerate HFs.
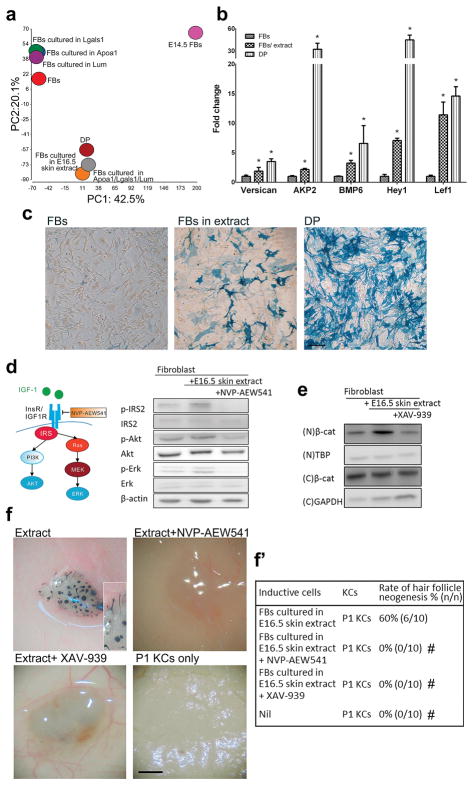
Fig. 6. Embryonic skin extract or 3-protein combination alters fibroblast gene expression to a dermal papilla like-profile and activates Wnt and Igf signaling.
Exposure to embryonic skin extract or a combination of the 3 proteins, Apoa1, Lgals1 and Lum, alters the gene expression to a HF dermal papilla fibroblast-like profile. (a) Principal component analysis of the transcriptomes of E14.5 dermal fibroblasts, adult DP cells and fibroblasts before and after exposure to selected proteins or E16.5 skin extract for 6 h. (b) Effect of a 6-hr exposure to E16.5 skin extract on the expression of the DP signature genes in cultured fibroblasts. *p < 0.05 with Student’s t-test, compared with fibroblasts (n = 3). (c) Alkaline phosphatase activity (blue color) of mouse adult fibroblasts cultured with rat E16.5 skin extract. Fibroblasts were originally negative for alkaline phosphatase activity (left panel) but had this activity after being cultured in the presence of the E16.5 skin extract for 3 days (central panel). Primary DP cells showed high alkaline phosphatase activity (right panel). FB: adult fibroblast; DP: dermal papilla cell. Bar: 100 μm. (d) Western blotting. The effect of E16.5 skin extract on insulin/Igf signaling in fibroblasts was inhibited by NVP-AEW541. (e) Western blotting. Enhanced Wnt signaling in fibroblasts by E16.5 skin extract was inhibited by XAV-939. N: nuclear; C: cytoplasmic. (f, f′) The effects of NVP-AEW541 and XAV-939 on HF induction by fibroblasts exposed to E16.5 skin extract. #p < 0.05 with Fisher’s test, compared with fibroblasts cultured with E16.5 skin extract only (n = 10). Bar: 500 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.7. Activation of Igf and Wnt signaling in fibroblasts after exposure to embryonic skin proteins is required for hair regeneration
To further analyze the transcriptomic data, the differentially expressed genes of fibroblasts exposed to 3-protein mix or each of the proteins alone was compared with those of fibroblasts exposed to E16.5 extract by SAS correlation analysis. The Spearman’s rank correlation coefficient (rs) for 3-protein mix and E16.5 extract, Apoa1 and E16.5 extract, Lgals1 and E16.5 extract and Lum and E16.5 extract was 0.320, 0.142, 0.111 and 0.116 (p < 0.0001), respectively. The data indicated that the transcriptomic profile of fibroblasts exposed to 3-protein mix is much closer to that of fibroblasts exposed to E16.5 extract than that of fibroblasts exposed to each protein alone. Moreover, the differentially expressed genes were analyzed by DAVID Bioinformatics Resources 6.7 software. We found that, after exposure to each of the 3 proteins, only 10 or less of the top 20 upregulated cellular processes matched the top 20 upregulated cellular processes of fibroblasts exposed to E16.5 extract (Table S3). In contrast, after exposure to Apoa1/Lgals1/Lum mix, 15 of the top 20 upregulated cellular processes matched those of fibroblasts exposed to E16.5 extract (Table S3). Of note, Wnt signaling and insulin-like growth factor (Igf) signaling were among the 15 shared pathways, but they were not included in the top 20 pathways upregulated by exposure to each protein alone. Wnt signaling has been demonstrated to be crucial for HF morphogenesis [12,48,53,54], and the role of Igf in HF development is not clear yet.
Consistent with the findings above, we observed enhanced phosphorylation of insulin receptor substrate 2 (Irs2) and the upregulation of Igf1 both at the transcript and protein levels after exposure to E16.5 extract (Fig. 6d). Inhibition of insulin/Igf signaling with NVP-AEW541, which is specific for Igf1 receptor kinase, ablated HF neogenesis (Fig. 6d, f and 6f′). In addition to Igf, adult fibroblasts treated with E16.5 extract activated canonical Wnt signaling evidenced by increased nuclear β-catenin (Fig. 6e). Inhibition of Wnt signaling with XAV-939, which stimulates β-catenin degradation, also suppressed HF neogenesis (Fig. 6e and f and f′). These results demonstrate that partial conversion of fibroblasts into a state with a DP-like property, along with Igf and Wnt activation, constitutes the core mechanism of extract-induced HF neogenesis. Compared with each of the 3 proteins alone, the Apoa1/Lgals1/Lum mix better recapitulate the effects of embryonic whole-skin extract on fibroblasts for HF neogenesis, including the stimulating effects on Igf and Wnt signaling.
4. Discussion
We show that the three core proteins, Apoa1, Lgals1, and Lum are necessary and sufficient to induce HF neogenesis from adult cells, and that additional factors can further enhance the efficiency. ApoA1 is the major protein component of plasma high-density lipoprotein particles [55]. Due to their function in lipid metabolism and transport, apolipoproteins are actively involved in tissue healing and regeneration where newly formed lipid membranes are required [[56], [57], [58]]. In addition to lipid metabolism, apolipoproteins also exhibit important functions in regulating morphogen signaling and epithelial-mesenchymal interaction during tissue development and regeneration [57,59,60]. Lgals1 belongs to the carbohydrate-binding galectin family with an affinity for beta-galactosides and is expressed in various tissues [61]. Lgals1 is not only present within the cells but also secreted into the extracellular matrices. Lgals1 modulates a variety of functions, including cell growth, cell migration, lineage differentiation, immunity and tissue development [[61], [62], [63], [64], [65], [66]]. It also promotes tissue regeneration [66,67]. Lum, a leucine rich proteoglycan and one of the extracellular matrix proteins, is widely present in many organs [68,69]. Extracellular matrix proteins, including proteoglycans, not only contribute to the shape and structural support for tissues but also regulate development, growth and healing via modulating cytokine activities [68,70]. Apart from its role in the organization of extracellular matrix, Lum also affects cell signaling, modulates the bioactivity of growth factors through binding and regulates tissue regeneration [69,[71], [72], [73], [74]]. Here we demonstrated that these 3 proteins collectively promote the competency of adult fibroblasts for HF neogenesis. In addition, we also demonstrated a novel way to induce HF neogenesis by transfecting adult fibroblasts with of mRNAs of these three genes, Apoa1/Lgals1/Lum.
For regeneration to take place, both the competency of keratinocytes and inductive cues are required [17,19,21]. In the past, adult HF regeneration was principally achieved by co-transplanting competent epithelial cells with inductive embryonic mesenchymal progenitors or specialized adult DP fibroblasts [5,[16], [17], [18], [19], [26], [27],26,27], which both are limited in availability and inductive abilities. Here, we uncovered a new approach that eliminates these limitations. Cell-free extract from the blastema of amputated limbs in adult salamanders also contains inductive proteins that are favorable for regeneration [28,75]. We expect that inductive proteins, analogous to the Apoa1/Lgals1/Lum mix we found in the skin, exist in other mammalian organs [27], and that their identification will open novel organ-specific regeneration protocols. Conversion of adult murine fibroblasts toward an HF-inductive state by rat E16.5 dermal extracts suggests that developmental signaling pathways are conserved [12], and can potentially be applied to humans. The extent to which this approach is applicable to human HF induction is to be determined. Recently, 3D printing has been applied to the generation of human skin equivalents [78]. Since embryonic protein factors were able to induce new HFs when they were incorporated into a dermal equivalent, combination of these protein factors with 3D printing technology might enable the creation of trichogenic skin equivalents. Because the development and regeneration of skin appendages exhibit regional difference [76,77], the context of extracellular proteins from different regions of developing skin can vary. Therefore, skin protein extracts from distinct body regions at specific developmental stages might be able to guide the neogenesis of different skin appendages, such as nails and sweat glands.
Examination of the temporal and spatial expression pattern showed that these three proteins are enriched in, but not limited to, the dermis of the perifolliculogenetic stage. In E16.5 embryonic rat skin, proteins of Apoa1 and Lum are also present in the epidermis, but Lgals1 is absent in the epidermis. Hence, this spatial distribution is consistent with the result that E16.5 dermal extract but not E16.5 epidermal extract is able to induce HF neogenesis (Fig. S1b and S1b′). Since these three proteins together are able to render adult non-hair fibroblasts competent for hair neogenesis, their enriched expression in the embryonic perifolliculogenetic dermis might help to create a favorable environment to enhance the robustness of HF morphogenesis.
Wnt signaling has been shown to be key to embryonic morphogenesis and postnatal HF neogenesis of HFs [12,13,48,53,54,79]. Our results suggest that the 3 proteins Apoa1/Lgals1/Lum together may help to augment Wnt signaling. Additionally, we showed that the 3 proteins Apoa1/Lgals1/Lum also promote Igf signaling and the activated Igf signaling is required for the conversion of fibroblasts to the competent state for HF neogenesis. The result supports the notion that Igf signaling may be involved in embryonic HF development [80]. Because the co-enrichment of the three proteins in the developmental dermis is restricted to a short timeframe, their effect on promoting HF development should subside after the HF morphogenetic stage. In addition to these positive environmental factors we found, other activating and inhibitory factors might also help to regulate the competency and time window for the development of HFs in the embryonic skin [12,79,81,82].
The skin extract-stimulated inductive ability of adult fibroblasts is lost after a washout period (Fig. 5b), suggesting that the gained inductive state is not permanent and can be reversed when the key environmental factors are removed. This contrasts that fact that, for bona fide DP cells, their cell fate and their characteristic HF inductive property can be stably maintained during cell expansion in vitro for up to several passages [17,35]. The loss of embryonic skin extract-induced inductive ability of adult fibroblasts after a washout period indicates the observed conversion toward an inductive DP-like state is transient and may not equal the profound and stable epigenetic cell fate reprogramming induced by transcription factors [83]. In HF neogenesis experiment utilizing inductive mesenchymal cells and in spontaneous HF neogenesis in big wounds [13,84], it has been shown that non-hair adult fibroblasts can contribute to and be committed to a DP fate during the induced morphogenetic epithelial-mesenchymal interaction, demonstrating the robustness and completeness of the natural cell reprogramming scheme. In our experiment, the skin extract-stimulated fibroblasts are likely to be finally reprogrammed and stabilized into a DP fate by a morphogenetic epithelial-mesenchymal interaction triggered by skin extract-stimulated fibroblasts. During this process, epithelial keratinocytes are also reprogrammed into a follicular fate by the epithelial-mesenchymal interaction. With our approach, adult fibroblasts are only stimulated toward the state necessary for reactivating HF morphogenesis. In this work, the fibroblasts were cultured in a serum-containing culture medium supplemented with embryonic protein factors. Whether serum proteins have a beneficial role in conferring trichogenic ability upon fibroblasts in vitro remains to be determined.
We also found that a certain threshold concentration of the proteins is required for HF neogenesis (Fig. S1c). In the in vivo conditions, as the concentration of delivered proteins or mRNAs decays below the threshold for HF regeneration due to either diffusion or degradation, the HF inductivity will be restricted to a narrow post-injection period. Hence, this can allow for a controllable window of time for HF neogenesis.
5. Conclusion
Regeneration of lost tissues in adults depends on the recapitulation of the developmental process of the respective organs. Using the HF regeneration as a model, we unveiled a novel method to re-elicit the developmental scheme for organ neogenesis. We demonstrated that defined extracellular proteins enriched in the embryonic developmental environment can reactivate organogenesis from adult cells. Three core proteins, Apoa1, Lgals1 and Lum, are necessary and sufficient for this regenerative process and that additional factors can further enhance its efficiency. Adult cells exposed to these pro-regeneration environmental proteins become competent to re-engage in organogenesis. The concealed regenerative potential of adult cells can thus be unleashed in an environment reconstituted by defined extracellular protein factors.
Supplementary Material
Acknowledgments
We thank the staff of the 8th Core Lab, Department of Medical Research, National Taiwan University Hospital for technical support and lab members in the S.J.L. lab for discussion. The mass spectrometry analyses using the Orbitrap Fusion mass spectrometers were performed by the Common Mass Spectrometry Facilities, Institute of Biological Chemistry, Academia Sinica. Analysis of RNA-sequencing data was performed by the Bioinformatics Core, Center of Genomic Medicine, National Taiwan University. This work was supported by the Taiwan Bio-Development Foundation (TBF; to S.J.L.), Taiwan Ministry of Science and Technology grants (103-2325-B-002-008, 103–2628-B002-004-MY3, 104-2627-M-002-012 and SPARK program to S.J.L.; 102-2314-B-002-046-MY3 to S.H.P.; 104-2113-M-001-005-MY3 to Y.J.C.), National Taiwan University Hospital grants (NTU-CDP-103R7879 and NTU-CDP-1104R7879 to S.H.P.), a National Taiwan University grant (104R7602D4 to S.J.L.), Asia-Pacific La Roche-Posay Foundation (Basic Research Award to S.J.L.), NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases grant (R01-AR067273 to M.V.P.) and Pew Charitable Trust (00029641 to M.V.P.).
References
- 1.Brockes JP. Amphibian limb regeneration: rebuilding a complex structure. Science. 1997;276:81–87. doi: 10.1126/science.276.5309.81. [DOI] [PubMed] [Google Scholar]
- 2.Johnson SL, Weston JA. Temperature-sensitive mutations that cause stage-specific defects in Zebrafish fin regeneration. Genetics. 1995;141:1583–1595. doi: 10.1093/genetics/141.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker RO, Chapin S, Sherry R. Regeneration of the ventricular myocardium in amphibians. Nature. 1974;248:145–147. doi: 10.1038/248145a0. [DOI] [PubMed] [Google Scholar]
- 4.Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. 2012;18:1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yen CM, Chan CC, Lin SJ. High-throughput reconstitution of epithelial-mesenchymal interaction in folliculoid microtissues by biomaterial-facilitated self-assembly of dissociated heterotypic adult cells. Biomaterials. 2010;31:4341–4352. doi: 10.1016/j.biomaterials.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, et al. Perfusion-decellularized matrix: using nature’s platform to engineer a bio-artificial heart. Nat Med. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 7.Lei M, Schumacher LJ, Lai YC, Juan WT, Yeh CY, Wu P, et al. Self-organization process in newborn skin organoid formation inspires strategy to restore hair regeneration of adult cells. Proc Natl Acad Sci U S A. 2017;114 doi: 10.1073/pnas.1700475114. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fan SM-Y, et al. Biomaterials. 2018;167:E7101–E7110. 121e131 129. [Google Scholar]
- 8.Segers VF, Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–942. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 9.Martin P. Wound healingeaiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 10.Veves A, Falanga V, Armstrong DG, Sabolinski ML. Apligraf Diabetic FootUlcer S. Graft skin, a human skin equivalent, is effective in the management of noninfected neuropathic diabetic foot ulcers: a prospective randomized multicenter clinical trial. Diabetes Care. 2001;24:290–295. doi: 10.2337/diacare.24.2.290. [DOI] [PubMed] [Google Scholar]
- 11.O’Connor NE, Mulliken JB, Banks-Schlegel S, Kehinde O, Green H. Grafting of burns with cultured epithelium prepared from autologous epidermal cells. Lancet. 1:4. [PubMed] [Google Scholar]
- 12.Schneider MR, Schmidt-Ullrich R, Paus R. The hair follicle as a dynamic miniorgan. Curr Biol CB. 2009;19:R132–R142. doi: 10.1016/j.cub.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Ito M, Yang Z, Andl T, Cui C, Kim N, Millar SE, et al. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447:316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- 14.Seifert AW, Kiama SG, Seifert MG, Goheen JR, Palmer TM, Maden M. Skin shedding and tissue regeneration in African spiny mice (Acomys) Nature. 2012;489:561–565. doi: 10.1038/nature11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kligman AM, Strauss JS. The formation of vellus hair follicles from human adult epidermis. J Invest Dermatol. 1956;27:19–23. doi: 10.1038/jid.1956.71. [DOI] [PubMed] [Google Scholar]
- 16.Oliver RF. The induction of hair follicle formation in the adult hooded rat by vibrissa dermal papillae. J Embryol Exp Morphol. 1970;23:219–236. [PubMed] [Google Scholar]
- 17.Jahoda CA, Reynolds AJ, Oliver RF. Induction of hair growth in ear wounds by cultured dermal papilla cells. J Invest Dermatol. 1993;101:584–590. doi: 10.1111/1523-1747.ep12366039. [DOI] [PubMed] [Google Scholar]
- 18.Huang YC, Chan CC, Lin WT, Chiu HY, Tsai RY, Tsai TH, et al. Scalable production of controllable dermal papilla spheroids on PVA surfaces and the effects of spheroid size on hair follicle regeneration. Biomaterials. 2013;34:442–451. doi: 10.1016/j.biomaterials.2012.09.083. [DOI] [PubMed] [Google Scholar]
- 19.Yang CC, Cotsarelis G. Review of hair follicle dermal cells. J Dermatol Sci. 2010;57:2–11. doi: 10.1016/j.jdermsci.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins CA, Chen JC, Cerise JE, Jahoda CA, Christiano AM. Microenvironmental reprogramming by three-dimensional culture enables dermal papilla cells to induce de novo human hair-follicle growth. Proc Natl Acad Sci U S A. 2013;110:19679–19688. doi: 10.1073/pnas.1309970110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinberg WC, Goodman LV, George C, Morgan DL, Ledbetter S, Yuspa SH, et al. Reconstitution of hair follicle development in vivo: determination of follicle formation, hair growth, and hair quality by dermal cells. J Invest Dermatol. 1993;100:229–236. doi: 10.1111/1523-1747.ep12468971. [DOI] [PubMed] [Google Scholar]
- 22.Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, et al. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 23.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Ferraris C, Bernard BA, Dhouailly D. Adult epidermal keratinocytes are endowed with pilosebaceous forming abilities. Int J Dev Biol. 1997;41:491–498. [PubMed] [Google Scholar]
- 25.Longaker MT, Adzick NS. The biology of fetal wound healing: a review. Plast Reconstr Surg. 1991;87:788–798. doi: 10.1097/00006534-199104000-00032. [DOI] [PubMed] [Google Scholar]
- 26.Toyoshima KE, Asakawa K, Ishibashi N, Toki H, Ogawa M, Hasegawa T, et al. Fully functional hair follicle regeneration through the rearrangement of stem cells and their niches. Nat Commun. 2012;3:784. doi: 10.1038/ncomms1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Badylak SF. Regenerative medicine and developmental biology: the role of the extracellular matrix. Anatomical record Part B. New anatomist. 2005;287:36–41. doi: 10.1002/ar.b.20081. [DOI] [PubMed] [Google Scholar]
- 28.McGann CJ, Odelberg SJ, Keating MT. Mammalian myotube dedifferentiation induced by newt regeneration extract. Proc Natl Acad Sci U S A. 2001;98:13699–13704. doi: 10.1073/pnas.221297398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barker TH. The role of ECM proteins and protein fragments in guiding cell behavior in regenerative medicine. Biomaterials. 2011;32:4211–4214. doi: 10.1016/j.biomaterials.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 30.Reynolds AJ, Lawrence C, Cserhalmi-Friedman PB, Christiano AM, Jahoda CA. Trans-gender induction of hair follicles. Nature. 1999;402:33–34. doi: 10.1038/46938. [DOI] [PubMed] [Google Scholar]
- 31.Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Z/AP, a double reporter for cre-mediated recombination. Dev Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- 32.Li YC, Lin MW, Yen MH, Fan SM, Wu JT, Young TH, et al. Programmable laser-assisted surface microfabrication on a poly(vinyl alcohol)-coated glass chip with self-changing cell adhesivity for heterotypic cell patterning. ACS Appl Mater Interfaces. 2015;7:22322–22332. doi: 10.1021/acsami.5b05978. [DOI] [PubMed] [Google Scholar]
- 33.Phua KKL, Liu Y, Sim SH. Non-linear enhancement of mRNA delivery efficiencies by influenza A derived NS1 protein engendering host gene inhibition property. Biomaterials. 2017;133:29–36. doi: 10.1016/j.biomaterials.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Zheng Y, Du X, Wang W, Boucher M, Parimoo S, Stenn K. Organogenesis from dissociated cells: generation of mature cycling hair follicles from skin-derived cells. J Invest Dermatol. 2005;124:867–876. doi: 10.1111/j.0022-202X.2005.23716.x. [DOI] [PubMed] [Google Scholar]
- 35.Huang WY, Huang YC, Huang KS, Chan CC, Chiu HY, Tsai RY, et al. Stress-induced premature senescence of dermal papilla cells compromises hair follicle epithelial-mesenchymal interaction. J Dermatol Sci. 2017;86:114–122. doi: 10.1016/j.jdermsci.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Huang WY, Lai SF, Chiu HY, Chang M, Plikus M, Chan CC, et al. Mobilizing transit-amplifying cell-derived ectopic progenitors prevents hair loss from chemotherapy or radiation therapy. Canc Res. 2017 doi: 10.1158/0008-5472.CAN-17-0667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR:ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with Top Hatand Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ihaka R, Gentleman RR. A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 40.Wickham H. RPackage Version 04 0. 2006. Ggplot: an Implementation of the Grammar of Graphics. [Google Scholar]
- 41.Lin SJ, Foley J, Jiang TX, Yeh CY, Wu P, Foley A, et al. Topology of feather melanocyte progenitor niche allows complex pigment patterns to emerge. Science. 2013;340:1442–1445. doi: 10.1126/science.1230374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan CC, Fan SM, Wang WH, Mu YF, Lin SJ. A two-stepped culture method for efficient production of trichogenic keratinocytes. Tissue Eng CMeth. 2015;21:1070–1079. doi: 10.1089/ten.tec.2015.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tam PP, Parameswaran M, Kinder SJ, Weinberger RP. The allocation of epiblast cells to the embryonic heart and other mesodermal lineages: the role of ingression and tissue movement during gastrulation. Development. 1997;124:1631–1642. doi: 10.1242/dev.124.9.1631. [DOI] [PubMed] [Google Scholar]
- 44.Jahoda CA, Horne KA, Oliver RF. Induction of hair growth by implantation of cultured dermal papilla cells. Nature. 1984;311:560–562. doi: 10.1038/311560a0. [DOI] [PubMed] [Google Scholar]
- 45.Tsai CF, Wang YT, Yen HY, Tsou CC, Ku WC, Lin PY, et al. Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics. Nat Commun. 2015;6:6622. doi: 10.1038/ncomms7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masuda T, Tomita M, Ishihama Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J Proteome Res. 2008;7:731–740. doi: 10.1021/pr700658q. [DOI] [PubMed] [Google Scholar]
- 47.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using stage-tips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 48.Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 49.Wang YT, Tsai CF, Hong TC, Tsou CC, Lin PY, Pan SH, et al. An informatics-assisted label-free quantitation strategy that depicts phospho-proteomic profiles in lung cancer cell invasion. J Proteome Res. 2010;9:5582–5597. doi: 10.1021/pr100394u. [DOI] [PubMed] [Google Scholar]
- 50.Tsou CC, Tsai CF, Tsui YH, Sudhir PR, Wang YT, Chen YJ, et al. IDEAL-Q, an automated tool for label-free quantitation analysis using an efficient peptide alignment approach and spectral data validation. Mol Cell Proteomics : MCP. 2010;9:131–144. doi: 10.1074/mcp.M900177-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiang Y, Zhang CQ, Huang K. Predicting glioblastoma prognosis networks using weighted gene co-expression network analysis on TCGA data. BMC Bioinf. 2012;13(Suppl 2):S12. doi: 10.1186/1471-2105-13-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rendl M, Lewis L, Fuchs E. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 2005;3:e331. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen D, Jarrell A, Guo C, Lang R, Atit R. Dermal beta-catenin activity in response to epidermal Wnt ligands is required for fibroblast proliferation and hair follicle initiation. Development. 2012;139:1522–1533. doi: 10.1242/dev.076463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Dev Cell. 2002;2:643–653. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- 55.Brouillette CG, Anantharamaiah GM, Engler JA, Borhani DW. Structural models of human apolipoprotein A-I: a critical analysis and review. Biochim Biophys Acta. 2001;1531:4–46. doi: 10.1016/s1388-1981(01)00081-6. [DOI] [PubMed] [Google Scholar]
- 56.van der Vliet HN, Sammels MG, Leegwater AC, Levels JH, Reitsma PH, Boers W, et al. Apolipoprotein A-V: a novel apolipoprotein associated with an early phase of liver regeneration. J Biol Chem. 2001;276:44512–44520. doi: 10.1074/jbc.M106888200. [DOI] [PubMed] [Google Scholar]
- 57.Monnot MJ, Babin PJ, Poleo G, Andre M, Laforest L, Ballagny C, et al. Epidermal expression of apolipoprotein E gene during fin and scale development and fin regeneration in zebrafish. Dev Dynam : Off Publ Am Ass Anatom. 1999;214:207–215. doi: 10.1002/(SICI)1097-0177(199903)214:3<207::AID-AJA4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 58.Boyles JK, Zoellner CD, Anderson LJ, Kosik LM, Pitas RE, Weisgraber KH, et al. A role for apolipoprotein E, apolipoprotein A-I, and low density lipoprotein receptors in cholesterol transport during regeneration and remyelination of the rat sciatic nerve. J Clin Invest. 1989;83:1015–1031. doi: 10.1172/JCI113943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim TH, Lee YH, Kim KH, Lee SH, Cha JY, Shin EK, et al. Role of lungapolipoprotein A-I in idiopathic pulmonary fibrosis: antiinflammatory and antifibrotic effect on experimental lung injury and fibrosis. Am J Respir Crit Care Med. 2010;182:633–642. doi: 10.1164/rccm.200905-0659OC. [DOI] [PubMed] [Google Scholar]
- 60.Yao Y, Shao ES, Jumabay M, Shahbazian A, Ji S, Bostrom KI. High-density lipoproteins affect endothelial BMP-signaling by modulating expression of the activin-like kinase receptor 1 and 2. Arterioscler Thromb Vasc Biol. 2008;28:2266–2274. doi: 10.1161/ATVBAHA.108.176958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Camby I, Le Mercier M, Lefranc F, Kiss R. Galectin-1: a small protein with major functions. Glycobiology. 2006;16:137R–57R. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]
- 62.Puche AC, Poirier F, Hair M, Bartlett PF, Key B. Role of galectin-1 in the developing mouse olfactory system. Dev Biol. 1996;179:274–287. doi: 10.1006/dbio.1996.0257. [DOI] [PubMed] [Google Scholar]
- 63.Andersen H, Jensen ON, Moiseeva EP, Eriksen EF. A proteome study of secreted prostatic factors affecting osteoblastic activity: galectin-1 is involved in differentiation of human bone marrow stromal cells. J Bone Min Res Off J Am Soc Bone Min Res. 2003;18:195–203. doi: 10.1359/jbmr.2003.18.2.195. [DOI] [PubMed] [Google Scholar]
- 64.Lutomski D, Fouillit M, Bourin P, Mellottee D, Denize N, Pontet M, et al. Externalization and binding of galectin-1 on cell surface of K562 cells upon erythroid differentiation. Glycobiology. 1997;7:1193–1199. doi: 10.1093/glycob/7.8.1193. [DOI] [PubMed] [Google Scholar]
- 65.Almkvist J, Karlsson A. Galectins as inflammatory mediators. Glycoconj J. 2002;19:575–581. doi: 10.1023/B:GLYC.0000014088.21242.e0. [DOI] [PubMed] [Google Scholar]
- 66.Georgiadis V, Stewart HJ, Pollard HJ, Tavsanoglu Y, Prasad R, Horwood J, et al. Lack of galectin-1 results in defects in myoblast fusion and muscle regeneration. Dev Dynam Off Publ Am Ass Anatom. 2007;236:1014–1024. doi: 10.1002/dvdy.21123. [DOI] [PubMed] [Google Scholar]
- 67.Chan J, O’Donoghue K, Gavina M, Torrente Y, Kennea N, Mehmet H, et al. Galectin-1 induces skeletal muscle differentiation in human fetal mesenchymal stem cells and increases muscle regeneration. Stem cells. 2006;24:1879–1891. doi: 10.1634/stemcells.2005-0564. [DOI] [PubMed] [Google Scholar]
- 68.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nikitovic D, Katonis P, Tsatsakis A, Karamanos NK, Tzanakakis GN. Lumican, a small leucine-rich proteoglycan. IUBMB Life. 2008;60:818–823. doi: 10.1002/iub.131. [DOI] [PubMed] [Google Scholar]
- 70.Kresse H, Schonherr E. Proteoglycans of the extracellular matrix and growth control. J Cell Physiol. 2001;189:266–274. doi: 10.1002/jcp.10030. [DOI] [PubMed] [Google Scholar]
- 71.Saika S, Miyamoto T, Tanaka S, Tanaka T, Ishida I, Ohnishi Y, et al. Responseof lens epithelial cells to injury: role of lumican in epithelial-mesenchymal transition. Investig Ophthalmol Vis Sci. 2003;44:2094–2102. doi: 10.1167/iovs.02-1059. [DOI] [PubMed] [Google Scholar]
- 72.Saika S, Shiraishi A, Liu CY, Funderburgh JL, Kao CW, Converse RL, et al. Role of lumican in the corneal epithelium during wound healing. J Biol Chem. 2000;275:2607–2612. doi: 10.1074/jbc.275.4.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nikitovic D, Berdiaki A, Zafiropoulos A, Katonis P, Tsatsakis A, Karamanos NK, et al. Lumican expression is positively correlated with the differentiation and negatively with the growth of human osteosarcoma cells. FEBS J. 2008;275:350–361. doi: 10.1111/j.1742-4658.2007.06205.x. [DOI] [PubMed] [Google Scholar]
- 74.Naito Z. Role of the small leucine-rich proteoglycan (SLRP) family in pathological lesions and cancer cell growth. J Nippon Med Sch Nippon Ika Daigakuzasshi. 2005;72:137–145. doi: 10.1272/jnms.72.137. [DOI] [PubMed] [Google Scholar]
- 75.Sugiura T, Wang H, Barsacchi R, Simon A, Tanaka EM. MARCKS-like protein is an initiating molecule in axolotl appendage regeneration. Nature. 2016;531:237–240. doi: 10.1038/nature16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu CP, Polak L, Keyes BE, Fuchs E. Spatiotemporal antagonism in mesenchymal-epithelial signaling in sweat versus hair fate decision. Science. 2016:354. doi: 10.1126/science.aah6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Q, Oh JW, Lee HL, Dhar A, Peng T, Ramos R, et al. A multi-scale model for hair follicles reveals heterogeneous domains driving rapid spatio-temporal hair growth patterning. eLife. 2017:6. doi: 10.7554/eLife.22772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim BS, Lee JS, Gao G, Cho DW. Direct 3D cell-printing of human skin with functional transwell system. Biofabrication. 2017;9:025034. doi: 10.1088/1758-5090/aa71c8. [DOI] [PubMed] [Google Scholar]
- 79.Sennett R, Rendl M. Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Semin Cell Dev Biol. 2012;23:917–927. doi: 10.1016/j.semcdb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 81.Botchkarev VA, Botchkareva NV, Sharov AA, Funa K, Huber O, Gilchrest BA. Modulation of BMP signaling by noggin is required for induction of the secondary (nontylotrich) hair follicles. J Invest Dermatol. 2002;118:3–10. doi: 10.1046/j.1523-1747.2002.01645.x. [DOI] [PubMed] [Google Scholar]
- 82.Botchkarev VA, Botchkareva NV, Roth W, Nakamura M, Chen LH, Herzog W, et al. Noggin is a mesenchymally derived stimulator of hair-follicle induction. Nat Cell Biol. 1999;1:158–164. doi: 10.1038/11078. [DOI] [PubMed] [Google Scholar]
- 83.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TCM. Wernig Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Collins CA, Jensen KB, MacRae EJ, Mansfield W, Watt FM. Polyclonal origin and hair induction ability of dermal papillae in neonatal and adult mouse back skin. Dev Biol. 2012;366:290–297. doi: 10.1016/j.ydbio.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.